Abstract
The adenomatous polyposis coli (APC) tumor suppressor protein is associated with the regulation of Wnt signaling; however, APC also controls other cellular processes including the regulation of cell adhesion and migration. The expression of full-length APC in SW480 colorectal cancer cells (SW480+APC) not only reduces Wnt signaling, but increases membrane E-cadherin and restores cell–cell adhesion. This report describes the effects of full-length, wild-type APC (fl-APC) on cell–cell adhesion genes and p120-catenin isoform switching in SW480 colon cancer cells: fl-APC increased the expression of genes implicated in cell–cell adhesion, whereas the expression of negative regulators of E-cadherin was decreased. Analysis of cell–cell adhesion-related proteins in SW480+APC cells revealed an increase in p120-catenin isoform 3A; similarly, depletion of APC altered the p120-catenin protein isoform profile. Expression of ESRP1 (epithelial splice regulatory protein 1) is increased in SW480+APC cells, and its depletion results in reversion to the p120-catenin isoform 1A phenotype and reduced cell–cell adhesion. The ESRP1 transcript is reduced in primary colorectal cancer, and its expression correlates with the level of APC. Pyrvinium pamoate, which inhibits Wnt signaling, promotes ESRP1 expression. We conclude that re-expression of APC restores the cell–cell adhesion gene and posttranscriptional regulatory programs leading to p120-catenin isoform switching and associated changes in cell–cell adhesion.
INTRODUCTION
Colorectal cancer (CRC) is a leading cause of cancer-related deaths worldwide (World Health Organization, 2018). More than 80% of CRCs arise from biallelic, truncating mutations in the tumor suppressor gene adenomatous polyposis coli (APC) (Kinzler and Vogelstein, 1996; Clevers and Nusse, 2012). APC is a multifunctional protein implicated in a range of cellular processes including cell migration, differentiation, and cell–cell adhesion and contributes to normal intestinal crypt homeostasis (Nathke et al., 1996; Faux et al., 2004; Sansom et al., 2004; Dow et al., 2015); truncation of APC stimulates increased crypt production (Paulsen et al., 2001). APC is best characterized for its role in the regulation of Wnt/β-catenin signaling (Clevers and Nusse, 2012). Wnt signaling is essential for normal embryonic development (Nusse and Varmus, 1992; Logan and Nusse, 2004) and stem cell maintenance (Reya and Clevers, 2005). APC mutations result in aberrant Wnt/β-catenin signaling and increased transcription of Wnt target genes (e.g., AXIN2 and myc) (Peifer and Polakis, 2000; Giles et al., 2003; Huang and He, 2008). SW480 is a CRC cell line where only truncated APC is present. We have shown previously that re-expression of full-length, wild-type APC (fl-APC) in SW480 CRC cells (SW480+APC) reduces Wnt/β-catenin signaling, increases membrane-associated E-cadherin, and restores cell–cell adhesion (Faux et al., 2004). However, neither the detailed mechanisms controlling APC induction of E-cadherin membrane association nor the mechanisms associated with the role of APC in increasing cell–cell adhesion have been identified.
E-cadherin is a transmembrane protein that mediates calcium-dependent cell–cell adhesion through homophilic interactions with its extracellular domains (Gul et al., 2017). The cytoplasmic tail of E-cadherin interacts with the β- and p120-catenins through binding to their armadillo repeats and indirectly with α-catenin, which connects the complex to the actin cytoskeleton (Ozawa et al., 1989; Takeichi, 2014). β- and p120-catenins are multifunctional proteins with roles in determining the strength of cell–cell adhesion junctions (Nelson, 2008; Pieters et al., 2012) as well as transcriptional signaling (Park et al., 2005; Clevers and Nusse, 2012; Lee et al., 2014). β-catenin is a central component in the canonical Wnt signaling pathway: Wnt stimulation increases β-catenin binding to the Tcf family of transcriptional regulators, consequentially driving Wnt target gene expression (Giles et al., 2003; Huang and He, 2008). p120-catenin is required for tuning the role of E-cadherin activity at cell–cell contacts (Yap et al., 1998; Ireton et al., 2002; Davis et al., 2003). At the cell junctions p120-catenin stabilizes E-cadherin (Davis et al., 2003) and reduces E-cadherin endocytosis by masking the endocytic dileucine motif (Nanes et al., 2012). p120-catenin also modulates RhoGTPase activity through its RhoA-binding sites (Anastasiadis et al., 2000) and activates Rac1 and consequentially cell motility (Noren et al., 2000), as well as participating in gene transcription by interacting with Kaiso (Park et al., 2005), thereby modulating the range of gene targets stimulated by Wnt/β-catenin.
p120-catenin is crucial for E-cadherin function in cell–cell adhesion, and as such it is essential for normal development (Pieters et al., 2016) and homeostasis in both the intestinal mucosa (Smalley-Freed et al., 2010) and the skin (Perez-Moreno et al., 2006). The p120-catenin protein is present as multiple isoforms (Pieters et al., 2012), which occur as a result of alternative splicing of the p120-catenin (CTNND1) gene (Keirsebilck et al., 1998). The p120-catenin isoform 1A is the longest isoform and contains nine armadillo domains in the central part of the molecule and an extended N-terminus (Pieters et al., 2012). p120-catenin isoforms 3A and 4 are missing part and all of the N-terminal region, respectively. The relative levels of p120-catenin 1A (long) and 3A (short) are different depending on the tissue and cell type (Mo and Reynolds, 1996; Ireton et al., 2002). In particular, there is a switch from the short 3A isoform to the longer 1A isoform when epithelial cells transition to mesenchymal cells (Ireton et al., 2002). Accordingly, short and long isoforms are reported to behave differently in human cancers, where the 1A isoform has been linked to increased cell migration and invasion (Seidel et al., 2004; Yanagisawa et al., 2008; Slorach et al., 2011; Zhang et al., 2014). The p120-catenin isoform 1A inhibits RhoA activity, thereby promoting cell invasiveness, whereas the shorter 3A isoform does not (Yanagisawa et al., 2008). The mechanisms by which the splicing of the p120 precursor mRNA have been investigated (Warzecha et al., 2009), but the relationship of the splicing to Wnt/β-catenin signaling and cell–cell adhesion are not well understood.
Here we demonstrate that restoration of full-length, wild-type APC (fl-APC) results in altered cell–cell adhesion, changes in gene expression profiles for cell adhesion components, and the redistribution of key cell–cell junctional adhesion proteins. p120-catenin mRNA levels are not altered at the transcriptional level, but the levels of the p120-catenin isoforms change because of changes to mRNA splicing. SW480+APC cells predominantly express the 3A short isoform, whereas the parental cells express both 3A (short) and 1A (long) isoforms. p120-catenin isoform switching is stimulated by an epithelial splice regulatory protein (ESRP1), which has increased expression in SW480+APC cells. We show that ESRP1 levels are reduced in primary CRC tissue and that inhibition of Wnt signaling further increases ESRP1 expression and consequentially the E-cadherin function in cell–cell adhesion junctions, suggesting a mechanism by which APC and Wnt signaling regulate the epithelial phenotype.
