Graphical abstract
Keywords: Pathogenic fungus, Pathogenicity, Fatty acid, Fungus-fungus interaction, Medicinal fungus
Abstract
Paecilomyces penicillatus is one of the pathogens of morels, which greatly affects the yield and quality of Morchella spp.. In the present study, we de novo assembled the genome sequence of the fungus P. penicillatus SAAS_ppe1. We analyzed the transcriptional profile of P. penicillatus SAAS_ppe1 infection of Morchella importuna at different stages (3 days and 6 days after infection) and the response of M. importuna using the transcriptome. The assembled genome sequence of P. penicillatus SAAS_ppe1 was 39.78 Mb in length (11 scaffolds; scaffold N50, 6.50 Mb), in which 99.7% of the expected genes were detected. A total of 7.48% and 19.83% clean transcriptional reads from the infected sites were mapped to the P. penicillatus genome at the early and late stages of infection, respectively. There were 3,943 genes differently expressed in P. penicillatus at different stages of infection, of which 24 genes had increased expression with the infection and infection stage, including diphthamide biosynthesis, aldehyde reductase, and NAD (P)H-hydrate epimerase (P < 0.05). Several genes had variable expression trends at different stages of infection, indicating P. penicillatus had diverse regulation patterns to infect M. importuna. GO function, involving cellular components, and KEGG pathways, involving glycerolipid metabolism, and plant-pathogen interaction were significantly enriched during infection by P. penicillatus. The expression of ten genes in M. importuna increased during the infection and infection stage, and these may regulate the response of M. importuna to P. penicillatus infection. This is the first comprehensive study on P. penicillatus infection mechanism and M. importuna response mechanism, which will lay a foundation for understanding the fungus-fungus interactions, gene functions, and variety breeding of pathogenic and edible fungi.
1. Introduction
The genus Paecilomyces was established by Bainier (1907) and was initially considered to be similar to Penicillium in morphology and physiology. Luangsa-Ard et al. [1] demonstrated that Paecilomyces was polyphyletic across two subclasses (Sordariomycetidae and Eurotiomycetidae) based on phylogenetic analysis of the 18S rRNA sequence. Inglis and Tigano [2] supported the polyphyly interpretation of the genus through phylogenetic analysis of the 5.8S rRNA and internal transcribed spacer (ITS1 and ITS2) sequences from selected Paecilomyces species. The diverse species of Paecilomyces inhabit a wide range of environments. Some Paecilomyces species are human pathogens that can cause onychomycosis [3], pneumonia [4], and rhinosinusitis [5]. Other Paecilomyces species cause diseases of apple [6], pistachio [7], and other trees [8]. P. hepiali is an endoparasitic fungus of Cordyceps sinensis and has various pharmacological activities [9], [10].
P. penicillatus (white mold) is a pathogen of morels (Pezizomycetes, Ascomycota) [11], which are valuable edible fungi [12], [13], [14]. Morel cultivation is important in China [12], [15], [16] and is a major income source for many growers. However, white mold is a serious disease affecting the cultivation of Morchella spp. [11]. White mold reduces production and quality of morels during cultivation, storage, and transportation [11]. White mold is highly adaptable, and has the ability of rapid colonization and transmission [11], [17], which made it difficult to control without affecting morel production. The pathogenic mechanism of P. penicillatus and the related resistance mechanism of morels have not been fully understood [17].
The availability of high-throughput DNA sequencing platforms, including Roche 454, SOLiD, Illumina, Ion Torrent, and PacBio, can aid the study of genomic features [18], evolution, environmental adaptation, pathogenicity [19], and secondary metabolite synthesis of pathogenic fungi. The genomes of four Paecilomyces species (P. penicillatus CCMJ2836 [17], P. variotii CBS144490 HYG1 [20], P. variotii CBS 101075 [20], and P. niveus CO7 [21]) have been published. However, different fungal strains varied greatly in genome size, gene number and gene sequence. In order to more accurately analyze the pathogenic mechanism of P. penicillatus and the resistance mechanism of Morchella spp., we further sequenced and assembled the P. penicillatus genome based on two sequencing platforms.
We sequenced the genome of P. penicillatus using Illumina and PacBio technologies. PacBio sequencing produces long read lengths (>10 Kb) and greatly improves the continuity of genome assembly. Transcription profiles of P. penicillatus infecting Morchella importuna (an important cultivated morel species in China) at different stages were detected by transcriptome sequencing to reveal the pathogenic mechanisms of Paecilomyces and the resistance mechanisms of M. importuna. This study is the first report on pathogenic mechanism of Paecilomyces and resistance mechanism of Morels from the transcriptional level, which will promote our understanding of the genomic feature, fungi-fungi interaction, pathogenicity, and genome evolution of P. penicillatus and M. importuna.
2. Materials and methods
2.1. Fungal strains and experimental treatment
P. penicillatus mycelia were obtained from the Institute of Plant Protection, Sichuan Academy of Agricultural Sciences (No. SAAS_ppe1) [22]. They were isolated from the fruiting bodies of M. importuna with white mold disease and verified by Koch’s law. The P. penicillatus mycelia were cultured on potato dextrose agar medium (PDA, potato, 200 g/L; glucose, 20 g/L; agar, 15 g/L) at 28 °C for 7 d. Morel cultivation strains were purchased from a local company in Chengdu. M. importuna was cultivated in a field in Pujiang, Chengdu, Sichuan province. When the fruiting bodies of M. importuna reached 7 cm in height, we inoculated them with cultured P. penicillatus mycelia. Four groups of samples were collected for transcriptome analysis, and each group contained three replicates (Fig. 1). The groups were pure cultured P. penicillatus mycelia in PDA (Ppe); healthy fruiting bodies of M. importuna (Mim); M. importuna pathogenic sites (1 cm2) infected by P. penicillatus for 3 days (early stage of infection) (MP1); M. importuna pathogenic sites (1 cm2) infected by P. penicillatus for 6 days (late stage of infection) (MP2).
Fig. 1.
Samples of Paecilomyces penicillatus and Morchella importuna collected in this study. a, pure cultured P. penicillatus mycelia in PDA medium; b, healthy fruiting body of M. importuna; c, M. importuna fruiting bodies infected by P. penicillatus for 3 days; d, M. importuna fruiting bodies infected by P. penicillatus for 6 days; e, field pictures of M. importuna infected by P. penicillatus; The white arrows in c and d show the disease sites, which were collected for transcriptome analysis.
2.2. DNA and RNA extraction
Pure cultured P. penicillatus mycelia were harvested for DNA extraction. Genome DNA of P. penicillatus was extracted using a fungal DNA kit (Omega Bio-Tek, Norcross, GA, USA) according to manufacturer’s instructions. The extracted DNA was detected using 1% agarose gel electrophoresis. DNA quality was evaluated using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), with the optical density ratio of 260/280 nm and 260/230 nm greater than 1.8 and 2.0, respectively. Samples from the four groups (Ppe, Mim, MP1, and MP2) were collected and frozen in liquid nitrogen for RNA extraction. Total RNA was extracted using TransZol reagent (TransGen Biotech, Beijing, China) according to manufacturer’s instructions. We used the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA) to evaluate RNA quality. Truseq Stranded Total RNA Sample Preparation Kit (Illumina) was used to prepare paired-end libraries for each RNA-seq sample. The obtained libraries were subjected to quality control steps and then submitted to the Illumina HiSeq 2500 platform for sequencing (Personal Biotechnology, Shanghai).
