Abstract
For ~30 years, two distinct groups of clinical isolates of Klebsiella pneumoniae have been recognized. Classical strains (cKp) are typically isolated from patients with some degree of immunocompromise and are not virulent in mouse models of infection whereas hypervirulent strains (hvKp) are associated with community acquired invasive infections and are highly virulent in mouse models of infection. Hyperproduction of capsule and a hypermucoviscous colony phenotype have been strongly associated with the hypervirulence of hvKp strains. Recent studies have begun to elucidate the relationship between capsule gene expression, hypermucoviscosity and hypervirulence. Additionally, genes associated with hyperproduction of capsule and hypermucoviscosity in hvKp strains have been identified in a few cKp isolates. However, it is not clear how the acquisition of these genes impacts the virulence of cKp isolates. A better understanding of the potential risks of these strains is particularly important given that many of them are resistant to multiple antibiotics, including carbapenems.
Introduction
For more than a century, Klebsiella pneumoniae has been recognized as a human pathogen responsible for a variety of different infections, yet bacterial factors contributing to these different disease presentations were not clear. However, two recent advances in the field have provided a path forward to formulate and focus questions for future mechanistic studies. The first is the differentiation of two groups ofstrains currently of clinical importance.Thesecondisthe application of wholegenomesequencing and bioinformatic analysis of large groups of K. pneumoniae strains. Several excellent reviews of K. pneumoniae diseases, epidemiology, virulence factors and pathogenesis have been published recently [1••,2•,3•], thus we will not go into the details of these aspects of K. pneumoniae biology here and instead will focus on the relationship between capsule, hypermucoviscosity and hypervirulence.
For the past 30 years, two different groups of K. pneumoniae of particular concern have circulated — hypervirulent (hvKp) strains that are sensitive to carbapenem and classical strains (cKp) that frequently are carbapenem resistant (CR) [1••,3•,4]. The cKp strains are typically associated with nosocomial infections or infection in a long-term care setting, suggesting some degree of immunocompromise is necessary for these strains to cause disease. For these CR-cKp strains, few treatment options are available and, although colistin has been used, strains that are also resistant to colistin are emerging [5]. Bioinformatic analyses of large strain sets have been instrumental in helping to define genes linked to hypervirulence (hv-associated genes); these include rmpA, rmpA2, iroBCDN, iutA, iucABCD and ybt genes [6,7,8•,9,10••,11]. These hv-associated genes are often encoded on a large, highly conserved virulence plasmid such as pLVPK, but some strains have the hv-associated genes encoded on the chromosome as part of an ICE element [12–14]. The hv-associated genes listed above encode three different siderophore systems: salmochelin (iroBCDN), aerobactin (iutA, iucABCD) and yersiniabactin (ybt genes). cKp strains produce only enterobactin and occasionally yersiniabactin. However, production of multiple siderophores is strongly associated with hypervirulence and studies in mice have demonstrated a contribution of these siderophores to pathogenesis [14–20]. rmpA (regulator of mucoidy protein A) and rmpA2 encode transcriptional regulators that seem to play similar roles enhancing production of capsule [7,8•,12,21–23] (see section on Rmp regulators for further discussion of rmpA/A2).
Strains of K. pneumoniae that are hypermucoviscous (HMV) were associated early on with hypervirulence; mutations that result in loss of this HMV phenotype also result in a reduction in virulence [8•,21,24•,25••,26,27]. The assay most often used to determine the HMV phenotype is a positive string test, where a loop touched to a colony will pull a ‘string’ of >5 mm (Box 1). However, the string test is not quantitative and can be influenced by culture conditions, age of the colony and user technique. The threshold to define a positive string test (5 mm) is quite low and known hvKp strains can yield ‘strings’ of >40 mm. In addition, a recent study aimed at identifying biomarkers that distinguish hvKp from cKp found that the string test was a less accurate predictor of hypervirulence than several genetic markers [28]. A more quantitative test sometimes used to assess the HMV phenotype is the sedimentation assay (Box 1). This assay has the potential to distinguish not only HMV from non-HMV positive strains, but also the potential to define HMV intermediate strains.
Box 1. Assays for capsule production and capsule-dependent virulence phenotypes.
Uronic acid (UA) content.
UA is a component of many capsules often in the form of glucuronic acid or galactouronic acid. It is most frequently determined by a colorometric assay.
Capsule (cps) gene expression.
Frequently monitored using lacZ or gfp transcriptional reporters to the three defined promoters in the cps locus (Figure 1). qRT-PCR is also used and is gaining popularity.
Mucoidy. A quick test for mucoidy is the string test, where a loop or stick is used to touch a colony on a plate and, as it is lifted away, a string connects the stick and the colony. To be considered hypermucoviscous (HMV), the string must reach at least 5 mm, however these strings can stretch for several cm! Another test is the sedimentation assay. HMV strains do not pellet well upon centrifugation. This is a visually satisfying assay as the resulting supernatant remains turbid, whereas the non-mucoid strains form tight pellets with cleared supernatants. This can be a somewhat quantifiable assay if the OD600 of the supernatant is measured and compared to the OD600 of the starting culture.
Serum resistance.
Capsule confers protection against complement-mediated killing. Viable bacteria are determined following exposure and compared to the input to assess the ability of a strain to resist killing. This assay is a convenient way to assess capsule-dependent virulence attributes in vitro.
Tissue culture models.
