Abstract
Many lysosome functions are determined by a lumenal pH of ∼5.0, including the activity of resident acid-activated hydrolases. Lysosome pH (pHlys) is often increased in neurodegenerative disorders and predicted to be decreased in cancers, making it a potential target for therapeutics to limit the progression of these diseases. Accurately measuring pHlys, however, is limited by currently used dyes that accumulate in multiple intracellular compartments and cannot be propagated in clonal cells for longitudinal studies or used for in vivo determinations. To resolve this limitation, we developed a genetically encoded ratiometric pHlys biosensor, pHLARE (pH Lysosomal Activity REporter), which localizes predominantly in lysosomes, has a dynamic range of pH 4.0 to 6.5, and can be stably expressed in cells. Using pHLARE we show decreased pHlys with inhibiting activity of the mammalian target of rapamycin complex 1 (mTORC1). Also, cancer cells from different tissue origins have a lower pHlys than untransformed cells, and stably expressing oncogenic RasV12 in untransformed cells is sufficient to decrease pHlys. pHLARE is a new tool to accurately measure pHlys for improved understanding of lysosome dynamics, which is increasingly considered a therapeutic target.
INTRODUCTION
Lysosomes function as catabolic hubs independently as well as downstream of autophagy and nutrient sensing by mammalian target of rapamycin complex 1 (mTORC1). Additionally, lysosomes contribute to trafficking of intracellular vesicles, plasma membrane repair, pathogen degradation, resistance to chemotherapies, and a broad range of homeostatic responses to environmental cues (Xu and Ren, 2015; Perera and Zoncu, 2016). The lumenal pH of lysosomes (pHlys) is a major determinant of many lysosome functions, including catabolism by lumenal acid-activated hydrolyases (Mindell, 2012), fusion with endosomes and cargo sorting (Marshansky and Futai, 2008; Scott and Gruenberg, 2011), and roles in Ca2+ homeostasis (Lee et al., 2015). Although pHlys in normal cells is thought to be tightly regulated at ∼5.0, it is increasingly recognized to be dysregulated in diseases. Dysregulated lysosomes are common in neurodegenerative disorders (Nixon, 2013). Although controversial, increased pHlys is suggested with neurodegeneration (Majumdar et al., 2007; Wolfe et al., 2013; Lee et al., 2015), which is predicted to attenuate activity of lumenal acid-activated hydrolases and decrease protein degradation leading to protein aggregation. Increased pHlys is also reported with diabetic nephropathy (Liu et al., 2015) and is a determinant in some pathologies of lysosomal storage diseases (Colacurcio and Nixon, 2016) and in osteopetrosis (Kornak et al., 2001). In contrast, decreased pHlys may occur in cancers compared with untransformed cells, based on changes in autophagosome activity (Kenific and Debnath, 2015), roles in multidrug resistance (Daniel et al, 2013; Zhitomirsky and Assaraf, 2016), and reversed pHlys and cytosolic pH dynamics (Liu et al., 2018), with the latter confirmed to be higher in most cancers (Webb et al., 2011; White et al., 2017b). A lower pHlys in cancer would be consistent with cancer cells having increased lysosomal catabolism of macromolecules (Perera and Zoncu, 2016), which likely contributes to metabolic reprogamming for increasing biomass to fuel rapid proliferation. Accordingly, targeting mechanisms of dysregulated pHlys dynamics is proposed as a therapeutic approach to limit some disease pathologies (Appelqvist et al., 2013; Wolfe et al., 2013; Fraldi et al., 2016; Piao and Amaravadi, 2016).
Accurately determining pHlys, however, is currently limited by the lack of selective and effective probes, particularly for longitudinal or in vivo studies. This is in contrast to a number of highly reliable reagents for accurately measuring cytosolic pH, including fluorescent dyes and genetically encoded biosensors (Grillo-Hill et al., 2014). Commonly used reagents for measuring pHlys have a number of caveats (Wolfe et al., 2013). Fluorescent dextran conjugates are taken up by endocytosis or macropinocytosis and accumulate in lysosomes but also other types of intracellular vesicles. Oregon green dextran conjugates are an improvement over the originally used fluorescein dextran that has a low signal in acidic compartments, photobleaches rapidly, and are ratiometric for quantitative measurements (Johnson et al., 2016). However, all dextran conjugates require >24 h pulse-chase labeling protocols through endo-lysosomal trafficking and cannot be propagated in cells (Nilsson et al., 2010; Wolfe et al, 2013; Johnson et al., 2016). Using the membrane permeant dyes LysoTracker and LysoSensor that accumulate in acidic compartments resolve caveats with uptake; however, they have considerable background fluorescence outside lysosomes. LysoTracker, however, is not ratiometric and hence allows only qualitative determinations. Lysosensor is ratiometric and it has the best dynamic range of dyes used to measure pHlys, but it is not suitable for measurements of more than a few minutes because it induces an increase in pHlys (Wolfe et al, 2013). Additionally, because LysoSensor and LysoTracker are substrates for P-glycoprotein and actively transported out of cells (Zhitomirsky et al., 2018), their use in determining the role of pHlys in resistance to cancer therapies is problematic. Nanoprobes that are pH sensitive have also been used for determining organelle pH, but lysosome targeting remains problematic, and a recently developed two-photon fluorophore conjugated to a lysosome targeting morpholine (Wang et al., 2018) achieves specificity, but cellular uptake is similar to dyes and nanoprobes and as reported is not quantitative.
For an improved method to accurately and selectively quantify pHlys in real-time over long time periods, we developed a genetically encoded and ratiometric pHlys biosensor designated pHLARE (pH Lysosome Activity REporter). Here, we describe the pHLARE design of the lysosomal-associated membrane protein 1 (LAMP1) tagged at lumenal and cytosolic domains with distinct fluorophores, validate that pHLARE is predominantly localized in lysosomes and has strong sensitivity with a broad dynamic range, and use pHLARE to show changes in pHlys with known pharmacological regulators, including a decrease with inhibiting mTORC1 activity. We also confirm a lower pHlys in clonal cancer cells from different tissue backgrounds and with different mutational signatures as well as epithelial cells expressing oncogenic RasV12.
RESULTS
pHLARE design and validation
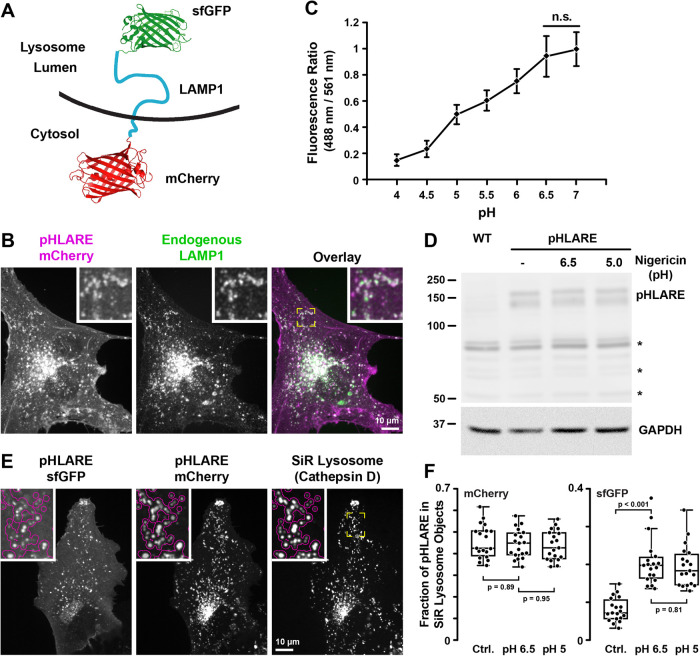
To selectively quantify pHlys, we generated pHLARE, a genetically encoded and ratiometric biosensor that can be stably expressed in cells. pHLARE encodes rat LAMP1, a lysosome transmembrane protein, tagged at the lumenal amino-terminus with superfolder GFP (sfGFP) (Figure 1A), a GFP variant having a pKa ∼5.9. We tested tagging the lumenal domain of LAMP1 with several fluorophores and found that sfGFP was optimal for dynamic range and limited photobleaching, as well as having increased thermostability and improved folding kinetics compared with EGFP (Pédelacq et al., 2006). The cytosolic pH probe pHluorin has a more neutral pKa of ∼7.0 (Grillo-Hill et al., 2014) that is too high for the low lysosome lumenal pH. mT-Sapphire and mCherry each have an appropriate pKa ∼4.5 (Shaner et al., 2004), but mT-Sapphire had a very low signal and substantial photobleaching. mCherry aggregated in the lysosome lumen and responded poorly for calibrating fluorescent signal to pH. In contrast, mCherry did not aggregate in the cytoplasm; hence, we tagged the cytoplasmic carboxyl-terminus of LAMP1 with mCherry (Figure 1A). The low pKa of mCherry makes it insensitive to changes in the cytosolic pH range and with sfGFP allows ratiometric analysis normalized for abundance of the biosensor. When expressed in human retinal pigment epithelial (RPE) cells, rat pHLARE localized predominantly in lysosomes, as indicated by colabeling with endogenous human LAMP1 (Figure 1B). By incubating cells in buffers at known pH values and containing the protonophore nigericin that allows intracellular membrane-bound compartments to equilibrate with extracellular pH, total cell pHLARE fluorescence had a ratiometric dynamic range between pH 4.0 and 6.5 (Figure 1C) and no apparent degradation of a predicted ∼150-kD band, as indicated by immunoblotting cell lysates with antibodies to RFP (Figure 1D). Two bands for pHLARE likely represent different levels of LAMP1 glycosylation.
