Abstract
Terpenes make up the largest class of natural products, with extensive chemical and structural diversity. Diterpenes, mostly isolated from plants and rarely prokaryotes, exhibit a variety of important biological activities and valuable applications, including providing antitumor and antibiotic pharmaceuticals. These natural products are constructed by terpene synthases, a class of enzymes that catalyze one of the most complex chemical reactions in biology: converting simple acyclic oligo-isoprenyl diphosphate substrates to complex polycyclic products via carbocation intermediates. Here we obtained the second ever crystal structure of a class II diterpene synthase from bacteria, tuberculosinol pyrophosphate synthase (i.e., Halimadienyl diphosphate synthase, MtHPS, or Rv3377c) from Mycobacterium tuberculosis (Mtb). This enzyme transforms (E,E,E)-geranylgeranyl diphosphate into tuberculosinol pyrophosphate (Halimadienyl diphosphate). Rv3377c is part of the Mtb diterpene pathway along with Rv3378c, which converts tuberculosinol pyrophosphate to 1-tuberculosinyl adenosine (1-TbAd). This pathway was shown to exist only in virulent Mycobacterium species, but not in closely related avirulent species, and was proposed to be involved in phagolysosome maturation arrest. To gain further insight into the reaction pathway and the mechanistically relevant enzyme substrate binding orientation, electronic structure calculation and docking studies of reaction intermediates were carried out. Results reveal a plausible binding mode of the substrate that can provide the information to guide future drug design and anti-infective therapies of this biosynthetic pathway.
Graphical Abstract
Terpenes and terpenoids comprise a class of natural products with more than 80000 members,1 whose structures are extremely varied and display a wide range of functions. These compounds range from essential metabolites, such as sterols, to unique secondary metabolites involved in communication and defense by various organisms. Terpene biosynthesis involves the isoprenyl diphosphate synthase-catalyzed condensation of isopentyl diphosphate and dimethylallyl diphosphate2 to form linear precursors such as geranyl diphosphate (C10), farnesyl diphosphate (C15), and geranylgeranyl diphosphate (C20). These linear substrates are then converted by terpene cyclases into complex, often polycyclic, monoterpenes, sesquiterpenes, and diterpenes, respectively.1,3,4 These terpenes are then functionalized, often by the addition of oxygen atoms, by additional biosynthetic enzymes.
While most known terpenes have been isolated from plants and fungi, various monoterpenes, sesquiterpenes, and diterpenes have been isolated from streptomycetes,5–9 Cyanobacteria,10 and Myxobacteria.11 Mycobacterium tuberculosis, the causative agent of tuberculosis, also contains a gene encoding a diterpene cyclase,12–14 and a broad lipidomics screen recently identified an abundant secreted diterpene, 1-tuberculosinyl adenosine (1-TbAd), as the end product of the Mtb diterpene pathway (see the details in the Supporting Information).15 Two genes, Rv3377c and Rv3378c, are necessary and sufficient to produce 1-TbAd in the nonpathogenic related mycobacterium Mycobacterium smegmatis.16 A previous study has shown that survival of Mtb in macrophages worsened with artificial damage to the Rv3377c gene.17 Rv3378c was thought to function as a pyrophosphatase,18 but the crystal structure revealed that it is a diverged cis-prenyl transferase that links adenosine and tuberculosinol pyrophosphate to form 1-TbAd15 (see the Supporting Information for the complete pathway mechanism). Purified Rv3377c, in the presence of Mg2+, rapidly converts geranylgeranyl diphosphate (GGPP) into tuberculosinol pyrophosphate (see Figure 1a for the reaction mechanism), indicating that Rv3377c is a class II diterpene cyclase enzyme.14,19 Class II terpene cyclases generate a carbocation by protonation of the C=C π-bond in the terminal isoprene unit, which involves a DXDD motif. These cyclases frequently have a multidomain architecture. In contrast, class I terpene cyclases promote the disconnection of a pyrophosphate to generate an allylic carbocation. These enzymes contain a DDXXD motif20 that binds the substrate diphosphate with the support of Mg2+ ions and are structurally related to isoprenyl diphosphate synthases.
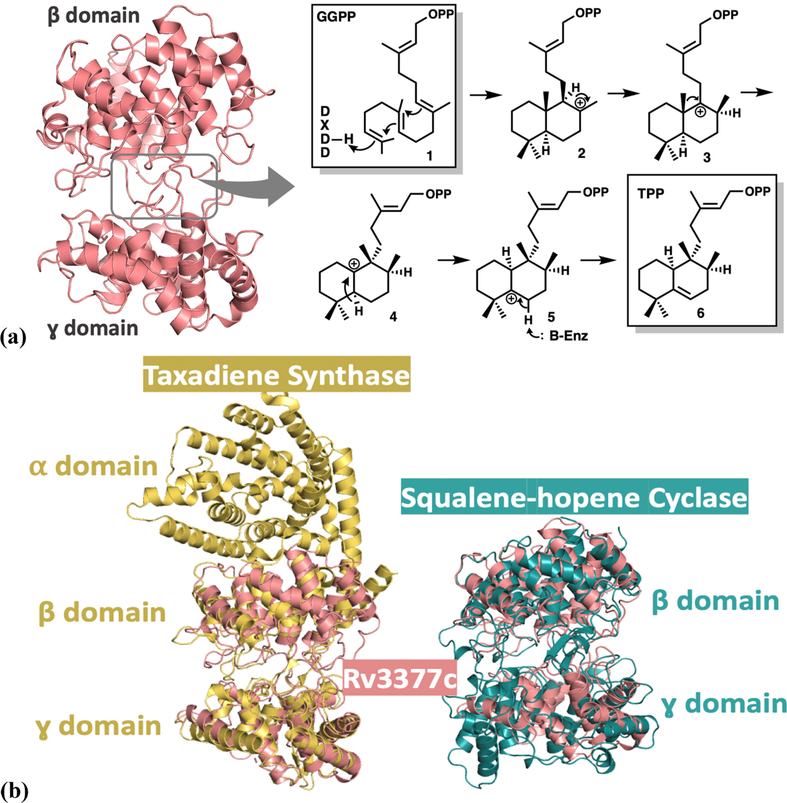
Figure 1.
