Abstract
State-of-the-art tissue-clearing methods provide subcellular-level optical access to intact tissues from individual organs and even to some entire mammals. When combined with light-sheet microscopy and automated approaches to image analysis, existing tissue-clearing methods can speed up and may reduce the cost of conventional histology by several orders of magnitude. In addition, tissue-clearing chemistry allows whole-organ antibody labelling, which can be applied even to thick human tissues. By combining the most powerful labelling, clearing, imaging and data-analysis tools, scientists are extracting structural and functional cellular and subcellular information on complex mammalian bodies and large human specimens at an accelerated pace. The rapid generation of terabyte-scale imaging data furthermore creates a high demand for efficient computational approaches that tackle challenges in large-scale data analysis and management. In this Review, we discuss how tissue-clearing methods could provide an unbiased, system-level view of mammalian bodies and human specimens and discuss future opportunities for the use of these methods in human neuroscience.
Histological techniques have been the standard procedure for investigating tissues for several decades; however, a complete understanding of biological mechanisms in health and disease requires an unbiased exploration of the whole organism, not just selected parts of tissue. This need is particularly evident in the context of the nervous system, which can be found throughout the body. Tissue-clearing methods now allow 3D imaging of intact tissues and even some entire organisms. Indeed, a century-old approach at rendering tissues transparent1 has been almost reinvented with recent developments in tissue-clearing reagents (TABLE 1) and protocols (Supplementary Table 1), efficient fluorescent labelling and rapid volumetric imaging by light-sheet microscopy2–14.
Table 1 |.
Tissue-clearing reagents used for whole-organ or body 3D imaging
| Chemical | Group | Role in tissue clearing | Cocktail or protocol | Refs |
|---|---|---|---|---|
| Methanol | Alcohol | Dehydration and permabilization | iDISCO, iDISCO+ | 4,8,26,36 |
| Ethanol | Alcohol | Dehydration | BABB | 14 |
| tert-Butanol | Alcohol | Dehydration | FluoClearBABB, uDISCO, PEGASOS | 42,169,170 |
| Tetrahydrofuran | Ether | Dehydration | 3DISCO, iDISCO, vDISCO | 4,8,28,33,36,171 |
| Benzyl alcohol | Aromatic alcohol | RI matching and delipidation (putative) | BABB, vDISCO | 14,28,172 |
| Benzyl benzoate | Aromatic ester | RI matching and delipidation (putative) | BABB, PEGASOS, vDISCO | 14,28,170,172 |
| Ethyl-3-phenylprop-2-enoate (ethyl cinnamate) | Aromatic ester | RI matching | Ethyl cinnamate method | 11 |
| Dibenzyl ether | Aromatic ether | RI matching and delipidation (putative) | 3DISCO, iDISCO, iDISCO+ | 4,7,8,26,33,36,171,173 |
| Diphenyl ether | Aromatic ether | RI matching and delipidation (putative) | uDISCO | 42 |
| Hexane | Alkane | Dehydration and delipidation? | BABB | 14 |
| Dichloromethane | Alkyl halide | Delipidation and dehydration | 3DISCO, uDISCO, iDISCO, iDISCO+ | 4,8,26,33,36,42 |
| Glycerol | Polyol | RI matching | Scale reagents, CLARITY, LUCID | 12,174–179 |
| 2,2′-Thiodiethanol | Thioalcohol | RI matching | Single treatment, CLARITY, LUCID, TOMEI | 10,161,177,178,180 |
| Sucrose | Oligosaccharide | RI matching | ScaleCUBIC2, UbasM-2 | 5,6,52,181 |
| D(–)-Fructose | Monosaccharide | RI matching | SeeDB, FRUIT | 13,165 |
| D(–)-Sorbitol | Monosaccharide | RI matching | sRIMS, ScaleS, STP tomography | 3,49,54 |
| Xylitol | Monosaccharide | Dehydration and RI matching | ClearSee | 182 |
| Methyl-β-cyclodextrin | Oligosaccharides | Delipidation (extracting cholesterol) | ScaleS, vDISCO | 28,49 |
| γ-Cyclodextrin | Oligosaccharides | Delipidation (extracting cholesterol) | ScaleS | 49 |
| Polyethylene glycol | Polyether | RI matching | ClearT2, PEGASOS | 163,170 |
| Dimethyl sulfoxide | Sulfoxide | RI matching | ScaleS, STP tomography | 49,50 |
| Urea | Urea | Hydration (putatively contributes to expansion and hence RI matching) and chemical penetration | Scale reagents, ScaleCUBIC reagents UbasM, FRUIT, ClearSee, vDISCO | 5,6,12,28,52,165,175,176,179,181–183 |
| 1,3-Dimethyl-2-imidazolidinone | Urea | Hydration (putatively contributes to expansion and hence RI matching) and chemical penetration | UbasM-1–UbasM-2 | 181 |
| Formamide | Amide | Putatively similar mechanism to urea | ClearT, ClearT2, RTF | 162,184 |
| N-Methylacetamide | Amide | RI matching | Ce3D | 185 |
| Nicotinamide | Aromatic amide | RI matching | CUBIC-R, CUBIC-R+ | 22,27 |
| N-Methylnicotinamide | Aromatic amide | RI matching | CUBIC-R, CUBIC-R+ | 22,27 |
| Antipyrine | Phenyl pyrazolone | RI matching and expansion | CUBIC-X, CUBIC-R, CUBIC-R+ | 17,22,27 |
| 5-(N-2,3-Dihydroxypropylacetamido)-2,4,6-triiodo-N,N′-bis(2,3-dihydroxypropyl) isophthalamide (iohexol) | Contrast reagent | RI matching | RIMS–SeeDB2–Ce3D | 3,52,185 |
| Diatrizoate acid | Contrast agent | RI matching | RI matching reagent in SWITCH protocol, FocusClear | 2,25,47 |
| Iodixanol | Contrast reagent | RI matching | RI matching reagent in SWITCH protocol | 25 |
| N-Acetyl-l-hydroxyproline (trans-1-acetyl-4-hydroxy-l-proline) | Amino acid | Relaxing collagen structure | ScaleS, vDISCO | 28,49 |
| N,N,N′,N′-Tetrakis (2-hydroxypropyl) ethylenediamine (Quadrol) | Amino alcohol | Delipidation, decolorization and RI matching | ScaleCUBIC-1, PACT-deCAL–Bone CLARITY, PEGASOS, vDISCO | 5,6,28,52,74,170 |
| Triethanolamine | Amino alcohol | Delipidation, decolorization and RI matching | ScaleCUBIC-2, RTF | 5,6,52,184 |
| N-Butyldiethanolamine | Amino alcohol | Delipidation, decolonization and pH adjustment | CUBIC-L, CUBIC-R+ | 22,27 |
| Meglumine or N-methyl-d-glucamine | Amino alcohol | Delipidation and RI matching | UbasM-1, RI matching reagent in SWITCH protocol | 25,181 |
| 1,3-Bis(aminomethyl) cyclohexane | Aliphatic amine | Delipidation, decolorization and pH adjustment | CUBIC-HL | 22 |
| Imidazole | Imidazole | Expansion and decalcification (with EDTA) | CUBIC-X, CUBIC-B | 17,22 |
| 1-Methylimidazole | Imidazole | Decolonization | CUBIC-P | 22 |
| Sodium dodecyl sulfate | Ionic detergent | Delipidation | CLARITY and related protocols (PACT, PARS, ACT, FACT, FASTClear and simplified CLARITY), SWITCH | 2,3,9,10,22,25,67,74,89,174,183,186–188 |
| Sodium dodecylbenzenesulfonate | Ionic detergent | Delipidation | CUBIC-HL | 22 |
| Sodium deoxycholate | Ionic detergent | Delipidation | ClearSee | 182 |
| Triton X-100 | Non-ionic detergent | Delipidation | Scale reagents, ScaleCUBIC-1, UbasM-1 (0.2%), Ce3D (0.1%), SUT, CUBIC-L, CUBIC-HL, vDISCO | 5,6,12,22,28,52,175,176,181,183,185 |
| Tween 20 | Non-ionic detergent | Delipidation | FocusClear | 47 |
| EDTA | EDTA | Decalcification | PACT-deCal, Bone CLARITY, CUBIC-B, PEGASOS, vDISCO | 9,22,28,74,170 |
BABB, benzyl alcohol and benzyl benzoate; Ce3D, clearing-enhanced 3D; EDTA, ethylenediaminetetraacetic acid; FACT, fast free-of-acrylamide clearing tissue; LUCID, illuminate cleared organs to identify target molecules; PEGASOS, polyethylene glycol-associated solvent system; RTF, rapid clearing method based on triethanolamine and formamide; RI, refractive index; RIMS, refractive index-matching solution; STP tomography, serial two-photon tomography; TOMEI, transparent plant organ method for imaging; UbasM, urea-based amino-sugar mixture.
Three major tissue-clearing approaches are currently available: hydrophobic, hydrophilic and hydrogel-based methods15,16 (FIG. 1); hydrophobic tissue-clearing and hydrophilic tissue-clearing methods are also referred as ‘solvent’ and ‘aqueous’ tissue-clearing methods, respectively. In general, these tissue-clearing methods, especially in highly efficient protocols, remove lipids (delipidation), pigments (decolourization) and calcium phosphate (decalcification) and aim to match the refractive indices (refractive index (RI) matching) of the specimens and the imaging media by reaching an almost complete level of transparency for intact organs and even entire adult rodent bodies. Although hydrophobic methods usually shrink the tissues and thereby allow the imaging of larger samples, hydrophilic and hydrogel-based methods, if the reagents have low osmolarity, can expand the specimens (expansion), which further increases the transparency and effective resolution (FIG. 1).
Fig. 1 |. Major tissue-clearing methods and their key features.
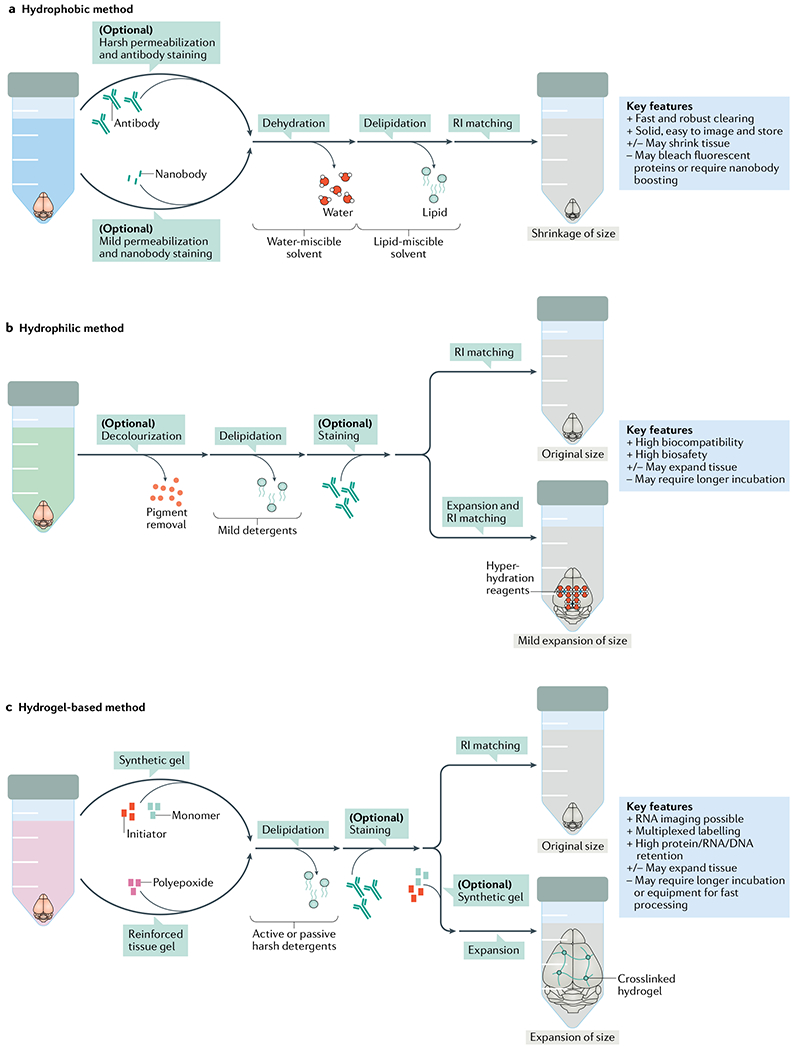
a | Hydrophobic methods rely on a complete dehydration of the tissue, followed by lipid extraction and refractive index (RI) matching using organic solvents. Flydrophobic methods are generally fast and can clear tissues fully. However, some hydrophobic methods can bleach the signal of fluorescent proteins rapidly. b | Hydrophilic methods are based on water-soluble solutions and are usually associated with higher biosafety and compatibility than hydrophobic methods. A potent hydrophilic method such as CUBIC starts with decolourization, which is followed by delipidation, RI matching and (optionally) expansion; some hydrophilic methods need longer incubation times for intact organs. c | Hydrogel embedding forms the third major category of tissue clearing. Hydrogel-based methods use either monomer and initiator molecules to make a synthetic gel or polyepoxide to make a reinforced tissue gel. Hydrogel-based methods can allow retainment of enough RNAs for assays such as fluorescence in situ hybridization and can be used to expand tissues several fold owing to the hydrogel mesh that glues the tissues. Some hydrogel-based methods need longer incubation times for intact organs.
