Abstract
Down syndrome (DS), caused by the trisomy of chromosome 21, is one of the common chromosomal disorders, the main clinical manifestations of which are delayed nervous development and intellectual disability. Long non-coding RNAs (lncRNAs) have critical roles in various biological processes, including cell growth, cell cycle regulation and differentiation. The roles of abnormally expressed lncRNAs have been previously reported; however, the biological functions and regulatory patterns of lncRNAs in DS have remained largely elusive. The aim of the present study was to perform a whole-genome-wide identification of lncRNAs and mRNAs associated with DS. In addition, global expression profiling analysis of DS-induced pluripotent stem cells was performed and differentially expressed (DE) lncRNAs and mRNAs were screened. Furthermore, the target genes and functions of the DE lncRNAs were predicted using Gene Ontology annotation and Kyoto Encyclopedia of Genes and Genomes signaling pathway enrichment analysis. The results revealed that the majority of the lncRNAs exerted functions in DS via cis-acting target genes. In addition, the results of the enrichment analysis indicated that these target genes were mainly involved in nervous and muscle development in DS. In conclusion, this integrative analysis using lncRNA and mRNA profiling provided novel insight into the pathogenesis of DS and it may promote the diagnosis and development of novel therapeutics for this disease.
Keywords: Down syndrome, long non-coding RNA, neurodevelopment, induced pluripotent stem cells
Introduction
Down syndrome (DS) or trisomy 21 is a chromosomal disorder that is caused by the total or partial trisomy of chromosome 21(1), which is associated with delayed nervous development, intellectual disability and a characteristic facial appearance. All affected individuals also experience cognitive delays and weak muscle tone (hypotonia) and they usually present with immune system problems (2). Furthermore, individuals with DS have been discovered to have an increased risk of developing Alzheimer's disease and leukemia (3).
Studies have demonstrated that neurological and immune abnormalities in DS commonly occur in the early developmental stage. At present, basic research on DS relies heavily on clinically obtained peripheral blood samples. However, these samples cannot meet the requirements of basic research, particularly when it comes to the study of early developmental mechanisms. Since 2006, induced pluripotent stem cells (iPSCs) have been considered an invaluable tool for disease modeling and regenerative therapies (4). iPSCs share most of the characteristics of human embryonic stem cells (ESCs); they are pluripotent and self-renew indefinitely, and have the potential to differentiate into any cell type in the body, including those pertinent to psychiatric disorders (5,6). Furthermore, iPSCs were indicated to have similar genetic and epigenetic features to those of ESCs (7,8). Therefore, iPSCs enable the investigation of the influence of the epigenetic status on the early developmental stage and provide a resource for stem cell-based therapies.
Long non-coding RNAs (lncRNAs) are non-protein coding transcripts that are >200 nucleotides long and are members of the family of ncRNAs. Although lncRNAs are not translated into protein, lncRNAs have been reported to serve a highly important regulatory role in the pathogenesis of numerous types of diseases (9). For instance, lncRNAs, as crucial gene antisense transcripts, have been indicated to drive the pathophysiology associated with Alzheimer's disease and regulators of oncogenes during the development of leukemia (10-12). Since patients with DS frequently develop Alzheimer's disease and leukemia, lncRNAs may be potential regulators in DS. A previous study by our group reported that lncRNAs were abnormally expressed in DS (13), suggesting that lncRNAs may be crucial factors in DS; however, the mechanistic roles of lncRNAs remain to be fully elucidated.
As an initial framework for understanding the function of lncRNAs, lncRNAs are typically divided into two types: Cis-acting and trans-acting, based on the local or distant regulation of gene expression, respectively (14,15). It has been suggested that the ability of lncRNA to regulate the expression levels of nearby genes in cis may be attributable to the mature lncRNA transcript or the fact that they do not rely on lncRNAs themselves to have a regulatory role (16). In trans-acting genes, lncRNAs have been indicated to interact with proteins and/or other RNA molecules to regulate the expression of target genes (16). Although an abnormal expression of lncRNAs has been observed in DS, the regulatory pattern of lncRNAs remains to be elucidated. To understand the potential function and regulatory pattern of lncRNAs in DS, the aim of the present study was to investigate the putative cis- or trans-acting targets of lncRNAs in DS and normal iPSCs. RNA sequencing (RNA-seq) was performed to analyze the genome-wide transcriptomic changes in DS. Subsequently, the regulatory mechanisms of differentially expressed (DE) lncRNAs were predicted and analyzed.