(a) Global structure of Rv3377c (PDB entry 6VPT), with dumbbell-shaped β (top) and γ (bottom) domains of the class II diterpene cyclases. Reaction mechanism of Rv3377c, which converts geranylgeranyl pyrophosphate (GGPP) to tuberculosinyl pyrophosphate (TPP). (b) Comparison of the structure of Rv3377c with closely related homologues. Overlay of Rv3377c (salmon) with taxadiene synthase (yellow, PDB entry 3P5P), which adopts an αβγ domain assembly (left). Overlay of Rv3377c (salmon) with squalene-hopene cyclase (green, PDB entry 2SQC) (right).
Recent crystal structures of plant and bacterial class II diterpene cyclases21–23 have revealed the domain architecture of these enzymes, the fold of each domain, the nature of the substrate binding sites, and the arrangement of catalytic residues in this protein family, although questions regarding the specific catalytically competent orientations of substrates remain. The plant enzymes share three domains (α, β, and γ) with the active site in a deep cleft between the β and γ domains.24–26 Although the β,γ-didomain architecture is widely observed, some class II cyclases have a single β-domain architecture.27 The catalytic acid in the characteristic DXDD motif shared by these enzymes is located at the bottom of the active-site cavity. The origin of product specificity for this widespread enzyme class remains a major unanswered question.
Compared to plant homologues, the sequence of Rv3377c is deeply diverged [the level of sequence identity is 23% between Rv3377c and taxadiene synthase from Pacific yew (see Figure 1b)] and Rv3377c is predicted to contain only the β and γ domains. To expand the structure–function correlates of this enzyme class, define the architecture of the substrate binding site, and promote inhibitor development, we determined the crystal structure of Mtb Rv3377c. This structure proved to be elusive for almost a decade after production of the initial crystals and was finally determined using Rosetta-based molecular replacement.
The fold of Rv3377c matches that of the β and γ domains of the plant enzymes and includes an interdomain loop with a conserved ion pair at the tip. The general acid that initiates cyclization interacts with H341 at the bottom of a large substrate binding cavity. Many aromatic residues line the cavity, as is expected for terpene synthases.1–3 These amino acids include two tryptophan residues that may switch rotamers as the reaction proceeds (vide infra).
Electronic structure calculations demonstrate that a reaction mechanism that involves cyclization and a series of hydride shifts and a methyl group migration is energetically viable. The results of extensive docking studies provide a model for the binding of intermediate carbocations and a template for structure-aided inhibitor design.
MATERIALS AND EXPERIMENTAL DETAILS
Amplification and Xloning.
An expression vector for full-length Rv3377c was created using the Gateway System (Invitrogen, Carlsbad, CA) from the genomic DNA of M. tuberculosis H37Rv (Colorado State University, Fort Collins, CO). A 1500 bp product was amplified using primers 3377F (5′-CTGGTGCCACGCGGTTCTCATATGGAGACTTTCAGGACT-3′) and 3377R (5′-GAAAGCTGGGTGAAGCTTTCATTGGTTACTCTCATC-3′), 5% DMSO, and TurboCX polymerase (Stratagene, La Jolla, CA). Polymerase chain reaction (PCR) products were run on a 1% agarose gel; the band was excised and purified using a QIAquick gel extraction kit (Qiagen, Venlo, The Netherlands). This PCR product was used as a template for a second amplification using primers GatewayTHF and GatewayR, 5% DMSO, and TurboCX polymerase, and the resulting product was purified as described above. The second PCR product was used in a recombination reaction with vector pDONR207 and BP clonase (Invitrogen). After treatment with proteinase K (Invitrogen), the recombination reaction was transformed into DH5α cells and correct recombination was selected on LB plates containing gentamicin. The plasmids from resulting colonies were sequenced and used to recombine the gene into destination vector pHMGWA, a thrombin-cleavable six-His-maltose binding protein tagged vector. The LR reaction mixture was transformed into DH5α and selected on LB plates containing 100 μg mL−1 ampicillin. Correct recombinants were confirmed by DNA sequencing.
Purification of Rv3377c.
BL21-CodonPlus(DE3) cells (Stratagene) harboring the Rv3377c expression plasmid were cultured in Terrific Broth containing 100 μg mL−1 ampicillin and 34 μg mL−1 chloramphenicol and grown at 37 °C to an OD600 of 0.8. The culture was shaken at 18 °C for 20 min; 100 μM isopropyl β-d-1-thiogalactopyranoside (final concentration) was added, and growth was continued at 18 °C for 18 h. Cells were harvested by centrifugation and resuspended in 0.3 M NaCl, 50 mM HEPES (pH 8.0), 2 mM MgCl2, 10% glycerol, 0.1% Triton X-100, and 0.5 mM tris(2-carboxyethyl)-phosphine hydrochloride (TCEP). After sonication on ice and centrifugation at 26000g for 1 h, the clarified cell lysate was loaded onto a 5 mL immobilized metal affinity chromatography (IMAC) column (Amersham-Pharmacia, Amersham) equilibrated with 0.1 M NiSO4. The resin was washed with 3 column volumes (CVs) of a buffer containing 25 mM imidazole. The protein was eluted in a buffer containing 250 mM imidazole, and the tag removed by thrombin cleavage. The untagged protein was dialyzed in 0.3 M NaCl, 50 mM HEPES (pH 8.0), 2 mM MgCl2, and 0.5 mM TCEP at room temperature overnight and loaded onto a 5 mL IMAC column equilibrated with 0.1 M NiSO4. The protein was eluted in 3 CVs of a buffer containing 25 mM imidazole and then concentrated to 5 mL before size exclusion chromatography [Sephacryl S-200 column (Amersham/GE)] in 150 mM NaCl, 20 mM Tris base (pH 7.5), and 0.5 mM TCEP. Purified protein was dialyzed into 50 mM NaCl, 20 mM Tris base (pH 7.5), and 0.5 mM TCEP and then concentrated to 6.0 mg mL−1. The protein was >90% pure as judged by sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Crystallization of Rv3377c.
Rv3377c was crystallized using the vapor diffusion method in 20% polyethylene glycol (PEG) 8000 and 0.1 M (cyclohexylamino)ethanesulfonic acid (CHES) (pH 9.5) with 2 mM MgCl2. Rod-shaped crystals were cryoprotected in a well solution with 20% glycerol and frozen in liquid nitrogen.
Structure Determination.
