Highlights
-
•
Penicillium polonicum, an endophyte of Ginkgo biloba, is a potent Taxol producer.
-
•
Taxol of P. polonicum has a strong anticancer effect against HEPG2 and MCF7.
-
•
The Taxol yield by P. polonicum was increased by 4.5 folds upon optimization with response surface methodology.
Keywords: Ginko biloba, Endophytic fungi, Penicillium polonicum, Taxol, Optimization, Placket-Burman design
Abstract
Twenty-eight fungal endophytes were recovered from the different parts of Ginkgo biloba and screened for their Taxol producing potency. Among these isolates, Penicillium polonicum AUMC14487 was reported as the potent Taxol producer (90.53 μg/l). The chemical identity of the extracted Taxol was verified from the TLC, HPLC, NMR, EDX, and FTIR analyses. The extracted Taxol displayed a strong antiproliferative activity against HEPG2 (IC50 4.06 μM) and MCF7 (IC50 6.07 μM). The yield of Taxol by P. polonicum was optimized by nutritional optimization with the Response Surface Methodology (RSM) using Plackett-Burman and Central Composite Designs. In addition to nutritional optimization, the effect of γ-irradiation of the spores of P. polonicum on its Taxol producing potency was evaluated. The yield of Taxol by P. polonicum was increased via nutritional optimization by response surface methodology with Plackett-Burman and FCCD designs, and γ-irradiation by about 4.5 folds, comparing to the control culture.
The yield of Taxol was increased by about 1.2 folds (401.2 μg/l) by γ -irradiation of the isolates at 0.5−0.75 kGy, comparing to the control cultures (332.2 μg/l).
The highest Taxol yield was obtained by growing P. polonicum on modified Czapek’s- Dox medium (sucrose 40.0 g/l, malt extract 20.0 g/l, peptone 2.0 g/l, K2PO4 2.0 g/l, KCl 1.0 g/l, NaNO3 2.0 g/l, MgSO4. 5H2O 1.0 g/l) of pH 7.0 at 30.0 °C for 7.0 days. From the FCCD design, sucrose, malt extract and incubation time being the highest significant variables medium components affecting the Taxol production by P. polonicum.
1. Introduction
Taxol is one of the most commercial anticancer drugs, with broad spectrum towards different types of cancer cell [[1], [2], [3]], it has been approved by USA FDA in 1992. The activity of Taxol elaborates from its unique specificity for binding with tubulin β-subunits heterodimer, promoting tubulin polymerization, disrupting the mitotic division of tumor cells [4,5]. The powerful activity of Taxol has been emphasized towards breast, lung, head, neck, uterine cancers and advanced forms of Kaposi’s sarcoma, as reviewed in [6,7]. Taxol has been firstly isolated and characterized from the bark of Yew tree Taxus brevifolia [8]. However, the tiny yield and vulnerability of this plant to unpredicted fluctuation with the ecological conditions were the major challenges for this source [9,10]. Semisynthetic process using 10-decaetylbaccatin III (DAB) intermediates extracted from the needles of Taxus baccata is the present technology for commercial Taxol production, however, the strong heterogeneity of DAB yield with environmental conditions of T. baccata is the major additional challenge [11].
Recently, exploring the Taxol producing potency of endophytic fungi isolated from the different medicinal plants raising the hope for using this technique as novel platform for commercial Taxol production [6,12,13]. The rationality of endophytic fungi as unique approach elaborates from their natural physiological identities such as fast growth, cost effective process, independent on climatic changes, feasibility of genetic manipulation, metabolic engineering and mass production [14]. Endophytic fungi are a group of fungi that internally colonize the different plant tissues without causing any apparent symptoms to their host plants [12,13,15]. Endophytic fungi especially inhabiting medicinal plants have been considered as a repertoire of plethora of novel secondary metabolites with the main chemical scaffold of terpenoids, steroids, quinones and polyketides [6,12,[16], [17], [18]]. Massive number of plants from different families were used as source for isolation of endophytic fungi and their Taxol producing potency were screened and determined as reviewed by [6,12,16,19]. Although, the promising implementation of the endophytic fungi for Taxol production, however, their lower reproducible yield and drastic loss of Taxol productivity with repeated subculturing and storage is the main practical challenge that hamper the commercial anticipation of these fungal for Taxol production [20,6,12,19]. Thus, searching for novel fungal endophytes from different medicinal plant hosts with promising, sustainable Taxol yield is the ultimate objective.
Ginkgo biloba has a great ethnopharmacological relevance, such as antibacterial, antioxidant, and anticancer, and anticardiovascular activities [21]. Several endophytic fungal isolates of G. biloba were collected and identified as potential producers to various flavonoids, alkaloids, terpenoids and miscellaneous compounds with diverse biological activities [21]. However, isolation and characterization of fungal endophytes with Taxol producing potency from G. biloba as family other than Taxaceae was the objective of this study. With the argument about the Taxol biosynthetic machineries, if it is derived from Taxus brevifolia “Taxol producing plant” via horizontal gene transfer, or naturally present on the genome of Taxol producing endophytic fungi [22,23], isolation of endophytic fungi with Taxol producing potency from family outside Taxaceae could be participate on unraveling this hypothesis. Thus, isolation of Taxol producing endophytic fungi from G. biloba in addition to assessment of the Taxol biosynthetic dependency on the plant host was the objective of the current study.
2. Material and methods
2.1. Isolation of the endophytic fungi dwelling Ginkgo biloba
Different parts of Ginkgo biloba (leaves, barks and twigs) were collected from Orman Garden (Cairo, Egypt) in spring 2017, and used as source for isolation of endophytic fungi. The different plant parts were surface sterilized with 70 % ethanol for 2 min, then by 2.5 % sodium hypochlorite for 2 min, then rinsing with sterile distilled water. The surface sterilized plant parts were sectioned into small pieces with sterile sharp blade, then placed on the surface of potato dextrose agar (PDA) with ampicillin (1 μg/mL) [24], incubated for 7 days at 30 °C. The effectiveness of surface sterilization of the plant parts was assessed by centrifuging the rinsing water, then 500 μl sterile water was added to the precipitate and plated into potato dextrose agar [[25], [26], [27]]. The developed hyphae of the endophytic fungal colonies were purified by subculturing on PDA and stored as slope and plate cultures at 4 °C. Control plate media were used to check the sterility of working area, while, positive control of plant parts without sterilization were used to check the epiphytic fungal flora.
2.2. Morphological and molecular identification of the recovered endophytic fungi
The recovered fungal isolates were identified to their species levels based on their micro and micro-morphological features by growing on Czapek’s-Dox and PDA media for 10 days at 30 °C, according to the references key [28,29]. The highest potential Taxol producing fungal isolates were further molecularly identified based on the sequence of internal transcribed spacer (ITS) flanking the 5.8S region (ITS1-5.8S-ITS2 rDNA) [16,30,31]. Fungal genomic DNA (gDNA) was extracted by pulverizing the mycelia (∼0.2 g) in liquid nitrogen then adding 1 ml cetyltrimethylammonium bromide (CTAB) extraction buffer (2% CTAB, 2% PVP40, 0.2 % 2-mercaptoethanol, 20 mM EDTA, 1.4 M NaCl in 100 mM Tris-HCl, pH 8.0). The gDNA was used as template for PCR with the primer set ITS4 5′-GGAAGTAAAAGTCGTAACAAGG-3′ and ITS5 5′-TCCTCCGCTTATT GATATGC-3′. The PCR reaction contains 10 μl of 2 × PCR master mixture (i-Taq™, Cat. No. 25027, iNTRON Biotech.), 2 μl of fungal gDNA, 1 μl of each primer (10 pmol/μl), completed to 20 μl with sterile distilled water [32]. The PCR was programed to initial denaturation at 94 °C for 2 min, denaturation at 94 °C for 30 s, annealing at 55 °C for 10 s, extension at 72 °C for 30 s for 35 cycles, and final extension at 72 °C for 2 min. The PCR amplicon was analyzed by 1.5 % agarose gel in 1 × TBE buffer (Ambion Cat# AM9864), using DNA ladder (1 kb Nex-gene Ladder, Puregene, Cat. # PG010-55DI) and visualized by gel documentation system (Vilber Lourmat, France). The amplicons were purified and sequenced by Applied Biosystems Sequencer, HiSQV Bases, Version 6.0 using the same primers set. The obtained ITS sequences were BLAST searched non-redundantly on the NCBI database. For the multiple sequence alignments, the sequences were imported into MEGA 6.0 software and aligned with Clustal W muscle algorithm [33] and the phylogenetic tree was constructed with neighbor-joining method of MEGA 6.0 with 1000 bootstrap replication [34].
2.3. Screening for Taxol producing endophytic fungi, Taxol extraction and quantification
The recovered endophytic fungi inhabiting Ginkgo biloba were screened for Taxol production using potato dextrose broth (PDB) (Cat# DF0549-17-9) [22]. The fungal isolates were grown on PDA media for 7 days at 30 °C. One plug of each fungal isolate was inoculated into 100 mL of the PDB per 250 mL Erlenmeyer flasks, the cultures were incubated at 30 ± 1 °C under shaking conditions for 15 days. After incubation, the cultures were filtered by sterile cheesecloth, the filtrates were amended with 0.2 % sodium bicarbonate for fatty acids precipitation. Taxol has been extracted with double volume of dichloromethane (DCM), and the organic phase was collected, solvent was evaporated to dryness, and the residues were re-dissolved in 3 mL of methanol [13,27,35]. Taxol was fractionated and identified by TLC analysis using Merck 1 mm (20 × 20 cm) pre-coated silica gel plates (TLC Silica gel 60 F254, Merck KGaA, Darmstadt, Germany). After running, Taxol was detected by UV illumination at 254 nm (Min-UVIS, DESAGA, Heidelberg, Germany) [13,27]. From the TLC silica gel plates, the putative spots of Taxol were scraped-off and dissolved in DCM, vortex vigorously for 10 min and centrifuged at 1000 rpm for 5 min. The precipitated silica particles were removed and the supernatant was taken for Taxol quantification and purity checking by HPLC (Agilent Technology, G1315D) of C18 reverse phase column (Eclipse Plus C18 4.6 × 150 mm, 3.5 μm, Cat.# 959963-902). The mobile phase was methanol/ acetonitrile/water (25:35:40, v/v/v) at flow rate 1.0 mL/min for 20 min [36]. Taxol fractions were scanned from 200 to 400 nm by photodiode array detector (DAD), their chemical identity and concentrations were confirmed from the retention time and absorption peak area comparing to authentic sample (Sigma-Aldrich, Cat. # T7402).