Capsule blocks adherence to cells in culture and prevents phagocytosis. Tissue culture assays are used to measure adherent and intracellular bacteria and can be performed with transformed cell lines or primary cells. Like the serum resistance assay, these assays are a relatively fast and inexpensive ways to assess capsule-dependent virulence properties in vitro.
Animal models.
There are several mouse models of Klebsiella infection used to study capsule mutants, including sepsis (intraperitoneal injection), pneumonia (intranasal or intratracheal inoculation), and gastrointestinal colonization (oral gavage). These models have been widely applied to assess mouse survival (single dose over several days), LD50 (multiple doses over several days), or bacterial burden (CFU/organ).
All K. pneumoniae strains produce an extracellular polysaccharide capsule required for virulence [1••] and more than 130 different capsule types have been identified in Klebsiella [11]. The hvKp strains tend to produce more capsule (as defined using a uronic acid [UA] assay [Box 1]) than cKp strains and early studies associated this hypercapsule production with the hv-associated gene rmpA (or the closely related gene rmpA2) [7,8•,12,21,22]. HvKp with mutations in rmpA, or other genes that affect capsule production in hvKp, typically lose the HMV phenotype and show a strong reduction in virulence when tested in mice [8•,23,24•]. Thus, the HMV phenotype became strongly associated with the amount of capsule production despite an early report suggesting HMV does not require excess capsule production [8•]. The recent report of a mutation in a gene encoding a transcriptional regulator (RmpC) that reduces expression of capsule (cps) genes and production of capsule, but does not affect the HMV phenotype, provides the clearest indication to date that the HMV phenotype does not require hyperproduction of capsule [25••]. However, mutations that prevent production of capsule also result in loss of HMV [21,24•,25••], underscoring a link between capsule and HMV.
In mouse models of infection (Box 1), hvKp strains are virulent even at low inoculation doses [24•,29–31] whereas cKp strains are not [32,33]. As mentioned above, hvKp strains typically have not been CR and CR-cKp have not been virulent in an immunocompetent host. However, in recent years CR-hvKp and CR-cKp that have acquired genes associated with hypervirulence (here designated as hv-CR-cKp) have been reported [34–40,41••,42••,43•]. In nearly all instances where tested, these isolates were considered to be HMV by the string test. To date, the majority of CR K. pneumoniae with hv-associated genes appear to be hvKp that have acquired a plasmid encoding a carbapenemase. Although virulence testing was not done in most of these reports, multi-locus sequence typing to determine strain sequence type (ST) and/or capsule type (K-type) indicated these strains come from ST or K-types strongly associated with hvKp. In reports of hv-CR-cKp, the hv-associated genes were encoded on pLVPK-like virulence plasmids that are typically found in hvKp. In most of these reports the function of the hv-associated genes or the effect of these genes on the virulence of these strains was not tested. However in cases where this was tested, acquisition of hv-associated genes was not always sufficient to confer virulence comparable to that of hvKp strains [41••,43•]. The strong association of HMV with capsule production and the association of HMV and capsule with virulence make understanding the regulation of cps expression and the link between cps expression and HMV important for determining what is required for a CR-cKp strain to become hypervirulent. Here we highlight what is currently known about HMV, cps expression and unanswered questions concerning HMV and virulence.
Strains used in Klebsiella capsule studies
In the field of K. pneumoniae capsule regulation, the majority of the research has been conducted using four hypervirulent strains. NTUH-K2044 [44] and CG43 [45] were both isolated from pyogenic liver abscesses. Strain Kp52145 is considered a reference strain for the K2 capsule type [46] and KPPR1S derivative of ATCC 43816 originated from a pneumonia patient [29,47]. Of these four strains, one (NTUH-K2044) is K1 capsule type while the other three are K2 capsule type, and they each are in different ST groups. CG43, Kp52145 and NTUH-K2044 each carry large virulence plasmids that confer hypervirulence, and NTUH-K2044 and KPPR1S carry a chromosomal ICEKp element that is similar to a portion of the virulence plasmid and encodes genes associated with hypervirulence. Common methods used to characterize capsule-associated phenotypes are described in Box 1.
Global regulators
Rcs phosphorelay
The Rcs phosphorelay is a complex signal transduction system found in many Gram-negative bacteria. It is comprised of RcsC (sensor kinase), RcsD (histidine phosphotransferase), RcsB (response regulator), RcsA (auxiliary protein), and RcsF (outer membrane lipoprotein) [48]. RcsB and RcsC were first identified by their role in colonic acid gene expression in Escherichia coli [49]. Soon thereafter, the Klebsiella homologs of RcsA and RcsB were found to function similarly when expressed in E. coli [50,51], and in K. pneumoniae [23,52]. It is now known that RcsB dimerizes with RcsA and binds a DNA sequence called the RcsAB box to activate expression of capsular gene expression in a number of bacterial organisms [48,53]. The direct regulation through binding of RcsAB to DNA has been shown for the upstream-most promoter of the three characterized capsule operons, located upstream of galF/ORF1 (Figure 1) [53]. In strains CG43 and KPPR1S, rcsB mutants have reduced cps expression, reduced UA levels, and pellet tightly [21,25••]. Similar trends have been reported on cps expression and UA levels in rcs mutants of strain NTUH-K2044 [54]. In addition, the KPPR1S ΔrcsB strain is attenuated in a murine pneumonia model [25••].
Figure 1.