One native X-ray data set was collected to 2.75 Å resolution at Lawrence Berkeley National Laboratory Advanced Light Source beamline 8.3.1 at 100 K and 11111 eV.
Molecular replacement models were prepared from eight template structures identified with hhsearch.28 Models were trimmed to aligned residues; non-identical side chains were truncated at Cγ, and molecular replacement was carried out in space group C2 using Phaser.29 One of the template structures, 3pya,30 an ent-copalyl diphosphate synthase, gave a very weak molecular replacement hit with a TFZ of 5.0 and an LLG of 39. Modeling building and refinement in MR-Rosetta31 dramatically improved the agreement with the crystal data, giving an LLG of 119; however, the resulting density map was noisy and suffered from model bias, and the model was difficult to improve. Further refinement in MR-Rosetta using this model as a starting point was unsuccessful, as was re-refinement of the original model in the new density map.
To allow further improvement of the model, a structural alignment was made between the MR-Rosetta refined model and all of the templates identified initially. This was done for the two domains of the protein separately; the process identified 3p5p25 as the template that best matched the N-terminal domain of the MR_Rosetta refined model and 2sqc32 as the template best matching the C-terminal domain. A hybrid model and sequence alignment was manually constructed from these two templates (superimposed on the MR-Rosetta model). MR-Rosetta was again run on this starting model, using the density map phased from the previous MR-Rosetta model. The resulting structure had a Phaser LLG of 412, and the model-phased map was readily interpretable by phenix.-autobuild,33 yielding R and Rfree values of 0.31 and 0.36, respectively.
Electronic Structure Calculation.
The electronic structure calculations [specifically, density functional theory (DFT)] were performed with Gaussian0934 with the mPW1PW91/6–31+G(d,p) functional/level of theory.33–39 The isoprenyl diphosphate tail of the carbocation structures were truncated in the gas phase calculation to avoid unnecessary sampling. Truncated carbocation intermediates 2–4 and transition state structures B–D were characterized in the manner described above (see Figure 2 for relative energies and Figure s4 for the description of the truncated structure). Intermediates 2–4 were then subjected to conformational search using Spartan 10 with the MMFF force field.35 All conformers were then optimized using ωB97X-D/6–31+G-(d,p).36 The conformers within 5 kcal/mol of the lowest-energy conformer were kept for each intermediate, resulting in two conformers of 2, two conformers of 3, four conformers of 4, and five conformers of 5. (Relative energies of the conformers are shown in Table s3. Structures are provided in the Supporting Information.)
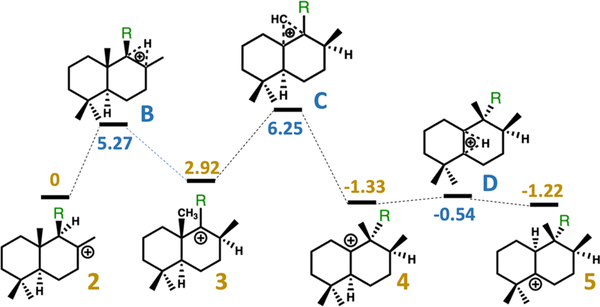
Figure 2.
Relative free energies of intermediates and transition state structures in kilocalories per mole, calculated with mPW1PW91/6–31+G(d,p) (see the Supporting Information for details). Yellow numbers are relative energies for minima, and blue numbers are relative energies for transition state structures. Note that the isoprenyl diphosphate tail was truncated to a methyl group for these calculations [R = CH3 rather than R = CH2CH2C(CH3)=CHCH2OPP was used].
Docking Simulation.
The optimized conformers of intermediates 2–5 were docked into the Rv3377c structure with Rosetta Molecular Modeling Suite47,48 using the Ref2015 scoring function.37 The crystal structure of Rv3377c was relaxed with the FastRelax38 procedure prior to docking simulations. During the docking simulation, a set of constraints was applied between Asp295 and the carbocation intermediates. A proton of Asp295 was donated to the substrate in the first step of the reaction (see Figure s4); therefore, distance, angle, and dihedral constraints were applied, and details about the constraints are summarized in Table s4. Then, 25000 docking simulations were performed for each intermediate. The resulting poses for each intermediate were then combined and filtered on the basis of the following criteria. (1) Constraint satisfaction. Only poses with a constraint score of ≤1 were kept for the next step filtering (a score of 0 means a perfect satisfaction of the constraints). (2) Total protein score. The lowest 25% poses were kept for the next step filtering. (3) Interface energy. Poses that were one standard deviation or lower from the mean interface energy were kept. Poses that passed the filtering undergo a pairwise root-mean-square deviation (RMSD) calculation of carbon atoms; i.e., RMSDs were calculated between each pose of intermediate 2 to 3, 2 to 4, and 2 to 5. Results of the RMSD calculation are summarized in Figure s5.
RESULTS AND DISCUSSION
Crystal Structure of Rv3377c.
Rv3377c was expressed in Escherichia coli, and the purified protein was crystallized in 20% polyethylene glycol (PEG) 8000 and 0.1 M (cyclohexylamino)ethanesulfonic acid (CHES) (pH 9.5) with 2 mM MgCl2. Despite repeated efforts over seven years and tests with mutant proteins suggested by the surface entropy reduction server,39 crystals were obtained only once. Native data were collected to 2.75 Å resolution, but the lack of a heavy-atom derivative or sufficiently related structures precluded crystallographic phase calculations. The structure was determined using molecular replacement with Rosetta-MR. The search model was a hybrid constructed from several different distant templates [Protein Data Bank (PDB) entries 2sqc32 and 3p5p25]. An initial solution with one molecule per asymmetric unit was obtained and refined using data processed in space group C2. The refined model (Rfree = 0.2392, and R = 0.2139) contains two molecules in the asymmetric unit. Residues 1, 103–113, 139–143, and 501 were not modeled due to a lack of electron density. Although 2.0 mM MgCl2 was added to the mother liquor of the crystal, with the intention of determining the catalytically important Mg2+ binding site, no bound Mg2+ ions can be interpreted in the electron density map of the crystal structure. Data collection and refinement statistics are given in the Supporting Information. The Rv3377c structure can be obtained from the Protein Data Bank (entry 6VPT).
