Abstract
The notion that topologically associating domains (TADs) are highly conserved across species is prevalent in the field of 3D genomics. But what exactly is meant by ‘highly conserved’, and what are the actual comparative data that support this notion? To address these questions, we performed a historical review of the relevant literature, and retraced numerous citation chains to reveal the primary data that were used as the basis for the widely accepted conclusion that TADs are highly conserved across evolution. A thorough review of the available evidence suggests the answer may be more complex than what is commonly presented.
Keywords: TADs, Topologically associating domains, 3D genome architecture, Gene regulation, Evolution, Hi-C
What are TADs?
Some of the most fascinating features to have emerged from research into 3D genome conformation are topologically associating domains (TADs). Originally discovered through the analysis of Hi-C (see Glossary) and 5C data [1–4], TADs appear on a chromatin contact map as large squares of enhanced contact frequency rising off the diagonal. Early studies reported that TADs are non-overlapping, self-interacting megabase-scale structures. More recent studies, using higher-resolution contact maps, as well as different inference algorithms, have revealed TAD structures at much smaller scales, and often nested within each other [5,6]. The precise nature of these features is still a matter of debate, with various definitions of TADs shifting as new algorithms arise and fresh discoveries are made about the mechanisms behind TAD formation (e.g. loop extrusion and compartmentalization) [6–13]. Previous studies have found relatively low concordance of TADs, defined by different algorithms and across various resolutions and parameters, further impeding a robust definition of these structures [14–16]. While efforts have been made to functionally distinguish various TADs at different scales [8,17–19], most studies, especially those that rely solely on Hi-C data, do not typically make these distinctions.
Regardless of the challenge of defining TADs, accumulating evidence suggests that TADs and other 3D structures may play a role in genome organization and function [5,17,20–23]. Studies assessing the direct transcriptional effects of TADs have found mixed results, with some locus-specific work suggesting a strong impact of TAD disruption on gene expression [24–27], while other genome-wide results imply only mild effects on expression [28–31]. Despite some uncertainty about the magnitude of regulatory changes induced by TAD disruptions, multiple independent lines of evidence suggest TADs may be functionally relevant. Genes located within the same TAD can have strongly correlated expression patterns and are often coregulated during cell differentiation [2,10,32]. TAD boundaries are strongly correlated with replication-timing domain boundaries [33], and are enriched for insulator elements such as CCCTC-binding factor (CTCF) [1,6]. Disruptions in normative TAD structures have also been implicated in a number of human pathologies [34–36]. Many believe that TADs putatively represent insulated neighborhoods, constraining the possible set of interactions between cis regulatory elements (CREs) and target genes [20,23,37].
The notion that TADs are highly conserved across species and cell types is prevalent. Determining TAD variability across cell types is important for understanding the extent to which the 3D genome structure affects differential gene regulation during development, enabling the regulatory and functional novelty observed across different cell lineages. In turn, assessing TAD conservation across evolution could help reveal the regulatory loci and mechanisms responsible for speciation and adaptation.
In this Opinion article, we focus on conservation of TADs across species. We do not discuss further the issues related to similarities and differences in TADs across cell types and tissues, which have been previously discussed [20,38–43]. Many studies state that TADs are conserved across species, but it is difficult to trace the origin of this claim. We set out to thoroughly review the evidence for evolutionary TAD conservation and found that, in fact, only a few studies have collected relevant data that provide direct evidence to support this notion.
Conservation in Context
In order to evaluate the evidence for conservation of TADs, it is important to consider what is implied by conservation of genetic and epigenetic features. At the level of a single feature – a single locus for example – conservation is easily defined when the state of the feature is identical across species. However, when studies refer to genome-wide conservation, they typically do not use a specific standard. When studies refer to a general property (for example, chromatin accessibility) as conserved, it implies that this property evolved under natural selection to maintain similarity across species. However, few studies formally test this hypothesis. In fact, for most functional genomic traits that are comparatively studied, a null model of ‘no selection’ is yet to be formulated.