The physical principles of these tissue-clearing methods, and how each tissue-clearing process (that is, delipidation, decolourization, decalcification, RI matching and expansion) contributes to tissue transparency, are beginning to be elucidated by considering the physical properties of organs. A recent study discovered that an organ can act as a polymer gel even in the absence of exogenous polymers17, which was originally predicted by Tanaka and colleagues via a bottom-up approach18,19. If an organ acts as a polymer gel, the RI of an organ (‘polymer gel’) can be described by both the RI and the volume of its components (‘monomers’) according to a theoretical formula called the Lorentz–Lorenz relation20,21. Therefore, delipidation and decalcification change the RI of an organ’s components and hence change the RI of the organ itself, whereas expansion can increase the volume of an organ’s components, also eventually reducing the RI of the organ. In either case, these processes allow the RI of an organ to be more easily matched with the RI of the medium and this results in minimization of light scattering. In addition, the minimization of light absorption, which also contributes to transparency of the specimens, can be achieved by decolourization (removing the pigments) of an organ. Recent comprehensive chemical profiling of hydrophilic tissue-clearing reagents provides a better understanding of the chemical principles of how each chemical functional group can contribute to tissue-clearing processes22.
In parallel to forging a better understanding of the principles underlying tissue-clearing methods, researchers are continuing to improve labelling and data-analysis tools to broaden the applications of tissue-clearing methods. Indeed, labelling options have expanded with the development of ex vivo deep-tissue labelling methods for whole mouse brain4,23–27 and whole mouse body6,27,28 and of in vivo systematic adeno-associated virus-based labelling of CNS and peripheral nervous system cells29,30. Once tissues are labelled and rendered transparent, light-sheet microscopy provides rapid 3D whole-brain imaging at subcellular resolution31. Some forms of light-sheet microscopy can already generate isotropic and high-resolution images with a substantially higher signal-to-noise ratio and a markedly higher resolution than for the images that can be generated by two-photon microscopy31. Furthermore, the development of powerful machine-learning algorithms will be essential to optimize the analysis of such large data sets, especially for image segmentation. The convergence of these diverse disciplines around the tissue-clearing methods will pave the way to unbiased 3D histological assessments, which should drastically accelerate the discovery of novel developmental, physiological and pathological mechanisms impacting whole organisms.
In this Review, we cover the landscape of rapidly emerging tissue-clearing methods and their related technologies — including labelling, light-sheet microscopy and data analysis and management — as well as applications of tissue clearing in neuroscience, especially in human studies. We then discuss the challenges and opportunities relating to the integration of these technologies to generate unbiased insights into human physiology and pathology.
Hydrophobic tissue clearing
Hydrophobic tissue-clearing approaches involve the use of organic solvents and often provide complete transparency of an intact specimen quickly32. For example, 3D imaging of solvent-cleared organs (3DISCO), developed by Ertürk and colleagues, can fully clear a whole adult mouse brain in 1–2 days7,33. Ethyl cinnamate has also been used in hydrophobic tissue clearing instead of organic solvents11. Because hydrophobic tissue-clearing methods are straightforward, only needing the sequential incubation of the specimen in different solutions, 3DISCO and its variants have already been widely used in imaging studies of neuronal circuits7,33, inflammation34, stem cells35,36 and cancer cells37–39, and in unsectioned rodent organs and human biopsy specimens40. DISCO-based methods have also been combined with deep-tissue immunolabelling approaches to study rodent embryos4,8, human embryos23, cancer biopsy specimens40,41, adult mouse brains4,26 and, more recently, whole mouse bodies (discussed later)28.
DISCO-based methods consist of an initial dehydration step to remove water — the major light scatterer in tissues (the RI of water is 1.33, whereas the RI of soft tissues is 1.44–1.56) — and a subsequent organic solvent immersion step to extract most of the lipids and increase the RI to match the average RI of biological tissues (the RI for both organic solvents and shrunk brain tissue is ~1.56)15. In most of the DISCO approaches (except immunolabelling-enabled DISCO+ (iDISCO+))34, dehydration leads to a marked shrinkage of the specimen32. Pan et al. developed the ultimate DISCO (uDISCO) method and shrank mouse bodies to about one third of their original size, which facilitated imaging of the entire bodies at cellular resolution by light-sheet microscopy42. The shrinkage of individual organs, especially CNS tissue, was isotropic; the bones also shrunk isotropically but to a lesser degree42. Through the use of uDISCO, the researchers visualized neuronal connectivity in the intact CNS of mice (that is, in the brain and spinal cord).
An important advantage of organic solvent-based clearing is the permanent preservation of the specimens, owing to the hardening of the cleared tissues. This allows multiple imaging sessions and long-term reanalysis of the samples, especially by immunolabelling methods, which can permanently stabilize the endogenous fluorescent signal28. Although clearing and imaging increasingly larger samples are valuable steps in the unbiased analysis of tissues, thorough labelling of large tissues with specific dyes and antibodies remains a challenge. Towards this goal, first, Tessier-Lavigne, Renier and colleagues developed iDISCO, which achieved immunolabelling of the adult mouse brain and allowed brain regions differentially activated during parenting behaviour to be uncovered26. iDISCO involves pretreatment of the specimen with solutions containing H2O2 and methanol to permeabilize mouse brain, a process that may also purge most of the epitopes for the antibodies26. In the future, development of new deep-tissue labelling approaches with full epitope preservation will be critical to broaden the applications of hydrophobic tissue-clearing methods.
To scale up the immunolabelling to whole adult mouse, Cai et al. recently introduced vDISCO (the ‘v’ refers to the variable domain of heavy-chain antibodies; that is, nanobodies), using high-pressure delivery of nanobodies for complete immunolabelling of the whole body with bright Atto dyes in the far-red region to overcome low signal intensity and autofluorescence of many tissues in the blue–green region28. This method amplified the fluorescent signal by two orders of magnitude and thereby allowed imaging of subcellular details and quantification of single cells deep in intact cleared mouse bodies through bones and muscles. vDISCO allowed the generation of the first whole-body neuronal projectome of adult mice, the study of neurodegeneration and inflammation throughout the body after CNS lesions and the co-discovery of skull–meninges connections, which seem to be critical for brain functions in physiological and pathological states28,43. As vDISCO enormously amplifies signal contrast even for dissected organs, in the future it will provide high-quality ground-truth data for machine learning-based algorithms aiming to map transgenically labelled mammalian brains11,44.
Hydrophilic tissue clearing
Hydrophilic tissue-clearing methods use water-soluble reagents for tissue clearing. Although the tissue-clearing performance of hydrophilic tissue-clearing methods was sometimes inferior to that of hydrophobic tissue-clearing methods, the former have obvious advantages, including high levels of biocompatibility, biosafety and preservation of protein function. Hydrophilic reagents usually form hydrogen bonds with components of tissues such as proteins as well as surrounding water molecules, which can help to preserve the 3D structure of tissue components and, thereby, the signal of fluorescent proteins. Moreover, hydrophilic reagents can be dissolved in water at a high concentration and can be used as an RI-matching medium to provide a high RI in the medium. Hydrophilic tissue-clearing methods also have a long history of about a quarter of a century (since 1995; BOX 1).
Box 1 |. History and recent development of hydrophilic tissue-clearing methods.
Beginning in 1995, the Russian physicist Tuchin and his colleagues conducted pioneering studies in hydrophilic tissue clearing. They tested a series of chemicals, including X-ray contrast agents (for example, trazograph), alcohols (for example, polyethylene glycol, glycerol and propylene glycol), sugars (for example, glucose and dextrans) and dimethyl sulfoxide (DMSO) to see if they could increase the refractive index of a refractive index-matching medium for biological samples such as human sclera145–152. Starting at a similar time, Chance and colleagues independently tested the effects of sugars (for example, mannitol, fructose, sucrose and glucose) and alcohols (for example, propanediol and methanol) on the optical properties of biological materials such as cells and tissues50,153 when they developed systems to measure the concentration of blood and the saturation of haemoglobin in tissues. Following these pioneering studies, Vargas and colleagues applied hydrophilic tissue-clearing methods involving glycerol, DMSO or glucose to hamster and rat skins154,155, and Wang and colleagues applied glycerol, propylene glycol, ethylene glycol, DMSO and oleic acid to skin tissues156–158. In 2005, Choi and colleagues compared the potentials of various reagents—including glycerol, trimethylolpropan, 1,3-butanediol, 1,4-butanediol, ethylene glycol, 2-methyl-l,3-propanediol, DMSO, linoleic acid, oleic acid and P-0062—with optically clear human skin159. Yeh and colleagues used sorbitol as a refractive index-matching component48, and Hell and colleagues discovered thiodiethanol as an refractive index-matching component160. Subsequently, Pavone et al. and Nemoto et al. developed tissue-clearing protocols10,161 that used thiodiethanol as a key refractive index-matching component. Mason and colleagues developed ClearT, which contains formamide as a key component162. Klein and colleagues developed a tissue-clearing protocol for adult mouse brain163 that uses sucrose as a key refractive index-matching component50. Jiang et al combined Scale and SeeDB protocols to construct FRUIT164, which uses urea12 and fructose13,50 as key tissue-clearing components. A series of X-ray contrast agents have frequently been used as refractive index-matching components since their introduction145. For example, the X-ray contrast agent iohexol was used as a key refractive index-matching component in a refractive index-matching solution3, whereas another X-ray contrast agent, iodixanol, was used in a SWITCH study25. See also the main text for the development and applications of FocusClear, Scale, SeeDB and CUBIC.
One of the first applications of hydrophilic tissue clearing to neuroscience was by Chiang and colleagues. They developed FocusClear, which includes an X-ray contrast agent (for example, diatrizoate acid) and a detergent (for example, Tween 20), and applied it to imaging of a whole cockroach brain45,46. More recently, Miyawaki and colleagues developed Scale12, which contains urea as a key tissue-clearing component, and can expand biological samples — for example, whole mouse embryos (embryonic day 13.5), infant mouse brains (postnatal day 15) and adult mouse brain slices (7–13 weeks old) — by hyperhydration47 and thereby reduce their RIs. In contrast to most hydrophobic tissue-clearing regents, Scale can more efficiently preserve fluorescent proteins12. This method has allowed imaging of yellow fluorescent protein-labelled Thy1-expressing neurons in infant and adult mice, green fluorescent protein-labelled neural stem cells in the adult mouse hippocampus and cell-cycle status of mouse embryos via detection of the Fucci-S/G2/M and Fucci-G1/G0 markers12. The researchers further developed Scale by combining urea with sorbitol48 to develop ScaleS49, which can be applied to adult, old and diseased mouse brain hemispheres. Imai and colleagues developed the See Deep Brain (SeeDB) protocol13, which uses fructose50 as its key RI-matching component, and used it to trace neural circuits in mouse olfactory bulbs because it does not expand or shrink biological samples and therefore preserves their morphology. They further refined this method by using an X-ray contrast agent with a higher RI, leading to the development of the SeeDB2 protocol, which has been applied to super-resolution imaging of neural circuits in mouse brains51.
Ueda and colleagues took a system-level approach to identifying potent hydrophilic tissue-clearing reagents by comprehensive chemical profiling. They discovered a series of amino alcohols that have both delipidation5 and decolonization6 capabilities and then developed a series of clear, unobstructed brain or body imaging cocktails and computational analysis (CUBIC) reagents for both delipidation and decolourization5,27,52. Profiling more than 1,600 hydrophilic chemicals, they also discovered a series of aromatic amides that can be used as potent RI-matching reagents22,27. By combination of the most advanced delipidation (CUBIC-L, where ‘L’ stands for ‘delipidation’) and RI-matching (CUBIC-R+, where ‘R’ stands for ‘RI matching’ and ‘+’ stands for a basic condition by adding an amino alcohol, N-butyldiethanolamine) reagents, the tissue-clearing performance of hydrophilic methods is now similar to or can even exceed that of hydrophobic methods without losing its advantages of biocompatibility, biosafety and preservation of protein function (for example, fluorescence)22. CUBIC has been applied to whole-brain imaging of immediate early gene expression induced by light exposure5 and drug administration53, whole-brain imaging of cancer metastasis27, whole-brain imaging of individual neurons54 and taste-sensing circuits in mouse brains55, and 3D brain imaging of glutaminergic synaptic connections56, hypothalamic neural subtypes57, layer-specific astrocyte morphology58 and visual projections of retinal ganglion cells59. CUBIC has been also applied to 3D imaging of other organs, including haematopoietic stem cells in bone marrow60, carcinoma in the lung61 and single-cell lineage tracing in the mammary gland62 and development of a heart63. Therefore, the peripheral nervous systems in these organs can be analysed in detail.
Delipidation in some hydrophilic tissue-clearing methods creates space for large substances such as antibodies to penetrate more rapidly and deeply into tissues, allowing 3D immunohistochemistry in large samples. CUBIC has been successfully used to achieve 3D immunohistochemistry for adult mouse brain, heart, lung, stomach and intestine5,6,22,27,52. Recently, CUBIC-L–CUBIC-R was combined with whole-brain 3D immunohistochemistry to image blood vessels in entire mouse brains27. CUBIC was also applied to 3D immunohistochemistry and imaging of neurons expressing vesicular acetylcholine transporter that innervated islets in a pancreas64. In addition to the enhanced penetration of antibodies owing to delipidation, weakening the interaction between an antibody and a tissue seems to accelerate the penetration of an antibody into a tissue. For example, the AbScale protocol, which uses the protein denaturant urea in antibody staining, achieved 3D immunohistochemistry of an adult mouse hemisphere49. An original Scale protocol uses urea for tissue clearing not for antibody staining. AbScale was used with brain-wide immunohistochemistry to image amyloid-β plaques, a hallmark pathological feature of Alzheimer disease, in mice27,49.
Hydrophilic methods can be also extended to expansion microscopy65. For example, the CUBIC-X protocol, which involves the use of an imidazole and an antipyrine as hyperhydrating reagents, can expand the adult mouse brain 10-fold in volume17 (the ‘X’ in CUBIC-X stands for ‘X-fold expansion). The use of the CUBIC-X protocol allows whole-brain cell profiling of adult mouse brains and allowed the development of a 3D single-cell-resolution mouse brain atlas (CUBIC-Atlas)17, which contains ~108 cells of an entire brain17,66 (FIG. 2). Such brain atlases could become widely used platforms for mapping cell activity (for example, the expression of immediate early genes), cell types (for example, specific subtypes of neurons and glia cells) and neural connectomes (for example, mapping the neural connections by adeno-associated virus and/or rabies virus).
