Graphical abstract
Keywords: Protein, Molecular dynamics, Amber, Charmm, Simulation, Protein dynamics, Peptide, Aggregation, Solubility, Molecular simulations, Water simulation, Force fields
Abstract
Continuous assessment of transferable forcefields for molecular simulations is essential to identify their weaknesses and direct improvement efforts. The latest efforts focused on better describing disordered proteins while retaining proper description of folded domains, important because forcefields of the previous generations produce overly compact disordered states. Such improvements should additionally alleviate the related problem of over-stabilized protein–protein interactions, which has been largely overlooked. Here we evaluated three state-of-the-art forcefields, current flagships of their respective developers, optimized for ordered and disordered proteins: CHARMM36m with its recommended corrected TIP3P* water, ff19SB with the recommended OPC water, and the 2019 a99SBdisp forcefield by D. E. Shaw Research with its modified TIP4P water; plus ff14SB with TIP3P as an example of the former generation of forcefields. Our evaluation entailed simulations of (i) multiple copies of a protein that is highly soluble yet undergoes weak dimerization, (ii) a disordered peptide with low, well-characterized alpha helical propensity, and (iii) a peptide known to form insoluble β-aggregates. Our results recapitulate ff14SB-TIP3P over-stabilizing aggregates and secondary structures and place a99SBdisp-TIP4PD at the other end i.e. predicting overly weak intermolecular interactions despite reasonably predicting secondary structure propensities. In-between, CHARMM36m-TIP3P* still over-stabilizes aggregates but predicts residue-wise alpha helical propensities in solution slightly better than ff19SB-OPC, while ff19SB-OPC poses the best prediction of weak dimerization of the soluble protein still predicting aggregation of the β-peptides. This independent assessment shows that the claimed forcefield improvements are real, but also that a right balance between noncovalent attraction and repulsion has not yet been reached. We thus propose developers to consider systems like those tested here in their forcefield tuning protocols. Last, the good performance of CHARMM36m-TIP3P* further shows that tuning 3-point water models might still be an alternative to the more costly 4-point models like OPC and TIP4PD.
1. Introduction
Molecular dynamics (MD) simulations using atomistic transferable forcefields are increasingly gaining practical utility in structural biology, assisting studies in drug discovery [1], [2], protein folding [3], [4], [5], protein structure prediction and refinement [3], [6], [7], intrinsically disordered peptides [8], unspecific interactions induced at high concentrations [9], [10], [11], [12], specific interactions between biomacromolecules [13], [14], [15] and with membranes [13], [14], [15], among some of the main applications. This has been possible thanks to the continuous efforts dedicated to benchmark and improve forcefields for molecular simulations and thanks to the improvements in commonplace hardware and molecular dynamics software, which enable today access to the multi-microsecond timescale through plain unbiased MD simulations in a few weeks or months of computing time for systems containing tens to low-hundred thousand atoms, and events occurring on even longer timescales by applying enhanced sampling methods. The role of continuously benchmarking simulations is of utmost importance, especially as the access to longer simulation timescales for larger systems unveils problems that remain undetected in shorter simulations and on smaller systems. One such important problem was realized in the last half decade: while the dynamic properties of single well-folded proteins simulated alone in solution were quite accurately reproduced by most forcefields, systems containing multiple copies of the same well-folded protein turned out to aggregation despite a real solution of the protein being perfectly stable. Moreover, these forcefields could not accurately describe the dynamical properties of disordered proteins, making them substantially more compact than what experimental measurements reported [16], [17], [18], [19], [20], [21]. The two problems seemed to be related to each other, most likely arising from problems in the balance of electrostatic forces and hydrophobic effects mediated by water properties which when balanced improve the resulting description. In particular, protein-water interactions were found to be too weak, thus favoring noncovalent protein–protein interactions leading to overstabilization of protein–protein complexes, protein aggregates, and disordered proteins.
In the last decade forcefield developers focused in improving the simulation of disordered peptides and proteins, first aiming only specifically at disordered systems [22], [23], [24], [25] and then, especially in the last 5 years, attempting to retain accuracy for folded proteins as well [26], [27], [28]. Recent reviews recap the strategies used by developers to improve simulations of disordered proteins and peptides [29], [30], [31]. Briefly, the main strategies adopted by forcefield developers involved fine-tuning the parameters for dihedral and non-covalent terms, adding specific corrections to backbone dihedrals to improve secondary structures attempting to reproduce disorder yet leaving place for secondary structure propensities, and adjusting water models to strengthen protein-water interactions. Dihedral angle corrections through grid-based energy additions from dihedral angle statistics (CMAP), and development of better water models, were in fact of key relevance. CMAP corrections were first introduced to CHARMM22 and turned out to improve description of disorder and secondary structure propensities, and to better capture the cooperativity of helix and hairpin formation yet retaining CHARMM22′s accuracy on folded proteins. Since then, similar approaches were adopted for the development of forcefields from CHARMM and also some Amber subfamilies, as summarized by Mu et al. [29]. While these dihedral corrections allowed mainly to improve the balance between secondary structures and disorder, the development of better water models that strengthen protein-water interactions was key for simulations to produce less compact ensembles of disordered proteins, more compatible with experimentally determined sizes. Water models most often used when tuning forcefields of the previous generations, mainly the 3-point TIP3P and SPC models, are well known from work in the previous decade to interact too softly with proteins, leading to the observed collapse of disordered ensembles and overly strong protein–protein interactions [20], [32].
Multiple studies have assessed the various forcefields from different generations upon their capacity to reproduce the behavior of disordered systems not used for forcefield tuning [17], [19], [21], [33], [34], [35], but studies on their ability to properly reproduce solubility and insolubility in the form of aggregation, fibrillation, etc. are scarce. Although the problem of aggregating proteins has not been explicitly tackled, one would expect it to be at least partially alleviated if it were truly of the same origin as the mainly water-blamed over-compaction of disordered states. This idea was suggested by some works and reviews but to date only limitedly tested [16.18,20], and totally overlooked by forcefield developers -perfectly reasonable because simulations of multiple proteins entail very large systems. Here we look into this problem by simulating systems that provide strong yes/no cues: protein solutions that should remain soluble or fibrillate -complemented by other subtler analyses.
Among the very large set of forcefields resulting from the combination of different optimization strategies and water models, in this work we have focused on three mainstream, transferable, nonpolarizable forcefields provided by the main forcefield developers as their “official” versions targeted to both disordered and folded proteins [26], [27], [28]. We tested these three forcefields for their capacity to keep a protein of high solubility in a soluble form, and an insoluble amyloid-prone peptide insoluble, while also keeping an eye on their capacity to properly describe disorder with secondary structure propensity. Specifically, we tested the latest official forcefields from the Amber, CHARMM and D. E. Shaw Research developers, each with the water model recommended and optimized to reproduce the structural dynamics of folded proteins, intrinsically disordered proteins, and folded proteins containing disordered regions. These forcefields are CHARMM36m with the recommended modified TIP3P (dubbed CHARMM36m-TIP3P* throughout the article) [26], amber99SB-disp with the recommended slightly modified version of TIP4P(D) (a99SBdisp-TIP4PD) [28], and ff19SB of the Amber family with the recommended OPC water model (ff19SB-OPC) [27]. We also tested ff14SB from the Amber family with the standard TIP3P water model (ff14SB-TIP3P) [36] as a representative of the previous generation of forcefields before the introduction of corrections tailored to better simulate disordered states. As far as we are aware of, the modern forcefields tested are the current “official” versions available from their developers as of August 2020, except for a further release from the D. E. Shaw group in 2020 that we could not accommodate to run but which we briefly mention in the discussion given its relevance to this work -as it was specifically devised for protein–protein interactions [37].