2.4. Spectroscopic analysis for Taxol authentication
The UV spectroscopic analyses of Taxol samples were measured at λ 227 nm (RIGOL, Ultra-3000 Series) comparing to authentic Taxol [13,30,35,37]. Blank media was used as negative baseline for all spectrophotometric analyses.
FT-IR spectrum of the purified Taxol samples was analyzed by JASCO FT-IR 3600 Spectrophotometer. The purified Taxol sample was grinded with KBr pellets, pressed into discs under vacuum, comparing to authentic one. The absorption was measured in the region 500 to 4000 cm−1 [3]. The chemical structure of extracted Taxol was confirmed from the H and C NMR spectroscopy (JEOL, ECA-500II, 500 MHz NMR) comparing to authentic Taxol. The samples were dissolved in CDCl3, chemical shifts are given in ppm (δ-scale) and the coupling constants are expressed in hertz (Hz).
2.5. Bioprocess optimization of the selected fungal isolates to maximize their Taxol yield
The chemical components of Taxol production medium by selected fungal isolate were optimized to maximize their yield of Taxol, using the different designs of Response Surface Methodology such as Placket-Burman design and Faced Central Composite design by the Design-Expert 7.0 statistical software package (Stat Ease Inc., Minneapolis, USA). Each experiment was run in three biological replicates and the mean values were considered. After incubation at the desired conditions, fungal biomass was filtrated, and the Taxol was extracted, purified and quantified by TLC and HPLC comparing to authentic sample as described above.
2.5.1. Placket-Burman design
Placket-Burman design has been used frequently for optimization of media component for fungal growth and production of desired compounds [38]. The optimum conditions for production of Taxol have been assessed in silico based on the media components that have been used in qualitative and quantitative screening [39]. Eleven factors have been included namely; sucrose, sodium nitrate, dipotassium phosphate, magnesium sulfate, potassium chloride, temperature, pH, incubation time, shaking speed, malt extract and peptone. The parameters were varied over two levels, the minimum and maximum ranges were selected. The statistical Design-Expert 7.0 was used to generate a set of 12 experimental designs. For each experiment, Taxol production was expressed by μg/l, trials were carried out in triplicates, and the average of Taxol yield was considered. Regression analysis of the trial data was conducted using statistical software. The effect of each variable was calculated [40] using the following equation
Where, E is the effect of a testing variable, M+ and M- are taxol concentration of trials at the parameter of higher and lower levels, respectively, and N is the number of experiments was carried out. The effect of each variable on the production was determining by calculating their respective E-values.
Where Tot high is the total responses at the high level, Tot low is the total responses at the low level, and No is the number of trials.
2.5.2. Faced central composite design
The most significant positive independent variables affecting Taxol production by the selected fungal isolates were optimized using FCCD to determine the individual and mutual interactions among the tested variables. FCCD is a statistical experimental design, in which each variable is represented by three different levels low (-1), medium (0), and high (+1) [41].
The linear, quadratic and interaction coefficients, regression model was illustrated by the following second-ordered polynomial equation,
Y, is the predicted response, β0 is the regression coefficient, βi is the linear coefficient, βii is the quadratic coefficient, Xi is the coded level of independent variable.
To determine the optimal levels of the variables for Taxol production, three-dimensional (3D) response surface curves were plotted to study the interaction between the various factors, and to determine the variable condition of each factor for maximum Taxol production. 3D graphs were carried out by holding three factors constants in an ideal level and plotting the obtained response (Taxol conc.) for varying levels of the other two factors.
2.6. Gamma irradiation effect on Taxol yield by the target fungal isolate
The selected potent fungal isolates were γ-irradiated with 60Cobalt source (Gamma cell 4000-A-India) at different doses (0.25–3.0 kGy) comparing to the control non-irradiated cultures. The dose rate was 1.2 kGy/h at the time of the experiments. The irradiated spores were inoculated into the optimal media under standard cultural conditions, comparing to the non-irradiated spores as control. The cultures were incubated for 15 days at 30.0 °C on a rotary shaker (120.0 rpm), filtered, and Taxol was extracted, purified and quantified by TLC and HPLC as described above.
2.7. Antimicrobial activity of Taxol against different microorganism
The antimicrobial activity of extracted fungal Taxol was assessed against different types of wound infectious bacterial isolates namely; Bacillus subtilis and Staphylococcus epidermidis (Gram positive) and Pseudomonas aeruginosa, Escherichia coli and Enterobacter agglomerans (Gram negative), in addition to the antifungal activity against Candida albicans. These organisms were freshly streaked on nutrient agar medium and incubated at 37 °C for 24 h. After incubation, bacteria were suspended in sterile peptone water to obtained standard inocula of ∼ 0.5 McFarland (1–1.5) ×108 CFU/mL with UV–vis spectrophotometer (at 600 nm). The examined Candida albicans inocula were set at (1–3) ×106 CFU/mL. The growth inhibition (mm) of microbial pathogens was assessed by agar disc diffusion method. Sterile standard antibiotic disks with the diameter of 6.0 mm were used to positive controls. Sterile paper discs (6.0 mm) were loaded with 20 μl of methanol as negative control and amoxicillin clavulanic acid (AMC) as positive control. Discs were loaded with different concentration of Taxol (0.1, 0.5 and 1.0 μg/ml). Three biological replicates were prepared. The plates were incubated at 37 °C for 24 h, and the resulting zones of inhibition were measured. Amoxicillin clavulanic acid (AMC); 30 μg/mL and Nystatin 100 μg/mL (6.0 mm diameter) were used to normalize the antimicrobial activity of Taxol. The inhibition Zone of growth was determined by a Vernier caliper (mm).
2.8. Antiproliferative activity
The antiproliferative activity of purified fungal Taxol has been evaluated against liver carcinoma (HPG2) and breast carcinoma (MCF7) (VACERA Institute, Cairo, Egypt). The viability of tumor cells was assessed by MTT assay [42]. The 96-well plate was seeded with 103 cells/ well, incubated overnight, then amended with different concentrations of fungal Taxol and re-incubated for 24 h. The MTT dye (20 μl) was added to each well, incubated for 2 h, and the developed formazan complex was dissolved by 100 μL DMSO, and the purple color was measured at λ570 nm. The IC50 value was expressed by the Taxol concentration reducing the growth of 50 % of initial number of tumor cells, normalizing to negative control.
2.9. Statistical analyses
The experiments were conducted in three biological replicates, and the results were expressed by mean ± STDV. The significance was calculated by one-way ANOVA with Fisher’s Least Significant Difference of post hoc test (https://www.easycalculation.com/statistics/fishers-lsd-calculator.php).
2.10. Fungal deposition
The ITS sequence of Penicillium polonicum was deposited at genbank with accession #MK128497.1, as well as, the isolate was physically deposited at Assiut University Mycological Centre (AUMC), Egypt, with accession # AUMC14487.
3. Results
3.1. Endophytic fungi of Ginkgo biloba; morphological, molecular identification and Taxol producing potency
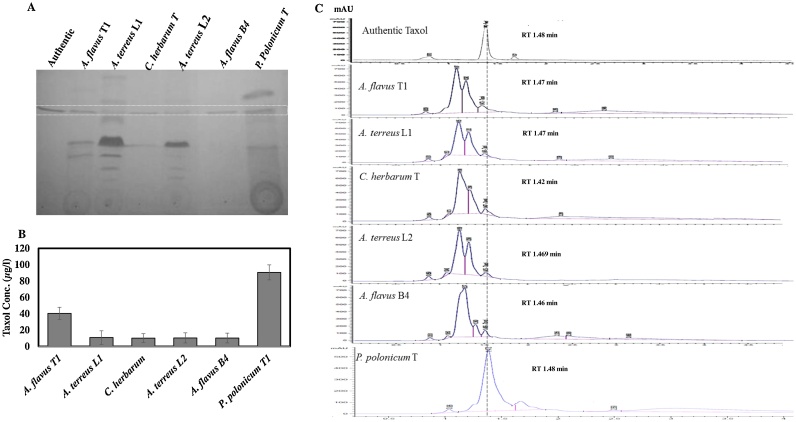
Twenty-eight endophyte fungal isolates were recovered from the barks, twigs and leaves of Ginkgo biloba on PDA agar medium (Table 1), these fungal isolates were allocated in twigs (8 fungal), barks (7 isolates) and leaves (13 f isolates) of plant. These fungal isolates were initially identified to the species level based on their morphological features according to the universal keys and were reported to belongs to four genera namely; Aspergillus, Penicillium, Fusarium and Cladosporium. Among these isolates, the prevalence of genus Aspergillus was reported to be > 75 %, followed by Fusarium that represented by 25 %, while Penicillium and Cladosporium were represented by about 7%. The genus Aspergillus was represented by five species namely; A. flavus (four isolates), A. oryzae (five isolates), A. niger (five isolates), A. fumigatus (three isolates), and A. terreus (four isolates). Two species of Penicillium (P. polonicum and P. egyptiacum) and one isolate of Cladosporium (C. herbarum) were recovered. The productivity of Taxol by the collected fungal isolates was assessed, by growing on PDB, incubation at the standard conditions, extraction and quantification of Taxol by TLC and HPLC. From the HPLC and TLC analyses (Fig. 1), the highest yield of Taxol has been reported by P. polonicum (90.53 μg/l), followed by A. flavus T1 (40.29 μg/l), while C. herabrum, A. terreus L1, A. terreus L4 and A. flavus L4 displayed a similar yield of Taxol (10.0–10.8 μg/l). However, the other fungal endophytes did not display any yield of Taxol based on the TLC and HPLC analyses. Among the recovered fungi, six isolates gave the highest potency for Taxol production, four of them were belong to genus Aspergillus (A. flavus T1, A. flavus L4, A. terreus L1, and A. terreus L4), in addition to P. polonicum and Cladosporium herbarum as revealed from the HPLC analysis. Aspergillus flavus T1, P. polonicum and C. harbarum have been isolated from the twigs, while, A. terreus L1, A. terreus L4 and A. flavus L4 have been isolated from leaves of G. biloba.