Without a formal test, what is typically meant when one concludes that a molecular trait is conserved? In most studies, conservation simply means ‘highly similar’ across species. While this is typically not a formal process, the degree of similarity, or variance, is evaluated and benchmarked based on other relevant comparisons. For example, if the level of observed variation in a trait is similar within and between species, the trait is typically deemed highly similar across species and therefore conserved. If variation between species is consistently low, regardless of the time, to the most recent common ancestors of the species, the trait is likely to be conserved. If different molecular features show a range of inter-species variability, the features with the lowest variance across species are assumed to be conserved. All of these examples point to ad-hoc definitions of conservation, but this does not mean that they are wrong.
Let us consider specific examples. Comparative studies have reported that genome-wide, the overlap of histone modification H3K4me3 locations in humans and chimpanzees is around 70% [44]. Remarkably, the genome-wide overlap of H3K4me3 locations in humans and mouse is also around 70% [45]. With these figures, not much can be said about the conservation of H3K4me3 locations in primates, but one can probably conclude with confidence that H3K4me3 locations are quite conserved between human and mouse. That said, the best genomic context for evaluating the degree of conservation in these comparisons may be other histone modifications, but even the minimal context provided here illustrates the importance of benchmarking similarity values in order to understand what they imply about conservation across species.
To date, comparative studies of chromatin conformation assessed TAD conservation based on overall similarity between species and the ad-hoc rationales discussed above, not a formal model of TAD evolution. With this in mind, we now turn to critically examine the existing evidence for evolutionary conservation of TADs.
Indirect evidence for conservation
The notion that TADs are highly conserved appears to be supported by a number of studies. One class of studies claiming TAD conservation, however, does not perform direct comparative assessment of TADs and boundaries across species. Instead, the indirect inference of TAD conservation is based on comparative functional genomic data that are independently associated with TADs.
One such example, which also happens to be one of the most commonly cited studies supporting TAD conservation, is Rudan et al. 2015 [46]. In this study, the authors collected comparative Hi-C data from liver cells of mouse, macaque, rabbit, and dog. They identified TADs in mouse and dog, but not in the other species. The authors’ conclusion of extensive conservation of chromosome structure was based on comparisons of placement and orientation of CTCF binding sites across species, as well as on inter-species comparison of inferred insulator activity at different distances from orthologous loci. Rudan et al. found that correlations of inferred insulator activity between mouse and every other species ranged from 0.34 to 0.61 (the authors did not report species pairwise comparisons that did not involve mouse). These correlation values may indicate some degree of conservation of 3D genome structure, but it is difficult to conclude from these analyses that TADs are indeed highly conserved across species. Moreover, Rudan et al.’s data collection was uneven across species, with ~275 million reads sequenced from mouse, ~150 million from rabbit, ~100 million from macaque, and ~550 million for dog. The large differences in read counts result in a difference in the power to infer insulator activity across species and hence complicate the interpretation of the reported results.
There are other widely cited studies which conclude that TADs are highly conserved based on indirect evidence. Harmston et al. 2017 [47], for instance, identified genomic regulatory blocks (GRBs, regions with a high density of conserved noncoding elements) in human, opossum, chicken, and spotted gar. They reported that GRBs are often quite conserved across species. Using previously collected Hi-C data from human and Drosophila, Harmston et al. have shown that GRBs often fall within TADs and/or have edges proximal to TAD boundaries in these two species. Based on these data, the authors concluded that TADs are generally conserved ancient features of the genome and that TAD boundaries are largely invariant between all the species in their study. However, the data reported by Harmston et al. shows that only about a third of the TADs were associated with GRBs; thus, even if one accepts the indirect inference based on GRBs as correct, up to two thirds of TADs may still not be conserved in these species, as no direct evidence for TAD conservation was presented in this study. Indeed, in their concluding statement, Harmston et al. are careful to note that only a subset of GRB-associated TADs appear to be ancient conserved structures. However, this paper is often cited as providing strong evidence for general TAD conservation across species.