Fig. 2 |. Whole-brain single-cell-resolution imaging and analysis.
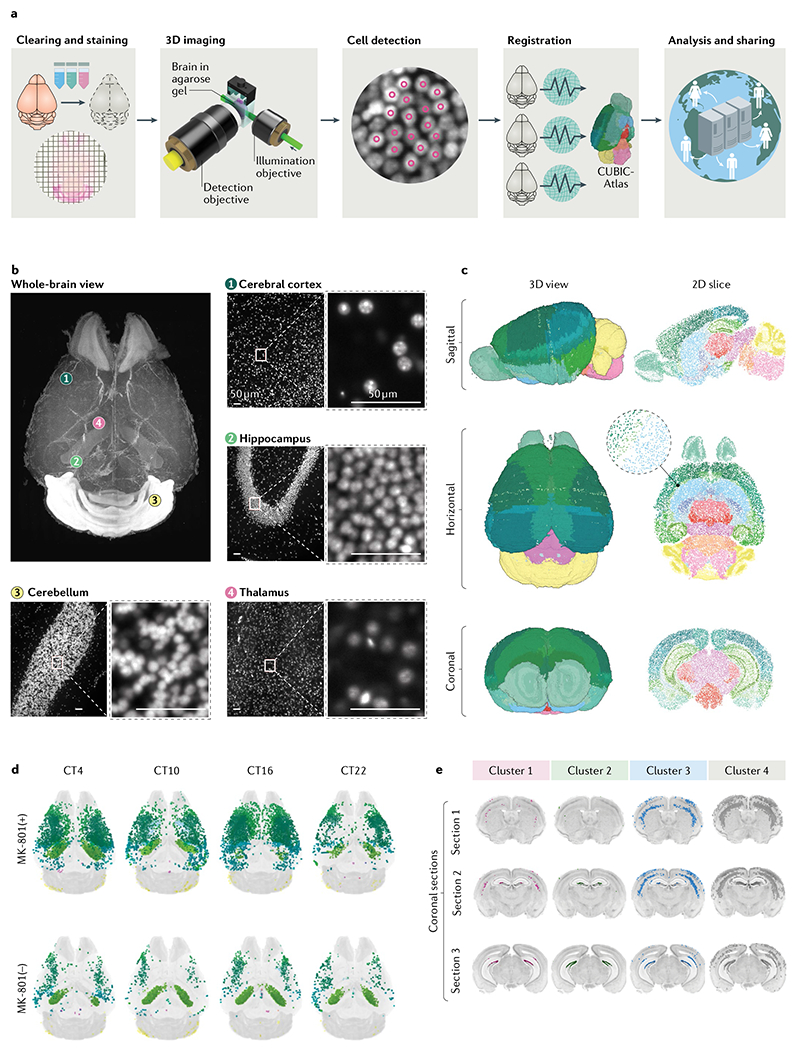
a | Tissue-clearing methods allow whole-brain profiling of cells. The brain data obtained can be registered in the 3D single-cell-resolution mouse brain atlas (CUBIC-Atlas) and shared by a worldwide research community via the Internet17. First, tissue clearing (and fluorescent staining if applicable) is applied to the specimen. Cleared brains are imaged using high-resolution light-sheet microscopy. A graphics processing unit-based high-speed cell counting program identifies all cells from the acquired images, rendering the whole tissue into an ensemble of cellular points (that is, a point cloud). Then, individual brains are registered onto the common brain coordinates to allow guantitative comparison and analysis. Finally, these data should be shared among researchers to allow collaborative and large-scale analysis. b | Images of an adult mouse brain obtained by the hydrophilic tissue-clearing and expansion protocol CUBIC-X and custom-made high-resolution light-sheet microscopy17. Nuclear staining using propidium iodide was applied to the cleared brain tissue. Magnified views of some of the representative brain regions, including cerebral cortex (view 1), hippocampus (view 2), cerebellum (view 3) and thalamus (view 4), are presented, along with the whole-brain overview. Scale bars indicate 50 μm after normalization of the sample expansion. c | The 3D single-cell-resolution mouse brain atlas CUBIC-Atlas. With use of nuclear staining images such as the ones shown in part b, all cell nuclei in a whole mouse brain were identified, totalling ~108 cells17,66. The colour of each cell represents anatomical annotations of each brain region obtained from the Allen Brain Atlas. d | The whole-brain neuronal activity profile with or without the long-term administration of a NMDA receptor inhibitor (MK-801) was quantified by imaging the destabilized fluorescent protein dVenus under the control of the Arc gene promoter by using the Arc–dVenus transgenic mouse53. Each Arc–dVenus mouse brain at different circadian times (CT) was mapped onto CUBIC-Atlas in a probabilistic manner to virtually reconstruct the time series17. Arc–dVenus-expressing cells are shown as dots, with their colours representing their anatomical areas. e | By clustering analysis of the data shown in part d, four distinct populations of cells were identified, exhibiting different activity patterns on MK-801 administration. Localization of each cluster is shown in coronal sections, revealing an inhomogeneous cellular population in the lower and upper dentate gyrus. Parts b–e were adapted from REF.17, Springer Nature Limited.
Hydrogel-based tissue clearing
To widen the applications of tissue-clearing technologies, Deisseroth, Chung and colleagues introduced a hydrogel-based tissue-clearing method called ‘cleared lipid-extracted acryl-hybridized rigid immunostaining/in situ hybridization-compatible tissue hydrogel’ (CLARITY), which secures a broad category of biomolecules at their physiological locations by covalently linking the molecules to an acryl-based hydrogel2 (FIG. 3). This unique hydrogel reinforcement allows complete and uniform removal of lipids from the tissue while minimizing structural damage and loss of biomolecules (10% protein loss in CLARITY versus 70% protein loss in formaldehyde-only fixed tissues)2. In CLARITY, both electrophoresis-driven and simple passive clearing can effectively remove lipids, which drastically increases the optical transparency and macromolecule permeability of the hydrogel–tissue hybrid2. The cleared intact organs can be imaged by fluorescence microscopy without loss of resolution. Small tissues can be readily stained with molecular probes (such as antibodies); however, the staining of large tissues requires longer incubation times as in all the tissue-processing methods that rely on passive diffusion.
Fig. 3 |. The SHIELD–MAP and AAV-based labelling system.
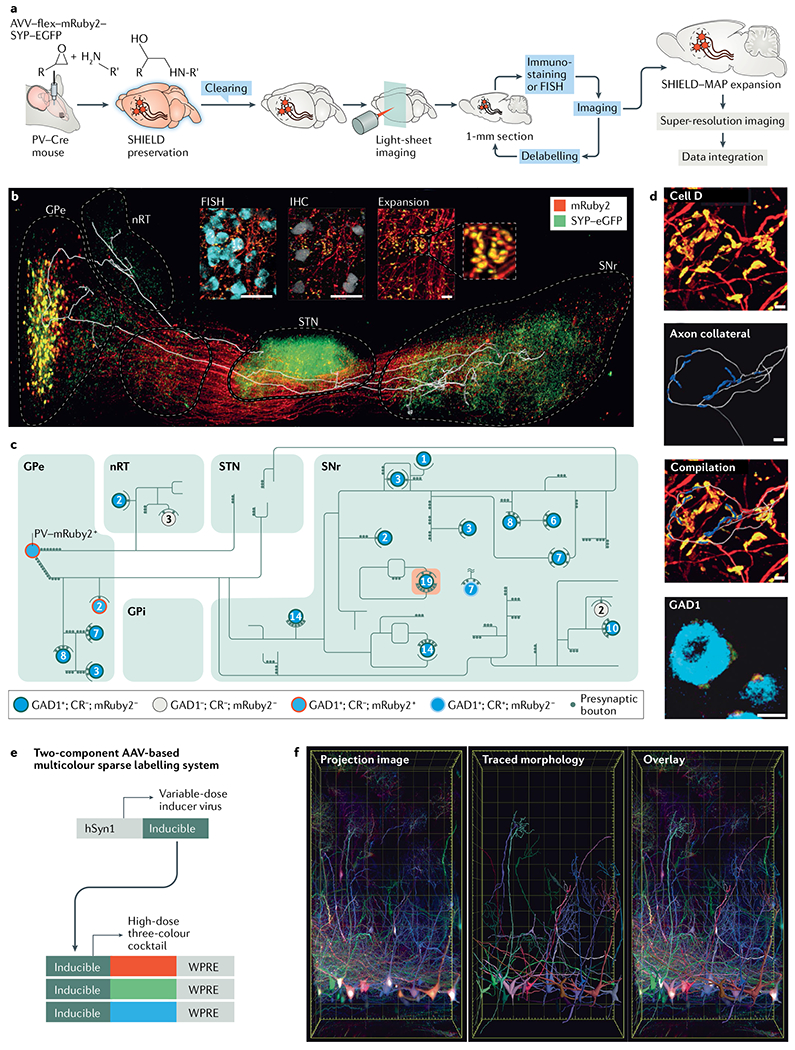
SHIELD (system-wide control of interaction time and kinetics of chemicals) combined with MAP (magnified analysis of proteome) allows integrated circuit mapping at single-cell resolution. a | The SHIELD–MAP pipeline. SHIELD allows fully integrated multiscale imaging of fluorescent protein-labelled circuits, mRNAs and proteins within the same mouse brain by simultaneously protecting the molecular and structural information within cleared tissues. b | The image shows a 3D rendering of fluorescent protein-labelled neuronal circuitry of parvalbumin (PV)-positive neurons in the globus pallidus externa (GPe) with an overlaid axon trace of a single labelled neuron. The inset shows example images from multi round staining and multiscale imaging. Scale bar 50 μm for the insets. c | Reconstructed axon arborization of the neuron and its downstream targets. Each circle represents a neuron. The number of putative axosomatic boutons is marked inside each circle. The colour of each circle provides molecular details for each neuron. d | Reconstructed putative axosomatic connectivity for ‘cell D’ (the cell highlighted in orange in part c). Ramified axons (grey) and enhanced green fluorescent protein (eCFP)-positive presynaptic boutons (blue) are segmented71. Scale bars 20 μm. e | Viral-assisted spectral tracing (VAST) can be used to label and visualize the 3D morphology and connectivity of cells in thick, cleared tissue blocks. Here, the schematic shows the two-component VAST labelling system30,80,122. A high dose of a three-vector cocktail (individually expressing a red, green or blue fluorescent protein) is coadministered along with a variable dose of an inducer vector that is required to turn on expression of the three proteins. To label specific cell populations, the expression of the fluorescent proteins can also be made Cre dependent. f | Projection image of the olfactory bulb where mitral cells are labelled by VAST (left). Sparse labelling of these cells allows tracing of their dendritic arbors (middle). An overlay of the traces and the projection image is shown in right-hand image. AAV, adeno-associated virus; CR, calretinin; FISH, fluorescence in situ hybridization; hSyn1, human synapsin 1 gene promoter; IHC, immunohistochemistry; GPi, globus pallidus interna; nRT, nucleus reticularis thalami; SNr, substantia nigra pars reticulata; STN,subthalamic nucleus; WPRE, Woodchuck hepatitis post-transcriptional regulatory element. Parts a–d are adapted from REF.71, Springer Nature Limited. Parts e and f are adapted from REF.80, Springer Nature Limited.
Variants of CLARITY3,9,67,68 have been developed to increase tissue permeability and probe penetration by decreasing gel density and the degree of crosslinking, such as the passive CLARITY technique (PACT)3,67. Weakening gel architecture, however, causes loss of tissue information. To accelerate molecular labelling without compromising tissue integrity, a new mode of transporting molecules into tissue was developed, termed ‘stochastic electrotransport’. Stochastic electrotransport generates electrophoretically driven diffusive random motion of a broad range of molecules (for example, antibodies, dyes and detergents) to rapidly deliver them into dense tissue gels and it allows uniform clearing and staining of intact mouse organs within 2 or 3 days (as opposed to weeks to months in methods that rely on the passive diffusion of such molecules)24.
The physicochemical properties of the acryl-based gel can be engineered to add functionalities to the tissue–gel hybrid. Boyden and colleagues hybridized tissue with an expandable hydrogel and then digested proteins using a proteinase to expand the construct isotropically for super-resolution imaging65. Chung and colleagues developed a technique called ‘magnified analysis of proteome’ (MAP) that eliminated the protein digestion step described above to preserve the 3D proteome of intact organs while allowing isotropic expansion69. In MAP, a highly dense hydrogel is synthesized in situ and then protein complexes are dissociated to achieve isotropic expansion of the 3D proteome at the organ level. The authors demonstrated its distinct utility for super-resolution imaging and reconstruction of neural projections. For better RNA detection, a recent study used 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide chemistry to anchor RNAs to polyacrylamide gel70.
These technological advances have allowed the brain-wide spatial mapping of biomolecules at subcellular resolution. However, the harsh chemical processing used in many tissue-clearing methods, such as detergent and organic solvent treatments, sometimes cause tissue damage. To address this limitation, a method termed ‘stabilization to harsh conditions via intramolecular epoxide linkages to prevent degradation’ (SHIELD) was developed to preserve the fluorescence of proteins, protein antigenicity, transcripts and tissue architecture in organ-scale transparent tissues. SHIELD uses polyepoxy chemicals to protect the physicochemical properties (for example, protein fluorescence and antigenicity) of biomolecules by forming intramolecular and intermolecular crosslinks71. To achieve uniform and controlled crosslinking in organ-scale tissues, polyepoxy chemicals are first dispersed in a buffer called ‘system-wide control of interaction time and kinetics of chemicals (SWITCH)-Off buffer’ that inhibits the fixation reaction25. Once the crosslinker has been uniformly dispersed, the reaction is turned on globally by moving the sample to a SWITCH-On buffer that enhances the fixation reaction. This simple strategy allowed scalable and uniform preservation of protein fluorescence, transcripts and proteins. SHIELD-preserved and delipidated tissue can withstand harsh antibody destaining conditions and therefore allows multiple rounds of staining and imaging of the same sample. Using SHIELD, the study authors demonstrated integrated mapping of neural circuits and their downstream targets at single-cell resolution as well as 3D molecular phenotyping of intact needle biopsy samples within only hours25 (FIG. 3). In addition, these technologies have allowed the study of 3D structure of cerebral organoids72 and network-specific amyloid progression73.
