Abstract
Purpose:
We combine SNR-efficient acquisition and model-based reconstruction strategies with newly available hardware instrumentation to achieve distortion-free in-vivo diffusion magnetic resonance imaging (dMRI) of the brain at submillimeter-isotropic resolution with high fidelity and sensitivity on a clinical 3T scanner.
Methods:
We propose Blip Up-Down Acquisition (BUDA) for multi-shot EPI using interleaved blip-up and -down phase encoding and incorporate B0 forward-modeling into structured low-rank reconstruction to enable distortion- and navigator-free dMRI. We further combine BUDA-EPI with an SNR-efficient Simultaneous Multi-Slab acquisition termed gSlider and dubbed the combined approach “gSlider BUDA-EPI”, to achieve high isotropic-resolution dMRI. To validate gSlider BUDA-EPI, whole-brain diffusion data at 860 and 780 μm datasets were acquired. Finally, to improve the conditioning and minimize noise penalty in BUDA reconstruction at very high resolutions where B0 inhomogeneity can have a detrimental effect, the level of B0 inhomogeneity was reduced by incorporating slab-by-slab dynamic shimming with a 32-channel AC/DC coil into the acquisition. Whole-brain 600μm diffusion data were then acquired with this combined approach of gSlider BUDA-EPI with dynamic shimming.
Results:
The results of 860 and 780μm datasets show high geometry fidelity with gSlider BUDA-EPI. With dynamic shimming, the BUDA reconstruction’s noise penalty was further alleviated. This enables whole-brain 600μm isotropic resolution diffusion imaging with high image-quality.
Conclusions:
gSlider BUDA-EPI enables high-quality distortion-free diffusion imaging across the whole-brain at sub-millimeter resolution, where the use of multi-coil dynamic B0 shimming further improve reconstruction performance, which can be particularly useful at very high resolutions.
Keywords: gSlider, distortion correction, shim array, high isotropic-resolution, diffusion-weighted imaging
Introduction
Diffusion magnetic resonance imaging (dMRI) with echo-planar imaging (EPI) is widely used for many neuroscientific and clinical applications. Submillimeter-isotropic-resolution dMRI has shown great potential for characterizing gray matter (1-3) as well as elucidate details of fine-scale brain structures such as the U-fibers in the white matter (4,5). However, the current resolution of in-vivo dMRI with EPI at 1.2-2.0mm-isotropic-resolution is too low to probe the fine detail structures of the brain, e.g. following the curvature of the U-fiber in white matter and resolving different layers of the gray matter (2,4,6). Compared to single-shot EPI (ss-EPI) acquisitions, multi-shot EPI (ms-EPI) (7,8) is a promising approach for high-resolution dMRI, as it can mitigate geometry distortions and T2* blurring. However, for submillimeter isotropic-resolution in-vivo dMRI, ms-EPI acquisitions are still limited by the following challenges: (i) artifacts arising from physiological noise-induced shot-to-shot phase variations, (ii) low signal-to-noise ratio (SNR), and (iii) remaining image distortions from B0 inhomogeneity and eddy currents. These issues and emerging mitigation approaches are outlined below.
(i) Combining multiple shots is prohibitively difficult in ms-EPI because of the shot-to-shot phase variations. These variations can be estimated and corrected using navigators (8,9), albeit at the cost of imaging efficiency and increased SAR. To overcome this issue, a range of navigator-free approaches have been explored which employ parallel imaging (10-12) with sparse or low-rank constraints (13,14), and deep learning (15,16) methods. In particular, Hankel structured low-rank constrained parallel imaging approaches (17-20) were recently proposed to enable navigator-free ms-EPI acquisition with higher in-plane acceleration and low geometric distortion.
(ii) Improving SNR is critical for high resolution dMRI. A number of acquisition approaches have been developed to boost SNR and reduce scan time in dMRI, by moving away from conventional 2D slice-by-slice imaging to more SNR-efficient simultaneous multi-slice (SMS) (21-24) and other volumetric acquisitions (25-27). With these approaches, a larger imaging volume is encoded during each EPI readout to provide better noise averaging to improve SNR. Among these approaches, three-dimensional dMRI with simultaneous multi-slab (SMSb) acquisition (28,29) is a promising strategy that enhance SNR with high acquisition efficiency for submillimeter-isotropic resolution dMRI. However, the shot-to-shot phase variations and the slab-boundary artifacts continues to be the key challenges for this strategy. To solve these issues, navigator-based and self-navigated methods (29,30) were proposed to correct the phase variations along both ky and kz dimensions, and slab-profile correction methods (31,32) were also proposed to mitigate the slab-boundary artifacts. Compared to these ‘Gz-gradient’ slab-encoded three-dimensional dMRI techniques, Generalized SLIce Dithered Enhanced Resolution (gSlider) (33) is another simultaneous multi-slab technique which differs in its use of ‘RF excitation’ rather than gradient-encoding to perform slab-encodings.
(iii) During the lengthy EPI readout, the magnetization is exposed to local field variations that stem from B0 inhomogeneity caused by tissue susceptibility interfaces inside the head as well as changes in chest cavity due to respiration. The local B0 inhomogeneity can lead to signal pileups, where multiple voxels overlap and cannot be disentangled without additional data acquisition. Geometric distortion scales badly with resolution since the time to acquire each line of k-space increases approximately linearly. This makes it imperative to reduce the geometric distortion in high resolution dMRI. To completely remove distortions from B0 inhomogeneity as well as eddy-current in dMRI and achieve high geometric fidelity, approaches that rely on additional EPI acquisitions with reversed phase encoding direction are typically employed. Since voxels stretched in one phase-encoding direction will be compressed in the other and vice versa, such information is employed in popular post-processing software packages, such as the FSL ‘TOPUP’ (34-36), to estimate and correct for distortions. Moreover, joint blip up/down parallel imaging reconstruction approaches using the hybrid-space SENSE reconstruction (37,38) have been proposed, which perform joint parallel imaging reconstruction of each pair of phase encoding acquisitions (blip up and down). Here field-map distortion information from TOPUP is directly incorporated into this reconstruction to generate distortion-free images.
In this work, to improve upon the hybrid-space SENSE approach, we first propose a Blip Up-Down Acquisition (BUDA) and joint parallel-imaging reconstruction with a structured low-rank constraint framework for distortion-free EPI (20). Second, we combine BUDA-EPI with gSlider, to enable high-isotropic-resolution dMRI with high-fidelity and SNR-efficiency. Finally, we improve the matrix conditioning and mitigate the noise penalty of the gSlider BUDA-EPI reconstruction by incorporating slab-by-slab dynamic shimming with a 32-channel AC/DC coil into the acquisition to reduce B0 inhomogeneity. Together, these methods enabled high-quality distortion-free whole-brain 600μm dMRI on a 3T clinical scanner.