Additional studies that are cited in support of TAD conservation, which did not perform direct comparisons of TADs across species, include Krefting et al. 2018 [48] and Lazar et al. 2018 [49]. Krefting et al. 2018 [48] considered TADs previously identified in humans [1,6] in the context of genomic rearrangement breakpoints identified in 13 species. They found enrichment for breakpoints at TAD boundaries and depletion within TAD bodies. Based on these observations, the authors made relatively strong claims about TAD stability across evolutionary timescales. Similarly, Lazar et al. 2018 [49] observed an enrichment for multiple species’ TAD boundaries at 67 genomic rearrangement breakpoints, identified between the human and gibbon genomes, ultimately concluding that TADs remain intact as functional units during evolution. Given that neither of these studies performed a direct comparison of TADs across species, it is difficult to conclude with confidence, based on their results, that TADs are highly conserved.
Direct but anecdotal evidence for conservation
The second class of studies that are widely cited as providing evidence for general TAD conservation considered only anecdotal evidence. These are studies that provide direct and strong evidence for conservation of TADs, but only in a small number of well-studied cases. It, thus, may be difficult to generalize from these studies and conclude with confidence that TADs are generally highly conserved across species.
Woltering et al. 2014 [50], for example, found that Hox loci across zebrafish and mouse tend to have similar TAD structure, and Gomez-Marin et al. 2015 found comparable TAD structures across a number of species at the Six loci [51]. Both of these studies, as well as a number of others [22,24,52], focus on loci that are highly conserved and/or thought to be critical for normal organismal development. Though these findings may underscore the functional importance of TADs, they do not provide strong enough evidence for broad and general TAD conservation. In particular, the focus on a subset of candidate loci that are more likely to contain conserved features may make it difficult to generalize these observations to a genome-wide scale.
Direct evidence for the conservation of TADs
A relatively small body of research studied TAD conservation by directly identifying TADs and boundaries in multiple species. Dixon et al. 2012 [1] collected Hi-C data and inferred TADs in human and mouse. This study was groundbreaking as it was one of the first to discover TADs and propose an algorithm to infer them from Hi-C contact maps (directionality index). This study is often cited as providing the first evidence that TADs are highly conserved between humans and mice. The authors collected 475 million sequencing reads from mouse Hi-C libraries but only 330 million reads from human. TAD boundaries were thought to be conserved if they had any overlap in the other species, with 76% of mouse boundaries found in humans but only 54% of human boundaries found in mice. If one considers the entire dataset of TAD boundaries, identified in both human and mouse (rather than the reported unilateral overlaps), ~31% of boundaries are shared between the two species.
These results were, and still are, interpreted as evidence for strong TAD conservation. To provide some context, we considered other functional annotations in human and mouse. There is about 60–75% overlap of loci marked by histone modification in humans and mouse [45], and between half to two-thirds of candidate regulatory regions are conserved in the two species [53]. Considering the observed proportion of overlapping TAD boundaries in human and mouse in this context, we believe that there is evidence for some level of conservation, but arguably, this cannot be considered strong enough evidence to support high conservation of TADs across species.
Another study that performed a direct comparative assessment of TADs is Rao et al. 2014 [6]. The authors collected ~6.5 billion Hi-C sequencing reads from human but only ~1.4 billion reads from mouse. The difference in read depth resulted in a striking difference in the power to infer TADs in the two species, with more than 9000 domains identified in human but only ~3000 domains found in mouse. The authors considered entire domains conserved if the center of a domain in one species was within 50 kb of an annotated domain in the other species (or within half the domain size, for domains smaller than 100 kb). Ultimately, Rao et al. 2014 reported that 45% of mouse domains (where they had considerably less power to identify TADs) were also present in human. Again, this points to some degree of conservation, but it is difficult to strongly conclude based on these data that TADs are highly conserved.
On the other hand…
There are a few studies which suggest that TADs may not be particularly conserved across species. Berthelot et al. 2015 [54] identified genomic rearrangement breakpoints in the genomes of human, mouse, dog, cow, horse, and a genomic reconstruction of the Boreoeutherian last common ancestor. The authors considered the overlap of rearrangement breakpoints with TADs that were previously identified in human [1]. Across several comparisons, the authors did not find evidence for a strong overlap of TAD boundaries and breakpoints, suggesting that TADs do not generally contribute to the locations of genomic rearrangements. Conversely, another similar study by Lazar et al. [49] observed strong overlap between TAD boundaries and genomic rearrangement breakpoints between the human and gibbon genomes. Both studies, however, used breakpoints rather than TADs as the basal set for comparisons, and hence represent indirect inferences addressing TAD conservation, making it difficult to generalize from their conclusions. Berthelot et al. [54] is nonetheless notable for interpreting the results of Dixon et al.’s [1] study as providing evidence for some TAD divergence between humans and mice. That the results of Dixon et al. 2012 can be interpreted by different groups, both as supporting conservation of TADs or lack thereof, highlights our notion that the foundation for the claim that TADs are highly conserved is not that strong.