Schematic of the capsule locus in KPPR1S. The three characterized promoters are indicated upstream of galF (also referred to as orf 1–2), wzi (orf 3–15) and manC (orf 16–17). Genes in dark blue are highly conserved among different capsule types; those in light blue are capsule type-specific, and those in dark blue hatches can vary depending on the sugar precursors that comprise the capsule. Orfs 12 and 13 appear to be unique to Klebsiella.
cAMP Receptor Protein (CRP)
CRP has been shown to repress cps expression in CG43 and NTUH-K2044 [55,56]. In CG43, expression from all three cps promoters was increased in the Δcrp strain, and CRP was shown to directly bind to the wzi and manC promoter regions. No CRP binding site was identified upstream of galF and binding of CRP to the galF promoter was not tested. CRP is known to repress rcsA expression, so it was speculated that the increased transcription from the galF promoter in the Δcrp strain was a consequence of increased levels of RcsA. Addition of glucose to the growth media resulted in increased cps expression whereas high cAMP levels (through mutation or direct addition to media) resulted in decreased cps expression. In NTUH-K2044, the Δcrp mutant had increased mucoviscosity and produced more UA than the parental strain. Mice inoculated by intraperitoneal injection with the Δcrp strain had an improved survival rate compared to those inoculated with the parental strain, suggesting this mutant is attenuated for virulence. However, the Δcrp strain has a growth defect and the crp mutation likely affects expression of a large number of genes. As one might predict that increased capsule production would enhance virulence, the attenuation of the Δcrp strain may, at least in part, be a consequence of the reduced growth rate and pleiotropic effects on gene expression.
Impact of iron levels on cps expression via Fur and IscR
The impact of iron homeostasis on cps gene expression has been investigated in strain CG43. Fur is an iron-responsive transcriptional regulator in the Enterobacteriaceae and was identified as playing a role in cps regulation when it was found to bind a region upstream of the rmpA promoter and repress its expression [21]. The Δfur strain was as resistant to sedimentation as the parental strain and produced more UA whereas a strain overproducing Fur formed tight pellets upon centrifugation and produced levels of UA comparable to the parental strain. A subsequent study showed that transcription of rcsA increased in the Δfur strain, which might also contribute to elevated cps gene expression [57]. This study included an analysis of cps regulation and showed elevated levels of expression from all three promoters in the Δfur mutant and demonstrated that the elevated cps expression requires RmpA and RcsA. Thus, the impact of Fur on capsule levels is likely an indirect action mediated via RmpA and RcsA.
IscR is another iron-responsive transcriptional regulator found to control cps expression. IscR was identified as a repressor of genes that encode iron-sulfur cluster assembly proteins, and it binds a [2Fe-2S] cluster [58]. An ΔiscR mutant in the K. pneumoniae strain CG43 had decreased UA levels, and slightly decreased cps expression from the galF and manC promoters [59]. An IscR box was identified upstream of galF and shown to be bound by IscR; this binding was dependent on the DNA sequence of the IscR box, and on the cysteine residues in IscR believed to coordinate the [2Fe-2S] cluster. These investigators further demonstrated that the ΔiscR mutant was more susceptible to killing by normal human serum, a phenotype associated with reduced capsule.
IscR is active when bound by Fe, and therefore activates cps expression when iron levels are sufficient. In contrast, Fur exerts its repressive activity upon cps expression (indirectly) when iron levels are sufficient and relieves this repression when iron levels are low. Thus, it seems that Klebsiella has acquired mechanisms to ensure capsule production remains at least at a basal level independent of the availability of iron, while still allowing for alterations in capsule production in response to extreme iron concentrations.
The Rmp regulators
RmpA and RmpA2
RmpA was identified as a regulator of the mucoidy phenotype in 1989 [8•]. It has a LuxR-type DNA-binding domain, but the N-terminal portion is unique to Klebsiella. The rmpA gene is located either on the chromosome or on a large virulence plasmid and is predominantly found only in strains that possess an HMV phenotype. The correlation between rmpA and HMV/hv is very high, and the presence of rmpA is among a set of genes proposed as biomarkers to identify potential hvKp strains [28,60]. RmpA has been shown to activate cps gene expression in numerous strains [7,21,25••], and to activate its own expression [25••], but no evidence of direct regulation has been published. RmpA was shown to interact with RcsB, and it is postulated that RmpA can substitute for RcsA, leading to activation of capsular genes in the absence of RcsA [21].
RmpA2 was initially found to regulate cps expression in the K. pneumoniae strain Chedid, a K2 capsule type strain [23]. The rmpA2 gene was given this name after sequencing revealed it to encode a longer protein than the originally cloned rmpA from Kp52145 [23]. Alignment of the coding sequences for RmpA and RmpA2 from several strains reveals that they share about 80% identity. Some strains, such as NTUH-K2044 and Kp52145, contain a truncated rmpA2 gene but it is unknown if these encode functional proteins. In CG43, the ΔrmpA2 strain is non-HMV but does not show much change in cps expression [21,22]. However, when rmpA2 is overexpressed, cps expression increased, so it may only impact transcription under certain conditions. To our knowledge, CG43 is the only strain known to have functional RmpA and RmpA2 proteins.
RmpC
In examining the role of RmpA in cps regulation in KPPR1S, it was noted that there was a neighboring gene predicted to encode another protein with a LuxR-type DNA-binding domain. Deletion of this gene, rmpC, resulted in reduced expression from cps promoters and reduced production of capsule similar to that observed for the rmpA mutant. However, unlike the rmpA mutant, this strain remains HMV [25••]. This is the first report of a mutant with reduced capsule that is still HMV. It was also determined to be co-transcribed with rmpA. The ΔrmpC strain is attenuated in the pneumonia model, but the phenotype is not as severe as mutants such as ΔrmpA and ΔkvrA that are non-HMV but have similar decreases in cps expression as the ΔrmpC mutant [25••]. Thus, it appears that HMV may be a more important virulence attribute than production of high levels of capsule.