RESULTS
Altered cell–cell adhesion transcript profiles in SW480 cells with restored APC expression
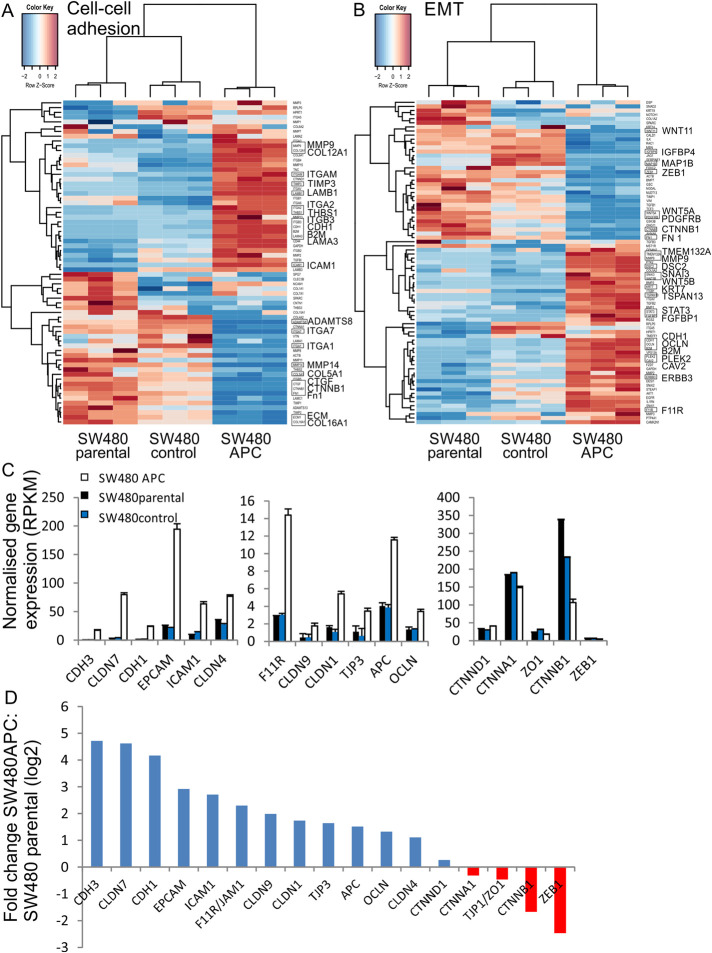
We have previously shown that expression of full-length APC protein in SW480 CRC cells (SW480+APC) reduces Wnt signaling, increases membrane β-catenin and E-cadherin, and restores cell–cell adhesion (Faux et al., 2004). The gene expression profiles that are altered by expression of wild-type APC have been determined by RNAseq analysis of SW480, SW480 control (empty vector-transfected control), and SW480+APC cells (King et al., 2016). The transcriptome sequencing identified a total of ∼25,130 genes, of which 1735 were significantly differentially expressed (King et al., 2016). Among the top differentially expressed genes were Wnt targets such as AXIN2, CCND2, and ID2 (DNA-binding protein inhibitor 2) and cell–cell adhesion genes, such as CDH1, CDH3, and MMP9 (Supplemental Table S1). Gene set enrichment analysis identified Wnt signaling, cell–cell adhesion, and epithelial-to-mesenchymal transition (EMT) as significant pathways induced by APC (King et al., 2016). Because re-expression of normal APC restored the cell–cell adhesion phenotype (Faux et al., 2004), we focused on changes in gene expression related to cell–cell adhesion. Hierarchical clustering of cell–cell adhesion and EMT gene expression profiles show differential expression in SW480+APC cells in comparison to SW480 and SW480 control cells (Figure 1, A and B). Our transcriptome analysis shows 22 cell–cell adhesion and 24 EMT genes as significantly differentially expressed (genes with an adjusted two-sided P value <0.05 and >2-fold change in expression) (Figure 1, A and B) (Supplemental Table S1). The analysis revealed that restoration of normal APC is associated with increased transcript expression of cell–cell adhesion genes, such as CDH1 (E-cadherin) and CDH3 (P-cadherin), intercellular adhesion molecule 1 (ICAM1), and epithelial cell adhesion molecule (EPCAM) as well as cell adhesion tight junction genes OCLN, CLDN 1, 4, 7,and 9 (Figure 1C). In contrast, there was no change in transcript level for CTNNA1, CTNND1, or tight junction gene ZO-1, whereas CTNNB1 and ZEB1 (Zinc Finger E-Box Binding Homeobox 1), a negative regulator of E-cadherin expression (Spaderna et al., 2006) were reduced at the transcript level in SW480+APC cells. The fold changes (log2) for significantly differentially expressed cell–cell adhesion genes are shown in Figure 1D. The reduction in the β-catenin transcripts in SW480+APC cells is consistent with changes in protein levels as reported previously (Faux et al., 2004). EMT is characterized by reduction in CDH1 and increased expression of cadherins such as CDH2 or CDH11 (Pal et al., 2018). While neither CDH2 or CDH11 is expressed, CDH23 expression is up-regulated in SW480 cells and CDH1 as well as CDH3 are rescued upon APC expression (Supplemental Table S1). Thus our transcriptome analysis suggests that expression of APC in SW480 cells results in specific gene expression changes and supports a role for APC in promoting cell–cell adhesion, possibly mediated through regulation of E-cadherin and/or Wnt signaling.
FIGURE 1:
Restoration of full-length APC promotes the expression of cell adhesion genes. (A, B) Heatmaps displaying hierarchical clustering for differentially expressed cell adhesion (A) and EMT (B) genes in SW480+APC compared with SW480 and SW480 control. The heatmap was drawn using log2 (+1 offset) expression values, mean centered and scaled by gene. Gene and sample dendrograms were generated using divisive hierarchical clustering (DIANA). (C) Selected cell–cell adhesion genes showing differential expression in SW480+APC, SW480 parental, and SW480 control cells expressed as normalized gene expression (RPKM). Mean ± SD (n = 3). (D) Log2 fold change in cell adhesion genes (from C) in SW480+APC compared with SW480 parental cells. Genes with an adjusted two-sided P value of less than 0.05 and showing a greater than twofold change in expression were considered differentially expressed. CTNND1, CTNNA1, and ZO1 are not differentially expressed.
p120-catenin subcellular distribution and isoform levels are altered upon APC expression
Given the changes in transcript for cell–cell adhesion genes, we investigated the subcellular distribution of cell–cell adhesion proteins (Figure 2A). As reported previously, β-catenin and E-cadherin redistribute to sites of cell–cell contact in SW480+APC cells, consistent with the altered morphology of the cells (Faux et al., 2004) (Figure 2A). The tight junction proteins ZO-1 and claudin-7 also demonstrate strong cell–cell contact staining in SW480+APC cells, in striking contrast to the control cells, which demonstrate diffuse cytoplasmic and some nuclear staining. We note that claudin-7 expression (CLDN7) is significantly increased at the transcript level in SW480+APC cells, whereas ZO-1 is not altered. p120-catenin (CTNND1) also demonstrates a strong redistribution to sites of cell–cell contact (Figure 2A) but as with ZO-1 is not differentially expressed at the transcript level (see Figure 1D).