Table 1.
Screening for Taxol producing endophytic fungi of Ginkgo biloba.
| Plant parts | Isolate No. | Fungal Isolate | TLC | HPLC (μg/L) |
|---|---|---|---|---|
| Twigs Barks | 1 | Aspergillus flavusT1 | ++ | 40.29 |
| 2 | Penicillium polonicum T1 | +++ | 90.53 | |
| 3 | Aspergillus oryzae 1 | + | – | |
| 4 | Aspergillus oryzae 2 | – | – | |
| 5 | Cladosporium herbarum | ++ | 10.02 | |
| 6 | Aspergillus fumigatus T1 | + | – | |
| 7 | Fusarium sp. 2 | – | – | |
| 8 | Aspergillus niger4 | + | – | |
| Bark | 9 | Aspergillus oryzae1 | – | – |
| 10 | Aspergillus terreus1 | – | – | |
| 11 | Fusarium sp. 1 | – | – | |
| 12 | Aspergillus flavus3 | + | – | |
| 13 | Aspergillus terreus3 | – | – | |
| 14 | Fusarium sp. 4 | – | – | |
| 15 | Aspergillus niger | + | – | |
| Leaves | 16 | Aspergillus terreusL1 | ++ | 10.76 |
| 17 | Aspergillus niger1 | + | – | |
| 18 | Aspergillus fumigatus1 | – | – | |
| 19 | Aspergillus terreus L4 | ++ | 10.34 | |
| 20 | Fusarium sp. 3 | – | – | |
| 21 | Penicillium egyptiacum | – | – | |
| 22 | Aspergillus niger5 | – | – | |
| 23 | Aspergillus flavus2 | + | – | |
| 24 | Aspergillus oryzae3 | – | – | |
| 25 | Aspergillus niger2 | + | – | |
| 26 | Aspergillus flavus L4 | ++ | 10.19 | |
| 27 | Aspergillus fumigatus3 | + | – | |
| 28 | Aspergillus oryzae5 | – | – |
Fig. 1.
Chromatographic analysis of the extracted Taxol and PCR screening for Taxol rate-limiting enzymes from the potent fungal endophytes of P. gracilior. The 24 fungal isolates were grown on PDB under standard cultural conditions, Taxol was extracted and checked by UV-absorbance at λ 227 nm and TLC analysis comparing to authentic Taxol. The highest Taxol producers (six isolates) were selected for further HPLC analysis and PCR amplification of Taxol biosynthetic marker genes. A, TLC chromatogram of extracted Taxol from the six fungal isolates (1, 59, 108, 4, 14 and 20). B, PCR amplicon of 10-deacetylbaccatin III-O-acetyltransferase (dbat). C, Overall Taxol yield from the selected fugal isolates from HPLC analysis. D, HPLC chromatogram from the selected six fungal isolates comparing to authentic Taxol. The retention time of authentic Taxol was 6.2 min, 1 μg/mL was injected into HPLC column giving peak area 1817.2 mAU that used for calculating the corresponding.
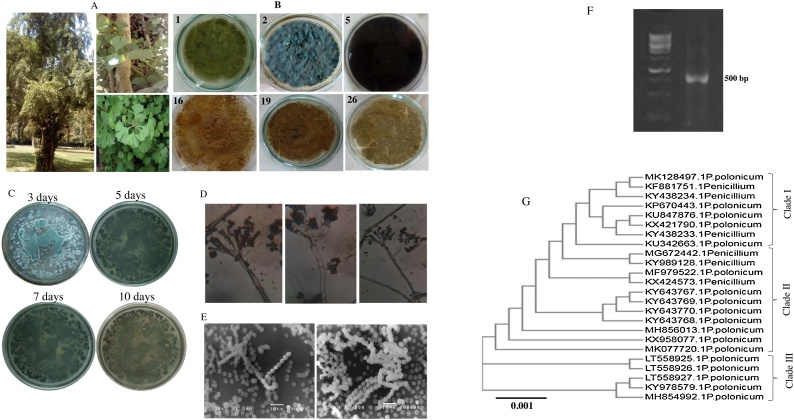
The morphological features of the potent six endophytic fungal isolates (A. flavus T1, P. polonicum, C. harbarum, A. terreus L1, A. terreus L4 and A. flavus L4) producing Taxol has been shown in Fig. 2. These fungal isolates were identified morphologically according to the universal morphological keys (Raper and Fennell, 1965; Pitt, 1980). P. polonicum growing optimally at 28 °C at Czapek’s-Dox agar, producing blue green conidia with a granular colony surface, with exudate droplets (Fig. 2). The conidiophores are terverticillate, slightly roughened on Czapek’s with definite phialides and metula, with smooth-walled and globose conidia (Frisvad, 1995) (Fig. 2). The colony reverse was cream yellow color pigment diffused into the agar medium. The microscopical and macroscopical features of this isolate typically follow the description of P. polonicum Westling by Frisvad and Samson [43]. Based on the Taxol yield, P. polonicum has been selected for further experimentation.
Fig. 2.
Taxol producing fungal endophytes of G. biloba. A; Morphological views of G. biloba twigs, cork and leaves. B; Potent Taxol producing endophytes; Aspergillus flavus (1), Penicillium polonicum (2), C. herbarum (5) from twigs; A. terreus 1 (16), A. terreus 2 (19) and A. flavus (26) from leaves. C; Microscopical views of isolate # 2 conidial heads (C) and conidiophores (D) at 1000x magnification by light microscope. Molecular identification of P. polonicum; PCR amplicon of ITS region of P. polonicum (E), normalizing to 250-10000bp ladder (Cat.#. SM0312), Phylogenetic analysis of P. polonicum by Maximum Likelihood method (F).
The morphological identification of P. polonicum has been validated by the molecular analysis of ITS region sequence (Cakir and Maden, 2015; Khalil et al., 209). Using gDNA as template, the PCR amplicon (∼550 bp) of P. polonicum was resolved, purified and sequenced. From the non-redundant blast search on NCBI database, the ITS sequence gave 99 % similarity with P. polonicum, with zero E values and 95 % query coverage. The isolate displayed 99 % similarity with P. polonicum isolates MH854992.1, NR103687.1, AF033475, MH862450.1, MH856411.1, NR119496.1, NR077206.1, JN942697.1, MH861768.1, MH856134.1, JN942742.1, NR138271.1, NR160219.1, MH862986.1 and MH856088.1 with zero E value and 95 % query coverage. The phylogenetic relatedness of P. polonicum with the database deposited isolates were constructed (Fig. 2). Based on the ITS sequence, three phylogenetic clades of P. polonicum were recovered with a strong sequence similarity as revealed from the root value 0.001, the target isolate belongs to P. polonicum clade I. Thus, from the microscopical and molecular analyses, the target isolate has been confirmed as P. polonicum, and deposited to genbank with accession number MK128497.1, as well as, the isolate has been deposited at Assiut University Mycological Centre (AUMC) under deposition # AUMC 14487.
3.2. Chemical identity, antimicrobial and anticancer activity of P. polonicum Taxol
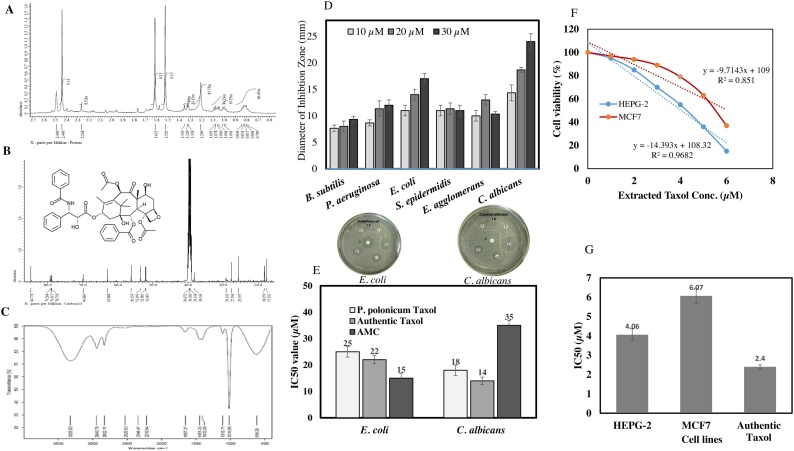
The chemical identity of P. polonicum Taxol has been validated by 1HNMR, 13CNMR, EDX and FT-IR analyses. After incubation of the fungal culture, Taxol was extracted by methylene chloride, initially checked by TLC and further purified by preparative TLC. Comparing to authentic Taxol, the putative Taxol samples were scrapped-off from the silica particles and further chemically analyzed. The resolved signals of 1HNMR and 13CNMR for P. polonicum extracted Taxol were identical to the standard Taxol, that distributed between 1.0 and 8.0 ppm (Fig. 3). Three proton signals were resolved at 1.0–3.0 ppm corresponding to methyl, acetate and acetylene groups, while the signals for aromatic moieties were resolved at 6.5–9.0 ppm. The FTIR spectra of P. polonicum Taxol were identical to authentic one. The purified Taxol had a representative peak at 3325.82 cm−1 that designated for the hydroxyl group (OH), peak at 2943.7 cm−1 that assigned to aliphatic CH stretch, peak at 1113.7 cm−1 corresponding to COO stretching frequency, and peak at 1019.8 cm−1 assigned for the aromatic C–H. The aliphatic C–H stretch was observed at 2943.7 cm−1, the C O stretch was positioned at 1657.3 cm−1, the peak at 1451.2 cm−1 due to NH stretching frequency. The carbonyl-oxygen stretching frequency was observed at 1413.1 and 1113.7 cm−1.