Rv3377c contains two helical domains that form a dumbbell shape and correspond to the β and γ domains of other class II diterpene cyclases (Figure 1a). Overlays of Rv3377c and close diterpene synthases taxadiene25 and squalene-hopene cyclase32 are shown in Figure 1b. The β and γ domains are conserved among different diterpene synthases as shown. Domain 1, or the β domain, forms an α6–α6 barrel of two concentric rings of parallel α chains and consists of α1 and α11–α20. The N-terminal and C-terminal helices are close to each other. Domain 2, or the γ domain, contains an α–α barrel consisting of helices α2–α10. The inner α helices of both domains are positioned so that the amino ends point toward an interdomain region, which consists of long loops from both domains and encloses a cavity of ∼738.4 Å3, measured by CAVER analyst with a 1.4 Å radius probe.40 An extended loop between the last two helices extends away from the β domain, packs against the interdomain connections, and inserts into a cleft in the γ domain. Lys476 at the tip of the loop forms an ion pair with Glu174 in the γ domain (see the Supporting Information for details).
Like other class II terpene cyclases, Rv3377c contains the QW motif, which has a general consensus of QX2–5GXW. There are two QW motifs in Rv3377c, and both are composed of QxxDGSWG. One QW motif is in the β domain, and the other is in the γ domain. Both motifs are on the outer surface of the protein, connecting two α helices, and the side chains of the Gln and Trp stack, as seen in other terpene cyclases1,3 (see the Supporting Information for details)
Electronic Structure Calculations.
Rv3377c catalyzes the conversion of GGPP to tuberculosinyl diphosphate. A reasonable mechanism for this transformation involves protonation and cyclization to form a carbocation with the relative configuration of copalyl diphosphate (CPP) (2 in Figure 2),14,19 followed by a sequence of 1,2-hydride shift (to form 3), 1,2-methyl group migration (to form 4), and another 1,2-hydride shift (to form 5). Deprotonation would then yield tuberculosinyl diphosphate. Dickschat and co-workers have shown through isotopic labeling studies that the relative configuration of the product is consistent with a chair/chair conformation of the precursor and initial protonation on the si face of the C=C π-bond of the terminal isoprene unit, i.e., the relative configuration of CPP.41
This putative mechanism was subjected to scrutiny using DFT calculations.42 All DFT calculations were performed with Gaussian09.34 All geometries were optimized using the mPW1PW91 method34,43 and the 6–31+G(d,p) basis set.44–48 All stationary points were characterized as minima or transition state structures using frequency calculations. Intrinsic reaction coordinate (IRC) calculations were used to confirm that transition state structures were connected directly to the intermediates shown.49,50 Relative free energies of intermediates and transition state structures along the reaction pathway are shown in Figure 2. Given the flexibility of the isoprenyl diphosphate tail in the absence of the enzyme, this group was truncated to a methyl group for DFT calculations, as shown in Figure 2.
Docking Simulations.
Conformers of each intermediate structure were identified with the MMFF force field using Spartan 10.35 For generating conformers of diphosphate-containing structures, fluorine was used in place of OPP, an approach analogous to that described previously for another terpene synthase (to avoid unnecessary sampling).51 All conformers generated were then fully optimized using Gaussian09 at the ωB97X-D/6–31+G(d,p) level of theory.36 Resulting structures within 5 kcal/mol of the lowest-energy conformer for each intermediate were kept and combined into conformational libraries for docking (see the Supporting Information for additional information).
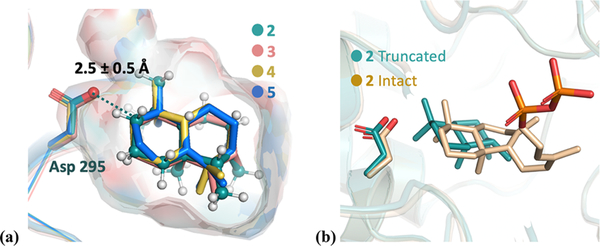
Docking simulations of intermediate structures into the Rv3377c protein structure were performed with the TerDockin approach51–53 using the Rosetta Molecular Modeling Suite.54,55 The crystal structure of Rv3377c was relaxed with the FastRelax38 procedure. The conformational libraries for each intermediate were then separately docked into the relaxed protein structure. Chemically meaningful constraints were applied during the docking simulations.51,53 Specifically, because the first step of the reaction mechanism is donation of a proton from Asp295 to the substrate to generate 2,1 the protonated alkyl carbon was constrained to Asp295 [distance of 2.5 ± 0.5 Å (see the Supporting Information for additional details)]. Then, 25000 docking runs were carried out for each intermediate; i.e., 100000 total docking runs were performed (all input and output files are attached as Supporting Information).
Results from docking runs for each structure were combined and then filtered on the basis of the satisfaction of the constraint, total protein energy (the lowest 25% was retained), and interface energy (only structures that were one standard deviation or lower from the mean were considered). The resulting docked structures were then aligned using TMalign,56 and RMSD calculations were performed on each carbon in the skeleton between intermediates. The resulting bound structures are shown in Figure 3a (see the Supporting Information for additional details about RMSD calculations). All intermediates are converged into a single binding mode. To confirm that docking truncated versions of carbocations does not lead to spurious results, docking of intact intermediate 2 was performed. As shown in Figure 3b, similar binding modes are predicted (see the Supporting Information for details). However, it has been observed that upon ligand binding there are often significant structural changes in the region interacting with the diphosphate with limited structural changes surrounding the core of the substrate. Therefore, as these changes in structure upon phosphate binding are likely beyond our current modeling capabilities, we emphasize here that our predictions about binding modes are relevant only to the bicyclic core of the substrate.
Figure 3.
(a) Overlay of lowest-RMSD structures for intermediates 2–5 (colored green, salmon, yellow, and blue, respectively). Asp295 is highlighted as sticks. The protons of intermediate 2 are colored white. The protonated carbon is highlighted as a dark blue sphere, which is a constraint to the Asp295 oxygen during the docking simulation. The surface indicates the active-site cavity. (b) Overlay of the lowest-energy docking result of intact 2 (beige) and truncated 2 (green, same structure as 2 in panel a).