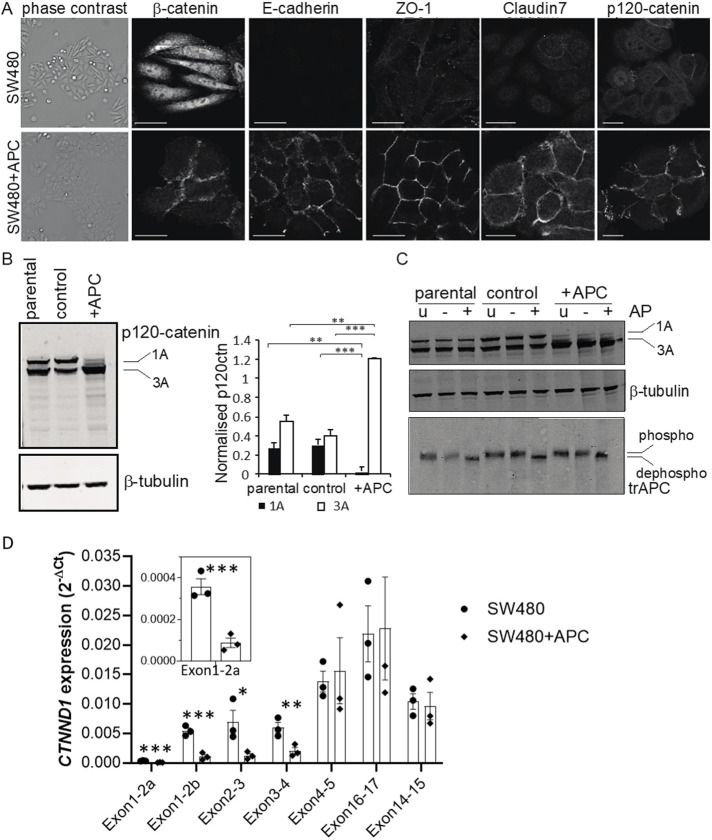
FIGURE 2:
Altered p120-catenin localization and isoform expression upon restoration of APC. (A) Cell adhesion proteins are localized to cell–cell contacts in SW480+APC but not SW480 parental cells. Phase contrast images (left-hand side) of SW480 parental and SW480+APC cells. Cells were immunostained with antibodies to β-catenin, E-cadherin, ZO-1, claudin-7, and p120-catenin. Shown are single confocal sections; scale bar 20 μm. (B) Immunoblot analysis of p120-catenin in SW480, SW480 control, and SW480+APC. p120-catenin migrates as two species in SW480 and SW480 control cells, but the faster migrating form predominates in SW480+APC cells. β-tubulin was used as a loading control. Quantified levels of normalized p120-catenin isoforms are shown on the right. Graph shows mean ± SEM, n = 5; **P < 0.005; ***P < 0.001, unpaired Student’s t test. (C) Altered migration of p120-catenin is not due to phosphorylation. SW480, SW480 control, and SW480+APC cells were untreated (u) or treated without (–) or with (+) Antarctic phosphatase (AP; 50 U) for 30 min at 37°C and proteins resolved by SDS–PAGE. Immunoblot analysis shows that phosphatase treatment did not alter the migration of p120-catenin and β-tubulin but resulted in the collapse of truncated APC to a faster migrating species (dephospho). (D) qRT-PCR analysis of CTNND1 mRNA expression with isoformspecific primers with extracts from SW480 and SW480+APC cells. SW480 cells express isoforms 1A and 3A, and SW480+APC cells express isoform 3A (mean ± SEM, n = 3; unpaired Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.005). Inset shows CTNND1 expression (exon2a).
There are two species of p120-catenin protein in SW480 parental and SW480 control cells, but in SW480+APC cells the faster migrating species predominates (Figure 2B). p120-catenin can be expressed as multiple isoforms due to alternative splicing (Keirsebilck et al., 1998) and four alternative transcriptional start sites. The migration pattern observed in SW480 cells is consistent with p120-catenin isoforms 1A and 3A (Pieters et al., 2012), the most common isoforms. In SW480 and SW480 control cells, isoforms 1A and 3A are equally distributed; however, in SW480+APC cells isoform 3A is increased by >2-fold (P < 0.005) and isoform 1A is barely detectable (Figure 2B). The increased expression of isoform 3 correlates with the increased membrane-associated E-cadherin and the pronounced epithelial morphology of the SW480+APC cells (Faux et al., 2004) (see Figure 2A). Thus, p120-catenin isoform expression is significantly altered upon expression of full-length APC.
To exclude that the aberrant migration of the p120-catenin species on SDS–PAGE was due to differences in phosphorylation, cell extracts were treated with phosphatase (Figure 2C). Phosphatase treatment did not affect the migration of p120-catenin species (Figure 2C, top panel). As a control, these analyses show that the truncated APC protein is phosphorylated in both the parental and SW480+APC cells (Figure 2C). Thus, altered migration of p120-catenin proteins is not due to phosphorylation. p120-catenin 1A and 3A isoform expression was also analyzed with quantitative-PCR using isoform-specific primers (Figure 2D; Supplemental Figure S1). CTNND1 mRNA expression was significantly higher with the primers that spanned the exon boundaries for CTNND1 1–2, 2–3, and 3–4 in SW480 compared with SW480+APC, that is, consistent with the expression of p120-catenin isoform 1A in SW480 but not SW480+APC cells (Figure 2D). In contrast, there was no difference in CTTND1 expression with the primers that detect both 1A and 3A isoforms, that is, those spanning exons 4–5 and 14–15 (Figure 2D). Expression of APC in SW480 CRC cells therefore results in a redistribution of cell–cell adhesion proteins to cell contacts, mirroring altered distributions in β-catenin and E-cadherin and altered p120-catenin isoform distribution. This suggests that relative levels p120-catenin isoforms may affect the adhesive and/or promigratory phenotype of SW480 cells.
APC promotes a switch to an epithelial p120-catenin isoform
Expression of full-length APC in SW480 cells results in altered Wnt and cell adhesion transcriptional signatures and is accompanied by redistribution of adhesion- and tight-junction proteins. To test whether APC regulates p120-catenin isoform levels, we depleted APC using small interfering RNA (siRNA). The truncated APC protein, present in SW480, SW480 control, and SW480+APC cells and full-length APC, present only in SW480+APC cells, is efficiently depleted at 72 h posttransfection (Figure 3A; Supplemental Figure S2A). Only low levels of full-length APC are present in the SW480+APC cells (Figure 3A and Faux et al., 2004). siRNA-mediated depletion of APC perturbs p120-catenin isoform expression whereby isoform 1 is increased upon APC depletion (2.7 ± 0.2-fold over mock control) (Figure 3, B and D). siRNA-mediated depletion of truncated APC protein also results in a modest but significant increase in p120-catenin isoform 1 in both SW480 parental and SW480 control cells (Figure 3), which suggests a residual function for the truncated APC protein that is likely to be Wnt-independent. While p120-catenin isoform 3A does not change markedly upon APC depletion, the ratio of p120-catenin isoforms 1A:3A is increased significantly in SW480+APC cells (3.16 ± 0.59-fold increase) in APC siRNA compared with mock control-treated cells (Supplemental Figure S2B). Depletion of APC results in a similar fold increase in β-catenin levels (2.6-± 0.7-fold) and a reduction of E-cadherin in SW480+APC cells (Figure 3C and Supplemental Figure S2C). The increase in p120-catenin isoform 1 following depletion of APC demonstrates that full-length APC controls p120-catenin isoform levels in SW480 cells.
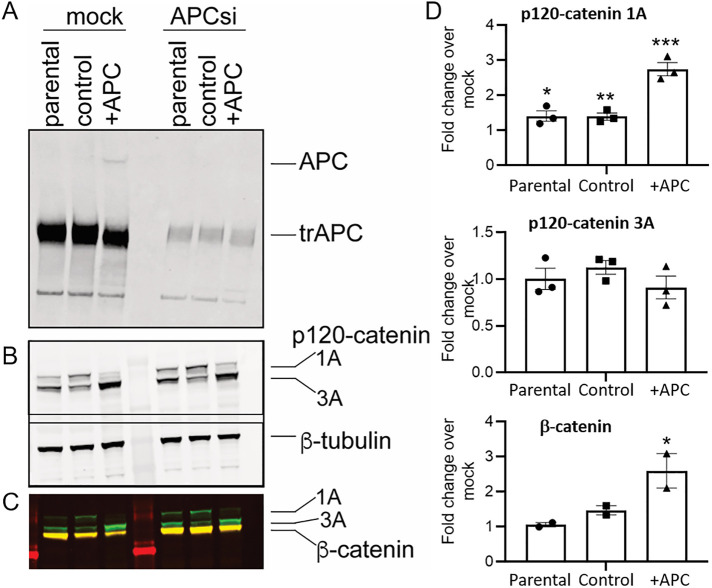
FIGURE 3:
p120-catenin isoform expression is dependent on APC. (A) Depletion of APC by siRNA at 72 h. APC levels were assessed by immunoblot analysis. Full-length APC is present in SW480+APC cells only; the truncated APC protein (trAPC) is indicated. (B) siRNA-mediated depletion perturbs p120-catenin isoform expression. SW480 parental, SW480 control, and SW480+APC cells were transfected with mock control and APC siRNA and p120-catenin levels assessed by immunoblot analysis. β-tubulin was used as a loading control. (C) β-catenin is reduced in mock-transfected SW480+APC cells compared with parental and control cells and increased in APC siRNA transfected cells. Immunoblots in B were probed with antibodies to β-catenin (yellow); p120-catenin is visible in green. (D) Quantitation of p120-catenin 1A (top) and 3A (middle) isoforms and β-catenin (bottom) normalized to β-tubulin in APC-depleted cells (72 h); graph shows fold change over mock control transfected cells, mean ± SEM (n = 3) for p120-catenin isoforms and mean ± SD (n = 2) for β-catenin, * P < 0.05; **P < 0.01, ***P < 0.005, unpaired Student’s t test.