On a brief description of the four force fields tested in this study, ff14SB was rebuilt from ff99SB with a new fit of all backbone and side chain dihedral parameters to improve the long-known deficiencies and biases of previous Amber force fields on secondary structures [36]. It is intended for use with standard TIP3P water and its parametrization was not much concerned with disordered proteins. With yet a new recalibration of backbone dihedrals and CMAP corrections plus additional adjustments, the most modern official Amber forcefield, ff19SB, performs better than ff14SB on disordered proteins when accompanied with the 4-point OPC water model. Much newer than the more established, computationally cheaper but arguably less accurate TIP3P water, the OPC model describes liquid water around room temperatures closer to experiment, and was shown to improve the description of some intrinsically disordered proteins even with older-generation forcefields like ff99SB [38]. The D. E. Shaw Research forcefield for folded and disordered proteins tested here was developed by tuning the torsional and van der Waals terms from amber99SB-ILDN and found to optimally describe disordered proteins if used together with a version of the 4-point TIP4P-D water model modified to slightly strengthen dispersive forces [28]. Coming from a separate family, CHARMM36m benefits from refined CMAPs relative to its predecessor to improve backbone dihedral dynamics and especially to remove its bias towards left-handed helices; plus improved salt bridge description between the guanidinium group of arginine and the different carboxylate groups. Moreover, its developers adjusted Lennard-Jones parameters of TIP3P water to strengthen protein-water interactions, and recommend this model for simulations with CHARMM36m as it produces less compact disordered states that better much the experimentally determined gyration radii [26].
We challenged the four forcefields against (i) solutions of multiple ubiquitin molecules that should remain soluble yet experience certain intermolecular contacts favored through one specific surface, (ii) an intrinsically disordered peptide of high solubility and slight helical propensity derived from huntingtin’s N-terminus (dubbed Htt-1-19) that was not employed for calibration of any of the tested forcefields, (iii) multiple copies of a peptide corresponding to residues 16–22 of Alzheimer-related amyloid-β peptide, highly insoluble and known to form β-amyloid fibers but setup as if in solution, and (iv) a portion of a β-fiber from the same protein (residues 16–40) of structure solved by X-ray diffraction. Whereas evaluation of disorder and secondary structure propensities in Htt-1-19 involves quantitative comparisons between global and residue-specific secondary structures observed in solution-state experiments and in the simulations, much like in other works tuning and evaluating forcefields, assessment of solubility was approached here through strong soluble/insoluble cues by working on extreme cases of (in)solubility. In our study, such extreme cases are represented by ubiquitin in the high-solubility end, and the amyloid-forming peptides in the other end.
Ubiquitin is a highly soluble protein, stable at millimolar concentrations for years at neutral and even slightly acidic pH [39] – it is in fact used as a standard for protein nuclear magnetic resonance (NMR). At such concentrations the average spacing between protein molecules is in the order of the size of the molecule itself; for example, at 10 mM the average surface-to-surface separation in a group of ubiquitin molecules would be close to the protein’s hydrodynamic radius of 11 Å. As a first approximation, then, it would be very likely that the protein molecules collide with each other often as they diffuse, giving place to transient interactions that distribute smoothly over the protein surface. Detailed NMR studies [40] have further revealed that at high concentrations ubiquitin engages dynamic interactions through a specific surface, an interaction limited to formation of dimers with a Kd of around 4.4 mM. Of course this dimerization is a highly dynamic equilibrium, i.e. the protein remains overall soluble. Moreover, the fact that homodimerization takes place faster than the chemical shift timescale indicates that the on and off rates are in the order of 106 s−1 or faster; thus, the transient binding and unbinding events that underly weak homodimerization occur multiple times per microsecond, a timescale accessible to modern atomistic simulations. In a previous work [16] we showed that forcefields of the previous amber99 generations could just not keep ubiquitin soluble at neutral pH, let alone predict reversible homodimerization: when 3 copies of ubiquitin were placed together and well-separated inside a box of TIP3P water at a concentration of around 5 mM, the molecules consistently aggregated with a few hundreds of nanoseconds leading to the incorrect prediction that such solution would not exist; moreover, the “aggregate” of protein molecules got increasingly more compact over time. Similar studies on other proteins of known high solubility have revealed the same problems, also for other forcefields contemporary of the amber99 family [18].
On the other end of the solubility spectrum, peptides 16–22 and 16–40 of the amyloid-β peptide are totally insoluble in water. They are so insoluble, that for experimentation they must be dissolved and handled in aqueous mixtures containing high mole fractions of acetonitrile, trifluoroacetic acids or other low-polarity solvents [41]. And even in such conditions, after a certain lag phase the peptides aggregate forming insoluble β-amyloid fibers. Simulations of amyloid aggregation starting from multiple copies of short peptides scattered inside a water box have been performed for years. Most such simulations show aggregation within timescales of tens to hundreds of nanoseconds, often producing β-rich structures as expected. However, the fact that forcefields tend to overstabilize interactions questions the validity of the observed aggregation pathways, timescales and structures. In fact, a recent study comparing several forcefields upon their capacity to predict aggregation of Aβ-16-22 showed quite different pathways, secondary structure contents and compactness [42]; and we are not aware of any simulations leading to perfect β-fibrils: in the best cases, simulations produce β-rich aggregates that look amorphous rather than as elongated fibrils -possibly a consequence of overly stabilizing interactions, but also possibly a realistic consequence of the high concentrations used in such simulations, as discussed by Strodel among other challenges for the simulation of amyloid aggregation processes [43].
We develop two more points before moving to the results section. First, we would like to distinguish among a set of related words and smoothen their meanings as used in theory/simulation and in experiments. Throughout this text, solubility and insolubility refer to the formation of a single homogenous phase as opposed to segregation into distinct phases. We call “aggregation” to the process by which a solution becomes two or more distinct phases, one of them much richer in protein, i.e. implying that the solute is insoluble, even if it may have been initially set as “soluble” upon preparation for MD simulation. On the contrary, dynamic interaction and dimerization imply interactions between defined numbers of molecules that are reversible in the timescale of an experiment or simulation, during which the system remains as a single phase i.e. soluble. “Aggregation” as used here is in principle irreversible, and encompasses processes like protein precipitation, crystallization, and fiber formation.
Second, we stress that the overall message of this work is neither on protein aggregation, nor on fiber formation or solubility calculations, but rather about forcefields overstabilizing interactions. To this aim we use the strong clear-cut cases of highly soluble vs. highly insoluble proteins/peptides to test the forcefields on a simple-to-judge basis. Besides, we compare the forcefields in their ability to reproduce dynamics of one peptide recently studied by us and others through NMR spectroscopies, not used in the tuning of any of the tested forcefields.
2. Results
We ran all the systems described throughout the text and summarized in Table 1 adding up to a total of around 28 µs for each force field: single 10 µs simulations for Htt-1-19 peptides, whose fast dynamics should be well covered in that timescale; three 2 µs replicas for systems containing 3 ubiquitin molecules, as this was enough to see strong aggregation in the cases where this happened; single 1 µs simulations for systems with 9 ubiquitin molecules as this was just to complement the 3-ubiquitin systems; single 2 µs simulations for 10 Aβ peptides as this was enough to see aggregation and where anyway the lack of replicas was compensated by the number of molecules; and three ~3 µs replicas started from an Aβ fiber. Table 1 provides more details on the systems, together with expected behaviors based on experimental knowledge of the systems and how they compare to what is observed in the simulations. We provide the coordinates of all simulated systems in the SI so that new forcefield developments can be tested directly.
Table 1.
Description of the systems, parameterization and trajectory lengths simulated in this work, together with a brief description of the key observations compared to the behavior expected based on experimental knowledge of the different systems.