Fig. 3.
Chemical analysis and antiproliferative activity of extracted Taxol from P. polonicum. Taxol was extracted, fractionated by TLC. The putative spots of Taxol were scraped-off from the TLC plates and spectroscopically analyzed. The spectra of 1HNMR (A), 13C NMR (B) and FT-IR analysis (C) for the extracted Taxol of P. polonicum. D, Antimicrobial activity of extracted Taxol against B. subtilis, S. epidermidis, P. aeruginosa, E. coli, E. agglomerans and C. albicans. E, The IC50 values of extracted Taxol towards E. coli and C. albicans. Antiproliferative activity of P. polonicum Taxol against liver tumor cells (HEPG2) and breast adenocarcinoma (MCF7) (F) and their IC50 values (G).
The antiproliferative activity of purified Taxol from P. polonicum was evaluated towards liver carcinoma (HPG2) and breast carcinoma (MCF7). From the viability assessment, the purified P. polonicum Taxol had IC50 values 4.06 μM and 6.07 μM towards HEPG2 and MCF7, respectively (Fig. 3). The IC50 values of Taxol from different sources was summarized in Table 2.
Table 2.
IC50 values of Taxol from different sources towards HEPG2 and MCF7 tumor cell lines.
| Taxol Source | Tumor cell line | IC50 (μM) | Reference |
|---|---|---|---|
| Penicillium polonicum | HEPG2 | 4.06 | This study |
| MCF7 | 6.07 | ||
| Aspergillus terreus | HEPG2 | 4.6 | [7] |
| MCF7 | 5.4 | ||
| Aspergillus flavipes | HEPG2 | 6.0 | [13] |
| MCF7 | 7.4 | ||
| Taxus brevifolia | HEPG2 | 600 | [12] |
| MCF7 | 762 | ||
| HEPG2 | 100 | ||
| HEPG2 | 610 | ||
| MCF7 | 590 | ||
| MCF7 | 2.5 | ||
| HeLa | 2.6 | ||
| A549 | 4.1 | ||
| HT-29 | 2.8 | ||
| Nodulisporium sylviforme | MCF7 | 12 | [55] |
| Cladosporium oxysporum | HCT15 | 430 | [58] |
The antimicrobial activity of the extracted Taxol from P. polonicum has been evaluated against various multidrug resistant bacteria such as Bacillus subtilis, Pseudomonas aeruginosa, Escherichia coli, Staphylococcus epidermidis, Enterobacter agglomerans in addition to fungal pathogen Candida albicans by the disc diffusion assay. Authentic Taxol and AMC were used as positive controls. From the results (Fig. 3), the antimicrobial activity of Taxol was noticeably fluctuated among the tested microbial isolates. Taxol had a significant antibacterial activity against E. coli, unlike to the mild activity towards B. subtilis, P. aeruginosa, S. epidermidis and E. agglomerans. Obviously, the extracted Taxol displayed a significant antifungal activity against C. albicans in a concentration dependent manner as revealed from the visual observation and diameter of inhibition zone with Taxol concentration, comparing to authentic Taxol (Fig. 3). The IC50 values of P. polonicum extracted Taxol towards E. coli and C. albicans were 25 and 18 μM, while, it was 22 and 14 μm for authentic Taxol, respectively. While, the IC50 value for E. coli and C. albicans were 15 and 35 μM, respectively (Table 3).
Table 3.
Antimicrobial activity of Taxol against different pathogenic microorganisms represented by diameter of inhibition zone (mm).
| Authentic Taxol | AMC | P. polonicum Taxol | |
|---|---|---|---|
| Bacillus subtilis | 8.6 ± 0.5 | 8.0 ± 1.00 | 11.33 ± 0.577 |
| Pseudomonas aeruginosa | 8.6 ± 0.5 | 6.0 ± 0.00 | 14.00 ± 1.00 |
| Escherichia coli | 11.0 ± 1.0 | 22.0 ± 1.0 | 22.00 ± 1.00 |
| Staphylococcus epidermidis | 9.5 ± 0.5 | 26.0 ± 1.0 | 11.00 ± 0.00 |
| Enterobacter agglomerans | 9.0 ± 0.0 | 19.0 ± 1.0 | 10.33 ± 0.577 |
| Candida albicans | 9.3 ± 1.5 | 6.00 ± 0.00 | 24.00 ± 1.52 |
The concentration of authentic Taxol and P. polonicum Taxol was is 20 μM.
3.3. Optimization of bioprocess variables using Response Surface Methodology
3.3.1. Plackett-Burman design screening for critical factors
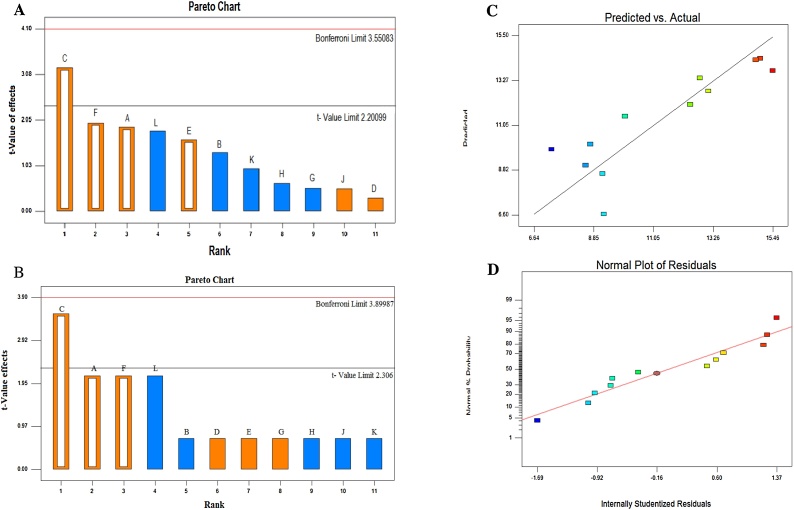
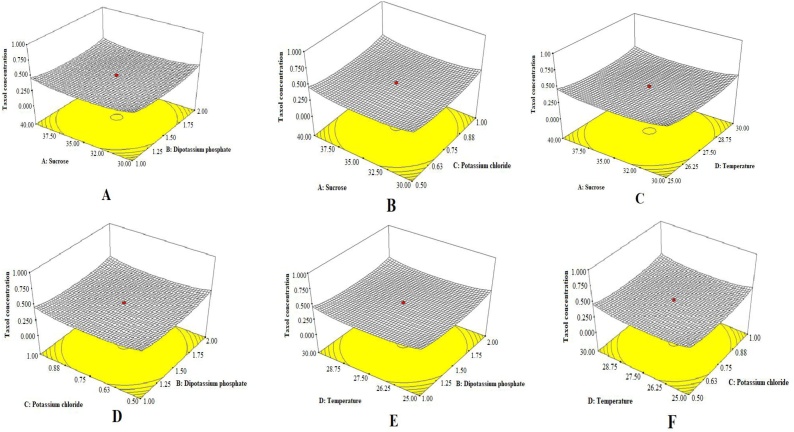
The media composition for Taxol production by P. polonicum were optimized by the statistical experimental design of Plackett-Burman. As revealed from the results (Fig. 4), sucrose, dipotassium phosphate, potassium chloride and temperature displayed a significant effect on Taxol production by P. polonicum. This design was used to draw up the media components and the cultural conditions that affect taxol production. The response was calculated as the concentration of Taxol by TLC and HPLC. All the experiments were performed in triplicates and the average concentration was considered. Taxol production response in the 12 runs experimental set-up was represented in Tables 4,5 . Statistical analysis of this design illustrates that the Model F-value of 5.16 implies their significant and Values of "p > F" fewer 0.050. The "Predicted R-Squared" of 0.2553 is in reasonable agreement with the "Adj. R- Squared" of 0.6018. "Adeq. Precision" measures the signal to noise ratio. A ratio greater than 4 is desirable. The "Adeq. Precision" ratio of 6.374 indicates an adequate signal [44]. This model can be used to navigate the design space as mentioned in Table 4. Based on parameter estimated and by applying multiple regression analysis on the experimental data, the test variables, and the response variable were relating by the following model equation: First-order model equation Terms of Coded Factors: Taxol Conc. =+1.81 + 0.31A+0.52C + 0.26E+0.32 F. The actual and predicted levels of Taxol by P. polonicum upon Plackett-Burman optimization bioprocess are summarized in Table 6 and Fig. 4. The highest actual (204 μg/l) and predicted (239 μg/l) yield of Taxol by P. polonicum upon factorial design by Plackett-Burman design was obtained at run # 9 at medium component; sucrose (40 g/l), malt extract (20 g/l), peptone (2 g/l), sodium nitrate (4 g/l), K2HPO4 (1 g/l), MgSO4 (1 g/l), KCl (1 g/l) of initial pH 6.0, and the cultures were incubated at 30 °C for 7 days at 120 rpm shaking speed. The effect of individual parameters studied in Plackett-Burman design was represented in Pareto Chart depicting the order of significance of variables involved in Taxol production (Fig. 4). Thus, the most significant variables affecting Taxol productivity by P. polonicum were dipotassium phosphate, temperature, and sucrose as revealed from the Plackett-Burman design.
Fig. 4.