Materials and methods
Sample preparation, RNA isolation and library preparation
Normal iPSCs and DS-iPSCs were generated from dermal fibroblast cells, which were purchased from the American Type Culture Collection (ACS-1011: Normal, newborn, male; ACS-1033: DS, newborn, male). For all experiments, three biological replicates were performed and ≥2 technical replicates were used within each biological replicate. Total RNA was extracted from the iPSCs using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA concentration and purity were measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). An Abiotin-labeled specific probe [Ribo-Zero™ ribosomal (r)RNA Removal kit] was used to remove the rRNA from the extracted total RNA. Subsequently, the complementary (c)DNA strand was synthesized using random primers and reverse transcriptase using the TruSeq Stranded kit (cat. no. PC-121-2001; Illumina, Inc.). The second-strand cDNA was subsequently synthesized and double-stranded cDNA was generated. The double-stranded cDNA underwent end-repair/dA-tail and adaptor ligation (17). The products were purified using magnetic beads to create the final cDNA library. High-throughput sequencing of the cDNA library was performed using Illumina HiSeq 2000 platform (Illumina, Inc.).
Sequencing and quality analysis
The clean reads were mapped to the reference genome sequence using HISAT tools (18) and the transcripts were assembled with StringTie (19). Following transcript reconstitution, Cuffcompare (20) was used to compare these transcripts with known mRNA and lncRNA to obtain information on their positional relationship. To distinguish the mRNAs and lncRNAs, three pieces of software [CPC (http://cpc.cbi.pku.edu.cn), txCdsPredic (http://hgdownload.soe.ucsc.edu/admin/exe/) and CNCI (http://www.bioinfo.org/software/cnci)] (20,21) were used to predict the coding ability of the transcripts and all transcripts met the following requirements: i) Fragments per kilo-base of exon per million fragments mapped (FPKM) ≥0.5; ii) coverage >1; iii) length >200. In addition, the estimation accuracy for genes with low expression was improved by Cuffmerge (22) and the combined transcripts were used as final results for subsequent analysis. Following the analysis of all transcripts annotated as mRNA or lncRNA, Bowtie2 was used to compare clean reads to the reference sequence (23), and the FPKM were then calculated as the expression levels of genes and transcripts using RSEM software (v1.3.1; http://deweylab.biostat.wisc.edu/rsem) (24). The results obtained were used for the subsequent analysis.
Identification of DE lncRNAs and mRNAs
The differential expression levels of lncRNAs and mRNAs were calculated based on the normalized FPKM using the DEGseq package (25). q<0.001 and |log2 fold change (FC)| >1 were set as the threshold values for significant differential expression.
lncRNA target gene prediction and enrichment analysis
To determine the correlation between lncRNAs and mRNAs, Spearman's rank correlation coefficient and Pearson's correlation coefficient were calculated and the strength and the direction of the correlations were thereby obtained, with Spearman_correlations and Pearson_correlations ≥0.6 considered to indicate a significant correlation. The sliding window strategy was used to search cis-acting target genes 10 kb upstream and 20 kb downstream of mRNAs (26). Above this range, RNAplex (http://www.tbi.univie.ac.at/~htafer/) was used to analyze the binding energy of lncRNA to mRNA (27). A binding energy of E-value ≤30 was considered to be trans-acting. To predict the cis-acting results, the different lncRNA-mRNA modules were counted, including lncRNAs located within 10 kb upstream or 20 kb downstream of mRNA, and the overlap between lncRNA and mRNA was determined. The overlap may be divided into different subclasses, including lnc-Overlap-mRNA and lnc-AntiOverlap-mRNA (28,29). The classification criteria are presented in Table SI. These cis target genes were subjected to Gene Ontology (GO; http://geneontology.org) functional term and Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/) signaling pathway enrichment analysis.