MarR-family regulators
KvrA
KvrA is the K. pneumoniae homolog of SlyA, a regulator of virulence in Salmonella. KvrA, along with KvrB, was identified as contributing to virulence through a screen that specifically targeted genes predicted to encode MarR-family regulators [24•]. The ΔkvrA mutant is essentially avirulent in the mouse pneumonia model, and was found to be non-HMV and to have reduced cps expression from the manC and galF promoters. Subsequently, KvrA was also shown to regulate expression from the rmpA promoter [25••], but cps gene expression in the ΔkvrA strain could not be restored by expression of rmpA on a plasmid. This suggests that KvrA may impact expression from the galF and manC promoters independently of its effect on rmpA. KvrA is found in both cKp and hvKp strains, but it did not appear to control cps expression in the classical strain KPNIH1, an ST258 isolate [24•]. However, a KPNIH1 kvrA mutant does have a colonization defect, suggesting there are other genes regulated by KvrA that potentially contribute to pathogenesis.
It is not yet known if KvrA directly binds to the rmpA or cps promoters. The function of this class of MarR-like regulators that includes SlyA and RovA involves derepressing H-NS-mediated gene silencing (reviewed in [61]). There is one report on the role of H-NS in a clinical isolate (K39, non-HMV) where cps expression, UA levels and mucosviscosity were elevated in an hns mutant [62]. It is therefore tempting to speculate that the role of KvrA in cps transcription is as a derepressor of H-NS-mediated silencing.
KvrB
KvrB is the K. pneumoniae homolog of EmrR. In E. coli, EmrR represses the EmrAB drug efflux pump [63]. The genes encoding EmrAB homologs are encoded downstream of KvrB, and the genetic context is the same as in E. coli, suggesting this locus functions similarly in K. pneumoniae. In addition, the EmrR homolog in an uropathogenic strain of E. coli, called MprA, is required for capsule expression [64]. A ΔkvrB mutant was attenuated in the mouse pneumonia model, but the colonization levels in the lungs and spleens were much higher than for other mutants such as ΔkvrA and ΔrcsB [24•,25••]. This ΔkvrB mutant is non-HMV and has reduced cps expression, both of which likely contribute to its virulence defect. In addition, like KvrA, KvrB was found to regulate the rmpA promoter. But unlike KvrA, the effect of KvrB on cps expression is likely to be solely through its action at the rmpA promoter as cps expression was restored in the ΔkvrB strain carrying rmpA on a plasmid [25••].
Other regulators
KvgAS and KvhAS two-component systems
Both cKp and hvKp strains encode homologs of the Evg/BvgAS two-component system, designated KvhAS [65]. The hvKp strains have a second pair of Evg/BvgAS homologs, designated KvgAS, and a neighboring gene encoding another response regulator named KvhR. In strain CG43, KvhR appears to play a role in both cps expression and HMV, and KvgA appears only to impact HMV. Loss of kvhA did not appear to affect either HMV or cps expression, but overexpressing kvhA did result in a reduction of both phenotypes. The exact nature of how these systems are involved in capsule and HMV has yet to be explored. The fact that KvhR and KvgA both impact HMV and are only found in hvKp suggests further investigation is warranted to understand their potential contribution to hypervirulence.
TraDIS screen
In a recent study, Dorman et al. utilized sedimentation properties to screen a transposon library for genes that contribute to altered capsule phenotypes [66•]. They found 19 transcriptional regulators, 13 of which were proposed to increase capsule production. Among the 19 were RmpA and RcsB, and they also identified H-NS, KvrA and KvrB as well as a number of other regulators with no previously known role in capsule synthesis. With all but RcsB, it remains to be determined which of these regulators acts directly at the capsule promoters.
Concluding remarks
So many regulators of capsule …
Summation of the literature on transcriptional regulation of K. pneumoniae capsule genes reveals there are numerous regulators that likely function to ensure cps expression is activated when needed (Figure 2). While most of these regulators have not yet been demonstrated to act directly at cps promoters, the abundance of regulators that contribute to cps regulation is an indicator of how critical capsule is to K. pneumoniae survival. Complicating analyses of capsule regulation is that it has been difficult to extrapolate results from one strain to another due to the variability in methods used and differences in regulatory gene content between strains (see Box 2). In addition, the role(s) of these regulators need to be evaluated in the context of other regulatory mutants to ascertain a number of critical factors, such as direct versus indirect regulation, whether they participate in a regulatory cascade, and whether there is cooperative regulation.
Figure 2.