Blackett-Burman design of the different variables for production of Taxol by P. polonicum. Pareto Charts showing the effect of individual factors on Taxol production where: C- Dipotassium phosphate, F- Temperature, A- Sucrose, L- Peptone, E- Potassium chloride, B- Sodium nitrate, K- Malt extract, H- Incubation time, G- pH, J- shaking speed, D- Magnesium sulfate (A, B). The normal probability plots of the variables for Taxol production by P. polonicum as determined by the first order polynomial equation. C, Plot of correlation between predicted and actual Taxol yield by P. polonicum. D, Normal probability plots of residuals.
Table 5.
Minimum and maximum ranges of the selected parameters in PBD for optimization of Taxol production.
| No. | Factors | Levels |
|
|---|---|---|---|
| Minimum (-1) | Maximum (1) | ||
| 1 | Sucrose (g /L) | 30.0 | 40.0 |
| 2 | Malt Extract (g /L) | 20.0 | 30.0 |
| 3 | Peptone (g /L) | 1.0 | 2.0 |
| 4 | Sodium nitrate (g /L) | 2.0 | 4.0 |
| 5 | Dipotassium phosphate (g /L) | 1.0 | 2.0 |
| 6 | Magnesium sulfate (g /L) | 0.5 | 1.0 |
| 7 | Potassium chloride (g /L) | 0.5 | 1.0 |
| 8 | Temperature | 25.0 | 30.0 |
| 9 | pH | 6.0 | 7.0 |
| 10 | Incubation time (days) | 7.0 | 12.0 |
| 11 | Shaking speed (rpm) | 120.0 | 150.0 |
Table 4.
Analysis of variance (ANOVA) for Taxol production.
| Source | Sum of Squares |
df | Mean Square | F Value | p-value Prob > F | Significance | R-Squared | Std. Dev. |
|---|---|---|---|---|---|---|---|---|
| Model | 6.40 | 4 | 1.60 | 5.16 | 0.0296 | Significant | 0.7466 | 0.56 |
| A- Sucrose. | 1.12 | 1 | 1.12 | 3.62 | 0.0989 | Significant | ||
| C-Dipotassium phosphate | 3.23 | 1 | 3.23 | 10.43 | 0.0145 | Significant | ||
| E- Potassium chloride | 0.81 | 1 | 0.81 | 2.60 | 0.1510 | Significant | ||
| F- Temperature | 1.24 | 1 | 1.24 | 3.98 | 0.0862 | Significant | ||
| Residual | 2.17 | 7 | 0.31 | |||||
| Cor Total | 8.57 | 11 |
Table 6.
Plackett-Burman design to evaluate factors affecting Taxol production by P. polonicum.
| Run | X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 | X9 | X10 | X11 | Taxol |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Experimental | Predicted | Residuals | ||||||||||||
| 1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 193.4 | 220.0 | −26.6 |
| 2 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 115.9 | 76.0 | 39.9 |
| 3 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | 126.6 | 100.0 | 26.6 |
| 4 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 165.7 | 171.0 | −5.3 |
| 5 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | 108.1 | 53.0 | 55.1 |
| 6 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | 197.0 | 225.0 | −28.0 |
| 7 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | 197.1 | 163.0 | 34.1 |
| 8 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | 69.7 | 84.0 | −14.3 |
| 9 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | −1 | −1 | −1 | 204.0 | 239.0 | −35.0 |
| 10 | 1 | 1 | 1 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 154.2 | 154.0 | 0.2 |
| 11 | −1 | −1 | −1 | 1 | −1 | 1 | 1 | −1 | 1 | 1 | 1 | 80.4 | 73.0 | 7.4 |
| 12 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | −1 | 30.6 | 85.0 | −54.4 |
3.3.2. Face Centered Central Composite Design (FCCD) bioprocess optimization
Face Centered Central Composite Design was adopted for the choice of optimal level of the significant variable that was identified based on the Plackett-Burman design. The most significant variables “dipotassium phosphate, potassium chloride, sucrose and temperature” have been selected for further optimization using FCCD design. The results of 30 runs of four variables with positive effects reveals the significant effect of the tested variables on the yield of Taxol by P. polonicum (Table 7). The yield of Taxol was increased by about 1.6 folds (332.08 μg/l) upon optimization with FCCD at the run #1 which the medium contains sucrose (40 g/l), dipotassium phosphate (2 g/l), potassium chloride (1 g/l), at incubation temperature 30 °C for 7 days incubation comparing to the basal medium at run #14 (204.1 μg/l). Upon FCCD optimization, the predicted Taxol yield was increased from 239 μg/l at run # 14 into 371.0 μg/l at run# 1, by about 1.55 folds increment. The three-dimensional response surface curve showing the effect of interactions of the most significant parameters; sucrose, dipotassium phosphate, potassium chloride and incubation temperature were shown in Fig. 5. The statistical analysis of the Linear Model design demonstrates that the F-value is 28.06 implying the significance of the model, and the values of "P > F" lower by about 0.050 reveals the significance of the model term. In this case sucrose, dipotassium phosphate, potassium chloride and temperature are significant model terms as shown in Table 8. The analysis revealed that the "Pred. R-Squared" of 0.7159 is in reasonable agreement with the "Adj. R-Squared" of 0.7887. Adeq. Precision" ratio of 19.736 measures the signal to noise ratio, as the ratio greater than four that being desirable, the Adeq. Precision ratio of 23.874 indicates the adequacy of signals. The results obtained from the CCD experiment were analyzed by ANOVA that yielded the following regression equation at the level of Taxol production: First-order model equation Terms of Coded Factors: Taxol Conc. =+2.17 + 0.28A+0.46B + 0.16C+0.24D.
Table 7.
Face Centered Central Composite design of the variables; sucrose, dipotassium phosphate, potassium chloride and temperature on the yield of Taxol by P. polonicum.
| Runs | Sucrose (g/L) |
Dipotassium phosphate (g/L) |
Potassium chloride (g/L) |
Temperature (°C) |
Taxol yield (μg/l) |
||
|---|---|---|---|---|---|---|---|
| Experimental | Predicted | Residuals | |||||
| 1 | 40 | 2 | 1 | 30 | 332.08 | 371.0 | −38.89 |
| 2 | 40 | 2 | 0.5 | 25 | 313.76 | 355.1 | −41.38 |
| 3 | 35 | 1.50 | 0.75 | 22.50 | 238.14 | 262.0 | −23.81 |
| 4 | 30 | 2 | 1 | 25 | 180.24 | 144.2 | 36.05 |
| 5 | 40 | 2 | 1 | 25 | 229.2 | 206.3 | 22.92 |
| 6 | 40 | 2 | 0.5 | 30 | 239.04 | 234.3 | 4.78 |
| 7 | 30 | 1 | 0.5 | 25 | 55.68 | 54.6 | 1.11 |
| 8 | 30 | 2 | 1 | 30 | 231.6 | 254.8 | −23.16 |
| 9 | 25 | 1.5 | 0.75 | 27.5 | 143.28 | 107.5 | 35.82 |
| 10 | 40 | 1 | 1 | 25 | 146.16 | 127.2 | 19.00 |
| 11 | 35 | 2.5 | 0.75 | 27.5 | 211.44 | 232.6 | −21.14 |
| 12 | 35 | 1.5 | 0.75 | 32.5 | 194.96 | 191.1 | 3.90 |
| 13 | 35 | 1.5 | 0.75 | 27.5 | 169.92 | 186.9 | −16.99 |
| 14 | 40 | 1 | 1 | 30 | 204.0 | 239.0 | −35.0 |
| 15 | 35 | 1.5 | 1.25 | 27.5 | 189.5 | 208.5 | −18.95 |
| 16 | 35 | 1.5 | 0.75 | 27.5 | 172.2 | 146.3 | 25.82 |
| 17 | 35 | 1.5 | 0.75 | 27.5 | 174.6 | 144.9 | 29.68 |
| 18 | 30 | 1 | 1 | 30 | 151.9 | 127.6 | 24.31 |
| 19 | 45 | 1.5 | 0.75 | 27.5 | 194.6 | 190.7 | 3.89 |
| 20 | 30 | 2 | 0.50 | 25 | 138.8 | 138.8 | 0.00 |
| 21 | 35 | 1.5 | 0.75 | 27.5 | 169.3 | 147.3 | 22.01 |
| 22 | 35 | 1.5 | 0.75 | 27.5 | 169.4 | 203.3 | −33.89 |
| 23 | 40 | 1 | 0.50 | 30 | 156.1 | 187.3 | −31.22 |
| 24 | 35 | 0.5 | 0.75 | 27.5 | 151.8 | 182.1 | −30.35 |
| 25 | 40 | 1 | 0.50 | 25 | 104.6 | 125.6 | −20.93 |
| 26 | 35 | 1.5 | 0.75 | 27.5 | 169.0 | 202.8 | −33.81 |
| 27 | 30 | 1 | 0.50 | 30 | 107.0 | 107.0 | 0.00 |
| 28 | 30 | 1 | 1 | 25 | 99.4 | 119.3 | −19.89 |
| 29 | 35 | 1.5 | 0.25 | 27.50 | 150.8 | 165.9 | −15.08 |
| 30 | 30 | 2 | 0.50 | 30 | 221.3 | 190.3 | 30.98 |
Fig. 5.
Three-dimensional response surface curve showing the effect of interactions of a) Sucrose and Dipotassium phosphate b) Sucrose and Potassium chloride c) Sucrose and Temperature d) Potassium chloride and Dipotassium phosphate e) Temperature and Dipotassium phosphate f) Temperature and Potassium chloride from the Plackett-Burman design.
Table 8.