2.3. Genome sequencing and assembly
We constructed two different gDNA libraries for the Illumina HiSeq system and PacBio system. Illumina sequencing libraries were constructed using a library construction kit (Illumina, San Diego, CA, USA) with 400 bp inserts. The PacBio libraries were constructed according to the manufacturers’ instructions of the PacBio system with a 20-kb-insert size. Generated gDNA libraries were subjected to Illumina Hiseq and PacBio RS II platforms for genomic sequencing by Personal Biotechnology (Shanghai, China). Long reads from the PacBio platform were used to de novo assemble the genome. Short reads from the Illumina platform were used to conduct a genome survey and base level correction after the assembly [23]. We used the Illumina sequencing data to estimate genome size with the K-mer-based method. The frequency of each K-mer from the short sequencing data was calculated using Jellyfish (v2.1.3) [24]. FALCON package [25] was used to assemble the long sequencing reads generated from the PacBio RS II platform into contigs. Using the PacBio long reads, we polished the assembled genome sequences through two rounds of polishing with Quiver. Pilon [26] was used to perform genome-wide base-level correction with the Illumina short sequencing reads. Then, we used the GapCloser program [27] to fill gaps and obtained a final draft genome.
2.4. Repetitive element and non-coding RNA annotation
Both homologous comparison and ab initio prediction were applied to annotate repeat elements in the P. penicillatus genome [28]. For homologous repeat annotation, RepeatMasker v4.0.5 [29] and RepeatProteinMask [29] were used for known repeat element types by searching against the Repbase database [30]. For ab initio repeat annotation, LTR_FINDER [31], RepeatScout [32], and RepeatModeler v1.0.4 (http://repeatmasker.org/RepeatModeler/) were used to construct a de novo repetitive element database, and the RepeatMasker [29] (http://repeatmasker.org/RMDownload.html) was used to annotate repeat elements with the database. The tRNA and rRNA genes in the P. penicillatus genome were predicted using tRNAscan-SE v1.3.1 [33] and RNAmmer v1.2 [34], respectively. Other non-coding RNAs in the P. penicillatus genome were annotated by the internal tool using Rfam database [35].
2.5. Protein-coding gene prediction and functional annotation
Exonerate software [36] was used to predict protein-coding genes in the P. penicillatus genome using protein sequences from closely related species. Augustus [37], GlimmerHMM [38], and SNAP [39] were used for ab initio prediction with the optimized parameters that were trained using high-quality proteins derived from the PASA gene models [40]. We combined gene models predicted by the two methods using EvidenceModeler [40]. Carbohydrate-active enzymes in the P. penicillatus genome were annotated using hmmscan v3.1b2 [41] against the Carbohydrate-Active enzymes Database [42] (http://www.cazy.org) with an E-value threshold of 1E-5. Public biological function databases of SwissProt [43], evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) [44], NR from NCBI, Gene Ontology (GO) [45], and Kyoto Encyclopedia of Genes and Genomes (KEGG) [46] were used for functional annotation of the predicted genes. Secondary metabolite encoding gene clusters were predicted using AntiSmash [47].
2.6. Analyses of RNA-Seq data
Data qualities were checked by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We used the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) program to filter out rRNA reads, short-fragment reads, sequencing adapters, and other low-quality reads from raw reads of the sequences. We mapped the remaining clean reads onto the P. penicillatus and M. importuna genomes using Bowtie2 software [48] based on a local alignment algorithm. We judge whether the RNA reads belongs to M. importuna or P. penicillatus according to the similarities between the reads and corresponding genome sequences. The expression levels were normalized by calculating the fragments per kilobase million reads (FPKM) value. DESeq R package (version 1.22.0) was used to calculate differentially expressed genes (DEGs). The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 as detected by DESeq were designated as differentially expressed [49]. GO enrichment analysis of DEGs was implemented by the GOseq R packages that were based on a Wallenius non-central hyper-geometric distribution [50]. KOBAS [51] was used to test the statistical significance of enrichment of DEGs in KEGG pathways [52].
2.7. Gene expression validation by RT-qPCR
To validate the gene expression calculated by transcriptome, we conducted RT-qPCR analysis of 19 genes identified by RNA-Seq. About 2 μg of total RNA from each sample was reverse-transcribed by reverse transcriptase M−MLV (TaKaRa). We designed gene-specific primers based on the gene sequences and used the 5.8S rRNA genes as endogenous loading controls for testing the validity of the template preparation [53]. At least three rounds of independent RT-PCR reactions were conducted to confirm the expression of each gene.
2.8. Data availability
The Illumina short-read sequencing data have been deposited to NCBI Sequence Read Archive (SRA) (Data Citation 1: NCBI Sequence Read Archive SRR9051197). The PacBio long-read sequencing data have been deposited to NCBI Sequence Read Archive (SRA) (Data Citation 2: NCBI Sequence Read Archive SRR9051168). The transcriptome data are available through the NCBI SRA (Data Citation 3: NCBI Sequence Read Archive SRR9682527-SRR9682529). The assembled genome version is available at GenBank (Data Citation 4: GenBank VCCP00000000.1). The annotation gff3 file of the assembled genome is available at Figshare (Data Citation 5: Figshare https://doi.org/10.6084/m9.figshare.8146202.v3). The reference genome of M. importuna can be found in JGI database (https://mycocosm.jgi.doe.gov/Morimp1/Morimp1.home.html) [16].
3. Results
3.1. Genome assembly of P. penicillatus
After removing adaptors in the raw reads, we obtained about 7.70 Gb (coverage of 187.65×) long sequencing subreads from the PacBio platform. The subreads N50 and the longest subreads were 10.64 kb and 63.81 kb, respectively. To improve the accuracy of reads for the P. penicillatus genome, approximately 4.05 Gb short sequencing reads were generated from the Illumina platform using paired-end sequencing libraries with insert lengths of 400 bp. Reads with residual adapter sequences and low-quality values (<20) were filtered out. Approximately 3.75 Gb (coverage of 91.25 × ) clean reads were obtained for the k-mer analysis and base correction of the genome (Fig. S1). We used the GapCloser program [27] to fill gaps and obtained a final 39.78 Mb draft genome. The P. penicillatus draft genome contained 11 scaffolds with a scaffold N50 length of 6.50 Mb (Table 1). All 11 scaffolds were over 10 kb, with the longest scaffold being 7.29 Mb. The GC content of the P. penicillatus genome was 44.19%, which was the lowest among the five Paecilomyces genomes. The single copy orthologs (BUSCO, version 3.0) [54] were used to evaluate the completeness of the assembled genome, and 99.7% of the 290 fungal BUSCO genes were identified in the final assembly. This indicated that the assembly of the P. penicillatus genome was of a high quality.
Table 1.