Methods
Multi-shot EPI with Blip Up-Down Acquisition (BUDA-EPI)
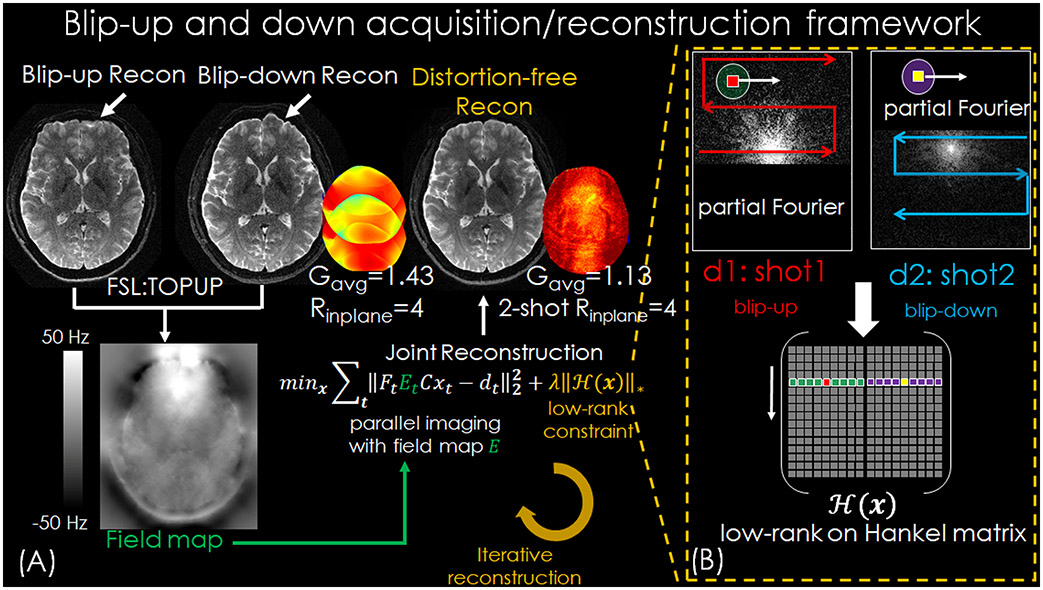
Figure 1 shows the acquisition and reconstruction framework of BUDA-EPI. In BUDA-EPI, two EPI-shots sample complementary subsets of k-space, one with a positive ky traversal (blip-up) and the other with a negative traversal (blip-down), to create opposing distortions (Figure 1(B)). The BUDA reconstruction includes the following steps: (i) individual SENSE reconstructions (39) with L1-wavelet constraint for blip-up and blip-down acquisitions, respectively. The L1-wavelet regularization is used to reduce noise and add spatial smoothing on the reconstructed images to help improve the subsequent B0 inhomogenity estimation. As the white arrows on the top-left of Figure 1(A) indicate, each individual shot from such reconstruction has significant geometric distortions along the phase-encoding direction. (ii) Estimating the field maps using SENSE reconstructed blip-up and blip-down images. The field map containing B0 and eddy-current effects can be extracted from this acquisition pair via FSL ‘TOPUP’(34,35,40). (iii) This field map is then incorporated into a joint-reconstruction with a structured low-rank constraint across the two shots to account for shot-to-shot phase variations, which can be expressed as:
| (1) |
where Ft is the undersampled Fourier operator in tth shot, Et is the estimated off-resonance information, C are the coil sensitivities estimated from a distortion-free gradient-echo pre-scan data using ESPIRiT (41), xt is the distortion-free image and dt are the k-space data for shot t. In the forward model, the distortion-free images are multiplied by coil sensitivities and distorted by the phase modulation of the off-resonance map in hybrid (x-ky) space as per (38). The constraint enforces a low-rank prior on the block-Hankel representation of the multi-shot data x, which is applied in k-space where small blocks of k-space data (e.g. kx × ky = 7 × 7) are concatenated from each shot in the column axis. This low-rank constraint with the rank regularization parameter λ (e.g. λ=1) exploits the similarity of the reconstructed images from the multi-shot data, where the reconstructed images across shots should have the same image magnitude, but differing smooth image phase from shot-to-shot phase variations. Subsequent to the low-rank reconstruction, the two multi-shot images xt for each diffusion direction are combined to produce a single image, where the background phase in each shot is estimated by spatial smoothing using a hamming filter with the filter window size equal to that of the acquired symmetric k-space region, and removed before the combination(42,43).
Figure 1.
(A) Reconstruction flowchart of BUDA-EPI. (B) Complementary k-space acquisition strategy is applied to each shot and the reconstructed multi-shot data are reshaped to the block-Hankel representation and enforced by the low rank prior in each iteration to provide robustness to shot-to-shot phase corruption.
Compared to reconstructing the Blip-up and -down EPI separately, joint BUDA-EPI reconstruction achieves improved reconstruction conditioning as illustrated by the marked reduction in g-factor noise penalty (Figure 1(A)). In the standard multi-shot interleave-EPI with no shot-to-shot phase variation, the total acceleration factor would be Rinplane =2 with two-shot acquisition. However, due to the shot-to-shot phase variations and the B0 inhomogeneity induced mismatches in the two blip up and down shots, the net total acceleration factor of the 2-shot BUDA data should be higher than Rinplane =2. Therefore, we refrain from using the total acceleration factor here but rather provide the acceleration factor of each acquisition shot while noting that the acquisition is performed across 2 shots as per Figure 1. Additionally, a partial Fourier (p.f.) acquisition where EPI shots sample complementary k-space sections is incorporated directly into the low-rank reconstruction (Figure 1(B)), so a short-TE can be attained to minimize T2-related signal losses.
Combining gSlider with BUDA-EPI (gSlider BUDA-EPI)
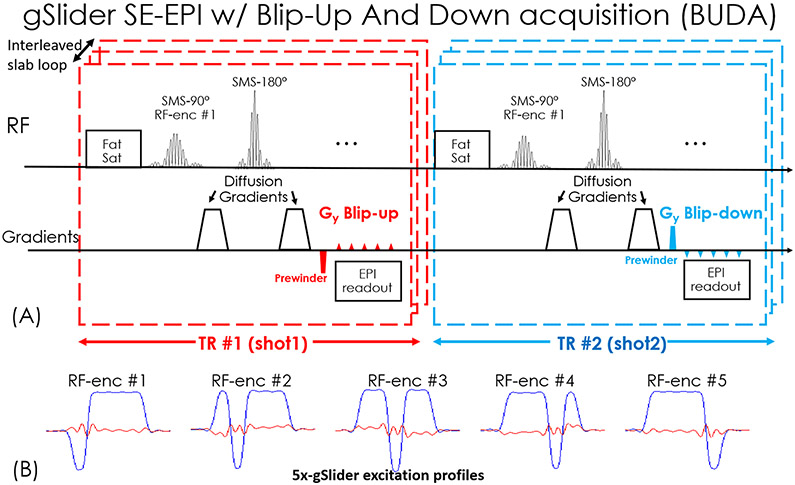
BUDA-EPI is incorporated into gSlider simultaneous multi-slab acquisition, where the blip-up and blip-down shots for each RF slab-encoding are acquired sequentially in two consecutive TRs (Figure 2(A)). Five RF-encoding pulses (the excitation profiles are shown in Figure 2(B)) are used for a 5x-gSlider slab-encoding; resulting in a total of 10 EPI-shots per slab across blip-up and -down encodings, all acquired together across 10 consecutive TRs, where each RF slab-encoding is designed to produce a slab-encoded volume with high image-SNR. Through this approach, high-quality dMRI has been achieved with self-navigation of shot-to-shot phase variations and bulk subject motion (44,45). The gSlider RF-excitations are performed across simultaneous multi-slab (SMSb) to acquire a large number of slices per EPI-shot, e.g. 10 simultaneous slices using gSlider RF-encoding of 5 sub-slices per slab and Multiband (MB) factor of 2 across two slabs. Here, blipped-CAIPI EPI readout is employed to achieve controlled-aliasing across the slabs to improve conditioning of the reconstruction (22). BUDA reconstruction is first performed for each RF-encoding volume using Equation 1, where extension in the forward model was made to allow for the reconstruction across simultaneously acquired slabs. Subsequently, the processed RF slab-encoded volumes are combined together in a forward model based reconstruction to resolve the sub-slices and create a high slice-resolution volume as per (33).