ANOVA for response surface linear model for the experiments with CCD.
| Source | Sum of Squares |
df | Mean Square | F Value | p-value Prob > F | Significance | R-Squared | Std. Dev. |
|---|---|---|---|---|---|---|---|---|
| Model | 9.02 | 4 | 2.26 | 28.06 | < 0.0001 | Significant | 0.8178 | 0.28 |
| A-Sucrose | 1.88 | 1 | 1.88 | 23.37 | < 0.0001 | Significant | ||
| B-Dipotassium phosphate | 5.15 | 1 | 5.15 | 64.03 | < 0.0001 | Significant | ||
| C-Potassium chloride | 0.60 | 1 | 0.60 | 7.47 | 0.0114 | Significant | ||
| D-Temperature | 1.40 | 1 | 1.40 | 17.36 | 0.0003 | Significant | ||
| Residual | 2.01 | 25 | 0.080 | |||||
| Lack of Fit | 2.01 | 20 | 0.10 | 133.8 | < 0.0001 | Significant | ||
| Pure Error | 3.749 | 5 | 7.498 | |||||
| Cor Total | 11.03 | 29 |
3.4. Effect of Gamma irradiation and types of medium on Taxol production by P. polonicum
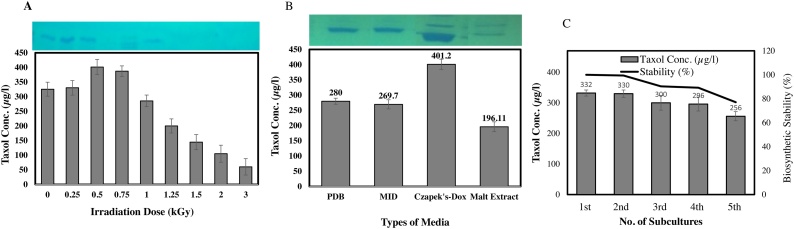
The yield of Taxol by P. polonicum has been further optimized with irradiation of the isolate with gamma-rays, in an endeavor to triggers the biosynthetic machinery of Taxol. The spores of P. polonicum was irradiated by gamma rays at different doses and grown on the optimized Czapek’s-Dox medium by response surface methodology with Plackett-Burman and FCCD designs. The cultures were incubated under standard conditions, and Taxol was extracted and quantified by TLC and HPLC. Noticeably, the yield of Taxol was increased by about 1.2 folds (401.2 μg/l) by γ -irradiation of the isolates at 0.5−0.75 kGy, comparing to the control cultures (332.2 μg/l) (Fig. 6). However, a significant reduction on the yield as well as fungal growth was observed upon further irradiation doses.
Fig. 6.
Effect of Gamma irradiation dose and types of media on Taxol yield by Penicillium polonicum. A, The fungal cultures were irradiated with different doses of gamma rays, Taxol was extracted and quantified. B, The isolate of P. polonicum was grown on different types of media regarding to the optimized Czapek’s-Dox medium as control, cultures were incubated at the desired conditions, and Taxol was extracted and quantified as described in Materials and Methods. The upper panel is the TLC plate with Taxol spots for each corresponding sample.
The influence of type of medium on Taxol production by P. polonicum has been assessed by growing the isolate on PDB, MID, modified Czapek’s-Dox and malt extract. The cultures were incubated, Taxol was extracted and quantified. From the results (Fig. 6), the highest Taxol yield by P. polonicum has recovered by growing on modified Czapek’s-Dox medium (401.2 μg/l), however, the yield of Taxol by the isolate upon growing on PDB, MID and Malt extract media was about 190−240 μg/l). Thus, upon nutritional optimization bioprocessing, and γ-irradiation, the yield of Taxol by P. polonicum has been strongly enhanced by about 4.4 folds (401.2 μg/l) comparing to the basal medium (90.5 μg/l).
3.5. Effect of subculturing of P. polonicum on its Taxol biosynthetic stability
The effect of subculturing of P. polonicum on their Taxol biosynthetic stability has been evaluated, by the growing the different generations on the modified Czapek’s-Dox media, incubated at standard conditions, Taxol was extracted and quantified as described above. From the results (Fig. 6), the biosynthetic stability of Taxol by P. polonicum was reduced by about 20 % at the fifth cultural generation, comparing to the original culture (zero culture).
4. Discussion
Fungal endophytes of with Taxol producing potency raised the hope for mass production of Taxol due to their fast growth, cost effective fermentation process, independence on climatic changes, resistance to shearing and feasibility of genetic manipulation [7,13,16,19,30,45,46]. However, the anticipation of fungi for industrial production of Taxol has been challenged by their lower reproducible yield and drastic loss of Taxol productivity with repeated subculturing [20,22]. Most of the endophytic Taxol-producing fungi were isolated from Taxus sp and Podocarpus sp which are belonging to family Taxaceae [6,12,20,22,35]. Exploring of the Taxol producing potency by endophytic fungi from plants outside Taxaceae family with sustainable Taxol productivity is the main objective by biotechnologists. Ginko biloba is one of the most traditionally recognized medicinal plants for its ethnopharmacological relevance such as anticancer activity, anti-inflammatory and antimicrobial activities [21], however, the Taxol producing potency of the endophytic fungi inhabiting this plant remain obscure. Thus, the objective of this study was to isolate and assess the Taxol producing potency of the endophytic fungi from G. biloba, as well as to assess the molecular biosynthetic stability of Taxol from the potent fungal isolate with storage and subcultures.
Twenty-eight endophyte fungal isolates were recovered from the different parts of Ginkgo biloba. Among the recovered fungi, the endophytic isolate P. polonicum gave the highest Taxol yield (90.53 μg/l), as authenticated from the TLC, UV- absorption and HPLC analysis. Similar screening paradigm for Taxol production has been reported for endophytes from Podocarpus gracilior [7] and other plants [13,16]. The Taxol yield by P. polonicum has been coincide with A. flavipes [13], A. terreus [27,37] an endophytes of P. gracilior, as well as for endophytes of Taxus spp such as A. candidus [47], Fusarium solani [48], A. niger [49] and A. fumigatus [50]. Consistently, the yield of Taxol by P. polonicum was coincident with A. flavipes “saprophytic isolate” [13]. The identity of P. polonicum the potent Taxol producing fungal isolate was confirmed from the molecular analysis of the ITS region, the sequence was deposited on Genbank with accession #MK128497.1, as well as at Assiut University Mycological Centre (AUMC) under deposition # AUMC14487. Similarly, the identity of P. polonicum was confirmed based on the sequence of ITS region (Khalil et al., 2019, Çakir and Maden, 2015). Visagie et al., 2014. The isolate had 99 % similarity with P. polonicum isolates MH854992, NR103687, AF033475, MH862450, MH856411, NR119496, NR077206, JN942697, MH861768, MH856134, JN942742, NR138271, with zero E. value and 95 % query coverage.
The chemical identity of P. polonicum Taxol has been validated by 1HNMR, 13CNMR, EDX and FT-IR analyses. The resolved signals of 1HNMR and 13CNMR for P. polonicum Taxol were identical to the standard Taxol, that distributed between 1.0 and 8.0 ppm. Three proton signals were resolved at 1.0–3.0 ppm corresponding to methyl, acetate and acetylene groups, while the signals for aromatic moieties were resolved at 6.5–9.0 ppm [35]. Consistently, for all Taxane scaffolds, signals for their side chains protons were resolved at 2.0–7.0 ppm, while those for benzoate (C2), phenyl (C3) and benzamide (C3) groups were resolved at 7.0 and 8.4 ppm [51,52]. The FTIR spectra of P. polonicum Taxol were identical to authentic one, represented by a peak at 3325.82 cm−1 for the hydroxyl group (OH), peak at 2943.7 cm−1 to the aliphatic CH stretch, peak at 1113.7 cm−1 to carboxyl stretching frequency, and peak at 1019.8 cm−1 for the aromatic C–H. The aliphatic C–H stretch was observed at 2943.7 cm−1, the C O stretch was positioned at 1657.3 cm−1, the peak at 1451.2 cm−1 due to NH stretching frequency.
From the antimicrobial profile, Taxol had a significant antifungal activity against C. albicans, with mild antibacterial activity against E. coli. The extracted Taxol displayed a significant antifungal activity against C. albicans in a concentration dependent manner. The IC50 values of P. polonicum extracted Taxol towards E. coli and C. albicans were 25 and 18 μM, while, it was 22 and 14 μm for authentic Taxol, respectively. The fungicidal activity of Taxol is one of the most characteristic biological features that has been observed towards a plethora of pathogenic fungi [53,23]. Taxol production by endophytes could be one of the main defense mechanisms of plant against different phytopathogens attacking the host plant. The antiproliferative activity of Taxol from P. polonicum has been evaluated towards various types of tumor including liver carcinoma (HPG2) and breast carcinoma (MCF7). From the viability assessment, P. polonicum Taxol had IC50 values 4.06 μM and 6.07 μM towards HEPG2 and MCF7, respectively. Similar IC50 values for Taxol purified from A. terreus [7], A. flavipes [13], N. sylviforme, C. oxysporum [51,54], T. brevifolia [47,55] towards HEPG2 and MCF7 tumor cells.
The Taxol yield by P. polonicum has been optimized by modification of the chemical components of Czapek’s-Dox as basal screening medium. The effect of eleven independent nutritional variables were assessed on productivity of Taxol by P. polonicum with the multiple response surface methodology. Plackett-Burman design and Centered Central Composite design have been used successfully for optimization of multiple variables by the microorganisms [44,56,57]. The predicted and actual yields of Taxol by P. polonicum as resolved from Plackett-Burman design were ranged from 30.6 and 239.0 μg/l, reflecting the significance of optimization process and efficiency of Plackett-Burman design. The highest actual Taxol yield by P. polonicum was 204.0 μg/l with the optimized variables sucrose (40 g/l), malt extract (20 g/l), peptone (2 g/l), NaNO3 (4 g/l), K2HPO4 (1 g/l), MgSO4.5H2O (1 g/l), KCl (1 g/l), temperature 30 °C, pH 6.0, and incubated for 7 days at 120 rpm shaking. From the normal probability plot, the points are arranged near the diagonal line, revealing the adequate independent distribution of the variables. Thus, the expected yield of Taxol by P. polonicum was fitted maximally with the experimental results. From the Plackett-Burman design, the most significant variables affecting Taxol yield were sucrose (p-value 0.0989), K2HPO4 (p-value 0.0145), KCl (p-value 0.151) and incubation temperature (p-value 0.0862). The highly significant variables affecting Taxol yield by P. polonicum was further optimized with the FCCD design, giving the maximum yield (332.08 μg/l) with 40 g/l sucrose, 2 g/LK2HPO4, 1 g/l KCl and 30 °C incubation temperature. From the FCCD, sucrose (p-value <0.0001) and K2HPO4 (p-value <0.0001) were the most significant variable affecting Taxol yield by P. polonicum as revealed from the highest f-value 23.37 and 64.03, respectively. Thus, using the surface response methodology bioprocessing with Plackett-Burman and FCCD designs, the yield of Taxol by P. polonicum was increased into 401.25 μg/l comparing to 90.53 μg/l of control, by about 4.5 folds increment.