FIGURE 1:
pHLARE localizes to lysosomes. (A) Schematic of pHLARE with rat LAMP1 tagged at the lumenal amino-terminus with sfGFP and at the cytoplasmic carboxyl-terminus with mCherry. (B) pHLARE stably expressed in human RPE cells, visualized with anti-mCherry antibodies, colocalizes with endogenous LAMP1, visualized with anti-human LAMP1 antibodies. (C) Fluorescence ratios of pHLARE in RPE cells in nigericin-containing buffers between pH 4.0 and 7.0. Data are means ± SEM of 15 cells from three separate cell preparations. Statistical analysis by Tukey–Kramer HSD indicates significant differences at all pH values except between pH 6.5 and 7.0. (D) Representative RFP immunoblot of three preparations of lysates from RPE cells, WT, and stably expressing pHLARE and untreated or treated with nigericin buffer at the indicated pH values for 5 min. The asterisks indicate nonspecific bands seen also in RPE WT cells not expressing pHLARE and GAPDH is used as a loading control. (E) Images of live RPE cells stably expressing pHLARE and stained with SiR-lysosome, a far-red pepstatin A that binds cathepsin D. The magenta outline in the insets at higher magnification indicate lysosomes detected in the SiR-lysosome channel showing overlap with pHLARE. (F) Analysis of the amount of pHLARE fluorescence signal associated with SiR-lysosome objects. As expected, the fluorescence distribution in the mCherry channel is insensitive to pH equilibration, while the SiR lysosome-associated sfGFP fluorescence increases. Statistical analysis by Tukey–Kramer HSD, n = 19 cells.
To develop methods to measure pHlys using pHLARE, we first analyzed how much pHLARE fluorescence signal originated from lysosomes in live cells by determining the fraction of pHLARE fluorescence within objects defined by SiR-lysosome, a highly specific membrane-permeant far-red-tagged pepstatin A that binds cathepsin D (Figure 1E). Nearly half of the pHLARE mCherry fluorescence signal was associated with lysosomes (Figure 1F) with the other half mostly localized to the cell membrane. Importantly, using the same analysis, the acidophilic dye LysoTracker was less specific with a substantial cytoplasm background and only a third of the total LysoTracker fluorescence signal originating from lysosomes (Supplemental Figure S1E). The distribution of the pHLARE mCherry lysosome signal remained the same in pH 7.0 and 5.0 nigericin buffers (Figure 1F) and the total fluorescence signal remained nearly unchanged (Supplemental Figure S1B), confirming pH independence of the mCherry channel within this calibration pH range. In contrast, the fraction of pHLARE sfGFP signal in lysosomes increased significantly following pH equilibration (Figure 1F) while the total pHLARE sfGFP fluorescence signal decreased nearly 10-fold at low pH (Supplemental Figure S1B). Object-based quantification of the overlap of pHLARE mCherry with SiR-lysosome showed high pH-independent colocalization of the mCherry channel with SiR-lysosome (86 ± 4% overlap) similar to colocalization between SiR-lysosome and LysoTracker (78 ± 6% overlap) (Supplemental Figure S1C). Together, these data demonstrate that pHLARE is at least as specific for lysosomes as LysoTracker, although a small contribution from pHLARE-positive endosomes that have not yet fully matured into cathepsin D containing lysosomes cannot be ruled out.
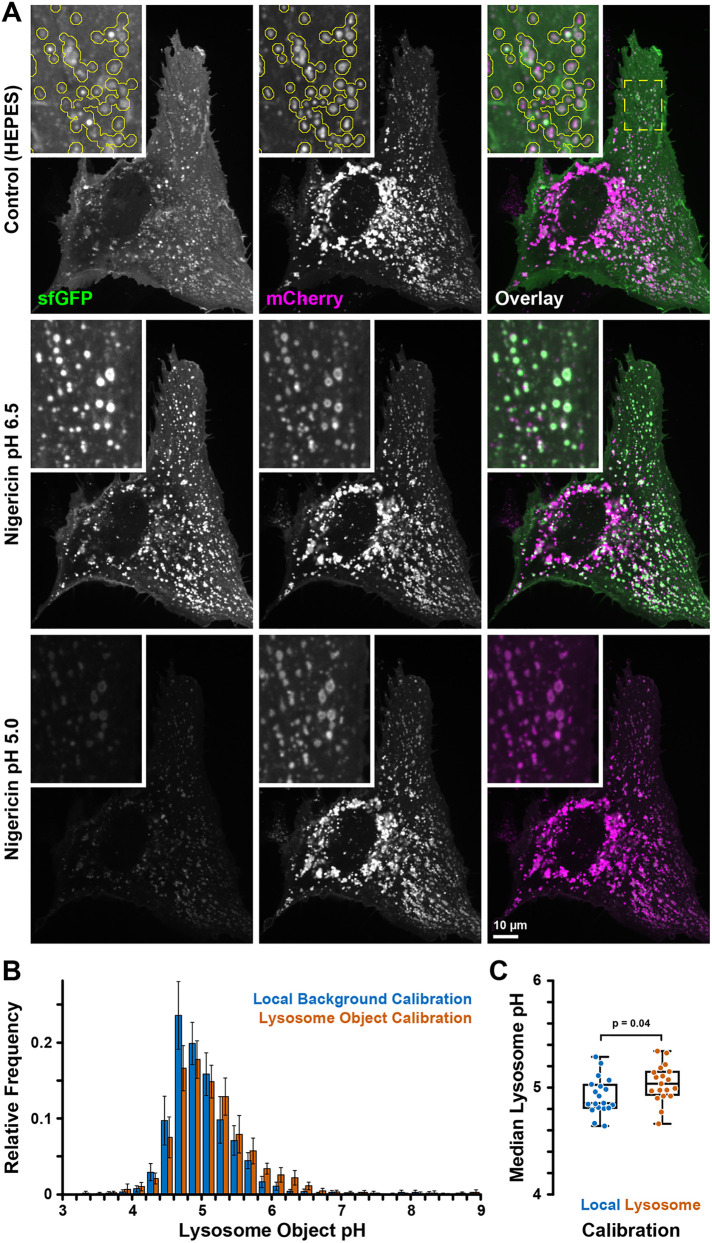
The fluorescence ratios of pHLARE were calibrated to pH with nigericin (Figure 2, A and B), similar to calibrating measurements of cytosolic pH (Grillo-Hill et al., 2014). After each experiment, cells are sequentially incubated with nigericin-containing buffers at higher (∼6.5) and lower (∼5.0) pH. Nigericin, a protonophore, equilibrates extracellular and intracellular pH (pHi), which provides fluorescence ratios at known pH values and calibration of pHLARE fluorescence ratios to absolute pHlys, which is determined for each cell. However, because lysosomes have differences in size, shape, aggregation, and localization, and approximately half of the whole cell pHLARE fluorescence signal does not originate from lysosomes, calibrating pHLARE fluorescence is more complex than calibrating fluorescent probes for cytosolic pH. We therefore developed an image analysis algorithm to objectively and reproducibly define lysosome objects in the pHlys-insensitive mCherry channel (Figure 2A). The average fluorescence intensities of these objects minus corresponding backgrounds defined as the contour around each lysosome object in the two pHLARE fluorescence channels were used to determine fluorescence ratios. The pH value for each lysosome object is calculated based on the nigericin calibration for each cell, as described above. It is important to note that during calibration, mCherry on the cytosolic side of pHLARE is exposed to low pH. To minimize skewing the calibration curve, to quantify pHlys, we use calibration lower pH values between 5.0 and 5.2 (Supplemental Figure S1B). Although we typically detected up to 100 lysosome objects per cell, lysosomes are often close to one another and therefore lysosome objects frequently contain many lysosomes, especially in central cell areas where lysosomes tend to cluster. While it may be possible to estimate the number of individual lysosomes based on the area of a lysosome cluster, we did not attempt this. In addition, because pHLARE fluorescence channels are acquired sequentially, a very small number of lysosomes that move substantially between the two image acquisitions will yield erroneous and unrealistic measurements (Figure 2B), and individual lysosome object pH values within a cell should be interpreted cautiously. To minimize the impact of such outliers, we use the median pH of all lysosome objects per cell to calculate a cell pHlys. Thus, the median pHlys is based on lysosome objects and not individual lysosomes. Because all lysosome objects are weighted equally, individual lysosomes and small clusters contribute more to the median cell pHlys. We were concerned that pHlys equilibration would be incomplete in nigericin due to potentially high lysosome buffering capacity and, therefore, tested two different calibration methods using either the local background around lysosomes to measure pHLARE fluorescence ratios in nigericin or the lysosome signal itself. We validated this analysis by comparing pHlys measurements in 19 RPE cells (Figure 2, B and C). With ∼0.1 pH units the difference between these two calibration methods was consistently small (4.92 ± 0.04 [mean and SEM]; lysosome signal calibration pHlys = 5.03 ± 0.04), although it reached statistical significance in this population of cells. For consistency, going forward we used the local background calibration. Fluorescence at the plasma membrane, which is excluded from the pHlys analysis by local background subtraction, could be due to lysosome exocytosis (Rodríguez et al., 1997; Jaiswal et al., 2002) or direct trafficking of LAMP1 to the plasma membrane (Rohrer et al., 1996). It should be noted that this plasma membrane signal is more obvious in the sfGFP channel because of reduced contrast with the quenched sfGFP signal in acidic lysosomes but is present in both pHLARE channels (Figure 1F). In general, less than 10 frames were acquired for steady-state pHlys and nigericin calibrations, during which time there was minimal photobleaching, determined at 488 and 561 nm excitation (Supplemental Figure S2).