ESRP1 controls p120-catenin isoform switching in SW480+APC cells
To gain mechanistic insight into how APC mediates alterations in p120-catenin isoform switching and the generation of the epithelial phenotype, we investigated genes that can induce a p120-catenin isoform switch. EMT genes such as c-FOS, SNAI1, SNAI2 (slug), SIP1/ZEB2, TWIST, and ZEPPO (Shapiro et al., 2011) have been associated with the p120-catenin isoform switch. Epithelial splice regulatory proteins (ESRP1 and ESRP2) coordinate an epithelial cell type–specific splicing program and have been shown to regulate splicing of several genes including p120-catenin (Warzecha et al., 2009). Indeed, we found that both ESRP1 and ESRP2 were significantly increased at the transcript level in SW480+APC cells (Figure 4A) (log2-fold change in SW480+APC compared with SW480 parental and SW480 control was 6.00 and 4.734 for ESRP1 and 1.3 and 1.178 for ESRP2, respectively). Moreover, siRNA-mediated depletion of APC resulted in a marked reduction in ESRP1 mRNA in SW480+APC cells, whereas Wnt target genes AXIN2 and ID2 mRNA levels increased, as expected (Figure 4B). In concordance with mRNA levels, ESRP1 and ESRP2 protein levels were also markedly increased in SW480APC compared with SW480 parental cells (Figure 4C). Depletion of ESRP1 resulted in reduced ESRP1 protein and mRNA in SW480+APC cells (Figure 4, C and D). ESRP2 siRNA treatment resulted in reduced ESRP2 protein (Figure 4C). We found that ESRP1 was also reduced upon ESRP2-mediated siRNA treatment but to a lesser extent than upon ESRP1-siRNA, and likewise, ESRP2 was reduced when ESRP1 was depleted (Figure 4C). Depletion of ESRP1 did not reverse the shift from 1A to 3A isoforms but did result in a greater than fourfold increase in the p120-catenin isoform 1 in SW480+APC cells (Figure 4, E and F). p120-catenin isoform 1 was also increased when ESRP2 was depleted (Figure 4, E and F). We tested the combination of ESRP1 and ESRP2 siRNAs to determine whether depletion of both ESRPs would further increase the levels of isoform 1A. However, co-knockdown of ESRP1 and 2 resulted in an increase similar to that of the knockdown of ESRP1 alone (Supplemental Figure S3), suggesting that ESRP1 is more likely to regulate p120-catenin isoform switching in SW480 cells. Interestingly, depletion of ZEB1 resulted in increased ESRP2 but not ESRP1 in SW480 cells, but the knockdown of ZEB1 did not affect p120-isoform distribution (Supplemental Figure S4).
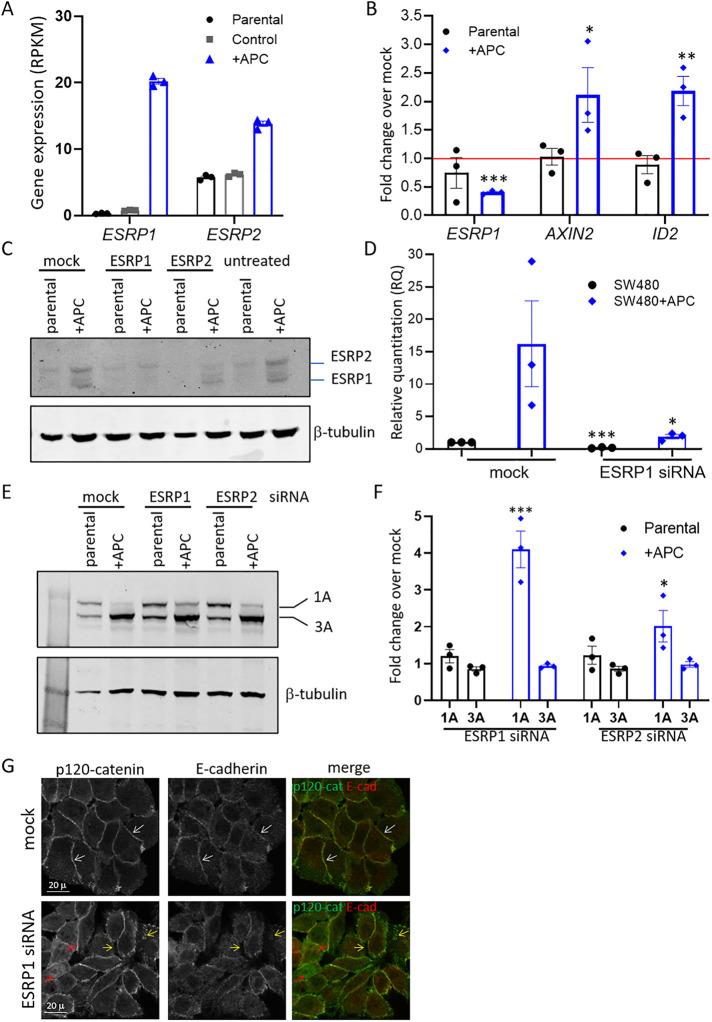
FIGURE 4:
ESRP1 is regulated by APC and promotes p120-catenin isoform switching in SW480+APC cells. (A) Increased expression of ESRP1 and ESRP2 transcript in SW480+APC cells from RNAseq analysis of SW480 parental, SW480 control, and SW480+APC (APC+) cells. Graph shows mean ± SD gene expression (RPKM) (n = 3). (B) siRNA-mediated depletion of APC results in reduced ESRP1 mRNA levels and increased Wnt target gene expression. qRT-PCR analysis of ESRP1, AXIN2, and ID2 mRNA in APC siRNA transfected SW480 parental and SW480+APC cells. Data are expressed as fold change over mock control transfected cells, mean ± SEM from three independent experiments, *P < 0.05, **P < 0.001, ***P < 0.005, unpaired Student’s t test. (C) Increased ESRP1 and ESRP2 in SW480+APC cells is reduced following siRNA-mediated depletion of ESRP1 and ESRP2, respectively. Immunoblot analysis of ESRP1/2. β-tubulin was used as a loading control. Shown is representative of three independent experiments. (D) siRNA-mediated depletion of ESRP1. SW480 parental and SW480+APC cells were transfected with siRNAs targeting ESRP1. Graph shows quantitation of ESRP1 mRNA assessed by qRT-PCR for mean ± SEM, n = 3. *P < 0.05, ***P < 0.005, unpaired Student’s t test. (E) Depletion of ESRP1 by siRNA results in increased p120-catenin isoform 1A in SW480+APC cells. Immunoblot analysis of p120-catenin. β-tubulin was used as a loading control. (F) Quantitation of p120-catenin isoforms 1A and 3A normalized to β-tubulin in ESRP1 and ESRP2- depleted cells expressed as fold change over mock-transfected cells. Shown is mean ± SEM, n = 3, *P < 0.05, ***P < 0.005, Student’s t test. (G) Depletion of ESRP1 by siRNA results in reduced cell–cell contact staining in SW480+APC cells. Mock or ESRP1 siRNA–treated cells were stained with antibodies to p120-catenin and E-cadherin. Shown are confocal sections of the fluorescence images for the p120-catenin (p120-cat) green and E-cadherin (Ecad) red, in merged image. Cell–cell junctions (white arrows), disrupted junctional staining (yellow arrows), and p120-catenin nuclear staining (red arrows) are indicated. Scale bars 20 μm.
