Abstract
The endoplasmic reticulum (ER) is one of the largest cytoplasmic organelles in eukaryotic cells and plays a role in many cellular processes, such as the production and quality control of secretory protein, lipid synthesis, and calcium homeostasis. The ER cannot be generated de novo, and thus its proper inheritance during cell division is paramount to the health and survival of the daughter cells. Although previous work has uncovered the cytoskeletal components involved, we still lack a comprehensive understanding of the intricate steps of and the cytoplasmic and membrane-bound components involved in ER inheritance. To directly address these issues, we utilized microfluidics and genetic analyses to show that before nuclear migration, early ER inheritance can be further divided into three distinctive steps. Moreover, we demonstrated that perturbing each of these steps affects the cell’s ability to mitigate ER stress. Thus, proper ER inheritance is essential to ensuring a healthy, functional cell.
INTRODUCTION
One of the hallmarks of eukaryotic cells is that their various biochemical activities are compartmentalized into membrane-bound organelles, which are fine-tuned for specific cell functions. During cell division, the inheritance of the genome, which is regulated by DNA cell cycle checkpoints, has to occur accurately and timely in order to faithfully transmit genetic information. The inheritance of most organelles relies on the directional transport of motor proteins on cytoskeletal tracks provided by either actin or microtubules. For instance, vacuoles and peroxisomes are transferred into the daughter cell by Myosin 2 on actin cables (Provance and Mercer, 1999; Fagarasanu et al., 2006; Peng and Weisman, 2008; Tang et al., 2019), while nuclear inheritance is dependent on microtubules (Du et al., 2004). The endoplasmic reticulum (ER) is morphologically distinct from other organelles; it consists of a continuous network of membrane tubules and sheets that undergo reorganization. The ER membrane is continuous with the outer nuclear membrane, which is classified as perinuclear ER (pnER) and is embedded with the nuclear pore complex. The cortical ER (cER) is juxtaposed with the plasma membrane (PM) and occasionally makes physical contacts with it. In higher eukaryotes, the interconnected ER tubules between the pnER and the cER consist of three-way junctions (Schwarz and Blower, 2016). In budding yeast, fewer ER tubules traverse the cytoplasm and connect the pnER and cER. Although a continuous luminal domain connects the pnER and cER (Luedeke et al., 2005), they are morphologically and functionally different. The pnER regulates materials entering and exiting the nucleus, while cER plays a role in lipid synthesis (Pichler et al., 2001; Tavassoli et al., 2013). Also, the inheritance of the pnER and the cER occurs via separate mechanisms at different stages of the cell cycle. In Saccharomyces cerevisiae, cells undergo closed mitosis in which the nuclear membrane (pnER) never breaks down and pnER inheritance takes place in G2/M phases of the cell cycle as part of nuclear migration, which is mediated by astral microtubules and the dynein–dynactin motor proteins that “walk” on microtubules by ATP hydrolysis, providing the force necessary to pull the nucleus into the daughter cell (Varshney and Sanyal, 2019).
The inheritance of cER in yeast is distinct from nuclear migration and may not be entirely dependent on the cytoskeleton. Disrupting the microtubules with nocodazole, a microtubule-depolymerizing agent, does not affect the delivery or maintenance of cER in the bud (Fehrenbacher et al., 2002). Although the actin cytoskeleton plays a role in maintaining the morphology and dynamics of cER, perturbing actin cables by either Latrunculin A treatment or temperature-sensitive mutants of the actin-encoding ACT1 gene does not completely abolish cER inheritance (Prinz et al., 2000; Fehrenbacher et al., 2002). Instead, cER inheritance occurs as early as bud emergence, when a pool of cER is observed to concentrate at the incipient bud site (Fehrenbacher et al., 2002). Because cER is associated with the PM, it may be passively inherited to the bud by following PM expansion during bud growth, but the precise mechanism is not understood. Nonetheless, many mutants have been identified to affect the structure of cER. One class of mutants are involved in ER-to-Golgi vesicular trafficking (Prinz et al., 2000), and another set of mutants were found to encode for proteins that form a tethering complex between cER and PM (Manford et al., 2012). Additionally, the integral ER proteins Aux1 and Ice2 have been implicated in the distribution of cER in the bud (Du et al., 2001; Estrada de Martin et al., 2005). The variety of mutants identified thus far likely suggests that the inheritance of cER is a multistep process that requires multiple redundant components.
More recently, the different stages of ER inheritance in yeast have been further revealed by an ultrastructural study that used electron microscopy tomography (EM) to reconstruct the ER in three-dimension (West et al., 2011). According to this study, in cells with small buds, an ER tubule that appears to have originated from the mother pnER is pointed toward the bud. Next, this ER tubule seems to anchor at the bud tip and spread into a “nexus” of ER in the bud cortex in close proximity to the PM. This peripheral ER in the bud contains both tubular and cisternal structures and is consistent with what we know as cER (West et al., 2011). Even though this study unveiled clear details of the ER structural modifications at different stages of bud growth, temporal information is lacking, and the presence of alternative routes to inherit the ER into the daughter cell remain elusive.
In addition to the ER inheritance stages under the normal growth condition, little was known about how the ER inheritance is impacted by the functional status of the ER. Previously, we discovered a cell-cycle checkpoint mechanism called the ER stress surveillance (ERSU) pathway that ensures that daughter cells inherit functional ER, and not ER that has been damaged by ER stress (Babour et al., 2010). Ultimately, the blocking of ER inheritance during ER stress leads to the cytokinesis block, leading to the halting of the cell cycle (Chao et al., 2019). Interestingly, we found that the ERSU pathway is independent of the well-known unfolded protein response (UPR) pathway (Walter and Ron, 2011) that reestablishes ER homeostasis. While the UPR is activated by IRE1, an ER transmembrane kinase/endoribonuclease, the ERSU does not utilize IRE1. Rather, ERSU is regulated by the mitogen activated protein (MAP) kinase Slt2. Thus, as ER function is replenished by the UPR, cells reenter the cell cycle and resume ER inheritance (Piña and Niwa, 2015).