Hydrogel-based methods are further applicable to the whole-body scale. Gradinaru and colleagues developed perfusion-assisted agent release in situ (PARS), which rendered rodent bodies transparent, allowing maps of central and peripheral nerves at target organs to be obtained3,68. Gradinaru and colleagues realized that the circulatory system (the vasculature) could be used to deliver clearing agents and labels instead of relying on passive diffusion, which is prohibitively slow for large organs or whole organisms. CLARITY-based methods2 can be adapted for bones74, and for magnified single-cell visualization while retaining fluorescent markers (see Supplementary Fig. 4 in REF.9).
Labelling
For CLARITY and other tissue-clearing methods to reach their full potential, it will be imperative to integrate data from post-mortem samples with markers of functionality. We cannot remove the lipids while keeping the brain alive, but we can at least store a short-term ‘memory of neuronal activity’ via transcriptional or biochemical changes (for example, Ca2+ influx) that can be evaluated after death and brain-wide. The history of activity in brain circuits can also be reported by changes in protein expression. As discussed earlier, hydrophobic, hydrophilic and hydrogel-based tissue-clearing methods are compatible with antibody staining of intact organs. The legacy of brain activity is also reported by changes in RNA transcripts, which can be detected by single-molecule fluorescence in situ hybridization and are indeed retained in PACT-cleared tissues3,75. More broadly, preserving the spatial relationships of tissues while accessing the transcriptome of selected cells is of crucial interest for many areas of biology and it motivated the development of methods for multicolour, multi-RNA imaging in deep tissue71. With use of single-molecule hybridization chain reaction, tissue hydrogel embedding and clearing by PACT and light-sheet microscopy, the detection of single-molecule mRNAs in approximately millimetre-thick brain slices is possible76. With rRNA labelling in PACT-cleared samples, researchers mapped the identity and growth rate of pathogens in sputum samples from individuals with cystic fibrosis77.
Tissue-clearing methods have been particularly useful in tracing long-range projections in the CNS and the peripheral nervous system. However, to maximize their impact, the clearing methods need to be complemented by labelling methods that can, for example, highlight the desired circuits with strong, morphology-filling markers, while preserving their genetic identity and be easy to use and highly customizable (that is, they minimize the need to generate or cross transgenic animals)78. This has been nicely applied to study the topography of dopaminergic projections in the mouse brain79.
As morphology reconstruction is difficult in data sets from densely labelled neurons, it would be desirable for the fraction of cells labelled to be easily controlled. Recruiting the power of recently engineered systemic adeno-associated viruses that can cross the blood–brain barrier29,80 has been beneficial in this respect, and methods are now available to label individual cells in the brain, peripheral nervous system and other organs with a wide range of hues using genetically encoded fluorescent proteins expressed by multiple viral vectors. Viral-assisted spectral tracing is a highly customizable multicolour labelling system with controlled labelling density that allows detailed studies of the 3D morphology of cells in intact, thick, cleared tissue samples (FIG. 3). The unique power of a labelling approach based on gene delivery is that in addition to it providing morphology markers, one can introduce state-altering genes and evaluate the effects on morphology of state-modified and state-unmodified cells within the same subject; coupled with tissue clearing, this will greatly enhance our understanding of physiology and pathology81.
Applications in humans
Our knowledge of the human body is primarily macroscopic and based on classic anatomical methods that were established centuries ago and that require the dissection and slicing of individual organs such as the brain. However, recent studies have showed that tissue clearing and light-sheet microscopy are starting to revolutionize not only the study of experimental animals but also the study of the anatomy of and disease diagnosis in humans. The surge of tissue-clearing innovation comes primarily from neurobiologists, who are facing a daunting task: deciphering the human connectome. The nervous system is by far the most complex organ, with probably hundreds of different cell types forming intricate circuits and networks, the organization of which is extremely difficult to understand from 2D slices. The combination of tissue clearing and light-sheet microscopy has already allowed us to image entire mouse brains and spinal cords and to better understand the extent of CNS deterioration in some animal models of neurological diseases8,26,82,83. The CUBIC approach has been combined with stimulated emission depletion microscopy to better visualize synaptic contacts onto the dendritic spines of pyramidal neurons in the mouse cortex84. The next challenge is to translate this knowledge to the analysis of the human nervous system and to develop applications that will improve the post-mortem pathological evaluation of brain disorders.
CLARITY-based clearing procedures have allowed successful imaging of cortical pyramidal neurons and interneurons85,86 and cerebellar Purkinje cells and granule cells87 in healthy human brain samples. Such procedures have also permitted imaging of pathological human brain samples, including visualization of tissue from an individual with autism2, α-synuclein inclusions, dopaminergic axons and substantia nigra in Parkinson disease88, Purkinje cells, mitochondria and vasculature in cerebellar ataxia87 and amyloid plaques in Alzheimer disease85. Other brain-clearing techniques, such as iDISCO82 and ScaleS49, have also been tried on Alzheimer disease brain samples; although confirmatory, these studies provided more direct evidence for an association between microglia and amyloid plaques, with these techniques also facilitating the detection of diffuse plaques. CUBIC27 has been used in whole-brain immunohistochemistry of blood vessels in cancer metastasis. Moreover, GABAergic interneurons were imaged in slices from hemimegalencephalic cortex that was cleared with 2,2′-thiodiethanol10, and neurons and vessels were imaged in spinal cord fragments that were cleared with ACT-PRESTO (active clarity technique–pressure-related efficient and stable transfer of macromolecules into organs)89. The clearing reagent OPTIClear90 is compatible with lipophilic dyes and has been used to label mossy fibre axons in adult human cerebellum.
Together, the studies mentioned above provide proof of concept for and support the feasibility of human brain clearing, but these and other studies illustrate the current technical limitations of this approach in humans. First, clearing was efficient on relatively thin brain slices or blocks of no more than a few hundred cubic micrometres2,25,49,82,85,87,88,90, corresponding at best to about 1/1,500 of the total human brain volume91. Good transparency sometimes required a significant extension of the clearing time, compared with a few days in rodents10. Second, formalin-fixed human brain tissue is highly autofluorescent, is rich in blood and contains lipofuscin-type pigments and neuromelanin, which are very difficult to clear. Third, immunostaining was successful only on samples thinner than a few hundred micrometres, and many antibodies failed to work following tissue-clearing treatment. Using single-chain variable fragments of conventional antibodies or nanobodies might be a good option to increase penetration into the brain samples92. Therefore, although the progress so far is promising, routine post-mortem 3D imaging of the entire human brain is still currently out of reach and remains a tantalizing challenge. Scalability of the clearing method will be essential, and there is also a need to develop light-sheet microscopes with much longer working distances and a larger field of view. Solvent-based clearing methods, which can drastically reduce the sample volume, might then be preferred. When optimized, human brain clearing will be a unique tool for correlating and validating in vivo 3D data obtained by MRI and diffusion tensor imaging. This will be essential to improve the diagnosis of disease.
Although 3D imaging of transparent adult human CNS is still in its infancy, tissue clearing has already provided a wealth of new information in the field of human embryology and allows easy access to 3D cytoarchitecture of millimetre-sized brain organoids92. Solvent-based DISCO clearing methods proved to be perfectly adapted to the transparentization of human embryos and fetuses at least up to 3 months of gestation23,93. Moreover, with use of this approach, whole-mount immunolabelling of centimetre-sized human specimens was performed with a large panel of antibodies23 (FIG. 4), and indicated how motor and sensory axons invade the hands, limbs, head and various organs and revealed an unexpected heterogeneity and stochasticity of sensory nerve branching pattern23. 3D imaging of DISCO-cleared human embryos also demonstrated that neurons secreting gonadotropin-releasing hormone migrate in two separate pathways and colonize many brain regions outside the hypothalamus93. The innervation of the embryonic pancreas was described following the use of CLARITY94. One can now revisit embryology and envisage building a comprehensive 3D cartography of human development. Indeed, tissue clearing will be extremely valuable to the Human Developmental Cell Atlas project95, one component of the Human Cell Atlas, an international initiative that aims to identify and map all cells of the human body.
Fig. 4 |. Towards a 3D developmental human cell atlas.
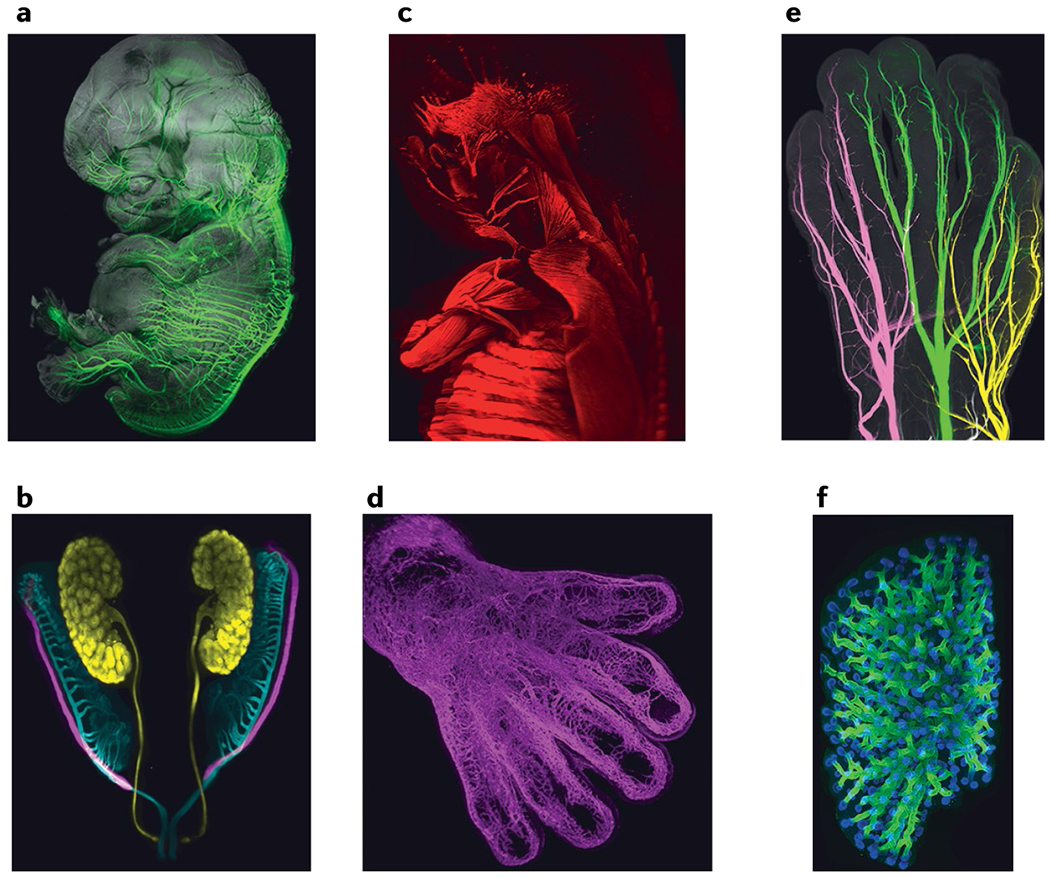
Solvent-based tissue clearing is particularly suitable for the analysis of human embryos as it allows visualization and mapping of immunolabelled cells in large and intact specimens. This provides unprecedented views of developing organs and human cells. a–d | Peripheral nerves (part a), Müllerian and Wolffian ducts and kidney in the urogenital system (part b), muscles in the back, arm and head (part c) and the vasculature in the hand (part d) of a human embryo at 8 weeks of gestation. e,f | The three sensory nerves of the hand (part e) and lung epithelial tubules (part f) in a fetus at 9.5 weeks of gestation. Parts a,b,d–f are adapted with permission from REF.23, Elsevier. Part c is adapted with permission from REF165, Company of Biologists doi:10.1242/dev.180349.
Understanding how tumour cells and viruses proliferate and spread within the body are major questions in oncology and virology, and they are attracting greater attention96 in neuroscience because there are tight interactions between nervous systems and the immune system97. Recent applications of tissue-clearing methods have helped follow at an unprecedented resolution the dissemination of HIV-infected human T lymphocytes in mouse lymphoid organs96 and human cancer cell lines in whole cleared mice27. These studies have already demonstrated that the cellular resolution in cleared organs significantly outperforms what is achieved with classic tumour detection methods such as bioluminescence. The cellular composition of cleared human tumour biopsy samples has also been described94,98–100. Therefore, once adopted by histopathologists, tissue clearing is expected to rapidly improve the analysis of tumour cell niches in their native context, diagnosis, staging of tumours and the validation of anticancer and antiviral therapies.
Light-sheet microscopy
To take full advantage of the clearing methods discussed so far and systematically extract structural information at the subcellular level, clearing methods require microscopes that are capable of rapid, high-resolution imaging of large volumes. Several powerful imaging methods have been developed or adapted for this purpose. Some of these efforts focused on pushing existing, well-established techniques, such as two-photon microscopy, to their performance limits. Notably, Chandrashekar, Myers and colleagues developed an entire imaging framework that integrates a fast, resonant-scanning two-photon microscope with a tissue vibratome to facilitate whole-brain imaging at high spatial resolution54. They achieved a spatial resolution of 0.45 μm laterally and 1.33 μm axially at a data acquisition rate of 1.6 ×106 μm3 per second (FIG. 5), which allowed the fully automated acquisition of entire mouse brains in approximately 1 week. To increase the speed and resolution even further, ongoing efforts are taking advantage of emerging imaging strategies. Light-sheet microscopy101–103 is a key approach to this end, as it promises particularly striking performance advances in both spatial and temporal regimes.
