Abstract
Phosphoinositide signaling lipids are essential for several cellular processes. The requirement for a phosphoinositide is conventionally studied by depleting the corresponding lipid kinase. However, there are very few reports on the impact of elevating phosphoinositides. That phosphoinositides are dynamically elevated in response to stimuli suggests that, in addition to being required, phosphoinositides drive downstream pathways. To test this hypothesis, we elevated the levels of phosphatidylinositol-3-phosphate (PI3P) by generating hyperactive alleles of the yeast phosphatidylinositol 3-kinase, Vps34. We find that hyperactive Vps34 drives certain pathways, including phosphatidylinositol-3,5-bisphosphate synthesis and retrograde transport from the vacuole. This demonstrates that PI3P is rate limiting in some pathways. Interestingly, hyperactive Vps34 does not affect endosomal sorting complexes required for transport (ESCRT) function. Thus, elevating PI3P does not always increase the rate of PI3P-dependent pathways. Elevating PI3P can also delay a pathway. Elevating PI3P slowed late steps in autophagy, in part by delaying the disassembly of autophagy proteins from mature autophagosomes as well as delaying fusion of autophagosomes with the vacuole. This latter defect is likely due to a more general defect in vacuole fusion, as assessed by changes in vacuole morphology. These studies suggest that stimulus-induced elevation of phosphoinositides provides a way for these stimuli to selectively regulate downstream processes.
INTRODUCTION
Phosphoinositide (PPI) lipids are signaling molecules that play critical roles in multiple cellular processes. It is assumed that generation of specific PPI species on membranes recruits distinct effector proteins that regulate downstream pathways (reviewed in Schink et al., 2016). PPI species are generated by phosphorylation at the 3, 4, and 5 positions of the inositol head group of phosphatidylinositol (PI) and the levels of these lipids are dynamically regulated by PPI lipid kinases and phosphatases in response to stimuli (Balla, 2013). For example, during hyperosmotic shock in Saccharomyces cerevisiae, there is a transient 15 to 20-fold elevation in phosphatidylinositol-3,5-bisphosphate (PI(3,5)P2) (Duex et al., 2006a). Most studies of how PPI lipids regulate downstream pathways use knockout or knockdown of PPI kinases to deplete a specific PPI lipid and thereby test the necessity for that PPI species in a process (Kihara et al., 2001). However, the dynamic regulation of PPI lipids suggests that in addition to being required for specific pathways, changes in PPI lipids may drive downstream processes. Here, we test this hypothesis directly by manipulating the levels of phosphatidylinositol 3-phosphate (PI3P), which is dynamically regulated in yeast (Duex et al., 2006a).
In yeast, the PI 3-kinase, Vps34, is the sole enzyme responsible for generating PI3P from PI (Schu et al., 1993). Vps34 functions within two large regulatory complexes (Kihara et al., 2001). Complex I is dedicated to autophagy, and complex II is required for several trafficking pathways in the endomembrane system. The pseudokinase Vps15 is absolutely required for Vps34 function and PI3P synthesis in yeast (Stack et al., 1993). In addition to Vps34 and Vps15, both complexes contain Vps30 (BECN1 in mammalian systems), which contains a BARA domain crucial for binding the PI 3-kinase complex to membranes (Huang et al., 2012; Rostislavleva et al., 2015). Furthermore, each complex contains complex-specific protein subunits, which specify the intracellular localization of the complexes (Obara et al., 2006). Complex I includes Atg14 and Atg38 (Araki et al., 2013), whereas complex II contains Vps38 (Kihara et al., 2001).
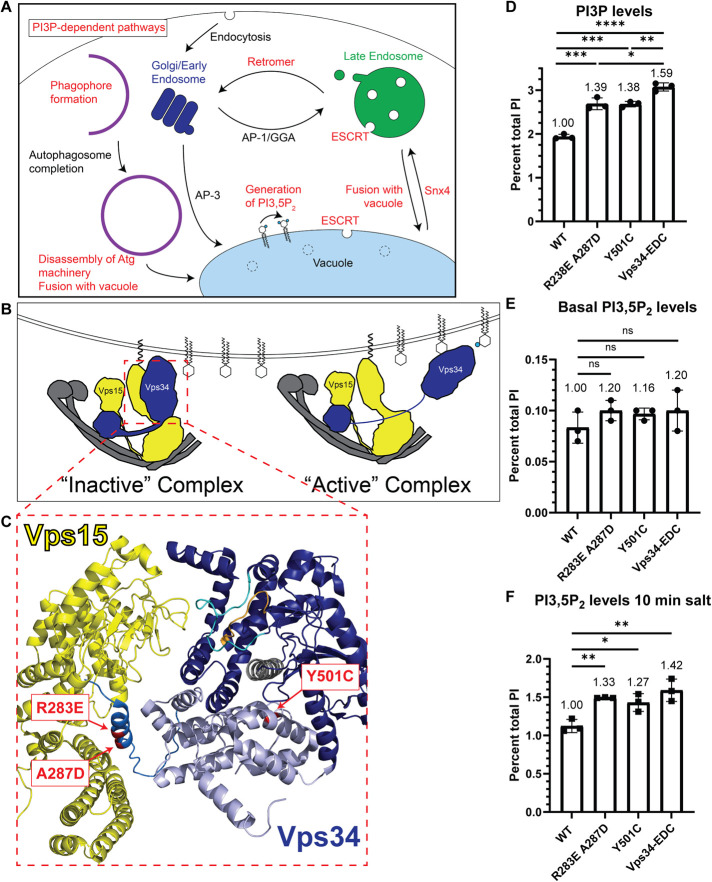
Several PI3P-dependent cellular processes have been identified in cells (Figure 1A). In yeast, synthesis of PI(3,5)P2 is achieved by phosphorylation of PI3P at the 5 position (Gary et al., 1998). Thus, Vps34 mutants that do not produce PI3P also do not produce PI(3,5)P2 (Dove et al., 1997).
FIGURE 1:
Generation of hyperactive Vps34 mutants. (A) Schematic indicating several PI3P-dependent intracellular trafficking pathways in yeast. PI3P serves as the substrate for PI(3,5)P2. PI3P is also required for Snx4-dependent retrograde transport from the vacuole and retromer-dependent retrograde transport from endosomes. ESCRT function at late endosomes and the vacuole is also dependent on PI3P. Furthermore, PI3P is required for phagophore formation during the initiation of autophagy, and then PI3P is removed before the disassembly of some autophagy proteins from the surface of mature autophagosomes. In addition, fusion of autophagosomes and other vesicles with the vacuole, as well as homotypic vacuole fusion (not depicted), are PI3P-dependent pathways. (B) Vps34 is proposed to be regulated in part via changes in contact of the Vps34 HELCAT domain with Vps15. In the inactive conformation, the Vps34 HELCAT domain contacts the Vps15 scaffold. During activation, the Vps34 HELCAT domain is proposed to alter the contact with Vps15 and allow Vps34 to access its PI substrate (Stjepanovic et al., 2017). (C) Crystal structure of the helical (light blue) and kinase (dark blue) domains of Vps34 and its contact with the pseudokinase domain of Vps15 (yellow; Rostislavleva et al., 2015). The three amino acid changes that comprise the Vps34-EDC hyperactive mutant are indicated (red). Two of these mutations, R283E and A287D, are on an alpha-helix N-terminal to the helical domain of Vps34 (neutral blue) and may hinder Vps34 HELCAT interaction with Vps15 and favor the active Vps34 conformation. The Y501C mutation is in the helical domain of Vps34 and faces the alpha-C helix (gray) of the kinase domain, nearby the activation (cyan) and catalytic (orange) loops of Vps34. (D–F) The Vps34 mutants R283E A287D and Y501C elevate PI3P levels. PI(3,5)P2 levels are also elevated during hyperosmotic shock. Combining the mutants to Vps34-EDC further elevated PI3P levels and PI(3,5)P2 levels. vps34Δ cells were transformed with a wild-type or mutant pRS416-Vps34 plasmid. PPI lipid levels were measured by metabolically labeling cells with myo-3H-inositol for 16 h. Prior to harvest, indicated cultures were exposed to 10 min of hyperosmotic shock. PPI lipid head groups were separated by anion exchange and HPLC. n = 3. Error bars indicate SD. Unpaired t test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
PI3P is also required for multiple retrograde transport pathways. Yeast retromer functions in retrograde transport of transmembrane sorting receptors from endosomes back to the Golgi (Seaman et al., 1997, 1998). The retromer requires PI3P, which recruits retromer sorting nexins Vps5 and Vps17 to membranes (Burda et al., 2002). These sorting nexins contain PI3P-binding PX domains as well as BAR domains (SNX-BAR; Yu and Lemmon, 2001). Recently, another SNX-BAR protein, Snx4, has been implicated in retrograde transport for retrieval of proteins from the vacuole to endosomes (Ma et al., 2017; Suzuki and Emr, 2018). PI3P is required for proper Snx4 localization and is necessary for Snx4-dependent retrograde transport (Nice et al., 2002; Suzuki and Emr, 2018).
Function of the endosomal sorting complexes required for transport (ESCRT) pathway also depends on PI3P. The ESCRT complex functions on both endosomes and the vacuole to target selected transmembrane proteins for degradation. The selected proteins are first ubiquitinated, which enables their recognition and binding to the ESCRT complex. Once the cargo proteins are bound, the ESCRT complex generates vesicles via inward budding into the lumen of the organelle, and the cargoes are ultimately degraded in the vacuole. ESCRT-0 functions by binding ubiquitinated ESCRT client proteins and recruiting ESCRT-I to membranes. Vps34 is required for proper localization of the ESCRT-0 subunit Vps27, which contains a PI3P-binding FYVE domain (Katzmann et al., 2003).