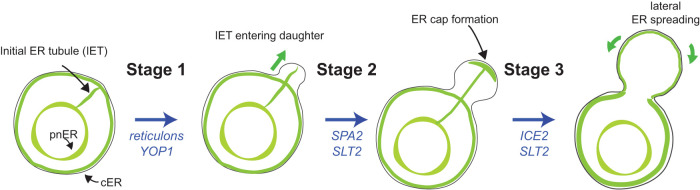
To further the understanding of the ER inheritance, we set out to investigate how the spatial–temporal and morphological changes of the ER are orchestrated during ER inheritance in live yeast cells upon employment of microfluidic chambers. Furthermore, we investigated how ER stress affects ER inheritance and also tested impacts of genetic mutants that have been reported to block ER inheritance. We found that ER inheritance into the daughter cell can be largely divided into three stages in live-cell analyses. In stage one, an ER tubule emanated from the mother pnER is oriented toward and extends into the incipient bud. In stage two, this ER tubule will be extended and ultimately anchored at the bud tip and form an enriched ER cap. Finally, in stage three, new ER tubules emerge from the ER cap and spreads into the bud periphery, forming cER. ER inheritance was blocked at one of these stages in different mutants, suggesting that these key stages are rate-limiting stages for the ER inheritance.
RESULTS AND DISCUSSION
The reticulons and Yop1 regulate the first stage of endoplasmic reticulum inheritance
We reported that ER stress inhibits cER inheritance into the daughter cell as part of the ERSU events (Babour et al., 2010; Piña and Niwa, 2015; Piña et al., 2016, 2018; Chao et al., 2019). Although ER inheritance during normal growth conditions has been described, ER inheritance during ER stress has not been investigated in detail. To further investigate, we performed live-cell time-lapse imaging to observe the sequence of events that lead to ER inheritance and its block with improved temporal resolution. We utilized a microfluidics chamber to simulate a native growth environment for a living cell culture (Figure 1A). The microchamber was coated with Con A to immobilize cells (Hansen et al., 2015) and was continuously perfused with either fresh medium or medium containing tunicamycin (Tm), a well-characterized ER stress inducer that inhibits N-glycosylation, so that cells accumulate nonglycosylated unfolded proteins in the ER lumen that activate both the unfolded protein response (UPR) and the ERSU cell cycle checkpoint.
FIGURE 1:
Rtn1, Rtn2, and Yop1 are required for the generation of an initial ER tubule (IET) from the mother pnER. (A) Schematic of the design of the microfluidics device used throughout this study. (B) WT cells expressing Pho88-GFP were immobilized in a ConA-coated microfluidics device, which was continuously perfused with fresh synthetic complete (SC) medium. All images were taken every 4 min from a single central focal plane using fluorescent microscopy. Yellow arrows point to the appearance of an IET in the mother (at 4 min) and in the bud (at 20 min) or an ER cap in the bud (at 24 min); red arrows indicate the approximate locations of those structures in the DIC image. Also shown are the tracings of ER fluorescence. All scale bars 2 µm. See also Supplemental Movies S1 (Pho88-GFP) and S2 (Sec61-GFP) and Supplemental Figures S1A and S3A. (C) Similar to B, but the medium contained 1µg/ ml tunicamycin (Tm). Yellow arrowheads indicate the absence of an expected IET or ER cap in the bud. See also Supplemental Movie S3. (D) Quantifications of buds containing an IET as visualized using Pho88-GFP in cells grown in SC or SC+Tm. Error bars indicate standard error (S.E.) and asterisk indicates p < 0.005. (E) Cartoon representations of early events of ER inheritance in cells under normal or ER stress conditions. (F) WT cells expressing Hmg1-GFP were immobilized on an agar pad containing YPD and imaged every 4 min. Images are deconvolved projections from a z-stack. Yellow arrows indicate an IET extending from mother pnER toward the bud; red arrows indicate the approximate locations of those structures in DIC images. (G) Similar to D but in rtn1∆rtn2∆yop1∆ cells. Yellow arrowheads indicate the absence of an expected IET. (H) Similar to F, quantifications of daughter cells containing an IET as visualized using Hmg1-GFP in the indicated strains. n > 50 for each strain. Error bars, S.E.; asterisk, p < 0.05.
We used Pho88-GFP, an integral membrane protein, as an ER marker to monitor the earliest stages of ER inheritance (Figure 1B; Supplemental Movie S1). The first clear event observed was a tubular structure emanating from the pnER and directed toward the incipient bud (Figure 1B, 4 min). This early tubular ER was also observed with a different ER membrane reporter, Sec61-GFP (Supplemental Figure S1A; Supplemental Movie S2). While a few ER tubules linking the cER and the pnER could be detected in the mother, only one tubule entered the bud (Figure 1B, 20 min). From here on, we will refer to this tubular ER structure as the “initial ER tubule (IET).”