Assembly information of five Paecilomyces genomes.
| Species |
|
P.penicillatus | Paecilomyces variotii | Paecilomyces variotii |
|
|---|---|---|---|---|---|
| Strain | SAAS_ppe1 | CCMJ2836 | CBS144490 HYG1 | CBS 101,075 | CO7 |
| Data storage database | GenBank | GenBank | JGI | JGI | JGI |
| Sequencing technology | PacBio RS II + Illumina HiSeq | PacBio Sequel | Illumina HiSeq | PacBio | MySeq |
| Genome size (Mbp) | 39.78 | 40.08 | 32.37 | 30.11 | 36.02 |
| BUSCO | 99.7% | 97.20% | 99.0% | 99.6% | |
| GC content (%) | 44.19 | 44.70 | 45.07 | 46.71 | 47.11 |
| Sequencing read coverage depth | 279x | 105 x | 1.0 x | 127.61x | 104x |
| Contigs | 11 | 51 | 158 | 86 | 586 |
| Scaffolds | 11 | 51 | 126 | 86 | 586 |
| Scaffold N50 (kb) | 6,504 | 2,595 | 642.74 | 1,732 | 185 |
| The largest Scaffolds (Mbp) | 7.29 | 4.09 | 2.10 | 4.97 | 0.71 |
| Predicted gene models | 8,619 | 9,454 | 9230 | 9270 | 10,584 |
| Carbohydrate-active enzymes | 385 | 299 | 322 | 319 | 435 |
3.2. Annotation of the P. penicillatus genome
A total of 8,619 protein-coding genes (PCGs) with a mean of 2.6 exons per gene were annotated in the P. penicillatus genome based on de novo prediction and a homology-based search (Table 1). The average length of PCGs identified in the P. penicillatus genome was 1,593 bp. There were 8,056 (96.48%) PCGs similar to the sequences in the NCBI nr database, 7,844 (93.94%) homologous to sequences in eggNOG, 5,759 (68.97%) mapped to GO, 5,089 (60.95%) classified in Swiss-Prot, and 3,276 (39.23%) assigned to KEGG (Fig. S2). We identified 50 secondary metabolite encoding gene clusters in the P. penicillatus genome using AntiSmash. A total of 6.07 Mb repetitive elements were identified in the P. penicillatus genome, accounting for 15.26% of the whole genome. A total of 574 tRNA and 143 rRNA genes were annotated in the P. penicillatus genome. We also predicted 32 other ncRNAs in the P. penicillatus genome. Comparative genomic analysis showed that the assembly of the P. penicillatus SAAS_ppe1 genome had a longer scaffold N50 value than the other four Paecilomyces genomes. All scaffolds were longer than 10 kb, indicating that the assembly of P. penicillatus SAAS_ppe1 was more complete than that of the other three Paecilomyces genomes (Table 1). P. penicillatus SAAS_ppe1 had a larger genome size than the other two Paecilomyces species (one species has two strains) but smaller than P. penicillatus CCMJ2836. The genome of P. penicillatus SAAS_ppe1 had the least number of gene models (Table 1). Carbohydrate-active genes in the P. penicillatus SAAS_ppe1 genome were more than that in P. penicillatus CCMJ2836, P. variotii CBS144490 HYG1, and P. variotii CBS 101,075 genomes, but less than that in P. niveus CO7 genome. However, the genes related to auxiliary activities (AAs), carbohydrate esterases (CEs), and polysaccharide lyases (PLs) were the most in the P. penicillatus SAAS_ppe1 genome among the five Paecilomyces genomes detected (Fig. S3).
3.3. Overview of the RNA-Seq data
To study the transcriptional regulation mechanism of P. penicillatus during infection of M. importuna, as well as the transcriptional response of M. importuna to infection by P. penicillatus, we detected the expression profiles of pure cultured P. penicillatus mycelia, healthy M. importuna fruiting bodies, and infected samples at different stages using an RNA-Seq technique. An average of 9.08 Gb of raw data (with an average of 60,135,493 raw reads) was generated for each sample, with a Q20 and Q30 of 97.43% and 93.50%, respectively (Table S1). After quality control, we generated an average of 9.01 Gb clean data (with an average of 59,820,429 clean reads) from the raw data; about 0.53% of the raw reads were discarded. All the clean reads were mapped to a corresponding reference genome. The average mapped rates were 78.39% and 95.49% of the samples Mim and Ppe to corresponding genomes, respectively. A total of 86.34% and 74.49% clean reads from the infected sites were mapped to the genome of M. importuna at early and late stages of infection, respectively. However, 7.48% and 19.83% clean reads from the infected sites were mapped to the P. penicillatus genome at early and late stages of infection, respectively. An average of 5.93% clean reads from the infected sites with mixed M. importuna fruiting bodies and P. penicillatus mycelia were not mapped to the two reference genomes.
3.4. Genes up-regulated or down-regulated with infection stage in P. penicillatus
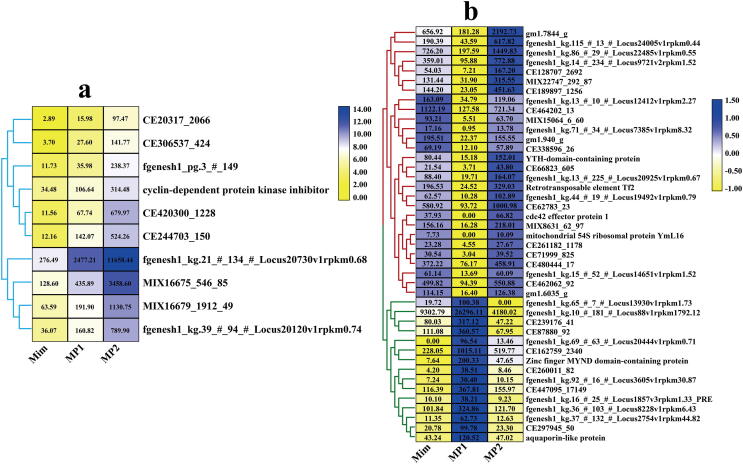
Of the 8,466 genes expressed in pure culture and infected mycelia of P. penicillatus, 3,943 genes were differently expressed between the P. penicillatus samples at different stages of infection (Fig. 2). Of the 3,943 differently expressed genes, the expression of 24 genes increased with the infection and infection stage (Fig. 3a), and they included diphthamide biosynthesis, aldehyde reductase, NAD(P)H-hydrate epimerase, and GPI anchored protein (P < 0.05). A total of 25 genes were down-regulated with the infection and infection stage (P < 0.05). The down-regulated genes were related to NAD-dependent epimerase/dehydratase, glyoxalase-like domain-containing protein, putative histidine--tRNA ligase, and acetyltransferase (Fig. 3b).
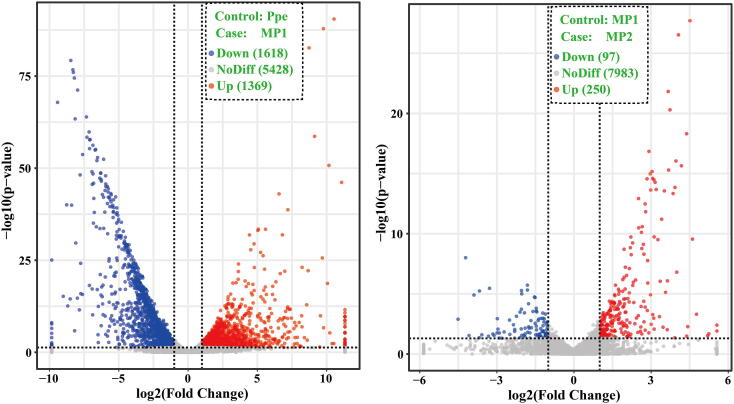
Fig. 2.