Figure 2.
(A) Sequence diagram of gSlider BUDA-EPI. (B) The designed 5x-gSlider excitation profiles.
To ensure high quality reconstruction, the effects on the slab profile from B1+ inhomogeneity and incomplete T1 recovery at short TRs were also incorporated into the reconstruction as per (45). The B1+ correction requires an extra low-resolution B1+ map, which was obtained using a vendor supplied Turbo-FLASH based B1+ mapping sequence (46). The scan time for this B1+ mapping at 3.4×3.4×5 mm3 resolution whole brain was ~20 seconds.
Our in-vivo study was approved by the institutional review board (IRB). To validate gSlider BUDA-EPI, whole-brain 1 mm isotropic resolution diffusion imaging data were acquired with a Siemens 32 channel head coil. The protocol used: FOV = 220×220×130 mm3 (26 slabs, 5 mm slab thickness), MB × Rinplane × gSlider= 2 × 4 × 5 per shot, CAIPI-shift =2, b = 1000 s/mm2, image matrix size = 220 × 220, TR/TE=3500/86 ms. Using 10 EPI shots, a distortion-free 1mm isotropic diffusion volume for each diffusion direction was acquired in 35 seconds. A matching T2-weighted 3D fast spin-echo (FSE) data was acquired to serve as a distortion-free reference.
To compare the reconstruction performance between hybrid-space SENSE (38) and BUDA method, g-factor was used as a guide for the conditioning of the overall reconstruction. For the hybrid-space SENSE reconstruction, we estimate the shot-to-shot phase differences from the individual blip-up and -down reconstructions, and then incorporate them into the g-factor calculation. To evaluate the Hankel low-rank regularization in the BUDA approach, the g-factor is calculated using the pseudo multiple replica method (47) with 200 repetitions of the Monte-Carlo simulation. In each Monte-Carlo repetition, independent and identically distributed complex gaussian noise is added to the raw k-space data of the pre-whitten coil channels. Each repetition runs the joint BUDA reconstruction and structured low-rank regularization with identical parameters used in the actual diffusion imaging reconstruciton. The square root of Rinplane increase in noise in the accelerated acquisition has been accounted for in the g-factor calculated by the pseudo multiple replica method.
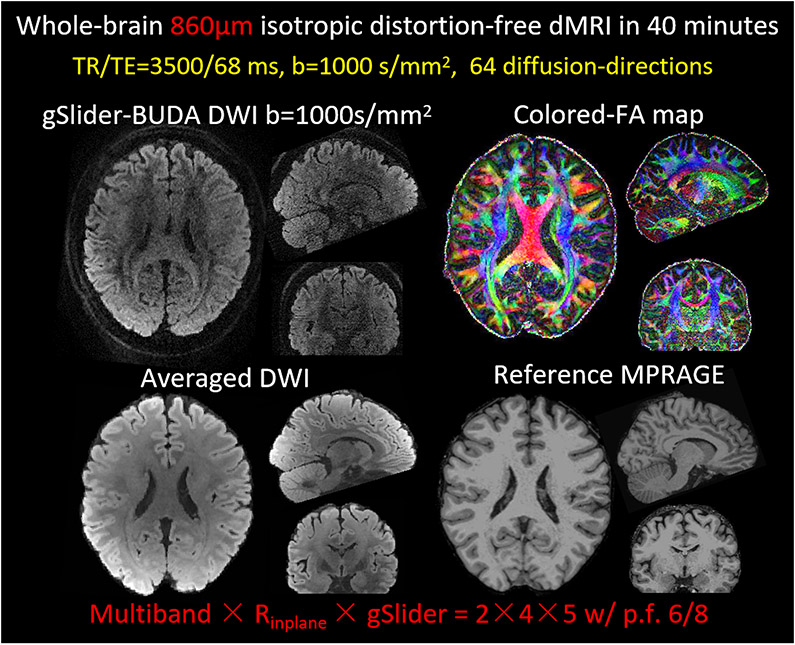
Whole brain 860μm data were acquired with the same 32 channel head coil using the following imaging parameters: FOV = 220 × 220 × 129mm3 (30 slabs, 4.3 mm slab thickness), image matrix size = 256 × 256, TR/TE = 3500/68ms. Five gSlider RF-encodings across blip up (phase-encoding direction: Anterior to Posterior) and down (phase-encoding direction: Posterior to Anterior) shots were collected at MB × Rinplane = 2 × 4, with CAIPI-shift of 2 and p.f. = 6/8. Sixty-four diffusion-directions with b =1000 s/mm2 were acquired, along with interspersed eight b=0 volumes. The total acquisition time was 40 minutes. Diffusion analysis and colored fractional anisotropy (FA) maps of the whole brain 860μm data were generated using the FSL software (34).
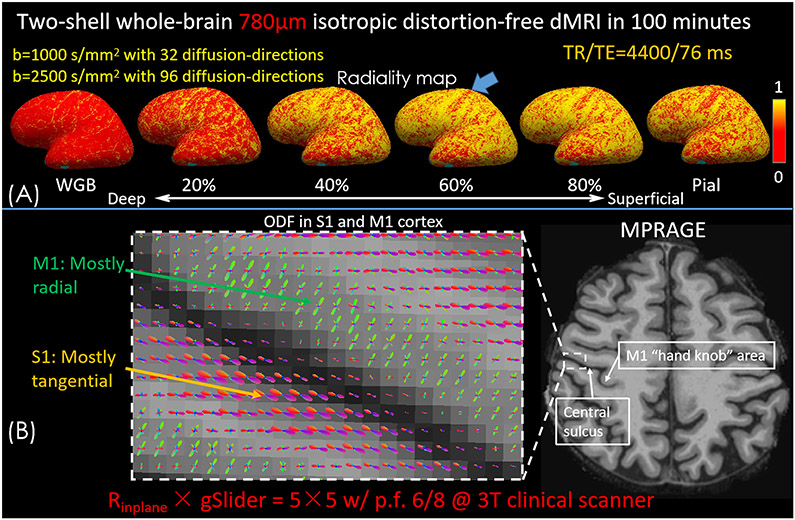
A longer gSlider BUDA-EPI acquisition was also performed at a higher spatial resolution to investigate the potential of gSlider BUDA-EPI in delineating gray-matter structures. Whole-brain 780 μm data were acquired across two shells using the same 32 channel head coil. The imaging parameters were FOV = 220 × 220 × 117mm3 (30 slabs, 3.9 mm slab thickness), image matrix size = 282×282, TR/TE = 4400/76 ms. Thirty-two diffusion-directions were acquired with b=1000s/mm2 and ninety-six diffusion-directions with b = 2500s/mm2. To reduce TE and T2* blurring Rinplane = 5 with p.f. = 6/8 was employed for each shot, where the missing p.f. data were recovered through BUDA low-rank reconstruction as described above. The use of Rinplane = 5 results in an effective echo-spacing of 0.196ms. 3D-MPRAGE was acquired along with the diffusion data for surface-based analysis. For cortical depth analysis, the intermediate surfaces between white and pial surfaces were generated by FreeSurfer (48) from the MPRAGE data. The diffusion principal eigenvectors were then aligned to the MPRAGE data using boundary-based co-registration (49) and projected to intermediate surfaces to calculate the radiality (50,51). For the acquired multi-shell data, orientation distribution function (ODF) was also estimated utilizing MRtrix software (52) multi-tissue-multi-shell CSD method (1,53).