FIGURE 4:
The effects of ER stress on the third stage of ER inheritance, involving the spreading of cER into the bud cortex. (A) Later time points (48 to 68 min) of the microfluidic experiment shown in Figure 1B. Yellow arrows indicate cER and red arrows indicate the approximate locations of cER in DIC images. See also Supplemental Movie S1, Supplemental Figure S3A, and Supplemental Movie S4. All scale bars 2 µm. (B) slt2∆ice2∆ cells expressing Pho88-GFP were imaged using microfluidics in SC media. Yellow arrowheads indicate incomplete ER spreading in the daughter cell; red arrows indicate those locations in DIC. Magnified views of the cER (Pho88-GFP) of slt2∆ice2∆ daughter cells were shown. See also Supplemental Figure S3C and Supplemental Movie S6. (C) WT and slt2∆ice2∆ cells expressing Pho88-GFP. Yellow arrows indicate cER and yellow arrowheads indicate missing cER; red arrows indicate those locations in DIC. Magnified views of the cER (Pho88-GFP) of either WT or slt2∆ice2∆ daughter cells are shown. (D) Quantifications of cER of cells shown in C (n > 50). The proportion of cell perimeter with associated cER was assessed in mothers and buds separately using Pho88-GFP as the ER marker. Error bars, S.E.; asterisks, p < 0.05. (E) WT and slt2∆ice2∆ cells expressing Tcb3-GFP. Yellow arrows indicate cER and yellow arrowheads indicate incomplete cER; red arrows indicate those locations in DIC images. Magnified views of the cER (Tcb3-GFP) of either WT or slt2∆ice2∆ daughter cells are shown. (F) Similar to C, but cells were treated with Tm. Yellow arrowheads indicate the expected ERSU-induced cER inheritance block phenotype; red arrows indicate those locations in DIC. Magnified views of the cER (Pho88-GFP) of either WT or slt2∆ice2∆ daughter cells were shown. See also Supplemental Figure S3D for quantitation. For one of the WT cell images, the position of the cell surface membrane is traced with a white dotted line. (G) Serial dilutions of WT and indicated mutants at mid-log phase were spotted onto SC media supplemented with 1 M sorbitol and with or without 0.1 µg/ml Tm. (H) Similar to G, but slt2∆ice2∆ cells containing the indicated plasmids were used. (I) Cartoon representations illustrating cER spreading from ER cap. Deleting SLT2 and ICE2 interferes with cER spreading.
We next investigated how ER stress might alter IET entry into daughter cells. This time, the microfluidics chamber was perfused with medium containing tunicamycin (Tm) to induce ER stress on wild-type (WT) cells expressing Pho88-GFP (Figure 1C; Supplemental Movie S3). We found that although the IET emanated from the mother pnER was still oriented correctly to the incipient bud neck, it did not enter the bud (Figure 1C, 0–20 min). Subsequently, as the daughter cell grew, little GFP accumulated in the bud (Figure 1C; ∼28–32 min). This persisted even when the bud became slightly larger (Figure 1C, 44 min). Quantitation indicated that IET entry into buds was significantly reduced when cells were under ER stress (Figure 1D). Taken together, these results showed that ER stress blocked IET entry into the daughter cell (Figure 1E), suggesting that components responsible for the early stages of the ER inheritance were sensitive to ER stress.
Previously, we identified Rtn1, Rtn2, and Yop1 as additional components of the ERSU pathway and that rtn1∆rtn2∆yop1∆ cells were unable to activate the ERSU response (Piña et al., 2016). Thus, we next investigated the details of ER inheritance in rtn1∆rtn2∆yop1∆ cells using a different ER reporter, Hmg1-GFP. In time-lapse studies, we noticed that IETs were often missing in the mother in rtn1∆rtn2∆yop1∆ cells (Figure 1, F and G; Supplemental Figure S1B). We also frequently failed to detect tubular ER in the buds of rtn1∆rtn2∆yop1∆ cells (Figure 1, G and H). In those cells that did contain an IET that originated from the mother pnER, the IET failed to orient correctly toward the bud (Figure 2A for WT vs. Figure 2, B and C for rtn1∆rtn2∆yop1∆ cells). Importantly, rtn1∆rtn2∆yop1∆ cells did not show contiguous cER with normal appearance (Figure 2A for WT vs. Figure 2, B and C for rtn1∆rtn2∆yop1∆ cells). This was expected, as Rtn1, Rtn2, and Yop1 play roles in establishing ER curvature (Voeltz et al., 2002). Nonetheless, we were surprised to find that a few rtn1∆rtn2∆yop1∆ daughter cells still inherited some ER (Figure 2, D and E), although the majority of these cells (∼80%) had abnormal tubules (Figure 2F), suggesting that ER inheritance was atypical in the absence of Rtn1, Rtn2, and Yop1. Instead of inheriting ER via an IET that emanated normally from the mother pnER, the cER of rtn1∆rtn2∆yop1∆ mother cell appeared to invade the daughter cell cortex and generated cER-like structures with severely altered morphologies (Figure 2, D and E, arrowheads). ER stress did not induce additional ER inheritance defects in rtn1∆rtn2∆yop1∆ cells (Figure 2, G and H, and Supplemental Figure S1, B and C; quantitation in Figure 2I).
FIGURE 2:
Abnormal cER inheritance in rtn1∆rtn2∆yop1∆ cells. (A) Visualization of an IET (ER was labeled with Hmg1-GFP) pointing toward the bud (yellow arrow) in a WTe cell. All scale bars 2 µm. (B–E) Time-lapse images of four different rtn1∆rtn2∆yop1∆ cells expressing Hmg1-GFP. Also shown are the tracings of ER in these cells to illustrate the four different types of ER inheritance defects observed. Yellow arrowheads indicate non-WT tubular structures that either did not enter the bud—B, C—or did not originate from the mother pnER—D, E. (F) Quantifications of the abnormal ER tubules observed in unstressed WT and rtn1∆rtn2∆yop1∆ cells (n > 50). Error bars, S.E.; asterisk, p < 0.05. (G, H) Sample images of WT or rtn1∆rtn2∆yop1∆ cells expressing Hmg1-GFP. Yellow arrowheads showed the absence of an IET in buds due to ER stress induction (Tm). (I) Quantifications of IET in mothers with an emerging bud in WT and rtn1∆rtn2∆yop1∆ cells (n > 50). Error bars, S.E.; asterisk, p < 0.05; n.s., not significant. (J) Serial dilutions of WT, rtn1∆rtn2∆yop1∆, and ire1∆ cells at mid-log phase were spotted onto SC media containing DMSO only or 0.5 µg/ml Tm.
