Abstract
Histone lysine demethylation modification is a critical epigenetic modification. Lysine demethylase 2A (KDM2A), a Jumonji C domain-containing demethylase, demethylates the dimethylated H3 lysine 36 (H3K36) residue and exerts little or no activity on monomethylated and trimethylated H3K36 residues. KDM2A expression is regulated by several factors, such as microRNAs, and the phosphorylation of KDM2A also plays a vital role in its function. KDM2A mainly recognizes the unmethylated region of CpG islands and subsequently demethylates histone H3K36 residues. In addition, KDM2A recognizes and binds to phosphorylated proteins, and promotes their ubiquitination and degradation. KDM2A plays an important role in chromosome remodeling and gene transcription, and is involved in cell proliferation and differentiation, cell metabolism, heterochromosomal homeostasis and gene stability. Notably, KDM2A is crucial for tumorigenesis and progression. In the present review, the documented biological functions of KDM2A in physiological and pathological processes are comprehensively summarized.
Keywords: demethylation, epigenetic, tumorigenesis, cell differentiation
1. Introduction
Histone is an integral part of the nucleosome, the basic unit of chromatin (1). The nucleosome core particle consists of ~147 bp of DNA wrapped around the histone octamer (H2A, H2B, H3 and H4) in two circles (2). The core particles of the nucleosome are connected to histone H1 via ~60 bp of connective DNA. The N-terminal ‘tail’ of the histones is an important target for several histone-modifying enzymes (2).
Histone methylation and demethylation are an important pair of histone epigenetic modifications, which play key roles in gene transcription regulation. Methylation occurs on the lysine and arginine residues of histones, including K9, K27 and K36 of histone H3, K20 of H4, R2, R17 of H3, and R3 of H4(3). The transcriptional regulation of histone lysine methylation is closely associated with lysine residue sites and methylation degree. The methylation of H3K4, H3K36 and H3K79 is often accompanied by the activation of gene transcription, while the methylation of H3K9, H3K27 and H4K20 inhibits gene transcription (3,4).
Histone lysine demethylases are mainly composed of lysine specific demethylase (LSD) and Jumonji C (JmjC)-domain-containing demethylases (JMJD) family demethylases (5). The LSD family consists of two members, lysine demethylase 1A (KDM1A)/LSD1 and KDM1B/LSD2, which remove the monomethyl and dimethyl (me1/me2) of histone lysine residues via amine oxidation reaction. In 2004, researchers at Harvard Medical School were the first to report that LSD1 removes dimethyl and monomethyl modifications of histone H3K4 in vitro, in the presence of the co-factor FAD and a proton nitrogen (6). In vivo, dimethylation and methylation of histone H3K9 can be removed to inhibit gene expression (6). LSD1 is a member of the monoamine oxidase family, and its action requires the participation of an extra proton on the ε-N atom (7). Thus, its demethylation is limited by the substrate and cannot be modified by the demethylation of trimethyllysine (7).
JMJD uses an oxygenase mechanism to remove monomethyl, dimethyl and trimethyl (me1/me2/me3) lysine residues. In 2006, researchers from the University of North Carolina at Chapel Hill reported that JmjC domain histone demethylase 1A (JHDM1A), also known as KDM2A, demethylated H3-methyl-K36 generating formaldehyde and succinate in the presence of Fe (II) and alpha-ketoglutarate (8,9).
According to sequence homology and structural similarity, JmjC domain demethylases are divided into seven subfamilies with different functions (KDM2-8) (9,10). As an important epigenetic modification, histone lysine demethylation regulates important physiological and pathological processes, such as tissue development and tumorigenesis (11). Notably, each subfamily of JMJD demethylase inhibits specific substrates of different histone lysine residues (Table I) (11).
Table I.
Main members of the JMJD family (11).
| JMJD family | Also known as | Gene localization | Substrate |
|---|---|---|---|
| JHDM1A | FBXL11/KDM2A | 11q31.1 | H3K36me2/1 |
| JHDM1B | FBXL10/KDM2B | 12q24.31 | H3K36 |
| JMJD1A | JHDM2A | 2p11.2 | H3K9me2 |
| JMJD2A | KDM4A | 1p34.1 | H3K9me3/2, H3K36me3/2 |
| JMJD2B | KIAA0876 | 19p13.3 | H3K9me3/2 |
| JMJD2C | GASC1 | 9p24.1 | H3K9me3/2 |
| JMJD2D | KDM4D | 11q21 | H3K9me3/2/1 |
| JMJD3 | KIAA0346 | 17p13.1 | H3K27me3/2 |
| JMJD6 | PSR | 17p25 | H3R2, H4R3 |
| JARID1A | RBP2 | 12p11 | H3K4me3/2 |
| JARID1B | PLU-1 | 1q32 | H3K4me3/2/1 |
| JARID1C | SMCX | Xp11.22-p11.21 | H3K4me3/2 |
| JARID1D | SMCY | Yq | H3K4me3/2 |
| UTX | KDM6A | Xp11.2 | H3K27me3/2 |
| UTY | UTY1 | Yq11 | H3K27 |
JMJD, Jumonji C (JmjC)-domain-containing demethylases.