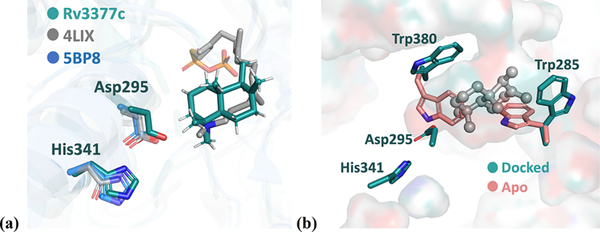
The crystal structure of Rv3377c shows that the active-site cavity sits at the interface of the β and γ domains, which agrees with structures of other class II diterpene synthases. The general acid, Asp295, is positioned deep in the cavity and forms hydrogen bonds to His341 and a potential water molecule. This corresponds to the general acid Asp313 and His359 in ent-copalyl diphosphate synthase (CPS) from Streptomyces platensis, the first bacterial class II diterpene synthase with a determined crystal structure.57 In the plant ent-copalyl diphosphate synthase from Arabidopsis thaliana, the general acid, Asp379, is hydrogen bonded with Asn425 instead.30 An overlay of these three structures is shown in Figure 4a. The binding mode of the co-crystallized substrate analogue in CPS agrees with that of docked intermediate 2 in Rv3377c. The hydrogen bonding orients the general acid Asp and facilitates the initial proton transfer. Packing against each other, Trp285 and Trp380 block access to the general acid. The arrangement of Trp285 and Trp380 after the docking suggests that they move to create space upon substrate binding, as shown in Figure 4b. Similarly, studies on ent-CPS suggested that the loop containing Trp380 undergoes a conformational change upon substrate analogue binding.21
Figure 4.
(a) Overlay of Rv3377c and CPS from a bacterium and a plant. The Rv3377c structure is colored green, bacterial CPS from S. platensis blue (PDB entry 5BP8), and plant CPS from A. thaliana (AtCPS) gray (PDB entry 4LIX). The gray sticks represent the (S)-15-aza-14,15-dihydrogeranylgeranyl thiolodiphosphate that is co-crystallized with 4LIX. The residues highlighted as sticks represent key catalytic residues: Asp295 and His341. (b) Comparison between the docked Rv3377c structure (green) with the apo crystal structure (salmon), with the gray sphere and stick representing the predicted binding mode of intermediate 2. On the basis of the modeling, both Trp380 and Trp285 are predicted to serve as a gate for binding.
Diterpene synthases bind a flexible substrate in an orientation relevant to product formation, trigger carbocation formation, shield reactive carbocation intermediates from premature quenching by water molecules, and allow inherent carbocation reactivity to be expressed.58–61 Elucidating how carbocations—minima, transition state structures, and species between them along rearrangement reaction coordinates—bind to terpene active sites is a frontier area of research.51,53,62–64 AtCPS shares the same substrate, GGPP, and first carbocation intermediate, 2, with Rv3377c. Thus, the sequence divergence of AtCPS and Rv3377c provides a unique opportunity to identify which residues mediate the formation of common cyclic intermediate 2 and which distinguish the following reaction pathways.
Identical residues are clustered around the pyrophosphate binding site, the DXDTT motif, and the bottom of the active-site cavity (see the Supporting Information for details). A reasonable hypothesis is that the shared residues mediate common steps in the AtCPS and Rv3377c reactions, while residue differences are responsible for the formation of the distinct products. In AtCPS, a base abstracts a proton from intermediate 2, resulting in C=C formation, but in Rv3377c, hydride and methyl group migrations occur before deprotonation. Consequently, an active-site base must be positioned differently.
The structure of the Rv3377c active site is compared to that of AtCPS in Figure 5. Previous studies of AtCPS have shown that a water molecule activated by ligation of a His/Asn pair could constitute the base that deprotonates the carbocation intermediate.65 In AtCPS, the H263F/N322L double mutation results in 75% of the product being (−)-kolavenyl diphosphate, consistent with this pair functioning as the base. In Rv3377c, these residues are replaced by hydrophobic residues, F171 and A217, allowing rearrangement to proceed past carbocation 2. Similar behavior was observed in OsCPS4 (syn-copalyl diphosphate synthase from Oryza sativa) where mutation of H501F leads to rearrangement instead of immediate quenching of the initial carbocation intermediate.66
Figure 5.
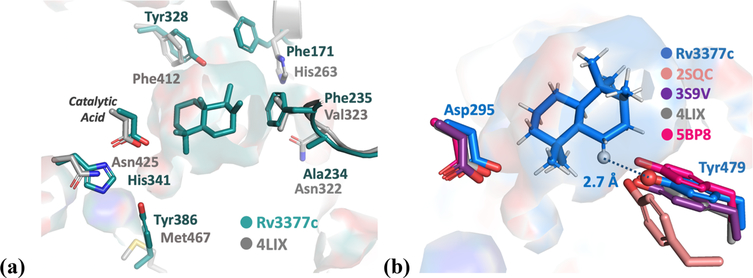
(a) Comparison of Rv3377c (green) and AtCPS (gray) active sites. (b) Tyr is conserved among class II diterpene synthases: blue for intermediate 5 docked into the Rv3377c structure, salmon for squalene-hopene synthase (PDB entry 2SQC), purple for abietadiene synthase from Abies grandis (AgAS, PDB entry 3S9V), gray for ent-copalyl diphosphate synthase from A. thaliana (AtCPS, PDB entry 4LIX), and pink for ent-copalyl diphosphate synthase from S. platensis (PDB entry 5BP8).
Additional residue differences between AtCPS and Rv3377c (within 6 Å of the ligand) are shown in Figure 5a. Changes to aromatic residues, including Tyr328, Phe235, and Tyr386, may lead to favorable interactions with the carbocation produced by rearrangement of 2.
On the basis of our docking results, the identity of a possible residue that may deprotonate carbocation 5 was identified. The oxygen atom of Tyr479 is predicted to be ∼2.7 Å from the proton that is ultimately removed. This group may function directly as a base or may activate a water molecule for deprotonation. The pKa of an O-protonated phenol is around −6, and the pKa of a typical carbocation lacking conjugation is less than −10.67 This indicates that the transfer of a proton from a carbocation to the tyrosine is energetically feasible. Similarly, a serine was reported to accept a proton from a carbocation intermediate in ent-kaurene synthase.68 This Tyr is conserved in class II diterpene synthases, as shown in Figure 5b. Previous studies suggested that the corresponding Tyr in AgAS might be the base for the deprotonation of the carbocation intermediate,22 while the equivalent Tyr in squalene-hopene cyclase (PDB entry 2SQC) was suggested to stabilize carbocations formed early along the polycyclization reaction coordinate.32
CONCLUSIONS
We describe the three-dimensional crystal structure of Rv3377c, the diterpene cyclase from M. tuberculosis. The results of docking calculations using this new structure suggest that the substrate need not move much during rearrangement (Figure 3a). Thus, given the energetics predicted for the enzyme-free rearrangement (Figure 2), upon carbocation formation, the conversion of 2 to 5 is likely extremely rapid and does not require active manipulation by the surrounding enzyme.58 The results shown in Figure 2 also indicate that 2–5 may all be in equilibrium, with the identity of the final product of Rv3377c determined by the position of the base that terminates the cascade reaction.69 On the basis of our docking results, we propose that Tyr479 is likely this base.