Finally, we assessed the functional significance of the abnormal ER inheritance in rtn1∆rtn2∆yop1∆ cells. In growth assays, we found that rtn1∆rtn2∆yop1∆ cells grew similarly to WT cells under normal conditions, but grew significantly worse on Tm-containing medium, although not to the same extent as the UPR- deficient ire1∆ cells (Figure 2J). These results suggested that proper formation and entry of the IET was critical for cell survival under ER stress.
The results so far indicated that the shape of ER may be an important determinant of IET formation. As shown previously by EM tomography, pnER consists of smooth ER sheets (West et al., 2011). So how might a tubule arise from a flat-membrane sheet structure? The reticulons and Yop1 contain a conserved protein domain that inserts into the ER membrane as hairpins (Voeltz et al., 2006), which may facilitate the generation of membrane curvature by forming a wedge on the cytoplasmic side of the membrane (Shibata et al., 2010). Inducing membrane curvatures by recruiting membrane shaping proteins is a common cellular strategy for generating new structures from the smooth surface of the ER, such as COPII vesicle formation at ER exit sites (Jensen and Schekman, 2011). Thus, the curvature-forming property of reticulons and Yop1 may be the mechanism responsible for IET formation from pnER. In support of this hypothesis, the ER in cells lacking reticulons is converted into large sheets (Voeltz et al., 2006), and we indeed observed that the ER tubules emanating from pnER were often poorly formed, if formed at all, in cells missing reticulons and Yop1. As a result, the buds in these cells had very little cER, suggesting that the shape of the ER is highly regulated and is important for ER inheritance. The growth defect of rtn1∆rtn2∆yop1∆ cells in the presence of ER stress highlighted their significance in formation of IET and ER inheritance.
Spa2 and Slt2 are required for the formation of an endoplasmic reticulum cap in the second stage of endoplasmic reticulum inheritance
After IET entered the daughter cell, we observed strong Pho88-GFP signals accumulating at the bud tip (Figure 1B, 28–44 min). We termed this rich ER deposit at the bud tip the “ER cap.” We also witnessed ER cap formation with other ER reporters, including Hmg1-GFP (Figure 1F) or Sec61–GFP (Supplemental Figure S1A). Moreover, the ER cap was only apparent in cells with small to medium buds (bud index of ∼0.15) and not in cells with very small (bud index ∼0.05) or medium to large buds (bud index ∼0.3; Supplemental Figure S2A). Thus, we hypothesized that the ER cap might have differentiated from the IET upon its binding to proteins at the bud tip. In support of this, previous studies have described that the ER is anchored at the bud tip before spreading to the cell cortex (Fehrenbacher et al., 2002; Wiederkehr et al., 2003).
Previously, we reported that the MAP kinase Slt2 is needed for activating the ERSU pathway (Babour et al., 2010). While Slt2 performs multiple cellular functions, we found that the population of Slt2 involved in ERSU is localized to the bud tip (Chao et al., 2019). Thus, we reasoned that identifying Slt2 interacting partners at the bud tip might be an avenue to uncover the component(s) that anchors the IET at the bud tip. Through searching genetic interactions annotated for SLT2 on the Saccharomyces Genome Database (SGD), we noticed that SPA2 has a negative genetic interaction with SLT2 (Costanzo et al., 2010). Spa2 is part of the polarisome complex and is localized to the bud tip early in the yeast cell cycle (Pruyne and Bretscher, 2000). Two yeast–two hybrid screens have demonstrated that Spa2 interacts directly with Slt2 (Sheu et al., 1998; van Drogen and Peter, 2002; Supplemental Figure S2B). Importantly, while reexamining our previously published DHFR screen for Slt2 (Chao et al., 2019), we noticed that Spa2 showed increased interactions with Slt2 during ER stress (Supplemental Figure S2C). In addition to these physical interaction data, other lines of evidence also suggest that Spa2 may function with Slt2 in ER inheritance under normal growth conditions (Loewen et al., 2007; Li et al., 2013). Thus, we investigated how Spa2 might function with Slt2 in the later stages of ER inheritance after IET entry into the daughter cell.
To confirm the protein–-protein interactions between Slt2 and Spa2, we used the protein-complementation assay with split-YFP as a reporter to visualize the direct binding of Slt2 and Spa2 in living cells (Michnick et al., 2007). In this assay, Slt2 was fused to one half of YFP, while Spa2 was fused to the complementary half of YFP. Physical interaction between Slt2 and Spa2 proteins brought both fragments of YFP into close proximity, allowing them to reconstitute and emit fluorescence. In addition to demonstrating the physical interaction between two proteins, this also revealed the subcellular site of the interaction. We found that YFP was localized to the tips of small buds, a location similar to that of the ER caps (Figure 3A). Furthermore, Spa2-GFP coimmunoprecipitated with a fraction of total cellular Slt2 (Supplemental Figure S2D). These results revealed that a pool of Slt2 localized to the tip of small buds interacted with Spa2 during polarized cell growth, consistent with published observations (Li et al., 2010).