Validation of DE lncRNAs and mRNAs
In order to verify the functions related to lncRNAs in DS-iPSCs and normal iPSCs, lncRNAs and mRNAs were selected based on the results of lncRNA target gene prediction and enrichment analysis. Subsequently, several lncRNA-mRNA pairs were screened using RT-qPCR to demonstrate the reliability of the analysis. RNA extraction was performed following using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized using random primers and the PrimeScript RT Reagent kit (cat. no. RR047A; Takara Bio, Inc.). RT-qPCR was performed with the One-Step TB Green PrimeScript RT-PCR Kit (cat. no. RR066A; Takara Bio, Inc.) following the manufacturer's instruction. RT-qPCR parameters were as follows: 95˚C for 2 min, 40 cycles of 95˚C for 5 sec and 60˚C for 34 min. Primers for the RT-qPCR were designed using Primer Bank (https://pga.mgh.harvard.edu/primerbank/) or Primer 5.0 software. The GAPDH gene was used to standardize the expression levels. The sequences of PCR were as follows: GAPDH forward, 5'-CGGAGTCAACGGATTTGGTCGTAT-3' and reverse, 5'-AGCCTTCTCCATGGTGGTGAAGAC-3'; NONHSAT009060.2 forward, 5'-CCTGGCTTCTGGTCAAAC-3' and reverse, 5'-AAGGCAACTCAGTCACTAACAC-3'; NONHSAT022318.2 forward, 5'-TCAGTTCAAGGCAACACTGC-3' and reverse 5'-AGGTGGCACTGACCATATCC-3'; ACTN3 forward, 5'-GTACCGCAACGTCAACGTG-3' and reverse, 5'-CGTAGTCGATGAGGTCAGGG-3'; and SRGAP2C-F forward, 5'-ATTGGGCAGCTGAGCATACA-3', and reverse, 5'-TTGGGTCCAGTAACGTATTCCA-3'. Each type of iPSCs included three samples and all reactions were performed three times for each sample. The results of the RT-qPCR for verification were analyzed using the 2-ΔΔCq method (30). Student's t-test was performed on RT-qPCR data and P<0.05 was considered significant.
Cytoplasmic and nuclear fractionation
The Minute (TM) Cytoplasmic and Nuclear Fractionation kits (Invent; cat. no. SC-003) were used for the separation of nuclear and plasma iPSCs. Concurrently, GAPDH was used as the internal reference gene for the nucleus and X-inactive specific transcript (XIST) was used as the internal reference for the cytoplasm (31). The expression location and relative expression levels of the lncRNAs were determined by RT-qPCR. RT-qPCR was performed as aforementioned.
Statistical analysis
A total of 48 samples were designed for the experiment (4 genes x2 samples x2 subcellular locations x3 technical repetitions). The RT-qPCR results for each gene were subjected to one-way ANOVA and for significance, Tukey's honestly significant differences test was performed using SPSS (version 16.0; SPSS, Inc.). For all tests, a P<0.05 was set for statistical significance.
Results
Identification of DE genes in iPSCs by RNA-Seq
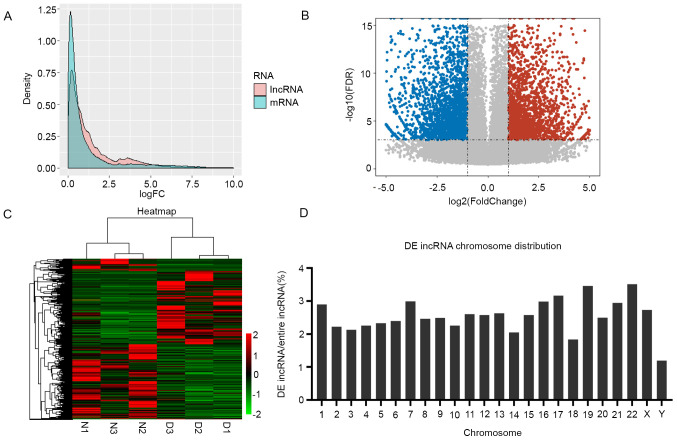
To identify lncRNAs and mRNAs expressed in DS, six cDNA libraries were constructed using DS-iPCs and normal iPSCs. A total of 763,789,996 raw reads were obtained. Following the removal of reads containing N ratios >10%, low-quality reads and adapter fragments, 699,869,860 clean reads were obtained. Following comparison of the data with the reference genome, 85,191 lncRNA transcripts and 40,726 mRNAs were identified using bioinformatics analysis. Compared to mRNAs, more lncRNAs were observed to be significantly DE (P<0.05; Fig. 1A), which suggested the functional importance of lncRNAs in DS. RNA-Seq analysis of lncRNA transcription (4,285 upregulated and 5,239 downregulated; Fig. 1B and C) and mRNA transcription (4,017 upregulated and 4,432 downregulated) demonstrated that differential gene expression was present in DS-iPSCs. All DE lncRNAs were well annotated with known chromosomal locations; DE lncRNAs were distributed across chromosomes without any locational preference (Fig. 1D).
Figure 1.