2. Structure and biological function of KDM2A
KDM2A belongs to the KDM2 family. Tsukada et al (8) discovered the first histone demethylase containing the JmjC domain using biological and chemical methods. The KDM2 family in the human genome includes two genes, KDM2A and KDM2B. The KDM2A gene is in 11q13.2, also known as FBXL11/JHDM1A/FBL7/CXXC8/FBL11/LILINA. The encoded protein belongs to the F-box protein family, which is characterized by the F-box containing 40 amino acid sequences, constituting one of the four subunits of the ubiquitin-protein ligase complex (12).
The KDM2A transcripts annotated on the NCBI website mainly have two types, and the longer isoform encoding protein consists of a JmjC domain, a CXXC-zinc finger (ZF-CXXC) domain, a plant homologous zinc finger (PHD) domain, an F-box-like domain and mitogenic exit network protein 1 (AMN1) (13). Conversely, the short-form KDM2A has no JmjC region, which is the catalytic core of demethylation (8). The ZF-CXXC domain specifically recognizes unmethylated CpG islands (14), and the recognition requires the participation of linker DNA. KDM2A binds to CpG islands and demethylates the dimethylated H3K36 residue, and exerts weak activity for monomethylated H3K36 residue (15,16). In addition to the two standard transcripts annotated on the NCBI websites, several KDM2A transcripts have been predicted and reported, such as the isoforms missing the N-terminal JmjC domain or the AMN1 domain. In addition, there are also significant functional differences between the subtypes (17). For example, the alternative isoform of KDM2A lacking the N-terminal demethylase domain can negatively regulate canonical Wnt signaling (12,17-20).
3. KDM2A expression and regulation
KDM2A is located in the nucleus and binds to unmethylated CpG DNA through the ZF-CxxC domain (14), which is essential for maintaining heterochromosomal homeostasis (21). KDM2A is extensively expressed in different tissues, with high expression levels in the brain, testis, ovaries and lungs (22). In addition, KDM2A is highly expressed in most tumors except prostate cancer (21,23-25). As an epigenetic regulator, the expression and biological function of KDM2A are affected by multiple external factors (26,27). In pathological processes, such as gastric cancer and glioblastoma, LINC00460, microRNA (miRNA/miR)-29b, miR-134-5p and miR-3666 directly bind to the KDM2A promoters to regulate KDM2A expression (24,28-31). Inflammation, hypoxia or reactive oxygen species production promote KDM2A expression (26,32), and upregulation of KDM2A induced by human papilloma virus (HPV)16E7 promotes tumorigenesis and progression of cervical cancer (33). Metformin activates the AMPK signaling pathway and decreases intracellular succinic acid levels, while activation of KDM2A decreases ribosomal RNA (rRNA) transcription (27). p300 can directly acetylate KDM2A at position K409, which in turn decreases demethylation of H3K36me2 and enhances the transcription of p21 and PUMA, thereby inhibiting the growth and metastasis of osteosarcoma (34). Mild glucose starvation induces KDM2A-mediated demethylation of H3K36me2 via the AMPK signaling pathway to decrease rRNA transcription and the proliferation of breast cancer cells (35). In non-small cell lung cancer, the carcinogen TPA activates cyclooxygenase-2 (COX-2) expression via KDM2A-mediated H3K36 dimethylation near the COX-2 promoter (36).
JmjC domain-containing histone lysine demethylases (KDM2-7) are important epigenetic regulators and potential targets for cancer (11). Thus, there is great interest to investigate and identify selective and therapeutic KDMs inhibitors (37). Understanding the structure of lysine demethylases and their modular synthetic approach has helped design and develop a series of highly selective KDM2/7 inhibitors (38,39). Some inhibitors exhibit antiproliferative activity, and so may be used as candidates for anticancer agents (38). Human immunodeficiency virus and HPV induce epigenetic alterations in host cells by altering the levels of H3K36 methylation within the promoter region of CTLA-4 and FOXP3, resulting in several diseases and different types of cancer (40,41). Histone demethylase inhibitors combined with checkpoint blockade may be used as a novel cancer treatment strategy (41-43). As an inhibitor of KDM2A, plant growth regulator Daminozide has been reported to significantly abrogate the effect of KDM2A on histone demethylation, and exhibits promising results as an anticancer therapeutic strategy (44,45).