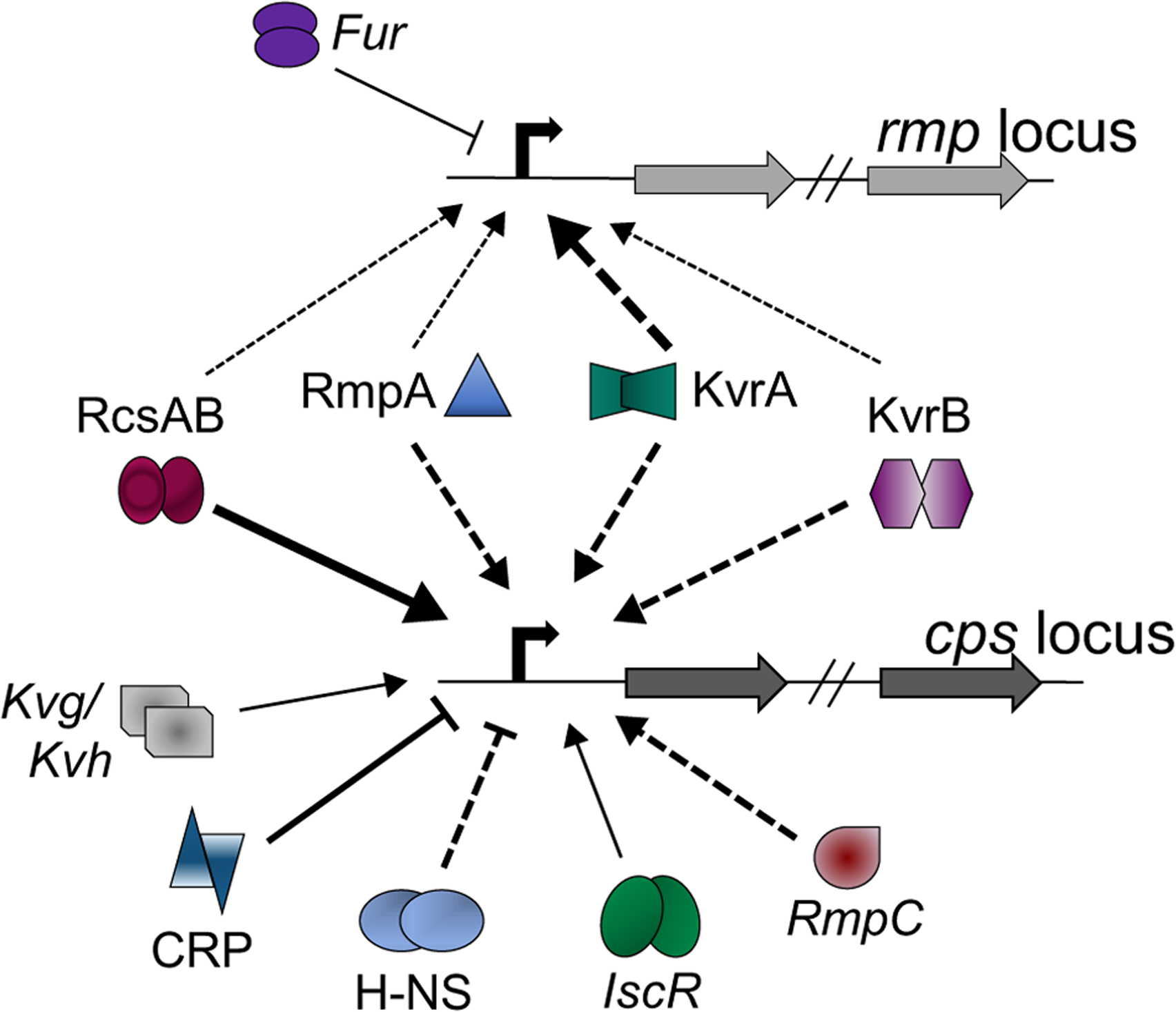
Complex matrix of capsule regulation. Each shape represents a different transcriptional regulator known to contribute in some way to the expression of capsule biosynthetic genes. If known or logically presumed, dimerization is indicated by doublets. Arrow thickness indicates the relative impact of the regulator: thin, small fold-change in mutant strain; thicker, larger fold-change in mutant strain. Arrows indicate activators, and T-shaped lines indicate repressors. Solid lines indicate that direct binding has been demonstrated for at least one of the cps promoters, and dashed lines indicate direct binding has not been tested. Italicized proteins have been characterized in only a single K. pneumoniae strain whereas the others have been characterized in at least 2 strains.
Box 2. A proposal to the Klebsiella community.
The methodology used to define capsule production, HMV and hypervirulence has varied significantly from one study to another, making direct comparisons between strains a challenge. In studies examining cps transcriptional regulation, some forms of capsule regulation (reporter or qRT-PCR) and quantification of UA levels are fairly constant between studies, but growth medium and conditions vary. In reports of newly isolated hv-CR-cKp strains, HMV is primarily assessed by the string test, and capsule levels and virulence are rarely examined. Assessments of HMV have ranged between qualitative (string test or images of centrifuged cultures) to quantitative (sedimentation assay). Virulence studies in mice have also been variable (see Box 1). While these types of variations are not unusual, as labs often favor a particular set of assays, this does make it difficult to extrapolate analyses between strains. This is further complicated by the recently recognized degree of genomic variation between strains of K. pneumoniae. As more strains are isolated that have both CR and hv-associated genes, there is a greater need for understanding the mechanisms that confer hypervirulence. Development and application of a consistent set of assays will expedite advancements and make it easier to assess the potential risks associated with these new isolates. Thus, we propose, at a minimum, the following assays for evaluating capsule production, hypermucoviscosity and hypervirulence:
Capsule should be quantified using the UA assay
HMV should be quantified using the sedimentation assay
Virulence should be measured in mice using a mouse model consistent with the origin of the strain (i.e., sepsis model for a blood isolate, pneumonia model for lung isolate, etc.)
In assessing potentially hypervirulent cKp isolates, some consideration should be given to determining the scale of virulence (i.e., is the strain more virulent than typical classical isolates? Has it become hypervirulent or does it have an intermediate virulence phenotype?).
But what really causes HMV and what is the link between HMV and hypervirulence?