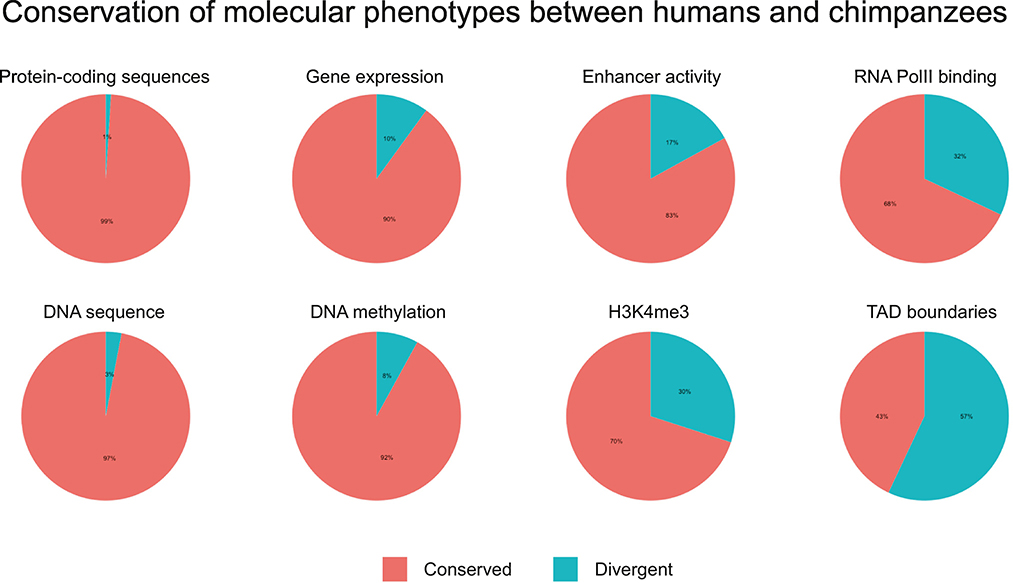
The notion that TADs may not be particularly conserved is also supported by another study, which directly inferred TADs in humans and chimpanzees [55]. An initial analysis found only ~43% of TADs conserved between these species, but across many different parameters (e.g. resolution, window size, genome assembly), and different downstream analysis decisions, no more than 78% of domains and 83% of TAD boundaries were found to be shared between humans and chimpanzees—a much lower percentage than what has been seen across these species for a number of other functional regulatory phenotypes (Figure 1). As the study noted, visual inspection of contact maps at algorithmically inferred divergent TADs sometimes suggested conservation, further highlighting the broader issue of inaccurate TAD inference.
Figure 1.
Conservation and divergence of different molecular phenotypes between human and chimpanzee. The proportion of features classified as conserved or divergent for quantitative traits such as gene expression and chromatin modification will depend on the power of the study (mostly based on the number of individuals sampled from each species). The data presented in this figure is based on studies that sampled similar number of individuals regardless of the phenotype studied (between 4–10 individuals from each species).
Other findings, particularly in plants, also suggest that TAD positions may not be conserved across species. Dong et al 2017 [56], for instance, collected Hi-C data from maize, tomato, sorghum, foxtail millet, and rice, and found relatively little conservation of TADs across these species. Xie et al. 2019 [21] assessed TAD conservation in two different mustard plants, Brassica rapa and Brassica oleracea, and reported that about 25% of all TADs are found in both species.