Additionally, PI3P is required for fusion of vesicles with the vacuole, and homotypic vacuole fusion. This is due in part to the direct binding of the SNARE Vam7 (Cheever et al., 2001; Fratti and Wickner, 2007) and the heterodimeric GEF subunits Mon1 and Ccz1 (Cabrera et al., 2014) to PI3P. Moreover, Vps34-dependent generation of PI3P is required for the vacuole association of many additional proteins required for vacuole fusion (Lawrence et al., 2014).
PI3P is also required for autophagy. Defects in PI 3-kinase complex I block autophagy in yeast (Kihara et al., 2001). Early in autophagy, PI3P recruits Atg18 to the phagophore (Dove et al., 2004; Obara et al., 2008) where it interacts with Atg2 and tethers preautophagosomal membranes to the endoplasmic reticulum, allowing Atg2 to transport lipids to promote autophagosome biogenesis (Rieter et al., 2013; Kotani et al., 2018; Valverde et al., 2019).
Following autophagosome formation, Ymr1, a myotubularin family protein and putative PI3P phosphatase (Taylor et al., 2000; Parrish et al., 2004) is crucial for autophagosome fusion with the vacuole. Deletion of YMR1 causes a failure of key autophagy machinery including Atg18 to dissociate from mature autophagosomes. This leads to accumulation of autophagosomes in the cytoplasm (Cebollero et al., 2012) and suggests that turnover of PI3P is crucial for this process. PI3P also has a positive role in a late step in autophagy. In vitro studies indicate that fusion of autophagosomes with the vacuole requires PI3P and likely acts by recruiting the Rab GTPase Ypt7, which in turn recruits the HOPS tethering complex (Bas et al., 2018). Thus, while PI3P is required for some steps in autophagy, impeding turnover of PI3P inhibits the resolution of autophagosomes.
Here, we report the generation and use of hyperactive mutations in Vps34 and the discovery that PI3P drives select pathways, including synthesis of PI(3,5)P2 during hyperosmotic shock and retrograde transport of Atg27. In these cases, the PI3P-dependent step is rate limiting. We also show that hyperactive Vps34 does not affect ESCRT function at endosomes or on the vacuole. Thus, elevating PI3P does not always increase the overall rate of a complex pathway. We also show that elevating PI3P can delay a pathway. Hyperactive Vps34 does not lead to an acceleration in the induction of autophagy, but inhibits late steps in autophagy, in part via a delay in disassembly of the autophagy machinery from the surface of mature autophagosomes and also a delay in fusion of autophagosomes with the vacuole. This latter defect is likely due to a more general defect in vacuole fusion, as evidenced by an increase in the number of vacuole lobes per cell, which is consistent with a defect in homotypic vacuole fusion. Overall, our studies suggest that stimulus-induced elevation of PI3P levels regulates some, but not all, PI3P-dependent membrane trafficking pathways and that phosphoinositide lipids are commonly rate limiting in pathways where they are required.
RESULTS AND DISCUSSION
Generation of hyperactive Vps34 mutants
To test the hypothesis that changes in PPI levels drive downstream processes, we devised a strategy for specifically elevating PI3P. PI3P was an attractive target because several downstream PI3P-dependent processes have been identified in yeast. Additionally, the existing 4.4 Å crystal structure of the yeast PI 3-kinase complex allowed us to predict amino acid changes that would increase PI3P levels (Rostislavleva et al., 2015). Moreover, the crystal structure also allowed us to map the location of amino acid changes responsible for the hyperactivity of the Vps34 mutants identified in our screen.
We found that overexpression of Vps34 alone or together with Vps15 caused a modest, 13%, increase in PI3P levels despite robust overexpression (Supplemental Figure S1, A and B). This result suggests that either overexpression of additional subunits of the Vps34 complex are required for increased Vps34 function or that Vps34 kinase activity is negatively regulated.
To achieve more robust elevation of PI3P levels, we tested whether point mutations in Vps34 would yield a hyperactive enzyme. Vps34 activity is proposed to be regulated in part via changes in the contact of the Vps34 helical and kinase domains (HELCAT) with the Vps15 pseudokinase domain (Figure 1B; Stjepanovic et al., 2017). In the inactive conformation, the Vps34 HELCAT domain contacts the Vps15 scaffold. During activation, the Vps34 HELCAT domain is proposed to alter its contact with Vps15 and allow Vps34 to access its PI substrate. Based on the structure of the PI 3-kinase complex, we introduced point mutations in Vps34 along its contact site with Vps15 (Supplemental Figure S1C). We tested eight Vps34 point mutants and identified two mutations (R283E and A287D) that robustly increased PI3P by 36 and 27%, respectively (Supplemental Figure S1D).
Combining R283E and A287D, but not other mutations along the Vps34 HELCAT-Vps15 interface, elevated PI3P levels by ∼40% (Supplemental Figure S1E). These two mutants are on an alpha-helix N-terminal to the helical domain of Vps34 and face Vps15 (Figure 1C). Identification of these hyperactive Vps34 mutants provides further evidence that altered contact between the Vps34 HELCAT domain and Vps15 promotes Vps34 kinase activity.
We also performed an unbiased genetic screen for hyperactive Vps34 mutants using a method similar to the one used to generate hyperactive mutations in the PI3P 5-kinase, Fab1 (Duex et al., 2006b; Lang et al., 2017). We generated a hypomorphic Vps34K759D allele, which has a mutation in the activation loop of Vps34, and found that it lowers PI3P levels to ∼20% of wild-type Vps34. We then performed random PCR mutagenesis on the C-terminal half of Vps34 K759D (Supplemental Figure S2A). Mutagenized plasmids were tested for their ability to rescue growth in vps34Δ cells grown on rapamycin at 33°C. From 22 independently isolated mutants, we identified nine unique point mutations. Five of these mutations elevate PI3P levels. The best of these mutations, Y501C, elevated PI3P ∼40% (Figure 1C and Supplemental Figure S2C). Interestingly, each of the five mutated residues is located on either the alpha-C helix of the Vps34 kinase domain or an adjacent helix of the helical domain (Supplemental Figure S2B). Conformational changes in the alpha-C helix are critical to regulating kinase function (Taylor et al., 2015). We hypothesize these hyperactive mutants favor an active conformation of the alpha-C helix. It is not known whether these are regulatory sites on the native enzyme. We determined that combining the R283E and A287D mutant with Y501C (Vps34-EDC) elevates PI3P by ∼60%, which is higher than either mutant alone (Figure 1D). The Vps34-EDC mutant does not change Vps34 protein levels (Supplemental Figure S3, A and B), nor does it change the localization of Vps34 within the cell as measured by the amount of Envy-Vps34 that colocalizes with the vacuole (Supplemental Figure S3, C and D). This suggests that the distribution of PI3P in the cell is most likely unchanged by the hyperactive mutant.
PI3P is the substrate for the PI3P 5-kinase Fab1, which generates PI(3,5)P2 (Gary et al., 1998). We tested whether elevation of PI3P via hyperactive Vps34 leads to an elevation of PI(3,5)P2. At basal conditions, no statistically significant increase in PI(3,5)P2 levels was detected (Figure 1E). However, when hyperosmotic shock was used to induce a transient elevation of PI(3,5)P2 (Duex et al., 2006a), the presence of hyperactive Vps34 mutants resulted in a further elevation of PI(3,5)P2 (Figure 1F). Note that while PI3P levels decrease during hyperosmotic shock, hyperactive Vps34 mutants still elevate PI3P above wild-type levels (Supplemental Figure S2D). Thus, hyperactive Vps34 drives elevation of PI(3,5)P2 during hyperosmotic shock. We ruled out several potential mechanisms by which hyperactive Vps34 might elevate PI(3,5)P2. We determined that hyperactive Vps34-EDC does not change the amount of Fab1-Envy that co-localizes with the vacuole (Supplemental Figure S4, A and B). Additionally, there was no change in the amount of Fab1 complex member Fig4 (Botelho et al., 2008) that colocalizes with the vacuole (Supplemental Figure S4, C and D). These results suggest that hyperactive Vps34 does not result in increased recruitment of the Fab1 complex to membranes. Moreover, the increase in PI(3,5)P2 is likely not due to inhibition of Fig4, the PI(3,5)P2 5-phosphatase (Gary et al., 2002). Catalytically dead Fig4 mutants exhibit a mild increase in PI(3,5)P2 under basal conditions, yet a decrease in PI(3,5)P2 at 10 min following hyperosmotic shock (Duex et al., 2006b; Strunk et al., 2020). Thus, the increase in PI(3,5)P2 caused by Vps34-EDC most likely occurs because of increased availability of PI3P, which may provide more substrate for Fab1 and/or activate Fab1. Note that the additional PI(3,5)P2 provided during hyperosmotic shock by hyperactive Vps34-EDC does not affect the growth of yeast cells following hyperosmotic shock (Supplemental Figure S4E).