Fig. 5 |. Resolution and speed of custom and commercial light-sheet microscopes.
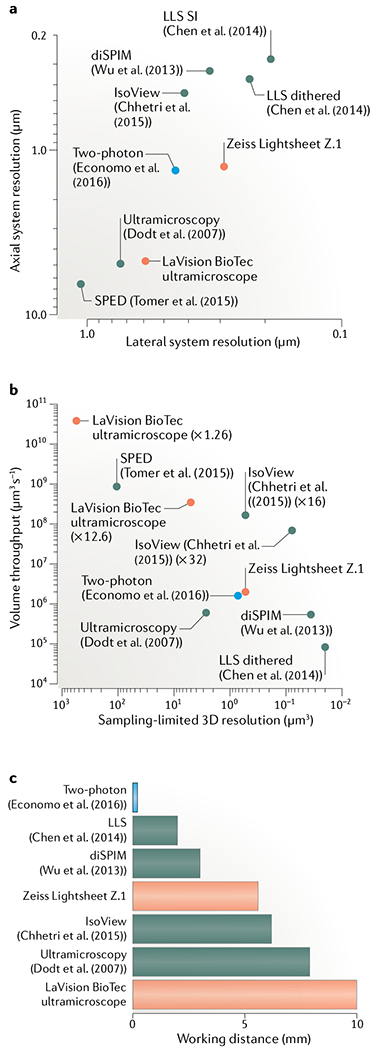
a | Lateral versus axial system resolution of custom14,105,110–112 (red) and commercial166,167 (green) light-sheet microscopes and state-of-the-art two-photon microscopes54 (blue). System resolution values168 representthe optical configurations and light-sheet properties reported in each study for the demonstration experiments shown in part b. The guantification disregards spatial sampling limitations inherent to the choice of detector. Resolution values are provided as the full width at half maximum (FWHM) size of the system point spread function. Lattice light-sheet (LLS) microscopy uses two different acquisition modes (dithered light sheet versus structured illumination (SI)), which affect the speed and resolution. b | Sampling-limited 3D resolution versus volume throughput for commercial light-sheet microscopes (green) and imaging experiments performed with custom light-sheet (black) and two-photon (blue) microscopes. 3D resolution is defined as , where dlat is the sampling-limited lateral FWHM size and dlat is the sampling-limited axial FWHM size of the point spread function. Lateral sampling can be adjusted by changing the magnification of the detection system, which typically affects both volume throughput and effective resolution (for examples, see ×1.26 versus ×12.6 magnification settings for the LaVision BioTec ultramicroscope or ×16 versus ×32 magnification settings for IsoView). Data points for custom light-sheet and two-photon microscopy refer to imaging of hippocampal dendrites14, Caenorhabditis elegans development111, cellular dynamics110, zebrafish nervous system105–112 and mouse brains54. Data points for commercial microscopes reflect the technical specifications provided by the manufacturer165,166. All techniques included in the plot have been successfully applied to the imaging of cleared tissues. c | Working distance of the detection systems for the methods shown in parts a and b. The design by Economo et al.54 does not require long-working-distance optics for imaging mouse brains since their microscope is integrated with a tissue vibratome. Chhetri et al.112 surrounded the specimen with four identical objectives for illumination and fluorescence detection, which allows an increase in the bidirectional working distance by a factor of 2 (compared with the native working distance of a single objective) in the special case of imaging transparent, cleared tissues. The maximum supported specimen size can in principle also be doubled in other light-sheet techniques at the expense of temporal resolution by rotating the specimen by 180° and acquiring volumetric data from this opposite view. Tomer et al.105 proposed using the majority of the detection objective’s working distance to introduce a block of high-refractive-index material in the detection path. The effective working distance (distance between the block and the in-focus region of the specimen) is thus typically not identical to the native working distance of the objective. For this reason, the latter technique is not included in this plot.
The central idea in light-sheet microscopy is to illuminate the specimen with a thin sheet of laser light from the side and acquire an image of the illuminated plane with a camera-based detection system. In the most common implementations of light-sheet microscopy, the wide-field detection arm is oriented at a right angle to the light-sheet illumination axis and separate objectives are used for illumination and detection. Compared with conventional microscopy, this approach offers two primary advantages for the imaging of large tissue volumes. First, the imaging speed is limited only by the acquisition rate of the camera and thus exceptionally high data rates of up to several hundred million voxels per second can be achieved with state-of-the-art sCMOS detectors. Second, photobleaching and phototoxic effects are kept to a minimum, since only the plane in the focus of the detection system is illuminated with laser light. The amount of energy any part of the sample is exposed to is thus constant and independent of the total size of the sample or imaging volume. By contrast, conventional or confocal microscopes, which use the same objective for illumination and detection, illuminate also out-of-focus regions in the imaging process (above and below the focal plane). In these latter methods, local light exposure thus increases linearly with the size of the imaging volume and can even result in complete bleaching of parts of the specimen before volume acquisition is complete.
Shortly after its modern reincarnation at the beginning of this century, light-sheet microscopy was first applied to the imaging of cleared tissues in 2007 by Dodt and colleagues14. To rapidly acquire volumetric image data from fluorescently labelled, excised hippocampi and even whole embryonic mouse brains, they developed a light-sheet microscope with low-magnification, long-working-distance detection optics and two opposing illumination arms. This approach offered a spatial resolution on the order of several μm axially and slightly less than 1 μm laterally (FIG. 5). Since this first report, many other studies have used light-sheet imaging for conceptually similar experiments. Initial efforts relied exclusively on custom-built systems67,104–108, but soon thereafter the first commercial products followed, such as the LaVision BioTec ultramicroscope5,6,8,23,33,42,52. These microscopes were quickly made compatible with a variety of clearing methods and a wide spectrum of biological specimens, including various organs2,5,7,33, whole rat brains107, chicken embryos106, human embryos23 and even entire adult mice3,6,42.
Imaging speed in light-sheet microscopy is generally limited by the frame rate of the camera, and most approaches use similar optics and similar types of static101 or scanned102 light sheets. Therefore, comparable spatial and temporal resolutions have been achieved with most systems developed and used to date. Importantly, however, in the past few years, new approaches have allowed fundamental advances in spatial resolution in light-sheet microscopes designed for live imaging applications. When a relatively large field of view is being imaged, the resolution in standard light-sheet microscopy is comparably low along the direction of the detection axis — typically several μm at best. This limitation arises from the relatively thick light sheets that are generated by conventional beam-shaping techniques. By contrast, high, spatially isotropic resolution of about a few hundred nm can now be achieved using Bessel beams109 or lattice light-sheets110, which present an alternative to conventionally used Gaussian beams and allow the illumination of exceptionally thin sections of a specimen (FIG. 5). Moreover, high resolution on the order of 300–400 nm in all spatial directions has also been realized by combining light-sheet microscopy with the concept of multiview imaging111–113. The key idea behind this latter approach is that the directions along which resolution is low and high, respectively, can be permuted by imaging the same specimen twice from two orthogonal views. When the complementary information from these views is combined by image processing, the low axial resolution observed in one given view of the specimen can thus simply be replaced by the much greater lateral resolution offered by the second, perpendicular view of the same specimen. This concept ofmultiview imaging, as it has been implemented in diSPIM111 and IsoView112 light-sheet microscopy, is a particularly promising candidate for high-resolution imaging of large volumes, since it can be implemented in a way that offers high spatial resolution over a large field of view at exceptionally high volume acquisition rates112. For example, in IsoView microscopy, this concept allows an isotropic spatial resolution of 400 nm at a volume throughput of more than 108 μm3 per second, while offering the long working distance needed for imaging large specimens without physical sectioning (FIG. 5). Irrespective of the choice of microscope implementation, the imaging of large samples furthermore benefits from automation strategies and computational techniques for adapting the microscope to the specimen’s optical properties. Although cleared specimens are exceptionally transparent, remaining spatial variability in the RI or small mismatches in the RIs of the specimen and the imaging medium can perturb the light path both in light-sheet illumination and in fluorescence detection. To ensure optimal overlap between light sheets and detection focal planes throughout the specimen volume — and thus optimal resolution, contrast and signal strength — adaptive imaging and autofocusing techniques have been developed104,114–116, which are conceptually compatible with most existing light-sheet microscope designs.
When one is imaging very large specimens in the millimetre or even centimetre range, it is important to note that even techniques with a relatively large field of view typically cannot fit the entire specimen within their native imaging volume. These limitations arise from a fundamental trade-off between field size and numerical aperture (and thus resolution) in the design of conventional optics, constraints related to the properties of high-end digital cameras, and from challenges associated with the creation of sufficiently thin light sheets over a large field of view. Thus, a typical workaround in existing approaches to high-resolution imaging of large samples, such as lattice light-sheet microscopy or IsoView light-sheet microscopy, is to rely on tiling strategies for imaging specimens as large as entire cleared and/or expanded nervous systems117,118. This approach largely decouples specimen size from spatial resolution, allowing state-of-the-art microscopes to produce images of optimal quality irrespective of whether the instrument is used to image single cells or an entire brain (assuming good performance of the tissue-clearing method). The two downsides of this approach are the limited data throughout, owing to the sequential nature of optical tiling, and the need for sophisticated computational methods for image processing (see the next section). Ongoing research efforts are therefore investigating other possible solutions to these bottlenecks, including the development of axially swept light-sheet microscopy119 and alternative optical strategies for generating light sheets120.
In the future, we expect to see powerful, new implementations of light-sheet microscopy that combine these latest breakthroughs in high-speed, high-resolution imaging with new large-aperture, long-working-distance detection optics developed by several major optics companies specifically for imaging large cleared tissues. Moreover, further advances in imaging speed will become possible by pursuing multiplexing strategies that parallelize the imaging process across large tissue volumes.
Data analysis and management
With the advent of powerful microscopes that allow high-speed, high-resolution imaging of large specimens, there is a need for new computational approaches that tackle downstream challenges in large-scale data analysis and management. For example, a raw recording of a single mouse brain obtained with high-resolution light microscopy typically comprises thousands of individual 3D subvolumes (also referred to as ‘tiles’) and tens of terabytes of image data. The tiling of image acquisition is usually necessary owing to the limited field of view of light microscopes, which, for specimens as large as a mouse brain, can capture only a small portion of the specimen volume at a time. Thus, translation of either specimen or optics and sequential acquisition of multiple subvolumes is required to eventually arrive at an image of the entire specimen. To efficiently store, process and extract biological meaning from the resulting image data, powerful computational methods are needed to facilitate the following: large-scale data management; multitile image registration and fusion; image analysis and mapping of image data to existing brain atlases; and visualization and interactive viewing of the 3D image data. Each of these challenges is the focus of ongoing efforts in the field, and a range of useful computational tools are now available.
In the realm of data management, recent efforts have led to the creation of several file formats designed to facilitate rapid compression and decompression of large-scale light microscopy image data. The KLB file format is a block-based, lossless compression format that provides JPEG2000-like compression factors while increasing write and read speeds fivefold (up to 500 and 1,000 MB per second, respectively)121. The block-based implementation furthermore offers fast access to local regions in large multidimensional images. KLB supports integration of other compression algorithms in the block-based container and requires only a CPU, thus allowing effective deployment on inexpensive, lower-end computers. By combining KLB with background masking, a 30-fold to 500-fold reduction in data size was achieved121. Similarly, the BigDataViewer122 Fiji plugin leverages the infrastructure of the ImageJ ecosystem123, namely the ImgLib2 library124, to provide seamless access to various block-based data formats, including KLB, hierarchical data format version 5 (HDF5), Imaris and CATMAID (collaborative annotation toolkit for massive amounts of image data). In addition, it provides a powerful caching scheme that ensures that data are transferred optimally. When coupled with the HDF5 container, for example, in the context of multiview reconstructions of light-sheet microscopy data125,126, BigDataViewer allows straightforward combination with arbitrary compression schemes. The reliance on ImgLib2, which is designed to write efficient image analysis algorithms in Java independent of the data type (8-bit, 16-bit, RGB and so on), dimensionality (1D to nD) and storage strategy (memory, local or remote file systems), makes it possible to deploy through BigDataViewer complex image processing pipelines on virtually limitless data. The B3D format offers a complementary approach to high-performance data compression using graphics processing unit-based compute unified device architecture (CUDA) processing127. Although B3D does not use a block-based architecture, it offers excellent write/read speeds on the order of 1,000 MB per second and includes lossless and lossy compression schemes. A reduction in storage requirements and a speed-up of computational processing of image data can also be achieved by using alternative representations of the information encoded in an image. For example, the adaptive particle representation adaptively resamples an image, guided by local information content and using local gain control, and stores the information as a set of particle cells with associated intensity values128. The developers of the adaptive particle representation showed that such a representation not only facilitates data compaction but also has the potential to speed up subsequent image processing steps, such as filtering or image segmentation. The challenge will be to adapt existing image processing solutions to this new data representation paradigm.