It should be noted that the existence of TADs in plants, worms, yeast, and other non-mammalian species is a matter of active debate [42]. While chromatin conformation capture experiments have revealed self-interacting TAD-like structures in many of these species, their characteristics and mechanisms of formation often differ substantially from those of mammalian TADs [19,57]. In many cases, these species lack homologs for insulator proteins thought to be essential for the formation of mammalian TADs (e.g. CTCF) [19]. Such differences may make it difficult to view inferences from these other species as compelling evidence against strong TAD conservation. More samples and deeply sequenced Hi-C libraries from these species, as well as a greater understanding of possible mechanisms of TAD-like feature formation, will be necessary to thoroughly assess conservation of TAD structures across all of evolution.
Concluding remarks
It is important to note that we are not taking a strong position ‘for’ or ‘against’ the notion of TAD conservation. Based on available evidence, we argue that there is currently no satisfying answer to the question of the degree of TAD conservation. While the results from certain studies suggest some degree of conservation, others often lead to much lower estimates. This, combined with ineffective study designs and variable analytical choices, further obscure the issue. Although many studies state that TADs are conserved across species, there are only sparse data supporting or refuting this claim. In our mind, there is no strong basis for the common and often unchallenged notion that TADs are highly conserved.
One of the largest factors affecting our ability to assess evolutionary TAD conservation is the lack of a gold standard for inferring TADs or for comparing them across species. As others have noted, TADs are variously and poorly defined, and it seems likely that stable TADs observed in Hi-C data represent statistical features that emerge from averaging more dynamic interactions across millions of cells [58] (see Outstanding Questions). The few studies that did directly compare TADs across species made somewhat arbitrary choices about how to call these features conserved. Until we arrive at a more precise definition of TADs that converges on robust metrics to infer them and assess their conservation, future studies hoping to assess 3D genome conservation across species should attempt to use a wide variety of TAD algorithms and parameters, as well as new interspecies Hi-C analytical methods to robustly assess 3D genome conservation [59,60]. TADs represent one intriguing feature of 3D genome architecture, with evolutionary conservation of other features (e.g. regulatory loops) being even less clear. In order to understand the overall regulatory dynamics, we must refine our understanding of TADs, and agree on how to infer and compare them across species and cell types.
Outstanding Questions.
What is the “ground truth” when it comes to TAD inference? Numerous inference algorithms have been proposed and applied, but which should be used widely, and why?
Are TADs identified at different scales (megabase, sub-megabase, etc.) representative of similar structures formed by similar processes, or should they be considered different features entirely? Can the field arrive at a consensus definition for TADs, mathematical and/or biological?
Are TADs defined based on bulk Hi-C data biologically meaningful to begin with? How might inferences made from millions of cells, be deconvoluted and reconciled with emerging evidence from single cells that TADs are dynamic structures?
What is the order of events with respect to 3D genome structure and transcription? Does the formation of TADs dictate possible sets of gene regulatory interactions, or do independent regulatory interactions themselves give rise to TAD structures? If both are the case, can any predictive distinction be drawn between TADs that dictate transcriptional activity vs. TADs that emerge as a result of transcriptional activity?
Are TADs largely shared across cell types within an organism, and does the extent of tissue sharing stratify by any particular facet of TADs? To what extent are differences in TADs across cell types also conserved across species?
What is the nature of TAD-like structures observed in non-mammalian species? How do these structures arise and what is their functional significance, particularly for species that lack homologs for insulator proteins implicated in TAD formation in mammals?
In the absence of a new unifying definition and inference method for TADs, what is the best method to compare them across development and evolution? For interspecies comparisons, how might we best model the process of TAD evolution in order to obtain a null model for neutrality against which to test conservation?
Highlights.
Topologically associating domains (TADs) are highly self-interacting regions in the genome, identified through analysis of chromosome conformation capture data.
TADs are often thought of as stable neighborhoods of gene regulation, constraining possible interactions between cis-regulatory elements and their target genes.
TADs are poorly defined from a biological perspective and have low concordance across different inference algorithms.
To date, comparative studies of TAD structures did not properly account for study design, technical, and analytical issues, but often concluded TADs are highly conserved across species.
A careful examination of the interspecies comparative data, available, to assess TAD conservation suggests that, while they certainly have some functional conservation, specific TAD structures and locations may not be especially conserved across evolutionary lineages.