We investigated junctional p120-catenin and E-cadherin staining in SW480+APC cells treated with ESRP1 siRNA. The SW480+APC cells show evident cell–cell contact staining with junctional markers such as E-cadherin, ZO1, and p120-catenin, consistent with an epithelial phenotype (see Figures 2A and 4G). p120-catenin junctional staining was disrupted upon ESRP1 depletion with less tightly packed cells and an increase in nuclear signal (Figure 4G). The increased nuclear signal in ESRP1-depleted cells resembles the p120-catenin distribution in the SW480 parental cells (Figure 2A). Similarly, E-cadherin staining at cell junctions was reduced. This is in contrast to mock-treated (control) cells, which display p120-catenin and E-cadherin at sites of cell adhesion (Figure 4G). Thus treatment with ESRP1 siRNAs partially reverted the epithelial phenotype and cell–cell junction morphology toward a more mesenchymal phenotype. Collectively these data show that restoring full-length APC expression in SW480 cells promotes ESRP1/2 expression leading to altered epithelial p120-catenin isoform expression and that ESRP1 depletion shifts the splicing program to partially mesenchymal.
p120-catenin isoform expression in colon cancer cells
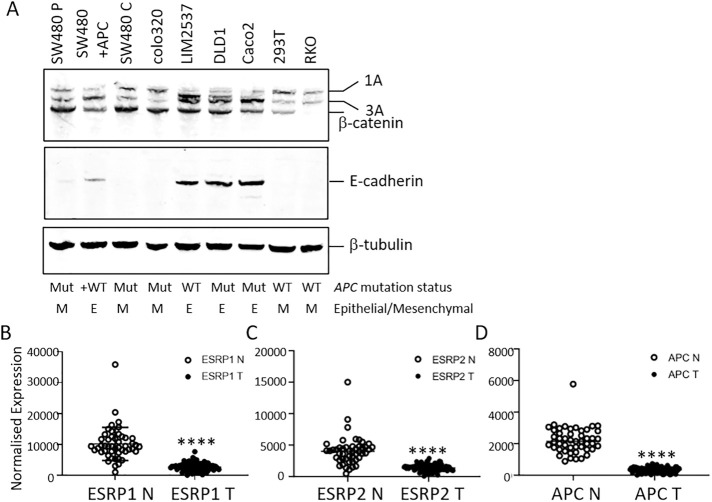
Expression of wild-type full-length APC in SW480 cells leads to increased expression of ESRP1 and ESPR2, suggesting that APC can regulate ESRP1/2 expression and thereby regulate isoform splicing, including p120-catenin. The corollary of this observation is that truncated APC is unable to regulate ESRP and p120-catenin isoform splicing. To investigate the contribution of APC to ESRP and isoform distribution, we investigated p120-catenin isoform expression in a panel of cell lines (Figure 5A). In cells with APC mutations (Colo320, DLD1, Caco2), p120-catenin isoform 1A is expressed: in Colo320 cells isoform 1A is the dominant isoform. In contrast, LIM2537 cells, which contain full-length APC, predominantly express isoform 3A, similar to SW480+APC cells (Figure 5A). This correlates with ESRP1 and ESRP2 transcript analysis in these cells (Wang et al., 2017) and supports our findings in SW480/SW480+APC cells. Both DLD1and Caco2 cells express a higher level of the epithelial 3A isoform compared with 1A (Figure 5A). These cells contain mutated APC but also demonstrate an epithelial morphology with intact cell adhesion junctions (Hidalgo et al., 1989; Tanaka et al., 2016). A defining feature of epithelial cells is E-cadherin expression at cell–cell junctions, which is absent in mesenchymal cells, and this correlates with ESRP and epithelial isoform expression (Warzecha et al., 2009, 2010; Baum and Georgiou, 2011). To assess whether the morphology of the cells was indicative of the isoform distribution, we analyzed E-cadherin expression in the cell lines (Figure 5A). E-cadherin is expressed in cells with an epithelial phenotype that also express higher levels of isoform 3A. Moreover, cells with a mesenchymal phenotype, including RKO and HEK293 (Buck et al., 2007; Warzecha et al., 2009; Ayinde et al., 2017), show a similar distribution of p120-catenin isoforms to SW480 cells and also express low levels of E-cadherin (Figure 5A) and ESRP (Warzecha et al., 2009, 2010; Wang et al., 2017). The distribution of p120-catenin isoforms therefore correlates well with E-cadherin expression and underscores the importance of ESRP and epithelial isoform expression in governing the epithelial phenotype.
FIGURE 5:
p120-catenin isoform distribution and ESRP1 expression in CRC. (A) p120-catenin isoform distribution in a panel of cell lines. Blots were probed with antibodies to p120-catenin, β-catenin, E-cadherin, and β-tubulin. The APC mutation status and epithelial/mesenchymal phenotype of each cell line are indicated. (B–D) ESRP1 (B), ESRP2 (C), and APC (D) transcript levels in normal colon (N) and CRC primary tumors (T). N n = 48, T n = 87; ****P < 0.0001, unpaired t test with Welch’s correlation.
ESRP1 and ESRP2 are reduced in primary CRC
We next explored whether the alterations in ESRP1 and ESRP2 observed in SW480 cells are representative of APC loss of heterozygosity (LOH) in CRCs. We interrogated RNA transcript data from 87 primary CRCs and 48 normal colon tissues (Wang et al., 2017) for expression of ESRP1, ESRP2, and APC (Figure 5, B–D). ESRP1 transcript levels were significantly higher in normal colon tissue in comparison to CRC tumor samples (Figure 5B), consistent with our findings in SW480+APC rescue cells compared with SW480 CRC cells. Similarly, ESRP2 showed a robust increase in normal tissue compared with CRC tumor tissue, albeit with lower overall expression (Figure 5C). As most CRCs (>70%) have truncating mutations in the APC gene and frequently demonstrate LOH or nonsense-mediated decay, APC transcript levels were significantly reduced in CRC tumor tissue in comparison to normal colon (Figure 5D). Moreover, we found significant correlation between ESRP1 and APC transcript levels in normal colon (Pearson r 0.6943, P < 0.0001) and in primary CRC tissue (Pearson r 0.4752, P < 0.0001). While the correlation between ESRP2 and APC was significant in normal colon (Pearson r 0.5649, P < 0.0001), there was no correlation with ESRP2 and APC in primary CRC samples (Pearson r 0.111, P = 0.3060). This may be reflected in the larger changes in ESRP1 compared with ESRP2 in SW480/SW480+APC cells. The reduced ESRP1 transcript in primary CRC and correlation with APC indicate a role for APC in regulation of ESRP1 and subsequent epithelial gene splicing events.
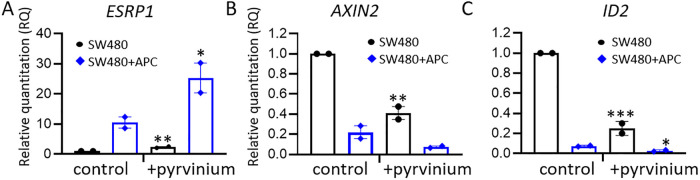
Wnt signaling inhibition promotes ESRP1 expression
To investigate the Wnt regulatory role, SW480 and SW480+APC cells were treated with the Wnt signaling inhibitor pyrvinium pamoate (Thorne et al., 2010) and the effect on the ESRP1 transcript level assessed. Wnt inhibition stimulated ESRP1 expression in both SW480 and SW480+APC cells (Figure 6A) with concomitant reductions in the expression of Wnt target genes, for example, AXIN2 and ID2 (Figure 6, B and C), but there was only a modest effect on the p120-catenin isoform distribution (Supplemental Figure S5). These results suggest that APC-mediated negative regulation of Wnt signaling promotes the expression of ESRP1 and consequentially p120-catenin switching to isotype 3A, which is associated with increased cell–cell adhesion and the epithelial phenotype (Figure 7).
FIGURE 6:
Inhibition of Wnt signaling promotes ESRP1 expression. SW480 parental and SW480+APC cells were treated with either vehicle control or Wnt inhibitor pyrvinium pamoate (1 μM, 18 h) and mRNA levels (RQ) assessed by qRT-PCR for ESRP1 (A), AXIN2 (B), and ID2 (C). Graphs show mean ± SEM, n = 2, *P < 0.05, **P < 0.01, ***P < 0.005, unpaired Student’s t test.