Hyperactive Vps34 increases retrograde transport of Atg27
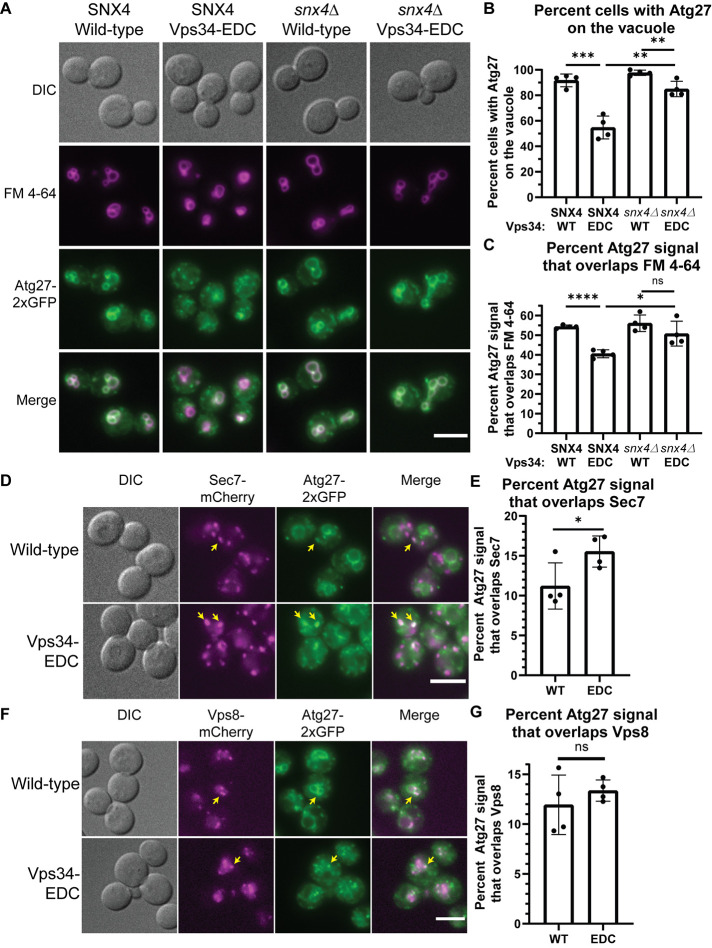
Atg27 is a cargo for PI3P-dependent Snx4- and retromer-dependent retrograde transport pathways. Following its synthesis, Atg27 is delivered from the Golgi to the vacuole via the AP-3 pathway (Segarra et al., 2015). From the vacuole, Atg27 undergoes retrograde transport to endosomes in a Snx4-dependent manner (Ma et al., 2017; Suzuki and Emr, 2018). From endosomes, Atg27 undergoes retrograde transport back to the Golgi via the retromer (Suzuki and Emr, 2018). Due to the cyclic nature of its transport, changes in Atg27 localization signify changes in the rates of each of these transport steps.
We tested whether hyperactive Vps34 increases the rate of retrograde transport of Atg27 from the vacuole to endosomes and/or Golgi. In wild-type cells, Atg27-2xGFP was primarily localized to the vacuole (Figure 2A) with a few Golgi (Figure 2D) and endosomal (Figure 2F) puncta. Notably, in the presence of Vps34-EDC, Atg27-2xGFP was more punctate (Figure 2A) and fewer hyperactive Vps34-EDC cells had Atg27-2xGFP visible on the vacuole (Figure 2B). Moreover, on a per cell basis, less Atg27-2xGFP colocalized with the vacuole (Figure 2C). Blocking retrograde transport from the vacuole by deletion of SNX4 suppressed Atg27-2xGFP traffic from the vacuole (Figure 2, A–C). These results suggest that hyperactive Vps34-EDC accelerates the Snx4 and/or retromer pathways.
FIGURE 2:
Hyperactive Vps34 increases retrograde transport of Atg27. (A–C) Hyperactive Vps34-EDC results in fewer cells that contain Atg27-2xGFP on the vacuole (FM 4-64, magenta; B) and in less total Atg27-2xGFP colocalization with FM 4-64 (C). Deletion of SNX4 results in the retention of Atg27-2xGFP on the vacuole. vps34Δ or vps34Δsnx4Δ cells with Atg27-2xGFP integrated at the endogenous locus were transformed with pRS416-Vps34 or pRS416-Vps34-EDC. After labeling with FM 4-64, cells were chased at 24°C for 3 h before imaging. DIC, differential interference contrast. Scale bar = 5 µm. Cells were scored as either having Atg27-2xGFP visible on the vacuole (colocalization with FM 4-64) or present only in puncta. The scorer was blinded to the genotype of the cells being quantified (B). To measure Atg27-2xGFP localization on a population basis, the Atg27-2xGFP signal that overlaps FM 4-64 was divided by total Atg27-2xGFP signal (C). Quantification of at least 40 cells per n; n = 4. Error bars indicate SD. Unpaired t test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. (D , E) Atg27-2xGFP partially colocalizes with the trans-Golgi (Sec7-mCherry). This colocalization increases in the presence of hyperactive Vps34-EDC. vps34Δ cells with Atg27-2xGFP and Sec7-mCherry integrated at the endogenous loci were transformed with pRS416-Vps34 or pRS416-Vps34-EDC. DIC, differential interference contrast. Scale bar = 5 µm. Examples of Atg27-2xGFP puncta that colocalize with Sec7-mCherry are indicated by yellow arrows. The Atg27-2xGFP signal that overlaps Sec7-mCherry was divided by total Atg27-2xGFP signal. Quantification of at least 40 cells per n, n = 4. Error bars indicate SD. Unpaired t test. * = p < 0.05. (F, G) Atg27-2xGFP partially colocalizes with endosomes (Vps8-mCherry). This colocalization is not statistically different for hyperactive Vps34-EDC compared with wild-type. vps34Δ cells with Atg27-2xGFP and Vps8-mCherry integrated at the endogenous loci were transformed with pRS416-Vps34 or pRS416-Vps34-EDC. DIC, differential interference contrast. Scale bar = 5 µm. Examples of Atg27-2xGFP puncta that colocalize with Vps8-mCherry are indicated by yellow arrows. The Atg27-2xGFP signal that overlaps Vps8-mCherry was divided by total Atg27-2xGFP signal. Quantification of at least 40 cells per n, n = 4. Error bars indicate SD. Unpaired t test. ns = p > 0.05.
The loss of Atg27-2xGFP localization to the vacuole is likely not caused by impairment of the AP-3–dependent anterograde delivery of Atg27 to the vacuole. We tested the localization of another AP-3 client protein Yck3 (Sun et al., 2004) and found that it was delivered to the vacuole in the presence of Vps34-EDC (Supplemental Figure S5A), indicating that Vps34-EDC does not cause a defect in AP-3 function.
We tested whether the additional Atg27-2xGFP puncta present in Vps34-EDC correspond to Golgi and/or endosomes. We found that expression of Vps34-EDC resulted in a higher percentage of the total cellular Atg27-2xGFP that colocalizes with the Golgi marker, Sec7-mCherry (Figure 2, D and E). There was no statistically significant difference between Vps34-EDC–expressing cells compared with wild-type with respect to the total cellular Atg27-2xGFP signal that colocalized with endosome marker Vps8-mCherry (Figure 2, F and G). Accumulation of Atg27-2xGFP in the Golgi is consistent with the hypothesis that hyperactive Vps34-EDC leads to increases in both Snx4- and retromer-dependent retrograde transport of Atg27.
We tested whether deletion of the retromer subunit VPS35 suppresses the increased Atg27-2xGFP localization to the Golgi caused by Vps34-EDC. Consistent with the retromer being required for accelerated retrograde transport of Atg27, in vps35∆ cells, there was no difference in Atg27-2xGFP localization to the Golgi between wild-type Vps34 and Vps34-EDC (Supplemental Figure S5, B and C). However, surprisingly, vps35∆ cells exhibited increased localization of Atg27-2xGFP to the Golgi compared with wild-type cells. This result was unexpected because a previous study reported that Atg27 accumulates on endosomes in vps35∆ cells with less Atg27 on Golgi (Suzuki and Emr, 2018), which fits with the view that the retromer functions in retrograde traffic of proteins from endosomes to the Golgi. However, this previous study is complicated by the fact that experiments were performed following 1 h of treatment with rapamycin. Because rapamycin inhibits protein synthesis in yeast (Barbet et al., 1996), we reasoned that rapamycin treatment may have masked a defect in AP-3-dependent anterograde delivery of Atg27 to the vacuole, leading to accumulation of Atg27 in the Golgi. However, deletion of VPS35 does not appear to affect AP-3 function, as another AP-3 client protein Yck3 is delivered to the vacuole in vps35∆ cells (Supplemental Figure S5D). Thus, under basal conditions, trafficking of Atg27 in vps35∆ cells is more complex than previously appreciated. Together, these results suggest that, in yeast, elevating PI3P drives retrograde transport from the vacuole, and leaves open the possibility that retrograde traffic from endosomes to the Golgi is accelerated as well.
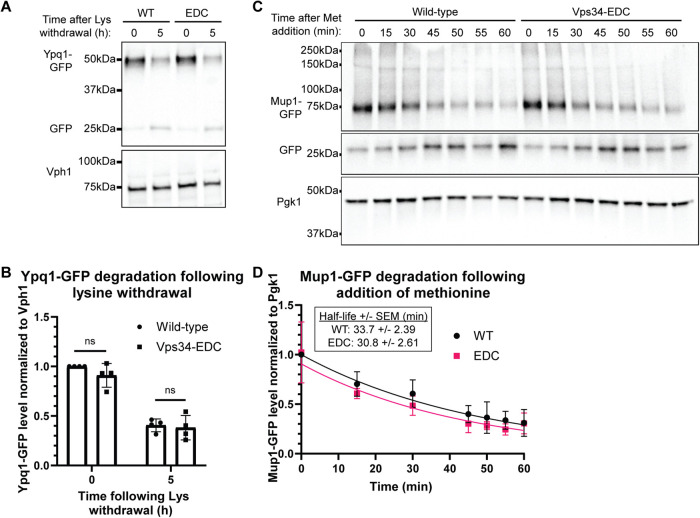
Hyperactive Vps34 does not affect ESCRT-dependent degradation of amino acid transporters Ypq1 or Mup1
ESCRT function is regulated by PI3P, and plays a crucial role in trafficking select transmembrane proteins to the vacuole for degradation, including the vacuolar cationic amino acid transporter Ypq1 (Li et al., 2015; Zhu et al., 2017). Following withdrawal of lysine, Ypq1 is ubiquitinated and internalized inside the vacuole by the ESCRT machinery where it is degraded.