gSlider BUDA-EPI with multi-coil dynamic B0 shimming
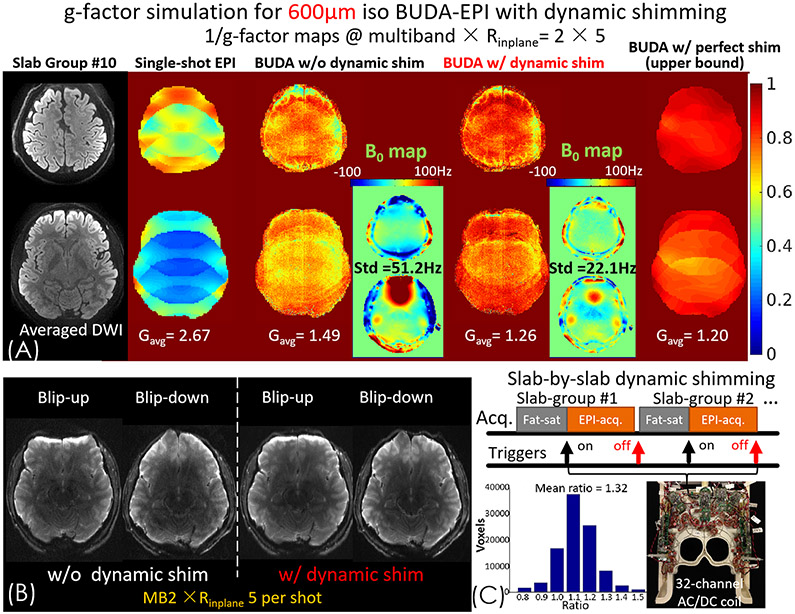
We further push the spatial resolution of gSlider BUDA-EPI to 600 μm isotropic using MB × Rinplane = 2 × 5 and p.f. = 5/8 to reduce the TE to 65ms and achieve an effective-echo-spacing of 0.256 ms. With the lengthened EPI readout and echo-spacing at this very high spatial resolution, the amount of phase accrual from B0 inhomogeneity across the readout will increase. The phase accrual from B0 inhomogeneity as a function of ky is of opposing signs for the blip-up and blip-down acquisitions. This acts to reduce the orthogonality/complementarity of the interleaved k-space encodings of the two BUDA shots which sample complementary ky data points. The increase in such phase accrual differences at high spatial resolution will therefore negatively impact the conditioning of BUDA reconstruction. To mitigate this issue, dynamic B0 shimming with a custom-build 32-channel integrated ΔB0/Rx array (AC/DC coil) (54) was used to reduce the B0 inhomogeneity and hence the level of distortion differences between the blip-up and-down data. Our previous study (45) showed that the use of dynamic B0 shimming method (55-57) with the AC/DC coil (54) could significantly mitigate the local field inhomogeneity by more than 50%, which when combined with advanced parallel imaging can enable dMRI acquisition with ss-EPI at ~8x distortion mitigation compared to the non-accelerated ss-EPI with standard 2nd order shim. This includes a factor of roughly 2x distortion reduction from improved B0 homogeneity achieved by the AC/DC coil compared to baseline global shimming with the scanner’s built-in 1st-2nd order spherical harmonic shims. This should improve the conditioning of BUDA reconstruction at 600 μm resolution.
With gSlider SMSb acquisition, dynamic shimming was performed across simultaneously acquired slabs (45). Two external triggers are added astride each EPI acquisition to enable slab-by-slab B0 shimming with the 32-channel AC/DC coil. To avoid poor performance in whole-brain fat suppression, the slab-by-slab shimming was turned off during fat saturation. To assess the performance gain from dynamic shimming, g-factor noise was calculated for acquisitions with and without dynamic shimming.
To optimize the performance of dynamic shimming for BUDA-EPI reconstruction, the shim target region is adjusted. The original shim optimization only targets brain tissue regions, which provides good shim performance in brain tissue areas at the cost of large B0 inhomogeneity in the skull areas. With parallel imaging acceleration, the FOV-folding/aliasing will cause the skull areas to fold into the brain tissue regions. As such, in the BUDA reconstruction there is a coupling of the problematic skull areas with large B0 inhomogeneity to the brain tissue areas, resulting in an increased in g-factor penalty in the brain regions. To avoid this issue, we expand the target shim optimization area to include the front and the back of the skulls that can alias into the brain, to ensure good shim in these regions as well.
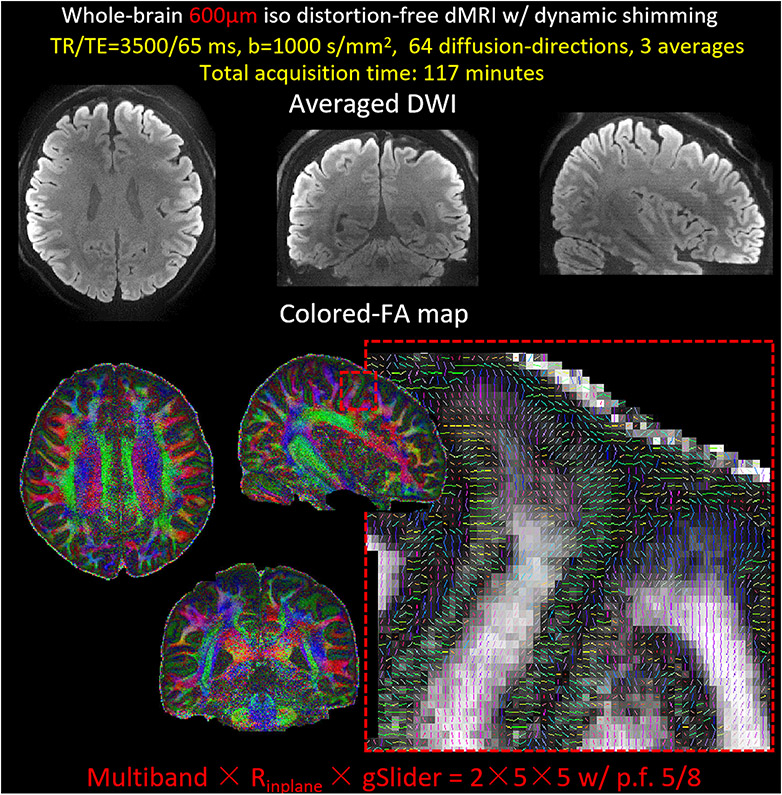
Using gSlider BUDA-EPI with dynamic shimming on the 32 channel AC/DC coil, whole-brain 600μm data were acquired with: FOV: 220 × 220 × 126mm3 (42 slab, 3.0mm slab thickness), image matrix size: 360 × 360, TR/TE = 3500/65ms, MB × Rinplane = 2 × 5 per shot with CAIPI-shift of 2 and p.f. = 5/8. Sixty-four diffusion-directions with b =1000 s/mm2 along with interspersed eight b=0 volumes, three averages. The total acquisition time is 117 minutes.
All in-vivo measurements were performed on a 3T commercial scanner (MAGNETOM Prisma, Siemens Healthineers, Erlangen, Germany). All image reconstructions were performed in MATLAB (MathWorks, Natick, MA).