The data generated on HMV have been from hvKp strains, but the primary focus has been on expression of capsule with less emphasis on the role of the transcriptional regulators in conferring HMV. This is in large part due to the assumption that the HMV phenotype is a consequence of capsule overproduction. However, we recently reported on a regulator that impacts cps expression but not HMV, indicating overproduction of capsule is not a requirement for HMV [25••]. Mutants that have a complete loss of capsule appear to be incapable of attaining HMV as a manC mutant overproducing RmpA remains non-HMV (personal observations). Thus, the HMV phenotype is likely to be due to a combination of capsule and something else. Although HMV is dependent on the presence of capsule, HMV and capsule need to be considered independently in addition to investigating the link that connects them.
Finally, more emphasis needs to be placed on quantifying HMV and assessing virulence of hv-CR-cKp clinical isolates (see Box 2). As more hv-CR-cKp strains are isolated, characterized and sequenced, the genetic data can be used to identify genes that correlate with HMV and hypervirulence. Genomic and phenotypic analyses will facilitate an understanding of what causes a strain to become HMV and hypervirulent. This information also will be essential in determining if the capacity to acquire HMV and/or hypervirulence is capsule type-dependent, and will be important for assessing the risks associated with cKp strains that acquire genes associated with HMV and hypervirulence in hvKp strains.
Acknowledgements
A special thanks to S. Falkow for providing exactly the right balance of support and independence for me to develop as a scientist (VLM). This work was supported by a grant from the National Institutes of Health (R21 AI132925) to V. Miller.
Footnotes
Conflict of interest
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.••.Paczosa MK, Mecsas J: Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 2016, 80:629–661. [DOI] [PMC free article] [PubMed] [Google Scholar]; Excellent, comprehensive review of K. pneumoniae biology and pathogenesis.
- 2.•.Martin RM, Bachman MA: Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 2018, 8:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]; Excellent review highlighting connections between virulence potential and the accessory genome.
- 3.•.Russo TA, Marr CM: Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 2019, 32 e00001–19. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive review of hvKp epidemiology, genomics and pathogenesis.
- 4.Sellick JA, Russo TA: Getting hypervirulent Klebsiella pneumoniae on the radar screen. Curr Opin in Infect Dis 2018, 31:341–346 10.1097/QCO.0000000000000464. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X et al. : Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016, 16:161–168. [DOI] [PubMed] [Google Scholar]
- 6.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE: Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genomics 2016, 2:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu C-R, Lin T-L, Chen Y-C, Chou H-C, Wang J-T: The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 2011, 157:3446–3457. [DOI] [PubMed] [Google Scholar]
- 8.•.Nassif X, Fournier J-M, Arondel J, Sansonetti PJ: Mucoid phenotype of Klebsiella pneumoniae is a plasmid-encoded virulence factor. Infect Immun 1989, 57:546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first report of rmpA and its role in the hypermucoviscous phenotype.
- 9.Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, Gottberg von A, Goossens H, Wagener MM, Benedi VJ: Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis 2007, 13:986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.••.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J et al. : Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 2015, 112: E3574–E3581. [DOI] [PMC free article] [PubMed] [Google Scholar]; Landmark paper providing genomic analyses of a large and diverse collection of K. pneumoniae isolates. This study demonstrated K. pneumoniae can be divided into three distinct species, that there is a large accessory genome and that there are a distinct set of genes associated with hvKp strains.
- 11.Follador R, Heinz E, Wyres KL, Ellington MJ, Kowarik M, Holt KE, Thomson NR: The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genomics 2016, 2:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y-T, Chang H-Y, Lai Y-C, Pan C-C, Tsai S-F, Peng H-L: Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 2004, 337:189–198. [DOI] [PubMed] [Google Scholar]
- 13.Lam MM, Wyres KL, Duchêne S, Wick RR, Judd LM, Gan Y-H, Hoh CH, Archuleta S, Molton JS, Kalimuddin S et al. : Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 2018, 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, Brisse S, Holt KE: Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genomics 2018, 12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh P-F, Lin T-L, Lee C-Z, Tsai S-F, Wang J-T: Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 2008, 197:1717–1727. [DOI] [PubMed] [Google Scholar]
- 16.Bachman MA, Miller VL, Weiser JN: Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog 2009, 5:e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bachman MA, Lenio S, Schmidt L, Oyler JE, Weiser JN: Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio 2012, 3 e00224–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachman MA, Oyler JE, Burns SH, Caza M, Lepine F, Dozois CM, Weiser JN: Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipcalin 2. Infect Immun 2011, 79:3309–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu W-L, Ko WC, Cheng K-C, Lee C-C, Lai C-C, Chuang Y-C: Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis 2008, 62:1–6. [DOI] [PubMed] [Google Scholar]
- 20.Russo TA, Olson R, MacDonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM: Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 2014, 82:2356–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL: RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 2010, 192:3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai Y-C, Peng H-L, Chang H-Y: RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol 2003, 185:788–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wacharotayankun R, Arakawa Y, Ohta M, Tanaka K, Akashi T, Mori M, Kato N: Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect Immun 1993, 61:3164–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.•.Palacios M, Miner TA, Frederick DR, Sepulveda VE, Quinn JD, Walker KA, Miller VL: Identification of two regulators of virulence that are conserved in Klebsiella pneumoniae classical and hypervirulent strains. mBio 2018, 9 e01443–18. [DOI] [PMC free article] [PubMed] [Google Scholar]; First study to identify conserved regulators of the MarR family (KvrA and KvrB) that affect virulence in both cKp and hvKp strains.