Transcriptional patterns of lncRNAs in DS-iPSCs. (A) Density plot demonstrating the FC values for the expression levels of lncRNA and protein-coding genes. Pink and blue represent the FC of lncRNAs and mRNAs, respectively. The differential expression of lncRNAs was more significant than that of mRNAs. (B) Volcano plot displaying the 4,285 upregulated and 5,239 downregulated lncRNAs. The horizontal line represents P<0.001 in log10 scale and the vertical lines represent the upregulation and downregulation by two-FC. The red and blue dots represent the upregulated and downregulated lncRNAs, respectively, and non-significantly DE genes are represented by black circles. (C) Heatmap indicating the DE lncRNAs in the DS and control samples. Each column represents one sample and each row one lncRNA. The relative expression levels of lncRNAs are depicted according to the color scale. Red indicates upregulation and green downregulation, whereas the three left columns represent the control samples and the three right columns the DS samples. The DE lncRNAs are clearly self-segregated into clusters, N1, N2 and N3 represent the three biological replications as normal control, and D1, D2 and D3 are represent the three biological replications from the DS patient. (D) Proportion of DE lncRNAs in each chromosome. The ordinate indicates the percentage of DE lncRNAs in each chromosome. lncRNA, long non-coding RNA; DS, Down syndrome; iPSCs, induced pluripotent stem cells; DE, differentially expressed; FDR, false discovery rate; FC, fold change.
Target gene prediction of DE lncRNAs
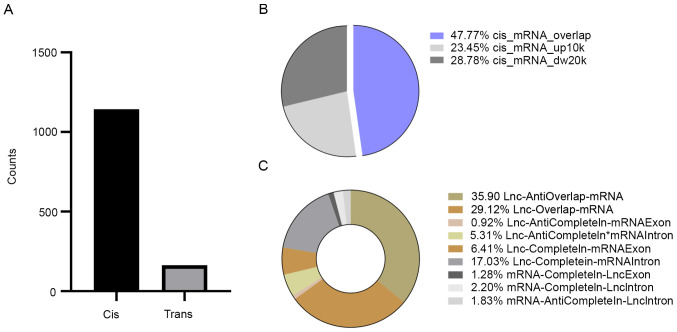
lncRNA expression levels have a significant impact on neighboring (cis) or distal (trans) protein-coding genes. LncRNA-mRNA interactions are useful to clarify the biological functions of DE lncRNAs. To determine the lncRNAs and their potential functions in DS, the potential interactions between the lncRNA and mRNA transcripts were investigated. The cis-/trans-acting target analysis was performed using the association of the expression profiles of DE-lncRNAs and DE-mRNAs. The mRNAs adjacent to the lncRNAs were screened as cis-acting target genes. Trans-acting was determined by calculating the binding energy. First, target genes were predicted in the whole genome and 30,118 lncRNA-mRNA pairs were obtained. The lncRNA-mRNA pairs were screened using the DE mRNAs and lncRNAs. In total, 1,305 lncRNA-mRNA pairs involved in cis and trans regulation were identified. Altogether, 1,143 cis-acting lncRNA-mRNA interactions involving 883 DE mRNAs and 162 trans-acting lncRNA-mRNA interactions involving 118 DE mRNAs were identified (Fig. 2A). Overall, the influence of cis-acting of lncRNAs was discovered to be more important than trans-acting, indicating that cis-acting lncRNAs may have a more significant role in DS (P<0.01).
Figure 2.
Statistical analysis of cis- and trans-acting RNAs. (A) Categorization of cis- and trans-regulatory divergence. The numbers of trans- and cis-acting lncRNA-mRNA pairs are provided, including 1,143 cis-acting and 162 trans-acting lncRNA-mRNA pairs. (B) Ratio of overlap 10 kb upstream and 20 kb downstream in cis-regulation. Each sector of the diagram indicates one of three categories that may be classified based on location divergence: Cis_mRNA_overlap (blue), cis_mRNA_up10k (red) and cis_mRNA_dw20k (gray). (C) The proportion of different classifications in overlapping lncRNAs with intronic/exon overlap was defined in more detail. lncRNA, long non-coding RNA.
In cis-acting, most interactions were overlapping, with 546 lncRNA-mRNA pairs identified; the remaining lncRNA-mRNA pairs were mapped 10 kb upstream (268 pairs) and in 20 kb downstream, with 329 lncRNA-mRNA pairs (Fig. 2B). It was revealed that >60% of the overlapping interaction were accounted for by the lncRNA-anti-overlap-mRNA (35.90%), lncRNA-overlap-mRNA (29.12%) and lncRNA-completein-mRNA intron (17.03%; Fig. 2C), which may be an important part of cis-acting in DS. The results indicated that cis-acting may not necessarily rely on a promoter or enhancer but also on the gene body.