4. Clinical significance of KDM2A in human cancers
KDM2A is abnormally expressed in different tumors, and it plays a vital role in tumorigenesis and progression (12). Wagner et al (46) demonstrated that KDM2A binds to the dual-specificity phosphatase 3 (DUSP3) gene promoter region and inhibits its expression, which in turn increases phosphorylation of ERK1/2 and promotes the occurrence and metastasis of non-small cell lung cancer. Another study reported similar findings for KDM2A in non-small cell lung cancer, with HDAC3 as the target gene (25). In addition, it has been reported that c-Fos recruits KDM2A to the COX-2 promoter region to promote the transcription of COX-2, and treatment with the carcinogen TPA promotes the recruitment process (36). Huang et al (47) demonstrated that KDM2A is highly expressed in gastric cancer tissues, which promotes the proliferation and metastasis of tumor cells by inhibiting programmed cell death protein 4 expression. Furthermore, KDM2A reverses epithelial-to-mesenchymal transition by regulating the PI3K signaling pathway, which promotes the progression of ovarian cancer (48). In colon cancer, LINC01278 upregulates KDM2A expression to promote cancer progression (49), and KDM2A expression is associated with cyclin D1 expression and cell proliferation (50). In breast cancer, studies have reported that KDM2A binds to the promoter region of genes, such as E2F1, which inhibits its transcriptional regulation and results in the invasion and metastasis of breast cancer cells (23,51). Conversely, another study has revealed a completely different transcriptional regulation mechanism in breast cancer. The results of this study demonstrated that KDM2A directly upregulates JAG1 expression, which affects breast cancer stem cell-related characteristics and angiogenesis (45). Subsequently, it has been reported that the short isoform of KDM2A (without the JmjC region) is crucial for promoting the tumorigenesis and progression of breast cancer, while the role of the long isoform remains unknown (17). High expression levels of KDM2A enhance cancer-associated fibroblasts (52), and combined expression of KDM2A and KDM2B may be associated with clinical prognosis in patients with breast cancer (53). Researchers at Harvard Medical School cloned four cDNA subtypes of KDM2A and demonstrated that only the KDM2A-N782 isoform (the longer isoform N-terminal 782 amino acids, including JmjC, CXXC and Ring domains, but not F-box and AMN1 domains) has a significant effect on promoting cell proliferation (19). Our previous study reported that KDM2A expression is significantly upregulated in hepatocellular carcinoma (HCC) tissues compared with adjacent normal tissues. In addition, high KDM2A expression is associated with poor prognosis and overall survival in patients with HCC (54). KDM2A augments stem cell-like characteristics via demethylation of histone H3K36 at promoters of stemness-associated transcription factors, such as OCT4, NANOG and SOX2(54).
In most cases, KDM2A promotes the progression of tumors. However, KDM2A knockdown in zebrafish has been reported to disrupt the transcriptome and result in high frequencies of spontaneous melanoma (55). In addition, interference with KDM2A expression in HT29 cells increases colony formation in soft agar (18). KDM2A affects the NF-κB pathway by demethylating the K218/K221 site of p65 in HT29 cells (56,57). In different types of tumors, KDM2A has different pro-oncogenic or anti-oncogenic effects, which may be associated with intertumor heterogeneity or the extensiveness of KDM2A downstream target genes and demethylation sites. In addition, multiple splicing forms of KDM2A play different roles in tumorigenesis and progression of different types of cancer (12).
5. Role of KDM2A in cell differentiation and development
KDM2A has been reported to play a vital role in the differentiation of stem cells and the development of embryos (19). Several studies have investigated the role of KDM2A in the proliferation and differentiation of stem cells from the apical papilla (SCAPs) (58,59). The transcription factor, BCOR, interacts with KDM2A to affect the expression of stem-related genes, such as SOX2(59). In addition, KDM2A abrogates the inhibition of p15INK4B and p27kip1, and demethylation of SFRP2 to regulate the differentiation and proliferation of SCAPs into adipocytes and chondrocytes (58-60). KDM2A exhibits synergistic effects with BCL6, inhibits cell proliferation and regulates osteogenic differentiation of mesenchymal stem cells via the epidermal growth factor epiregulin, EREG (61). Researchers cloned four cDNA isoforms of KDM2A and demonstrated that transfection with the transcript variant of KDM2A-N782 containing N-terminal 782AA can significantly promote keratinocytes proliferation (19). Notably, KDM2A promotes the vitamin C-dependent reprogramming process of adult cells, accelerates the cell cycle and inhibits resting senescence, and plays an important role in the process of induced pluripotent stem cells (62).
Researchers analyzed KDM2A expression in the teeth of mice during embryonic and postnatal stages via reverse transcription-quantitative PCR and immunohistochemistry analyses. The results demonstrated that KDM2A may play essential roles in cell proliferation and tooth differentiation of mice (63). In addition, KDM2A knockdown exhibited severe embryonic lethality, accompanied by severe growth defects and weight loss (64). KDM2A knockdown affects the expression of cell cycle regulatory factors, such as p21Cip1, which in turn decreases cell proliferation and increases apoptosis (64). KDM2A plays a vital role in maintaining embryonic stem cells by affecting the methylation levels of H3K36me2 and H3K4me3, and regulating the expression of germ cell-related genes (65). In the xenopus model, the stability of nuclear β-catenin depends on its methylation/demethylation, and the lysine demethylase Kdm2a/b specifically demethylates and degrades non-phosphorylated β-catenin in the nucleus to regulate the canonical Wnt signaling pathway, which plays a decisive role in the formation of the anterior-posterior body axis in xenopus embryos (66).