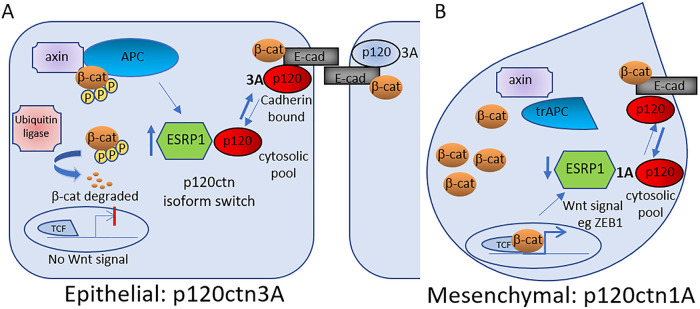
FIGURE 7:
Model for APC regulation of p120-catenin isoform splicing and cell–cell adhesion. Intact APC inhibits Wnt/β-catenin signaling and can promote ESRP1, which controls p120-catenin isoform splicing and contributes to maintaining cell–cell adhesion and an epithelial phenotype (A). Truncated APC leads to increased Wnt/β-catenin signaling and reduced ESRP1 and isoform switching and a mesenchymal cell type (B).
DISCUSSION
We have identified that APC is a modulator of cell–cell adhesion through altered expression of cell–cell adhesion genes and the master epithelial splice regulator ESRP1. We had previously shown that expression of full-length APC in SW480 cells eradicates tumor growth and re-establishes cadherin-mediated cell–cell adhesion (Faux et al., 2004). Here we show a novel role for APC in promoting ESRP1 expression, which in turn modifies the splicing of p120-catenin. Expression of the epithelial p120-catenin isoform promotes cell–cell adhesiveness by stabilization of E-cadherin, allowing reactivation of E-cadherin function (Ireton et al., 2002; Davis et al., 2003). We propose that ESRP1 gene expression is regulated by APC and that the lack of ESRP1-mediated splicing of p120-catenin reduces cell–cell adhesion in CRC.
In this report we show that in addition to regulating Wnt signaling genes (King et al. 2016), APC regulates the expression of EMT-associated genes. These alterations could either be a consequence of Wnt signaling (Brembeck et al., 2006) or a direct effect of APC on cytoskeletal-dependent processes (McCartney and Nathke, 2008; Nelson and Nathke, 2013). The APC-induced changes in cell–cell adhesion may not be a direct outcome of effects of Wnt signaling. A Wnt-independent function is suggested by the increase in p120-catenin 1A following APC depletion even in cells containing only truncated APC. The master splicing regulator, ESRP1, has not previously been reported as a Wnt target gene (Nusse and Clevers, 2017) and yet is among the most abundantly increased genes when full-length APC is expressed in SW480 cells. It is intriguing that ESRP1 expression is increased further by inhibition of Wnt signaling, but this did not translate into detectable changes in p120-catenin isoform switching. ESRP1 regulates splicing of p120-catenin, which functions to stablize E-cadherin at cell junctions (Ireton et al., 2002; Warzecha et al., 2009). Thus, APC expression effectively reduces Wnt signaling but also has effects on cell–cell adhesion and EMT, underscoring the impact of APC mutation on a range of cellular processes important in tumorigenesis (Hankey et al., 2018).
Despite efficient depletion of ESRP1 (>80%) and reversion to p120-catenin isoform 1A, we detected only modest decreases in isoform 3A and E-cadherin. Similarly, Warzecha et al. (2010) did not detect a change in E-cadherin upon ESRP1 knockdown, and ESRP1 overexpression results in an attenuated response (Shapiro et al., 2011; Horiguchi et al., 2012). We cannot rule out the possibility that other genes, such as transcriptional repressor ZEPPO, are involved in repression of E-cadherin and p120-catenin isoform regulation (Slorach et al., 2011). However, the striking increase in ESRP1 expression reported here, along with its function in isoform splicing (Warzecha et al., 2009, 2010; Lamouille et al., 2014; Pradella et al., 2017), point to ESRP1 as a key mechanism for promoting p120-catenin isoform splicing in the SW480+APC cells.
In addition to modulation of ESRP1 by re-expression of APC or APC depletion, we show striking decreases in ESRP1 and 2 in primary CRC and detected that ESRP1 can be increased directly by inhibition of Wnt signaling. ESRP2 expression was also increased in SW480+APC cells but to a lesser extent than ESRP1; the depletion of ESRP2 resulted in only a modest increase in the level of the p120-catenin isoform 1A, and the combination of ESRP1 and ESRP2 siRNA did not result in any further increase. Significantly, increased expression of ESRP1 and ESRP2 has been shown to correlate with epithelial splice signatures and favorable CRC outcomes (Deloria et al., 2016; Mager et al., 2017). ESRP1 expression is reduced in a number of other cancers, including head and neck (Ishii et al., 2014), breast (Fici et al., 2017), ovarian (Jeong et al., 2017), pancreas (Ueda et al., 2014), and lung (Walser et al., 2018), in which it is recognized as a favorable prognostic factor for patients with metastatic disease. The transcriptional repressors SNAI and ZEB1 as well as the miR200 family have been implicated in regulation of ESRP expression (Haraguchi et al., 2016; Larsen et al., 2016; Walser et al., 2018). Indeed the high levels of ESRP1 with correspondingly low ZEB1 in SW480+APC cells are consistent with direct regulation of ESRP1 by ZEB1; however, knockdown of ZEB1 resulted in an increased level of ESRP2 but not ESRP1. Wnt inhibition did modulate ESRP1 expression further, indicating that ESRP1 can be controlled by a Wnt-ZEB1-axis. Our findings implicate APC and Wnt signaling in EMT processes through alterations in transcription as well as isoform splicing of cell–cell adhesion genes.
In conclusion, the restoration of full-length APC in SW480 colon cancer cells that express only truncated APC returns the cells to an epithelial phenotype with functional cell–cell adhesion junctions. In addition to the well-characterized changes in β-catenin and Wnt signaling targets, we report significant changes in cell–cell adhesion and other regulatory signatures. In particular, we identified ESRP1 as a key gene involved in the regulation of p120-catenin isoform splicing. These data imply that the alteration in p120-catenin splicing, upon re-expression of full-length APC, contributes to E-cadherin–mediated cell–cell adhesion. While APC’s role as a negative regulator of Wnt signaling is important, the impact of APC on Wnt-independent pathways such as cell–cell adhesion signaling needs to be considered. The switch from an epithelial to a mesenchymal-type tumor cell can involve significant changes in many signaling and regulatory pathways, including the levels of ESRP1 and ESRP2 (Warzecha et al., 2009). ESRP1 is not usually considered a Wnt target gene (Cong et al., 2004) and does not appear to be influenced directly by changes in cell–cell signaling. Presumably, complex responses to changes in kinase activity (e.g., CK1 and GSK3) and location will have Wnt-independent effects on cell morphology, metabolism, and motility. The levels of cytoplasmic β-catenin and the interactions of β-catenin with proteins in the nucleus, cytoplasm, and cell membrane change with tyrosine kinase activation and cell–cell adhesion. The consequential effects of the APC-induced p120-catenin isoform switch on E-cadherin turnover and cell–cell adhesion may be separate from Wnt-driven events but significant contributors to the suppression of the tumor phenotype.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Antibodies
The following antibodies were used: anti–E-cadherin (HECD1 #ab1416; Abcam; 24E10 #3195; Cell Signaling; #610182; BD Transduction Laboratories), anti–claudin-7 (#349100; Invitrogen), anti–p120-catenin (#610134; BD Transduction Laboratories), anti-ZO1 (#61096; BD Transduction Laboratories), anti–β-tubulin (AA2 #T8328; Sigma Aldrich), anti-APC (H290; Santa Cruz), anti–β-catenin (#610153; BD Transduction Laboratories), and anti-ESRP1/2 (Rockland). Antibodies were used at 1:1000 for immunoblot analysis and 1:200 for immunostaining, unless otherwise stated. Secondary antibodies for immunoblots were Alexa488 goat anti-mouse/rabbit (#A-11001 and #A-11035; Thermo Scientific) and Alexa546 goat anti-mouse/rabbit (#A-11030 and #A-11035; Thermo Scientific), used at 1:10,000.