Using Western blot analysis, we tested whether hyperactive Vps34-EDC accelerates ESCRT-dependent degradation of Ypq1 following withdrawal of lysine by measuring Ypq1-GFP levels normalized to Vph1. We found that degradation of Ypq1-GFP was not affected by Vps34-EDC (Figure 3, A and B), suggesting that elevating PI3P does not affect ESCRT function in the degradation of Ypq1.
FIGURE 3:
Hyperactive Vps34 does not affect ESCRT-dependent degradation of amino acid transporters Ypq1 or Mup1. (A, B) ESCRT-dependent internalization of Ypq1-GFP following the withdrawal of lysine is not statistically significantly different for hyperactive Vps34-EDC compared with wild-type. vps34Δ cells were transformed with pRS416-Vps34 or pRS416-Vps34-EDC and pRS414-Ypq1-GFP. To induce Ypq1-GFP internalization, cells were transferred to media lacking lysine. Samples were collected at 0 and 5 h following lysine withdrawal. Ypq1-GFP protein levels were analyzed via Western blot using anti-GFP antibody, and were normalized to Vph1. Levels were then normalized to wild-type at the zero time point. Representative of n = 4. Error bars indicate SD. Unpaired t test. ns = p > 0.05. (C, D) ESCRT-dependent internalization of Mup1-GFP following the addition of methionine is not statistically significantly different for hyperactive Vps34-EDC compared with wild-type. vps34Δ cells were transformed with pRS416-Vps34 or pRS416-Vps34-EDC and pRS414-Mup1-GFP. Cells were grown in media lacking methionine. To induce Mup1-GFP internalization, cells were transferred to media containing methionine. Samples were collected at the indicated time points following methionine addition. Mup1-GFP protein levels were analyzed via Western blot using anti-GFP antibody, and normalized to Pgk1. Levels were then normalized to wild-type at the zero time point. Representative of n = 4. Error bars indicate SD. The degradation rate of Mup1-GFP was determined using a linear mixed effects model. The logarithmic transformation of Mup1-GFP levels was modeled as a linear function of time and allowed to vary by genotype. Replicate-specific intercepts were included to account for residual correlation between protein levels within the same replicate. The difference in degradation rates between genotypes was not statistically significant (a 95% confidence interval of the genotype by time interaction contained 0). The equation generated by the linear mixed effects model is plotted (D). The half-life of Mup1-GFP following readdition of methionine was calculated to be 33.7 min (SEM 2.39 min) with wild-type Vps34, and 30.8 min (SEM 2.61 min) with Vps34-EDC.
We also tested a second ESCRT substrate, the plasma membrane methionine transporter Mup1 (Teis et al., 2008). Mup1 accumulates on the plasma membrane when cells are starved for methionine. When methionine is reintroduced to cells, Mup1 is ubiquitinated, endocytosed, delivered to endosomes, and internalized by ESCRT into multivesicular bodies, which then fuse with the vacuole where Mup1 is degraded (Menant et al., 2006; Teis et al., 2008). Thus, to test ESCRT function, we measured Mup1-GFP levels normalized to Pgk1 following readdition of methionine.
Similar to what was observed with Ypq1, there were no statistically significant differences in the rate of Mup1 degradation between wild-type and hyperactive Vps34-EDC following readdition of methionine (Figure 3, C and D). The half-life of Mup1-GFP following readdition of methionine was calculated to be 33.7 min (SEM 2.39 min) with wild-type Vps34, and 30.8 min (SEM 2.61 min) with Vps34-EDC. This experiment indicates that elevating PI3P does not affect ESCRT function in the degradation of Mup1. Overall, in contrast to the retrograde transport of Atg27, elevating PI3P does not accelerate ESCRT function on the vacuole or endosomes, demonstrating that elevating PI3P does not always increase the overall rate of a complex pathway.
Hyperactive Vps34 inhibits a late step in autophagy
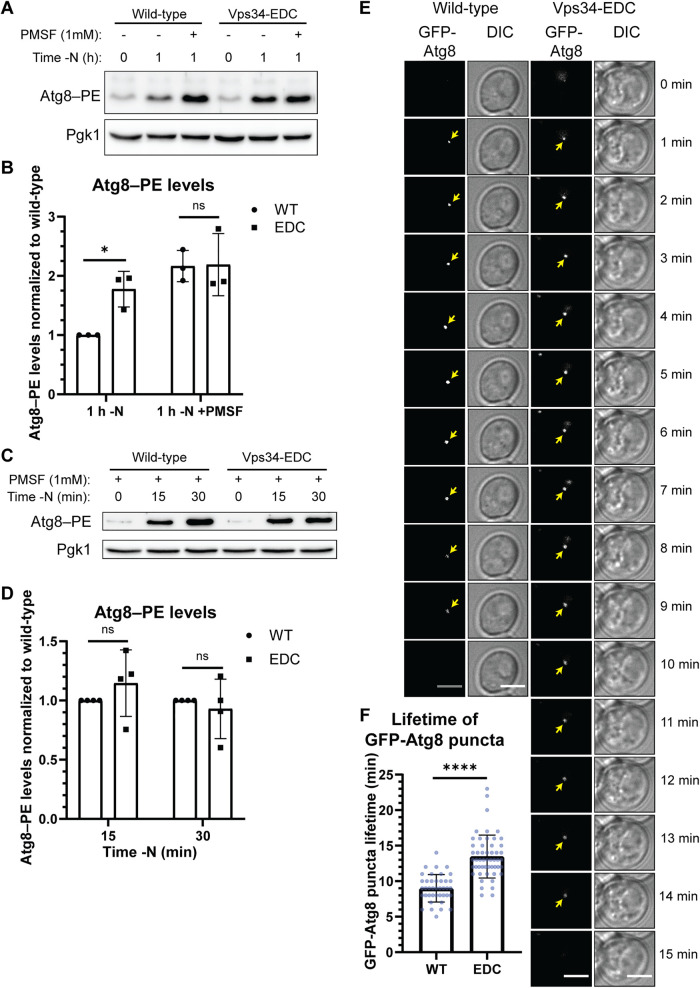
PI3P is involved in multiple steps of autophagy, including autophagosome biogenesis, disassembly of the autophagy machinery from the surface of mature autophagosomes, and the fusion of mature autophagosomes with the vacuole. As part of our analysis, we investigated each of these steps. A combination of autophagy induction and flux was assessed by determining the levels of lipidated Atg8 (Atg8–PE; Klionsky et al., 2016). Atg8 (LC3 and GABARAP subfamilies in mammals) is a ubiquitin-like protein. The expression of ATG8 is strongly induced after autophagy induction, following which Atg8 is conjugated to phosphatidylethanolamine (PE), on the phagophore membrane (Ichimura et al., 2000). Following phagophore expansion and closure, and before autophagosome–vacuole fusion, Atg8–PE on the cytoplasmic surface of the resulting autophagosome is cleaved from the membrane. The lumenal surface of the autophagosome, however, contains Atg8–PE that is trapped within the autophagosome. This pool of Atg8–PE can be monitored as autophagic cargo because, following fusion with the vacuole, this pool of Atg8–PE is degraded in the vacuolar lumen. Consequently, the steady-state level of Atg8–PE after autophagy induction is a function of both its generation and its degradation in the vacuole.
Treating cells with the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) blocks the vacuolar degradation of Atg8–PE, facilitating the measurement of autophagy induction independent of Atg8–PE turnover. Following 1 h of autophagy stimulation by nitrogen starvation, no difference in Atg8–PE levels could be detected between strains expressing Vps34-WT and Vps34-EDC in the presence of PMSF (Figure 4, A and B), suggesting that the extent of autophagy induction was unchanged by hyperactive Vps34-EDC. Similarly, in the presence of PMSF, no difference in Atg8–PE levels could be detected between strains expressing Vps34-WT and Vps34-EDC at either 15 or 30 min following nitrogen starvation (Figure 4, C and D). These results suggest that autophagy initiation requires, but is not driven by PI3P.