6. Biological functions of KDM2A in other physiological and pathological processes
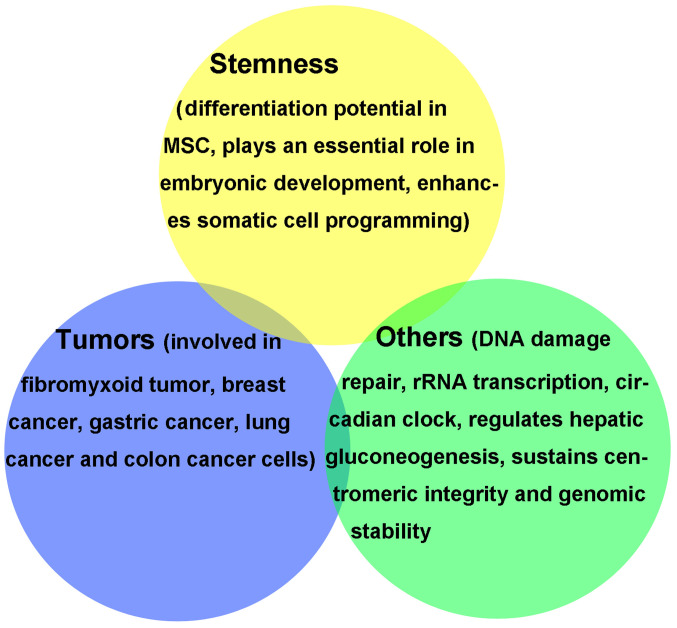
In addition to its involvement in embryonic development and stem cell differentiation, KDM2A is also associated with tumorigenesis and progression. KDM2A regulates several physiological and pathological processes (Fig. 1) (12). Age-associated DNA methylation changes in blood leukocytes during early childhood may reflect epigenetic maturation, since histone modifiers and chromatin remodeling factors, such as KDM2A, are locus susceptible and play key roles in leukocyte biology (67). JmjC domain-containing histone lysine demethylases (KDM2-7) are known to be altered with aging, which could be associated with the regulation of the aging process and age-related diseases (68). Pan et al (69) demonstrated that KDM2A negatively regulates gluconeogenesis-related genes in vivo, which silences KDM2A expression and accelerates the synthesis of liver glycogen. Overexpression of KDM2A decreases blood sugar levels and this regulatory effect is achieved by the demethylation of H3K36 in the C/EBPa promoter region by KDM2A (69). Frescas et al (21) reported that KDM2A is involved in regulating the expression of small non-coding RNAs that are encoded by the clusters of satellite repeats at the centromere and are essential for maintaining heterochromatin homeostasis. The transcription of rRNA genes is a rate-limiting step in ribosomal synthesis, which changes in response to environmental stimuli (26,27,70). It has been demonstrated that the demethylase KDM2A containing the JmjC domain remarkably decreases rDNA transcription in response to starvation, which is accompanied by the demethylation of H3K36me2 in the rDNA promoter (71). In addition, KDM2A binds to the unmethylated CpG sequence of rDNA promoter under starvation stress via the CXXC-ZF domain, and demethylates H3K36me2 in the rDNA promoter to decrease the transcription of rDNA (70-72). Reischl and Kramer (73) demonstrated that KDM2A is an important circadian clock regulator. KDM2A directly binds to the promoter regions of CLOCK/BMAL1-regulated genes via a CXXC-ZF motif and regulates the expression of clock genes, such as Nr1d1, which plays a key role in maintaining the mammalian circadian rhythms (73). ATM protein (ataxia-telangiectasia mutated) is an important signal molecule in DNA repair (74); however, its specific molecular mechanism remains unclear. It has been reported that following DNA damage, ATM specifically phosphorylates the KDM2A serine 632 site, which decreases the chromosome binding capacity of KDM2A and increases the degree of dimethylation of H3K36 at the DNA damage site (75). This in turn recruits the MRE11 complex to the injury site, which interacts with BRCT2 to induce repair and cell survival (75). KDM2A is also recruited to DNA double-strand breaks and interacts with 53BP1 to ensure the stability of the genome (76). Downregulated KDM2A expression increases H3K36me2 at DNA damage sites to inhibit transcription and promote repair (77).
Figure 1.
Biological function of lysine demethylase 2A in pathological and physiological processes. MSC, mesenchymal stem cell; rRNA, ribosomal RNA.
7. Conclusions
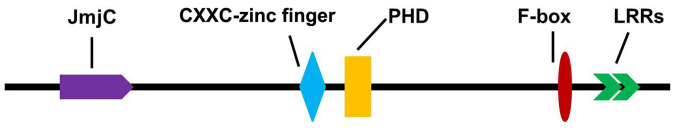
KDM2A belongs to the JMJD family, which consists of a JmjC domain, a PHD zinc finger structure, LRR-AMN1 and F-box, CXXC-zinc finger structure domain and other components (Fig. 2) (8). As a histone demethylase, KDM2A specifically demethylates both monomethylated and dimethylated lysine-36 of histone H3, and is an important epigenetic modification (13). KDM2A also contains SKP1-cullin-F-box, a subunit of the ubiquitin protein ligase complex, which recognizes and binds to some phosphorylated proteins and promotes their ubiquitination and degradation (76).