Functional analysis of the DE lncRNAs in DS
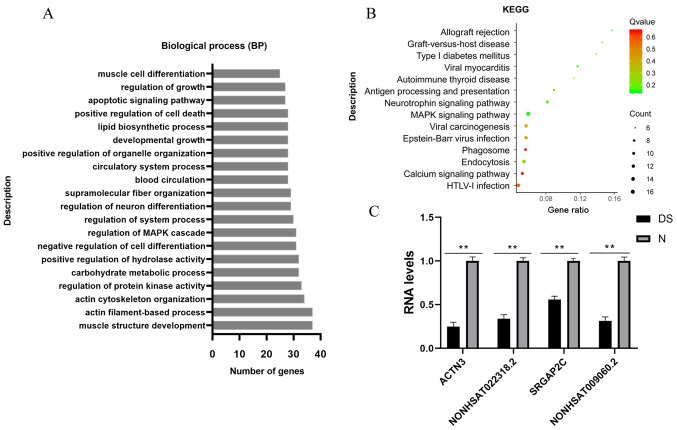
To gain further insight into the function of identified lncRNAs, GO functional term enrichment and KEGG signaling pathway enrichment analysis were used to understand the biological functions and signaling pathways of the target genes of each lncRNA module. The biological pathways of the targets and their upstream/downstream relationship may help determine the potential molecular mechanisms of lncRNAs and contribute to the design experiments to further verify the pathogenesis of lncRNA in DS (32). To further investigate the potential function of DE lncRNAs, GO functional term enrichment analysis and KEGG signaling pathway enrichment analysis were also performed. A total of 397 DE cis-acting target genes were obtained following the conversion of the transcript ID into the gene ID and the removal of duplicates. To further reveal the biological pathway information of the potential targets of these DE lncRNAs in DS, those DE protein-coding genes were subjected to GO functional term enrichment analysis. Representative GO terms significantly enriched by DE mRNAs were identified based on a P-value (P≤0.01, manually removing redundant functional terms). Functional analysis demonstrated that cis target genes of lncRNAs were enriched in 183 GO terms, which were involved in nervous and muscle system development (Fig. 3A). The corresponding potential target genes were subjected to KEGG signaling pathway enrichment analysis. The results revealed that these DE target genes were able to be mapped to 23 signaling pathways; among these signaling pathways, 15 significantly enriched KEGG signaling pathways were determined (P<0.05; Fig. 3B), including the ‘Neurotrophin signaling pathway’ and ‘MAPK signaling pathway’. These results suggested that DE lncRNAs may act through their target genes to regulate the nervous, muscle and immune systems during the early development of DS. Therefore, these results indicated that lncRNAs may function in cis mode on neighboring DS genes to influence nervous and muscle development. To further validate the present results, two lncRNA-mRNA pairs were selected for RT-qPCR verification. Detailed lncRNA-mRNA information was provided in Table SII. lncRNAs with a significant differential expression [logFC ≥|2|; false discovery rates (q) ≤0.001, (False discovery rates were calculated using the q-value conversion algorithm)] and their target genes that were enriched in neural and muscle biological pathways were selected for verification. The DS phenotype may be used to associate enrichment results with specific genes. Therefore, two mRNAs were selected: SLIT-ROBO Rho GTPase activating protein 2C (SRGAP2C) and actin-α3 (ACTN3), which were enriched in the biological process term of nerve and muscle development. Significant differences were discovered in both the expression levels of the mRNAs and corresponding lncRNAs (logFC ≥|2|, P≤0.01; Fig. 3C).
Figure 3.
Target genes were screened by enrichment and quantitative analysis. (A) Analysis of the top 20 overrepresented GO terms of lncRNAs, including GO:0061061, muscle structure development; GO:0042692, muscle cell differentiation; GO:0051961, negative regulation of nervous system development; and GO:0045664, regulation of neuron differentiation. (B) KEGG signaling pathway enrichment analysis of the corresponding cis potential target genes of lncRNAs in DS. The enrichment factor was calculated by the number of enriched genes and the P-value. (C) Validation of the two pairs of lncRNA-mRNA using RT-qPCR. Relative expression levels of selected mRNAs SRGAP2C and ACTN3 and lncRNAs NONHSAT009060.2 and NONHSAT022318.2 in DS (n=3), as compared with the controls (n=3). The results of the reverse RT-qPCR analysis were consistent with the RNA-sequencing data. Bar graphs represent the mean ± standard deviation. All reactions were repeated three times for each mRNA or lncRNA. GAPDH was used as the internal control. **P<0.01 vs. normal controls. BP, biological process; GO, Gene Ontology; lncRNA, long non-coding RNA; KEGG, Kyoto Encyclopedia of Genes and Genomes; DS, Down syndrome; N, normal control; ACTN3, actin-α3; SRGAP2C, SLIT-ROBO Rho GTPase activating protein 2C.