Results
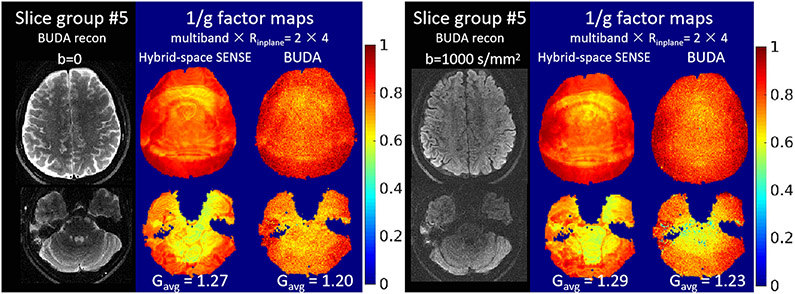
Figure 3 shows the g-factor results of the hybrid-space SENSE and the BUDA reconstructions for cases with no shot-to-shot phase variation (b=0) and with shot-to-shot phase variations (b=1000 s/mm2) at MB × Rinplane = 2 × 4 accelerations. For the b=0 data, where there is minimal shot-to-shot phase variation, the averaged g-factors of BUDA vs hybrid-space SENSE are 1.20 vs 1.27, which demonstrate a small but noticable improvement in SNR from the structured low-rank constraint. For the diffusion-weighted data, the shot-to-shot phase variation reduces the orthogonality of the blip-up and blip-down EPI encodings that samples complementary k-space locations, which will lead to an increase in g-factor. In this case, both the BUDA and the hybrid-space SENSE methods have increased g-factor penalty when compared to the b=0 case, but with the BUDA method (Gavg =1.23) still retaining a lower g-factor penalty when compared to the hybrid-space SENSE method (Gavg =1.29). Since the physiological noise induced shot-to-shot phase-difference varies across different shots, we also validate the performance of hybrid-space SENSE and BUDA reconstrucitons with different shot-to-shot phase variations. The 1/g-factor maps of three different shot-to-shot cases across different diffusion-directions are shown in Supporting Information Figure S1. From the figure it can be seen that the g-factor maps vary across different shot-to-shot phase variations. From the figure it can be seen that the g-factor maps vary across different shot-to-shot phase variations. Nonetheless, in all cases, the proposed BUDA method shows reduced g-factor penalty compared to the hybrid-space SENSE method.
Figure 3.
The reconstruction results and 1/g-factor maps of the b=0 and 1000 s/mm2 EPI data, with MB × Rinplane = 2 × 4 accelerations.
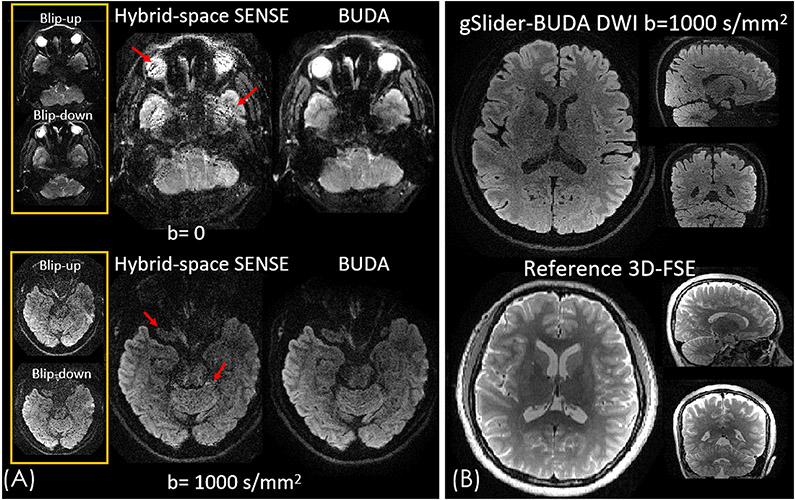
Figure 4 (A) shows the comparison between the proposed BUDA and hybrid-space SENSE reconstructions for an acquisition at MB × Rinplane = 2 × 4. Both hybrid-space SENSE and proposed BUDA incorporate field maps into the forward model of the joint reconstruction to correct the geometric distortion. However, as the red arrows highlighted in Figure 4(A), the residual artifacts in the hybrid-SENSE reconstruction were eliminated in BUDA reconstruction, demonstrating that the proposed BUDA has better reconstruction performance with reduced residual artifacts compared to the hybrid-space SENSE method at high acceleration factors. Figure 4(B) shows the single-average single diffusion-direction DWI reconstructed from 35 seconds of gSlider BUDA-EPI acquisition, showing the high SNR of the proposed method at 1mm isotropic resolution. Compared to the reference T2-weighted 3D-FSE images, the whole-brain diffusion volume reconstructed by the proposed method yielded images in three orthogonal views, which closely matched those of the reference images.
Figure 4.
(A) Comparison between hybrid-space SENSE and BUDA reconstructions for an acquisition at MB × Rinplane = 2 × 4 (B) The diffusion weighted volume reconstructed by gSlider-BUDA method. The diffusion weighted volume retains high geometric fidelity that closely resembled that of the reference 3D-FSE images.
Figure 5 shows the high-fidelity whole-brain diffusion-weighted images (DWI), colored-FA maps and averaged DWIs in three orthogonal views from gSlider BUDA-EPI at 860μm isotropic resolution. As can be seen from the averaged DWI volume, the reconstructed volumetric results retain high geometric fidelity with the reference MPRAGE images.
Figure 5.
Three orthogonal views of i) whole-brain single-direction diffusion-weighted images (DWI), ii) colored-FA maps and iii) 64-diffusion-direction averaged DWI from gSlider BUDA-EPI at 860μm isotropic resolution. As can be seen from the averaged DWI volume, the diffusion data retains high geometric fidelity with the reference MPRAGE.
Figure 6 shows the radiality maps and the ODF acquired from the two-shell whole brain 780μm gSlider BUDA-EPI data. Figure 6(A) shows the radiality maps across the different cortical depth on the inflated brain surface. In general, consistent with previous studies (50,51), radiality is low at the white-gray boundary (WGB), higher at the middle cortical depths and lower at the pial surface. Low radiality can be observed in the somatosensory cortex (S1) (indicated by blue arrow) at all cortical depths which points to the S1 area consisting mostly of tangential fibers. Figure 6(B) shows the zoom-in views of the ODF in S1 and primary motor cortex (M1). We found a primarily radial orientation in M1 and tangential fibers to the local cortical surface orientation in S1 as expected.
Figure 6.
(A) Radiality maps at the surface of the white-gray boundary (WGB) surface, the surfaces of 20%, 40%, 60% and 80% of cortical thickness into gray matter, and the pial surface on the inflated brain surface. (B) ODF results in primary motor cortex (M1) and somatosensory cortex (S1) obtained from the whole brain 780μm dMRI data with two shells.