Volcano plot of differentially expressed genes (DEGs) between samples (M. importuna). The X-axis represents the fold change of DEGs in different experimental groups; -log10 (p-value) in the Y-axis represents DEGs that were more significant as the value increased. The dots represent the genes; the grey dots indicate non-differentially expressed genes; the red dots represent up-regulated differentially expressed genes; the blue dots represent down-regulated differentially expressed genes. Ppe, pure cultured P. penicillatus mycelia in PDA medium; MP1, M. importuna fruiting bodies infected by P. penicillatus for 3 days; MP2, M. importuna fruiting bodies infected by P. penicillatus for 6 days. Each treatment had three biological replicates. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Genes up- (a) or down- (b) regulated with infection and infection stages in P. penicillatus. The black number in the color block indicates the fragments per kilobase million reads (FPKM) value of different genes. The gene expression level increased with color from yellow to blue. Ppe, pure cultured P. penicillatus mycelia in PDA medium; MP1, M. importuna fruiting bodies infected by P. penicillatus for 3 days; MP2, M. importuna fruiting bodies infected by P. penicillatus for 6 days. Each treatment had three biological duplicates. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Genes differentially expressed with infection stage in P. penicillatus
A total of 47 genes were first up-regulated and then down-regulated with infection and the infection stage in P. penicillatus (P < 0.05). These were related to fumitremorgin C monooxygenase-like protein, glycosyltransferase family 31 protein, ferric-chelate reductase, concanava A-like lectin/glucanase, and cupin 2 domain-containing protein (Fig. 4a). A total of 125 genes, including mannitol-1-phosphate 5-dehydrogenase, phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, glucose repressible protein Grg1, reductase, peptidase activity, and peptidase inhibitor I78 (Fig. 4b and 4c), were first down-regulated and then up-regulated with the infection and infection stage in P. penicillatus (P < 0.05).
Fig. 4.
Genes differentially expressed with infection and infection stage in P. penicillatus. a, gene first up-regulated and then down-regulated with the infection and infection stage in P. penicillatus; b and c, gene first down-regulated and then up-regulated with the infection and infection stage in P. penicillatus; the gene expression level increased with color from yellow to blue. Ppe, pure cultured P. penicillatus mycelia in PDA medium; MP1, M. importuna fruiting bodies infected by P. penicillatus for 3 days; MP2, M. importuna fruiting bodies infected by P. penicillatus for 6 days. Each treatment had three biological duplicates. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. Functional enrichment analysis in P. penicillatus
GO functional enrichment analysis showed that when the fruiting body of M. importuna was infected by P. penicillatus at the early stage, four GO terms, related to membrane, membrane part, intrinsic component of membrane, and integral component of membrane, were significantly enriched (P < 0.05). The four GO terms could be assigned to the cellular component category (Table S2). Only one GO term was significantly enriched in P. penicillatus samples at the late infection stage compared with samples at the early infection stage (P < 0.05). This was related to carbohydrate metabolic process in the category of biological processes.
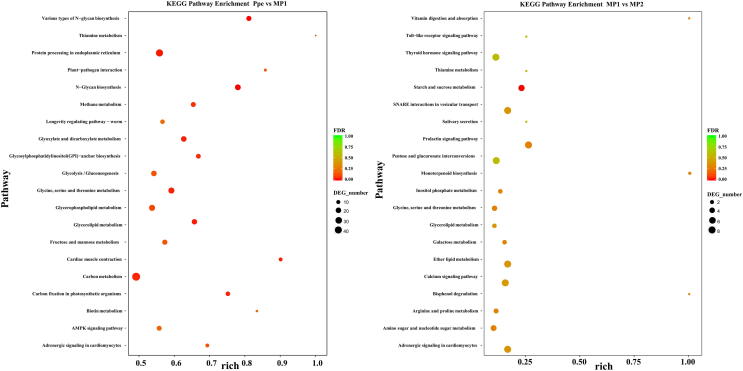
KEGG pathway analysis showed that 35 pathways were significantly enriched between the pure culture and the early infection stage of P. penicillatus mycelia (P < 0.05). These were involved in “N-Glycan biosynthesis,” “protein processing in endoplasmic reticulum,” “cardiac muscle contraction,” and “glycerolipid metabolism” (Fig. 5). Metabolism pathways relating to “glyoxylate and dicarboxylate metabolism” and “carbon fixation in photosynthetic organisms,” and organismal systems relating to “plant-pathogen interaction,” and “adrenergic signaling in cardiomyocytes” were also over-expressed at the early infection stage of P. penicillatus (P < 0.05). Ten KEGG pathways were significantly enriched in P. penicillatus mycelia at the late infection stage compared with those at the early stage (P < 0.05). These were related to starch and sucrose metabolism, amino sugar and nucleotide sugar metabolism, and galactose metabolism in the metabolism category, prolactin signaling pathway, and vitamin digestion and absorption in the category of organismal systems.
Fig. 5.
The mostly enriched KEGG pathways in different samples (P. penicillatus). Y-axis represents different KEGG categories; X-axis represents the rich factor. The size of the dot indicates the number of differentially expressed genes (DEGs) involved in the pathway; Color bars on the right represent the P-value of the KEGG pathway. Ppe, pure cultured P. penicillatus mycelia in PDA medium; MP1, M. importuna fruiting bodies infected by P. penicillatus for 3 days; MP2, M. importuna fruiting bodies infected by P. penicillatus for 6 days. Each treatment had three biological duplicates.
3.7. Genes up- or down-regulated with infection stage in M. Importuna
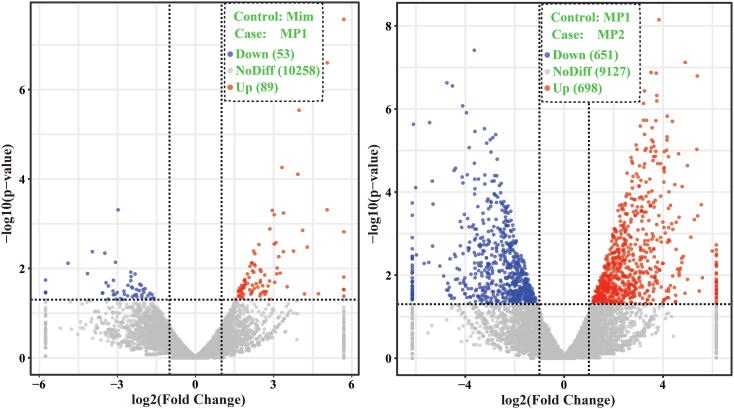
A total of 10,955 genes were expressed in healthy and diseased M. importuna fruiting bodies, of which 1,576 genes were significantly differently expressed between healthy and diseased M. importuna fruiting bodies at different stages (P < 0.05) (Fig. 6). There were 142 genes differently expressed between healthy M. importuna fruiting bodies and diseased M. importuna fruiting bodies at the early stage, and 1,349 genes differently expressed between diseased M. importuna fruiting bodies at the early and the late stages (P < 0.05). Of these differently expressed genes, the expression of 10 genes increased with the infection and infection stage (P < 0.05). These included cyclin-dependent protein kinase inhibitor and nine genes with unknown functions (Fig. 7a). No gene was always down-regulated with the infection and infection stage.