Supplementary Material
ACKNOWLEDGMENTS
L.M.P. started this work in 2005 as part of her Ph.D. thesis with T.A. Due to technical difficulties obtaining protein crystals, this work could not be completed in a timely manner. T.A. passed away in March 2014, and although he was unable to take part in writing the manuscript, his contributions to it remain. A great scientist and mentor, T.A. always pursued how structure can inform function and mechanism. This work would not have been possible without his guidance, support, and tenacity. This one’s for you, Tom. You are missed. Y.Z. acknowledges V. Yarov-Yarovoy for his feedback on this project.
Funding
The authors gratefully acknowledge support from the National Science Foundation (Grants 1827246, 1805510, 1627539, CHE-1565933, and CHE-030089 for computer time via the XSEDE) and the National Institutes of Health (Grants 2R01GM076324 and P42ES004699), the University of California—Davis, and the Rosetta Commons.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00762.
Supplemental methods and information (PDF)
Crystal structure files, docking input and output files, and electronic structural calculation files (ZIP)
Accession Codes
Protein Data Bank entry 6VPT.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.0c00762
Contributor Information
Yue Zhang, Department of Chemistry, University of California-Davis, Davis, California 95616, United States.
Lisa M. Prach, Department of Molecular and Cell Biology, University of California, Berkeley, California 94720, United States
Terrence E. O’Brien, Department of Chemistry, University of California-Davis, Davis, California 95616, United States.
Frank DiMaio, Department of Biochemistry, University of Washington, Seattle, Washington 98195, United States.
Daniil M. Prigozhin, Molecular Biophysics and Integrated Bioimaging Division, Lawrence Berkeley National Laboratory, Berkeley, California 94720, United States
Jacob E. Corn, Department of Biology, ETH Zurich, 8093 Zurich, Switzerland
Tom Alber, Department of Molecular & Cell Biology and QB3 Institute, University of California, Berkeley, California 94720, United States.
Justin B. Siegel, Department of Chemistry, Department of Biochemistry and Molecular Medicine, and Genome Center, University of California-Davis, Davis, California 95616, United States
Dean J. Tantillo, Department of Chemistry, University of California-Davis, Davis, California 95616, United States.
REFERENCES
- (1).Christianson DW (2017) Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 117, 11570–11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Poulter CD, Craig J, and Rr G (2008) Farnesyl Pyrophosphate. Encycl. Genet. Genomics, Proteomics Informatics 253, 672–672. [Google Scholar]
- (3).Christianson DW (2006) Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. 106, 3412–3442. [DOI] [PubMed] [Google Scholar]
- (4).Sacchettini JC, and Poulter CD (1997) Creating Isoprenoid Diversity. Science 277, 1788–1789. [DOI] [PubMed] [Google Scholar]
- (5).Gerber NN (1969) A Volatile Metabolite of Actinomycetes, 2-Methylisoborneol. J. Antibiot. 22, 508–509. [DOI] [PubMed] [Google Scholar]
- (6).Dickschat JS, Wenzel SC, Bode HB, Müller R, and Schulz S (2004) Biosynthesis of volatiles by the myxobacterium Myxococcus xanthus. ChemBioChem 5, 778–787. [DOI] [PubMed] [Google Scholar]
- (7).Dickschat JS, Martens T, Brinkhoff T, Simon M, and Schulz S (2005) Volatiles released by a Streptomyces species isolated from the North Sea. Chem. Biodiversity 2, 837–865. [DOI] [PubMed] [Google Scholar]
- (8).Schulz S, and Dickschat JS (2007) Bacterial volatiles: The smell of small organisms. Nat. Prod. Rep. 24, 814–842. [DOI] [PubMed] [Google Scholar]
- (9).Yamada Y, Kuzuyama T, Komatsu M, Shin-ya K, Omura S, Cane DE, and Ikeda H (2015) Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. U. S. A. 112, 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Izaguirre G, Hwang CJ, Krasner SW, and McGuire MJ (1982) Geosmin and 2-methylisoborneol from cyanobacteria in three water supply systems. Appl. Environ. Microbiol. 43, 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Dickschat JS, Nawrath T, Thiel V, Kunze B, Müller R, and Schulz S (2007) Biosynthesis of the off-flavor 2-methylisoborneol by the myxobacterium Nannocystis exedens. Angew. Chem., Int. Ed. 46, 8287–8290. [DOI] [PubMed] [Google Scholar]
- (12).Mann FM, Xu M, Chen X, Fulton DB, Russell DG, and Peters RJ (2009) Edaxadiene: A new bioactive diterpene from Mycobacterium tuberculosis. J. Am. Chem. Soc. 131, 17526–17527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Mann FM, Prisic S, Hu H, Xu M, Coates RM, and Peters RJ (2009) Characterization and inhibition of a class II diterpene cyclase from Mycobacterium tuberculosis. Implications for tuberculosis. J. Biol. Chem. 284, 23574–23579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Nakano C, and Hoshino T (2009) Characterization of the Rv3377c gene product, a type-B diterpene cyclase, from the Mycobacterium tuberculosis H37 genome. ChemBioChem 10, 2060–2071. [DOI] [PubMed] [Google Scholar]
- (15).Layre E, Lee HJ, Young DC, Jezek Martinot A, Buter J, Minnaard AJ, Annand JW, Fortune SM, Snider BB, Matsunaga I, Rubin EJ, Alber T, and Moody DB (2014) Molecular profiling of Mycobacterium tuberculosis identifies tuberculosinyl nucleoside products of the virulence-associated enzyme Rv3378c. Proc. Natl. Acad. Sci. U. S. A. 111, 2978–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Prach L, Kirby J, Keasling JD, and Alber T (2010) Diterpene production in Mycobacterium tuberculosis. FEBS J. 277, 3588–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Pethe K, Swenson DL, Alonso S, Anderson J, Wang C, and Russell DG (2004) Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc. Natl. Acad. Sci. U. S. A. 101, 13642–13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Nakano C, Ootsuka T, Takayama K, Mitsui T, Sato T, and Hoshino T (2011) Characterization of the Rv3378c gene product, a new diterpene synthasefor producing tuberculosinol and (13R, S)-isotuberculosinol (nosyberkol), from the mycobacterium tuberculosis H37Rv genome. Biosci., Biotechnol., Biochem. 75, 75–81. [DOI] [PubMed] [Google Scholar]