Figure 7(A) shows the 1/g-factor map comparisons of a representative slice-group of single-shot EPI, BUDA without and with dynamic shim, and with a perfect shim (no B0 inhomogeneity) at 600um isotropic resolution with MB × Rinplane = 2 × 5. It can be seen that the distortion differences between blip-up and blip-down shots are much mitigated using dynamic shimming (Figure 7(B)). Compared to the averaged g-factor of single-shot EPI (Gavg =2.67), the joint reconstruction of complementary k-space encoding from the two BUDA acquisition shots significantly reduces g-factor (Gavg =1.49). However, high g-factor persist in areas of large B0 inhomogeneity, where phase accrual from B0 inhomogeneity disturbs the orthogonality of the k-space encodings in the two BUDA shots. The g-factor result for BUDA with perfect shim (no B0 inhomogeneity) on the far-right of the figure is an idealized case, which represents the upper bound of the BUDA reconstruction performance (effectively MB × Rinplane = 2 × 2.5, Gavg =1.20). With optimized dynamic shimming, B0-variation is reduced by >50% as shown in the B0 maps (standard deviation of B0-variation is 22.1 Hz with dynamic shimming vs. 51.2 Hz without dynamic shimming in this slice-group), which results in 32% g-factor improvement (Figure 7(C)), approaching performance of the perfect shim case (Gavg =1.26 for BUDA with dynamic shim vs. Gavg =1.20 for BUDA with perfect shim). While one representative slice-pair is shown here, similar g-factor gains are achieved over the whole brain (The whole brain Gavg = 1.18 with dynamic shim vs. Gavg = 1.41 without dynamic shim).
Figure 7.
(A) 1/g-factor maps of single-shot EPI, BUDA-EPI without and with dynamic shim, and with perfect shim (no B0 inhomogeneity); all at MB × Rinplane= 2 × 5. (B) Using the 32-channel AC/DC coil with slab-by-slab dynamic shimming, the distortion difference between blip-up and blip-down shots are mitigated and hence (C) the average g-factor of BUDA reconstruction improves by 32% compared to no dynamic shimming.
Figure 8 shows the averaged DWIs from 64 diffusion-encoding directions and colored-FA maps of the 600 μm isotropic data acquired using gSlider BUDA-EPI with dynamic shimming. The zoomed-in figure shows a sagittal view of colored-FA map, displaying the high-resolution details from this dataset.
Figure 8.
The averaged diffusion-weighted images and the colored-FA maps from 64-diffusion-direction whole-brain 600μm gSlider BUDA-EPI with dynamic shimming. The zoom-in sagittal view of a slice of colored FA maps, displaying the high-resolution capability of the 600μm data.
Discussion
In this work, a highly efficient multi-shot EPI acquisition/reconstruction framework was proposed for distortion-free, submillimeter-isotropic-resolution dMRI. With interleaved blip up-down acquisition, the field maps were estimated and then incorporated into the forward model of joint parallel imaging reconstruction with Hankel structured low-rank constraint to obtain distortion-free images. Combining two reverse-polarity shots during parallel imaging and incorporating a field map into the forward model removes distortion, resolves voxel pile ups and enables high acceleration factors per shot with good joint reconstruction performance. Reverse polarity acquisition in BUDA offers a “smart partial Fourier” approach where the missing portion of k-space is also flipped, so that the two-shots provide complementary information and help prevent loss of resolution.
Previous studies such as hybrid-space SENSE (37,38) performs joint reconstruction of the 2-shots by including their phase difference and field map into the forward model to correct both shot-to-shot phase variations and geometric distortion. Such joint reconstruction was shown to significantly reduce the g-factor noise penalty when compared to reconstructions on the blip-up and blip-down acquisitions separately. Nonetheless, at high parallel imaging accelerations, navigation-free reconstructions with this approach can result in image artifacts and noise amplification due to the poor SNR of diffusion-weighted images and the inaccurate estimation of the shot-to-shot phase difference. Rather than stitching the shots together in k-space, in the BUDA method the blip-up and down shots are constrained to be similar using implicit structured low-rank constraint (18). The combination of k-space data across the two shots through such a low-rank constraint obviates the need for an explicit estimation of the shot-to-shot phase variations while also provides the g-factor reduction benefit of a joint reconstruction as demonstrated via the pseudo multiple replica g-factor calculation. We note that the same structured low-rank constraint has also been utilized in the MUSSELS method (17) to reconstruct interleaved EPI with good reconstruction performance.
The BUDA method is combined with gSlider simultaneous multislab acquisition to boost SNR and achieve distortion-free dMRI with high-SNR efficiency. There are some limitations in the proposed gSlider BUDA-EPI framework, which provide opportunities for future research. In particular, motion artifacts pose a challenge in dMRI, as acquisitions are typically long and span a large number of diffusion directions. For BUDA-EPI, motion corruptions between the blip-up and –down shots can result in poor B0 map estimation and image artifacts in the joint reconstruction. In our dMRI BUDA-EPI, the blip–up and –down shots for each diffusion direction are acquired in consecutive TRs to minimize the time between these acquisitions. Nonetheless, motion can still occur between these acquisitions. Preliminary development of a motion-robust reconstruction for BUDA-EPI has already been pursued (58), which uses the initial SENSE reconstructed images to estimate motion parameters and updated field-maps, which are incorporated into to the forward model of the joint reconstruction. This approach can also be combined with the motion-corrected gSlider (mc-gSlider) reconstruction (44) to provide motion-robustness across RF encodings.
Compared to the conventional ms-EPI which reduces image distortion, BUDA corrects for the image distortions completely but causes additional noise penalty in the reconstruction at high B0 inhomogeneity areas. This noise penalty increases with resolution as the echo-spacing and the EPI readout duration increase to cause larger phase accumulation due to B0 inhomogeneity. To mitigate this issue, in this work, slab-by-slab dynamic shimming was used to reduce the B0 inhomogeneity level, which was shown to improve the conditioning of reconstruction. To further improve the conditioning of reconstructions, adding saturation bands to saturate out the front and back part of the skull regions could also be used to get rid of the large B0 inhomogeniety issue in the problematic skull regions. This could be used together with the optimized shim procedure so that it would allow us to retain most of the shim performance when compared to the brain-only dynamic shimming. Future work will explore the use of ultra-high performance gradients such as in (59,60) to reduce the echo-spacing, as well as the use of higher channel count AC/DC shim array for further B0 inhomogeneity mitigation. These improvements should further reduce the noise penalty and allow for higher per shot acceleration still that would be necessary to limit T2* blurring as we push for higher spatial resolution. Note also that the B0 mitigation performance of the AC/DC coil array will degrade at higher multi-band factors as it is more difficult to achieve high quality shimming simultaneously across a larger number of slices (55). A higher channel count AC/DC coil array will also mitigate this issue. Nonetheless, with the gSlider-BUDA acquisition, the desired TR of 3.5s can already be achieved with MB 2 for our 600 μm acquisition. This TR of 3.5s was used to provide a good balance in the trade-off between SNR efficiency vs. spin-history and motion sensitivity issues(45). Therefore, we don’t need higher MB factors to further reduce TR. Moreover, the use of more shots/segmentations in the BUDA acquisition could offer an alternative or be used synergistically with ultra-high-performance gradient and shim array to achieve even higher per shot acceleration. This represents an interesting avenue to explore in future work but would also come at a cost of increased scan time and motion sensitivity, as well as more difficult joint reconstruction.
As we push forward with our encoding capability to achieve higher and higher isotropic resolution dMRI, significant decrease in voxel size will lead to a proportionate decrease in SNR. To achieve high SNR-efficiency, gSlider simultaneous multislab acquisition was employed in this work, but SNR remains a challenge for 600μm whole-brain dMRI. To reduce noise, incorporation of recent diffusion denoising methods (61,62) can be used to enhance SNR, while also provide the capability for faster acquisition through random sub-sampling of RF-encodings in the gSlider acquisition (61).
Conclusion
In this work, we developed gSlider BUDA-EPI with dynamic B0 shimming to achieve distortion-free submillimeter-isotropic-resolution dMRI in vivo. Such approach should aid in the push towards in vivo dMRI at the mesoscale to study fine-scale structures of the brain.