Fig. 6.
Volcano plot of differentially expressed genes (DEGs) between samples (M. importuna). The X-axis represents the fold change of DEGs in different experimental groups; -log10 (p-value) in the Y-axis represents DEGs that were more significant as the value increased. The dots represent the genes; the grey dots indicate non differentially expressed genes; the red dots represent up-regulated differentially expressed genes; the blue dots represent down-regulated differentially expressed genes. Mim, healthy fruiting bodies of M. importuna; MP1, M. importuna fruiting bodies infected by P. penicillatus for 3 days; MP2, M. importuna fruiting bodies infected by P. penicillatus for 6 days. Each treatment had three biological duplicates. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Genes differentially expressed with the infection and infection stage in M. importuna. a, genes up-regulated with infection and infection stages in M. importuna; b, the upper part of genes was first down-regulated and then up-regulated with the infection and infection stage, and the lower part of the genes was first up-regulated and then down-regulated with the infection and infection stage in M. importuna; the black number in the color block indicates the fragments per kilobase million reads (FPKM) value of different genes. The gene expression level increased with color from yellow to blue. Mim, healthy fruiting bodies of M. importuna; MP1, M. importuna fruiting bodies infected by P. penicillatus for 3 days; MP2, M. importuna fruiting bodies infected by P. penicillatus for 6 days. Each treatment had three biological duplicates. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.8. Genes differentially expressed with infection stage in M. Importuna
A total of 15 genes were first up-regulated and then down-regulated with the infection and infection stage in M. importuna (P < 0.05), and they were related to zinc finger MYND domain-containing protein, aquaporin-like protein, and 13 genes with unknown functions (Fig. 7b). Twenty-eight genes, including YTH-domain-containing protein, retrotransposable element Tf2, cdc42 effector protein, mitochondrial 54S ribosomal protein YmL16, and 24 hypothetical proteins were first down-regulated and then up-regulated with the infection and infection stage in M. importuna (P < 0.05).
3.9. Functional enrichment analysis in M. Importuna
GO functional enrichment analysis showed that no GO term was significantly enriched between healthy and diseased M. importuna fruiting bodies at different stages. KEGG pathway analysis showed that nine pathways were significantly enriched between the healthy and diseased M. importuna fruiting bodies at the early stage (P < 0.05) (Fig. 8). Organismal systems pathways relating to “longevity regulating pathway,” “PPAR signaling pathway,” and “GABAergic synapse,” metabolism pathway relating to “fatty acid metabolism,” “fatty acid biosynthesis,” and “biosynthesis of unsaturated fatty acids,” genetic information processing pathway relating to “fanconi anemia,” and environmental information processing pathway relating to “FoxO signaling” were over-expressed at the early infection stage of M. importuna (P < 0.05). Four KEGG pathways were significantly enriched between diseased M. importuna fruiting bodies at the early and late stages (P < 0.05). These were related to glycan degradation, carbon metabolism, and pentose phosphate pathway in the category of metabolism and mineral absorption in the organismal systems category.
Fig. 8.
The mostly enriched KEGG pathways in different samples (M. importuna). Y-axis represents different KEGG categories; X-axis represents the rich factor. The size of the dot indicates the number of differentially expressed genes (DEGs) involved in the pathway; the color bars on the right represent the P-value of KEGG pathway. Mim, healthy fruiting bodies of M. importuna; MP1, M. importuna fruiting bodies infected by P. penicillatus for 3 days; MP2, M. importuna fruiting bodies infected by P. penicillatus for 6 days. Each treatment contains three biological duplicates.
3.10. Validation of the DEG results by qPCR analysis
We validated gene expression profiles obtained by transcriptome analysis using qPCR analysis. This confirmed the expression levels of 19 selected genes, with 5.8S rRNA serving as the reference gene (Table S3). All the 19 identified genes were successfully amplified and resulted in a single band. Three genes were up-regulated in M. importuna at the early infection stage compared with healthy M. importuna fruiting bodies. Five genes exhibited higher expression levels in diseased M. importuna fruiting bodies at the late infection stage compared to the early infection stage. Five genes were down-regulated and one was up-regulated in P. penicillatus at the early infection stage compared to pure cultured P. penicillatus mycelia. Six genes exhibited higher expression levels in P. penicillatus at the late infection stage than in the early infection stage. The selected genes used for qPCR validation were significantly differentially expressed as calculated by DEG analysis. This supported the reliability of the transcriptome analysis (Table S4).
4. Discussion
P. penicillatus is a pathogenic fungus that greatly reduces the yield and quality of morels [11]. However, the pathogenicity mechanism of P. penicillatus has not been fully understood. Fungal genomes and mitogenomes are variable in size, structure, intron and gene number [55], [56], [57], [78]. In order to more accurately analyze the pathogenic mechanism of P. penicillatus and the resistance mechanism of Morchella spp., we further sequenced and assembled the P. penicillatus SAAS_ppe1 genome to improve the integrity of genome assembly. We obtained the genome of P. penicillatus SAAS_ppe1 based on PacBio and Illumina sequencings. The genome of P. penicillatus SAAS_ppe1 had a longer scaffold (N50) than the other four Paecilomyces species, and all its scaffolds were greater than 10 kb long. BUSCO assessment results showed that 99.7% of the 290 fungal BUSCO genes exist in the assembled genome of P. penicillatus SAAS_ppe1, which was higher than the other four Paecilomyces genomes. The combined application of PacBio and Illumina sequencing methods produced a P. penicillatus genome that was of higher quality than other species in the genus [58], [59]. The other four Paecilomyces genomes were assembled based on only one sequencing method. A high-quality P. penicillatus genome enabled analysis of its pathogenicity mechanism. We were able to successfully distinguish the transcripts of P. penicillatus and M. importuna in the pathogenic sites, and analyze pathogenic mechanisms and transcriptional response mechanisms. In addition, compared with the other Paecilomyces species, P. penicillatus has fewer gene models and more carbohydrate-active genes belonging to auxiliary activities (AAs), carbohydrate esterases (CEs), and polysaccharide lyases (PLs). The increased number of carbohydrate-active genes may contribute to the strong adaptation and infection abilities of P. penicillatus [60], [61].