FIGURE 4:
Hyperactive Vps34 inhibits a late step in autophagy. (A, B) Comparison of Atg8–PE protein levels in the absence and presence of PMSF, a protease inhibitor, suggests that Vps34-EDC inhibits a late step in autophagy, but does not change the rate of autophagy induction. During autophagy, the levels of Atg8–PE are determined by both covalent attachment of phosphatidylethanolamine (PE) to Atg8 and by degradation of Atg8–PE once autophagosomes fuse with the vacuole. When degradation of Atg8–PE is inhibited by the serine protease inhibitor, PMSF, there was no difference in Atg8–PE levels in Vps34-WT compared with Vps34-EDC, suggesting that autophagy induction is unchanged by Vps34-EDC. However, in the absence of PMSF, Atg8–PE protein levels are elevated by hyperactive Vps34-EDC following 1 h of nitrogen starvation, suggesting that Vps34-EDC inhibits a late step in autophagy. vps34Δ cells were transformed with pRS416-Vps34 or pRS416-Vps34-EDC. Atg8–PE protein levels were analyzed via Western blot using anti-Atg8 antibody. Atg8–PE levels were normalized to Pgk1. Levels were further normalized to wild-type at 1 h of nitrogen starvation. Representative of n = 3. Error bars indicate SD. Unpaired t test. ns = p > 0.05, * = p < 0.05. (C, D) Following 15 and 30 min of nitrogen starvation, Atg8–PE protein levels are unchanged between hyperactive Vps34-EDC and wild-type when degradation of Atg8–PE is inhibited by the protease inhibitor, PMSF. This result suggests that Vps34-EDC does not affect autophagy induction. vps34Δ cells were transformed with pRS416-Vps34 or pRS416-Vps34-EDC. Atg8–PE protein levels were analyzed via Western blot using anti-Atg8 antibody. Atg8–PE levels were normalized to Pgk1. Levels were further normalized to wild-type at 15 and 30 min following nitrogen starvation. Representative of n = 4. Error bars indicate SD. Unpaired t test. ns = p > 0.05. (E, F) Hyperactive Vps34-EDC results in an increase in the lifetime of GFP-Atg8 puncta in cells. vps34Δ cells were cotransformed with pRS414-GFP-Atg8 and pRS416-Vps34 or pRS416-Vps34-EDC. Cells were imaged every minute for 26 min following 30 min of nitrogen starvation. Twenty z-slices that were 0.2 µm apart were acquired at each time point. Single z-slice. DIC, differential interference contrast. Scale bar = 3.5 µm. GFP-Atg8 puncta were tracked over time with the lifetime of each puncta calculated as the time between the first and last frames the puncta were visible. Only GFP-Atg8 puncta that could be followed unambiguously over their lifetime were analyzed. For wild-type, n = 40. For Vps34-EDC, n = 52. Error bars indicate SD. Unpaired t test. **** = p < 0.0001.
As a second independent measure of the induction of autophagy, we investigated the transcriptional up-regulation of critical autophagy genes, ATG1, ATG7, and ATG9, whose expression is elevated during nitrogen starvation (Delorme-Axford and Klionsky, 2018). We found no differences in mRNA levels between Vps34-WT and Vps34-EDC following 30 min of nitrogen starvation (Supplemental Figure S6A), suggesting that hyperactive Vps34 does not affect the transcription of autophagy genes.
Whereas the levels of Atg8–PE are very similar between Vps34-WT and Vps34-EDC following starvation in the presence of PMSF, comparing Atg8–PE levels in the absence of PMSF reveals that Vps34-EDC partially blocks autophagy flux. In the absence of PMSF, degradation of Atg8–PE can occur after autophagosome fusion with the vacuole. Following 1 h of nitrogen starvation in the absence of PMSF, Atg8–PE protein levels were markedly lower in Vps34-WT compared with Vps34-EDC (Figure 4, A and B). The observation that no difference in Atg8–PE levels was detected between Vps34-WT and Vps34-EDC in the presence of PMSF, but that Atg8–PE was lower in Vps34-WT in the absence of PMSF, indicates that hyperactive Vps34 partially inhibits vacuolar delivery and degradation of Atg8–PE.
To further characterize the defect in a late step of autophagy, we examined the dynamics of autophagosome biogenesis using GFP-tagged Atg8 as a marker. Hyperactive Vps34-EDC results in an increased number of GFP-Atg8 puncta in cells following 30 min of nitrogen starvation (Supplemental Figure S6, B and C). This result is consistent with either an increase in the induction of autophagy and/or with the inhibition of a late step in autophagy. Using time-lapse microscopy of GFP-Atg8 puncta, we measured the lifetime of GFP-Atg8 puncta in strains expressing either Vps34-WT or Vps34-EDC. GFP-Atg8 puncta in the presence of Vps34-WT persisted for 9 min, consistent with previous studies (Cebollero et al., 2012). In contrast, GFP-Atg8 puncta in the presence of Vps34-EDC persisted for 13.5 min (Figure 4, E and F). These results suggest that the resolution of autophagosomes is delayed by hyperactive Vps34. Further evidence of this inhibition is observed by measuring the degradation of GFP-Atg8 via the appearance of proteolytically resistant free GFP by immunoblot analysis. Expression of hyperactive Vps34-EDC resulted in a reduction in the appearance of free GFP following 1 and 2 h of nitrogen starvation (Figure 5, A and B), indicating a reduction in autophagy flux.
FIGURE 5:
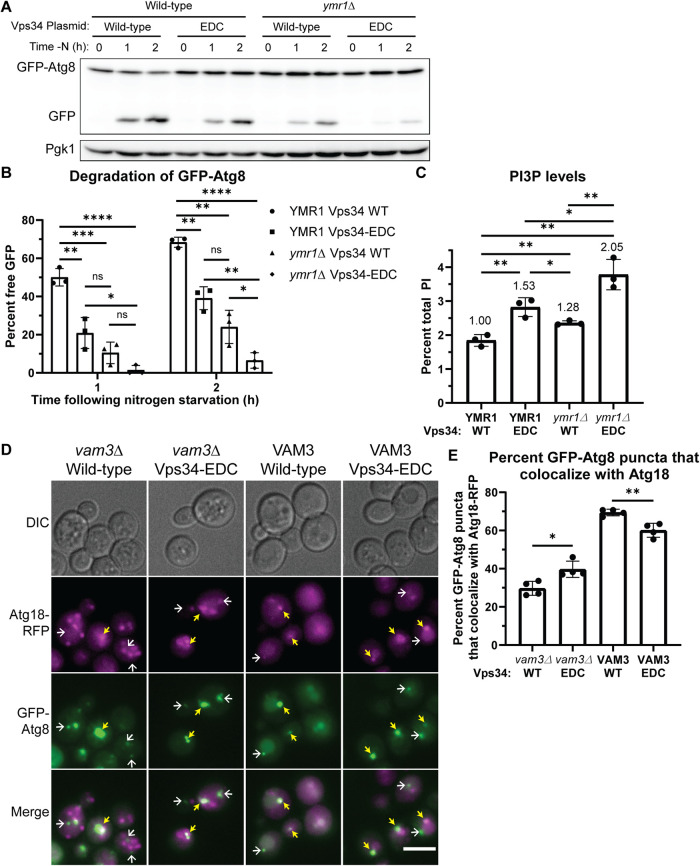
Hyperactive Vps34 inhibits a late step in autophagy. (A, B) The Vps34-EDC mutant or deletion of YMR1 inhibits degradation of GFP-Atg8 following nitrogen starvation. Combining Vps34-EDC with a deletion of YMR1 further inhibits degradation of GFP-Atg8, indicating a block in autophagy flux. vps34Δ or vps34Δ ymr1Δ cells were cotransformed with pRS416-Vps34 or pRS416-Vps34-EDC and copper inducible pRS414-pCup1-GFP-Atg8. No exogenous copper was added during this experiment. GFP-Atg8 and GFP protein levels were analyzed via Western blot using anti-GFP antibody. Free GFP levels were divided by the sum of GFP-Atg8 and free GFP and expressed as a percent. Representative of n = 3. Error bars indicate SD. Unpaired t test. ns = p > 0.05, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001. (C) The Vps34-EDC mutant and deletion of the yeast myotubularin gene YMR1 elevated PI3P levels. Combining Vps34-EDC with deletion of YMR1 further elevated PI3P levels. vps34Δ or vps34Δ ymr1Δ cells were transformed with pRS416-Vps34 or pRS416-Vps34-EDC. PPI lipid levels were measured by metabolically labeling cells with myo-3H-inositol for 16 h, harvesting cells, and separating PPI lipid head groups by anion exchange and HPLC. n = 3. Error bars indicate SD. Unpaired t test. * = p < 0.05, ** = p < 0.01. (D, E) Hyperactive Vps34-EDC leads to a defect in disassembly of the autophagy machinery from the surface of mature autophagosomes as well as a defect in fusion of autophagosomes with the vacuole. In the presence of Vam3, Vps34-EDC caused a small but statistically significant decrease in the percentage of GFP-Atg8 puncta that colocalize with Atg18-RFP after 1 h of nitrogen starvation. This suggests that Vps34-EDC leads to a minor defect in fusion of autophagosomes with the vacuole. However, in vam3Δ cells where fusion of autophagosomes with the vacuole is completely blocked, hyperactive Vps34-EDC leads to a small but statistically significant increase in the percentage of GFP-Atg8 puncta that colocalize with Atg18-RFP after 1 h of nitrogen starvation. This result suggests that Vps34-EDC also leads to a minor defect in the dissociation of key autophagy machinery from mature autophagosomes. vps34Δ or vps34Δ vam3Δ cells with Atg18-RFP integrated at the endogenous locus were transformed with pRS413-GFP-Atg8 and pRS416-Vps34 or pRS416-Vps34-EDC. Cells were imaged after 1 h of nitrogen starvation. DIC, differential interference contrast. Scale bar = 5 µm. Individual GFP-Atg8 puncta were scored on whether or not they colocalized with Atg18-RFP puncta. The scorer was blinded to the genotype of the cells being quantified. Cells without visible Atg18-RFP were excluded from quantification. Examples of GFP-Atg8 puncta that colocalize with Atg18-RFP are indicated by yellow arrows. Examples of GFP-Atg8 puncta that do not colocalize with Atg18-RFP are indicated by white arrows. Quantification of 100 cells per n, n = 4. Error bars indicate SD. Unpaired t test. ** = p < 0.01, **** = p < 0.0001.