Figure 2.
Structure of lysine demethylase 2A. JmjC, Jumonji C; PHD, plant homologous zinc finger.
The methylation and demethylation of histones has always been relatively balanced, affecting gene regulation and several biological functions. In addition to affecting chromosome structure, the level of histone methylation also acts as a scaffolding molecule to recognize and bind certain transcription factors and affect transcription regulation (3). Abnormal KDM2A expression and an imbalance of target gene methylation result in tumorigenesis and the progression of different types of cancer (12).
In conclusion, KDM2A is closely associated with physiological processes, such as stem cell differentiation, cell rhythm and metabolism, as well as heterosomal stability and DNA damage repair.
Acknowledgements
Not applicable.
Funding Statement
Funding: The present review was supported by the Medicine and Health Science Technology Development Program of Shandong province (grant no. 2019WS198) and the Jinan Clinical Medical Science Technology Innovation Program (grant no. 201704080).
Availability of data and materials
Not applicable.
Authors' contributions
LL and QL conceived the present review and performed the literature review. LL drafted the initial manuscript. JL prepared the table and revised the manuscript for important intellectual content. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 a resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 3.Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49(e324) doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Fang Y, Liao G, Yu B. Targeting histone lysine demethylase LSD1/KDM1A as a new avenue for cancer therapy. Curr Top Med Chem. 2019;19:889–891. doi: 10.2174/156802661911190725094910. [DOI] [PubMed] [Google Scholar]
- 8.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 9.Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 11.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vacik T, Ladinovic D, Raska I. KDM2A/B lysine demethylases and their alternative isoforms in development and disease. Nucleus. 2018;9:431–441. doi: 10.1080/19491034.2018.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlotorynski E. Protein folding: Structure-function analysis of KDM2A. Nat Rev Mol Cell Biol. 2014;15(630) doi: 10.1038/nrm3870. [DOI] [PubMed] [Google Scholar]
- 14.Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou JC, Blackledge NP, Farcas AM, Klose RJ. Recognition of CpG island chromatin by KDM2A requires direct and specific interaction with linker DNA. Mol Cell Biol. 2012;32:479–489. doi: 10.1128/MCB.06332-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iuchi S, Paulo JA. Lysine-specific demethylase 2A enhances binding of various nuclear factors to CpG-rich genomic DNAs by action of its CXXC-PHD domain. Sci Rep. 2019;9(5496) doi: 10.1038/s41598-019-41896-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Liu L, Holowatyj A, Jiang Y, Yang ZQ. Integrated genomic and functional analyses of histone demethylases identify oncogenic KDM2A isoform in breast cancer. Mol Carcinog. 2016;55:977–990. doi: 10.1002/mc.22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Z, Cheung P, Kuo AJ, Yukl ET, Wilmot CM, Gozani O, Patel DJ. A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes Dev. 2014;28:1758–1771. doi: 10.1101/gad.246561.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iuchi S, Green H. Lysine-specific demethylase 2A (KDM2A) normalizes human embryonic stem cell derived keratinocytes. Proc Natl Acad Sci USA. 2012;109:9442–9447. doi: 10.1073/pnas.1206176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladinovic D, Pinkas D, Šopin T, Raška O, Liška F, Raška I, Vacík T. Alternative isoforms of KDM2A and KDM2B lysine demethylases negatively regulate canonical Wnt signaling. PLoS One. 2020;15(e236612) doi: 10.1371/journal.pone.0236612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frescas D, Guardavaccaro D, Kuchay SM, Kato H, Poleshko A, Basrur V, Elenitoba-Johnson KS, Katz RA, Pagano M. KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle. 2008;7:3539–3547. doi: 10.4161/cc.7.22.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:63–70. doi: 10.1093/dnares/6.1.63. [DOI] [PubMed] [Google Scholar]
- 23.Chen JY, Luo CW, Lai YS, Wu CC, Hung WC. Lysine demethylase KDM2A inhibits TET2 to promote DNA methylation and silencing of tumor suppressor genes in breast cancer. Oncogenesis. 2017;6(e369) doi: 10.1038/oncsis.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong Y, Zou S, Yang F, Xu X, Bu W, Jia J, Liu Z. RUNX3-mediated up-regulation of miR-29b suppresses the proliferation and migration of gastric cancer cells by targeting KDM2A. Cancer Lett. 2016;381:138–148. doi: 10.1016/j.canlet.2016.07.038. [DOI] [PubMed] [Google Scholar]