Supplementary Material
Supporting Information Figure S1. The 1/g-factor maps of hybrid-space SENSE and BUDA reconstructions with three diffusion-directions. The physiological noise induced shot-to-shot phase-difference varies across three diffusion-directions.
Acknowledgement
This work was supported in part by NIH research grants: R01EB020613, R01MH116173, R01EB016695, U01EB025162, U01MH093765, R01EB028797, U24EB028984, R00EB021349 and U01EB026996.
References
- 1.Leuze CWU, Anwander A, Bazin PL, et al. Layer-specific intracortical connectivity revealed with diffusion MRI. Cereb. Cortex 2014;24:328–339 doi: 10.1093/cercor/bhs311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assaf Y Imaging laminar structures in the gray matter with diffusion MRI. Neuroimage 2019;197:677–688 doi: 10.1016/j.neuroimage.2017.12.096. [DOI] [PubMed] [Google Scholar]
- 3.Ganepola T, Nagy Z, Ghosh A, Papadopoulo T, Alexander DC, Sereno MI. Using diffusion MRI to discriminate areas of cortical grey matter. Neuroimage 2018;182:456–468 doi: 10.1016/j.neuroimage.2017.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song AW, Chang HC, Petty C, Guidon A, Chen NK. Improved delineation of short cortical association fibers and gray/white matter boundary using whole-brain three-dimensional diffusion tensor imaging at submillimeter spatial resolution. Brain Connect. 2014;4:636–640 doi: 10.1089/brain.2014.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianciardi M, Toschi N, Edlow BL, et al. Toward an In Vivo Neuroimaging Template of Human Brainstem Nuclei of the Ascending Arousal, Autonomic, and Motor Systems. Brain Connect. 2015;5:597–607 doi: 10.1089/brain.2015.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl. Acad. Sci. U. S. A 2000;97:11050–11055 doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen N kuei, Guidon A, Chang HC, Song AW. A robust multi-shot scan strategy for high-resolution diffusion weighted MRI enabled by multiplexed sensitivity-encoding (MUSE). Neuroimage 2013;72:41–47 doi: 10.1016/j.neuroimage.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holdsworth SJ, Skare S, Newbould RD, Guzmann R, Blevins NH, Bammer R. Readout-segmented EPI for rapid high resolution diffusion imaging at 3T. Eur. J. Radiol 2008;65:36–46 doi: 10.1016/j.ejrad.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong HK, Gore JC, Anderson AW. High-resolution human diffusion tensor imaging using 2-D navigated multishot SENSE EPI at 7 T. Magn. Reson. Med 2013;69:793–802 doi: 10.1002/mrm.24320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu ML, Chang HC, Chung HW, Truong TK, Bashir MR, Chen NK. POCS-based reconstruction of multiplexed sensitivity encoded MRI (POCSMUSE): A general algorithm for reducing motion-related artifacts. Magn. Reson. Med 2015;74:1336–1348 doi: 10.1002/mrm.25527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z, Ma X, Truong TK, Song AW, Guo H. Phase-updated regularized SENSE for navigator-free multishot diffusion imaging. Magn. Reson. Med 2017;78:172–181 doi: 10.1002/mrm.26361. [DOI] [PubMed] [Google Scholar]
- 12.Liao C, Manhard MK, Bilgic B, et al. Phase-matched virtual coil reconstruction for highly accelerated diffusion echo-planar imaging. Neuroimage 2019;194:291–302 doi: 10.1016/j.neuroimage.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Liao C, Chen Y, Cao X, et al. Efficient parallel reconstruction for high resolution multishot spiral diffusion data with low rank constraint. Magn. Reson. Med 2017;77:1359–1366 doi: 10.1002/mrm.26199. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y, Levine EG, Tian Q, et al. Motion-robust reconstruction of multishot diffusion-weighted images without phase estimation through locally low-rank regularization. Magn. Reson. Med 2018. doi: 10.1002/mrm.27488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bilgic B, Chatnuntawech I, Manhard MK, et al. Highly accelerated multishot echo planar imaging through synergistic machine learning and joint reconstruction. Magn. Reson. Med 2019:mrm.27813 doi: 10.1002/mrm.27813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal HK, Mani MP, Jacob M. Modl-mussels: Model-based deep learning for multishot sensitivity-encoded diffusion mri. IEEE Trans. Med. Imaging 2020;39:1268–1277 doi: 10.1109/TMI.2019.2946501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mani M, Jacob M, Kelley D, Magnotta V. Multi-shot sensitivity-encoded diffusion data recovery using structured low-rank matrix completion (MUSSELS). Magn. Reson. Med 2017;78:494–507 doi: 10.1002/mrm.26382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haldar JP. Low-Rank Modeling of Local k-Space Neighborhoods (LORAKS) for Constrained MRI. IEEE Trans. Med. Imaging 2014;33:668–681 doi: 10.1109/TMI.2013.2293974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bilgic B, Kim TH, Liao C, et al. Improving parallel imaging by jointly reconstructing multi-contrast data. Magn. Reson. Med 2018;80:619–632 doi: 10.1002/mrm.27076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao C, Cao X, Cho J, Zhang Z, Setsompop K, Bilgic B. Highly efficient MRI through multi-shot echo planar imaging. In: Lu YM, Papadakis M, Van De Ville D, editors. Wavelets and Sparsity XVIII. Vol. 11138. SPIE; 2019. p. 43. doi: 10.1117/12.2527183. [DOI] [Google Scholar]
- 21.Setsompop K, Cohen-Adad J, Gagoski BA, et al. Improving diffusion MRI using simultaneous multi-slice echo planar imaging. Neuroimage 2012;63:569–580 doi: 10.1016/j.neuroimage.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn. Reson. Med 2012;67:1210–1224 doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moeller S, Yacoub E, Olman CA, et al. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain FMRI. Magn. Reson. Med 2010;63:1144–1153 doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feinberg DA, Moeller S, Smith SM, et al. Multiplexed Echo Planar Imaging for Sub-Second Whole Brain FMRI and Fast Diffusion Imaging Valdes-Sosa PA, editor. PLoS One 2010;5:e15710 doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engström M, Skare S. Diffusion-weighted 3D multislab echo planar imaging for high signal-to-noise ratio efficiency and isotropic image resolution. Magn. Reson. Med 2013;70:1507–1514 doi: 10.1002/mrm.24594. [DOI] [PubMed] [Google Scholar]
- 26.Wu W, Poser BA, Douaud G, et al. High-resolution diffusion MRI at 7T using a three-dimensional multi-slab acquisition. Neuroimage 2016;143:1–14 doi: 10.1016/J.NEUROIMAGE.2016.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang H-C, Sundman M, Petit L, et al. Human brain diffusion tensor imaging at submillimeter isotropic resolution on a 3Tesla clinical MRI scanner. Neuroimage 2015;118:667–75 doi: 10.1016/j.neuroimage.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruce IP, Chang H-C, Petty C, Chen N-K, Song AW. 3D-MB-MUSE: A robust 3D multi-slab, multi-band and multi-shot reconstruction approach for ultrahigh resolution diffusion MRI. Neuroimage 2017;159:46–56 doi: 10.1016/J.NEUROIMAGE.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang HC, Hui ES, Chiu PW, Liu X, Chen NK. Phase correction for three-dimensional (3D) diffusion-weighted interleaved EPI using 3D multiplexed sensitivity encoding and reconstruction (3D-MUSER). Magn. Reson. Med 2018;79:2702–2712 doi: 10.1002/mrm.26944. [DOI] [PubMed] [Google Scholar]