| System | Parameterization(protein-water) | Replicas × length | Expected behavior (italics) vs. observations in the simulations |
|---|---|---|---|
| Single Htt-1-19 peptide in 100 mM KCl at neutral pH | Soluble and disordered but with ~10–30% helical propensity according to circular dichroism spectra that peaks at residues 14–18[45]. | ||
| ff14SB-TIP3P | 1 × ~ 10 µs | Helix propensity too high (45%), highest for residues 3–12. | |
| CHARMM36m-TIP3P* | 1 × ~ 10 µs | 22% helix propensity, highest for residues 9–15. | |
| a99SBdisp-TIP4PD | 1 × ~ 10 µs | 28% helix propensity, highest for residues 3–14. | |
| ff19SB-OPC | 1 × ~ 10 µs | 29% helix propensity, highest for residues 4–11. | |
| 3 ubiquitin molecules around 5 mM, in 100 mM KCl at neutral pH | Soluble but undergoing weak dimerization through a favored interface (residues 4–12, 42–51 and 62–71), exchanging in the microsecond timescale[40]. | ||
| ff14SB-TIP3P | 3 × ~ 1.7 µs | Aggregation into dimers and then trimers that get increasingly more compact. | |
| CHARMM36m-TIP3P* | 3 × ~ 2 µs | Aggregation into dimers and then trimers but less compact than with ff14SB-TIP3P. | |
| a99SBdisp-TIP4PD | 3 × ~ 1.9 µs | Soluble with a few short-lived encounters that do not match the NMR data; no trimers. | |
| ff19SB-OPC | 3 × ~ 2.2 us | Dimers lasting for tens to few hundreds nanoseconds, contacts most consistent with NMR data yet not perfect; no trimers. | |
| 9 ubiquitin molecules around 5 mM, in 100 mM KCl at neutral pH | Soluble but undergoing weak dimerization through a favored interface (residues 4–12, 42–51 and 62–71), exchanging in the microsecond timescale[40]. | ||
| ff14SB-TIP3P | 1 × ~ 0.95 µs | Aggregates form and grow bigger and more compact. | |
| CHARMM36m-TIP3P* | 1 × ~ 0.88 µs | Aggregates form and grow bigger, but less compact than ff14SB-TIP3P. | |
| a99SBdisp-TIP4PD | 1 × ~ 1.1 µs | Highly soluble, only collisions through no preferred surface. | |
| ff19SB-OPC | 1 × ~ 1.1 us | Some dimers form that then grow into bigger aggregates, but slower than C36m. | |
| 10 Aβ 16–22 molecules at around 10 mM, in 50 mM KCl at neutral pH | Totally insoluble in water. Forms β-rich fibrils and amorphous aggregates depending on conditions and concentration[41]. | ||
| ff14SB-TIP3P | 1 × ~ 1.8 µs | Aggregate of large β content | |
| CHARMM36m-TIP3P* | 1 × ~ 1.8 µs | Aggregate of large β content | |
| a99SBdisp-TIP4PD | 1 × ~ 2 µs | Remains soluble, some reversible formation of different pairs of strands | |
| ff19SB-OPC | 1 × 1.95 us | Largely soluble, with small aggregates of β content | |
| Fiber stretch of 10 Aβ-40 strands from PDB 2LNQ, in 50 mM KCl at neutral pH | Totally insoluble fiber that does not dissociate in water[51]. | ||
| ff14SB-TIP3P | 3 × ~ 3 µs | Rigid, max Cα RMSD 5–6 Å | |
| CHARMM36m-TIP3P* | 3 × ~ 3 µs | Less rigid than ff14SB, max Cα RMSD 8–10 Å. | |
| a99SBdisp-TIP4PD | 3 × ~ 3 µs | Similar to CHARMM36m, max Cα RMSD 8–10 Å. | |
| ff19SB-OPC | 3 × ~ 3 us | As rigid as ff14SB-TIP3P, max Cα RMSD 5–6 Å | |
2.1. Testing solutions of multiple ubiquitin molecules
Akin to our previous work [16] but with state-of-the-art transferable protein forcefields as of 2019–2020, our first test involved two systems containing ~5–6 mM solutions of ubiquitin [40], more precisely 3 or 9 protein molecules in solvent boxes of size ~100 × 100 × 100 and ~135 × 135 × 135 Å3. At pH 7 ubiquitin bears no net charge (pI 6.8 from sequence and from experimental measurements) yet it is highly soluble up to 5–10 mM concentrations, and highly stable at mildly acidic to mildly basic pH. Although long-recognized as a monomeric protein, analytical ultracentrifugation and NMR experiments revealed that it undergoes dynamic dimerization with a weak dissociation constant of 4.4 mM [40]. Chemical shift mapping and paramagnetic relaxation enhancement NMR experiments indicate that this noncovalent dimerization takes place through an expansion of the slightly hydrophobic surface patch centered at Ile44, that ubiquitin uses to recognize many of its binding partners: residues 4–12, 42–51 and 62–71 (while crystallographic complexes of covalent ubiquitin dimers show that they are stabilized by noncovalent contacts through surfaces that involve mainly residues around the same surface patch: 8, 9, 24, 32–54, 59, 68 and 70–75). The NMR data also informs that the binding/unbinding equilibrium takes place in the fast regime of chemical shifts, i.e. faster than the microsecond timescale. This implies that microsecond-long simulations should display, on top of unspecific transient interactions and contacts produced by random collisions, a good number of binding and unbinding events favored through the residues identified by NMR to drive the weak noncovalent dimerization. In assessing our simulations, we therefore looked at two main cues: whether ubiquitin remains soluble monomeric, forms dimers, trimers, or higher-order aggregates when more molecules are available; and whether the interfaces that mediate any interactions are enriched in those residues identified by NMR or not. Of note, by working at concentrations slightly above the Kd and given the fast kinetics of the process, a perfect simulation should readily reveal dimers and even possibly experience some unbinding events where the dimers resolve into monomers; while trimers or any other bigger complex are not expected.
With each forcefield, we ran the 3-molecule systems in three independent replicas and the 9-molecule systems once. From a visual inspection of the trajectories with 3 protein molecules it is evident that they aggregate together strongly with ff14SB-TIP3P (example in Fig. 1A), less so with CHARMM36m-TIP3P*, and even less so or not at all with the other two modern forcefields. For a more quantitative assessment we computed the distances between the centers of mass of all three possible pairs of proteins over time (Fig. 1B). The numbers confirm faster protein–protein complexation happening with ff14SB-TIP3P than with CHARMM36m-TIP3P*, both showing quite strong binding to an extent that these simulations would predict ubiquitin to be insoluble. Meanwhile, the other two forcefields do not show aggregation of the three molecules, at least in the timescale simulated. ff19SB-OPC shows only dimer formation including swaps i.e. events in which the free molecule binds the dimer releasing the opposing monomer (see for example replica 3 at around 1400 ns). With the a99SBdisp-TIP4PD simulation the proteins seem to only quickly collide with each other without forming any complexes except for the last 200 ns of replica 2 and two short periods of replica 3, and never forming any trimers in the simulated timescale.
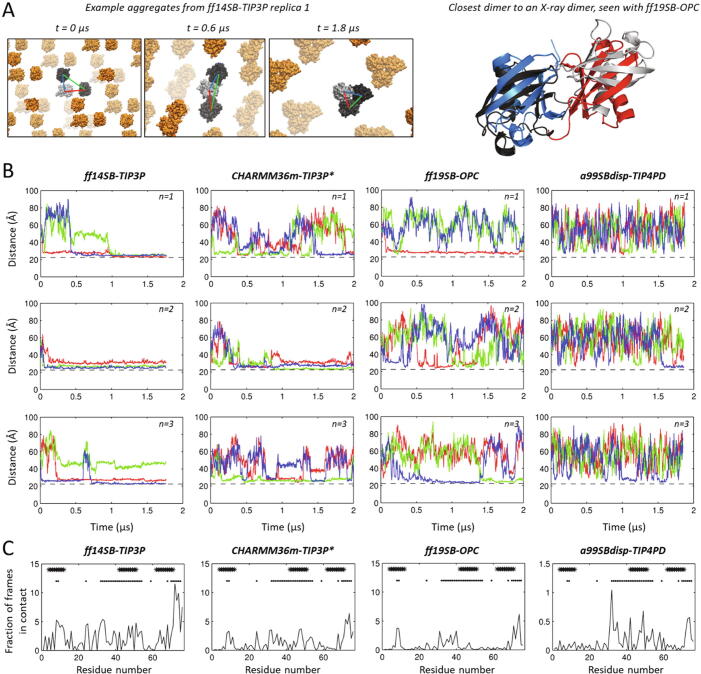
Fig. 1.