In an endeavor to maximize the yield of Taxol by P. polonicum, the fungal spores were irradiated by γ-rays, and the spores were grown on the modified Czapek’s-Dox medium, Taxol was extracted and quantified. Upon γ-rays irradiation, the Taxol yield by P. polonicum was increased by about 1.2 folds comparing to the control cultures, suggesting the induction of biosynthetic gene cluster of Taxol. Similar results have been reported for Taxol biosynthesis by Fusarium maire and Nodulisporium sylviforme that increased by about 8.2 and 2.5 folds upon irradiation [58,6,12].
The molecular biosynthetic stability of Taxol by P. polonium has been evaluated with the fungal subculturing. The Taxol yield by P. polonicum was reduced by about 20 % upon subculturing till the 5th subcultural generation. Attenuation of the biosynthetic machinery of Taxol by the fungi is the common metabolic practical challenge that limits the anticipation of fungi to be an industrial platform for commercial production of Taxol [6,12,14,22]. Noticeably, the attenuation of the biosynthetic potency of Taxol from P. polonicum have the same pattern to other endophytic fungi isolated from family Taxaceae.
In conclusion, the endophytic fungi inhabiting G. biloba were recovered and identified to the species level based on their morphological and molecular traits. The isolate P. polonicum was recovered from twigs of G. biloba and recognized as potent Taxol producer, based on the metabolic and molecular biomarker for Taxol synthesis. The yield of Taxol by P. polonicum was maximized via nutritional optimization of the isolate by response surface methodology with Plackett-Burman and FCCD designs, in addition to γ-irradiation by about 4.5 folds, comparing to the control isolate. The biosynthetic potency of Taxol by P. polonicum is relatively more stable with the fungal multiple subculturing, comparing to other endophytes. Convincingly, the biosynthetic machinery of Taxol from endophytes inhabiting plants outside Taxaceae family, could be have an unique molecular regulators and expressing signals independent on their host plants.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgment
We appreciate the financial support from Academy of Scientific Research and Technology, Egypt, to Ashraf S.A. El-Sayed.
Contributor Information
Gamal M. El-Sherbiny, Email: gamalelsherbiny1970@yahoo.com.
Ashraf S. El-Sayed, Email: ash.elsayed@gmail.com.
References
- 1.Straubinger R.M., Sharma A., Murray M., Mayhew E. Novel Taxol formulations: taxol-containing liposomes. J. Natl. Cancer Inst. Monographs. 1993:69–78. [PubMed] [Google Scholar]
- 2.Brown D. In: Itokawa H., Lee K.-H., editors. Vol. 2003. 2003. Preclinical and clinical studies of the taxanes. (Taxus – The Genus Taxus). [Google Scholar]
- 3.Kumaran R.S., Muthumary J., Hur B.K. Production of taxol from Phyllosticta spinarum, an endophytic fungus of Cupressus sp. Eng. Life Sci. 2008;8:438–446. [Google Scholar]
- 4.Cragg G.M., Schepartz S.A., Suffness M., Grever M.R. The taxol supply crisis. New NCI policies for handling the large-scale production of novel natural product anticancer and anti-HIV agents. J. Nat. Prod. 1993;56:1657–1668. doi: 10.1021/np50100a001. [DOI] [PubMed] [Google Scholar]
- 5.Goldspiel B.R. Clinical overview of the taxanes. Pharmacotherapy. 1997;17:110S–125S. doi: 10.1002/j.1875-9114.1997.tb03813.x. [DOI] [PubMed] [Google Scholar]
- 6.El-Sayed A.S.A., Abdel-Ghany S.E., Ali G.S. Genome editing approaches: manipulating of lovastatin and taxol synthesis of filamentous fungi by CRISPR/Cas9 system. Appl. Microbiol. Biotechnol. 2017;101:3953–3976. doi: 10.1007/s00253-017-8263-z. [DOI] [PubMed] [Google Scholar]
- 7.El-Sayed A.S.A., Safan S., Mohamed N.Z., Shaban L., Ali G.S., Sitohy M.Z. Induction of Taxol biosynthesis by Aspergillus terreus, endophyte of Podocarpus gracilior Pilger, upon intimate interaction with the plant endogenous microbes. Process. Biochem. 2018;71:31–40. doi: 10.1016/j.procbio.2018.04.020. [DOI] [Google Scholar]
- 8.Wani M.C., Taylor H.L., Wall M.E., Coggon P., McPhail A.T. Plant antitumor agents. VI. The isolation and strcture of taxol, a novel antileukemic and antitumo agent from Taxus bretvifolia. J. Am. Chem. Soc. 1971:2325–2327. doi: 10.1021/ja00738a045. [DOI] [PubMed] [Google Scholar]
- 9.Malik S., Cusidó R.M., Mirjalili M.H., Moyano E., Palazón J., Bonfill M. Production of the anticancer drug taxol in Taxus baccata suspension cultures: a review. Process. Biochem. 2011;46:23–34. doi: 10.1016/j.procbio.2010.09.004. [DOI] [Google Scholar]
- 10.Wang C., Wang Y., Wang Y., Fan M., Luo F., Qian Z. Characterization, pharmacokinetics and disposition of novel nanoscale preparations of paclitaxel. Int. J. Pharm. 2011;414:251–259. doi: 10.1016/j.ijpharm.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Thomas P.A., Polwart A. Taxus baccata L. J. Ecol. 2003;91:489–524. doi: 10.1046/j.1365-2745.2003.00783.x. [DOI] [Google Scholar]
- 12.El-Sayed A.S.A., El-Sayed M.T., Rady A.M., Zein N., Enan G., Shindia A., El-Hefnawy S., Sitohy M., Sitohy B. Exploiting the biosynthetic potency of taxol from fungal endophytes of conifers plants; genome mining and metabolic manipulation. Molecules. 2020;25:3000. doi: 10.3390/molecules25133000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Sayed A.S.A., Ali D.M.I., Yassin M.A., Zayed R.A., Ali G.S. Sterol inhibitor “Fluconazole” enhance the Taxol yield and molecular expression of its encoding genes cluster from Aspergillus flavipes. Process. Biochem. 2019;76:55–67. [Google Scholar]
- 14.Stierle A., Strobel G., Stierle D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science. 1993;260:214–216. doi: 10.1126/science.8097061. [DOI] [PubMed] [Google Scholar]
- 15.El-Sayed A.S.A., Moustafa A.H., Hussein H.A., El-Sheikh A.A., El-Shafey S.N., Fathy N.A.M., Enan G.A. Potential insecticidal activity of Sarocladium strictum, an endophyte of Cynanchum acutum, against Spodoptera littoralis, a polyphagous insect pest. Biocatal. Agric. Biotechnol. 2020;24 doi: 10.1016/j.bcab.2020.101524. [DOI] [Google Scholar]
- 16.El-Sayed A.S.A., Akbar A., Iqrar I., Ali R., Norman D., Brennan M., Ali G.S. A glucanolytic Pseudomonas sp. associated with Smilax bona-nox L. displays strong activity against Phytophthora parasitica. Microbiol. Res. 2018;207:140–152. doi: 10.1016/j.micres.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 17.Gond S.K., Kharwar R.N., White J.F. Will fungi be the new source of the blockbuster drug taxol? Fungal Biol. Rev. 2014;28:77–84. [Google Scholar]
- 18.Vasundhara M., Kumar A., Reddy M.S. Molecular approaches to screen bioactive compounds from endophytic fungi. Front. Microbiol. 2016;7:1–12. doi: 10.3389/fmicb.2016.01774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Sayed A.S.A., Shindia A.A., AbouZaid A.A., Yassin A.M., Shad Ali G., Sitohy M.Z. Biochemical characterization of peptidylarginine deiminase-like orthologs from thermotolerant Emericella dentata and Aspergillus nidulans. Enzyme Microb. Technol. 2019;124:41–53. doi: 10.1016/j.enzmictec.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Staniek A., Woerdenbag H., Kayser O. Taxomyces andreanae: a presumed paclitaxel producer demystified? Planta Med. 2009;75:1561–1566. doi: 10.1055/s-0029-1186181. [DOI] [PubMed] [Google Scholar]
- 21.Yuan Z., Tian Y., He F., Zhou H. Endophytes from Ginkgo biloba and their secondary metabolites. Chin. Med. 2019;14(51):1–40. doi: 10.1186/s13020-019-0271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinig U., Scholz S., Jennewein S. Getting to the bottom of Taxol biosynthesis by fungi. Fungal Divers. 2013;60:161–170. doi: 10.1007/s13225-013-0228-7. [DOI] [Google Scholar]
- 23.Kusari S., Singh S., Jayabaskaran C. Rethinking production of Taxol® (paclitaxel) using endophyte biotechnology. Trends Biotechnol. 2014;32:304–311. doi: 10.1016/j.tibtech.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 24.El-Sayed A.S., Khalaf S.A., Aziz H.A. Characterization of homocysteine γ-lyase from submerged and solid cultures of Aspergillus fumigatus ASH (JX006238) J. Microbiol. Biotechnol. 2013;23:499–510. doi: 10.4014/jmb.1208.08070. [DOI] [PubMed] [Google Scholar]