- (19).Nakano C, Okamura T, Sato T, Dairi T, and Hoshino T (2005) Mycobacterium tuberculosis H37Rv3377c encodes the diterpene cyclase for producing the halimane skeleton. Chem. Commun, 1016–1018. [DOI] [PubMed] [Google Scholar]
- (20).Ashby MN, and Edwards PA (1990) Elucidation of the deficiency in two yeast coenzyme Q mutants. Characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J. Biol. Chem. 265, 13157–13164. [PubMed] [Google Scholar]
- (21).KÖksal M, Hu H, Coates RM, Peters RJ, and Christianson DW (2011) Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat. Chem. Biol. 7, 431–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhou K, Gao Y, Hoy JA, Mann FM, Honzatko RB, and Peters RJ (2012) Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis. J. Biol. Chem. 287, 6840–6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Janke R, Görner C, Hirte M, Brück T, and Loll B (2014) The first structure of a bacterial diterpene cyclase: CotB2. Acta Crystallogr., Sect. D: Biol. Crystallogr. 70, 1528–1537. [DOI] [PubMed] [Google Scholar]
- (24).Cao R, Zhang Y, Mann FM, Huang C, Mukkamala D, Hudock MP, Mead ME, Prisic S, Wang K, Lin FY, Chang TK, Peters RJ, and Oldfield E (2010) Diterpene cyclases and the nature of the isoprene fold. Proteins: Struct., Funct., Genet. 78, 2417–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Köksal M, Jin Y, Coates RM, Croteau R, and Christianson DW (2011) Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 469, 116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Gao Y, Honzatko RB, and Peters RJ (2012) Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat. Prod. Rep. 29, 1153–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Moosmann P, Ecker F, Leopold-Messer S, Cahn JKB, Dieterich CL, Groll M, and Piel J (2020) A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds. Nat. Chem. 12, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Söding J (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960. [DOI] [PubMed] [Google Scholar]
- (29).McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Köksal M, Potter K, Peters RJ, and Christianson DW (2014) 1.55 Å-Resolution Structure of Ent-Copalyl Diphosphate Synthase and Exploration of General Acid Function By Site-Directed Mutagenesis. Biochim. Biophys. Acta, Gen. Subj. 1840, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Dimaio F, Terwilliger TC, Read RJ, Wlodawer A, Oberdorfer G, Wagner U, Valkov E, Alon A, Fass D, Axelrod HL, Das D, Vorobiev SM, Iwaï H, Pokkuluri PR, and Baker D (2011) Improved molecular replacement by density- and energy-guided protein structure optimization. Nature 473, 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wendt KU, Lenhart A, and Schulz GE (1999) The structure of the membrane protein squalene-hopene cyclase at 2.0 Å resolution. J. Mol. Biol. 286, 175–187. [DOI] [PubMed] [Google Scholar]
- (33).Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, and Adams PD (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr., Sect. D: Biol. Crystallogr. 64, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr., Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Keith T, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, and Fox DJ (2013) Gaussian 09, rev. D.01, Gaussian Inc. [Google Scholar]
- (35).Shao Y, Molnar LF, Jung Y, Kussmann J, Ochsenfeld C, Brown ST, Gilbert ATB, Slipchenko LV, Levchenko SV, O’Neill DP, DiStasio RA, Lochan RC, Wang T, Beran GJO, Besley NA, Herbert JM, Yeh Lin C, Van Voorhis T, Hung Chien S, Sodt A, Steele RP, Rassolov VA, Maslen PE, Korambath PP, Adamson RD, Austin B, Baker J, Byrd EFC, Dachsel H, Doerksen RJ, Dreuw A, Dunietz BD, Dutoi AD, Furlani TR, Gwaltney SR, Heyden A, Hirata S, Hsu CP, Kedziora G, Khalliulin RZ, Klunzinger P, Lee AM, Lee MS, Liang W, Lotan I, Nair N, Peters B, Proynov EI, Pieniazek PA, Min Rhee Y, Ritchie J, Rosta E, David Sherrill C, Simmonett AC, Subotnik JE, Lee Woodcock H, Zhang W, Bell AT, Chakraborty AK, Chipman DM, Keil FJ, Warshel A, Hehre WJ, Schaefer HF, Kong J, Krylov AI, Gill PMW, and Head-Gordon M (2006) Advances in methods and algorithms in a modern quantum chemistry program package. Phys. Chem. Chem. Phys. 8, 3172–3191. [DOI] [PubMed] [Google Scholar]
- (36).Chai J. Da, and Head-Gordon M (2008) Long-range corrected hybrid density functionals with damped atom-atom dispersion corrections. Phys. Chem. Chem. Phys. 10, 6615–6620. [DOI] [PubMed] [Google Scholar]
- (37).Park H, Bradley P, Greisen P, Liu Y, Mulligan VK, Kim DE, Baker D, and Dimaio F (2016) Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. J. Chem. Theory Comput. 12, 6201–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Conway P, Tyka MD, DiMaio F, Konerding DE, and Baker D (2014) Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 23, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Goldschmidt L, Cooper DR, Derewenda ZS, and Eisenberg D (2007) Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 16, 1569–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, and Damborsky J (2012) CAVER 3.0: A Tool for the Analysis of Transport Pathways in Dynamic Protein Structures. PLoS Comput. Biol. 8, e1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Citron CA, Rabe P, Barra L, Nakano C, Hoshino T, and Dickschat JS (2014) Synthesis of isotopically labelled oligoprenyl diphosphates and their application in mechanistic investigations of terpene cyclases. Eur. J. Org. Chem. 2014, 7684–7691. [Google Scholar]