Nuclear and cytoplasmic location of the target lncRNAs
The subcellular localization of an lncRNA is closely associated with its biological mechanism; thus, a correlation analysis between the localization and expression levels of lncRNAs in iPSCs was performed. In addition, nuclear fractionation analysis was performed and the expression levels of NONHSAT009060.2 and NONHSAT022318.2 in the nucleus and cytoplasm were analyzed. Nuclear segregation was assayed using XIST, while the cytoplasmic segregation was analyzed using GAPDH as the housekeeping gene. The results clearly suggested that NONHSAT009060.2 and NONHSAT022318.2 were expressed in both the cytoplasm and nucleus, with NONHSAT009060.2 exhibiting a moderate preference for the nucleus (Fig. S1A and B). Since lncRNAs located in the nucleus play a major role in transcriptional regulation and lncRNAs located in the cytoplasm mainly play a role in post-transcriptional regulation (Fig. S1), this indicated that the expression assessment and identification of the lncRNAs was valid. Since the subcellular localization of a lncRNA is not dependent on the cell type (33), the model is applicable to all human cells.
Discussion
DS is a complex syndrome mediated by numerous genes (34). Epigenetic modifications, such as DNA methylation and lncRNAs, are necessary mechanisms to regulate gene expression levels. A diverse range of mechanisms have been reported for the lncRNA-mediated epigenetic modulation of gene expression. Previous studies have indicated that lncRNA may mainly serve through cis- and trans-regulatory functions to regulate the expression levels of target genes (35,36), which is the major regulatory pattern of lncRNA in higher organisms.
A previous study by our group indicated that lncRNA expression profiles were significantly dysregulated in DS-iPSCs (13). The present study also showed that the differential expression levels of lncRNAs were more significant than that of mRNAs, indicating that lncRNAs play an important role in the regulation process; however, the exact manner in which lncRNAs are involved in the pathogenesis of DS has remained elusive. To further investigate the roles of epigenetic alterations on biological processes and signaling pathways in DS, a comprehensive analysis of lncRNA expression levels and potential regulatory patterns in normal iPSCs and DS-iPSCs was performed. The results revealed that the cis-acting role of lncRNAs had an important impact in DS. The cis-acting pattern was classified according to the positional relationship between lncRNAs and mRNAs. The present classification results demonstrated that ~1/2 of the lncRNAs-mRNAs involved in cis-acting belonged to lncRNAs with overlapping mRNAs and the lncRNAs and mRNAs exhibited the highest antisense overlap. Previous studies have also demonstrated that lncRNAs regulated the coding gene expression levels through the genomic position effect (37). Antisense lncRNAs transcribed against and overlapping with the protein-coding genes have been indicated to regulate their protein-coding counterparts through multiple mechanisms (38). Several antisense lncRNAs have been suggested to regulate the shear and degradation of mRNA through complementary pairing with mRNA following transcription, affecting the expression levels of mRNA (39,40). The present results also validated the results of a previous study indicating that antisense transcription may be far more extensive than previously anticipated (41). It should be noted that cis-acting antisense intronic RNAs have a regulatory function (42), which may stabilize protein-coding transcripts or regulate their alternative splicing. The most abundant wholly intronic antisense RNAs are transcribed from introns of genes related to the regulation of transcription (43). Cis-acting antisense intronic lncRNAs were also observed in the present data, which provided a clue to their functional relevance. This observation offers a novel avenue for future studies.
To precisely clarify the function of DE target genes, GO term enrichment and KEGG signaling pathway enrichment analysis was performed to investigate the potential biological signaling pathways and functions involved. The results of the present study revealed that the cis-acting targets of DE lncRNAs were mainly enriched in the biological process term ‘nervous and muscle growth and development’, which may be associated with the mental impairment and low muscle tone of patients with DS. The functions of lncRNAs have remained to be fully elucidated and the present study provided comprehensive novel insight into the regulatory patterns of mRNAs and lncRNAs in DS by analyzing expression profiles.