Tests on a concentrated ubiquitin solution. (A) Representative structures discussed in the text. On the left, example progression of aggregate formation from the starting system in replica 1 of the simulation with ff14SB-TIP3P, showing each protein of the cell in shades of grey and proteins of the periodic cells in orange. On the right, the closest match to an experimental ubiquitin-ubiquitin dimer (blue and red, PDB 1AAR) in the simulations with ff19SB-OPC (proteins from the simulation shown in grey), fit either to Cα atoms of residues 1–72 of both proteins (left) or of only one protein (right, with the proteins separated for clarity, the cyan and magenta proteins are aligned). (B) Interprotein distances (i.e. distance between centers of mass) over time for the 3 possible pairs of ubiquitin molecules, in the 3 independent replicas of 3 ubiquitin molecules parameterized with the 4 tested forcefields. Red = distance between protein molecules 1 and 2, green = distance between molecules 1 and 3, blue = distance between molecules 2 and 3, as depicted on the left in panel A. Each row of plots is an independent replica (n = 1,2,3); then from left to right: ff14SB-TIP3P, CHARMM36m-TIP3P*, ff19SB-OPC and a99SBdisp-TIP4PD. In these plots, the dashed line is twice the gyration radius of ubiquitin i.e. 22 Å). (C) Fraction of frames in which each residue established contacts with residues of other ubiquitin molecules, averaged from all replicas for each forcefield. Asterisks denote the residues involved in dimerization according to the NMR study by Liu et al, while dots denote residues involved in crystal contacts in X-ray structures of covalent ubiquitin dimers. Note that for a99SBdisp-TIP4PD the y axis is 10 times smaller than in the other plots. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Qualitatively, what is observed with ff19SB-OPC and a99SBdisp-TIP4PD would in principle reflect reality better than the aggregation observed with ff14SB and CHARMM36m, but orders-of-magnitude longer simulations would be required to unbiasedly characterize the thermodynamics and kinetics of the dimerization equilibrium with each parametrization and thus better conclude which one reproduces a real solution of concentrated ubiquitin the best. We can however further compare the forcefields by using the information about residues known to be engaged in weak noncovalent dimerization from NMR experiments and in covalent ubiquitin dimers as crystallized, by comparing directly the residues engaged in contact in the trajectories and in the structures and NMR data (averaged over all replicas for each forcefield in Fig. 1C). We clarify here that the NMR data only points at which residues are engaged in the surface that mediates the noncovalent dimerization, but contains no information about the specific pairs of residues in contact. This is why in Fig. 1C we compare the NMR-identified residues just with the number of contacts experienced by each residue of each protein (with any residue of the other proteins) in the simulations. What we observed upon such comparison is that although none of the forcefields captures the regions involved in dimerization very well, the residue-wise contact profiles produced by ff19SB-OPC and a99SBdisp-TIP4PD seem best, especially those produced by ff19SB-OPC which peak at residues 10, 30–40 and 65-end thus partially overlapping with the NMR-derived data for noncovalent dimerization and the noncovalent contacts observed in some X-ray structures of covalently linked ubiquitin dimers. Interestingly, simulations with ff19SB-OPC explore a few times a dimeric arrangement within ~7 Å Cα RMSD (covering both proteins) of the covalent ubiquitin dimer in PDB 1AAR, which is among the covalent complexes that best match the NMR data for noncovalent dimerization [40] (Fig. 1A, right). In turn, for CHARMM36m-TIP3P* and ff14SB-TIP3P the aggregates end up involving far too many residues in contact and no clear preferred binding poses. Moreover, the dimers that form with these two forcefields eventually evolve into trimeric clusters that get increasingly more compact behaving as mere aggregates, as illustrated from replica 1 of the simulation with ff14SB-TIP3P in Fig. 1A.
The simulations on systems containing 9 ubiquitin molecules reveal similar outcomes, making more patent the strong inter-protein binding with ff14SB-TIP3P and with CHARMM36m-TIP3P*, showing also some aggregation for ff19SB-OPC that was not clear for the simulations with 3 proteins. The effects are also evident in plots of the total solvent-accessible surface areas vs. time, such that ff14SB-TIP3P seems to favor the most compact aggregates followed by CHARMM36m-TIP3P*, then ff19SB-OPC and finally a99SBdisp-TIP4PD where the proteins remain fully soluble and monomeric (Fig. S1).
Overall, as conclusion of these tests on concentrated ubiquitin solutions, the two forcefields with finely tuned 4-point water molecules (ff99SB-OPC and a99SBdisp-TIP4PD) seem to better reproduce a real solution of concentrated ubiquitin. Of them, a99SBdisp-TIP4PD keeps the protein more stable in solution, showing no preferred interactions through any surface although within this timescale and at the employed concentrations reversible dimer formation should happen. Meanwhile, the residues involved in protein–protein contacts in the simulations with ff99SB-OPC are in better agreement with the identities of residues engaged in actual interactions as determined by NMR for weak noncovalent dimerization and in X-ray structures of covalent ubiquitin dimers. Thus, for this system the ff99SB-OPC simulations seem to reproduce reality the best.
2.2. Testing a disordered peptide with localized helical propensity
As briefly reviewed in the introduction, the main goal of recent forcefield optimization efforts including those that led to CHARMM36m-TIP3P*, ff19SB-OPC and a99SBdisp-TIP4PD tested in this work, was not to properly account for solubility in multiple-protein systems as tested above for ubiquitin but rather to properly account for structural dynamics of isolated, highly disordered, soluble polypeptides -which being related to the protein–protein interaction problem, could have simultaneously solved both. We tested the four forcefields against this specific issue by simulating a disordered peptide not used in any of the parameter optimization protocols of the tested forcefields: the first 19 amino acids of huntingtin’s exon 1, i.e. the 17 fully conserved N-terminal amino acids followed by the first two glutamine residues of the glutamine expansion, dubbed here Htt-1-19. This peptide has a net charge of +1 at pH 7; it is highly soluble and intrinsically disordered but with a slight alpha helical propensity quantified at around 10–30% from circular dichroism (CD) spectra at pH 7, that gets further stabilized in presence of trifluoroethanol [44], [45]. Furthermore, 13C chemical shift propensities for exon 1 and the Htt-1-19 peptide at pH 7 disclose residual alpha helical propensity around residues ~5–19, with a maximum at around residues 15–18 [45], [46], [47] at acidic and neutral pH (Fig. 2A). Meanwhile, crystallization of a protein fusion of huntingtin’s exon 1 to maltose binding protein (MBP) enforces a fully helical conformation as revealed by X-ray diffraction [48].
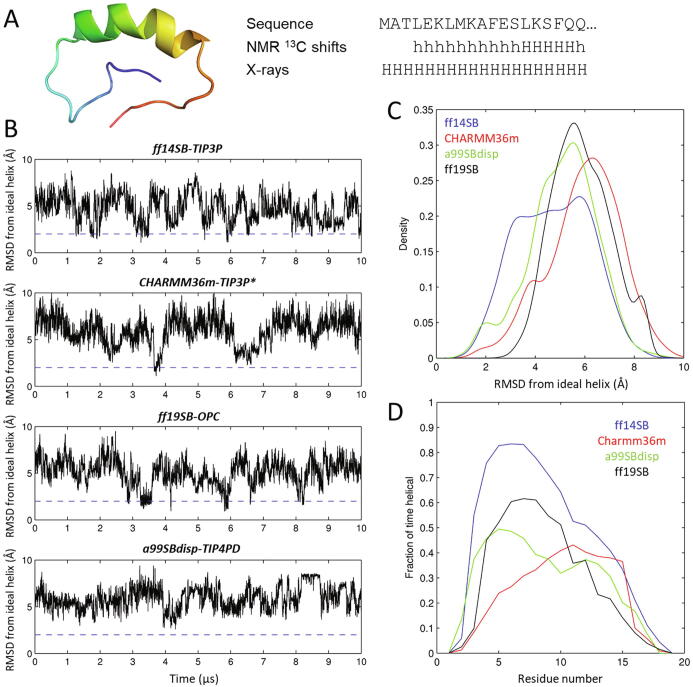
Fig. 2.
Tests on Huntingtin’s Nt19 peptide. (A) Model of Huntingtin segment Nt17Q17 computed from 13C chemical shifts at neutral pH by Baias et al. [46], showing on the right the sequence of the Htt-1-19 peptide under study and the helical propensities derived for it from 13C chemical shifts [45] and from an X-ray structure of huntingtin’s exon 1 fused to maltose binding protein (PDB ID 3IO6), where “h” and “H” mean low and helical propensity, respectively. (B) RMSD from an ideal helical conformation over 10 μs of unbiased MD simulation of Htt-1-19. The dashed line is set at 2 Å RMSD, under which the peptide is essentially fully helical. (C) Histogram of RMSD densities from the data in panel B. (D) Fractional helicity per residue averaged throughout each of the four simulations. The total average helical content is 45% for ff14SB-TIP3P, 23% for CHARMM36m-TIP3P*, 28% for a99SBdisp-TIP4PD and 29% for ff19SB-OPC.