FIGURE 2:
Cell-based pHLARE pH calibration. (A) pHLARE-expressing RPE cell in NaHCO3-containing buffer (described for pHi measurements) and in nigericin buffers of two different calibration pH values showing sfGFP (green) and mCherry (magenta) fluorescence. The three images are scaled identically showing the increase of sfGFP signal in lysosomes at near neutral pH equilibration and the dramatic overall decrease of sfGFP fluorescence at low pH. Insets show the indicated region at higher magnification. The yellow outline in the top row indicates lysosome objects detected in the mCherry channel. Note that lysosome size is near the resolution limit of optical microscopy and pHLARE lysosome membrane localization is only resolved in larger lysosomes. (B) Frequency histogram of lysosome object pH distribution in RPE cells obtained with the two different calibration methods as indicated. Bars are the average of n = 19 cells and error bars indicate 95% confidence interval. (C) Box plot of the median pHlys of the same set of cells. Statistical analysis by unpaired t test.
Using pHLARE, we determined steady-state pHlys in untransformed cells from different species and tissue origins. Human RPE and MCF10A mammary epithelial cells, and canine MDCK kidney epithelial cells (Figure 3A) and human pancreatic ductal epithelial (HPDE) cells (Figure 6A) had a mean cell pHlys between 5.0 and 5.4. These values are similar to the pHlys of 5.1 to 5.2 in HeLa cells determined using cresyl violet (Ostrowski et al., 2016) or Oregon green dextran (Johnson et al., 2016) but substantially higher than the ∼4.3 in keratinocytes determined using FITC–conjugated dextran and flow cytometry (Nilsson et al., 2010). For MCF10A cells there was no difference in pHlys when pHLARE was transiently or stably expressed in cells (Supplemental Figure S2B). In contrast, the mean cell pHlys in mouse NIH 3T3 fibroblasts and hamster CCL39 lung fibroblasts was ∼6.0–6.1 and significantly higher compared with RPE epithelial cells (Figure 3A). These values for fibroblasts are also higher than that of ∼4.9 in mouse Swiss 3T3 fibroblasts determined by fluorescence–conjugated dextran (Lin et al., 2003), in embryonic fibroblasts determined using LysoSensor (Wolfe et al., 2013), and the ∼4.4 in human skin fibroblasts determined using LysoSensor (Coffey et al., 2014). The distinct difference in pHlys of epithelial cells compared with fibroblasts is unclear and warrants further investigation to determine a possible functional significance.
FIGURE 3:
Cell-specific pHlys. (A) Average steady-state pHlys in RPE cells stably expressing pHLARE (n = 74 cells) and in MCF10A (23 cells), MDCK (n = 17 cells), NIH-3T3 (15 cells), and CCL39 (n = 11 cells) transiently expressing pHLARE and maintained in growth medium. Box plots show median, first, and third quartile, with whiskers extending to observations within 1.5 times the interquartile range, and all individual data points include data for individual cells obtained from three to seven separate cell preparations. Statistical analysis by Tukey–Kramer HSD test. (B) Steady-state pHlys of lysosomes within 3 μm of the nuclear membrane (perinuclear) and 3 μm of the distal margin of membrane protrusions in RPE cells, indicated by hatched lines. Data are expressed as described in A and obtained from six separate cell preparations. (C–E) Lysosome motility, including mean speed (C), maximum speed (D), and displacement (E) of RPE control cells and RPE-pHLARE cells. Plots indicate mean and SEM with Student’s t test from three separate cell preparations.
FIGURE 6:
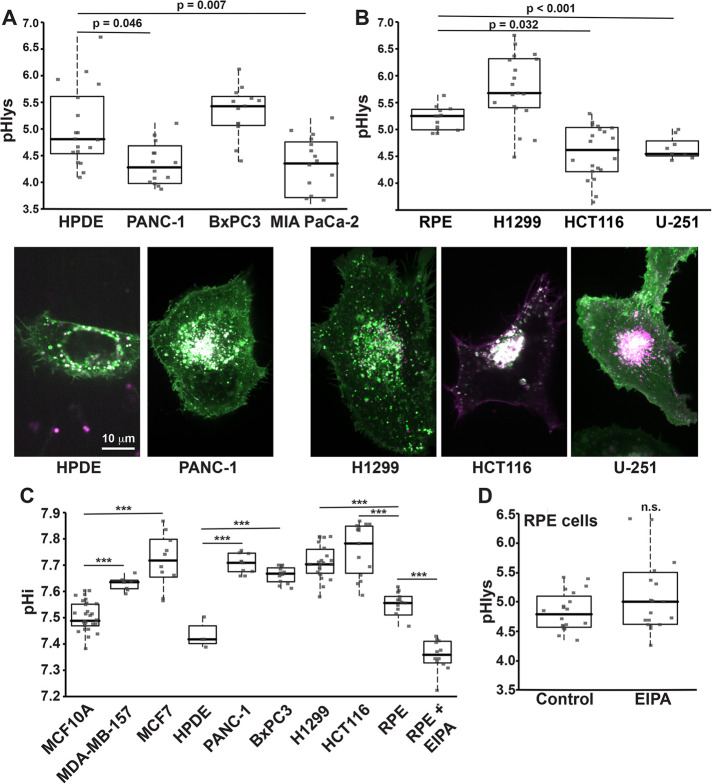
pHlys and pHi in cancer and untransformed cells. (A) Images of pHLARE and average pHlys of human untransformed HPDE cells (n = 19 cells) and pancreatic adenocarcinoma PANC-1 (n = 17 cells), BxPC3 (n = 13 cells), and MIA PaCa-2 (n = 20 cells) cells. (B) Images of pHLARE and average pHlys of human RPE untransformed cells (n = 12 cells), H1299 lung cancer cells (n = 19 cells), HCT116 colon cancer cells (n = 22 cells), and U-251 glioblastoma cells (n = 9 cells). (C) Steady-state pHi of the indicated cell types determined in a NaHCO3-containing buffer. ***p < 0.001. (D) Average pHlys of RPE cells in the absence (Control; n = 20 cells) and presence of EIPA (10 μM, 18 h; n = 19 cells). Box plots show median, first, and third quartile, with whiskers extending to observations within 1.5 times the interquartile range and all individual data points. Statistical analysis by Tukey–Kramer HSD test, with data obtained from three to five separate cell preparations.
The values described above are averages for all lysosomes in the indicated cell types; however, recent findings suggest that pHlys within each cell may be heterogeneous (Johnson et al., 2016). In many cell types, lysosomes have a predominantly perinuclear localization, but smaller populations are scattered within the cytosol and are also peripheral near the plasma membrane. Perinuclear lysosomes are suggested to have a lower lumenal pH than peripheral lysosomes (Johnson et al., 2016), although this was not quantitatively determined but rather predicted from qualitative differences in the fluorescence of Alexa Fluor–conjugated dextran and also measured in a HEPES buffer in the absence of HCO3-. We found that in RPE cells with clear membrane protrusions, lysosomes within 3 μm of the distal membrane have an average pHlys of 5.48 ± 0.12 (mean ± SEM) that is significantly higher than the pHlys 4.80 ± 0.12 of perinuclear-clustered lysosomes in RPE cells (Figure 3B), indicating that spatially distinct lysosomes have different steady-state lumenal pH values. However, spatially heterogeneous pHlys may depend on cell type and conditions. For human osteocarcoma cells, lowering extracellular pH of the medium causes a redistribution of lysosomes from predominantly perinuclear to predominantly peripheral (Walton et al., 2018). We confirmed that pHLARE expression did not change lysosome motility. Using spinning disk confocal microscopy and live-cell tracking every 3 s for 5 min LysoTracker loaded in wild-type (WT) RPE cells and RPE cells expressing pHLARE, we found no difference in mean speed (Figure 3C), maximum speed (Figure 3D), or displacement (Figure 3E).