The present study also provided a novel perspective for pathological processes in DS, namely that cis-acting lncRNAs have a significant role in controlling/regulating the expression levels of the target genes and thus modulating the clinical phenotype of DS. For instance, SRGAP2C was discovered to act antagonistically on these proteins during the development of cortical neurons, regulating the development of excitatory and inhibitory synapses of the cortical pyramidal neurons in vivo, protracting the maturation and increasing the density of excitatory and inhibitory synapses and indirectly increasing neuronal migration. SRGAP2C has also been implicated in cognition, learning and memory (44). Nerve disorders in patients with DS are currently among the most critical research hotspots. The results of the present study suggested that the abnormal expression levels of neurodevelopment-related genes caused by dysregulated lncRNA expression may be an important cause of nervous system damage occurring during the early developmental stage. ACTN3 is primarily expressed in skeletal muscle and functions as a structural component of the sarcomeric Z line, which has been indicated to be associated with vertical craniofacial skeletal patterns (45). In addition, previous studies have reported that the lack of ACTN3 led to decreased muscle strength (46). It is well-known that craniofacial dysplasia frequently occurs in patients with DS (47) and that the downregulation of ACTN3 in the early developmental stage may be a possible factor for the onset of these clinical symptoms in DS.
In conclusion, in the present study, bioinformatics analysis was performed to preliminarily predict the functions of lncRNAs and their interactions with mRNAs in DS. Further studies should be performed to determine the interactions between lncRNAs and the target genes mentioned above. Elucidation of the precise transcriptional regulatory role of lncRNAs in DS may help understand the pathogenesis of DS and promote the diagnosis and development of novel therapeutics for this disease.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: This work was supported by grants from the National Key Research and Development Program of China (grant nos. 2019YFA0801402, 2016YFC1000503 and 2016YFC0905102), the National Natural Science Foundation of China (grant nos. 81971421 and 81471485), Key Disciplines of Top Priority in Shanghai (grant no. 2017ZZ02019), Shanghai Key Clinical Specialty Project (grant no. shslczdzk05705) and the Experimental Animals Project of Shanghai Municipality (grant nos. 18140901600 and 18140901601).
Availability of data and materials
The sequencing data of all samples were deposited in the Sequence Read Archive (SRA), under accession no. SRP289173. The sequencing data set is comprised of 6 samples, three replicates for each of the two groups: The normal control group and DS group, including the following:
Normal control 1 (N1) https://www.ncbi.nlm.nih.gov/sra/SRX9382487
Normal control 2 (N2) https://www.ncbi.nlm.nih.gov/sra/SRX9382488
Normal control 3 (N3) https://www.ncbi.nlm.nih.gov/sra/SRX9382489
DS 1 (D1) https://www.ncbi.nlm.nih.gov/sra/SRX9382484
Authors' contributions
WM, as the first author, contributed to the experimental design, drafting of the manuscript and the analysis and interpretation of the data for the study. YL completed the cell culture experiments. WM and HM performed RT-qPCR. ZR reviewed and edited the manuscript. JY and ZR are the corresponding authors and contributed to the experimental conception and design, the critical revision of the manuscript and the final approval of the manuscript to be published. WM and JY were responsible for checking and approving the authenticity of the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Patterson D. Molecular genetic analysis of Down syndrome. Hum Genet. 2009;126:195–214. doi: 10.1007/s00439-009-0696-8. [DOI] [PubMed] [Google Scholar]
- 2.Korenberg JR, Chen XN, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C, et al. Down syndrome phenotypes: The consequences of chromosomal imbalance. Proc Natl Acad Sci USA. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weijerman ME, de Winter JP. Clinical practice. The care of children with Down syndrome. Eur J Pediatr. 2010;169:1445–1452. doi: 10.1007/s00431-010-1253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Bilic J, Izpisua Belmonte JC. Concise review: Induced pluripotent stem cells versus embryonic stem cells: Close enough or yet too far apart? Stem Cells. 2012;30:33–41. doi: 10.1002/stem.700. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann A, Ziller M, Spengler D. Progress in iPSC- based modeling of psychiatric disorders. Int J Mol Sci. 2019;20(4896) doi: 10.3390/ijms20194896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–906. doi: 10.1042/BST20120020. [DOI] [PubMed] [Google Scholar]