We obtained the transcriptional profiles of a pure culture of P. penicillatus mycelia, healthy fruiting bodies of M. importuna, and mixed transcripts of P. penicillatus and M. importuna at early and late infection stages, from pathogenic sites. During the early stage of infection, 86.34% and 7.48% of the mixed transcripts were mapped to the genome of M. importuna and P. penicillatus, respectively. While in the late stage of infection, 74.49% and 19.83% of the mixed transcripts were mapped to the genome of M. importuna and P. penicillatus, respectively. These results showed that M. importuna was dominant in the pathogenic site, but its biomass and transcripts decreased over time, indicating that healthy M. importuna tissues were gradually infected by P. penicillatus. The biomass of P. penicillatus gradually increased with the infection and infection stage. Differentially expressed gene analysis showed that 24 and 25 genes were up-regulated and down-regulated, respectively, with the infection and infection stage, indicating that these genes are involved in the process of P. penicillatus infection. The up-regulated genes included diphthamide biosynthesis, aldehyde reductase, NAD (P) h-hydroxy epimerase, and GPI anchored protein. The biological function of diphthamide is not well understood. Previous studies showed that diphthamide modification plays an important role in regulating the sensitivity of cells to bacterial toxins and maintaining translation fidelity [62], [63]. Our results suggest that diphthamide plays a role in P. penicillatus infection. Aldehyde reductase is a member of the aldo–keto reductase superfamily, which catalyzes the NADPH-dependent reduction of a variety of aldehydes to their corresponding alcohols [64], [65]. NAD (P) h-hydroxy epimerase can help repair NAD(P)H hydrates [66]. GPI anchored proteins act as membrane anchors of many eukaryotic cell surface proteins [67]. These genes are important in the physiological regulation of eukaryotes. Our study provides the first information on the regulatory roles of these genes in the process of P. penicillatus infection.
We found 47 genes that were first up-regulated and then down-regulated with the infection and infection stage in P. penicillatus. These genes were related to glycosyltransferase family 31 proteins and ferric-chelate reductase. Glycosyltransferases are important proteins for the synthesis of complex carbohydrates [68]. Ferric-chelate reductase participates in the synthesis of sesquiterpenoids with important biological activities in fungi [69]. A total of 125 genes were first down-regulated and then up-regulated with the infection and infection stage in P. penicillatus, including mannitol-1-phosphate 5-dehydrogenase and glucose repressible protein Grg1. Mannitol-1-phosphate 5-dehydrogenase participates in the mannitol cycle for NADPH regeneration [70]. Glucose repressible protein Grg1 helps fungi utilize a wide range of carbon sources except for glucose [71]. GO functional enrichment showed that, in the early infection stage, cellular component function was significantly enriched in P. penicillatus, showing its important role in early infection of P. penicillatus. In the late infection stage, carbohydrate metabolic process was over expressed in P. penicillatus. These results show that P. penicillatus has diverse regulation patterns that it uses to infect M. importuna in different infection stages. KEGG pathways, including “glycerolipid metabolism” and “plant-pathogen interaction” were over-expressed at the early infection stage of P. penicillatus. Glycerolipid metabolism helps regulate lipid synthesis and metabolism and influences stress responses [72]. Plant-pathogen interactions can play an important role in the process of pathogens infecting plants [73]. Our study revealed that metabolic pathways have an important regulatory role in the early stage of fungus-fungus interactions. Ten KEGG pathways were significantly enriched in P. penicillatus mycelia at the late infection stage compared to the early infection stage. These pathways were related to amino sugar and nucleotide sugar metabolism and galactose metabolism, which are involved in the regulation of cell morphology and plant-fungus interactions [74], [75].
M. importuna is cultivated in China and other countries [76], [77]. However, the yield of M. importuna is reduced by pathogens. P. penicillatus is an important pathogen of M. importuna that can cause stalk and cap rot [11]. However, the transcriptional response mechanism of M. importuna to pathogens was previously unknown. We found that the expression levels of 10 genes increased with the infection and infection stage in M. importuna and included cyclin-dependent protein kinase inhibitor and nine genes with unknown functions. Fifteen genes were first up-regulated and then down-regulated with the infection and infection stage in M. importuna. Twenty-eight genes were first down-regulated and then up-regulated with the infection and infection stage in M. importuna. The results indicated that these genes had roles in regulating the response of M. importuna to P. penicillatus infection. Fatty acid biosynthesis and metabolism pathways were significantly enriched at the early infection stage of M. importuna. Fatty acid biosynthesis and metabolism plays an important role in the plant-fungus interaction [73]. We found that this pathway participated in the fungus-fungus interaction. The functions of many proteins from M. importuna are not understood. This study shows the transcriptional response mechanism of M. importuna and the data will also be useful in the functional analysis of M. importuna proteins.
Funding
This research was funded by the Foundation Program of the Financial & Innovational Capacity Building Project of Sichuan (2018QNJJ-013 &2016GYSH-014).
Author Contributions
Conceived and designed experiments: C.C., R.F., J.W., and Q.L.. Analyzed the data: C.C., X.L., and X.C.. Wrote and review the paper: C.C., and Q.L.. Funding acquisition and project administration: D.L..
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.04.065.
Contributor Information
Qiang Li, Email: leeq110@126.com.
Daihua Lu, Email: 453831354@qq.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Quality control of the Paecilomyces penicillatus genome sequencing data. (a) Base quality using Phred scores for the data from Illumina sequencing. In the box plots, the whiskers represent the range between the 10% and 90% quantiles; the yellow boxes show the range between the upper and lower quartiles; the red lines represent median. The blue line indicates the mean quality; (b) Sequence content across all bases; the X-axis represents the base position in reads (5’–>3’), and the Y-axis represents the proportion of a base in reads; (c) Distribution of read-over-read quality; (d) Read length distribution of PacBio sequencing after filtering.
Supplementary figure 2.
Functional annotation of the protein-coding genes in the Paecilomyces penicillatus genome against five databases.
Supplementary figure 3.
CAZy function classification of the five Paecilomyces genomes.