To explore whether yet higher elevation of PI3P levels would further reduce autophagy flux, we enhanced the elevation of PI3P by combining the Vps34-EDC mutant with a knockout of the YMR1 gene, encoding a putative PI3P phosphatase. We found that the Vps34-EDC ymr1Δ mutant elevated PI3P by 105%, compared with 53% by Vps34-EDC alone and 28% by a ymr1Δ mutant alone (Figure 5C). Deletion of YMR1 in our strain background more robustly elevated PI3P levels than previously reported (Parrish et al., 2004), and the increase in PI3P levels caused by YMR1 deletion was more evident in the presence of Vps34-EDC. Notably, the double Vps34-EDC ymr1Δ mutant inhibited autophagy flux considerably more than either mutation alone as measured by degradation of GFP-Atg8 by immunoblot analysis (Figure 5, A and B). These results strongly indicate an inverse correlation between autophagy flux and an increase in cellular PI3P. Furthermore, the turnover of PI3P on autophagosomes may be critical for the successful completion of a late step in autophagy.
Before the fusion of mature autophagosomes with the vacuole, some autophagy proteins, including the PI3P-binding protein Atg18, must be removed from the autophagosome surface (Cebollero et al., 2012). Removal of proteins from the surface of autophagosomes likely requires depletion of PI3P via the PI3P phosphatase Ymr1 (Cebollero et al., 2012). Conversely, in vitro studies suggest that PI3P is required for the fusion of the autophagosome with the vacuole (Bas et al., 2018). Thus, we tested whether the expression of hyperactive Vps34-EDC had an impact on the displacement of Atg18 from the autophagosome surface and/or the fusion of autophagosomes with the vacuole.
To specifically test displacement of Atg18 from the autophagosome surface independent of autophagosome–vacuole fusion, we blocked autophagosome–vacuole fusion by deleting the vacuolar t-SNARE Vam3 (Cebollero et al., 2012). We assessed the disassembly of Atg18 after 1 h of nitrogen starvation, by determining the percentage of GFP-Atg8–positive autophagosomes that colocalized with surface Atg18-RFP. In vam3Δ cells, hyperactive Vps34-EDC leads to a 10% increase in the percentage of GFP-Atg8 puncta that colocalize with Atg18-RFP (Figure 5, D and E). This result suggests that hyperactive Vps34-EDC leads to a defect in the dissociation of autophagy proteins from mature autophagosomes.
We also tested whether there was a defect in autophagosome fusion with the vacuole in a strain with wild-type Vam3. After 1 h of nitrogen starvation we measured the percentage of GFP-Atg8–positive autophagosomes that colocalize with Atg18-RFP. In this experiment, both disassembly of Atg18-RFP from the autophagosome surface and autophagosome–vacuole fusion can occur. A defect in autophagosome–vacuole fusion will result in the persistence of GFP-Atg8 puncta that do not colocalize with Atg18-RFP, and thus a decrease in the colocalization between GFP-Atg8 and Atg18-RFP. Indeed, we found that hyperactive Vps34-EDC caused a 9% decrease in the percentage of GFP-Atg8 puncta that colocalize with Atg18-RFP (Figure 5, D and E). Note that the 10% defect in disassembly of Atg18-RFP from autophagosomes that was observed in the vam3Δ cells likely also occurs in the wild-type VAM3 strain. Thus, this result suggests that Vps34-EDC leads to approximately a 20% defect in fusion of autophagosomes with the vacuole. Additionally, the total number of GFP-Atg8 puncta observed in vam3Δ cells did not change between wild-type and hyperactive Vps34-EDC (Supplemental Figure S6D). Because autophagosomes are not turned over in vam3Δ cells, this result provides further evidence that hyperactive Vps34-EDC does not affect the induction of autophagy, indicating that differences in autophagy induction are not responsible for changes in the colocalization of GFP-Atg8 and Atg18-RFP. Note that Atg18 protein levels were not different between wild-type and Vps34-EDC under nutrient-rich conditions or following nitrogen starvation (Supplemental Figure S6, E and F).
Together, our results suggest that whereas hyperactive Vps34-EDC has no effect on the induction of autophagy, it results in a decrease in autophagic flux in part via defects in both disassembly of the autophagy machinery from the surface of mature autophagosomes and fusion of autophagosomes with the vacuole. However, the 10% defect in disassembly of autophagy proteins from autophagosomes and the 20% defect in autophagosome–vacuole fusion do not fully account for the observed defect in autophagic flux. Autophagic flux, as measured by Atg8–PE levels, lifetime of Atg8 puncta, or degradation of GFP-Atg8 indicated a defect in the range of 50–70%. Thus, there are likely additional steps in the resolution of autophagy that are affected by hyperactive Vps34-EDC. Interestingly, despite inhibiting autophagy, hyperactive Vps34-EDC does not affect the survival of yeast cells during long-term nitrogen starvation (Supplemental Figure S6G), which suggests that Vps34-EDC cells maintain sufficient levels of autophagy function to allow for survival during chronic nitrogen starvation.
The finding that hyperactive Vps34-EDC causes a partial defect in autophagosome fusion with the vacuole raises the possibility that there is a more global defect in fusion with the vacuole. A defect in vacuole–vacuole fusion would result in a more fragmented vacuole morphology. To assess homotypic vacuole fusion, we counted the number of vacuole lobes per cell, finding a small but significant 12% increase in vacuole lobes (Figure 6, A and B). This difference was predominantly due to an ∼40% decrease in the number of cells with a single vacuole (Figure 6C). While an increase in the number of vacuole lobes per cell could also be caused by an increase in vacuole fission, a modest impairment of fusion fits closely with the observed defect in autophagosome fusion with the vacuole.
FIGURE 6:
Hyperactive Vps34-EDC may lead to a modest decrease in homotypic vacuole fusion. (A–C) Hyperactive Vps34-EDC results in a 12% increase in the average number of vacuole lobes in a cell (B), which occurs primarily due to a decrease in the number of cells with 1 vacuole lobe (C). vps34Δ cells were transformed with pRS416-Vps34 or pRS416-Vps34-EDC. After labeling with FM 4-64, cells were chased at 24°C for 3 h before imaging. DIC, differential interference contrast. Scale bar = 5 µm. The number of vacuole lobes was counted by a scorer who was blinded to the genotype of the cells being quantified. Quantification of at least 100 cells per n, n = 4. Error bars indicate SD. Unpaired t test (B). Two-way ANOVA and Dunnett’s post-hoc test (C). ns = p > 0.05, * = p < 0.05, ** = p < 0.01.
Conclusions
Most studies of PPI lipids use knockdown, knockout, or drug inhibition of PPI kinases to test whether a specific PPI is essential for a pathway. However, this approach does not indicate whether a PPI lipid is a key regulator of the pathway of interest. The ability to generate mutations that robustly elevate a PPI lipid provides an effective tool to elucidate the regulatory roles of PPI lipids. Previously, our lab generated hyperactive mutations in the PI3P 5-kinase, Fab1, by screening for mutants that rescued Fab1 function in strains where key Fab1 activators were knocked out (Duex et al., 2006b; Lang et al., 2017). These mutants led to mechanistic insights into the regulation of Fab1 (Lang et al., 2017). In this study, instead of knocking out genes encoding Vps34 regulators, we performed a screen based on rescuing growth of a hypomorphic Vps34 allele that was mutated near the active site of the enzyme. The mutants identified in this screen were concentrated near the active site of the kinase and most likely help favor an active conformation of the kinase, but may not be regulatory sites on the native enzyme. These two different screening strategies appear to return mutations that provide insight into different aspects of PPI kinase function and regulation. These differences should guide the design of future screens for hyperactive PPI kinases. A screen that avoids crippling the active site of the kinase is more likely to yield mechanistic insights into the regulation of a PPI kinase. When regulators of PPI kinases are not known or when the sole goal of a hyperactive mutant is to determine downstream effects of elevating a PPI lipid, a screening strategy based on a hypomorphic PPI kinase allele is straightforward because catalytic site residues can be readily identified.
The major advantage of the Vps34-EDC mutant was that it enabled specific and direct testing of the downstream effects of elevating PI3P. It should be noted that in these experiments, the Vps34-EDC plasmid is constitutively expressed and thus PI3P is constitutively elevated. This is in contrast with wild-type cells where stimulus-induced changes in phosphoinositides occur on the order of minutes (Duex et al., 2006a). Cells may respond differently to chronic versus acute elevation of phosphoinositides. We determined that elevation of PI3P drives some pathways, including synthesis of PI(3,5)P2 during hyperosmotic shock, as well as retrograde transport of Atg27. These findings demonstrate that elevation of PI3P drives some downstream processes and that in these cases, the PI3P-dependent step is rate limiting. In contrast, hyperactive Vps34 does not affect ESCRT function, demonstrating that elevating PI3P does not always increase the overall rate of a complex pathway. Much like ESCRT function, induction of autophagy is not accelerated by hyperactive Vps34, though, conversely, autophagic flux is lowered by hyperactive Vps34, in part by delaying disassembly of the autophagy machinery from the surface of mature autophagosomes as well as a delay in fusion of autophagosomes with the vacuole. These results provide evidence that PI3P can also inhibit specific steps within a pathway. Thus, these studies suggest that stimulus-induced elevation of PI3P can positively or negatively regulate some PI3P-dependent membrane trafficking pathways, and that phosphoinositide lipids are commonly rate limiting in pathways where they are required. Moreover, these findings suggest that stimulus-induced elevation of PPI lipids provides a way for stimuli to selectively regulate pathways.