Transforming the raw image data of a large, cleared specimen into a spatially coherent, high-resolution image volume that is suitable for data visualization and analysis involves several critical steps. The individual image tiles need to be precisely aligned and fused, and an additional multiview deconvolution step may be needed if complementary views of the specimen were recorded with the light microscope. BigStitcher is scalable and efficient software that facilitates such computations for terabyte-scale image data of large specimens, such as entire mammalian brains or entire invertebrate nervous systems117. This tool leverages the power of ImgLib2 and BigDataViewer and is conveniently available as a plugin for the widely used image processing package Fiji129. Once these initial image processing steps are complete and a high-quality image volume of the complete specimen has been reconstructed, downstream analyses and data mining frequently involve mapping and comparison of the image data with an anatomical reference atlas, image segmentation, annotation of the image data, and interactive data visualization. For the mapping of image volumes to each other or to a common anatomical atlas, several tools have been developed130–132, but they currently cannot process very large data volumes. Bigwarp133 allows manual, interactive, landmark-based deformable image alignment on arbitrarily large images as it uses BigDataViewer for visualization and navigation and a thin plate spline implemented in Java to build a deformation from point correspondences134. For image segmentation, powerful approaches to image analysis based on machine learning are quickly gaining momentum. The iLastik toolkit has been widely adopted as a user-friendly framework for interactive image classification and segmentation with an intuitive graphical user interface135. Familiarity with machine learning algorithms or expertise in image processing is not required to use this toolkit, as the user communicates with the software simply by labelling a small subset of the image data, thus visually indicating the desired classification scheme. iLastik then learns from these user-provided labels to build classifiers that are suitable for automated processing of large data sets. Real-time feedback furthermore allows the user to iteratively refine segmentation results and update the respective classifiers.
Other software tools support the investigation of structure–function relationships in the mouse brain. For example, an open-source framework consisting of the software packages WholeBrain and OpenBrainMap136 allows integration of anatomical, molecular and functional light-microscopy image data. This framework uses information and definitions from the mouse reference atlas generated by the Allen Institute137,138, and it includes tools for mapping labelled neurons into a versatile, standardized brain atlas, statistical data analysis and visualization and sharing of data through an interactive Web interface. CATMAID139 is another useful software framework that provides efficient access to large-scale electron and light microscopy image data sets as well as an infrastructure for collaborative annotation of such data through a decentralized Web interface. These design principles have been enabling not only for applications in neuroscience, and the field of connectomics in particular140, but also for large-scale projects in other areas of the life sciences, such as whole-tissue cell tracking in developmental biology141. Finally, powerful methods designed to specifically address the need for interactive 3D visualization of large images contribute another essential facet to the spectrum of open-source tools for handling high-resolution images of large, cleared specimens. BigDataViewer122 and TeraFly142 allow 3D viewing of large image stacks and are designed to be very responsive to user manipulation of the specimen’s coordinate system, such as translation, rotation and zoom. BigDataViewer is smoothly integrated in the Fiji ecosystem and plays a central role in advanced software suites such as BigStitcher, BigWarp and MaMuT for manual segmentation and tracking of cells in multiview light-sheet microscopy data143. TeraFly is designed in a particularly memory-efficient way and provides true 3D rendering capabilities, such as real-time alpha blending, as well as a ‘Virtual Finger’ feature that maps user input from the 2D plane of the computer screen to the 3D location in the data set corresponding to the selected biological structure.
Future objectives
Classical histology is typically performed on only a few selected thin slices. However, analysis of selected tissue sections is prone to inevitable biases because one may miss important biomedical information that is located elsewhere. By contrast, 3D histology on intact transparent specimens provides a much greater amount of information and thus a greater level of insight into anatomy and pathology. Moreover, it can speed up and reduce the cost of histology by several-thousand-fold compared with classical histology approaches.
Although the efficacy of tissue-clearing methods has improved considerably, we still lack a rigorous understanding of the physical and chemical principles underlying tissue-clearing processes. Moreover, we still need more robust labelling, imaging and data analysis tools to broaden the applications of tissue-clearing methods. In particular, we need the following: rapid and homogeneous protein and RNA labelling methods for whole organs of rodents and primates, large human tissues and, even, whole adult rodent and primate bodies; light-sheet microscopes that are capable of imaging tissues as large as organs and bodies of adult rodents and primates with isotropic subcellular resolution (less than 1 μm); and faster and more accurate algorithms for stitching, atlas registration and structure recognition in data sets larger than tens of terabytes. Furthermore, a standard atlas of organs and bodies of rodent and primates is required to quantitatively compare protein and RNA expression in different individuals. It would be ideal if new methods such as multiplexed robust fluorescence in situ hybridization, which allows imaging of dozens of mRNAs on brain sections, could also be combined with tissue-clearing methods144. We also expect that tissue-clearing methods will be combined with other prominent technologies, including single-cell RNA sequencing and mass spectrometry, to obtain spatial and temporal information on the quality and quantity of RNAs and proteins over entire organs and bodies.
In the future, tissue-clearing methods will significantly impact the drug development process by introducing unbiased readouts of entire organs and bodies in treated versus control groups, thereby assessing whether drug candidates for given neurological diseases can cross the blood-brain barrier and whether they can bind to desired targets in the brain and spinal cord at the level of single cells. Until now, targeting the amyloid peptides in the plaques by specific antibodies has been a major endeavour in an effort to develop an effective treatment for Alzheimer disease, but clinical trials of this approach have failed to show any efficacy. We speculate that one of the major contributing reasons for the failure in translational research is the lack of technologies that truly assess the targeting of such drugs at single-cell resolution in the whole body of preclinical animal models. Therefore, monitoring biodistributions and actions of drugs at single-cell resolution will be critical to comprehensively understand underlying cellular mechanisms and thereby design effective drugs. In this regard, nanotechnology will also greatly facilitate targeted drug delivery in many fields of biomedical science, including neuroscience. The effects of nanoparticle-based drugs are currently assessed by coarse imaging methods such as bioluminescence, which can detect the signal only when they are of millimetre size, significantly jeopardizing the use of nanoparticles to conduct nanoscale functions. Tissue-clearing methods will be a key technology in the development of drug-carrying nanoparticles, as such methods will allow identification of these agents in the whole rodent and primate bodies at single-cell resolution.
3D histopathology provided by tissue-clearing methods will also significantly scale up the investigation of human tissues in both preclinical and clinical arenas. For example, labelling and imaging whole human biopsy samples from patients with cancer (several thousand times larger than in classical histology) at single-cell resolution will provide much faster and more accurate diagnosis and staging of the tumours. Finally, clearing, labelling and imaging of adult human tissues on the order of centimetres will be one of the primary technologies to reveal the cellular structure of whole organs and eventually map circuits in whole human brains.
Supplementary Material
Acknowledgements
The authors thank E. A. Susaki for help in compiling Table 1 and Supplementary Table 1 for current tissue-clearing protocols and reagents, K. Matsumoto and Y. Shinohara for drawing the chemical structures in the supplementary information, T. Mano for contributing to the CUBIC figure, R. Cai and C. Pan for contributing to the uDISCO figure, S. R. Kumar, G. M. Coughlin, R. Challis and C. Challis for contributing to the viral-assisted spectral tracing figure and Y.-G. Park, C. H. Sohn, T. Ku, V. Lilascharoen and B. K. Lim for contributing to the SHIELD figure. The authors also gratefully acknowledge grant support from Brain/MINDS, the Basic Science and Platform Technology Program for Innovative Biological Medicine (AMED/MEXT), the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research (S)) and the Human Frontier Science Program Research Grant Program (HFSP RGP0019/2018) (H.R.U.), the Munich Cluster for Systems Neurology (SyNergy), the Fritz Thyssen Stiftung and the Deutsche Forschungsgemeinschaft (A.E.), the David and Lucile Packard Foundation (Packard Fellowship), the McKnight Foundation, the US National Institutes of Health (NIH) (1-DP2-ES027992; U01MH117072), the NCSOFT Cultural Foundation and the Koreaan Institute for Basic Science (IBS-R026-D1) (K.C.), the NIH BRAIN Initiative, the NIH Office of the Director and the US National Science Foundation (NeuroNex) (V.G.), LABEX LIFESENSES (reference ANR-10-LABX-65) managed by the French Agence National de la Recherche within the Investissements d’Avenir programme under reference ANR-11-IDEX-0004-02 (A.C.), the European Regional Development Fund in the framework of the Czech IT4Innovations National Supercomputing Center path to exascale project, project number CZ.02.1.01/0.0/0.0/16_013/0001791, within the Czech Research, Development and Education Operational Programme (P.T.) and the Howard Hughes Medical Institute (P.J.K.).
Glossary
- Tissue clearing
A method to make a biological specimen transparent by minimization of light scattering and light absorption by the biological specimen
- Hydrophobic tissue clearing
One of three major tissue-clearing methods; it uses hydrophobic (water-immiscible) reagents. it is also referred to as ‘solvent tissue clearing’
- Hydrophilic tissue clearing
One of three major tissue-clearing methods; it uses hydrophilic (water-miscible) reagents. it is also referred to as ‘aqueous tissue clearing’ but does not always involve the use of water.
- Refractive index
(RI). The ratio of the speed of light in a vacuum to its speed in a specified medium. The RI of a vacuum is 1 by definition, whereas the RI of water is ~1.33
- Light-sheet microscopy
A technique that allows fast, high-resolution imaging of large biological specimens with low light exposure by rapidly acquiring images of thin optical sections illuminated by laser light sheets
- Passive CLARITY technique
(PACT). A technique that allows flexible hydrogel formulation and clearing without the need to use electrophoresis
- Refractive index-matching solution
A solution that is compatible with the passive CLARITY technique, perfusion-assisted agent release in situ and CLARITY It provides high-resolution imaging at depth by further reducing light scattering in both cleared and uncleared samples
- Perfusion-assisted agent release in situ
(PARS). A method that allows whole-rodent clearing and labelling. It uses the intrinsic circulatory system (the vasculature) to deliver clearing agents and labels, instead of relying on passive diffusion, which can be prohibitively slow far large organs or whole organisms
- Hydrogel-based tissue clearing
One of three major tissue-clearing methods; it crosslinks biological specimens to make a synthetic hydrogel
- sCMOS detectors
Scientific-grade CMOS-based cameras that offer a large sensor area, high pixel count, low noise, high frame rate, high dynamic range and high quantum yield, all of which are highly desirable properties for detectors used in widefield fluorescence light microscopy
- Hierarchical data format version 5
(HDF5). A hierarchical data format and versatile data model to manage and represent extremely large and complex data objects
- Compute unified device architecture
(CUDA). A parallel computing platform and programming model developed by Nvidia for general computing on its own graphics processing units
- Adaptive particle representation
A content-adaptive representation of fluorescence microscopy images that overcomes storage and processing bottlenecks in big microscopy image data
Footnotes
Competing interests
H.R.U. is a co-inventor on a patent applications covering the CUBIC reagents (PCT/JP2014/070618 (pending), patent applicant is RIKEN, other co-inventors are E. A. Susaki and K. Tainaka; PCT/JP2017/016410 (pending), patent applicant is RIKEN, other co-inventors are K. Tainaka and T. Murakami) and a co-founder of CUBICStars Inc. A.E. is the applicant and the inventor on a patent application for technologies relating to vDISCO clearing (PCT/EP2018/063098 (pending)). K.C. is the inventor or a co-inventor on patents and patent applications for CLARITY (PCT/US2013/031066 (active), patent applicant is Stanford University, co-inventor is K. A. Deisseroth), stochastic electrotransport (PCT/US2015/024297 (active), patent applicant is MIT), SHIELD (PCT/US2016/064538 (pending), applicant is Massachusetts Institute of Technology (MIT), other co-inventors are E. Murray and J. H. Cho), SWITCH (PCT/US2016/064538 (pending), applicant is MIT, other co-inventors are E. Murray and J. H. Cho) and MAP (PCT/US2017/030285 (pending), applicant is MIT, other co-inventors are T. Ku, J. M. Swaney and J. Y. Park) and a co-founder of LifeCanvas Technologies. V.G. is a co-inventor on patent applications covering PACT and PARS (PCT/US2014/048985 (active), applicant is California Institute of Technology, other co-inventors are V. Gradinaru and B. Yang) and adeno-associated virus (US14/485,024 (active), applicant is California Institute of Technology, other co-inventors are B. E. Deverman, P. H. Patterson and V. Gradinaru) technologies. P.J.K. is an inventor or co-inventor on patents and patent applications covering multiview imaging (US14/049,470 (active), applicant is Howard Hughes Medical Institute) and adaptive light-sheet microscopy (PCT/US2017/038970 (pending), applicant is Howard Hughes Medical Institute, other co-inventors are R. K. Chhetri and L. A. Royer). P.T. and A.C. declare no competing interests.
Peer review information
Nature Reviews Neuroscience thanks S. Gentleman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41583-019-0250-1.
References
- 1.Spalteholz W Über das Durchsichtigmachen von Menschlichen und Tierischen Präparaten. (S. Hirzel, 1914). [Google Scholar]
- 2.Chung K et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article is the first to demonstrate in situ synthesis of a hydrogel and its fusion with tissue via covalent bonds to form a cleared tissue–hydrogel hybrid.
- 3.Yang B et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article first reports clearing of adult rodents via the vasculature; RNA detection in cleared tissues with single-molecule resolution by single-molecule fluorescence in situ hybridization and that specific formulations of tissue–hydrogel hybrids can significantly expand.
- 4.Renier N et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014. [DOI] [PubMed] [Google Scholar]
- 5.Susaki EA et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014. [DOI] [PubMed] [Google Scholar]; This article first reports on the delipidation activity of amino alcohols and the high-performance hydrophilic tissue-clearing method CUBIC and reports tissue clearing of the marmoset brain.
- 6.Tainaka K et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924 (2014. [DOI] [PubMed] [Google Scholar]; This article first demonstrates that decolourization of haem is possible in mild chemical conditions by amino alcohols, and reports whole-body imaging of the adult mouse.
- 7.Ertürk A et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat. Med 18, 166–171 (2012. [DOI] [PubMed] [Google Scholar]; This work demonstrates the application of an organic solvent-clearing method (3DISCO) on the intact adult mouse CNS to study neurodegeneration and regeneration.
- 8.Belle M et al. A simple method for 3D analysis of immunolabeled axonal tracts in a transparent nervous system. Cell Rep. 9, 1191–1201 (2014. [DOI] [PubMed] [Google Scholar]; This work is the first to combine whole-mount immunostaining, 3DISCO clearing and light-sheet microscopy to analyse axon guidance defects in mutant mice.