- 30.Moeller S, Ramanna S, Lenglet C, et al. Self-navigation for 3D multishot EPI with data-reference. Magn. Reson. Med 2020:mrm.28231 doi: 10.1002/mrm.28231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van AT, Aksoy M, Holdsworth SJ, Kopeinigg D, Vos SB, Bammer R. Slab profile encoding (PEN) for minimizing slab boundary artifact in three-dimensional diffusion-weighted multislab acquisition. Magn. Reson. Med 2015;73:605–613 doi: 10.1002/mrm.25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu W, Koopmans PJ, Frost R, Miller KL. Reducing slab boundary artifacts in three-dimensional multislab diffusion MRI using nonlinear inversion for slab profile encoding (NPEN). Magn. Reson. Med 2016;76:1183–1195 doi: 10.1002/mrm.26027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Setsompop K, Fan Q, Stockmann J, et al. High-resolution in vivo diffusion imaging of the human brain with generalized slice dithered enhanced resolution: Simultaneous multislice (gSlider-SMS). Magn. Reson. Med 2018;79:141–151 doi: 10.1002/mrm.26653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. Fsl. Neuroimage 2012;62:782–790 doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Andersson JLR, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: Application to diffusion tensor imaging. Neuroimage 2003;20:870–888 doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 36.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. In: NeuroImage. Vol. 23. Academic Press; 2004. pp. S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 37.Zhu K, Dougherty RF, Wu H, et al. Hybrid-Space SENSE Reconstruction for Simultaneous Multi-Slice MRI. IEEE Trans. Med. Imaging 2016;35:1824–1836 doi: 10.1109/TMI.2016.2531635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahneisen B, Aksoy M, Maclaren J, Wuerslin C, Bammer R. Extended hybrid-space SENSE for EPI: Off-resonance and eddy current corrected joint interleaved blip-up/down reconstruction. Neuroimage 2017;153:97–108 doi: 10.1016/J.NEUROIMAGE.2017.03.052. [DOI] [PubMed] [Google Scholar]
- 39.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magn. Reson. Med 1999;42:952–962 doi: . [DOI] [PubMed] [Google Scholar]
- 40.Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. In: NeuroImage. Vol. 23. ; 2004. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 41.Uecker M, Lai P, Murphy MJ, et al. ESPIRiT - An eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magn. Reson. Med 2014;71:990–1001 doi: 10.1002/mrm.24751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holdsworth SJ, Aksoy M, Newbould RD, et al. Diffusion tensor imaging (DTI) with retrospective motion correction for large-scale pediatric imaging. J. Magn. Reson. Imaging 2012;36:961–971 doi: 10.1002/jmri.23710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eichner C, Cauley SF, Cohen-Adad J, et al. Real diffusion-weighted MRI enabling true signal averaging and increased diffusion contrast. Neuroimage 2015;122:373–384 doi: 10.1016/j.neuroimage.2015.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang F, Bilgic B, Dong Z, et al. Motion-robust sub-millimeter isotropic diffusion imaging through motion corrected generalized slice dithered enhanced resolution (MC-gSlider) acquisition. Magn. Reson. Med 2018;80:1891–1906 doi: 10.1002/mrm.27196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao C, Stockmann J, Tian Q, et al. High-fidelity, high-isotropic-resolution diffusion imaging through gSlider acquisition with B1+ and T1 corrections and integrated ΔB0/Rx shim array. Magn. Reson. Med 2020;83:56–67 doi: 10.1002/mrm.27899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung S, Kim D, Breton E, Axel L. Rapid B1+ mapping using a preconditioning RF pulse with TurboFLASH readout. Magn. Reson. Med. 2010;64:n/a–n/a doi: 10.1002/mrm.22423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive quantification of signal-to-noise ratio and g-factor for image-based and k-space-based parallel imaging reconstructions. Magn. Reson. Med 2008;60:895–907 doi: 10.1002/mrm.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischl B FreeSurfer. Neuroimage 2012;62:774–781 doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. Neuroimage 2009;48:63–72 doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McNab JA, Polimeni JR, Wang R, et al. Surface based analysis of diffusion orientation for identifying architectonic domains in the in vivo human cortex. Neuroimage 2013;69:87–100 doi: 10.1016/j.neuroimage.2012.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan Q, Nummenmaa A, Polimeni JR, et al. HIgh b-value and high Resolution Integrated Diffusion (HIBRID) imaging. Neuroimage 2017;150:162–176 doi: 10.1016/j.neuroimage.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tournier JD, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol 2012;22:53–66 doi: 10.1002/ima.22005. [DOI] [Google Scholar]
- 53.Jeurissen B, Tournier JD, Dhollander T, Connelly A, Sijbers J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 2014;103:411–426 doi: 10.1016/j.neuroimage.2014.07.061. [DOI] [PubMed] [Google Scholar]
- 54.Stockmann JP, Witzel T, Keil B, et al. A 32-channel combined RF and B0shim array for 3T brain imaging. Magn. Reson. Med 2016;75:441–451 doi: 10.1002/mrm.25587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stockmann JP, Wald LL. In vivo B0 field shimming methods for MRI at 7 T. Neuroimage 2018;168:71–87 doi: 10.1016/J.NEUROIMAGE.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Juchem C, Nixon TW, McIntyre S, Boer VO, Rothman DL, de Graaf RA. Dynamic multi-coil shimming of the human brain at 7 T. J. Magn. Reson 2011;212:280–288 doi: 10.1016/J.JMR.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou J, Stockmann JP, Arango N, et al. An orthogonal shim coil for 3T brain imaging. Magn. Reson. Med 2020;83:1499–1511 doi: 10.1002/mrm.28010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao X, Liao C, Zhang Z, et al. MOCO-BUDA: motion-corrected blip-up/down acquisition with joint reconstruction for motion-robust and distortion-free diffusion MRI of brain. In: ISMRM 28th Annual Meeting. Sydney, Austrialia; 2020. p. 0465. [Google Scholar]
- 59.Foo TKF, Laskaris E, Vermilyea M, et al. Lightweight, compact, and high-performance 3T MR system for imaging the brain and extremities. Magn. Reson. Med 2018;80:2232–2245 doi: 10.1002/mrm.27175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilm BJ, Hennel F, Roesler MB, Weiger M, Pruessmann KP. Minimizing the echo time in diffusion imaging using spiral readouts and a head gradient system. Magn. Reson. Med 2020:mrm.28346 doi: 10.1002/mrm.28346. [DOI] [PubMed] [Google Scholar]
- 61.Ramos-Llordén G, Ning L, Liao C, et al. High-fidelity, accelerated whole-brain submillimeter in vivo diffusion MRI using gSlider-spherical ridgelets (gSlider-SR). Magn. Reson. Med 2020:mrm.28232 doi: 10.1002/mrm.28232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haldar JP, Liu Y, Liao C, Fan Q, Setsompop K. Fast submillimeter diffusion MRI using gSlider-SMS and SNR-enhancing joint reconstruction. Magn. Reson. Med 2020;84:762–776 doi: 10.1002/mrm.28172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1. The 1/g-factor maps of hybrid-space SENSE and BUDA reconstructions with three diffusion-directions. The physiological noise induced shot-to-shot phase-difference varies across three diffusion-directions.