- 25.Xiong Z.-Q., Yang Y.-Y., Zhao N., Wang Y. Diversity of endophytic fungi and screening of fungal paclitaxel producer from Anglojap yew, Taxus x media. BMC Microbiol. 2013;13:71. doi: 10.1186/1471-2180-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Sayed A.S.A., Ruff L.E., Ghany S.E.A., Ali G.S., Esener S. Molecular and spectroscopic characterization of Aspergillus flavipes and Pseudomonas putida L-methionine γ-lyase in vitro. Appl. Biochem. Biotechnol. 2017;181:1513–1532. doi: 10.1007/s12010-016-2299-x. [DOI] [PubMed] [Google Scholar]
- 27.El-Sayed A.S.A., Safan S., Mohamed N.Z., Shaban L., Ali G.S., Sitohy M.Z. Induction of Taxol biosynthesis by Aspergillus terreus, endophyte of Podocarpus gracilior Pilger, upon intimate interaction with the plant endogenous microbes. Process. Biochem. 2018;71:31–40. [Google Scholar]
- 28.Raper K.B., Fennell D.I. Williams and Wilkins; 1965. The Genus Aspergillus. [Google Scholar]
- 29.Domsch K., Gams W., Anderson T. Vol. 1. 1980. (Compendium of Soil Fungi). [Google Scholar]
- 30.El-Sayed A.S.A., Yassin M.A., Ali G.S. Transcriptional and proteomic profiling of Aspergillus flavipes in response to sulfur starvation. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Sayed A.S.A., Yassin M.A., Khalaf S.A., El-Batrik M., Ali G.S., Esener S. Biochemical and pharmacokinetic properties of PEGylated cystathionine γ-lyase from aspergillus carneus KF723837. J. Mol. Microbiol. Biotechnol. 2015;25:301–310. doi: 10.1159/000437331. [DOI] [PubMed] [Google Scholar]
- 32.Kebeish R., El-Sayed A., Fahmy H., Abdel-Ghany A. Molecular cloning, biochemical characterization, and antitumor properties of a novel L-asparaginase from Synechococcus elongatus PCC6803. Biochemistry (Moscow) 2016;81 doi: 10.1134/S000629791610014X. [DOI] [PubMed] [Google Scholar]
- 33.Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 35.El-Sayed A.S.A., Fathalla M., Yassin M.A., Zein N., Morsy S., Sitohy M., Sitohy B. Conjugation of Aspergillus flavipes taxol with porphyrin increases the anticancer activity of taxol and ameliorates its cytotoxic effects. Molecules. 2020;25:1–13. doi: 10.3390/molecules25020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nims E., Dubois C.P., Roberts S.C., Walker E.L. Expression profiling of genes involved in paclitaxel biosynthesis for targeted metabolic engineering. Metab. Eng. 2006;8:385–394. doi: 10.1016/j.ymben.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 37.El-Sayed A.S.A., Mohamed N.Z., Safan S., Yassin M.A., Shaban L., Shindia A.A., Shad Ali G., Sitohy M.Z. Restoring the Taxol biosynthetic machinery of Aspergillus terreus by Podocarpus gracilior Pilger microbiome, with retrieving the ribosome biogenesis proteins of WD40 superfamily. Sci. Rep. 2019;9:11534. doi: 10.1038/s41598-019-47816-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurme S.T., Surwase S.N., Jadhav J.P., Author C. 2020. Maximizing the L-DOPA Production Through Response Surface Methodology B.Y. Newly Isolated Bacterium Aeromonas sp. SNS. [DOI] [Google Scholar]
- 39.Sai-Ut S., Benjakul S., Sumpavapol P., Kishimura H. Optimization of gelatinolytic enzyme production by B. amyloliquefaciens sp. H11 through Plackett–Burman design and response surface methodology. Int. Aquat. Res. 2014;6:59. [Google Scholar]
- 40.Trust B. In: Biometrika Trust The Design of Optimum Multifactorial Experiments. Plackett R.L., Burman J.P., editors. Oxford University Press on behalf of Biometrika Trust Stable; 1946. [Google Scholar]
- 41.Elazazy M.S., El-Hamshary M., Sakr M., Al-Easa H.S. Plackett-Burman and Box-Behnken designs as chemometric tools for micro-determination of L-Ornithine. Spectrochimica Acta Part A: Mol. Biomol. Spectrosc. 2018;193:397–406. doi: 10.1016/j.saa.2017.12.044. [DOI] [PubMed] [Google Scholar]
- 42.Cory A.H., Owen T.C., Barltrop J.A., Cory J.G. Use of an aqueous soluble tetrazolium/ formazan assay for cell growth assays in culture. Cancer Commun. 1991:207–212. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
- 43.Frisvad J.C., Samson R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 2004;49:1–173. [Google Scholar]
- 44.El-Naggar N.E.A., Moawad H., El-Shweihy N.M., El-Ewasy S.M., Elsehemy I.A., Abdelwahed N.A.M. Process development for scale-up production of a therapeutic L-asparaginase by Streptomyces brollosae NEAE-115 from shake flasks to bioreactor. Sci. Rep. 2019;9:1–18. doi: 10.1038/s41598-019-49709-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Sayed A.S., Shindia A.A. Characterization and immobilization of purified Aspergillus flavipesl-methioninase: continuous production of methanethiol. J. Appl. Microbiol. 2011;111:54–69. doi: 10.1111/j.1365-2672.2011.05027.x. [DOI] [PubMed] [Google Scholar]
- 46.El-Sayed A.S.A., Hassan M.N., Nada H.M.S. Purification, immobilization, and biochemical characterization of l-arginine deiminase from thermophilic Aspergillus fumigatus KJ434941: anticancer activity in vitro. Biotechnol. Prog. 2015;31 doi: 10.1002/btpr.2045. [DOI] [PubMed] [Google Scholar]
- 47.P. Zhang, P.-P. Zhou, L.-J. Yu, An Endophytic Taxol-Producing Fungus from Taxus media, Cladosporium cladosporioides MD2, (n.d.). 10.1007/s00284-008-9270-1. [DOI] [PubMed]
- 48.Deng B.W., Liu K.H., Chen W.Q., Ding X.W., Xie X.C. Fusarium solani, Tax-3, a new endophytic taxol-producing fungus from Taxus chinensis. World J. Microbiol. Biotechnol. 2009;25:139–143. doi: 10.1007/s11274-008-9876-2. [DOI] [Google Scholar]
- 49.Zhao K., Ping W., Li Q., Hao S., Zhao L., Gao T., Zhou D. Aspergillus niger var. taxi, a new species variant of taxol-producing fungus isolated from Taxus cuspidata in China. J. Appl. Microbiol. 2009;107:1202–1207. doi: 10.1111/j.1365-2672.2009.04305.x. [DOI] [PubMed] [Google Scholar]
- 50.Sun D., Ran X., Wang J. [Isolation and identification of a taxol-producing endophytic fungus from Podocarpus] Acta Microbiol. Sin. 2008;48:589–595. [PubMed] [Google Scholar]
- 51.Chmurny G.N., Hilton B.D., Brobst S., Look S.A., Witherup K.M., Beutler J.A. 1H- and 13C-NMR Assignments for Taxol, 7-epi-Taxol, and Cephalomannine. J. Nat. Prod. 1992;55:414–423. doi: 10.1021/np50082a002. [DOI] [PubMed] [Google Scholar]
- 52.Zhang P., Zhou P.-P., Yu L.-J. An endophytic taxol-producing fungus from Taxus x media, Aspergillus candidus MD3. FEMS Microbiol Lett. 2009;293(2):155–159. doi: 10.1111/j.1574-6968.2009.01481.x. [DOI] [PubMed] [Google Scholar]
- 53.Kusari S., Lamshöft M., Spiteller M. Aspergillus fumigatus Fresenius, an endophytic fungus from Juniperus communis L. Horstmann as a novel source of the anticancer pro-drug deoxypodophyllotoxin. J. Appl. Microbiol. 2009;107:1019–1030. doi: 10.1111/j.1365-2672.2009.04285.x. [DOI] [PubMed] [Google Scholar]
- 54.Rangarajulu Senthil Kumaran R., Choi Y., Lee S., Jeon H., Jung H., Kim H. Isolation of taxol, an anticancer drug produced by the endophytic fungus, Phoma betae. Afr. J. Biotechnol. 2012;11:950–960. doi: 10.5897/AJB11.1937. [DOI] [Google Scholar]
- 55.Wang X., Wang C., Sun Y.-T., Sun C.-Z., Zhang Y., Wang X.-H., Zhao K. Taxol produced from endophytic Fungi Induces apoptosis in human breast, cervical and ovarian cancer cells. Asian Pacific J. Cancer Prev. 2015;16:125–131. doi: 10.7314/apjcp.2015.16.1.125. [DOI] [PubMed] [Google Scholar]
- 56.Cao W., Gong G., Liu X., Hu W., Li Z., Liu H., Li Y. 2014. Optimization of Epothilone B Production by Sorangium cellulosum Using Multiple Steps of the Response Surface Methodolog Y. [DOI] [Google Scholar]
- 57.Martorell M.M., Pajot H.F., Rovati J.I., Figueroa L.I.C. Optimization of culture medium composition for manganese peroxidase and tyrosinase production during Reactive Black 5 decolourization by the yeast Trichosporon akiyoshidainum. Yeast. 2012;29:137–144. doi: 10.1002/yea.2896. [DOI] [PubMed] [Google Scholar]
- 58.Zhou X., Zhu H., Liu L., Lin J., Tang K. A review: recent advances and future prospects of taxol-producing endophytic fungi. Appl. Microbiol. Biotechnol. 2010;86:1707–1717. doi: 10.1007/s00253-010-2546-y. [DOI] [PubMed] [Google Scholar]