pHLARE measures dynamic changes in pHlys
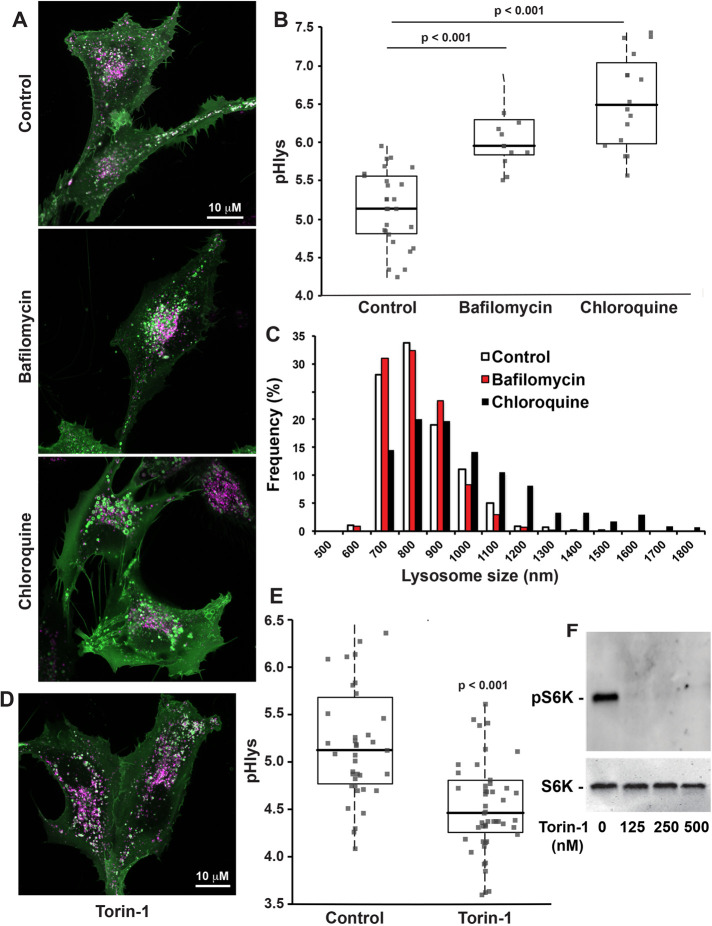
We used pHLARE to confirm increased pHlys when RPE cells were incubated with NH4Cl, both 30 mM for 15 min and 5 mM for 18 h (Supplemental Figure S3, A and B), which increases the pH of membrane-bound compartments as NH3 entering compartments complex with H+ (White et al., 2017a). We also confirmed increased pHlys in RPE cells treated for 60 min with bafilomycin (100 nM), a V-ATPase inhibitor, and with chloroquine (100 µM), which neutralizes acidic intracellular compartments (Figure 4, A and B). In control RPE cells, lysosome size was within the reported range of 100–1000 nm (Appelqvist et al., 2013; Bandyopadhyay et al., 2014; Xu and Ren, 2015), and the majority of lysosomes had a mean size of 784 ± 133 nm. With chloroquine but not with bafilomycin, however, lysosomes were larger with a mean size of 934 ± 244 nm and there were fewer lysosomes smaller than 600 nm and more lysosomes larger than 1000 nm (Figure 4, A and C). Lysosome size can increase as a result of accumulated undigested material. However, the increased pHlys with bafilomycin and chloroquine were similar; hence, both presumably decreased catabolism but not an increase in lysosome size, which suggests a different mechanism for increased size with chloroquine. A previous report (Gallagher et al., 2017) found that lysosome swelling with chloroquine is dependent on glucose in the medium but also that increased pHlys with chloroquine is independent of lysosomal swelling. Additionally, possible mechanisms include swelling from chloroquine accumulating in the lumen or activation of the transcription factor EB (TFEB) pathway that increases lysosome biogenesis in response to lysosomal storage dysfunction (Sardiello et al., 2009).
FIGURE 4:
pHLARE measures dynamic changes in pHlys. (A, B) RPE cells stably expressing pHLARE were untreated (control, n = 27 cells) or treated with 100 nM of the V-ATPase inhibitor Bafilomycin A1 (n = 15 cells) or 100 μM chloroquine (n = 15 cells) for 2 h before acquiring images, which were used to calculate pHlys. (C) Lysosome size with indicated conditions determined by measuring more than 1,000 lysosomes in four cell preparations for controls and three cell preparations with bafilomycin and chloroquine treatment. (D, E) pHlys of RPE cells stably expressing pHLARE untreated (control, n = 39 cells) or treated with 250 nM Torin-1 (n = 50 cells) for 2 h before acquiring images. (F) Representative immunoblot of lysates from two separate RPE cell preparations untreated or treated with the indicated concentrations of Torin-1 and probed with antibodies to total and phosphorylated S6K. Box plots show median, first, and third quartile, with whiskers extending to observations within 1.5 times the interquartile range and all individual data points. Statistical analysis by Tukey–Kramer HSD test, with data obtained from three separate cell preparations in B, C, and F and four separate cell preparations in D.
Lysosome functions in nutrient sensing and responses are in part determined by mTORC1, which when active is associated with the lysosomal membrane. Inside (lysosome) to outside (cytoplasm) signaling regulates mTOCR1 activity (Settembre et al., 2013; Rebsamen et al., 2015; Wang et al., 2015). Whether mTORC1 activity regulates pHlys, however, has received limited attention. Pharmacologically inhibiting mTOCR1 increases autophagy and decreases pHlys; however, the latter was determined qualitatively by increased LysoTracker and LysoSensor staining but not quantitatively by calibrating the fluorescence signal to pH (Zhou et al., 2013). We used RPE cells stably expressing pHLARE and quantitative measurements to show that Torin-1, an ATP-competitive inhibitor of mTORC1 activity (Thoreen et al., 2009), decreased pHlys from 5.22 ± 0.10 in controls to 4.63 ± 0.09 (mean ± SEM, 4 cell preparations) (Figure 4, D and E). We confirmed that Torin-1 decreased phosphorylation of ribosomal protein S6 kinase beta-1 (S6K1) (Figure 4F), an mTORC1 substrate, and treated RPE cells with 250 nM Torin-1 for 2 h, as previously described (Thoreen et al., 2012). Decreased pHlys with inhibiting mTORC1 activity is consistent with the established role of active mTORC1 in stimulating biosynthetic pathways and inhibiting cellular catabolism (Saxton and Sabatini, 2017). Accordingly, our findings suggest that active mTORC1 might increase pHlys to limit catabolism. However, pHlys was not different in RPE cells maintained for 24 h in medium containing 0.2% fetal bovine serum (FBS) compared with cells maintained in growth medium containing 10% FBS (Supplemental Figure S3C). Previous findings indicate that acute (2–6 h) serum deprivation markedly changes lysosome morphology from circular to tubular and increases pHlys, although the latter was determined qualitatively but not quantitatively using LysoTracker, and by 24 h lysosome morphology and lumenal pH was restored to steady-state conditions by a feedback loop involving increased mTOR activity (Yu et al., 2010). Our findings suggest that pHlys dynamics over longer periods may be more responsive to signals related to nutrient deprivation and not general growth conditions.
Cancer cells with decreased pHlys
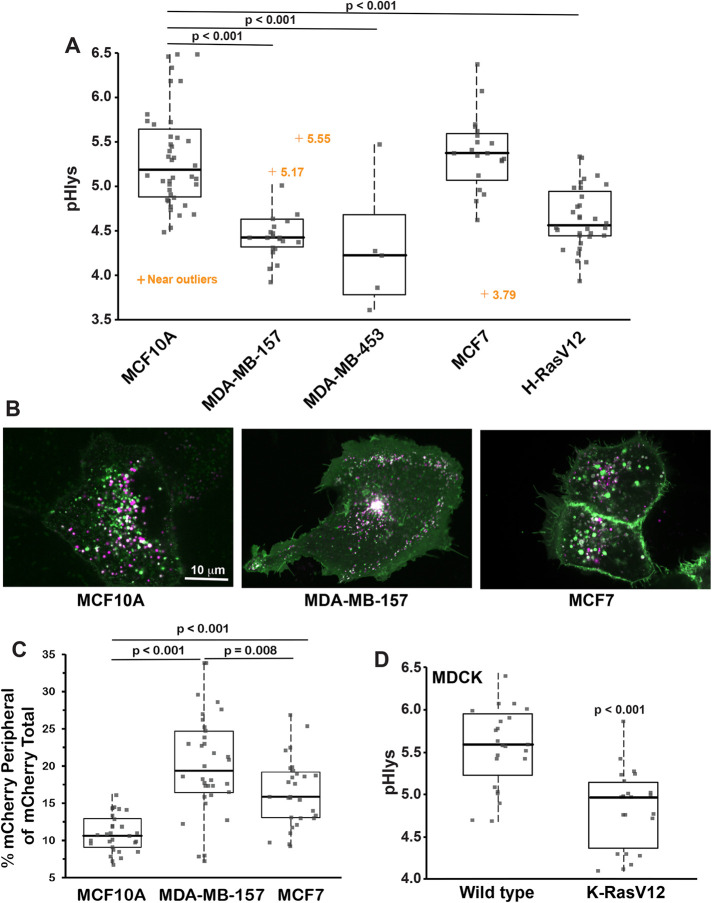
Cancer progression and metastasis are associated with striking changes in lysosomes, including their volume, composition, cellular distribution, and lumenal enzyme activities (Appelqvist et al., 2013; Perera and Bardeesy, 2015; Hamalisto and Jaattela, 2016). Despite many of these properties being determined by pHlys, there are limited data on pHlys in cancer compared with tissue-matched untransformed cells (Nilsson et al., 2010). We used pHLARE to measure steady-state pHlys in clonal human cancer cells from different tissue origins and with different mutational signatures. The pHlys of MDA-MB-157 and MDA-MB-453 basal, triple negative, and invasive breast cancer cells (4.56 ± 0.11 and 4.31 ± 0.28, respectively, mean ± SEM) was significantly lower than in immortalized but not transformed MCF10A mammary epithelial (5.36 ± 0.09) (Figure 5, A and B). In contrast, the pHlys of MCF7 lumenal, estrogen receptor- and progesterone receptor-positive, and benign breast cancer cells (5.34 ± 0.12) was not different than MCF10A cells (Figure 5, A and B), which was also reported (Altan et al., 1999). In MDA-MB-157 but not MCF7 cells, there were more peripherally localized lysosomes compared with MCF10A cells (Figure 5C), determined by mCherry fluorescence within 3 μm of the plasma membrane, which is consistent with the increased invasiveness of basal breast cancer cells (Neve et al., 2006). In contrast with RPE cells, however, in MDA-MB-157 cells there was no difference in pHlys in peripheral compared with perinuclear lysosomes. Additionally, we did not see an increase in the number of peripheral lysosomes in in pancreatic (Figure 6A) or other cancer cells (Figure 6B).