- 25.••.Walker KA, Miner TA, Palacios M, Trzilova D, Frederick DR, Broberg CA, Sepulveda VE, Quinn JD, Miller VL: A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio 2019, 10 e00089–19. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that rmpA is the first gene of an operon that encodes an additional regulator of cps expression, RmpC. A mutation in rmpC results in reduced cps gene expression and reduced capsule but has wildtype levels of hypermucoviscosity. This is the first characterized mutant demonstrating that hyperproduction of capsule is not a requirement for hypermucoviscosity.
- 26.Yu W-L, Ko WC, Cheng K-C, Lee H-C, Ke D-S, Lee CC, Fung CP, Chuang Y-C: Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. J Exp Med 2006, 42:1351–1358. [DOI] [PubMed] [Google Scholar]
- 27.Fang C-T, Chuang Y-P, Shun C-T, Chang S-C, Wang J-T: A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 2004, 199:697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR et al. : Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 2018, 56 e00776–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawlor MS, Hsu J, Rick PD, Miller VL: Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol 2005, 58:1054–1073. [DOI] [PubMed] [Google Scholar]
- 30.Cortés G, Borrell N, de Astorza B, Gómez C, Sauleda J, Albertí S: Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect Immun 2002, 70:2583–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawlor MS, Handley SA, Miller VL: Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect Immun 2006, 74:5402–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong H, Carter RA, Leiner IM, Tang Y-W, Chen L, Kreiswirth BN, Pamer EG: Distinct contributions of neutrophils and CCR2 +monocytes to pulmonary clearance of different Klebsiella pneumoniae strains. Infect Immun 2015, 83:3418–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen DA, Hilliard JK, Tiemann KM, Todd EM, Morley SC, Hunstad DA: Klebsiella pneumoniae FimK promotes virulence in murine pneumonia. J Infect Dis 2016, 213:649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cejas D, Fernández Canigia L, Rincón Cruz G, Elena AX, Maldonado I, Gutkind GO, Radice MA: First isolate of KPC-2-producing Klebsiella pneumonaie sequence type 23 from the Americas. J Clin Microbiol 2014, 52:3483–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J: Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis 2015, 37:107–112. [DOI] [PubMed] [Google Scholar]
- 36.Turton JF, Payne Z, Coward A, Hopkins KL, Turton JA, Doumith M, Woodford N: Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-ST23 and “non-hypervirulent” types ST147, ST15 and ST383. J Med Microbiol 2018, 67:118–128. [DOI] [PubMed] [Google Scholar]
- 37.Liu B-T, Su W-Q: Whole genome sequencing of NDM-1-producing serotype K1 ST23 hypervirulent Klebsiella pneumoniae in China. J Med Microbiol 2019, 68:866–873 10.1099/jmm.0.000996. [DOI] [PubMed] [Google Scholar]
- 38.Karlsson M, Stanton RA, Ansari U, McAllister G, Chan MY, Sula E, Grass JE, Duffy N, Anacker ML, Witwer ML et al. : Identification of a carbapenemase-producing hypervirulent Klebsiella pneumoniae isolate, United States. Antimicrob Agents Chemother 2019, 63 10.1128/AAC.00519-19e00519-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harada S, Aoki K, Ishii Y, Ohno Y, Nakamura A, Komatsu M, Tateda K: Emergence of IMP-Producing Hypervirulent Klebsiella pneumoniae Carrying a pLVPK-Like Virulence Plasmid. 2019. 10.1016/j.ijantimicag.2019.05.007. [DOI] [PubMed]
- 40.Wei D-D, Wan L-G, Deng Q, Liu Y: Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in mainland China. Diagn Microbiol Infect Dis 2016, 85:192–194. [DOI] [PubMed] [Google Scholar]
- 41.••.Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, Wang Q, Wang X, Chen H, Li H et al. : Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 2015, 71:553–560. [DOI] [PubMed] [Google Scholar]; An early report of CR-hvKp as well as CR-cKp isolates with hv-associated genes; these hv-CR-cKp strains were not virulent in mouse models of sepsis underscoring the need for more detailed phenotypic characterization of putative hv-CR-cKp strains.
- 42.••.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Wai-Chi Chan E, Shu L, Yu J, Zhang R et al. : A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 2018, 18:37–46. [DOI] [PubMed] [Google Scholar]; This report describes cKp isolates with some hv-associated genes encoded on a large plasmid with regions homologous to pLVPK. These strains had a resistance phenotype in PMNs similar to hvKp.
- 43.•.Huang Y-H, Chou S-H, Liang S-W, Ni C-E, Lin Y-T, Huang Y-W, Yang T-C: Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother 2018, 73:2039–2046. [DOI] [PubMed] [Google Scholar]; This report describes a cKp strain with intermediate virulence in a mouse model of sepsis. This strain contains a novel plasmid encoding many hv-associated genes.