MATERIALS AND METHODS
Yeast strains, plasmids, and media
Yeast cultures were grown in yeast extract peptone dextrose (YEPD) containing 1% yeast extract, 2% peptone, and 2% dextrose or synthetic complete (SC) media lacking the indicated amino acid(s) at 24°C unless specified. For nitrogen starvation, yeast strains were cultured in SD-N medium containing 0.19% yeast nitrogen base, 2% glucose, and vitamins, and lacking amino acids and ammonium sulfate (Formedium). Yeast strains and plasmids are listed in Supplemental Tables S1 and S2, respectively.
Phosphoinositide lipid labeling and quantification
Yeast myo-3H-inositol labeling and total cellular phosphoinositide extraction, deacylation, and measurements were performed as described (Bonangelino et al., 2002; Duex et al., 2006a). Briefly, cells were grown in the appropriate SC media to midlog phase, washed with SC media lacking inositol, and used to inoculate 5 ml of SC media lacking inositol and containing 50 μCi of myo-3H-inositol. Cells were grown for 16 h shaking at 24°C, harvested by centrifugation, and resuspended in 100 μl inositol-free media. For hyperosmotic shock, 100 μl of inositol-free media with 1.8 M NaCl was added to the sample for 10 min. For basal conditions, 100 μl of inositol-free media was added to the 100-μl sample. Cells were then killed via addition of ice-cold 4.5% perchloric acid. Cells were lysed using a minibeadbeater for 2 min, then immediately put on ice for 2 min. This was repeated two more times for a total lysis time of 6 min. Cell extracts were centrifuged at 16,000 × g for 10 min at room temperature. Pellets were washed with 1 ml of 100 mM EDTA, pH 8.0, then resuspended in 50 μl distilled deionized water (ddH2O). Samples were deacylated with 1 ml methylamine reagent for 1 h at 55°C, then dried in a speed vac concentrator. Pellets were resuspended in 300 μl of ddH2O, mixed with 300 μl of a 20:4:1 mixture of butanol/ethyl ether/formic acid ethyl ester, vortexed, and centrifuged for 2 min at 16,000 × g. The lower aqueous phase was then transferred to a fresh microcentrifuge tube. This sample extraction was repeated, and samples were dried in a speed vac concentrator. Dried samples were resuspended in 60 μl ddH2O and analyzed by high-performance liquid chromatography (HPLC) using a SAX anion exchange column. Buffer A (ddH2O) and buffer B (1 M (NH4)2HPO4, pH 3.8) are used to generate the following gradients run at a 1 ml/min flow rate: 1% buffer B for 5 min, 1–20% buffer B for 44 min, 20–50% buffer B for 3.75 min, and 50% buffer B for 8 min. To quantify scintillation counts from each sample, the raw counts in each peak were expressed as a percentage of total phosphatidylinositol-related species, calculated from summation of the counts of the five glycero-inositol peaks present in yeast (PI, PI3P, PI4P, PI(3,5)P2, and PI(4,5)P2). Background scintillation counts were calculated from adjacent regions and subtracted from all peaks.
Screen for hyperactive Vps34 mutants
A schematic of our screen is shown in Supplemental Figure S2A. A pRS416-Vps34-K759D plasmid was gapped by digesting with XhoI restriction enzyme. The gapped plasmid was purified by agarose electrophoresis followed by DNA purification (Qiagen). Primers AM30 (5-GAATTCACTATTGTGGATGCCGTATCTTCG-3) and AM31 (5-GGGTAACGCCAGGGTTTTCC-3) were used to PCR amplify the gapped region along with ∼100 bases upstream of and downstream from the XhoI restriction sites using error-prone Taq DNA polymerase (Invitrogen). Gapped plasmid backbone and mutated PCR product were cotransformed into vps34∆ cells. Two days following cotransformation, colonies were replica plated to plates containing 10 nM rapamycin and grown at 33°C. From an estimated 12,000 colonies, mutant Vps34 plasmids were isolated from 59 single colonies that rescued growth on rapamycin at 33°C and amplified in Escherichia coli. When mutant plasmids were retransformed into vps34∆ cells, 33 of the 59 plasmids rescued growth on rapamycin at 33°C. Sanger sequencing of those 33 plasmids revealed that 11 plasmids contained a mutation at the D759 locus. The 22 remaining independent mutants indicated nine unique point mutations. Of those nine mutations, changes at five of them elevated PI3P levels.
Fluorescence microscopy
Yeast cells were grown in the appropriate SC media to midlog phase. Live cell images were obtained on a DeltaVision Restoration system (Applied Precision) using an inverted epifluorescence microscope (IX-71; Olympus) with a charge-coupled device camera (Cool-SNAP HQ; Photometrics) and processed in Fiji. When vacuoles were visualized, cells were labeled with 12 μg FM 4-64 in 250 μl media for 1 h, then washed twice and grown in 5 ml fresh SC media for one doubling time (2–3 h; Vida and Emr, 1995).
Quantification of fluorescence microscopy images
Quantification of the number of vacuole lobes, the number of cells with Atg27-2xGFP visible on the vacuole, the number of GFP-Atg8 puncta, and the colocalization of GFP-Atg8 puncta with Atg18-RFP puncta was performed by a scorer who was blinded to the genotype of the cells being quantified.
The amount of Envy-Vps34, Fab1-Envy, or Fig4-Envy that colocalized with the vacuole was measured using Fiji. Background fluorescence was subtracted using a 5.0 pixel rolling ball radius in the green channel and a 10.0 pixel rolling ball radius in the FM 4-64 channel. FM 4-64 images were thresholded such that pixels with signal present were set to 1 and pixels without signal present were set to 0 using the Otsu method. Total signal that colocalizes with the vacuole was calculated by multiplying the background subtracted green channel image by the FM 4-64 thresholded image and determining the integrated intensity of the resulting image. The total colocalized green signal was divided by the number of cells in the quantified images to give the green signal intensity on the vacuole per cell.
Colocalization of Atg27-2xGFP with FM 4-64, Sec7-mCherry, or Vps8-mCherry was measured using Fiji. Background fluorescence was subtracted using a 5.0 pixel rolling ball radius and images were thresholded such that pixels with signal present were set to 1 and pixels without signal present were set to 0. The Otsu method was used, except for Vps8-mCherry where the Renyi entropy method was used. Total Atg27-2xGFP signal was calculated by multiplying the background subtracted Atg27-2xGFP image by the Atg27-2xGFP thresholded image and determining the integrated intensity of the resulting image. The Atg27-2xGFP signal that colocalized with FM 4-64, Sec7-mCherry, or Vps8-mCherry was calculated by multiplying the Atg27-2xGFP thresholded image and the FM 4-64/Sec7-mCherry/Vps8-mCherry thresholded image. The resulting image was multiplied by the background subtracted Atg27-2xGFP image and integrated intensity of the resulting image was determined. Colocalized Atg27-2xGFP signal was divided by total Atg27-2xGFP signal to calculate percent overlap.
Western blot analysis
Yeast cells were grown in the appropriate SC media to midlog phase. Before harvesting, cells were treated in the appropriate experiment-specific conditions. For Ypq1-GFP and Mup1-GFP degradation assays, cells were washed twice in lysine-free SC media before resuspending in lysine-free SC media or resuspended in SC media containing methionine, respectively (Zhu et al., 2017). For Atg8–PE and GFP-Atg8 assays, cells were shifted to nitrogen-starvation media (-N) for autophagy induction. One OD of cells was harvested, and 100 μl urea lysis buffer was added (1% SDS, 8 M urea, 10 mM Tris, pH 6.8, 10 mM EDTA, 0.01% bromophenol blue, 0.2% β-mercaptoethanol, Roche complete protease inhibitor cocktail). One-half volume of 0.5 mm Zirconia glass beads was added and tubes were vortexed in a microtube mixer for 10 min at 4°C. Supernatants were transferred to fresh tubes and samples were heated for 10 min at 75°C and run on an SDS polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane at 60 V for 16 h. Membranes were blocked in 5% milk before incubation with the indicated primary antibody. Membranes were washed three times for 5 min in Tris-buffered saline containing 0.1% TWEEN-20, incubated in secondary antibodies, washed again, developed with ECL prime (GE Healthcare; Pgk1) or Clarity Max (Bio-Rad; other antibodies), imaged on a Bio-Rad ChemiDoc imager, and quantified using FIJI. For immunoblot analyses, the following antibodies were used: mouse anti-GFP (1:1000; Roche), mouse anti-Myc (1:1000; clone 9E10 EMD; Millipore), mouse anti-Pgk1 (1:10,000; Invitrogen), mouse anti-Vph1 (1:1000; Abcam), rabbit anti-Atg8 (described previously; Huang et al., 2000), mouse anti-Dpm1 (1:5000; ThermoFisher), mouse anti-HA (1;1000; Covance MMS-101P).
Analysis of Mup1-GFP degradation
Mup1-GFP levels were normalized to Pgk1. A linear mixed-effects model was used to assess the degradation rate of Mup1-GFP. The logarithmic transformation of Mup1-GFP levels was modeled as a linear function of time and allowed to vary by genotype. Replicate-specific intercepts were included to account for residual correlation between protein levels within the same replicate. A 95% confidence interval of the genotype by time interaction was used to determine whether there were statistically significant differences between wild-type and hyperactive Vps34-EDC.