- 25.Dhar SS, Alam H, Li N, Wagner KW, Chung J, Ahn YW, Lee MG. Transcriptional repression of histone deacetylase 3 by the histone demethylase KDM2A is coupled to tumorigenicity of lung cancer cells. J Biol Chem. 2014;289:7483–7496. doi: 10.1074/jbc.M113.521625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka Y, Obinata H, Konishi A, Yamagiwa N, Tsuneoka M. Production of ROS by Gallic acid activates KDM2A to reduce rRNA transcription. Cells. 2020;9(2266) doi: 10.3390/cells9102266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y, Konishi A, Obinata H, Tsuneoka M. Metformin activates KDM2A to reduce rRNA transcription and cell proliferation by dual regulation of AMPK activity and intracellular succinate level. Sci Rep. 2019;9(18694) doi: 10.1038/s41598-019-55075-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F, Liang S, Liu X, Han L, Wang J, Du Q. LINC00460 modulates KDM2A to promote cell proliferation and migration by targeting miR-342-3p in gastric cancer. Onco Targets Ther. 2018;11:6383–6394. doi: 10.2147/OTT.S169307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shou T, Yang H, Lv J, Liu D, Sun X. MicroRNA3666 suppresses the growth and migration of glioblastoma cells by targeting KDM2A. Mol Med Rep. 2019;19:1049–1055. doi: 10.3892/mmr.2018.9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Wei C, Zhang Z, Jin Q, Xiao X. MiR-134-5p regulates myocardial apoptosis and angiogenesis by directly targeting KDM2A After myocardial infarction. Int Heart J. 2020;61:815–821. doi: 10.1536/ihj.19-468. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, Chen X, Jiang J, Wan X, Wang Y, Xu P. Epigallocatechin gallate reverses gastric cancer by regulating the long noncoding RNA LINC00511/miR-29b/KDM2A axis. Biochim Biophys Acta Mol Basis Dis. 2020;1866(165856) doi: 10.1016/j.bbadis.2020.165856. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, Li G, Han N, Zhang X, Cao Y, Cao Y, Fan Z. Secreted frizzled-related protein 2 promotes the osteo/odontogenic differentiation and paracrine potentials of stem cells from apical papilla under inflammation and hypoxia conditions. Cell Prolif. 2020;53(e12694) doi: 10.1111/cpr.12694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ou R, Zhu L, Zhao L, Li W, Tao F, Lu Y, He Q, Li J, Ren Y, Xu Y. HPV16 E7-induced upregulation of KDM2A promotes cervical cancer progression by regulating miR-132-radixin pathway. J Cell Physiol. 2019;234:2659–2671. doi: 10.1002/jcp.27080. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Sun B, Zhang Q, Dong H, Zhang J. p300 Acetylates JHDM1A to inhibit osteosarcoma carcinogenesis. Artif Cells Nanomed Biotechnol. 2019;47:2891–2899. doi: 10.1080/21691401.2019.1638790. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka Y, Yano H, Ogasawara S, Yoshioka S, Imamura H, Okamoto K, Tsuneoka M. Mild glucose starvation induces KDM2A-mediated H3K36me2 demethylation through AMPK To reduce rRNA transcription and cell proliferation. Mol Cell Biol. 2015;35:4170–4184. doi: 10.1128/MCB.00579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu S, Yang Y, Du Y, Cao LL, Li M, Shen C, Hou T, Zhao Y, Wang H, Deng D, et al. The transcription factor c-Fos coordinates with histone lysine-specific demethylase 2A to activate the expression of cyclooxygenase-2. Oncotarget. 2015;6:34704–34717. doi: 10.18632/oncotarget.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAllister TE, England KS, Hopkinson RJ, Brennan PE, Kawamura A, Schofield CJ. Recent progress in histone demethylase inhibitors. J Med Chem. 2016;59:1308–1329. doi: 10.1021/acs.jmedchem.5b01758. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki T, Ozasa H, Itoh Y, Zhan P, Sawada H, Mino K, Walport L, Ohkubo R, Kawamura A, Yonezawa M, et al. Identification of the KDM2/7 histone lysine demethylase subfamily inhibitor and its antiproliferative activity. J Med Chem. 2013;56:7222–7231. doi: 10.1021/jm400624b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerken PA, Wolstenhulme JR, Tumber A, Hatch SB, Zhang Y, Müller S, Chandler SA, Mair B, Li F, Nijman SMB, et al. Discovery of a highly selective cell-active inhibitor of the histone lysine demethylases KDM2/7. Angew Chem Int Ed Engl. 2017;56:15555–15559. doi: 10.1002/anie.201706788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Q, Chen L, Song Y, Ma L, Xiao P, Chen L, Zhen H, Han R, Chen X, Sun S, et al. Induction of co-inhibitory molecule CTLA-4 by human papillomavirus E7 protein through downregulation of histone methyltransferase JHDM1B expression. Virology. 2019;538:111–118. doi: 10.1016/j.virol.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Abdel-Hameed EA, Ji H, Shata MT. HIV-induced epigenetic alterations in host cells. Adv Exp Med Biol. 2016;879:27–38. doi: 10.1007/978-3-319-24738-0_2. [DOI] [PubMed] [Google Scholar]