Starting from a fully extended conformation set up at pH 7, we simulated the peptide for 10 µs with each forcefield. We note that although 1H-15N NOE values and secondary 13C shifts reveal some local secondary structuring, the 15N relaxation data and the sharp NMR signals imply a very dynamic nature [45], [46]. Thus, 10 µs simulations should be long enough to capture some formation and rupture of local secondary structures, as we indeed observe. In fact, none of these trajectories gets stuck in any specific conformation. In order to make a more quantitative comparison, we recurred to a fully helical structure of the peptide as a reference to compare against. This seemed the most appropriate choice because our simulated peptide does not correspond to the exact same constructs studied by NMR; also because the only solution-state NMR model (shown in Fig. 2A) is based on 13C chemical shifts only hence it is of limited accuracy and it does not reflect the full dynamic nature of the peptide; and also because the strongly helical structure observed in the X-ray structure indicates that a fully helical structure is available within the conformational landscape of the peptide. By comparing the conformations adopted by the peptide against a fully helical structure taken as reference, more intelligible than a random disordered peptide, we can better compare the residue-averaged and global time-averaged helical propensities to estimates from solution-state CD (for global helix propensity) and 13C NMR chemical shifts (for residue-wise propensities).
Simulations with all 4 forcefields show some interconversions to conformations within 2–3 Å RMSD of a fully helical state (Fig. 2B,C); however, ff14SB-TIP3P favors this state substantially more than the other forcefields. Calculation of local secondary structure for each residue throughout each simulation (Fig. 2D) reveals 22% helix propensity for CHARMM36m-TIP3P*, 28% for a99SBdisp-TIP4PD and 29% for ff19SB-OPC, all lower than that for ff14SB-TIP3P (45%). Inspection per residue shows that the amber-based forcefields predict a maximal helical propensity between residues 4 and 10 with the a99SBdisp-TIP4PD and ff99SB-OPC profiles looking like dampened versions of ff14SB-TIP3P, whereas the per-residue helicity profile for CHARMM36m-TIP3P* is shifted to the C-terminus with highest helical propensity between residues 9 and 15. Although all 3 modern forcefields capture roughly the right helical content as determined by CD, the profile over sequence observed for CHARMM36m-TIP3P* is more consistent with the residue-wise propensities estimated from 13C chemical shifts which show stronger alpha helical propensity towards the C-terminus.
2.3. Testing a highly insoluble fragment from Alzheimer-related amyloid-β peptide
We then challenged the forcefields to reproduce the opposite of what we tested with ubiquitin: the insolubility of peptides derived from the Alzheimer-related amyloid-β (Aβ) peptide. The full Aβ peptide is released upon proteolysis of the amyloid precursor protein as a version containing 39–43 residues [49]; it is somewhat soluble in water buffer but forms fibers after a lag phase whose duration depends on multiple factors, while some of its segments are totally insoluble, i.e. water solutions of them cannot be prepared. Specifically, we here tested the four forcefields on two systems containing multiple copies of two segments of the Aβ peptide known to rapidly form β-rich aggregates upon attempt to dissolution in water at neutral pH [50]. These peptides are in fact so insoluble in water, that their experimental handling requires the use of large mole fractions of other solvents [41]. And even in such solvents, the peptides undergo aggregation within seconds to a few hours forming fibers if at micromolar concentrations or amorphous aggregates if at millimolar concentrations, in both cases very rich in β-sheets that are easily evidenced by CD, infrared spectroscopies, etc.
One set of simulations consists in 10 molecules of the Aβ-derived KLVFFAE segment initiated from random positions, i.e. well spread in solution, in a box that renders them ~10 mM in concentration. We ran this system in single replica with each forcefield, but we note that the presence of 10 molecules compensates for the lack of multiple independent runs, and as we detail below we observe quite neat similarities and differences among forcefields, that look significant at least in the timescale tested and possibly beyond too. The other set of simulations are setup from an X-ray structure of a portion of amyloid fiber formed by Aβ 26–40, of sequence QKLVFFAENVGSNKGAIIGLMVGGVV which includes the smaller KLVFFAE peptide that makes the first system. This structure of this piece of β-amyloid fiber was determined by X-ray diffraction (PDB ID 2LNQ [51]). The unit cell contains 5 polypeptides of this same sequence, each turning at the GSN triplet and folding on itself to form a hydrophobic interior and all stacking head-to-tail forming a quite planar arrangement that repeats itself along the crystal. We ran this system in triplicate with each forcefield, each for at least 3 µs.
On challenging the forcefields with these systems we expect the 10 mM solution of KLVFFAE to lead to aggregation with formation of β structures, and the amyloid Aβ 26–40 fiber to remain insoluble and well structured.
In the simulations starting from a solution of 10 KLVFFAE peptides (at pH 7 where the peptide is neutral) both CHARMM36m-TIP3P* and ff14SB-TIP3P led to β-rich aggregates (example in Fig. 3A for CHARMM36m-TIP3P*) within a few hundred nanoseconds, while a99SBdisp-TIP4PD and ff19SB-OPC did not, their peptides remaining fully disordered and with only some β structuring even after ~2 μs of simulation. Aggregation can be judged more objectively from the resulting drop in the total solvent-accessible surface area of the peptides (Fig. 3A), which is slightly stronger for ff14SB-TIP3P than for both CHARMM36m-TIP3P* and virtually null for the other two forcefields, although the simulation with ff19SB-OPC does feature some small drops in SASA due to some small fast-resolved aggregates. Overall this trend is analogous to that of aggregation propensity reported above for ubiquitin solutions. Coarsely, ff14SB-TIP3P and CHARMM36m-TIP3P* would pose that aggregation takes place within a microsecond timescale, which cannot be compared to a number in solution because this exact experiment cannot practically be carried out due to the extreme insolubility of the peptide. However, the main conclusion derived from these simulations, i.e. that the peptide is essentially insoluble, would be correct. For ff19SB-OPC and a99SBdisp-TIP4PD the conclusion is less clear, because it could well occur that the peptides aggregate in a longer timescale; in such case the simulations would be predicting a lag phase that does exist for many other amyloid-prone but less insoluble peptides like for example the full Aβ peptide itself.
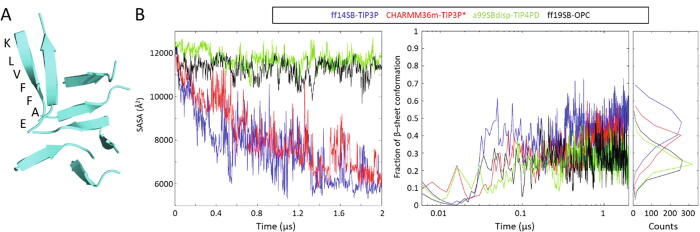
Fig. 3.
Tests on solutions of 10 randomly spread, extended KLVFFAE peptides. (A) Example β-aggregate, in this case observed at the end of the simulation with CHARMM36m-TIP3P*, with the amino acid sequence shown in one peptide. (B) Solvent-Accessible Surface Area (SASA) over time, fraction of residues in β conformation over time, and fractional β content averaged throughout the trajectories. In the three plots, blue is ff14SB-TIP3P, red is CHARMM36m-TIP3P*, green is a99SBdisp-TIP4PD and black is ff99SB-OPC. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Notably and consistently with the known nature of KLVFFAE aggregates, the peptides aggregated with ff14SB-TIP3P and CHARMM36m-TIP3P* adopt substantial β-sheet conformation (40–50%, Fig. 3B). In the simulations with a99SBdisp-TIP4PD and ff19SB-OPC, where peptides did not aggregate in the simulated timescale, there is also some 20% β structuring. In all cases the β content is stabilized by around 200 ns, but fluctuates largely especially with ff14SB-TIP3P (Fig. 3B, right). Overall, regarding the capacity to quickly predict the strong insolubility and β-rich structure of the KLVFFAE peptide CHARMM36m-TIP3P* and ff14SB-TIP3P would rank similarly, both better than the other two forcefields.
In all four simulations started from the structure of a piece of fiber of Aβ 16–40 (Fig. 4A, also at pH 7 where each fibril has a net charge of +1) its constituent peptides remain stack to the fiber over 3 μs of simulation, indicating insolubility in this timescale. However, RMSD profiles from the starting structure and β-sheet contents over time (Fig. 4B) show that the systems parameterized with ff14SB-TIP3P and ff19SB-OPC are somewhat more stable than those parametrized with a99SBdisp-TIP4PD and CHARMM36-TIP3P*. Although this could suggest that the peptides are beginning to dissolve, the contacts are still extensive and testing this hypothesis would require much longer simulations. It is interesting that the resulting trend is not exactly the same as in the other cases, because here ff19SB-OPC behaves closer to ff14SB-TIP3P, both possibly better than the other two forcefields by making the fibril stable.