- 9.Treweek JB et al. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc 10, 1860–1896 (2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costantini I et al. A versatile clearing agent for multi-modal brain imaging. Sci. Rep 5, 9808 (2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klingberg A et al. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using light-sheet microscopy. J. Am. Soc. Nephrol 28, 452–459 (2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hama H et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci 14, 1481–1488 (2011. [DOI] [PubMed] [Google Scholar]
- 13.Ke M-T, Fujimoto S & Imai T SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci 16, 1154–1161 (2013. [DOI] [PubMed] [Google Scholar]
- 14.Dodt HU et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007. [DOI] [PubMed] [Google Scholar]
- 15.Susaki EA & Ueda HR Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem. Biol 23, 137–157 (2016. [DOI] [PubMed] [Google Scholar]
- 16.Tainaka K, Kuno A, Kubota SI, Murakami T & Ueda HR Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu. Rev. Cell Dev. Biol 32, 713–741 (2016. [DOI] [PubMed] [Google Scholar]
- 17.Murakami TC et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci 21, 625–637 (2018. [DOI] [PubMed] [Google Scholar]; This article first demonstrates the polymer-gel property of a fixed mouse brain in the absence of exogenous polymers, reports expansion microscopy using hydrophilic reagents and reports the generation of a 3D single-cell-resolution mouse brain atlas, the CUBIC-Atlas.
- 18.Amiya T& Tanaka T Phase transitions in crosslinked gels of natural polymers. Macromolecules 20, 1162–1164 (1987. [Google Scholar]
- 19.Shibayama M& Tanaka T in Responsive Gels: Volume Transitions I 1–62 (Springer, 1993. [Google Scholar]
- 20.Lorentz HA Ueber die Beziehung zwischen der Fortpflanzungsgeschwindigkeit des Lichtes und der Körperdichte. Ann. Phys 9, 641–665 (1880. [Google Scholar]
- 21.Lorenz L Ueber die Refractionsconstante. Ann. Phys 11,70–103 (1880. [Google Scholar]
- 22.Tainaka K et al. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep 24, 2196–2210 (2018. [DOI] [PubMed] [Google Scholar]; This article first reports the chemical profiling of hydrophilic tissue-clearing reagents and proposes chemical principles of hydrophilic tissue-clearing methods.
- 23.Belle M et al. Tridimensional visualization and analysis of early human development. Cell 169, 161–173 e112 (2017. [DOI] [PubMed] [Google Scholar]; This study uses 3D imaging of cleared human embryos and fetuses to provide the first comprehensive description of the development of the human peripheral nervous system and the innervation pattern of many organs in intact specimens.
- 24.Kim SY et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl Acad. Sci. USA 112, E6274–E6283 (2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray E et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article is the first to demonstrate that controlling the chemical interaction time and kinetics achieves more uniform and scalable processing of large-scale tissues.
- 26.Renier N et al. Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubota SI et al. Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep 20, 236–250 (2017. [DOI] [PubMed] [Google Scholar]
- 28.Cai R et al. Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull–meninges connections. Nat. Neurosci 22, 317–327 (2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that vDISCO boosted fluorescent protein signal in intact transparent mice, thereby allowing detection of widespread CNS trauma effects, and reveals short vascular connections between the skull marrow and brain meninges.
- 29.Deverman BE et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol 34, 204–209 (2016. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first article to report efficient crossing of the blood–brain barrier in adult rodents by an engineered adeno-associated virus vector able to package the cargo of choice. A Cre-enzyme-based viral-vector screening method is introduced and it is demonstrated how whole-body tissue clearing can facilitate transduction maps of systemically delivered genes.
- 30.Bedbrook CN, Deverman BE& Gradinaru V Viral strategies for targeting the central and peripheral nervous systems. Annu. Rev. Neurosci 41, 323–348 (2018. [DOI] [PubMed] [Google Scholar]
- 31.Keller PJ & Ahrens MB Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 85, 462–483 (2015. [DOI] [PubMed] [Google Scholar]
- 32.Vigouroux RJ, Belle M & Chédotal A Neuroscience in the third dimension: shedding new light on the brain with tissue clearing. Mol. Brain 10, 33 (2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ertürk A et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc 7, 1983–1995 (2012. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z et al. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 528, 225–230 (2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acar M et al. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526, 126–130 (2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espinosa-Medina I et al. Parasympathetic ganglia derive from Schwann cell precursors. Science 345, 87–90 (2014. [DOI] [PubMed] [Google Scholar]
- 37.Oshimori N, Oristian D & Fuchs E TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 (2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garofalo S et al. Enriched environment reduces glioma growth through immune and non-immune mechanisms in mice. Nat. Commun 6, 6623 (2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Neubeck B et al. An inhibitory antibody targeting carbonic anhydrase XII abrogates chemoresistance and significantly reduces lung metastases in an orthotopic breast cancer model in vivo. Int. J. Cancer 143, 2065–2075 (2018. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka N et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat. Biomed. Eng 1,796–806 (2017. [DOI] [PubMed] [Google Scholar]
- 41.Garvalov BK & Ertürk A Seeing whole-tumour heterogeneity. Nat. Biomed. Eng 1,772–774 (2017. [DOI] [PubMed] [Google Scholar]
- 42.Pan C et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867 (2016. [DOI] [PubMed] [Google Scholar]
- 43.Herisson F et al. Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration. Nat. Neurosci 21, 1209–1217 (2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan C et al. Deep learning reveals cancer metastasis and therapeutic antibody targeting in whole body. Cell 179, 1661–1676.e19 (2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; This works describes a novel method to detect and quantify cancer metastasis and antibody drug targeting at cellular resolution in the entire mouse using whole-body clearing and deep learning.
- 45.Chiang AS et al. Three-dimensional mapping of brain neuropils in the cockroach, Diploptera punctata. J. Comp. Neurol 440, 1–11 (2001. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y-C & Chiang A-S High-resolution confocal imaging and three-dimensional rendering. Methods 30, 86–93 (2003. [DOI] [PubMed] [Google Scholar]
- 47.Richardson DS & Lichtman JW Clarifying Tissue Clearing. Cell 162, 246–257 (2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirshburg J, Choi B, Nelson JS & Yeh AT Correlation between collagen solubility and skin optical clearing using sugars. Lasers Surg. Med 39, 140–144 (2007. [DOI] [PubMed] [Google Scholar]
- 49.Hama H et al. ScaleS: an optical clearing palette for biological imaging. Nat. Neurosci 18, 1518–1529 (2015. [DOI] [PubMed] [Google Scholar]
- 50.Chance B, Liu H, Kitai T & Zhang Y Effects of solutes on optical properties of biological materials: models, cells, and tissues. Anal. Biochem 227, 351–362 (1995. [DOI] [PubMed] [Google Scholar]
- 51.Ke M-T et al. Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep 14, 2718–2732 (2016. [DOI] [PubMed] [Google Scholar]
- 52.Susaki EA et al. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc 10, 1709–1727 (2015. [DOI] [PubMed] [Google Scholar]
- 53.Tatsuki F et al. Involvement of Ca2+-dependent hyperpolarization in sleep duration in mammals. Neuron 90, 70–85 (2016. [DOI] [PubMed] [Google Scholar]
- 54.Economo MN et al. A platform for brain-wide imaging and reconstruction of individual neurons. eLife 5, e10566 (2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L et al. The coding of valence and identity in the mammalian taste system. Nature 558, 127–131 (2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Justus D et al. Glutamatergic synaptic integration of locomotion speed via septoentorhinal projections. Nat. Neurosci 20, 16–19 (2017. [DOI] [PubMed] [Google Scholar]
- 57.Romanov RA et al. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci 20, 176–188 (2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lanjakornsiripan D et al. Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat. Commun 9, 1623 (2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rousso DL et al. Two pairs of on and off retinal ganglion cells are defined by intersectional patterns of transcription factor expression. Cell Rep 15, 1930–1944 (2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen JY et al. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227 (2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cuccarese MF et al. Heterogeneity of macrophage infiltration and therapeutic response in lung carcinoma revealed by 3D organ imaging. Nat. Commun 8, 14293 (2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis FM et al. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat. Commun 7, 13053 (2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J et al. Single-cell lineage tracing reveals that oriented cell division contributes to trabecular morphogenesis and regional specification. Cell Rep 15, 158–170 (2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto J et al. Neuronal signals regulate obesity induced beta-cell proliferation by FoxM1 dependent mechanism. Nat. Commun 8, 1930 (2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen F, Tillberg PW & Boyden ES Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article first reports the concept of expansion microscopy based on an exogenous hydrogel polymer.
- 66.Matsumoto K et al. Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat. Protoc 14, 3506–3537 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Tomer R, Ye L, Hsueh B & Deisseroth K Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc 9, 1682–1697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gradinaru V, Treweek J, Overton K & Deisseroth K Hydrogel-tissue chemistry: principles and applications. Annu. Rev. Biophys 47, 355–376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ku T et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol 34, 973–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A & Deisseroth K Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell 164, 792–804 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park Y-G et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat. Biotechnol 37, 73–83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article is the first to demonstrate protection of tissue and biomolecular properties against harsh stress using polyfunctional crosslinkers.
- 72.Renner M et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J 36, 1316–1329 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Canter RG et al. 3D mapping reveals network-specific amyloid progression and subcortical susceptibility in mice. Commun. Biol 2, 360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Greenbaum A et al. Bone CLARITY: clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow. Sci. Transl Med 9, eaah6518 (2017). [DOI] [PubMed] [Google Scholar]; This article reports clearing of whole bone with preserved marrow and imaging endogenous fluorescence with single-cell resolution throughout the marrow by light-sheet microscopy.
- 75.Greenbaum A, Jang MJ, Challis C & Gradinaru V Q&A: how can advances in tissue clearing and optogenetics contribute to our understanding of normal and diseased biology? BMC Biol 15, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shah S et al. Single-molecule RNA detection at depth via hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DePas WH et al. Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio 7, e00796–e00816 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Treweek JB & Gradinaru V Extracting structural and functional features of widely distributed biological circuits with single cell resolution via tissue clearing and delivery vectors. Curr. Opin. Biotechnol 40, 193–207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Menegas W et al. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife 4, e10032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article reports novel viral vectors to deliver cargo to neurons throughout the brain and body via the bloodstream, and a complementary vector toolbox for sparse stochastic labelling for morphological assessment. These delivery methods for Golgi-like genetic labelling can complement tissue clearing and microscopy methods to yield refined maps of the nervous system across the brain and body.
- 81.Robinson JE & Gradinaru V Dopaminergic dysfunction in neurodevelopmental disorders: recent advances and synergistic technologies to aid basic research. Curr. Opin. Neurobiol 48, 17–29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liebmann T et al. Three-dimensional study of Alzheimer’s disease hallmarks using the iDISCO clearing method. Cell Rep 16, 1138–1152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Welniarz Q et al. Non cell-autonomous role of DCC in the guidance of the corticospinal tract at the midline. Sci. Rep 7, 410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hruska M, Henderson N, Le Marchand SJ, Jafri H & Dalva MB Synaptic nanomodules underlie the organization and plasticity of spine synapses. Nat. Neurosci 21, 671–682 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ando K et al. Inside Alzheimer brain with CLARITY: senile plaques, neurofibrillary tangles and axons in 3-D. Acta Neuropathol 128, 457–459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morawski M et al. Developing 3D microscopy with CLARITY on human brain tissue: towards a tool for informing and validating MRI-based histology. Neuroimage 182, 417–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Phillips J et al. Development of passive CLARITY and immunofluorescent labelling of multiple proteins in human cerebellum: understanding mechanisms of neurodegeneration in mitochondrial disease. Sci. Rep 6, 26013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu AKL et al. Bringing CLARITY to the human brain: visualization of Lewy pathology in three dimensions. Neuropathol. Appl. Neurobiol 42, 573–587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee E et al. ACT-PRESTO: rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci. Rep 6, 18631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lai HM et al. Next generation histology methods for three-dimensional imaging of fresh and archival human brain tissues. Nat. Commun 9, 1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Allen JS, Damasio H & Grabowski TJ Normal neuroanatomical variation in the human brain: an MRI-volumetric study. Am. J. Phys. Anthropol 118, 341–358 (2002). [DOI] [PubMed] [Google Scholar]
- 92.Birey F et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Casoni F et al. Development of the neurons controlling fertility in humans: new insights from 3D imaging and transparent fetal brains. Development 143, 3969–3981 (2016). [DOI] [PubMed] [Google Scholar]
- 94.Hsueh B et al. Pathways to clinical CLARITY: volumetric analysis of irregular, soft, and heterogeneous tissues in development and disease. Sci. Rep 7, 5899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Behjati S, Lindsay S, Teichmann SA & Haniffa M Mapping human development at single-cell resolution. Development 145, dev152561 (2018). [DOI] [PubMed] [Google Scholar]
- 96.Kieffer C, Ladinsky MS, Ninh A, Galimidi RP & Bjorkman PJ Longitudinal imaging of HIV-1 spread in humanized mice with parallel 3D immunofluorescence and electron tomography. eLife 6, e23282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dantzer R, O’Connor JC, Freund GG, Johnson RW & Kelley KW From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci 9, 46–56 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Glaser AK et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng 1, 0084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nojima S et al. CUBIC pathology: three-dimensional imaging for pathological diagnosis. Sci. Rep 7, 9269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Royen ME et al. Three-dimensional microscopic analysis of clinical prostate specimens. Histopathology 69, 985–992 (2016). [DOI] [PubMed] [Google Scholar]
- 101.Huisken J, Swoger J, Del Bene F, Wittbrodt J & Stelzer EH Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004). [DOI] [PubMed] [Google Scholar]
- 102.Keller PJ, Schmidt AD, Wittbrodt J & Stelzer EHK Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008). [DOI] [PubMed] [Google Scholar]
- 103.Voie AH, Burns DH & Spelman FA Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J. Microsc 170, 229–236 (1993). [DOI] [PubMed] [Google Scholar]
- 104.Ryan DP et al. Automatic and adaptive heterogeneous refractive index compensation for light-sheet microscopy. Nat. Commun 8, 612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tomer R et al. SPED light sheet microscopy: fast mapping of biological system structure and function. Cell 163, 1796–1806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gómez-Gaviro MV et al. Optimized CUBIC protocol for 3D imaging of chicken embryos at single-cell resolution. Development 44, 2092–2097 (2017). [DOI] [PubMed] [Google Scholar]
- 107.Stefaniuk M et al. Light-sheet microscopy imaging of a whole cleared rat brain with Thy1-GFP transgene. Sci. Rep 6, 28209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Niedworok CJ et al. Charting monosynaptic connectivity maps by two-color light-sheet fluorescence microscopy. Cell Rep 2, 1375–1386 (2012). [DOI] [PubMed] [Google Scholar]
- 109.Planchon TA et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8, 417–423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen BC et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes lattice light-sheet microscopy, which uses very thin laser light sheets for fluorescence generation and introduces a light-efficient concept for high-resolution imaging of a field of view of approximately 100 μm.