FIGURE 3:
The impact of ER stress on the second stage of ER inheritance, involving the formation of an ER cap. (A) Split-YFP PCA between Slt2 and Spa2. Yellow arrows indicate localization of the interaction at bud tips; red arrows indicate the approximate locations of those interactions in DIC images. All scale bars 2 µm. (B) slt2∆spa2∆ cells expressing Pho88-GFP were imaged using microfluidics in SC media. Yellow arrowhead indicates the absence of an ER cap, and red arrow indicates a bud tip. See also Supplemental Figure S3B and Supplemental Movie S5 (slt2∆spa2∆) vs. Supplemental Movie S3 (WT). (C) WT and slt2∆spa2 cells expressing Pho88-GFP. Yellow arrows indicate ER cap and cER in the daughter cells; yellow arrowheads indicate the absence of an ER cap; red arrows indicate the approximate locations of the interactions in DIC images. (D) Quantifications of ER cap formation for cells in C (n > 50) by measuring ER fluorescence ratios between bud tip and bud periphery and expressed as percentages. Error bars, S.E.; asterisk, p < 0.05. (E) Similar to C, but medium contained 1 µg/ml Tm to induce ER stress. Yellow arrowheads indicate the absence of cER and red arrows show those locations in DIC images. (F) Quantitation of ER inheritance in small-budded WT and slt2∆spa2∆ daughter cells grown with or without Tm. Only cells whose bud-to-mother size ratios were less than 0.3 were quantified. Error bars, S.E.; asterisks, p < 0.05; n.s., not significant. (G) WT and slt2∆spa2∆ cells expressing Tcb3-GFP. Yellow arrows indicate normal cER distribution and yellow arrowheads indicate defects in cER; red arrows indicate those locations in DIC images. Magnified views of the cER (Tcb3-GFP) of either WT or slt2∆spa2∆ daughter cells were shown. (H) Serial dilutions of WT and indicated mutants at mid-log phase were spotted onto SC media supplemented with 1 M sorbitol, and with or without 0.1 µg/ml Tm. (I) Similar to H, except that slt2∆spa2∆ cells were transformed with a pRS315 plasmid containing no insert or the indicated alleles of SLT2. (J) Cartoon representations showing the normal process of IET anchoring followed by the formation of ER cap. Deleting SLT2 and SPA2 interferes with both processes.
To dissect the functional significance of Spa2 in normal growth and under ER stress, we monitored ER inheritance in slt2∆spa2∆ cells. During ER stress, ER inheritance is normally blocked in WT cells early in the cell cycle (Babour et al., 2010; Piña and Niwa, 2015; Piña et al., 2018). Thus, evaluating the effects of ER stress in the later stages of ER inheritance, such as the anchoring of IET to the bud tip or the lateral expansion of cER from the bud tip, is rather difficult in WT cells. However, in slt2∆ cells, ER is inherited even under ER stress (but this ultimately leads to cell death), thus providing us with an opportunity to investigate the role of Spa2 after the ER anchoring step. To this end, we generated slt2∆spa2∆ cells carrying Pho88-GFP and performed microfluidics experiments. During normal growth, slt2∆spa2∆ cells showed nearly normal ER inheritance, but ER cap formation was noticeably missing when compared with WT (Figure 3B vs Figure 1B and Supplemental Figure S3A vs. Supplemental Figure S3B). Quantifications confirmed that ER cap formation was significantly reduced in slt2∆spa2∆ (Figure 3, C and D; Supplemental Figure S3A vs. Supplemental Figure S3B; Supplemental Movie S4 vs. Supplemental Figure S5). During ER stress, due to the lack of SLT2, cells were not able to initiate the ERSU response and ER inheritance was not blocked in slt2∆spa2∆, resulting in some ER signals being detected in the daughter cells (Figure 3, E and F).
To further characterize the nature of the ER observed in slt2∆spa2∆ cells, we utilized Tcb3-GFP, an integral ER protein that is also a part of the ER–PM contact site complex (Toulmay and Prinz, 2012). Tcb3-GFP allows the visualization of only the cER rather than total ER. Indeed, we found that in WT cells, Tcb3-GFP signal was abundant in the daughter cell cortex (Figure 3G, yellow arrows). However, in slt2∆spa2∆ cells, Tcb3-GFP signal was very patchy or fragmented (Figure 3G, yellow arrowheads). These results were consistent with our earlier findings that slt2∆spa2∆ cells did not inherit complete cER (Figure 3F). Interestingly, we noticed that the cER defects in slt2∆spa2∆ were more apparent in medium-budded cells, after the stage of ER cap formation (Figure 1B, 40 min onward; Supplemental Figure S1A, 40 min onward; Supplemental Figure S3A, 8 min onward), suggesting that the lack of an ER cap in the daughter cells of slt2∆spa2∆ may be associated with their cER defects.
To test if Spa2-dependent ER cap formation played a role in the ERSU pathway, and to investigate the significance of the Slt2–Spa2 interaction, we examined the growth of slt2∆spa2∆ cells. The growth of the double mutant was reduced as compared with that of either slt2∆ or spa2∆ cells, even under normal conditions, providing further support for a negative genetic interaction between SLT2 and SPA2 (Figure 3H). It should be noted that we added an osmotic stabilizer (sorbitol) to the media for this growth assay, since Slt2 is also involved in the cell wall integrity (CWI) response. Importantly, this growth difference was further exacerbated in the presence of ER stress (Figure 3H). Previously, we reported that both Slt2’s kinase activity and its autophosphorylation are critical for the ERSU pathway (Babour et al., 2010). Here, we found that either the autophosphorylation mutant (T190AY192F) or the kinase dead mutant (K54R) of Slt2 was capable of rescuing the growth sensitivity under both normal and ER stress growth conditions (Figure 3I). The extent of rescue by either allelic variants of Slt2 was similar to that of WT Slt2 regardless of ER stress induction, suggesting that Spa2 likely interacted with Slt2 outside of these residues. Taken together, the genetic and physical interactions between Slt2 and Spa2 indicated that they functioned together at the bud tip to facilitate the formation of the ER cap (Figure 3K).