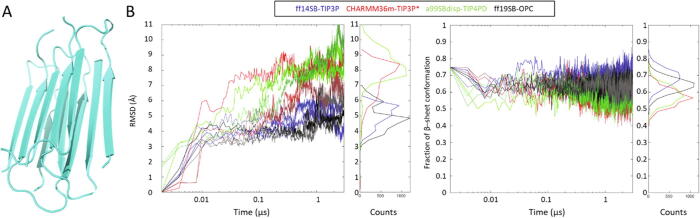
Fig. 4.
Tests on an amyloid aggregate. (A) View of the starting structure used in the simulations, from PDB ID 2LNQ [51]. (B) Overall Cα RMSD from the starting structure. (C) Overall β content against time. In both plots, ff14SB-TIP3P is in blue, CHARMM36m-TIP3P* in red, a99SBdisp-TIP4PD in green and ff19SB-OPC in black. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Summarizing the results in this section, it appears that ff14SB-TIP3P and CHARMM36-TIP3P* both greatly favor formation of amorphous β-rich aggregates, while a99SBdisp-TIP4PD shows much higher solubility and disorder in the simulated timescale. Although we cannot rule out that a99SBdisp-TIP4PD could produce β-rich aggregates in longer simulations, this seems unlikely, and the lack of any aggregation parallels our observations on the ubiquitin systems and even the latest observations from its developers on the much longer timescales they achieve with their specialized computer for MD simulations [37]. Meanwhile, ff19SB-OPC seems closer to a99SBdisp-TIP4PD than to the other forcefields considering that it did not show strong binding nor any aggregation of the Aβ peptides, although there were some transient interactions, but it seems closer to ff14SB-TIP3P regarding its capacity to keep the amyloid aggregate insoluble and stable. It is clear that much longer simulation lengths are required to draw more solid conclusions about the performance of these forcefields on these systems, especially to know if a99SBdisp-TIP4PD and CHARMM36-TIP3P* would eventually fully dissolve the Aβ fiber in much longer simulations and if ff19SB-OPC would eventually result in aggregation of the KLVFFAE peptides.
3. Discussion
We have here challenged the latest transferable nonpolarizable forcefields for atomistic molecular simulations from leader developers against a series of problems that involve (i) keeping soluble proteins in solution state with the additional difficulty of predicting weak dimerization, yet (ii) making insoluble peptides aggregate into β-rich structures and keeping a β-fiber insoluble, and (iii) properly keeping a disordered peptide not too compact yet reproduce its helical propensity. Although the tested forcefields were only optimized with the latter kind of goal in mind, on which all three modern forcefields improve relative to the older taken as reference, we do see some evidence of improved behavior of multiprotein systems too, especially for ff19SB-OPC and CHARMM36m-TIP3P*.
The results from all our tests can be summarized by ordering the forcefields by their tendency to make proteins and peptides more or less compact, either through strong binding and eventual aggregation (ubiquitin and Aβ peptides) or secondary structure stabilization (Htt-1-19). The resulting order is ff14SB-TIP3P as the most compaction-favoring forcefield, followed by CHARMM36m-TIP3P*, then ff19SB-OPC. and finally a99SBdisp-TIP4PD as the forcefield that totally disfavors aggregation. Within this series, ff14SB-TIP3P and a99SBdisp-TIP4PD represent two opposite extremes, both bad as the former overstabilizes ubiquitin aggregates and the α-helix propensity of the huntingtin peptide too much and the latter misses the expected intermolecular interactions between ubiquitin molecules and even between peptides expected to aggregate very rapidly as they are totally insoluble in water. In-between, both CHARMM36m-TIP3P* and ff19SB-OPC predict a correct extent of α-helical propensity for the huntingtin peptide (slightly better for CHARMM36m-TIP3P*) and β-strand propensity for the Aβ peptides. a99SBdisp-TIP4PD reproduces Htt-1-19 disorder and helical propensity similarly to ff19SB-OPC. CHARMM36m-TIP3P* over-stabilizes ubiquitin aggregates too much, whereas ff19SB-OPC keeps ubiquitin soluble with just some contacts that somewhat match those expected from NMR data but it does not lead to aggregates of the Aβ peptides at least in the timescale of the simulation, although it does keep the amyloid fibril in a compact state and it does produce some β-strand dimers in the aggregation simulation that could potentially result in β-aggregates in longer trajectories.
Regarding a99SBdisp-TIP4PD, although we cannot exclude that results could have been better in longer timescales, recent tests carried out by its developers showed that this forcefield cannot keep together some protein–protein complexes known to be of high affinity [37], thus attesting in favor of our conclusion that it fails to capture interprotein interactions. That same study includes the development of a new optimization forcefield supposed to improve protein–protein complexation while preserving good description of ordered and disordered proteins isolated in solution. Even such ad hoc optimization, despite improving from a99SBdisp-TIP4PD, still does not properly explain affinities, as the authors found. It is thus clear that the problem still deserves attention, the positive point being that at least this group of forcefield developers are now incorporating multi-protein systems in their targets for improvement. It is important though that observables for single proteins, either ordered, disordered or mixed, be kept under the eye, because our findings suggest that the optimal balance between stabilizing and destabilizing interactions requires exquisite tuning, and that optimizations that look good enough for disordered proteins are peptides may still be insufficient to properly describe multiprotein systems.
Importantly, all four forcefields could correctly capture the secondary structures preferred by the two peptides involved in the studies. Huntingtin’s peptide 1–19 is disordered with a substantial helical propensity that all forcefields captured, with ff19SB-OPC, CHARMM36m-TIP3P* and a99SBdisp-TIP4PD even correctly capturing the ~20% population estimated by circular dichroism (overly stabilized by ff14SB-TIP3P, as expected). Likewise, the short Aβ peptide aggregated in all four cases into β-sheets, also consistent with the secondary structure it adopts in fibrils and even in amorphous aggregates. Since short peptides have intrinsically fast dynamics, often in the microsecond timescale, current simulation power allows sampling their multiple conformational transitions thus coming closer to fulfillment of the ergodic hypothesis. A more extensive benchmark specifically tailored to predicting the secondary structures of peptides through atomistic simulations may then be worth in the near future.
A further important note concerns the water models. The good performance of CHARMM36m-TIP3P*, very comparable to that of ff19SB with OPC, shows that 3-point water models might still be good alternatives to the more costly 4-point models like OPC and TIP4PD. However, 4-point models are only recently being tested extensively, so their role in improving the description of multi-protein and disordered states may not have been fully exploited yet. A related point is that of polarizable forcefields like Drude [52] and Amoeba [53], which as far as we know have not been benchmarked in independent tests but could hold the key to complete descriptions of biomolecular physics especially regarding correct modulation of hydrogen bonding and charge-pair interactions, interactions of strong polarization origin such as cation-π interactions, and water-mediated effects [54], [55], [56].
An important limitation in most if not all MD simulation studies is the limited timescale of sampling relative to the actual timescale of relevant molecular motions. Enhanced-sampling simulations help to cover this gap, but they can introduce artifacts especially if the collective variables are not properly chosen or scanned [57], hence the preference for long unbiased simulations and replicas over forcing reaction coordinates. In our microsecond-timescale simulations, the predictions of ubiquitin and KLVFFAE peptide being quite insoluble are neat and aggregation looks irreversible when it happens; moreover, these cases involve assemblies with more than 2 molecules which are known to not exist (or in the best case be very scarcely populated). Similarly, a timescale of 10 µs should be sufficient to sample the fast dynamics of the short Htt-1-19 peptides, and in fact the simulations show multiple transitions between disordered and helix-like states. The number of transitions is per se not sufficient to compute free energies, but stresses the main differences quite clearly. Last, the case of the amyloid fiber is the only one where our results are most sensitive to simulation length and number of replicas, hence less conclusive. In this and all cases, a 10X longer simulation timescale could probably better discern the accuracies of the different forcefields, a timescale probably achievable within the next decade. Another caveat in our simulations of multiple ubiquitin and KLVFFAE peptides is that although the starting orientations are randomized, there is no way to ensure that these orientations do not introduce any artifacts or biases. Ideally, one should begin our simulations with a huge number of different configurations that sample the different relative orientations and distances, but this is of course intractable. On the good side, we note that the molecules are far enough to initially diffuse as if they were isolated in solution and differently in the different replicas, before engaging in contacts.