Acknowledgments
We apologize to the authors of relevant studies whose work was not addressed due to space limitations. We thank Natalia Gonzales, Jasmin Zohren, Sergey Kolchenko, Daniel Ibrahim, and Carlos Bustamante, for useful discussions, comments, and/or edits to the manuscript. YG is supported by NIH grant R35GM131726. IEE is supported by the Genetics and Gene Regulation Training Grant (T32 GM07197). The authors have no conflicts of interest to declare.
Glossary
- 5C
Chromosome conformation capture carbon copy, an experimental technique to assay contact probability between pairs of loci within a given genomic region (typically a megabase or less)
- Cis regulatory elements (CREs)
DNA sequences, typically non-coding, whose role is to affect the level of gene expression. CREs include enhancers, promoters, silencers, and insulators. CREs often contain transcription factor binding sites
- Compartmentalization
Segmentation of chromatin into megabase-scale compartments with different transcriptional and epigenetic properties: “A” compartments with actively transcribed genes and active histone marks, and “B” compartments with inactive genes and repressive histone marks. Sometimes further sub-divided into smaller compartments with various combinations of active and repressive epigenetic properties
- CCCTC-Binding Factor (CTCF)
Transcription factor involved in 3D genome architecture, thought to act as an insulator reducing physical interactions between sequences on either side of it. CTCF binding sites are frequently found in a convergent orientation at the boundaries of a TAD, and putatively halt the action of loop extrusion
- Genomic rearrangement breakpoints
Genomic locations representing a break in synteny blocks across genomes of different species; orthologous locations not consecutive or not oriented similarly across species
- Hi-C
High-throughput chromosome conformation capture, an experimental technique to assay contact probabilities between pairs of loci genome-wide
- Loop extrusion
Putative mechanism driving chromatin loop formation, whereby factors such as cohesin extrude a DNA loop until stalling at a TAD boundary, often due to interactions with proteins such as CTCF
- Regulatory loops
Specific chromatin loops that enable interactions between cis regulatory elements and promoters, often thought to impact regulation of gene expression
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485: 376–380. doi: 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nora EPP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485: 381–5. doi: 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48: 471–84. doi: 10.1016/j.molcel.2012.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. Three-dimensional folding and functional organization principles of the drosophila genome. Cell. 2012;148: 458–472. doi: 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 5.Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 2013;9: e1004018. doi: 10.1371/journal.pgen.1004018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao S, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159: 1665–1680. doi: 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippova D, Patro R, Duggal G, Kingsford C. Identification of alternative topological domains in chromatin. Algorithms Mol Biol. 2014;9: 14. doi: 10.1186/1748-7188-9-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon JR, Gorkin DU, Ren B. Chromatin domains: The unit of chromosome organization. Mol Cell. 2016;62: 668–680. doi: 10.1016/j.molcel.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153: 1281–95. doi: 10.1016/j.cell.2013.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan Y, Mariani L, Barozzi I, Schulz EG, Blüthgen N, Stadler M, et al. Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res. 2017;27: 479–490. doi: 10.1101/gr.212803.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci. 2018;115: 201717730. doi: 10.1073/pnas.1717730115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15: 2038–49. doi: 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci. 2015;112: E6456–E6465. doi: 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dali R, Blanchette M. A critical assessment of topologically associating domain prediction tools. Nucleic Acids Res. 2017;45: 2994–3005. doi: 10.1093/nar/gkx145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forcato M, Nicoletti C, Pal K, Livi CM, Ferrari F, Bicciato S. Comparison of computational methods for Hi-C data analysis. Nat Methods. 