Ice2 functions with Slt2 in the third stage of endoplasmic reticulum inheritance
In our microfluidics experiment with WT cells, we observed that at later time points, the ER cap was less prominent, and cER became more evenly distributed in the bud periphery (Figure 4A, beyond 48 min; Supplemental Figure S3A, after 20 min; Supplemental Movies S1, S4, and S7). This suggested that as the bud grew, ER proliferated from the ER cap to decorate the bud cortex, ultimately forming the PM-associated reticular cER network that is frequently described in the literature. For instance, a recent study used EM tomography to show that tubular ER in a small bud branches out from a central nexus toward the bud periphery and makes contacts with the PM, eventually establishing cER (West et al., 2011). In addition, another study used time-lapse microscopy to show that tubular ER in the bud undergoes dynamic branching during growth (Costanzo et al., 2010). Based on previously published and our current observations, we hypothesized that the dynamic remodeling of the ER cap, and the proliferation and spreading of ER into the bud cortex to establish cER is an essential step in ER inheritance that could be affected by ER stress.
We searched for candidates that might function with Slt2 in the later stages of ER inheritance, in particular ER spreading. Because SPA2 showed a negative genetic interaction with SLT2 and played a role in capturing tubular ER at the bud tip to form an ER cap, we reasoned that component(s) that interact negatively with SLT2 may include gene(s) involved in spreading ER from the ER cap. We noticed that ICE2 negatively interacts with SLT2 in an unbiased whole genome survey (Estrada de Martin et al., 2005). Ice2 is an ER transmembrane protein, and ice2∆ cells have mild defects in the distribution of cER (Babour et al., 2010). Thus, we wondered if the genetic interaction between ICE2 and SLT2 might suggest that ICE2 functions in spreading ER from the ER cap. To test this idea, we used live-cell imaging to follow ER inheritance in slt2∆ice2∆ cells, with a focus on the later stages (Figure 4B; Supplemental Figure S3, A and C; Supplemental Movie S6). Even though we observed normal ER tubule anchoring to the bud tip as well as the formation of an ER cap, we noticed that ER spreading in the bud was incomplete (Figure 4B, starting at the 20-min mark; Supplemental Figure S3C, after 32 min). We discovered that while WT cells have ∼80% of their cell periphery covered with cER, slt2∆ice2∆ cells only had ∼16% cER in the bud and ∼35% in the mother (representative images, Figure 4C; quantification, Figure 4D). In comparison, ∆ice2 single mutant had 39% cER in the bud and 50% in the mother (Figure 4D). Using Tcb3-GFP, we confirmed that slt2∆ice2∆ indeed had incomplete cER in the bud, representing severe defects in ER spreading (Figure 4E).
Preventing cER spreading at the bud cortex enhances endoplasmic reticulum stress surveillance activation
To further test the significance of the cER spreading step in the ERSU pathway, we examined the impact of ER stress on cER formation in slt2∆ice2∆ cells. As expected, WT cells blocked cER inheritance by the daughter cells (Figure 4F, left). In slt2∆ice2∆ cells, we observed only small amounts of fragmented cER in the daughter cells (Figure 4F, right; quantitation in Supplemental Figure S3D). Furthermore, slt2∆ice2∆ cells grew better than slt2∆ cells under ER stress (Figure 4G). Thus, the ER spreading defect induced by the additional deletion of ICE2 in slt2∆ cells (e.g., slt2∆ice2∆) may have rescued the cells’ sensitivity to ER stress. These results were in agreement with our previous results that preventing cER inheritance in slt2∆ cells by using a low dose of Latrunculin B, an actin temperature-sensitive mutation (act1-1), or MYO4 deletion rescued their growth sensitivity to ER stress (Babour et al., 2010). Interestingly, we found that Slt2’s kinase function was required for rescuing the growth defects of slt2∆ice2∆ cells under normal or ER stress conditions. The hypersensitivity of slt2∆ice2∆ cell to ER stress could only be rescued with the WT Slt2, as neither K54R nor T190A Y192F mutant alleles of SLT2 rescued the growth of slt2∆ice2∆ cells (Figure 4H). This finding was in contrast to our slt2∆spa2∆ data (Figure 3I), suggesting that the kinase function and the phosphorylation of Slt2 were required for its role in cER spreading, even though they are dispensable for ER cap formation. Taken together, these data suggest that SLT2 and ICE2 function together in the late stage of ER inheritance, namely ER spreading (Figure 4I).
In conclusion, we can now further divide early ER inheritance into three stages: 1) forming an ER tubule (IET) from the mother pnER and delivering it into the new bud; 2) anchoring the IET to the bud tip and forming an ER cap; and 3) spreading ER from the ER cap to the bud cortex to form cER (Figure 5). In the first stage, reticulons and Yop1 were important for the formation and orientation of the IET. Following IET entry into the daughter cell, Slt2 functioned with the polarisome Spa2 at the bud tip to regulate the formation of the ER cap. On the other hand, Slt2 functioned with the cER inheritance factor Ice2 to release ER from the ER cap, so that ER could spread to the bud periphery to form cER. Our findings here revealed more specific roles for Slt2 in each of the three ER inheritance stages, further suggesting that ER inheritance is likely regulated throughout the cell cycle via requisite checkpoints, which may be similar in principle to the morphogenesis check point mediated by the Swe1 kinase (Zarzov et al., 1996). In fact, Slt2 is found to undergo periods of activation via phosphorylation throughout the cell cycle, peaking at the polarized growth phase (Xu et al., 2009; Roberts et al., 2010). This indicates that Slt2 is likely most active during its required functions in stages 2 and 3 of early ER inheritance; by interacting with different partners such as Spa2 and Ice2, Slt2 may regulate these two distinct but tightly connected steps. Taken together, Slt2 may have emerged to become a crucial regulator that couples ER function to general cell-cycle progression, a critical property that warrants further study.
FIGURE 5:
Key ER inheritance stages. Cartoon drawings that illustrate the different stages of ER inheritance. Stage 1, which is mediated by the reticulons and Yop1, involves the extension of an IET from the mother pnER to the daughter cell. Stage 2, which is regulated by Slt2 and Spa2, involves the formation of an ER cap at the bud tip. Stage 3, which is mediated by Slt2 and Ice2, involves spreading ER from the ER cap to the bud cortex.