FIGURE 5:
Differences in pHlys in breast cancer cells and with oncogene transformation. (A) The average pHlys of human untransformed MCF10A mammary epithelial cells (n = 36 cells), breast cancer MDA-MB-157 (n = 24 cells), MDA-MB-453 (n = 6 cells), and MCF7 (n = 21) cells, and MCF10A cells stably expressing H-RasV12 (n = 24 cells). (B, C) Representative images of MCF10A, MDA-MB-157, and MCF7 cells expressing pHLARE (B) and quantification of lysosomes within 3 μm of the plasma membrane indicated by mCherry fluorescence. (C, D) The average pHlys in canine MDCK epithelial cells WT (n = 23) and stably expressing K-RasV12 (n = 23 cells). Box plots show median, first, and third quartile, with whiskers extending to observations within 1.5 times the interquartile range and all individual data points. Statistical analysis by Tukey–Kramer HSD test, with data obtained from three to five separate preparations of all cells except for two preparations of MDA-MB-453.
These breast cancer cell lines have different mutational signatures. To determine whether a single activated oncogene is sufficient to change pHlys, we used MCF10A cells stably expressing H-RasV12. We previously reported that these cells have a higher cytosolic pH (pHi) compared with WT MCF10A cells (Grillo-Hill et al., 2015), which we confirmed in the current study (Supplemental Figure S4A). We found that pHlys was significantly lower (4.67 ± 0.07) compared with WT MCF10A cells (5.36 ± 0.09) (Figure 5A). To further test the effect of oncogenic Ras, we found that canine MDCK cells stably expressing K-RasV12, which have a higher pHi (Supplemental Figure S4B), also have a significantly lower pHlys (4.84 ± 0.10) compared with the pHlys of WT MDCK cells (5.58 ± 0.10) (Figure 5D). These data indicate that a lower pHlys can be induced by a single activated oncogene and does not require the complex mutational signature of a cancer cell.
For clonal pancreatic ductal carcinoma cells, the pHlys of PANC-1 (4.39 ± 0.19) and MIA PaCa-2 (4.24 ± 0.17) but not BxPC3 (5.33 ± 0.14) was significantly lower than normal HPDE cells (5.12 ± 0.18) (Figure 6A). Additionally, the pHlys of HCT116 colon cancer cells (4.64 ± 0.12) and U-251 human glioblastoma cells (4.65 ± 0.19) but not H1299 lung cancer cells (5.46 ± 0.13) was significantly lower than RPE cells (Figure 6B). Although we compared pHlys of these cancer cells with RPE cells and not tissue-matched untransformed cells, the lower pHlys was not significantly different comparing MDA-MB-157, MDA-MB-453, PANC-1, HCT116, and U-251 cells. Additionally, all pHlys measurements were made with cells in growth medium containing FBS, suggesting that the lower pHlys in the indicated cancer cells was not a result of limited nutrient availability. However, distinct from MDA-MB-157 cells (Figure 5B), other cancer cells with a decreased pHlys did not have a notable increase in the abundance of peripherally localized lysosomes (Figure 6, A and B).
An unresolved question is whether pHlys and pHi dynamics are related. The higher pHi of cancer cells is decreased with loss of Stat3, which also increases pHlys by attenuating V-ATPase activity (Liu et al., 2018). Whether the effects of Stat3 on pHi and pHlys are mediated by similar or distinct mechanisms, however, remains undetermined. We found that all cancer cells included in our study had a significantly higher pHi compared with untransformed cells (Figure 6C), including those not having a lower pHlys such as pancreatic BxPC3 cells and lung H1299 cells. Additionally, treating RPE cells with EIPA (10 μM; 18 h), which inhibits H+ efflux by the plasma membrane Na-H exchanger NHE1, decreased pHi (Figure 6C) but did not change pHlys (Figure 6D). These data suggest that pHi and pHlys are not directly related and that their regulation by oncogenes is likely through distinct mechanisms.
DISCUSSION
We report the design, validation, and use of pHLARE, a new genetically encoded pHlys biosensor. As a ratiometric biosensor localized predominantly in lysosomes with a broad dynamic range, pHLARE resolves a number of limitations of currently used fluorescent dyes for measuring pHlys, including improved organelle specificity, quantitative determinations, and stability with the ease of being stably expressed in cells. The lumenal pH of each organelle is different, requiring the use of organelle-specific pH indicators, and is crucial for organelle function, including the pH-dependent function of proteins that reflect adaptations to the cellular compartment where they are localized (Chan and Warwicker, 2009). The steady-state pHlys of ∼5.0, the lowest of any cellular organelle, is particularly critical for the activity of lumenal acid-activated hydrolases, but recent findings suggest that pHlys may also contribute to Ca2+ homeostasis (Lee et al., 2015) as well as the localization of lysosomes in cells (Johnson et al., 2016), with the latter suggested to be important for cancer metastasis (Tu et al., 2008; Hamalisto and Jaattela, 2016; Pu et al., 2016). pHLARE was developed by fusing sfGFP and mCherry to lumenal and cytosolic domains, respectively, of LAMP1. The predominant localization of pHLARE in lysosomes was confirmed by colocalization with endogenous LAMP1 and with SiR-lysosome, a fluorescent pepstatin A that binds cathepsin D. Although LAMP1 is predominantly localized in organelles considered to be lysosomes or at the transition of late-endosomes to lysosomes, recent findings indicate that LAMP1 is not exclusively in degradative lysosomes containing detectable hydrolases (Cheng et al., 2018; Yap et al., 2018). Hydrolase-deficient lysosomes raise questions on catabolic-independent functions requiring a low lumenal pH that use of pHLARE could contribute to resolving.
A number of diseases, including neurodegenerative disorders and cancer, have dysregulated lysosome functions in catabolism, nutrient sensing, and trafficking (reviewed in Kallunki et al., 2012; Appelqvist et al., 2013; Perera and Zoncu 2016). Despite many of these functions being regulated by pHlys and whether pHlys is also dysregulated in these diseases, the determinants and consequences of dysregulated pHlys remain incompletely understood. As a new tool, pHLARE could contribute to resolving a number of current questions on the role of lysosomes in disease pathologies. A constitutively increased pHlys is reported to occur with neurodegenerative disorders (Majumdar et al., 2007; Wolfe et al., 2013; Lee et al., 2015) and in diabetic nephropathy (Liu et al., 2015). The predicted consequence of increased pHlys in these diseases is decreased activity of lumenal acid-activated hydrolases and catabolism of proteins and macromolecules.
Whether pHlys is dysregulated in cancer cells, which rely on increased catabolism of macromolecules to generate biomass for rapid proliferation, remains unresolved. To our knowledge, only one report compared pHlys in tissue-matched normal and cancer cells and found that clonal head and neck cancer cells have a higher pHlys than untransformed keratinocytes (Nilsson et al., 2010). However, in this previous study pHlys was determined after flow cytometry of cells loaded with FITC–conjugated dextran, and flow cytometry requires disrupting cell–matrix adhesions and using cells in suspension, which could change steady-state pHlys. We used pHLARE in adherent cells maintained in growth medium to show that human MDA-MB-157 and MDA-MB-453 breast cancer cells and PANC-1 pancreatic cancer cells had a significantly lower pHlys compared with tissue-matched untransformed MCF10A and HPDE cells, respectively. We also observed a lower pHlys in human HCT166 colon cancer cells and U-251 glioblastoma cells and in human mammary epithelial cells expressing oncogenic H- and K-RasV12. However, the pHlys of human MCF7 breast cancer cells, BxPC3 pancreatic cancer cells, and H1299 lung cancer cells was not different compared with untransformed cells. Additionally, clonal HeLa cervical cancer cells have a normallike pHlys of 5.1–5.2 (Ostrowski et al., 2016; Johnson et al., 2016). The causes and consequences of these differences in pHlys between distinct cancer cells remain to be determined. Although MCF7 cells are estrogen receptor- and progesterone receptor- positive and MCA-MB-157 and MDA-MB-453 are triple negative, their different pHlys is likely not determined by their mutational signature, as suggested by a single oncogene, RasV12, inducing a lower pHlys. However, different pHlys values could be related to invasiveness. MDA-MB-157 cells are basal-type and more mesenchymal and invasive than lumenal MCF7 cells that have tight cell–cell junctions (Neve et al., 2006). Also, PANC-1 cells are more invasive and migrate as single cells compared with BxPC3 cells that have tighter cell–cell contacts and migrate as collective cell sheets (Deer et al., 2010). Lysosomes can contribute to cell invasion by the exocytic release of lysosomal hydrolases degrading the extracellular matrix (Tu et al., 2008; Hamalisto and Jaattela, 2016; Pu et al., 2016). Although we found that MDA-MB-157 have significantly more pHLARE within 3 μm of the plasma membrane compared with MCF7 and MCF10A cells, we did not observe increased peripheral lysosomes in other cancer cells types that had a lower pHlys. We also did not observe a correlation between pHi and pHlys in cancer or untransformed RPE cells.