2017. doi: 10.1038/nmeth.4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zufferey M, Tavernari D, Oricchio E, Ciriello G. Comparison of computational methods for the identification of topologically associating domains. Genome Biol. 2018;19: 217. doi: 10.1186/s13059-018-1596-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beagan JA, Phillips-Cremins JE. On the existence and functionality of topologically associating domains. Nat Genet. 2020;52: 8–16. doi: 10.1038/s41588-019-0561-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sikorska N, Sexton T. Defining functionally relevant spatial chromatin domains: it’s a TAD complicated. J Mol Biol. 2019;432: 653–664. doi: 10.1016/j.jmb.2019.12.006 [DOI] [PubMed] [Google Scholar]
- 19.Szabo Q, Bantignies F, Cavalli G. Principles of genome folding into topologically associating domains. Sci Adv. 2019;5: eaaw1668. doi: 10.1126/sciadv.aaw1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrey G, Mundlos S. The three-dimensional genome: regulating gene expression during pluripotency and development. Development. 2017;144: 3646–3658. doi: 10.1242/dev.148304 [DOI] [PubMed] [Google Scholar]
- 21.Xie T, Zhang F-G, Zhang H-Y, Wang X-T, Hu J-H, Wu X-M. Biased gene retention during diploidization in Brassica linked to three-dimensional genome organization. Nat Plants. 2019;5: 822–832. doi: 10.1038/s41477-019-0479-8 [DOI] [PubMed] [Google Scholar]
- 22.Smith EM, Lajoie BR, Jain G, Dekker J. Invariant TAD boundaries constrain cell-type-specific looping interactions between promoters and distal elements around the CFTR locus. Am J Hum Genet. 2016;98: 185–201. doi: 10.1016/j.ajhg.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, Schwarzer W, et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24: 390–400. doi: 10.1101/gr.163519.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161: 1012–25. doi: 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hnisz D, Weintraub AS, Day DS, Valton A-L, Bak RO, Li CH, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351: 1454–1458. doi: 10.1126/science.aad9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, et al. CRISPR inversion of CTCF sites alters genome topology and Enhancer/Promoter function. Cell. 2015;162: 900–10. doi: 10.1016/j.cell.2015.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laugsch M, Bartusel M, Rehimi R, Alirzayeva H, Karaolidou A, Crispatzu G, et al. Modeling the pathological long-range regulatory effects of human structural variation with patient-specific hiPSCs. Cell Stem Cell. 2019;24: 736–752.e12. doi: 10.1016/j.stem.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 28.Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet. 2019; 1–11. doi: 10.1038/s41588-019-0462-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuin J, Dixon JR, Reijden MIJA, van der, Ye Z, Kolovos P, Brouwer RWW, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci. 2014;111: 996–1001. doi: 10.1073/pnas.1317788111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, et al. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol Cell. 2019;76: 473–484.e7. doi: 10.1016/j.molcel.2019.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao SSP, Huang S-C, Hilaire BGS, Engreitz JM, Perez EM, Kieffer-Kwon K-R, et al. Cohesin loss eliminates all loop domains. Cell. 2017;171: 305–320.e24. doi: 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramírez F, Bhardwaj V, Arrigoni L, Lam K, Grüning BA, Villaveces J, et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9: 189. doi: 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515: 402–5. doi: 10.1038/nature13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lupiáñez DG, Spielmann M, Mundlos S. Breaking TADs: How alterations of chromatin domains result in disease. Trends Genet. 2016;32: 225–237. doi: 10.1016/j.tig.2016.01.003 [DOI] [PubMed] [Google Scholar]
- 35.Ibn-Salem J, Köhler S, Love MI, Chung H-R, Huang N, Hurles ME, et al. Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome Biol. 2014;15: 423. doi: 10.1186/s13059-014-0423-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, Schöpflin R, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538: 265–269. doi: 10.1038/nature19800 [DOI] [PubMed] [Google Scholar]
- 37.Sexton T, Cavalli G. The role of chromosome domains in shaping the functional genome. Cell. 2015;160: 1049–1059. doi: 10.1016/j.cell.2015.02.040 [DOI] [PubMed] [Google Scholar]