- 10.Ng SY, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Emmrich S, Streltsov A, Schmidt F, Thangapandi VR, Reinhardt D, Klusmann JH. LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol Cancer. 2014;13(171) doi: 10.1186/1476-4598-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ, Wahlestedt C, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu JJ, Liu YN, Ren ZR, Yan JB. Dysfunctions of mitochondria in close association with strong perturbation of long noncoding RNAs expression in down syndrome. Int J Biochem Cell Biol. 2017;92:115–120. doi: 10.1016/j.biocel.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 14.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 16.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, Lai S, Rich JN, Inguva A, Shannon KM, et al. AMPK/FIS1-mediated mitophagy is required for self-renewal of human AML stem cells. Cell Stem Cell. 2018;23:86–100.e6. doi: 10.1016/j.stem.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(323) doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 26.Narzisi G, O'Rawe JA, Iossifov I, Fang H, Lee YH, Wang Z, Wu Y, Lyon GJ, Wigler M, Schatz MC. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat Methods. 2014;11:1033–1036. doi: 10.1038/nmeth.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tafer H, Amman F, Eggenhofer F, Stadler PF, Hofacker IL. Fast accessibility-based prediction of RNA-RNA interactions. Bioinformatics. 2011;27:1934–1940. doi: 10.1093/bioinformatics/btr281. [DOI] [PubMed] [Google Scholar]
- 28.Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–214. doi: 10.1016/j.neuroscience.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11(59) doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Q, Wan Q, Jiang Y, Liu J, Qiang L, Sun L. A landscape of murine long non-coding RNAs reveals the leading transcriptome alterations in adipose tissue during aging. Cell Rep. 2020;31(107694) doi: 10.1016/j.celrep.2020.107694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gudenas BL, Wang L. Prediction of LncRNA subcellular localization with deep learning from sequence features. Sci Rep. 2018;8(16385) doi: 10.1038/s41598-018-34708-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Letourneau A, Santoni FA, Bonilla X, Sailani MR, Gonzalez D, Kind J, Chevalier C, Thurman R, Sandstrom RS, Hibaoui Y, et al. Domains of genome-wide gene expression dysregulation in Down's syndrome. Nature. 2014;508:345–350. doi: 10.1038/nature13200. [DOI] [PubMed] [Google Scholar]
- 35.Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, et al. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016;18:637–652. doi: 10.1016/j.stem.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 36.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 37.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magistri M, Faghihi MA, St Laurent G III, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: Focus on natural antisense transcripts. Trends Genet. 2012;28:389–396. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paralkar VR, Taborda CC, Huang P, Yao Y, Kossenkov AV, Prasad R, Luan J, Davies JOJ, Hughes JR, Hardison RC, et al. Unlinking an lncRNA from its associated cis element. Mol Cell. 2016;62:104–110. doi: 10.1016/j.molcel.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 42.Yan MD, Hong CC, Lai GM, Cheng AL, Lin YW, Chuang SE. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet. 2005;14:1465–1474. doi: 10.1093/hmg/ddi156. [DOI] [PubMed] [Google Scholar]
- 43.Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, El-Jundi TA, da Silva AM, Reis EM, Verjovski-Almeida S. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8(R43) doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fossati M, Pizzarelli R, Schmidt ER, Kupferman JV, Stroebel D, Polleux F, Charrier C. SRGAP2 and its human-specific paralog co-regulate the development of excitatory and inhibitory synapses. Neuron. 2016;91:356–369. doi: 10.1016/j.neuron.2016.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cunha A, Nelson-Filho P, Marañón-Vásquez GA, Ramos AGC, Dantas B, Sebastiani AM, Silvério F, Omori MA, Rodrigues AS, Teixeira EC, et al. Genetic variants in ACTN3 and MYO1H are associated with sagittal and vertical craniofacial skeletal patterns. Arch Oral Biol. 2019;97:85–90. doi: 10.1016/j.archoralbio.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 46.Clarkson PM, Devaney JM, Dressman HG, Thompson PD, Hubal MJ, Urso M, Price TB, Angelopoulos TJ, Gordon PM, Moyna NM, et al. ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. J Appl Physiol (1985) 2005;99:154–163. doi: 10.1152/japplphysiol.01139.2004. [DOI] [PubMed] [Google Scholar]
- 47.van Marrewijk DJ, van Stiphout MA, Reuland-Bosma W, Bronkhorst EM, Ongkosuwito EM. The relationship between craniofacial development and hypodontia in patients with Down syndrome. Eur J Orthod. 2016;38:178–183. doi: 10.1093/ejo/cjv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data of all samples were deposited in the Sequence Read Archive (SRA), under accession no. SRP289173. The sequencing data set is comprised of 6 samples, three replicates for each of the two groups: The normal control group and DS group, including the following:
Normal control 1 (N1) https://www.ncbi.nlm.nih.gov/sra/SRX9382487
Normal control 2 (N2) https://www.ncbi.nlm.nih.gov/sra/SRX9382488
Normal control 3 (N3) https://www.ncbi.nlm.nih.gov/sra/SRX9382489
DS 1 (D1) https://www.ncbi.nlm.nih.gov/sra/SRX9382484