- 44.Wu K-M, Li L-H, Yan J-J, Tsao N, Liao T-L, Tsai H-C, Fung C-P, Chen H-J, Liu Y-M, Wang J-T et al. : Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 2009, 191:4492–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y-T, Chang H-Y, Lai Y-C, Pan C-C, Tsai S-F, Peng H-L: Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 2004, 337:189–198. [DOI] [PubMed] [Google Scholar]
- 46.Lery L, Frangeul L, Passet V, Frangeul, Almeida AS, Bialek-Davenet S, Barbe V, Bengoechea JA, Sansonetti P, Brisse S et al. : Comparative analysis of Klebsiella pneumoniae genomes identifies a phopholipase D family protein as a novel virulence factor. BMC Biol 2014, 12:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broberg CA, Wu W, Cavaloci JD, Miller VL, Bachman MA: Complete genome sequence of Klebsiella pneumoniae strain ATCC 43816 KPPR1, a rifampin-resistant mutant commonly used in animal, genetic, and molecular biology studies. Genome Announc 2014, 2 e00924–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wall E, Majdalani N, Gottesman S: The complex Rcs regulatory cascade. Annu Rev Microbiol 2018, 72:111–139. [DOI] [PubMed] [Google Scholar]
- 49.Gottesman S, Trisler P, Torres-Cabassa A: Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: Characterization of three regulatory genes. J Bacteriol 1985, 162:1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allen PM, Fisher D, Saunders JR, Hart CA: The role of capsular polysaccharide K21b of Klebsiella and of the structurally related colanic-acid polysaccharide of Escherichia coli in resistance to phagocytosis and serum killing. J Med Microbiol 1987, 24:363–370. [DOI] [PubMed] [Google Scholar]
- 51.Wacharotayankun R, Arakawa Y, Ohta M, Hasegawa T, Mori M, Horii T, Kato N: Involvement of rcsB in Klebsiella K2 capsule synthesis in Escherichia coli K-12. J Bacteriol 1992, 174:1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCallum KL, Whitfield C: The rcsA gene of Klebsiella pneumoniae O1:K20 is involved in expression of the serotype-specific K (capsular) antigen. Infect Immun 1991, 59:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wehland M, Bernhard F: The RcsAB box: characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J Biol Chem 2000, 275:7013–7020. [DOI] [PubMed] [Google Scholar]
- 54.Peng D, Li X, Liu P, Zhou X, Luo M, Su K, Chen S, Zhang Z, He Q, Qiu J et al. : Transcriptional regulation of galF by RcsAB affects capsular polysaccharide formation in Klebsiella pneumoniae NTUH-K2044. Microbiol Res 2018, 216:70–78. [DOI] [PubMed] [Google Scholar]
- 55.Lin C-T, Chen Y-C, Jinn T-R, Wu C-C, Hong Y-M, Wu W-H: Role of the cAMP-dependent carbon catabolite repression in capsular polysaccharide biosynthesis in Klebsiella pneumoniae. PLoS One 2013, 8:e54430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ou Q, Fan J, Duan D, Xu L, Wang J, Zhou D, Yang H, Li B: Involvement of cAMP receptor protein in biofilm formation, fimbria production, capsular polysaccharide biosynthesis and lethality in mouse of Klebsiella pneumoniae serotype K1 causing pyogenic liver abscess. J Med Microbiol 2017, 66:1–7. [DOI] [PubMed] [Google Scholar]
- 57.Lin C-T, Wu C-C, Chen Y-S, Lai Y-C, Chi C, Lin J-C, Chen Y, Peng H-L: Fur regulation of the capsular polysaccharide biosynthesis and iron-acquisition systems in Klebsiella pneumoniae CG43. Microbiology 2011, 157:419–429. [DOI] [PubMed] [Google Scholar]
- 58.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ: IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci U S A 2001, 98:14895–14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu C-C, Wang C-K, Chen Y-C, Lin T-H, Jinn T-R, Lin C-T: IscR regulation of capsular polysaccharide biosynthesis and iron-acquisition systems in Klebsiella pneumoniae CG43. PLoS One 2014, 9:e107812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu F, Lv J, Niu S, Du H, Tang Y-W, Pitout JDD, Bonomo RA, Kreiswirth BN, Chen L: Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (Sequence Type 258 [ST258] and ST11) and hypervirulent (ST23, ST65, ST86, and ST375) strains. J Clin Microbiol 2018, 56 e00731–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoebel DM, Free A, Dorman CJ: Anti-silencing: overcoming H-NS-mediated repression of transcription in gram-negative enteric bacteria. Microbiology 2008, 154:2533–2545. [DOI] [PubMed] [Google Scholar]
- 62.Ares MA, Fernández-Vázquez JL, Rosales-Reyes R, Jarillo-Quijada MD, Bargen von K, Torres J, González-y-Merchand JA, Alcántar-Curiel MD, la Cruz De MA: H-NS nucleoid protein controls virulence features of Klebsiella pneumoniae by regulating the expression of type 3 pili and the capsule polysaccharide. Front Cell Infect Microbiol 2016, 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lomovskaya O, Lewis K, Matin A: EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol 1995, 177:2328–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goh KGK, Phan M-D, Forde BM, Chong TM, Yin W-F, Chan K-G, Ulett GC, Sweet MJ, Beatson SA, Schembri MA: Genome-wide discovery of genes required for capsule production by uropathogenic Escherichia coli. mBio 2017, 8 e01558–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin C-T, Huang T-Y, Liang W-C, Peng H-L: Homologous response regulators KvgA, KvhA and KvhR regulate the synthesis of capsular polysaccharide in Klebsiella pneumoniae CG43 in a coordinated canner. J Biochem 2006, 140:429–438. [DOI] [PubMed] [Google Scholar]
- 66.•.Dorman MJ, Feltwell T, Goulding DA, Parkhill J, Short FL: The capsule regulatory network of Klebsiella pneumoniae defined by density-TraDISort. mBio 2018, 9 e01863–18. [DOI] [PMC free article] [PubMed] [Google Scholar]; This report describes an innovative method for screening a large set of transposon mutants for alterations in mucoviscosity. Many genes known to impact mucoviscosity as well as novel genes were identified.