- 38.Sauerwald N, Singhal A, Kingsford C. Analysis of the structural variability of topologically associated domains as revealed by Hi-C. NAR Genomics Bioinforma. 2019;2. doi: 10.1093/nargab/lqz008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauerwald N, Kingsford C. Quantifying the similarity of topological domains across normal and cancer human cell types. Bioinformatics. 2018;34: i475–i483. doi: 10.1093/bioinformatics/bty265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17: 2042–2059. doi: 10.1016/j.celrep.2016.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng H, Xie W. The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol. 2019;20: 535–550. doi: 10.1038/s41580-019-0132-4 [DOI] [PubMed] [Google Scholar]
- 42.Bonev B, Genetics C-G. Organization and function of the 3 D genome. 2016. doi: 10.1038/nrg.2016.112 [DOI] [Google Scholar]
- 43.Cubeñas-Potts C, Corces VG. Topologically associating domains: An invariant framework or a dynamic scaffold? Nucl Austin Tex. 2015;6: 430–4. doi: 10.1080/19491034.2015.1096467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cain CE, Blekhman R, Marioni JC, Gilad Y. Gene expression differences among primates are associated with changes in a histone epigenetic modification. Genetics. 2011;187: 1225–34. doi: 10.1534/genetics.110.126177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woo YH, Li W-H. Evolutionary conservation of histone modifications in mammals. Mol Biol Evol. 2012;29: 1757–67. doi: 10.1093/molbev/mss022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudan MV, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10: 1297–309. doi: 10.1016/j.celrep.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harmston N, Ing-Simmons E, Tan G, Perry M, Merkenschlager M, Lenhard B. Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat Commun. 2017;8: 441. doi: 10.1038/s41467-017-00524-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krefting J, Andrade-Navarro MA, Ibn-Salem J. Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol. 2018;16: 87. doi: 10.1186/s12915-018-0556-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazar NH, Nevonen KA, O’Connell B, McCann C, O’Neill RJ, Green RE, et al. Epigenetic maintenance of topological domains in the highly rearranged gibbon genome. Genome Res. 2018;28: 983–997. doi: 10.1101/gr.233874.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woltering JM, Noordermeer D, Leleu M, Duboule D. Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits. PLoS Biol. 2014;12: e1001773. doi: 10.1371/journal.pbio.1001773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gómez-Marín C, Tena JJ, Acemel RD, López-Mayorga M, Naranjo S, de la Calle-Mustienes E, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci. 2015;112: 7542–7547. doi: 10.1073/pnas.1505463112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galupa R, Nora EP, Worsley-Hunt R, Picard C, Gard C, van Bemmel JG, et al. A conserved noncoding locus regulates random monoallelic xist expression across a topological boundary. Mol Cell. 2020;77: 352–367.e8. doi: 10.1016/j.molcel.2019.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515: 355–64. doi: 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berthelot C, Muffato M, Abecassis J, Roest Crollius H. The 3D organization of chromatin explains evolutionary fragile genomic regions. Cell Rep. 2015;10: 1913–1924. doi: 10.1016/j.celrep.2015.02.046 [DOI] [PubMed] [Google Scholar]
- 55.Eres IE, Luo K, Hsiao CJ, Blake LE, Gilad Y. Reorganization of 3D genome structure may contribute to gene regulatory evolution in primates. PLOS Genet. 2019;15: e1008278. doi: 10.1371/journal.pgen.1008278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong P, Tu X, Chu P-Y, Lü P, Zhu N, Grierson D, et al. 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol Plant. 2017;10: 1497–1509. doi: 10.1016/j.molp.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 57.Acemel RD, Maeso I, Gómez-Skarmeta JL. Topologically associated domains: a successful scaffold for the evolution of gene regulation in animals. Wiley Interdiscip Rev Dev Biol. 2017;6: e265. doi: 10.1002/wdev.265 [DOI] [PubMed] [Google Scholar]
- 58.de Wit E. TADs as the caller calls them. J Mol Biol. 2019;432: 638–642. doi: 10.1016/j.jmb.2019.09.026 [DOI] [PubMed] [Google Scholar]
- 59.Yang Y, Zhang Y, Ren B, Dixon JR, Ma J. Comparing 3D genome organization in multiple species using phylo-hmrf. Cell Syst. 2019;8: 494–505.e14. doi: 10.1016/j.cels.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nuriddinov M, Fishman V. C-InterSecture—a computational tool for interspecies comparison of genome architecture. Bioinformatics. 2019;35: 4912–4921. doi: 10.1093/bioinformatics/btz415 [DOI] [PubMed] [Google Scholar]