Hyperosmotic shock growth assay
Yeast cells were grown to midlog phase in SC media and then diluted to equal concentrations. An equal volume of SC media or SC media with 1.8 M NaCl was added to the culture to begin the time course. ODs were measured immediately following the addition of SC media or SC media with 1.8 M NaCl, and then every 4 h for 24 h. The time zero OD measurement for each sample was normalized to 1. A natural logarithmic transformation was applied to the normalized ODs so that exponential growth is represented linearly.
Real-time quantitative PCR
Yeast were cultured in the appropriate SC media to midlog phase and then shifted to nitrogen-starvation media (-N) for 30 min for autophagy induction. Cells were collected and flash-frozen in liquid nitrogen. Total RNA was extracted using an RNA extraction kit (Clontech; Nucleo Spin RNA; 740955.250). Reverse transcription was carried out using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems/Thermo Fisher Scientific; 4368814). For each sample, 1 μg RNA was used for cDNA synthesis. Real-time quantitative PCR (RT-qPCR) was performed using the Radiant SYBR Green Lo-ROX qPCR kit (Alkali Scientific) in a CFX Connect (Bio-Rad; 1855201) real-time PCR machine. For all RT-qPCR experiments, melting curves were run after the PCR cycles to verify primer specificity. Relative gene expression was calculated using the 2–ΔΔCT method and normalized as indicated (Livak and Schmittgen, 2001).
Nitrogen starvation survival assay
Yeast cells were grown to midlog phase in SC media. Equal numbers of cells were collected and rinsed twice in nitrogen-starvation media before resuspending in nitrogen-starvation media. Following 1 d and 14 d of nitrogen starvation, equal volumes of culture were serially diluted 1:5 and spotted on SC plates. Plates were imaged following 3 d of yeast growth.
Supplementary Material
Acknowledgments
We thank members of the Weisman laboratory for their insightful discussions. We thank Christopher Burd for the Atg27-2xGFP strain used to make yeast strains LWY19264, LWY19252, LWY19306, LWY19358, LWY19518, and LWY19521. We thank Ajit Joglekar for discussions of quantification of Atg27-2xGFP colocalization. We thank Ming Li for pRS416-Ypq1-GFP used to make pRS414-Ypq1-GFP. We thank Mara Duncan for pRS416-Mup1-GFP used to make pRS414-Mup1-GFP. We thank Joseph Dickens of Consulting for Statistics, Computing, and Analytics Research at the University of Michigan for his help performing statistical analysis on the degradation of Mup1-GFP. We thank Fulvio Reggiori for pRS416-Atg18-GFP.
This work was supported by National Institutes of Health Grants no. R01-NS099340 and no. R01-GM062261 (to L.S.W.), Grant no. GM131919 (to D.J.K.), mCubed 3.0, University of Michigan (to L.S.W.), and an LSI Cubed Collaborative Grant from the Life Sciences Institute, University of Michigan (to N.S. and V.L.). N.S. was supported in part by NIGMS Grant no. T32-GM007315 and a Rackham Warner Lambert Fellowship from the Program in Cellular and Molecular Biology, University of Michigan.
Abbreviations used:
- ESCRT
endosomal sorting complexes required for transport
- HELCAT
helical and kinase domains
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- (PI(3,5)P 2)
phosphatidylinositol-3,5-bisphosphate
- PI3P
phosphatidylinositol 3-phosphate
- PMSF
phenylmethylsulfonyl fluoride
- PPI
phosphoinositide
- SC
synthetic complete.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-03-0191) on November 25, 2020.
REFERENCES
- Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y (2013). Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol 203, 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 93, 1019–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN (1996). TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7, 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bas L, Papinski D, Licheva M, Torggler R, Rohringer S, Schuschnig M, Kraft C (2018). Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J Cell Biol 217, 3656–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS (2002). Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol 156, 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho RJ, Efe JA, Teis D, Emr SD (2008). Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell 19, 4273–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P, Padilla SM, Sarkar S, Emr SD (2002). Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci 115, 3889–3900. [DOI] [PubMed] [Google Scholar]
- Cabrera M, Nordmann M, Perz A, Schmedt D, Gerondopoulos A, Barr F, Piehler J, Engelbrecht-Vandre S, Ungermann C (2014). The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J Cell Sci 127, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E, van der Vaart A, Zhao M, Rieter E, Klionsky DJ, Helms JB, Reggiori F (2012). Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol 22, 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M (2001). Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol 3, 613–618. [DOI] [PubMed] [Google Scholar]
- Delorme-Axford E, Klionsky DJ (2018). Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae. J Biol Chem 293, 5396–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH (1997). Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390, 187–192. [DOI] [PubMed] [Google Scholar]
- Dove SK, Piper RC, McEwen RK, Yu JW, King MC, Hughes DC, Thuring J, Holmes AB, Cooke FT, Michell RH, et al. (2004). Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J 23, 1922–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS (2006a). Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell 5, 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Tang F, Weisman LS (2006b). The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J Cell Biol 172, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti RA, Wickner W (2007). Distinct targeting and fusion functions of the PX and SNARE domains of yeast vacuolar Vam7p. J Biol Chem 282, 13133–13138. [DOI] [PubMed] [Google Scholar]
- Gary JD, Sato TK, Stefan CJ, Bonangelino CJ, Weisman LS, Emr SD (2002). Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol Biol Cell 13, 1238–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD (1998). Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol 143, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, et al. (2012). Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res 22, 473–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WP, Scott SV, Kim J, Klionsky DJ (2000). The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem 275, 5845–5851. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD (2003). Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol 162, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Kirisako H, Koizumi M, Ohsumi Y, Nakatogawa H (2018). The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci USA 115, 10363–10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang MJ, Strunk BS, Azad N, Petersen JL, Weisman LS (2017). An intramolecular interaction within the lipid kinase Fab1 regulates cellular phosphatidylinositol 3,5-bisphosphate lipid levels. Mol Biol Cell 28, 858–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence G, Brown CC, Flood BA, Karunakaran S, Cabrera M, Nordmann M, Ungermann C, Fratti RA (2014). Dynamic association of the PI3P-interacting Mon1-Ccz1 GEF with vacuoles is controlled through its phosphorylation by the type 1 casein kinase Yck3. Mol Biol Cell 25, 1608–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Rong Y, Chuang YS, Peng D, Emr SD (2015). Ubiquitin-dependent lysosomal membrane protein sorting and degradation. Mol Cell 57, 467–478. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma M, Burd CG, Chi RJ (2017). Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic 18, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menant A, Barbey R, Thomas D (2006). Substrate-mediated remodeling of methionine transport by multiple ubiquitin-dependent mechanisms in yeast cells. EMBO J 25, 4436–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ (2002). Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem 277, 30198–30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y (2008). The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem 283, 23972–23980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y (2006). Assortment of phosphatidylinositol 3-kinase complexes—Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell 17, 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish WR, Stefan CJ, Emr SD (2004). Essential role for the myotubularin-related phosphatase Ymr1p and the synaptojanin-like phosphatases Sjl2p and Sjl3p in regulation of phosphatidylinositol 3-phosphate in yeast. Mol Biol Cell 15, 3567–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieter E, Vinke F, Bakula D, Cebollero E, Ungermann C, Proikas-Cezanne T, Reggiori F (2013). Atg18 function in autophagy is regulated by specific sites within its beta-propeller. J Cell Sci 126, 593–604. [DOI] [PubMed] [Google Scholar]
- Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, et al. (2015). Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350, aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink KO, Tan KW, Stenmark H (2016). Phosphoinositides in control of membrane dynamics. Annu Rev Cell Dev Biol 32, 143–171. [DOI] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD (1993). Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88–91. [DOI] [PubMed] [Google Scholar]
- Seaman MN, Marcusson EG, Cereghino JL, Emr SD (1997). Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol 137, 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, McCaffery JM, Emr SD (1998). A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol 142, 665–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra VA, Boettner DR, Lemmon SK (2015). Atg27 tyrosine sorting motif is important for its trafficking and Atg9 localization. Traffic 16, 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Herman PK, Schu PV, Emr SD (1993). A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J 12, 2195–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stjepanovic G, Baskaran S, Lin MG, Hurley JH (2017). Vps34 kinase domain dynamics regulate the autophagic PI 3-kinase complex. Mol Cell 67, 528–534.e523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Steinfeld N, Lee S, Jin N, Munoz-Rivera C, Meeks G, Thomas A, Akemann C, Mapp AK, MacGurn JA, et al. (2020). Roles for a lipid phosphatase in the activation of its opposing lipid kinase. Mol Biol Cell 31, 1835–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Chen L, Cao W, Roth AF, Davis NG (2004). The yeast casein kinase Yck3p is palmitoylated, then sorted to the vacuolar membrane with AP-3-dependent recognition of a YXXΦ adaptin sorting signal. Mol Biol Cell 15, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SW, Emr SD (2018). Membrane protein recycling from the vacuole/lysosome membrane. J Cell Biol 217, 1623–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GS, Maehama T, Dixon JE (2000). Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci USA 97, 8910–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Shaw AS, Kannan N, Kornev AP (2015). Integration of signaling in the kinome: Architecture and regulation of the αC helix. Biochim Biophys Acta 1854, 1567–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD (2008). Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell 15, 578–589. [DOI] [PubMed] [Google Scholar]
- Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ (2019). ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 218, 1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Lemmon MA (2001). All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem 276, 44179–44184. [DOI] [PubMed] [Google Scholar]
- Zhu L, Jorgensen JR, Li M, Chuang YS, Emr SD (2017). ESCRTs function directly on the lysosome membrane to downregulate ubiquitinated lysosomal membrane proteins. Elife 6, e26403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.