Our study of ER inheritance in yeast may contribute to the understanding of how ER is distributed even in nondividing cells such as neurons. To integrate diverse electrical signals, neurons have highly branched dendrites, which have numerous membrane protrusions called dendritic spines that serve as postsynaptic terminals. How ER reaches distant dendritic spines hundreds of micrometers away from the cell body is not well understood, but is an important question to answer since dendritic spine dynamics are associated with learning and memory (Cui-Wang et al., 2012). Nevertheless, not all dendritic spines contain ER (Mostafavi et al., 2008). How, then, do neurons regulate the delivery of ER to dendritic spines? Even though neurons do not undergo cell division once differentiated, the formation of dendritic spines shares similarities with asymmetric cell division in yeast. Therefore, our findings may serve as basic starting points for interrogating the complex arrangements of ER in neurons.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions
All yeast strains, except for rtn1∆rtn2∆yop1∆ (MNY2703) and the accompanying WT (MNY1037), were on the basis of S288C, and grown at 30°C in synthetic-defined (SD) or complete (SC) medium. Single-deletion strains with the KanMX marker were obtained from the yeast haploid deletion collection (BY4741, MAT a, Thermo Scientific). Other single-deletion strains with the NatR marker were done by one-step homologous recombination of PCR-generated fragments in haploids using the p4339 plasmid in BY7092 strain (both from the Boone lab, University of Toronto). GFP tagging of endogenous proteins was done by the PCR method using the pKT128 (SpHIS5) plasmid (Tavassoli et al., 2013) in the BY7043 background (Chao et al., 2014). Double-deletion strains were generated by mating and tetrad dissection. PCA strains were generated as previously described (Kim et al., 2008). Plasmids p2188, p2190, and p2193 were kind gifts of D. Levin (Chao et al., 2014). pHVF1CT and pUVF2CT were gifts from C. Loewen (Chao et al., 2014).
Spot assays
Tenfold serial dilutions of log phase cells were carried out using a pin-frogger on agar plates containing SD media, and supplemented with 1M sorbitol where indicated. All assays were performed at 30°C for 24–48 h and repeated independently at least two times.
Microscopy
Cells were visualized using a Zeiss Axiovert 200M fluorescent microscope with a 100 × 1.3 NA objective, unless otherwise stated. Deconvolution of images was done using Axiovision software (Zeiss).
Microfluidics microscopy
Microfluidic devices were fabricated using polydimethyl siloxane (PDMS) following published procedures (Hansen et al., 2015), at the UCSD Biodynamics Laboratory. After cells were introduced into the device as published, the device was mounted on the microscope via a custom-made stage adaptor. Tubings (BTPE-90, Instech Laboratories, Inc. Plymouth Meeting, PA, USA) were attached to the device via custom-made connectors, and SC medium were perfused through the channels in the device by gravity flow. Single plane images were taken every four minutes using Axiovision software on a Zeiss microscope, and analyzed using ImageJ software (NIH).
Time-lapse microscopy on agarose pads
Time-lapse images of WT (MNY1037) and rtn1∆rtn2∆yop1∆ (MNY2703) strains (Figures 1 and 2) were collected on agarose (1.6%) pads containing 0.5X YPD (1% peptone, 0.5% yeast extract, 2% dextrose). Images were collected on a DeltaVision system (Applied Precision) consisting of an inverted epifluorescence microscope (IX71, Olympus) equipped with a temperature chamber maintained at 30°C for the duration of the time-lapse. Images are z-projections of deconvolved z-stacks. For Supplemental Figures S3 and S4, imaging was carried out on the Zeiss Axio Observer fluorescent microscope with a 100 × 1.3 NA objective on Agarose pads containing complete synthetic yeast medium. Deconvolution and z-projections were performed with Zen 2.3 Pro software (Zeiss).
Image quantification
All quantifications were done using ImageJ. To enable direct comparisons, WT and mutant cells were imaged on the same day with identical microscope settings. Bud-to-mother size ratios were determined by measuring the areas occupied by bud and mother cells on the corresponding transmission images. Small budded cells were classified as bud area less than 1/3 that of the mother, and medium budded cells had bud area greater than 1/3. To quantify ER fluorescence, a surface plot was generated from the fluorescent images, and the peak fluorescent intensities were estimated from the plot. Quantification of cER in WT and slt2∆ice2∆ cells was done by measuring the distance occupied by the ER fragments in the cell cortex, and dividing by the circumference of the bud or mother cell. All error bars represent SEM.
Acknowledgments
This work was funded by NIH RO1 Grant GM087415 (M.N.) and Cancer Research Coordinating Committee Grant CRR-20-632388 to M.N. J.C. was supported by a UCSD Molecular Biology Cancer postdoctoral fellowship and F.P. was supported by an NIH 5T32AI007469-20UCSD/LIAI allergy postdoctoral training grant.
Abbreviations used:
- ER
endoplasmic reticulum
- cER
cortical ER
- ConA
concavanine A
- DHFR
dihydrofolate reductase
- EM
electron microscopy
- ERSU
ER stress surveillance
- GFP
green fluorescence protein
- IET
initial ER tubule
- MAP
mitogen activated protein
- PM
plasma membrane
- pnER
perinuclear ER
- SC
synthetic complete
- SGD
saccharomyces genome database
- UPR
unfolded protein response
- WT
wild type
- YFP
yellow fluorescence protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-08-0558).