To conclude, we acknowledge that the forcefield improvements claimed by all developers are evident in our results, with a positive prospect as developments slowly converge to the right balance between sources of attractive and repulsive forces. Overall, in our tests CHARMM36m-TIP3P* and ff19SB-OPC seem to emerge as the most recommendable forcefields, with CHARMM36m-TIP3P* having the advantage that it entails much smaller number of particles for a given size of simulation box as it uses a 3-point water model. Of course, any conclusion recommending a forcefield over others is very limited and biased, due the small number of systems tested. Besides, certain tasks might be better suited to more specific forcefields; for example, one may want to stick to one of the forcefields specific for intrinsically disordered proteins for simulation of single purely disordered systems, and in fact some are still better than for example CHARMM36m for this [29]; while a project involving nucleic acids or membranes may require specific forcefields for these molecules that may have only been tested together with other specific forcefields for proteins.
As a closing remark we pose again that in order to advance forcefield evolution developers need to simultaneously consider not only single folded, disordered and mixed proteins, but also multiprotein systems expected to either remain soluble or aggregate. We provide in the SI the starting coordinates for all systems simulated here, so that they can be used for testing new forcefields in future works.
4. Methods
Systems to simulate concentrated ubiquitin solutions were prepared by randomly placing 3 or 9 ubiquitin molecules from PDB ID 1UBQ in a box of size ~100 × 100 × 100 Å3 (for 3 protein molecules, reaching 5 mM concentration) or ~135 × 135 × 135 Å3 (for 9 protein molecules, reaching 6 mM concentration). For Htt-1-19, the peptide was initially built in a fully extended conformation (64 Å long) and placed along the largest dimension of a box of size 100 × 70 × 50 Å3; then after some compression happening in <100 ns of simulation, a new, smaller system was prepared to extend the simulations as presented. The system with 10 Aβ peptides was setup by randomly placing 10 copies of a peptide built in extended conformation, inside a box of size 110 × 115 × 115 Å3 (10 mM). The Aβ fiber system was set from PDB 2QLN with minimum 20 Å spacing to the edges of the solvent box. All systems were prepared with standard protonation states corresponding to pH 7, and neutralized to 100 mM KCl. For parametrization, ff14SB-TIP3P systems were prepared with the Amber18 package and ff19SB-OPC systems with the Amber20 package; CHARMM36m-TIP3P* systems were built with CHARMM-GUI as of august 2018; and a99SBdisp-TIP4PD systems were built with Gromacs 2018 using the parameters provided by D.E. Shaw Research. Simulations with ff14SB were run with Amber18, those with ff19SB using Amber20 provided by the developers, and simulations with CHARMM36m and a99SBdisp were run with Gromacs 2018. All simulations were carried out in NPT conditions at 300 K and 1 atm, with 2 fs integration timestep, no special mass repartitioning, using SHAKE to constrain bond lengths involving hydrogen atoms, and 12 Å cutoff for nonbonded interactions switching from 10 Å with PME treatment of electrostatics. Before each NPT production simulation, we minimized the systems and equilibrated them in NVT from 0 to 300 K over 1 ns of simulation with CA atoms constrained followed by 1 ns of unrestrained simulation. All analyses were carried out with in-house VMD and Matlab scripts.
CRediT authorship contribution statement
Luciano A. Abriata: Conceptualization, Methodology, Investigation, Original and revised. Matteo Dal Peraro: Conceptualization, Original and revised.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We acknowledge the Swiss National Supercomputing Centre (CSCS) for access to computer time through grant CSCS-2018 to LAA and MDP.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.04.050.
Contributor Information
Luciano A. Abriata, Email: luciano.abriata@epfl.ch.
Matteo Dal Peraro, Email: matteo.dalperaro@epfl.ch.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Liu X., Shi D., Zhou S., Liu H., Liu H., Yao X. Molecular dynamics simulations and novel drug discovery. Expert Opin Drug Discov. 2018;13(1):23–37. doi: 10.1080/17460441.2018.1403419. [DOI] [PubMed] [Google Scholar]
- 2.Nunes-Alves A., Kokh D.B., Wade R.C. Recent progress in molecular simulation methods for drug binding kinetics. Curr Opin Struct Biol. 2020;64:126–133. doi: 10.1016/j.sbi.2020.06.022. [DOI] [PubMed] [Google Scholar]
- 3.Geng H., Chen F., Ye J., Jiang F. Applications of molecular dynamics simulation in structure prediction of peptides and proteins. Comput Struct Biotechnol J. 2019;17:1162–1170. doi: 10.1016/j.csbj.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferina J., Daggett V. Visualizing protein folding and unfolding. J Mol Biol. 2019;431:1540–1564. doi: 10.1016/j.jmb.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 5.Lindorff-Larsen K., Piana S., Dror R.O., Shaw D.E. How fast-folding proteins fold. Science. 2011;334(6055):517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 6.Heo L., Arbour C.F., Feig M. Driven to near-experimental accuracy by refinement via molecular dynamics simulations. Proteins Struct. Funct. Bioinforma. 2019;87:1263–1275. doi: 10.1002/prot.25759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heo L., Feig M. Experimental accuracy in protein structure refinement via molecular dynamics simulations. Proc Natl Acad Sci. 2018;115:13276–13281. doi: 10.1073/pnas.1811364115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastore A. The role of post-translational modifications on the energy landscape of Huntingtin N-terminus. Front. Mol. Biosci. 2019;6:95. doi: 10.3389/fmolb.2019.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abriata L.A., Spiga E., Peraro M.D. Molecular effects of concentrated solutes on protein hydration, dynamics, and electrostatics. Biophys J. 2016;111:743–755. doi: 10.1016/j.bpj.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spiga E., Abriata L.A., Piazza F., Dal Peraro M. Dissecting the effects of concentrated carbohydrate solutions on protein diffusion, hydration, and internal dynamics. J Phys Chem B. 2014;118:5310–5321. doi: 10.1021/jp4126705. [DOI] [PubMed] [Google Scholar]
- 11.Feig M., Sugita Y. Variable interactions between protein crowders and biomolecular solutes are important in understanding cellular crowding. J Phys Chem B. 2012;116(1):599–605. doi: 10.1021/jp209302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada R., Sugita Y., Feig M. Protein crowding affects hydration structure and dynamics. J Am Chem Soc. 2012;134(10):4842–4849. doi: 10.1021/ja211115q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marrink S.J. Computational modeling of realistic cell membranes. Chem Rev. 2019;119:6184–6226. doi: 10.1021/acs.chemrev.8b00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abriata L.A., Albanesi D., Dal Peraro M., de Mendoza D. Signal sensing and transduction by histidine kinases as unveiled through studies on a temperature sensor. Acc Chem Res. 2017;50:1359–1366. doi: 10.1021/acs.accounts.6b00593. [DOI] [PubMed] [Google Scholar]
- 15.Saita E. A coiled coil switch mediates cold sensing by the thermosensory protein DesK. Mol Microbiol. 2015;98:258–271. doi: 10.1111/mmi.13118. [DOI] [PubMed] [Google Scholar]
- 16.Abriata L.A., Dal Peraro M. Assessing the potential of atomistic molecular dynamics simulations to probe reversible protein-protein recognition and binding. Sci Rep. 2015;5:10549. doi: 10.1038/srep10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palazzesi F., Prakash M.K., Bonomi M., Barducci A. Accuracy of current all-atom force-fields in modeling protein disordered states. J Chem Theory Comput. 2015;11:2–7. doi: 10.1021/ct500718s. [DOI] [PubMed] [Google Scholar]
- 18.Petrov D., Zagrovic B., Dunbrack R.L. Are current atomistic force fields accurate enough to study proteins in crowded environments? PLoS Comput Biol. 2014;10(5) doi: 10.1371/journal.pcbi.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriques J., Cragnell C., Skepö M. Molecular dynamics simulations of intrinsically disordered proteins: force field evaluation and comparison with experiment. J Chem Theory Comput. 2015;11:3420–3431. doi: 10.1021/ct501178z. [DOI] [PubMed] [Google Scholar]