- 42.Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, Li Y, Chen H, Yang H, Hsu PH, et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell. 2018;174:549–563.e19. doi: 10.1016/j.cell.2018.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boehm D, Ott M. Host methyltransferases and demethylases: Potential new epigenetic targets for HIV cure strategies and beyond. AIDS Res Hum Retroviruses. 2017;33 (Suppl):S8–S22. doi: 10.1089/aid.2017.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose NR, Woon EC, Tumber A, Walport LJ, Chowdhury R, Li XS, King ON, Lejeune C, Ng SS, Krojer T, et al. Plant growth regulator daminozide is a selective inhibitor of human KDM2/7 histone demethylases. J Med Chem. 2012;55:6639–6643. doi: 10.1021/jm300677j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen JY, Li CF, Chu PY, Lai YS, Chen CH, Jiang SS, Hou MF, Hung WC. Lysine demethylase 2A promotes stemness and angiogenesis of breast cancer by upregulating Jagged1. Oncotarget. 2016;7:27689–27710. doi: 10.18632/oncotarget.8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner KW, Alam H, Dhar SS, Giri U, Li N, Wei Y, Giri D, Cascone T, Kim JH, Ye Y, et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest. 2013;123:5231–5246. doi: 10.1172/JCI68642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Liu Y, Yu L, Chen J, Hou J, Cui L, Ma D, Lu W. Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumour Biol. 2015;36:271–278. doi: 10.1007/s13277-014-2630-5. [DOI] [PubMed] [Google Scholar]
- 48.Lu DH, Yang J, Gao LK, Min J, Tang JM, Hu M, Li Y, Li ST, Chen J, Hong L. Lysine demethylase 2A promotes the progression of ovarian cancer by regulating the PI3K pathway and reversing epithelialmesenchymal transition. Oncol Rep. 2019;41:917–927. doi: 10.3892/or.2018.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xi C, Ye NY, Wang YB. LncRNA LINC01278 accelerates colorectal cancer progression via miR-134-5p/KDM2A axis. Eur Rev Med Pharmacol Sci. 2020;24:10526–10534. doi: 10.26355/eurrev_202010_23405. [DOI] [PubMed] [Google Scholar]
- 50.Cao LL, Du C, Liu H, Pei L, Qin L, Jia M, Wang H. doi: 10.1177/1724600818764069. Lysine-specific demethylase 2A expression is associated with cell growth and cyclin D1 expression in colorectal adenocarcinoma. Int J Biol Markers: Apr 1, 2018 (Epub ahead of print). doi: 10.1177/1724600818764069. [DOI] [PubMed] [Google Scholar]
- 51.Rizwani W, Schaal C, Kunigal S, Coppola D, Chellappan S. Mammalian lysine histone demethylase KDM2A regulates E2F1-mediated gene transcription in breast cancer cells. PLoS One. 2014;9(e100888) doi: 10.1371/journal.pone.0100888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen JY, Li CF, Lai YS, Hung WC. Lysine demethylase 2A expression in cancer-associated fibroblasts promotes breast tumour growth. Br J Cancer. 2020;124:484–493. doi: 10.1038/s41416-020-01112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Nicola I, Guerrieri AN, Penzo M, Ceccarelli C, De Leo A, Trerè D, Montanaro L. Combined expression levels of KDM2A and KDM2B correlate with nucleolar size and prognosis in primary breast carcinomas. Histol Histopathol. 2020;35:1181–1187. doi: 10.14670/HH-18-248. [DOI] [PubMed] [Google Scholar]
- 54.Lin Q, Wu Z, Yue X, Yu X, Wang Z, Song X, Xu L, He Y, Ge Y, Tan S, et al. ZHX2 restricts hepatocellular carcinoma by suppressing stem cell-like traits through KDM2A-mediated H3K36 demethylation. Ebiomedicine. 2020;53(102676) doi: 10.1016/j.ebiom.2020.102676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scahill CM, Digby Z, Sealy IM, Wojciechowska S, White RJ, Collins JE, Stemple DL, Bartke T, Mathers ME, Patton EE, Busch-Nentwich EM. Loss of the chromatin modifier Kdm2aa causes BrafV600E-independent spontaneous melanoma in zebrafish. PLoS Genet. 2017;13(e1006959) doi: 10.1371/journal.pgen.1006959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y, Gudkov AV, Stark GR. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci USA. 2009;106:16339–16344. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, Stark GR. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao R, Dong R, Du J, Ma P, Wang S, Fan Z. Depletion of histone demethylase KDM2A inhibited cell proliferation of stem cells from apical papilla by de-repression of p15INK4B and p27Kip1. Mol Cell Biochem. 2013;379:115–122. doi: 10.1007/s11010-013-1633-7. [DOI] [PubMed] [Google Scholar]