References
- 1.Luangsa-ard J.J., Hywel-Jones N.L., Samson R.A. The polyphyletic nature of Paecilomyces sensu lato based on 18S-generated rDNA phylogeny. Mycologia. 2004;96(4):773–780. doi: 10.1080/15572536.2005.11832925. [DOI] [PubMed] [Google Scholar]
- 2.Inglis P.W., Tigano M.S. Identification and taxonomy of some entomopathogenic Paecilomyces spp. (Ascomycota) isolates using rDNA-ITS Sequences. Genetics and Molecular Biology. 2006;29(1):132–136. [Google Scholar]
- 3.Fletcher C.L., Hay R.J., Midgley G., Moore M. Onychomycosis caused by infection with Paecilomyces lilacinus. Br J Dermatol. 1998;139:1133–1135. doi: 10.1046/j.1365-2133.1998.2576s.x. [DOI] [PubMed] [Google Scholar]
- 4.Feldman R., Cockerham L., Buchan B.W., Lu Z., Huang A.M. Treatment of Paecilomyces variotii pneumonia with posaconazole: case report and literature review. Mycoses. 2016;59(12):746–750. doi: 10.1111/myc.12525. [DOI] [PubMed] [Google Scholar]
- 5.Swami T., Pannu S., Kumar M., Gupta G. Chronic invasive fungal rhinosinusitis by Paecilomyces variotii: A rare case report. Indian J Med Microbiol. 2016;34(1):103–106. doi: 10.4103/0255-0857.174126. [DOI] [PubMed] [Google Scholar]
- 6.Biango-Daniels M.N., Hodge K.T. Paecilomyces Rot: A New Apple Disease. Plant Dis. 2018;102(8):1581–1587. doi: 10.1094/PDIS-12-17-1896-RE. [DOI] [PubMed] [Google Scholar]
- 7.Heidarian R., Fotouhifar K.-B., Debets A.J.M., Aanen D.K., Yurkov A.M. Phylogeny of Paecilomyces, the causal agent of pistachio and some other trees dieback disease in Iran. PLoS ONE. 2018;13(7):e0200794. doi: 10.1371/journal.pone.0200794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biango-Daniels M.N., Ayer K.M., Cox K.D., Hodge K.T. Paecilomyces niveus: Pathogenicity in the Orchard and Sensitivity to Three Fungicides. Plant Dis. 2019;103(1):125–131. doi: 10.1094/PDIS-04-18-0695-RE. [DOI] [PubMed] [Google Scholar]
- 9.Pang F., Wang L., Jin Y.u., Guo L., Song L., Liu G. Transcriptome analysis of Paecilomyces hepiali at different growth stages and culture additives to reveal putative genes in cordycepin biosynthesis. Genomics. 2018;110(3):162–170. doi: 10.1016/j.ygeno.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Li LZ, Liu YG, Teng LR, Lu JH, et al. (2016) Investigations on the antifatigue and antihypoxic effects of Paecilomyces hepiali extract. Mol Med Rep 13: 1861-1868. [DOI] [PubMed]
- 11.He X.L., Peng W.H., Miao R.Y., Tang J., Chen Y. White mold on cultivated morels caused by Paecilomyces penicillatus. FEMS Microbiol Lett. 2017;364 doi: 10.1093/femsle/fnx037. [DOI] [PubMed] [Google Scholar]
- 12.Li W., Cai Z.-N., Mehmood S., Liang L.-L., Liu Y.u., Zhang H.-Y. Anti-inflammatory effects of Morchella esculenta polysaccharide and its derivatives in fine particulate matter-treated NR8383 cells. Int J Biol Macromol. 2019;129:904–915. doi: 10.1016/j.ijbiomac.2019.02.088. [DOI] [PubMed] [Google Scholar]
- 13.Xiong C., Luo Q., Huang W.-L., Li Q., Chen C., Chen Z.-Q. The potential neuritogenic activity of aqueous extracts from Morchella importuna in rat pheochromocytoma cells. Food Sci Biotechnol. 2017;26(6):1685–1692. doi: 10.1007/s10068-017-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He P., Wang K.e., Cai Y., Hu X., Zheng Y., Zhang J. Involvement of autophagy and apoptosis and lipid accumulation in sclerotial morphogenesis of Morchella importuna. Micron. 2018;109:34–40. doi: 10.1016/j.micron.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Meng X., Che C., Zhang J., Gong Z., Si M., Yang G.e. Structural characterization and immunomodulating activities of polysaccharides from a newly collected wild Morchella sextelata. Int J Biol Macromol. 2019;129:608–614. doi: 10.1016/j.ijbiomac.2019.01.226. [DOI] [PubMed] [Google Scholar]
- 16.Tan H., Kohler A., Miao R., Liu T., Zhang Q., Zhang B.o. Multi-omic analyses of exogenous nutrient bag decomposition by the black morel Morchella importuna reveal sustained carbon acquisition and transferring. Environ Microbiol. 2019;21(10):3909–3926. doi: 10.1111/1462-2920.14741. [DOI] [PubMed] [Google Scholar]
- 17.Wang X., Peng J., Sun L., Bonito G., Guo Y. Genome Sequencing of Paecilomyces Penicillatus Provides Insights into Its Phylogenetic Placement and Mycoparasitism Mechanisms on Morel Mushrooms. Pathogens. 2020;9 doi: 10.3390/pathogens9100834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan K., Wang C., Liu W., Liu K., Jia B., Hao L. Genome sequence and transcriptomic profiles of a marine bacterium, Pseudoalteromonas agarivorans Hao 2018. Sci Data. 2019;6(1) doi: 10.1038/s41597-019-0012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X., Liu W., Zheng B. Complete genome sequencing of Comamonas kerstersii 8943, a causative agent for peritonitis. Sci Data. 2018;5(1) doi: 10.1038/sdata.2018.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urquhart A.S., Mondo S.J., Mäkelä M.R., Hane J.K., Wiebenga A.d., He G. Genomic and Genetic Insights Into a Cosmopolitan Fungus, Paecilomyces variotii (Eurotiales) Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.0305810.3389/fmicb.2018.03058.s00110.3389/fmicb.2018.03058.s002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biango-Daniels M.N., Wang T.W., Hodge K.T. Draft Genome Sequence of the Patulin-Producing Fungus Paecilomyces niveus Strain CO7. Genome Announc. 2018;6(25) doi: 10.1128/genomeA.00556-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C., Li Q., Chen H., Fu R., Wang J., Chen X. The complete mitochondrial genome of Paecilomyces penicillatus (Hypocreales: Sordariomycetes) Mitochondrial DNA Part B. 2019;4(1):199–200. [Google Scholar]
- 23.Liu H.-P., Xiao S.-J., Wu N., Wang D.i., Liu Y.-C., Zhou C.-W. The sequence and de novo assembly of Oxygymnocypris stewartii genome. Sci Data. 2019;6(1) doi: 10.1038/sdata.2019.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcais G, Kingsford C (2011) A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764-770. [DOI] [PMC free article] [PubMed]
- 25.De Landtsheer S, Trairatphisan P, Lucarelli P, Sauter T (2017) FALCON: a toolbox for the fast contextualization of logical networks. Bioinformatics 33: 3431-3436. [DOI] [PMC free article] [PubMed]
- 26.Pendleton M., Sebra R., Pang A.W.C., Ummat A., Franzen O., Rausch T. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods. 2015;12(8):780–786. doi: 10.1038/nmeth.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1(1) doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W., Yan H.-J., Chen S.-Y., Li Z.-Z., Yi J., Niu L.-L. The sequence and de novo assembly of hog deer genome. Sci Data. 2019;6(1) doi: 10.1038/sdata.2018.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tempel S. Using and understanding RepeatMasker. Methods Mol Biol. 2012;859:29–51. doi: 10.1007/978-1-61779-603-6_2. [DOI] [PubMed] [Google Scholar]
- 30.Bao W., Kojima K.K., Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucl Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price A.L., Jones N.C., Pevzner P.A. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 33.Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, et al. (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35: 3100-3108. [DOI] [PMC free article] [PubMed]
- 35.Griffiths-Jones S (2005) Annotating non-coding RNAs with Rfam. Curr Protoc Bioinformatics Chapter 12: Unit 12 15. [DOI] [PubMed]
- 36.Slater G.S., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinf. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanke M., Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucl Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majoros W.H., Pertea M., Salzberg S.L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20(16):2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 39.Korf I. Gene finding in novel genomes. BMC Bioinf. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, et al. (2018) HMMER web server: 2018 update. Nucleic Acids Res 46: W200-W204. [DOI] [PMC free article] [PubMed]