Differences in pHlys between cancer cells could also reflect drug resistance, as lysosomes contribute to resistance by sequestering and degrading chemotherapeutics (Piao and Amaravadi, 2016; Zhitomirsky and Assaraf, 2016). We favor the possibility that differences in pHlys are related to cancer cell-specific metabolic requirements and cellular mechanisms for generating biomass, according to the view that lysosomes contribute to metabolic reprogramming in some cancer cells along with changes in glycolysis and mitochondrial respiration (Perera and Bardeesy, 2015). Supporting the prediction that a lower pHlys may function as an alternative metabolic reprogramming mechanism, BxPC3 cells but not PANC-1 cells rely on a classically defined metabolic reprogramming of increased glycolysis and decreased mitochondrial respiration (Kovalenko et al., 2016; Tataranni et al., 2017), and inhibiting glycolysis decreases proliferation of BxPC3 cells but not PANC-1 cells (Tataranni et al., 2017, Nishi et al., 2018).
As a new reagent to more accurately measure pHlys that can be propagated in cells and used over long time periods, pHLARE will be invaluable for many objectives. First is to identify resident lysosome proteins regulating pHlys. The V-ATPase is currently the only well-accepted regulator of pHlys, with roles for a number of lysosome-localized transporters and pumps being controversial (Xu and Ren, 2015). Endogenous regulators of pHlys could be targets for therapeutics to restore dysregulated pHlys in diseases such as neurodegeneration and cancer. Second is to determine lysosome exocytosis, particularly in the context of cancer metastasis, by measuring increased sfGFP fluorescence on exposure to the extracellular space. Third, because pHLARE is genetically encoded it can be expressed in animal models to study lysosome dynamics in diseases and also pHlys dynamics during development, which remain unknown.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cloning and DNA constructs
Mammalian expression plasmids encoding rat LAMP1 (mCherry-Lysosomes-20, Addgene; 55073), mApple-LAMP1-pHluorin (Addgene; 54918), and sfGFP (Addgene; 54579) were obtained from the Michael Davidson collection at University of California, San Francisco (UCSF). Gibson assembly was used to generate pHLARE. Briefly, PCR was used to amplify the prolactin endoplasmic reticulum (ER) signal peptide (MDSKGSSQKGSRLLLLLVVSNLLLCQGVVS) from mApple-LAMP1-pHluorin, sfGFP from sfGFP-C1, and rat LAMP1 without the ER signal peptide (amino acids 22–407) from mCherry-Lysosome-20. The PCR inserts were assembled with restriction enzyme-digested pmCherry-N1 backbone in Gibson Assembly Master Mix (New England Biolabs; E2611S) as per manufacturer recommendations, and the assembled plasmids transformed into NEB5α cells (New England Biolabs; C3019). DNA sequencing was performed to verify the integrity of the construct.
Cell culture and transfection
All cell lines were maintained at 37°C and 5% CO2 except MDA-MB-157 and MDA-MB-453, which were maintained at 37°C and atmospheric CO2. Media obtained from Thermo Fisher Scientific were supplemented with penicillin–streptomycin. RPE cells, obtained from Sophie Dumont (UCSF), and MCF7 cells, obtained from the UCSF Cell Culture Facility, were maintained in DMEM/F21 supplemented with 10% heat-inactivated FBS (Atlanta Biological) and 2 mM L-glutamine. MDCK cells, obtained from the UCSF Cell Culture Facility; MDCK cells stably expressing K-RasV12, obtained from Zev Gartner (UCSF) and previously described (Schoenenberger et al., 1991); PANC-1 and MIA PaCa-2 cells, obtained from Rushika Perera (UCSF); and NIH-3T3 cells, obtained from Jeroen Roose (UCSF), were maintained in DMEM-H21 medium supplemented with 10% FBS. CCL39 cells, obtained from ATCC, were maintained in DMEM-H21 medium supplemented with 5% FBS. BxPC3 cells, obtained from Rushika Perera (UCSF), and H1299 cells, obtained from Torsten Wittmann (UCSF), were maintained in RPMI medium supplemented with 10% FBS. HCT 116 cells, obtained from Bert Vogelstein (Johns Hopkins University), were maintained in McCoy’s 5a medium supplemented with 10% FBS, and U-251 cells, obtained from Robert Judson-Torres (University of Utah), were maintained in EMEM supplemented with 2 mM glutamine, 1% nonessential amino acids, 1 mM sodium pyruvate, and 10% FBS. MDA-MB-157 cells, obtained from the J. M. Bishop Lab (UCSF), and MDA-MB-453 cells, obtained from ATCC, were maintained in Leibovitz L15 medium supplemented with 15% FBS. HDPE cells, obtained from Rushika Perera (UCSF), were maintained in keratinocyte medium supplemented with 10% FBS. MCF10A cells, WT, and stably expressing RasV12 were obtained from Jay Debnath (UCSF) and maintained and studied in 2D cultures as previously described (Debnath et al., 2003; Grillo-Hill et al., 2015). Mycoplasma contamination was tested 2×/year using media obtained from cells maintained for 48 h in the absence of penicillin–streptomycin by using a PCR Mycoplasma Detection Kit (abm #G238). Cell lines were authenticated commercially by IDEXX BioAnalytics.
Heterologous expression of proteins was achieved by transfecting cells with 1 μg of plasmid DNA/35 mm using FuGENE HD (Promega; E2311), as previously described (Webb et al., 2015). For transient expression of pHLARE, cells were washed 24 h after transfecting and imaged after an additional 24 h. For stable expression of pHLARE, 24 h after transfection cells were sorted for sfGFP and mCherry fluorescence on a FACS Aria II (BD Biosciences) at the UCSF Parnassus Flow Cytometry Core.
Immunolabeling and immunoblotting
For immunolabeling, RPE cells stably expressing pHLARE were plated at a density of ∼40 × 104 cells/well of a 6-well plate containing 100-mm glass coverslips and were maintained in complete growth media in a 37°C/5% CO2 incubator. Cells plated for 48 h were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, permeabilized in 0.1% Triton X-100 in PBS for 10 min, and incubated with blocking buffer (3% bovine serum albumin, 1% nonimmune goat serum, and 1% cold water fish gelatin) for 1 h. After washing in PBS, cells were incubated overnight at 4°C with primary antibodies to mCherry (Thermo Fisher Scientific; M11217) and human LAMP1 (Cell Signaling Technologies; 9091) diluted in blocking buffer. After 24 h, cells were washed with PBS and incubated with Alexa Fluor 568 goat anti-mouse (Thermo Fisher Scientific; A-11077) and Alexa Fluor 647 goat anti-rabbit (Thermo Fisher Scientific; A-21244) for 1 h. Coverslips were washed 3× in PBS, with Hoechst 33342 (Thermo Fisher Scientific; 62249) included in the second wash, and mounted on glass sides in ProLong gold antifade mountant (Thermo Fisher Scientific; P36930). Where indicated, RPE cells in growth medium were incubated with far-red SiR-lysosome (Cytoskeleton) for 60 min and washed before imaging live cells expressing pHLARE. Images were acquired by spinning disk confocal microscopy as described for imaging pHLARE.
For immunoblotting, proteins in total cell lysates were separated by SDS–PAGE and transferred to polyvinylidene difluoride membranes. The membranes were incubated with antibodies to RFP (Abcam 62341) or total and phosphorylated S6K1 (Cell Signaling Technologies; 9292 and 9205, respectively) for 24 h at 4°C. After washing, membranes were incubated with peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) for 1 h at RT, and bound antibodies were developed by enhanced chemiluminescence using SuperSignal West Femto (Thermo Fisher Scientific) and imaged using an Alpha Innotech FluorChem Q (Alpha Innotech).