- (42).Tantillo DJ (2011) Biosynthesis via carbocations: Theoretical studies on terpene formation. Nat. Prod. Rep. 28, 1035–1053. [DOI] [PubMed] [Google Scholar]
- (43).Matsuda SPT, Wilson WK, and Xiong Q (2006) Mechanistic insights into triterpene synthesis from quantum mechanical calculations. Detection of systematic errors in B3LYP cyclization energies. Org. Biomol. Chem. 4, 530–543. [DOI] [PubMed] [Google Scholar]
- (44).Becke AD (1993) A new mixing of Hartree-Fock and local density-functional theories. J. Chem. Phys. 98, 1372–1377. [Google Scholar]
- (45).Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652. [Google Scholar]
- (46).Lee C, Yang W, and Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B: Condens. Matter Mater. Phys. 37, 785–789. [DOI] [PubMed] [Google Scholar]
- (47).Stephens PJ, Devlin FJ, Chabalowski CF, and Frisch MJ (1994) Ab Initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 98, 11623–11627. [Google Scholar]
- (48).Tirado-Rives J, and Jorgensen WL (2008) Performance of B3LYP density functional methods for a large set of organic molecules. J. Chem. Theory Comput. 4, 297–306. [DOI] [PubMed] [Google Scholar]
- (49).Gonzalez C, and Schlegel HB (1990) Reaction path following in mass-weighted internal coordinates. J. Phys. Chem. 94, 5523–5527. [Google Scholar]
- (50).Fukui K (1981) The Path of Chemical Reactions - The IRC Approach. Acc. Chem. Res. 14, 363–368. [Google Scholar]
- (51).O’Brien TE, Bertolani SJ, Zhang Y, Siegel JB, and Tantillo DJ (2018) Predicting Productive Binding Modes for Substrates and Carbocation Intermediates in Terpene Synthases - Bornyl Diphosphate Synthase As a Representative Case. ACS Catal. 8, 3322–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Das S, Shimshi M, Raz K, Nitoker Eliaz N, Mhashal AR, Ansbacher T, and Major DT (2019) EnzyDock: Protein–Ligand Docking of Multiple Reactive States along a Reaction Coordinate in Enzymes. J. Chem. Theory Comput. 15, 5116–5134. [DOI] [PubMed] [Google Scholar]
- (53).O’Brien TE, Bertolani SJ, Tantillo DJ, and Siegel JB (2016) Mechanistically informed predictions of binding modes for carbocation intermediates of a sesquiterpene synthase reaction. Chem. Sci. 7, 4009–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Lange OF, and Baker D (2012) Resolution-adapted recombination of structural features significantly improves sampling in restraint-guided structure calculation. Proteins: Struct., Funct., Genet. 80, 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Alford RF, Leaver-fay A, Jeliazkov JR, O’Meara MJ, Dimaio FP, Park H, Shapovalov MV, Renfrew PD, Mulligan K, Kappel K, Labonte JW, Pacella MS, Bonneau R, et al. (2017) J. Chem. Theory Comput. 13, 3031–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Zhang Y, and Skolnick J (2005) TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Rudolf JD, Dong L-B, Cao H, Hatzos-Skintges C, Osipiuk J, Endres M, Chang CY, Ma M, Babnigg G, Joachimiak A, Phillips GN, and Shen B (2016) Structure of the ent-Copalyl Diphosphate Synthase PtmT2 from Streptomyces platensis CB00739, a Bacterial Type II Diterpene Synthase. J. Am. Chem. Soc. 138, 10905–10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Tantillo DJ (2017) Importance of Inherent Substrate Reactivity in Enzyme-Promoted Carbocation Cyclization/Rearrangements. Angew. Chem., Int. Ed. 56, 10040–10045. [DOI] [PubMed] [Google Scholar]
- (59).Cane DE (1990) Enzymatic Formation of Sesquiterpenes. Chem. Rev. 90, 1089–1103. [Google Scholar]
- (60).Pemberton RP, Ho KC, and Tantillo DJ (2015) Modulation of inherent dynamical tendencies of the bisabolyl cation via preorganization in epi-isozizaene synthase. Chem. Sci. 6, 2347–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Hammer SC, Syrén PO, and Hauer B (2016) Substrate Pre-Folding and Water Molecule Organization Matters for Terpene Cyclase Catalyzed Conversion of Unnatural Substrates. ChemistrySelect 1, 3589–3593. [Google Scholar]
- (62).Freud Y, Ansbacher T, and Major DT (2017) Catalytic Control in the Facile Proton Transfer in Taxadiene Synthase. ACS Catal. 7, 7653–7657. [Google Scholar]
- (63).Major DT, and Weitman M (2012) Electrostatically guided dynamics-the root of fidelity in a promiscuous terpene synthase? J. Am. Chem. Soc. 134, 19454–19462. [DOI] [PubMed] [Google Scholar]
- (64).Driller R, Janke S, Fuchs M, Warner E, Mhashal AR, Major DT, Christmann M, Brück T, and Loll B (2018) Towards a comprehensive understanding of the structural dynamics of a bacterial diterpene synthase during catalysis. Nat. Commun. 9, 3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Potter KC, Zi J, Hong YJ, Schulte S, Malchow B, Tantillo DJ, and Peters RJ (2016) Blocking Deprotonation with Retention of Aromaticity in a Plant ent-Copalyl Diphosphate Synthase Leads to Product Rearrangement. Angew. Chem., Int. Ed. 55, 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Potter KC, Jia M, Hong YJ, Tantillo D, and Peters RJ (2016) Product Rearrangement from Altering a Single Residue in the Rice syn-Copalyl Diphosphate Synthase. Org. Lett. 18, 1060–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).McCormack AC, More O’Ferrall RA, O’Donoghue AMC, and Rao SN (2002) Protonated benzofuran, anthracene, naphthalene, benzene, ethene, and ethyne: Measurements and estimates of pKa and pKR. J. Am. Chem. Soc. 124, 8575–8583. [DOI] [PubMed] [Google Scholar]
- (68).Jia M, Zhang Y, Siegel JB, Tantillo DJ, and Peters RJ (2019) Switching on a Nontraditional Enzymatic Base - Deprotonation by Serine in the ent-Kaurene Synthase from Bradyrhizobium japonicum. ACS Catal. 9, 8867–8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Hong YJ, and Tantillo DJ (2011) The taxadiene-forming carbocation cascade. J. Am. Chem. Soc. 133, 18249–18256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.