- 20.Best R.B., Zheng W., Mittal J. Balanced protein–water interactions improve properties of disordered proteins and non-specific protein association. J Chem Theory Comput. 2014;10:5113–5124. doi: 10.1021/ct500569b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman M.U., Rehman A.U., Liu H., Chen H.-F. Comparison and evaluation of force fields for intrinsically disordered proteins. J Chem Inf Model. 2020;60:4912–4923. doi: 10.1021/acs.jcim.0c00762. [DOI] [PubMed] [Google Scholar]
- 22.Yang S., Liu H., Zhang Y., Lu H., Chen H. Residue-specific force field improving the sample of intrinsically disordered proteins and folded proteins. J Chem Inf Model. 2019;59:4793–4805. doi: 10.1021/acs.jcim.9b00647. [DOI] [PubMed] [Google Scholar]
- 23.Wang W., Ye W., Jiang C., Luo R., Chen H.-F. New force field on modeling intrinsically disordered proteins. Chem Biol Drug Des. 2014;84:253–269. doi: 10.1111/cbdd.12314. [DOI] [PubMed] [Google Scholar]
- 24.Song D., Wang W., Ye W., Ji D., Luo R., Chen H.-F. Wiley Online Library; 2017. ff14IDPs force field improving the conformation sampling of intrinsically disordered proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song D., Luo R., Chen H.-F. The IDP-specific force field ff14IDPSFF improves the conformer sampling of intrinsically disordered proteins. J Chem Inf Model. 2017;57:1166–1178. doi: 10.1021/acs.jcim.7b00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian C. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J Chem Theory Comput. 2019;16:528–552. doi: 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- 28.Robustelli P., Piana S., Shaw D.E. Developing a molecular dynamics force field for both folded and disordered protein states. Proc Natl Acad Sci. 2018;115:E4758–E4766. doi: 10.1073/pnas.1800690115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mu J., Liu H., Zhang J., Luo R., Chen H.-F. Recent force field strategies for intrinsically disordered proteins. J Chem Inf Model. 2021;61(3):1037–1047. doi: 10.1021/acs.jcim.0c01175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J., MacKerell A.D., Jr Force field development and simulations of intrinsically disordered proteins. Curr Opin Struct Biol. 2018;48:40–48. doi: 10.1016/j.sbi.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W. Recent advances in atomic molecular dynamics simulation of intrinsically disordered proteins. Phys Chem Chem Phys. 2021;23(2):777–784. doi: 10.1039/d0cp05818a. [DOI] [PubMed] [Google Scholar]
- 32.Piana S., Donchev A.G., Robustelli P., Shaw D.E. Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J Phys Chem B. 2015;119:5113–5123. doi: 10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- 33.Wang A. Quality of force fields and sampling methods in simulating pepX peptides: a case study for intrinsically disordered proteins. Phys Chem Chem Phys. 2021;23:2430–2437. doi: 10.1039/d0cp05484d. [DOI] [PubMed] [Google Scholar]
- 34.Rauscher S. Structural ensembles of intrinsically disordered proteins depend strongly on force field: a comparison to experiment. J Chem Theory Comput. 2015;11:5513–5524. doi: 10.1021/acs.jctc.5b00736. [DOI] [PubMed] [Google Scholar]
- 35.Nerenberg P.S., Head-Gordon T. New developments in force fields for biomolecular simulations. Curr Opin Struct Biol. 2018;49:129–138. doi: 10.1016/j.sbi.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Maier J.A. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piana S., Robustelli P., Tan D., Chen S., Shaw D.E. Development of a force field for the simulation of single-chain proteins and protein-protein complexes. J Chem Theory Comput. 2020;16:2494–2507. doi: 10.1021/acs.jctc.9b00251. [DOI] [PubMed] [Google Scholar]
- 38.Shabane P.S., Izadi S., Onufriev A.V. General purpose water model can improve atomistic simulations of intrinsically disordered proteins. J Chem Theory Comput. 2019;15:2620–2634. doi: 10.1021/acs.jctc.8b01123. [DOI] [PubMed] [Google Scholar]
- 39.Wintrode P.L., Makhatadze G.I., Privalov P.L. Thermodynamics of ubiquitin unfolding. Proteins Struct Funct Bioinforma. 1994;18(3):246–253. doi: 10.1002/prot.340180305. [DOI] [PubMed] [Google Scholar]
- 40.Liu Z., Zhang W.-P., Xing Q., Ren X., Liu M., Tang C. Noncovalent dimerization of ubiquitin. Angew Chem Int Ed Engl. 2012;51(2):469–472. doi: 10.1002/anie.201106190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao K., Wang J., Zhou P., Wang C., Xu H., Zhao X. Self-assembly of short Aβ (16–22) peptides: effect of terminal capping and the role of electrostatic interaction. Langmuir. 2011;27(6):2723–2730. doi: 10.1021/la1034273. [DOI] [PubMed] [Google Scholar]
- 42.Samantray S., Yin F., Kav B., Strodel B. Different force fields give rise to different amyloid aggregation pathways in molecular dynamics simulations. J Chem Inf Model. 2020;60:6462–6475. doi: 10.1021/acs.jcim.0c01063. [DOI] [PubMed] [Google Scholar]
- 43.Strodel B. Amyloid aggregation simulations: challenges, advances and perspectives. Curr Opin Struct Biol. 2021;67:145–152. doi: 10.1016/j.sbi.2020.10.019. [DOI] [PubMed] [Google Scholar]
- 44.Chiki A. Mutant Exon1 Huntingtin aggregation is regulated by T3 phosphorylation-induced structural changes and crosstalk between T3 phosphorylation and acetylation at K6. Angew Chem Int Ed Engl. 2017;56:5202–5207. doi: 10.1002/anie.201611750. [DOI] [PubMed] [Google Scholar]
- 45.Chiki A. Site-specific phosphorylation of Huntingtin exon 1 recombinant proteins enabled by the discovery of novel kinases. Chembiochem Eur J Chem Biol. 2020 doi: 10.1002/cbic.202000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baias M. Structure and dynamics of the huntingtin Exon-1 N-terminus: a solution NMR perspective. J Am Chem Soc. 2017;139:1168–1176. doi: 10.1021/jacs.6b10893. [DOI] [PubMed] [Google Scholar]
- 47.Newcombe E.A. Tadpole-like conformations of huntingtin exon 1 are characterized by conformational heterogeneity that persists regardless of polyglutamine length. J Mol Biol. 2018;430:1442–1458. doi: 10.1016/j.jmb.2018.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim M. Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues. Prion. 2013;7(3):221–228. doi: 10.4161/pri.23807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Checler F. Processing of the β-amyloid precursor protein and its regulation in Alzheimer’s disease. J Neurochem. 1995;65(4):1431–1444. doi: 10.1046/j.1471-4159.1995.65041431.x. [DOI] [PubMed] [Google Scholar]
- 50.Petkova A.T. Self-propagating, molecular-level polymorphism in Alzheimer’s ß-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 51.Qiang W., Yau W.-M., Luo Y., Mattson M.P., Tycko R. Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc Natl Acad Sci. 2012;109:4443–4448. doi: 10.1073/pnas.1111305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin F.-Y. Further optimization and validation of the classical drude polarizable protein force field. J Chem Theory Comput. 2020;16:3221–3239. doi: 10.1021/acs.jctc.0c00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y. Polarizable atomic multipole-based AMOEBA force field for proteins. J Chem Theory Comput. 2013;9:4046–4063. doi: 10.1021/ct4003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin F.-Y., MacKerell A.D., Jr. Improved modeling of cation-π and anion-ring interactions using the drude polarizable empirical force field for proteins. J Comput Chem. 2020;41:439–448. doi: 10.1002/jcc.26067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jing Z. Polarizable force fields for biomolecular simulations: recent advances and applications. Annu Rev Biophys. 2019;48:371–394. doi: 10.1146/annurev-biophys-070317-033349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inakollu V.S., Geerke D.P., Rowley C.N., Yu H. Polarisable force fields: what do they add in biomolecular simulations? Curr Opin Struct Biol. 2020;61:182–190. doi: 10.1016/j.sbi.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 57.Pan A.C., Weinreich T.M., Shan Y., Scarpazza D.P., Shaw D.E. Assessing the accuracy of two enhanced sampling methods using EGFR kinase transition pathways: the influence of collective variable choice. J Chem Theory Comput. 2014;10:2860–2865. doi: 10.1021/ct500223p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.