Microscopy and pH measurements
For live-cell imaging of pHLARE, cells were plated on 35-mm MatTek dishes (MatTek Corporation; P35G-1.5-10-C). Fluorescent images were acquired using a customized spinning disk confocal (Yokogawa CSU-X1) on a Nikon Ti-E microscope with a 60× Plan TIRF 1.49 NA objective equipped with a Photometrics cMYO cooled CCD camera, largely as described (Stehbens et al., 2012). Image acquisition settings that caused less than 5% photobleaching were used all experiments. Conditions for experimentally changing pHlys included incubating cells in growth medium and prior to imaging with NH4Cl (30 mM for 15 min or 5 mM for 18 h), or 100 nM bafilomycin A1 (Sigma-Aldrich; B1793), 100 μM chloroquine (Sigma-Aldrich; C6628), or 250 nM Torin-1 (Tocris Bioscience; 4247) for 2 h.
Fluorescence ratios of pHLARE were first acquired in cell culture medium and then cells were sequentially incubated in a potassium-phosphate buffer (50 mM potassium phosphate, 80 mM potassium chloride, 1 mM magnesium chloride) containing 20 μM nigericin free acid (Thermo Fisher Scientific; N1495) at pH 6.5 and 5.0. Cells were incubated in nigericin buffers for 5 min before imaging to equilibrate extracellular and pHi. Nigericin incubations to calibrate fluorescence ratios to pHlys were included at the end of each acquisition to obtain a calibration conversion formula for each imaging experiment and for each cell. Minimal photobleaching during the number of frames for image acquisition, including nigericin calibrations, was determined at 488 and 561 nm excitation (Supplemental Figure S2).
Steady-state pHi was measured as previously described (Grillo-Hill et al., 2015; White et al., 2017a). In brief, cells plated for 48 h in 24-well dishes were washed and incubated for 15 min in a NaHCO3-containing buffer containing 1 μM of the pH-sensitive dye BCECF. BCECF fluorescence ratios, measured on a SpectraMax fluorescence plate reader (Molecular Devices), were calibrated to pHi by treating cells at the end of each measurement in a potassium phosphate buffer containing the protonophore nigericin (10 μM) at pH 6.5 and 7.5, as previously described (Grillo-Hill et al., 2014).
Lysosome tracking
For lysosome tracking experiments, live RPE cells, WT, and stably expressing pHLARE, were incubated with 50 nM Lysotracker Red DND-99 (Molecular Probes Cat. No. L-7528) for 1 h at 37°C. Imaging was then performed without further washing of cells using a spinning disk confocal microscope as described above. Images were recorded every 3 s over 5 min (101 frames in total) with 100 ms exposure time at 561 nm excitation. Movies were analyzed with Fiji software using the TrackMate v3.5.1 plugin for recognition and tracking as described (Tinevez et al., 2017). Settings were as follows: pixel width: 0.094 μm; pixel height: 0.094 μm; voxel depth: 1 μm; crop settings: not applied; select a detector: DoG detector with estimated blob size: 1.5 μm; threshold: 50; median filter: no; subpixel localization: yes; initial thresholding: none; select view: HyperStack Displayer; set filters on spots: none; select a tracker: simple LAP tracker; linking max distance: 8 μm; gap-closing max distance: 8 μm; gap-closing max frame gap: 2. Filter settings on tracks were as follows: Max number of gaps: 2; duration: 50; displacement: 4; mean quality: 500. More than 500 tracks per movie were analyzed with respect to track displacement and mean speed. Results were recorded in Microsoft Excel and bulk statistics were analyzed and displayed as box plots in GraphPad Prism 6 software.
pHLARE image analysis
To reproducibly define lysosome objects and corresponding local background regions that were then used to calculate fluorescence ratios and pHlys values, we implemented an image analysis algorithm in NIS Elements software based on mCherry fluorescence intensity thresholding and morphometric manipulation of the resulting binary images. Briefly, the mCherry channel was thresholded at an intensity three times above the nonlysosomal cell membrane pHLARE background. The thresholded binary was smoothened and dilated such that each lysosome area included sufficient surrounding background, and a corresponding local background was defined as the outer 1-pixel contour of each lysosome object. Fluorescent intensity measurements from both pHLARE channels of all lysosome and corresponding local background objects were exported into Microsoft Excel for further analysis.
The pHLARE fluorescence ratio r for each lysosome object was calculated as
in which n is the index for each lysosome object, Ilys is the fluorescence intensity of the lysosome object, and Ibkg is the fluorescence intensity of the corresponding local background. The lysosome pH was calculated in two different ways, using either the mean local background signal or the mean lysosome signal minus camera offset of all lysosome objects to calibrate the pHLARE fluorescence ratio based on the fluorescence intensities in the nigericin images. pH values for each lysosome object were then calculated assuming a linear relationship between pH and pHLARE fluorescence ratio. To reduce the impact of measurement error outliers, the cellular lysosome pH was calculated as the median of all individual lysosome object pH values per cell. An NIS Elements Macro “pHLARE_analysis.mac” performing the image operations semiautomatically, which also allows masking the cell of interest in the measurement and nigericin images, an example Excel worksheet for the pHlys calculations “pHLARE_analysis_worksheet.xlsx,” an example dataset “RPE_cell.zip,” as well as further step-by-step instructions on how to utilize the macro and excel worksheet, are included in the Supplemental Information.
To determine spatially localized pHlys in RPE cells, we used the analysis pipeline described above to measure lysosomes selectively within 3 µm of the distal margin of membrane protrusions and 3 µm of the nuclear membrane, and in MDA-MB-157 and MCF10A cells, we measured the total mCherry fluorescence of pHLARE within 3 µm of the plasma membrane relative to the total mCherry fluorescence of the cell. To determine lysosome size, we also used the analysis program described above that includes the area of lysosome objects. To evaluate the size of single lysosomes and not lysosome clusters, we used only the area of objects with a shape value between 0.9 and 1.0.
Object-based analysis of pHLARE overlap with SiR-lysosome was done with MATLAB using the adaptive thresholding function to define objects above the surrounding background fluorescence. Single pixels were removed and objects smoothed before calculating the fraction of area overlap.
Data presentation and statistical analysis
Box-and-whisker plots were generated using Analyse-It for Microsoft Excel and show median, first, and third quartile, observations within 1.5 times the interquartile range, and all individual data points. Significance of multiple comparisons was calculated by Tukey–Kramer honest significant difference (HSD) test in Analyse-It for Microsoft Excel or by Student’s paired t test. Figures were assembled in Adobe Illustrator CS5.
Supplementary Material
Acknowledgments
We gratefully acknowledge the contributions of technical assistance and expertise from Andreas Ettinger (Helmholtz Center Munich, Germany) and cell lines acquired from the individuals indicated above. This work was supported by National Institutes of Health Grants NS107480 and S10RR026758 (T.W.), CA197855 (D.L.B.), University of California Cancer Research Coordinating Committee (D.L.B.), and by funding from the Paul G. Allen Family Foundation (T.W. and D.L.B.). The authors also note that during the review of our paper, a genetically encoded ratiometric pHlys biosensor was reported (Ponsford et al., 2020. Autophagy 9,1–19) that also uses LAMP1 but with a different design of fluorophores.
Abbreviations used:
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- HPDE
human pancreatic ductal epithelial
- HSD
honest significant difference
- LAMP1
lysosomal-associated membrane protein 1
- mTORC1
mammalian target of rapamycin complex 1
- PBS
phosphate-buffered saline
- pHi
intracellular pH
- pHLARE
pH lysosome activity reporter
- pHlys
lysosome pH
- RPE
retinal pigment epithelial
- sfGFP
superfolder GFP
- S6K1
ribosomal protein S6 kinase beta-1
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-06-0383) on November 25, 2020.