- 42.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucl Acids Res. 2014;42(D1):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The UniProt C. UniProt: the universal protein knowledgebase. Nucl Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller J., Szklarczyk D., Julien P., Letunic I., Roth A. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucl Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blin K., Shaw S., Steinke K., Villebro R., Ziemert N. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucl Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q., Huang W., Xiong C., Zhao J. Transcriptome analysis reveals the role of nitric oxide in Pleurotus eryngii responses to Cd(2+) stress. Chemosphere. 2018;201:294–302. doi: 10.1016/j.chemosphere.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21(19):3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 52.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M. KEGG for linking genomes to life and the environment. Nucl Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zarivi O., Cesare P., Ragnelli A.M., Aimola P., Leonardi M. Validation of reference genes for quantitative real-time PCR in Perigord black truffle (Tuber melanosporum) developmental stages. Phytochemistry. 2015;116:78–86. doi: 10.1016/j.phytochem.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 54.Seppey M., Manni M., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 55.Cheng J., Luo Q., Ren Y., Luo Z., Liao W., Wang X.u. Panorama of intron dynamics and gene rearrangements in the phylum Basidiomycota as revealed by the complete mitochondrial genome of Turbinellus floccosus. Appl Microbiol Biotechnol. 2021;105(5):2017–2032. doi: 10.1007/s00253-021-11153-w. [DOI] [PubMed] [Google Scholar]
- 56.Li Q., Wu P., Li L., Feng H., Tu W., Bao Z. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int J Biol Macromol. 2021;172:560–572. doi: 10.1016/j.ijbiomac.2021.01.087. [DOI] [PubMed] [Google Scholar]
- 57.Li H., Wu S., Ma X., Chen W., Zhang J., Duan S. The genome sequences of 90 mushrooms. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-28303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhoads A., Au K.F. PacBio sequencing and its applications. Genomics Proteomics & Bioinformatics. 2015;13(5):278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Au K.F., Underwood J.G., Lee L., Wong W.H. Improving PacBio long read accuracy by short read alignment. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0046679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zerillo M.M., Adhikari B.N., Hamilton J.P., Buell C.R., Lévesque C.A., Tisserat N. Carbohydrate-active enzymes in pythium and their role in plant cell wall and storage polysaccharide degradation. PLoS ONE. 2013;8(9):e72572. doi: 10.1371/journal.pone.0072572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brouwer H., Coutinho P.M., Henrissat B., de Vries R.P. Carbohydrate-related enzymes of important Phytophthora plant pathogens. Fungal Genet Biol. 2014;72:192–200. doi: 10.1016/j.fgb.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 62.Su X., Lin Z., Lin H. The biosynthesis and biological function of diphthamide. Crit Rev Biochem Mol Biol. 2013;48(6):515–521. doi: 10.3109/10409238.2013.831023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaffrath R., Abdel-Fattah W., Klassen R., Stark M.J. The diphthamide modification pathway from Saccharomyces cerevisiae–revisited. Mol Microbiol. 2014;94:1213–1226. doi: 10.1111/mmi.12845. [DOI] [PubMed] [Google Scholar]
- 64.Rees-Milton K.J., Jia Z., Green N.C., Bhatia M., El-Kabbani O., Flynn T.G. Aldehyde reductase: the role of C-terminal residues in defining substrate and cofactor specificities. Arch Biochem Biophys. 1998;355(2):137–144. doi: 10.1006/abbi.1998.0721. [DOI] [PubMed] [Google Scholar]
- 65.El-Kabbani O., Judge K., Ginell S.L., Myles D.A.A., DeLucas L.J., Flynn T.G. Structure of porcine aldehyde reductase holoenzyme. Nat Struct Biol. 1995;2(8):687–692. doi: 10.1038/nsb0895-687. [DOI] [PubMed] [Google Scholar]
- 66.Niehaus TD, Elbadawi-Sidhu M, Huang L, Prunetti L, Gregory JF, 3rd, et al. (2018) Evidence that the metabolite repair enzyme NAD(P)HX epimerase has a moonlighting function. Biosci Rep 38. [DOI] [PMC free article] [PubMed]
- 67.Kinoshita T., Fujita M. Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J Lipid Res. 2016;57:6–24. doi: 10.1194/jlr.R063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McArthur J.B., Chen X. Glycosyltransferase engineering for carbohydrate synthesis. Biochem Soc Trans. 2016;44:129–142. doi: 10.1042/BST20150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Umezawa S, Akao H, Kubota M, Kino K (2020) Chemoenzymatic oxygenation method for sesquiterpenoid synthesis based on Fe-chelate and ferric-chelate reductase. Biosci Biotechnol Biochem 84: 780-788. [DOI] [PubMed]
- 70.Chase T., Jr. Mannitol-1-phosphate dehydrogenase of Escherichia coli. Chemical properties and binding of substrates. Biochem J. 1986;239:435–443. doi: 10.1042/bj2390435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kayikci O., Nielsen J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15 doi: 10.1093/femsyr/fov068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Henry S.A., Gaspar M.L., Jesch S.A. The response to inositol: regulation of glycerolipid metabolism and stress response signaling in yeast. Chem Phys Lipids. 2014;180:23–43. doi: 10.1016/j.chemphyslip.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beccaccioli M., Reverberi M., Scala V. Fungal lipids: biosynthesis and signalling during plant-pathogen interaction. Front Biosci (Landmark Ed) 2019;24:172–185. doi: 10.2741/4712. [DOI] [PubMed] [Google Scholar]
- 74.Lee D.-K., Ahn S., Cho H.Y., Yun H.Y., Park J.H., Lim J. Metabolic response induced by parasitic plant-fungus interactions hinder amino sugar and nucleotide sugar metabolism in the host. Sci Rep. 2016;6(1) doi: 10.1038/srep37434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gopinath R.K., Leu J.-Y. Hsp90 mediates the crosstalk between galactose metabolism and cell morphology pathways in yeast. Curr Genet. 2017;63(1):23–27. doi: 10.1007/s00294-016-0614-2. [DOI] [PubMed] [Google Scholar]
- 76.He P., Yu M., Wang K.e., Cai Y., Li B., Liu W. Interspecific hybridization between cultivated morels Morchella importuna and Morchella sextelata by PEG-induced double inactivated protoplast fusion. World J Microbiol Biotechnol. 2020;36(4) doi: 10.1007/s11274-020-02835-0. [DOI] [PubMed] [Google Scholar]
- 77.Liu Q., Ma H., Zhang Y.a., Dong C. Artificial cultivation of true morels: current state, issues and perspectives. Crit Rev Biotechnol. 2018;38(2):259–271. doi: 10.1080/07388551.2017.1333082. [DOI] [PubMed] [Google Scholar]
- 78.Liu Q., Ren Y., Shi X., Peng L., Zhao J. Comparative mitochondrial genome analysis of two ectomycorrhizal fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the subphylum Agaricomycotina. Int J Mol Sci. 2019;20:5167. doi: 10.3390/ijms20205167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina short-read sequencing data have been deposited to NCBI Sequence Read Archive (SRA) (Data Citation 1: NCBI Sequence Read Archive SRR9051197). The PacBio long-read sequencing data have been deposited to NCBI Sequence Read Archive (SRA) (Data Citation 2: NCBI Sequence Read Archive SRR9051168). The transcriptome data are available through the NCBI SRA (Data Citation 3: NCBI Sequence Read Archive SRR9682527-SRR9682529). The assembled genome version is available at GenBank (Data Citation 4: GenBank VCCP00000000.1). The annotation gff3 file of the assembled genome is available at Figshare (Data Citation 5: Figshare https://doi.org/10.6084/m9.figshare.8146202.v3). The reference genome of M. importuna can be found in JGI database (https://mycocosm.jgi.doe.gov/Morimp1/Morimp1.home.html) [16].