REFERENCES
- Babour A, Bicknell AA, Tourtellotte J, Niwa M (2010). A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell 142, 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JT, Piña F, Onishi M, Cohen Y, Lai Y-S, Schuldiner M, Niwa M (2019). Transfer of the septin ring to cytokinetic remnants in ER stress directs age-sensitive cell-cycle re-entry. Dev Cell 1–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JT, Wong AKO, Tavassoli S, Young BP, Chruscicki A, Fang NN, Howe LJ, Mayor T, Foster LJ, Loewen CJR (2014). Polarization of the endoplasmic reticulum by ER-septin tethering. Cell 158, 620–632. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, et al. (2010). The genetic landscape of a cell. Science 327, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui-Wang T, Hanus C, Cui T, Helton T, Bourne J, Watson D, Harris KM, Ehlers MD (2012). Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell 148, 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Ferro-Novick S, Novick P (2004). Dynamics and inheritance of the endoplasmic reticulum. J Cell Sci 117, 2871–2878. [DOI] [PubMed] [Google Scholar]
- Du Y, Pypaert M, Novick P, Ferro-Novick S (2001). Aux1p/Swa2p is required for cortical endoplasmic reticulum inheritance in Saccharomyces cerevisiae. Mol Biol Cell 12, 2614–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada de Martin P, Du Y, Novick P, Ferro-Novick S (2005). Ice2p is important for the distribution and structure of the cortical ER network in Saccharomyces cerevisiae. J Cell Sci 118, 65–77. [DOI] [PubMed] [Google Scholar]
- Fagarasanu A, Fagarasanu M, Eitzen GA, Aitchison JD, Rachubinski RA (2006). The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev Cell 10, 587–600. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher K, Davis D, Wu M, Boldogh I, Pon L (2002). Endoplasmic reticulum dynamics, inheritance, and cytoskeletal interactions in budding yeast. 13, 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Hao N, O’Shea EK (2015). High-throughput microfluidics to control and measure signaling dynamics in single yeast cells. Nat Protoc 10, 1181–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen D, Schekman R (2011). COPII-mediated vesicle formation at a glance. J Cell Sci 124, 1–4. [DOI] [PubMed] [Google Scholar]
- Kim KY, Truman AW, Levin DE (2008). Yeast Mpk1 mitogen-activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol Cell Biol 28, 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Du Y, Siegel S, Ferro-Novick S, Novick P (2010). Activation of the mitogen-activated protein kinase, Slt2p, at bud tips blocks a late stage of endoplasmic reticulum inheritance in Saccharomyces cerevisiae. 21, 1772–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ferro-Novick S, Novick P (2013). Different polarisome components play distinct roles in Slt2p-regulated cortical ER inheritance in Saccharomyces cerevisiae. 24, 3145–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJR, Young BP, Tavassoli S, Levine TP (2007). Inheritance of cortical ER in yeast is required for normal septin organization. J Cell Biol 179, 467–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedeke C, Frei S, Sbalzarini I, Schwarz H, Spang A, Barral Y (2005). Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. 169, 897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD (2012). ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell 23, 1129–1140. [DOI] [PubMed] [Google Scholar]
- Michnick S, Ear P, Manderson E, Remy I, Stefan E (2007). Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov 6, 569–582. [DOI] [PubMed] [Google Scholar]
- Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q (2008). GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol 9(Suppl 1), S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Weisman LS (2008). The cyclin-dependent kinase Cdk1 directly regulates vacuole inheritance. Dev Cell 15, 478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler H, Gaigg B, Hrastnik C, Achleitner G, Kohlwein S, Zellnig G, Perktold A, Daum G (2001). A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur J Biochem 268, 2351–2361. [DOI] [PubMed] [Google Scholar]
- Piña FJ, Niwa M (2015). The ER Stress Surveillance (ERSU) pathway regulates daughter cell ER protein aggregate inheritance. Elife 4, 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña FJ, Fleming T, Pogliano K, Niwa M (2016). Reticulons regulate the ER inheritance block during ER stress. Dev Cell 37, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña F, Yagisawa F, Obara K, Gregerson JD, Kihara A, Niwa M (2018). Sphingolipids activate the endoplasmic reticulum stress surveillance pathway. J Cell Biol 217, 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA (2000). Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol 150, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provance DW, Mercer JA (1999). Myosin-V: head to tail. Cell Mol Life Sci 56, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A (2000). Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci 113 (Pt 3), 365–375. [DOI] [PubMed] [Google Scholar]
- Roberts TF, Tschida KA, Klein ME, Mooney R (2010). Rapid spine stabilization and synaptic enhancement at the onset of behavioural learning. Nature 463, 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Blower MD (2016). The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci 73, 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M (1998). Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol 18, 4053–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA (2010). Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Li Y, Yu C, Wei Z (2019). Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2. J Biol Chem 294, 5896–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli S, Young BP, Cox RC, Prinz WA, de Kroon AIPM, Loewen CJR (2013). Plasma membrane–endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep 14, 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA (2012). A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci 125, 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F, Peter M (2002). Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr Biol 12, 1698–1703. [DOI] [PubMed] [Google Scholar]
- Varshney N, Sanyal K (2019). Nuclear migration in budding yeasts: position before division. Curr Genet 65, 1341–1346. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Rolls MM, Rapoport TA (2002). Structural organization of the endoplasmic reticulum. EMBO Rep 3, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- West M, Zurek N, Hoenger A, Voeltz GK (2011). A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. 193, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A, Du Y, Pypaert M, Ferro-Novick S, Novick P (2003). Sec3p is needed for the spatial regulation of secretion and for the inheritance of the cortical endoplasmic reticulum. Mol Biol Cell 14, 4770–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Yu X, Perlik AJ, Tobin WF, Zweig JA, Tennant K, Jones T, Zuo Y (2009). Rapid formation and selective stabilization of synapses for enduring motor memories. Nature 462, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzov P, Mazzoni C, Mann C (1996). The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J 15, 83–91. [PMC free article] [PubMed] [Google Scholar]