- 111.Wu YC et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat. Biotechnol 31, 1032–1038 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes diSPIM light-sheet microscopy, which rapidly acquires two orthogonal views of a specimen with a field of view of several hundred micrometres and hence combines high imaging speeds with a high, spatially isotropic resolution of 330 nm.
- 112.Chhetri RK et al. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat. Methods 12, 1171–1178 (2015). [DOI] [PubMed] [Google Scholar]; By allowing simultaneous orthogonal four-view imaging with a field of view approaching a 1 mm, IsoView light-sheet microscopy achieves a volume throughput of more than 108 μm3 per second and a high, spatially isotropic resolution of 400 nm.
- 113.Swoger J, Verveer P, Greger K, Huisken J & Stelzer EH Multi-view image fusion improves resolution in three-dimensional microscopy. Opt. Express 15, 8029–8042 (2007). [DOI] [PubMed] [Google Scholar]
- 114.Royer LA et al. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol 34, 1267–1278 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Royer LA, Lemon WC, Chhetri RK & Keller PJ A practical guide to adaptive light-sheet microscopy. Nat. Protoc 13, 2462–2500 (2018). [DOI] [PubMed] [Google Scholar]
- 116.Silvestri L et al. RAPID: real-time image-based autofocus for all wide-field optical microscopy systems. Preprint at bioRxiv 10.1101/170555 (2017). [DOI] [Google Scholar]
- 117.Hörl D et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods 16, 870–874 (2019). [DOI] [PubMed] [Google Scholar]; This artcle describes BigStitcher, which offers highly customizable stitching of arbitrarily large image mosaics and multiview selective plane illumination microscopy data building on the BigDataViewer infrastructure in Fiji.
- 118.Gao R et al. Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363, eaau8302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dean KM, Roudot P, Welf ES, Danuser G & Fiolka R Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophys. J 108, 2807–2815 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pende M et al. High-resolution ultramicroscopy of the developing and adult nervous system in optically cleared Drosophila melanogaster. Nat. Commun 9, 4731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Amat F et al. Efficient processing and analysis of large-scale light-sheet microscopy data. Nat. Protoc 10, 1679–1696 (2015). [DOI] [PubMed] [Google Scholar]
- 122.Pietzsch T, Saalfeld S, Preibisch S & Tomancak P BigDataViewer: visualization and processing for large image data sets. Nat. Methods 12, 481–483 (2015). [DOI] [PubMed] [Google Scholar]; This article describes BigDataViewer, which provides a well-engineered software framework to navigate and compute on arbitrarily large image data sets that is integrated into the popular Fiji ecosystem.
- 123.Schindelin J, Rueden CT, Hiner MC & Eliceiri KW The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev 82, 518–529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pietzsch T, Preibisch S, Tomancak P & Saalfeld S ImgLib2—generic image processing in Java. Bioinformatics 28, 3009–3011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Preibisch S, Saalfeld S, Schindelin J & Tomancak P Software for bead-based registration of selective plane illumination microscopy data. Nat. Methods 7, 418–419 (2010). [DOI] [PubMed] [Google Scholar]
- 126.Preibisch S et al. Efficient Bayesian-based multiview deconvolution. Nat. Methods 11, 645–648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Balazs B, Deschamps J, Albert M, Ries J & Hufnagel L A real-time compression library for microscopy images. Preprint at bioRxiv 10.1101/164624 (2017). [DOI] [Google Scholar]
- 128.Cheeseman BL, Gunther U, Susik M, Gonciarz K & Sbalzarini IF Forget pixels: adaptive particle representation of fluorescence microscopy images. Nat. Commun 9, 5160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tomer R, Denes AS, Tessmar-Raible K & Arendt D Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell 142, 800–809 (2010). [DOI] [PubMed] [Google Scholar]
- 131.Heckscher ES et al. Atlas-builder software and the eNeuro atlas: resources for developmental biology and neuroscience. Development 141,2524–2532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ronneberger O et al. ViBE-Z: a framework for 3D virtual colocalization analysis in zebrafish larval brains. Nat. Methods 9, 735–742 (2012). [DOI] [PubMed] [Google Scholar]
- 133.Bogovic JA, Hanslovsky P, Wong A & Saalfeld S in 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) 1123–1126 (IEEE, 2016). [Google Scholar]
- 134.Zheng Z et al. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell 174, 730–743.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sommer CS, Kothe C, Hamprecht U & Ilastik FA in 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 230–233 (IEEE, 2011). [Google Scholar]
- 136.Fürth D et al. An interactive framework for whole-brain maps at cellular resolution. Nat. Neurosci 21, 139–149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oh SW et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lein ES et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 139.Saalfeld S, Cardona A, Hartenstein V & Tomancak P CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25, 1984–1986 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schneider-Mizell CM et al. Quantitative neuroanatomy for connectomics in Drosophila. eLife 5, e12059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article describes the first applications of crowdsourcing of connectome tracing through the Web-based CATMAID framework.
- 141.Amat F et al. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods 11, 951–958 (2014). [DOI] [PubMed] [Google Scholar]
- 142.Bria A & Iannello G TeraStitcher - a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinforma 13, 316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wolff C et al. Multi-view light-sheet imaging and tracking with the MaMuT software reveals the cell lineage of a direct developing arthropod limb. eLife 7, e34410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Moffitt JR et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bakutkin VV, Maksimova IL, Semyonova TN, Tuchin VV & Kon IL in Ophthalmic Technologies V 137–142 (SPIE, 1995). [Google Scholar]
- 146.Zimnyakov DA, Tuchin VV, Michin AA, Kon IL & Serov AN in Ophthalmic Technologies VI 233–243 (SPIE, 1996). [Google Scholar]
- 147.Tuchin VV et al. in Photon Propagation in Tissues II 118–143 (SPIE, 1996). [Google Scholar]
- 148.Tuchin VV et al. Light propagation in tissues with controlled optical properties. J. Biomed. Opt 2, 401–417 (1997). [DOI] [PubMed] [Google Scholar]
- 149.Bashkatov AN et al. in Ophthalmic Technologies IX 311–320 (SPIE, 1999). [Google Scholar]
- 150.Tuchin VV et al. Optics of living tissues with controlled scattering properties. Proc. SPIE 3863, 10–21 (1999). [Google Scholar]
- 151.Tuchin VV, Xu X & Wang RK Dynamic optical coherence tomography in studies of optical clearing, sedimentation, and aggregation of immersed blood. Appl. Opt 41, 258–271 (2002). [DOI] [PubMed] [Google Scholar]
- 152.Xu X, Wang RK, Elder JB & Tuchin VV Effect of dextran-induced changes in refractive index and aggregation on optical properties of whole blood. Phys. Med. Biol 48, 1205–1221 (2003). [DOI] [PubMed] [Google Scholar]
- 153.Liu H, Beauvoit B, Kimura M & Chance B Dependence of tissue optical properties on solute-induced changes in refractive index and osmolarity. J. Biomed. Opt 1, 200–211 (1996). [DOI] [PubMed] [Google Scholar]
- 154.Vargas O, Chan EK, Barton JK, Rylander HG & Welch AJ Use of an agent to reduce scattering in skin. Lasers Surg. Med 24, 133–141 (1999). [DOI] [PubMed] [Google Scholar]
- 155.Vargas G, Chan KF, Thomsen SL & Welch AJ Use of osmotically active agents to alter optical properties of tissue: effects on the detected fluorescence signal measured through skin. Lasers Surg. Med 29, 213–220 (2001). [DOI] [PubMed] [Google Scholar]
- 156.Wang RK, Xu X, Tuchin VV & Elder JB Concurrent enhancement of imaging depth and contrast for optical coherence tomography by hyperosmotic agents. JOSA B 18, 948–953 (2001). [Google Scholar]
- 157.Xu X & Wang RK The role of water desorption on optical clearing of biotissue: studied with near infrared reflectance spectroscopy. Med. Phys 30, 1246–1253 (2003). [DOI] [PubMed] [Google Scholar]
- 158.Jiang J & Wang RK Comparing the synergistic effects of oleic acid and dimethyl sulfoxide as vehicles for optical clearing of skin tissue in vitro. Phys. Med. Biol 49, 5283–5294 (2004). [DOI] [PubMed] [Google Scholar]
- 159.Choi B et al. Determination of chemical agent optical clearing potential using in vitro human skin. Lasers Surg. Med 36, 72–75 (2005). [DOI] [PubMed] [Google Scholar]
- 160.Staudt T, Lang MC, Medda R, Engelhardt J & Hell SW 2,2′-Thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc. Res. Tech 70, 1–9 (2007). [DOI] [PubMed] [Google Scholar]
- 161.Aoyagi Y, Kawakami R, Osanai H, Hibi T & Nemoto T A rapid optical clearing protocol using 2,2′-thiodiethanol for microscopic observation of fixed mouse brain. PLOS ONE 10, e0116280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kuwajima T et al. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 140, 1364–1368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tsai PS et al. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels. J. Neurosci 29, 14553–14570 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hou B et al. Scalable and DiI-compatible optical clearance of the mammalian brain. Front Neuroanat 9, 19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Diogo R, Siomava N & Gitton Y Development of human limb muscles based on whole-mount immunostaining and the links between ontogeny and evolution. Development 146, dev180349 (2019). [DOI] [PubMed] [Google Scholar]
- 166.LaVision BioTec. The UltraMicroscope setup. LaVision BioTec https://www.lavisionbiotec.com/products/UltraMicroscope/specification.html (2019).
- 167.Zeiss. Lightsheet Z1. Zeiss https://www.zeiss.com/microscopy/us/products/imaging-systems/lightsheet-z-1.html#downloads (2019).
- 168.Engelbrecht CJ & Stelzer EH Resolution enhancement in a light-sheet-based microscope (SPIM). Opt. Lett 31, 1477–1479 (2006). [DOI] [PubMed] [Google Scholar]
- 169.Schwarz MK et al. Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains. PLOS ONE 10, e0124650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jing D et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res 28, 803–818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Becker K, Jährling N, Saghafi S, Weiler R & Dodt HU Chemical clearing and dehydration of GFP expressing mouse brains. PLOS ONE 7, e33916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Scott GD, Blum ED, Fryer AD & Jacoby DB Tissue optical clearing, three-dimensional imaging, and computer morphometry in whole mouse lungs and human airways. Am. J. Respir. Cell. Mol. Biol 51, 43–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ertürk A, Lafkas D & Chalouni C Imaging cleared intact biological systems at a cellular level by 3DISCO. J. Vis. Exp 10.3791/5138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Epp JR et al. Optimization of CLARITY for clearing whole-brain and other intact organs. eNeuro 2, ENEURO.0022-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Abe T et al. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development 140, 237–246 (2013). [DOI] [PubMed] [Google Scholar]
- 176.Breuss M et al. The expression of tubb2b undergoes a developmental transition in murine cortical neurons. J. Comp. Neurol 523, 2161–2186 (2015). [DOI] [PubMed] [Google Scholar]
- 177.Sekitani T et al. Ultraflexible organic amplifier with biocompatible gel electrodes. Nat. Commun 7, 11425 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mizutani H et al. Transparency-enhancing technology allows three-dimensional assessment of gastrointestinal mucosa: a porcine model. Pathol. Int 68, 102–108 (2018). [DOI] [PubMed] [Google Scholar]
- 179.Warner CA et al. An optical clearing technique for plant tissues allowing deep imaging and compatible with fluorescence microscopy. Plant. Physiol 166, 1684–1687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hasegawa J et al. Three-dimensional imaging of plant organs using a simple and rapid transparency technique. Plant Cell Physiol 57, 462–472 (2016). [DOI] [PubMed] [Google Scholar]
- 181.Chen L et al. UbasM: an effective balanced optical clearing method for intact biomedical imaging. Sci. Rep 7, 12218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kurihara D, Mizuta Y, Sato Y & Higashiyama T ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development 142, 4168–4179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang Z et al. Imaging transparent intact cardiac tissue with single-cell resolution. Biomed. Opt. Express 9, 423–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yu T et al. RTF: a rapid and versatile tissue optical clearing method. Sci. Rep 8, 1964 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li W, Germain RN & Gerner MY Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl Acad. Sci. USA 114, E7321–E7330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lai HM et al. Rationalisation and validation of an acrylamide-free procedure in three-dimensional histological imaging. PLOS ONE 11, e0158628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Xu N et al. Fast free-of-acrylamide clearing tissue (FACT)—an optimized new protocol for rapid, high-resolution imaging of three-dimensional brain tissue. Sci. Rep 7, 9895 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Perbellini F et al. Free-of-Acrylamide SDS-based tissue clearing (FASTClear) for three dimensional visualization of myocardial tissue. Sci. Rep 7, 5188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