- 59.Dong R, Yao R, Du J, Wang S, Fan Z. Depletion of histone demethylase KDM2A enhanced the adipogenic and chondrogenic differentiation potentials of stem cells from apical papilla. Exp Cell Res. 2013;319:2874–2882. doi: 10.1016/j.yexcr.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 60.Yu G, Wang J, Lin X, Diao S, Cao Y, Dong R, Wang L, Wang S, Fan Z. Demethylation of SFRP2 by histone demethylase KDM2A regulated osteo-/dentinogenic differentiation of stem cells of the apical papilla. Cell Prolif. 2016;49:330–340. doi: 10.1111/cpr.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du J, Ma Y, Ma P, Wang S, Fan Z. Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem Cells. 2013;31:126–136. doi: 10.1002/stem.1255. [DOI] [PubMed] [Google Scholar]
- 62.Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, Pei D. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Yi Q, Cao Y, Liu OS, Lu YQ, Wang JS, Wang SL, Yao R, Fan ZP. Spatial and temporal expression of histone demethylase, Kdm2a, during murine molar development. Biotech Histochem. 2016;91:137–144. doi: 10.3109/10520295.2015.1106586. [DOI] [PubMed] [Google Scholar]
- 64.Kawakami E, Tokunaga A, Ozawa M, Sakamoto R, Yoshida N. The histone demethylase Fbxl11/Kdm2a plays an essential role in embryonic development by repressing cell-cycle regulators. Mech Dev. 2015;135:31–42. doi: 10.1016/j.mod.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 65.Fu E, Shen J, Dong Z, Zhang W, Zhang Y, Chen F, Cheng Z, Zhao X, Shuai L, Lu X. Histone demethylase Kdm2a regulates germ cell genes and endogenous retroviruses in embryonic stem cells. Epigenomics. 2019;11:751–766. doi: 10.2217/epi-2018-0126. [DOI] [PubMed] [Google Scholar]
- 66.Lu L, Gao Y, Zhang Z, Cao Q, Zhang X, Zou J, Cao Y. Kdm2a/b lysine demethylases regulate canonical Wnt signaling by modulating the stability of nuclear β-catenin. Dev Cell. 2015;33:660–674. doi: 10.1016/j.devcel.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 67.Acevedo N, Reinius LE, Vitezic M, Fortino V, Söderhäll C, Honkanen H, Veijola R, Simell O, Toppari J, Ilonen J, et al. Age-associated DNA methylation changes in immune genes, histone modifiers and chromatin remodeling factors within 5 years after birth in human blood leukocytes. Clin Epigenetics. 2015;7(34) doi: 10.1186/s13148-015-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salminen A, Kauppinen A, Kaarniranta K. 2-Oxoglutarate-dependent dioxygenases are sensors of energy metabolism, oxygen availability, and iron homeostasis: Potential role in the regulation of aging process. Cell Mol Life Sci. 2015;72:3897–3914. doi: 10.1007/s00018-015-1978-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pan D, Mao C, Zou T, Yao AY, Cooper MP, Boyartchuk V, Wang YX. The histone demethylase Jhdm1a regulates hepatic gluconeogenesis. PLoS Genet. 2012;8(e1002761) doi: 10.1371/journal.pgen.1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka Y, Okamoto K, Teye K, Umata T, Yamagiwa N, Suto Y, Zhang Y, Tsuneoka M. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J. 2010;29:1510–1522. doi: 10.1038/emboj.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka Y, Umata T, Okamoto K, Obuse C, Tsuneoka M. CxxC-ZF domain is needed for KDM2A to demethylate histone in rDNA promoter in response to starvation. Cell Struct Funct. 2014;39:79–92. doi: 10.1247/csf.13022. [DOI] [PubMed] [Google Scholar]
- 72.Tsuneoka M, Tanaka Y, Okamoto K. A CxxC domain that binds to unmethylated CpG is required for KDM2A to control rDNA transcription. Yakugaku Zasshi. 2015;135:11–21. doi: 10.1248/yakushi.14-00202-2. (In Japanese) [DOI] [PubMed] [Google Scholar]
- 73.Reischl S, Kramer A. Fbxl11 is a novel negative element of the mammalian circadian clock. J Biol Rhythms. 2015;30:291–301. doi: 10.1177/0748730415587407. [DOI] [PubMed] [Google Scholar]
- 74.Jin MH, Oh DY. ATM in DNA repair in cancer. Pharmacol Ther. 2019;203(107391) doi: 10.1016/j.pharmthera.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 75.Cao LL, Wei F, Du Y, Song B, Wang D, Shen C, Lu X, Cao Z, Yang Q, Gao Y, et al. ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene. 2016;35(402) doi: 10.1038/onc.2015.311. [DOI] [PubMed] [Google Scholar]
- 76.Bueno M, Baldascini M, Richard S, Lowndes NF. Recruitment of lysine demethylase 2A to DNA double strand breaks and its interaction with 53BP1 ensures genome stability. Oncotarget. 2018;9:15915–15930. doi: 10.18632/oncotarget.24636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rezazadeh S, Yang D, Biashad SA, Firsanov D, Takasugi M, Gilbert M, Tombline G, Bhanu NV, Garcia BA, Seluanov A, Gorbunova V. SIRT6 mono-ADP ribosylates KDM2A to locally increase H3K36me2 at DNA damage sites to inhibit transcription and promote repair. Aging (Albany NY) 2020;12:11165–11184. doi: 10.18632/aging.103567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.