REFERENCES
- Altan N, Chen Y, Schindler M, Simon SM (1999). Tamoxifen inhibits acidification in cells independent of the estrogen receptor. Proc Natl Acad Sci USA 96, 4432–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelqvist H, Wäster P, Kågedal K, Öllinger K (2013). The lysosome: from waste bag to potential therapeutic target. J Mol Cell Biol 5, 214–26. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay D, Cyphersmith A, Zapata JA, Kim YJ, Payne CK (2014). Lysosome transport as a function of lysosome diameter. PLoS One 9, e86847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng XT, Xie YX, Zhou B, Huang N, Farfel-Becker T, Sheng ZH (2018). Characterization of LAMP-1 labeled nondegradative lysosomal and endocytic compartments in neurons. J Cell Biol 217, 3127–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey EE, Beckel JM, Laties AM, Mitchell CH (2014). Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience 263, 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colacurcio DJ, Nixon RA (2016). Disorders of lysosomal acidification - The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res Rev 32, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P, Warwicker J (2009). Evidence for the adaptation of protein pH-dependence to subcellular pH. BMC Biol 7, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel C, Bell C, Burton C, Harguindey S, Reshkin SJ, Rauch C (2013). The role of proton dynamics in the development and maintenance of multidrug resistance in cancer. Biochim Biophys Acta 1832, 606–617. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS (2003). Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268. [DOI] [PubMed] [Google Scholar]
- Deer EL, Gonzalez-Hernandez J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ (2010). Phenotype and genotype of pancreatic cancer cell lines. Pancreas 39, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraldi A, Klein AD, Medina DL, Settembre C (2016). Brain disorders due to lysosomal dysfunction. Annu Rev Neurosci 39, 277–295. [DOI] [PubMed] [Google Scholar]
- Gallagher LE, Radhi OA, Abdullah MO, McCluskey AG, Boyd M, Chan EYW (2017). Lysosomotropism depends on glucose: a chloroquine resistance mechanism. Cell Death Dis 8, e3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo-Hill BK, Webb BA, Barber DL (2014). Ratiometric imaging of pH probes. Methods Cell Biol 123, 429–448. [DOI] [PubMed] [Google Scholar]
- Grillo-Hill BK, Choi C, Jimenez-Vidal M, Barber DL (2015). Increased H+ efflux is sufficient to induce dysplasia and necessary for viability with oncogene expression. eLife 4, e03270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamalisto S, Jaattela M (2016). Lysosomes in cancer-living on the edge (of the cell). Curr Opin Cell Biol 39, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal JK, Andrews NW, Simon SM (2002). Membrane proximal lysosomes are the major vesicles responsible for calcium-dependent exocytosis in nonsecretory cells. J Cell Biol 159, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, Ostrowski P, Jaumouille V, Grinstein S (2016). The position of lysosomes within the cell determines their luminal pH. J Cell Biol 212, 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki T, Olsen OD, Jaattela M (2012). Cancer-associated lysosomal changes: friends or foes? Oncogene 32, 1995–2004. [DOI] [PubMed] [Google Scholar]
- Kenific CM, Debnath J (2015). Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol 25, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornak U, Kasper D, Bösl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ (2001). Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell 104, 205–215. [DOI] [PubMed] [Google Scholar]
- Kovalenko I, Glasauer A, Schöckel L, Sauter DR, Ehrmann A, Sohler F, Hägebarth A, Novak I, Christian S (2016). Identification of KCa3.1 Channel as a novel regulator of oxidative phosphorylation in a subset of pancreatic carcinoma cell lines. PLoS One 11, e0160658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, McBrayer MK, Wolfe DM, Haslett LJ, Kumar A, Sato Y, Lie PP, Mohan P, Coffey EE, Kompella U, et al (2015). Presenilin 1 maintains lysosomal Ca2+ homeostasis via TRPML1 by regulating vATPase-mediated lysosome acidification. Cell Rep 12, 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HJ, Herman P, Lakowicz JR (2003). Fluorescence lifetime-resolved pH imaging of living cells. Cytometry A 52, 77-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Palmfeldt J, Lin L, Colaço A, Clemmensen KKB, Huang J, Xu F, Liu X, Maeda K, Luo Y, Jäättelä M (2018). STAT3 associates with vacuolar H+-ATPase and regulates cytosolic and lysosomal pH. Cell Res 28, 996–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WJ, Shen TT, Chen RH, Wu HL, Wang YJ, Deng JK, Chen QH, Pan Q, Huang CM, Fu CM, et al. (2015). Autophagy-lysosome pathway in renal tubular epithelial cells is disrupted by advanced glycation end products in diabetic nephropathy. J Biol Chem 290, 20499–20510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Cruz D, Asamoah N, Buxbaum A, Sohar I, Lobel P, Maxfield FR (2007). Activation of microglia acidifies lysosomes and leads to degradation of Alzheimer amyloid fibrils. Mol Biol Cell 18, 1490–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshansky V, Futai M (2008). The V-type Hfl-ATPase in vesicular trafficking: Targeting, regulation and function. Curr Opin Cell Biol 20, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA (2012). Lysosomal acidification mechanisms. Annu Rev Physiol 74, 69–86. [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark NB, Coppe JP, Tong F, Speed T, et al. (2006). A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson C, Roberg K, Grafström RC, Ollinger K (2010). Intrinsic differences in cisplatin sensitivity of head and neck cancer cell lines: Correlation to lysosomal pH. Head Neck 32, 1185–1194. [DOI] [PubMed] [Google Scholar]
- Nishi K, Suzuki M, Yamamoto N, Matsumoto A, Iwase Y, Yamasaki K, Otagiri M, Yumita N (2018). Glutamine deprivation enhances acetyl-CoA carboxylase inhibitor-induced death of human pancreatic cancer cells. Anticancer Res 38, 6683–6689. [DOI] [PubMed] [Google Scholar]
- Nixon RA (2013). The role of autophagy in neurodegenerative disease. Nat Med 19, 983–997. [DOI] [PubMed] [Google Scholar]
- Ostrowski PP, Fairn GD, Grinstein S, Johnson DE (2016). Cresyl violet: a superior fluorescent lysosomal marker. Traffic 17, 1313–1321. [DOI] [PubMed] [Google Scholar]
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS (2006). Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24, 79–88. [DOI] [PubMed] [Google Scholar]
- Perera RM, Bardeesy N (2015). Pancreatic cancer metabolism: Breaking it down to build it back up. Cancer Discov 5, 1247–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, Zoncu R (2016). The lysosome as a regulatory hub. Annu Rev Cell Dev Biol 32, 223–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S, Amaravadi RK (2016). Targeting the lysosome in cancer. Ann NY Acad Sci 1371, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Guardia CM, Keren-Kaplan T, Bonifacino JS (2016). Mechanisms and functions of lysosome positioning. J Cell Sci 129, 4329–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen M, Pochini L, Stasyk T, de Araujo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, et al. (2015). SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez A, Webster P, Ortego J, Andrews NW (1997). Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J Cell Biol 137, 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer J, Schweizer A, Russell D, Kornfeld S (1996). The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J Cell Biol 132, 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM (2017). mTOR signaling in growth, metabolism and disease. Cell 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenenberger CA, Zuk A, Kendall D, Matlin KS (1991). Multilayering and loss of apical polarity in MDCK cells transformed with viral K-ras. J Cell Biol 112, 873–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CC, Gruenberg J (2011). Ion flux and the function of endosomes and lysosomes: pH is just the start: the flux of ions across endosomal membranes influences endosome function not only through regulation of the luminal pH. Bioessays 33, 103–110. [DOI] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, et al. (2013). TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 15, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Stehbens S, Pemble H, Murrow L, Wittmann T (2012). Imaging intracellular protein dynamics by spinning disk confocal microscopy. Methods Enzymol 504, 293–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataranni T, Agriesti F, Ruggieri V, Mazzoccoli C, Simeon V, Laurenzana I, Scrima R, Pazienza V, Capitanio N, Piccoli C (2017). Rewiring carbohydrate catabolism differentially affects survival of pancreatic cancer cell lines with diverse metabolic profiles. Oncotarget 8, 41265–41281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284, 8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GG, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90. [DOI] [PubMed] [Google Scholar]
- Tu C, Ortega-Cava CF, Chen G, Fernandes ND, Cavallo-Medved D, Sloane BF, Band V, Band H (2008). Lysosomal cathepsin B participates in the podosome-mediated extracellular matrix degradation and invasion via secreted lysosomes in v-Src fibroblasts. Cancer Res 68, 9147–9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton ZE, Patel CH, Brooks RC, Yu Y, Ibrahim-Hashim A, Riddle M, Porcu A, Jiang T, Ecker BL, Tameire F, et al. (2018). Acid suspends the circadian clock in hypoxia through inhibition of mTOR. Cell 174, 72–87.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Dong B, Kong X, Zhang N, Song W, Lin W (2018). Dual site-controlled two-photon fluorescent probe for the imaging of lysosomal pH in living cells. Luminescence 33, 1275–1280. [DOI] [PubMed] [Google Scholar]
- Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. (2015). Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BA, Chimenti M, Jacobson MP, Barber DL (2011). Dysregulated pH: a perfect storm for cancer progression. Nature Cancer Rev 11, 671–677. [DOI] [PubMed] [Google Scholar]
- Webb BA, Forouhar F, Szu FE, Seetharaman J, Tong L, Barber DL (2015). Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations. Nature 523, 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Garrido Ruiz RG, Szpiech ZA, Strauli NB, Hernandez RD, Jacobson JP, Barber DL (2017a). Cancer-associated arginine to histidine mutations confer a gain in pH sensing to mutant proteins. Sci Signaling 10, pii: eaam9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Grillo-Hill BK, Barber DL (2017b). Dysregulated pH dynamics enables cancer cell behaviors. J Cell Sci 130, 663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe DM, Lee JH, Kumar A, Lee S, Orenstein SJ, Nixon RA (2013). Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci 37, 1949–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Ren D (2015). Lysosomal physiology. Annu Rev Physiol 77, 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap CC, Digilio L, McMahon LP, Garcia ADR, Winckler B (2018). Degradation of dendritic cargos requires Rab7-dependent transport to somatic lysosomes. J Cell Biol 217, 3141–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. (2010). Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhitomirsky B, Assaraf YG (2016). Lysosomes as mediators of drug resistance in cancer. Drug Resist Updat 24, 23–33. [DOI] [PubMed] [Google Scholar]
- Zhitomirsky B, Farber H, Assaraf YG (2018). LysoTracker and MitoTracker Red are transport substrates of P-glycoprotein: implications for anticancer drug design evading multidrug resistance. J Cell Mol Med 22, 2131–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Tan SH, Nicolas V, Bauvy C, Yang ND, Shang J, Y. X, Codogno P, Shen HM (2013). Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res 23, 508–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.