Cell culture conditions
SW480 cells (from the American Type Culture Collection, SW480 control, and SW480+APC were authenticated (King et al., 2016) and were cultured in RPMI supplemented with 1.08% thioglycerol, 50 mg/ml hydrocortisone, 100 U/ml insulin, and 10% fetal calf serum (FCS) plus 1.5 mg/ml G418 for the control and +APC cells. Colo320, HEK293, RKO, DLD1, Caco2, and LIM2537 cells were cultured in DME supplemented with 10% FCS. Cells were tested for mycoplasma every 4–6 mo. All cell lines were authenticated by STR (short tandem repeat) profiling analysis at the Australian Genome Research Facility (AGRF) (Parkville, VIC, Australia) using the GenePrint 10 System (Promega) (Wang et al., 2017).
RNAseq analysis
RNA from SW480, SW480+APC, and SW480 control cells were sequenced and analyzed as described (King et al., 2016). Differential gene expression analysis for cell adhesion and EMT-specific genes was conducted using read counts with the Bioconductor edgeR package as described (King et al., 2016).
Cell based assays
Cells were cultured and transfected with siRNA using Lipofectamine 2000 (Thermo Fisher). The following siRNA were used: APC (ON-TARGETplus L-03869), ESRP1 (ON-TARGETplus L-20672-01), ESRP2 (ON-TARGETplus L-014523), and control ON-TARGETplus nontargeting pool D-001810-10) (all from Dharmacon). Cells were harvested for immunoblot analysis or immunostained for confocal microscopy acquired with an Olympus confocal microscope. Cells were counterstained with fluorescently labeled 4’,6-diamidino-2-phenylindole (DAPI) (#10236276001; Roche Diagnostics). For immunoblot analysis, the levels of protein were quantified using densitometry normalized to β-tubulin.
Quantitative real-time PCR analysis
mRNA was extracted and purified (Illustra RNAspin Mini Kit #25-0500-70; GE LifeSciences) as per the manufacturer’s instructions. Reverse transcription was performed using the High Capacity cDNA Reverse Transcription Kit (#4368814 AB; Applied Biosystems). Real-time PCR amplification was achieved using PowerSYBR Green PCR Master Mix (#4367659; Applied Biosystems) and Taqman Fast Advanced Master Mix (ThermoFisherScientific Applied Biosystems; 4444557) reagents. The SYBR green primers were as follows: AXIN2: (#QT00037639; Qiagen); Primer sequences used: GAPDH FWD: CAATGACCCCTTCATTGACC, REV: TGATGACAAGCTTCCCGTTC; ESRP1 FWD: TGCTGTTCTGGAAAGTCGTG, REV: CTTTGCAGTCCTCCGTCAGT; ID2 FWD: TCAGCCTGCATCACCAGAGA, REV: CTGCAAGGACAGGATGCTGATA. The Taqman probes were CTNND1 (HS00931672_m1; HS00931681_m1; HS00931673_m1; HS00609741_m1; HS00609742_m1; HS00931670_m1; HS00931671_m1), and GAPDH (HS99999905_m1). SYBR green samples were analyzed on a 7300 Real-Time PCR system (Applied Biosystems) using SDS software version 4.0 (Applied Biosystems). The Taqman samples were run on a Viia7 Real-Time PCR System (ThermoFisherScientific). Target gene expression is expressed relative to the housekeeping gene expression (GAPDH) and is presented in arbitrary units (2(-ΔCt)).
Statistical analysis
Statistical analyses were performed using a Student’s t test, unless otherwise described. Data presented graphically are the means ± SEM for three independent experiments unless otherwise stated.
Supplementary Material
Acknowledgments
This work was supported by National Health and Medical Research Council (NH&MRC) Program Grant #487922. The funding body had no role in the design of the study, collection, analysis, or interpretation of data or in writing the manuscript. This work was also supported by the Walter and Eliza Hall Institute. We thank the members of the Burgess laboratory for critical evaluation of the data.
Abbreviations used:
- APC
adenomatous polyposis coli
- CDH1
E-cadherin
- CDH3
P-cadherin
- CRC
colorectal cancer
- CTNND1
catenin delta (p120-catenin)
- EMT
epithelial-to-mesenchymal transition
- ESRP1
epithelial splice regulatory protein 1
- ID2
DNA-binding protein inhibitor 2
- ZEB1
Zinc Finger E-Box Binding Homeobox 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-05-0321) on November 25, 2020.
REFERENCES
- Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford HC, Zheng Y, Reynolds AB (2000). Inhibition of RhoA by p120 catenin. Nat Cell Biol 2, 637–644. [DOI] [PubMed] [Google Scholar]
- Ayinde O, Wang Z, Griffin M (2017). Tissue transglutaminase induces epithelial-mesenchymal-transition and the acquisition of stem cell like characteristics in colorectal cancer cells. Oncotarget 8, 20025–20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B, Georgiou M (2011). Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol 192, 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Rosario M, Birchmeier W (2006). Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev 16, 51–59. [DOI] [PubMed] [Google Scholar]
- Buck E, Eyzaguirre A, Barr S, Thompson S, Sennello R, Young D, Iwata KK, Gibson NW, Cagnoni P, Haley JD (2007). Loss of homotypic cell adhesion by epithelial-mesenchymal transition or mutation limits sensitivity to epidermal growth factor receptor inhibition. Mol Cancer Ther 6, 532–541. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R (2012). Wnt/beta-catenin signaling and disease. Cell 149, 1192–1205. [DOI] [PubMed] [Google Scholar]
- Cong F, Schweizer L, Varmus H (2004). Casein kinase Iepsilon modulates the signaling specificities of dishevelled. Mol Cell Biol 24, 2000–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB (2003). A core function for p120-catenin in cadherin turnover. J Cell Biol 163, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloria AJ, Hoflmayer D, Kienzl P, Lopatecka J, Sampl S, Klimpfinger M, Braunschmid T, Bastian F, Lu L, Marian B, et al. (2016). Epithelial splicing regulatory protein 1 and 2 paralogues correlate with splice signatures and favorable outcome in human colorectal cancer. Oncotarget 7, 73800–73816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, Lowe SW (2015). Apc restoration promotes cellular differentiation and reestablishes Crypt homeostasis in colorectal cancer. Cell 161, 1539–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faux MC, Ross JL, Meeker C, Johns T, Ji H, Simpson RJ, Layton MJ, Burgess AW (2004). Restoration of full-length adenomatous polyposis coli (APC) protein in a colon cancer cell line enhances cell adhesion. J Cell Sci 117, 427–439. [DOI] [PubMed] [Google Scholar]
- Fici P, Gallerani G, Morel AP, Mercatali L, Ibrahim T, Scarpi E, Amadori D, Puisieux A, Rigaud M, Fabbri F (2017). Splicing factor ratio as an index of epithelial-mesenchymal transition and tumor aggressiveness in breast cancer. Oncotarget 8, 2423–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles RH, van Es JH, Clevers H (2003). Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta 1653, 1–24. [DOI] [PubMed] [Google Scholar]
- Gul IS, Hulpiau P, Saeys Y, van Roy F (2017). Evolution and diversity of cadherins and catenins. Exp Cell Res 358, 3–9. [DOI] [PubMed] [Google Scholar]
- Hankey W, Frankel WL, Groden J (2018). Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: implications for therapeutic targeting. Cancer Metastasis Rev 37, 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Kondo M, Uchikawa R, Kobayashi K, Hiramatsu H, Kobayashi K, Chit UW, Shimizu T, Iba H (2016). Dynamics and plasticity of the epithelial to mesenchymal transition induced by miR-200 family inhibition. Sci Rep 6, 21117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo IJ, Raub TJ, Borchardt RT (1989). Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 96, 736–749. [PubMed] [Google Scholar]
- Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, Fujii H, Yamaguchi A, Miyazawa K, Miyazono K, Saitoh M (2012). TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene 31, 3190–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, He X (2008). Wnt/beta-catenin signaling: new (and old) players and new insights. Curr Opin Cell Biol 20, 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, Anastasiadis PZ, Matrisian L, Bundy LM, Sealy L, et al. (2002). A novel role for p120 catenin in E-cadherin function. J Cell Biol 159, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S, Motizuki M, Masuyama K, Miyazawa K (2014). Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem 289, 27386–27399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HM, Han J, Lee SH, Park HJ, Lee HJ, Choi JS, Lee YM, Choi YL, Shin YK, Kwon MJ (2017). ESRP1 is overexpressed in ovarian cancer and promotes switching from mesenchymal to epithelial phenotype in ovarian cancer cells. Oncogenesis 6, e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirsebilck A, Bonne S, Staes K, van Hengel J, Nollet F, Reynolds A, van Roy F (1998). Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics 50, 129–146. [DOI] [PubMed] [Google Scholar]
- King LE, Love CG, Sieber OM, Faux MC, Burgess AW (2016). Differential RNA-seq analysis comparing APC-defective and APC-restored SW480 colorectal cancer cells. Genom Data 7, 293–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B (1996). Lessons from hereditary colorectal cancer. Cell 87, 159–170. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15, 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, Dospoy PD, Augustyn A, Hight SK, Sato M, et al. (2016). ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest 126, 3219–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Ji H, Furuta Y, Park JI, McCrea PD (2014). p120-catenin regulates REST and CoREST, and modulates mouse embryonic stem cell differentiation. J Cell Sci 127, 4037–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Nusse R (2004). The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20, 781–810. [DOI] [PubMed] [Google Scholar]
- Mager LF, Koelzer VH, Stuber R, Thoo L, Keller I, Koeck I, Langenegger M, Simillion C, Pfister SP, Faderl M, et al. (2017). The ESRP1-GPR137 axis contributes to intestinal pathogenesis. eLife 6, ee28366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney BM, Nathke IS (2008). Cell regulation by the Apc protein Apc as master regulator of epithelia. Curr Opin Cell Biol 20, 186–193. [DOI] [PubMed] [Google Scholar]
- Mo YY, Reynolds AB (1996). Identification of murine p120 isoforms and heterogeneous expression of p120cas isoforms in human tumor cell lines. Cancer Res 56, 2633–2640. [PubMed] [Google Scholar]
- Nanes BA, Chiasson-MacKenzie C, Lowery AM, Ishiyama N, Faundez V, Ikura M, Vincent PA, Kowalczyk AP (2012). p120-catenin binding masks an endocytic signal conserved in classical cadherins. J Cell Biol 199, 365–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathke IS, Adams CL, Polakis P, Sellin JH, Nelson WJ (1996). The adenomatous polyposis coli tumor suppressor protein localizes to plasma membrane sites involved in active cell migration. J Cell Biol 134, 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S, Nathke IS (2013). Interactions and functions of the adenomatous polyposis coli (APC) protein at a glance. J Cell Sci 126, 873–877. [DOI] [PubMed] [Google Scholar]
- Nelson WJ (2008). Regulation of cell-cell adhesion by the cadherin-catenin complex. Biochem Soc Trans 36, 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Liu BP, Burridge K, Kreft B (2000). p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J Cell Biol 150, 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R, Clevers H (2017). Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999. [DOI] [PubMed] [Google Scholar]
- Nusse R, Varmus HE (1992). Wnt genes. Cell 69, 1073–1087. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, Kemler R (1989). The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J 8, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Bhattacharya S, Kalyan G, Hazra S (2018). Cadherin profiling for therapeutic interventions in epithelial mesenchymal transition (EMT) and tumorigenesis. Exp Cell Res 368, 137–146. [DOI] [PubMed] [Google Scholar]
- Park JI, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, Barton MC, Deroo T, Vleminckx K, Moon RT, McCrea PD (2005). Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev Cell 8, 843–854. [DOI] [PubMed] [Google Scholar]
- Paulsen JE, Steffensen IL, Loberg EM, Husoy T, Namork E, Alexander J (2001). Qualitative and quantitative relationship between dysplastic aberrant crypt foci and tumorigenesis in the Min/+ mouse colon. Cancer Res 61, 5010–5015. [PubMed] [Google Scholar]
- Peifer M, Polakis P (2000). Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science 287, 1606–1609. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E (2006). p120-catenin mediates inflammatory responses in the skin. Cell 124, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters T, Goossens S, Haenebalcke L, Andries V, Stryjewska A, De Rycke R, Lemeire K, Hochepied T, Huylebroeck D, Berx G, et al. (2016). p120 catenin-mediated stabilization of e-cadherin is essential for primitive endoderm specification. PLoS Genet 12, e1006243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters T, van Roy F, van Hengel J (2012). Functions of p120ctn isoforms in cell-cell adhesion and intracellular signaling. Front Biosci (Landmark Ed) 17, 1669–1694. [DOI] [PubMed] [Google Scholar]
- Pradella D, Naro C, Sette C, Ghigna C (2017). EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer 16, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Clevers H (2005). Wnt signalling in stem cells and cancer. Nature 434, 843–850. [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, et al. (2004). Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev 18, 1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel B, Braeg S, Adler G, Wedlich D, Menke A (2004). E- and N-cadherin differ with respect to their associated p120ctn isoforms and their ability to suppress invasive growth in pancreatic cancer cells. Oncogene 23, 5532–5542. [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB (2011). An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7, e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slorach EM, Chou J, Werb Z (2011). Zeppo1 is a novel metastasis promoter that represses E-cadherin expression and regulates p120-catenin isoform expression and localization. Genes Dev 25, 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley-Freed WG, Efimov A, Burnett PE, Short SP, Davis MA, Gumucio DL, Washington MK, Coffey RJ, Reynolds AB (2010). p120-catenin is essential for maintenance of barrier function and intestinal homeostasis in mice. J Clin Invest 120, 1824–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, Jung A, Kirchner T, Brabletz T (2006). A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology 131, 830–840. [DOI] [PubMed] [Google Scholar]
- Takeichi M (2014). Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol 15, 397–410. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kobayashi W, Haraguchi M, Ishihata K, Nakamura N, Ozawa M (2016). Snail1 expression in human colon cancer DLD-1 cells confers invasive properties without N-cadherin expression. Biochem Biophys Rep 8, 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS, Jernigan KK, Meyers KC, Hang BI, Waterson AG, et al. (2010). Small-molecule inhibition of Wnt signaling through activation of casein kinase 1alpha. Nat Chem Biol 6, 829–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J, Matsuda Y, Yamahatsu K, Uchida E, Naito Z, Korc M, Ishiwata T (2014). Epithelial splicing regulatory protein 1 is a favorable prognostic factor in pancreatic cancer that attenuates pancreatic metastases. Oncogene 33, 4485–4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser TC, Jing Z, Tran LM, Lin YQ, Yakobian N, Wang G, Krysan K, Zhu LX, Sharma S, Lee MH, et al. (2018). Silencing the Snail-dependent RNA splice regulator ESRP1 drives malignant transformation of human pulmonary epithelial cells. Cancer Res 78, 1986–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mouradov D, Wang X, Jorissen RN, Chambers MC, Zimmerman LJ, Vasaikar S, Love CG, Li S, Lowes K, et al. (2017). Colorectal cancer cell line proteomes are representative of primary tumors and predict drug sensitivity. Gastroenterology 153, 1082–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP (2010). An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J 29, 3286–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP (2009). ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, Anastasiadis PZ (2008). A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J Biol Chem 283, 18344–18354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Niessen CM, Gumbiner BM (1998). The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J Cell Biol 141, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao Y, Jiang G, Zhang X, Zhao H, Wu J, Xu K, Wang E (2014). Impact of p120-catenin isoforms 1A and 3A on epithelial mesenchymal transition of lung cancer cells expressing E-cadherin in different subcellular locations. PLoS One 9, e88064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.