Abstract
Existing tumor markers for testicular germ cell tumor (TGCT) cannot detect the presence of pure teratoma. Serum miRNAs have strong performance detecting other subtypes of TGCT. Previous reports suggest high levels of miR-375 expression in teratoma tissue. The purpose of this study was to explore the role of serum miRNA, including miR-375, in detecting the presence of teratoma at postchemotherapy retroperitoneal lymph node dissection (PC-RPLND).
We prospectively collected presurgical serum from 40 TGCT patients undergoing PC-RPLND (21 with teratoma at RPLND and 19 with no evidence of disease). We examined the utility of serum miR-375-3p and miR-375-5p by quantitative polymerase chain reaction, and searched for other putative serum miRNAs with small RNA sequencing. The area under the receiver operating characteristic curve (AUC) and univariate analyses were utilized to evaluate test characteristics and predictors of teratoma.
Both serum miR-375-3p and miR-375-5p exhibited poor performance (miR-375-3p: 86% sensitivity, 32% specificity, AUC: 0.506; miR-375-5p: 55% sensitivity, 67% specificity, AUC: 0.556). Teratoma at orchiectomy was the only predictor of PC-RPLND teratoma. Small RNA sequencing identified three potentially discriminatory miRNAs, but further validation demonstrated no utility. Our results confirm prior reports that serum miR-375 cannot predict teratoma, and suggest that there may not exist a predictive serum miRNA for teratoma.
Patient summary
We found that serum miR-375 cannot detect the presence of teratoma at postchemotherapy retroperitoneal lymph node dissection (PC-RPLND). We are also unable to find any other serum miRNAs predictive of pure teratoma at PC-RPLND. Hence, the lack of a reliable circulating marker of teratoma remains a critical clinical need.
Keywords: Retroperitoneal lymph node dissection, MicroRNA, Teratoma, Serum biomarker
The serum tumor markers (STMs) lactate dehydrogenase, beta-human chorionic gonadotropin, and alpha-fetoprotein are a mainstay in the diagnosis and management of testicular germ cell tumors (TGCTs). Recent work has revealed that serum miRNAs exhibit greatly improved performance over these STMs in detecting viable TGCT, but neither conventional STMs nor TGCT-associated serum miRNA is sensitive to pure teratoma [1]. Teratoma is a common finding at postchemotherapy retroperitoneal lymph node dissection (PC-RPLND), as 25–40% of resected lymph nodes will contain teratoma and 40–50% will exhibit no signs of residual disease [2]. Therefore, nearly half of patients receiving PC-RPLND will do so unnecessarily. A circulating marker sensitive to pure teratoma could prevent these unnecessary operations. Until recently, potential targets for the detection of teratoma were elusive. A recent genomic and epigenomic analysis of testicular tumors demonstrated a promising high level of expression of miR-375 among teratomas [3]. We first reported that serum miR-375-3p was uninformative for teratoma in a small cohort [4], and other recent studies investigating the utility of circulating miR-375-3p have confirmed that it is not predictive of teratoma [5]. We therefore set out to search for other serum miRNAs predictive of pure teratoma at PC-RPLND.
Following institutional review board approval, we collected serum from 40 patients with ≥1-cm-diameter retroperitoneal mass immediately prior to bilateral full-template PC-RPLND between 2016 and 2019. The pathology of resected nodes was examined by experienced genitourinary pathologists and classified as postpubertal-type teratoma (21 cases total: 19 pure postpubertal teratoma, one teratoma with yolk sac elements, and one teratoma with yolk sac and embryonal carcinoma elements) or control (19 cases total: seven benign, 11 fibrosis, and one necrosis). Baseline characteristics of this population are described in Supplementary Table 1. There were no significant differences between the groups concerning age, clinical stage, or conventional STM levels (Supplementary Table 2). We examined miR-375-3p (primary strand) and miR-375-5p (passenger strand) levels in the serum using an extraction and quantitative polymerase chain reaction (qPCR) workflow similar to those described previously for TGCT serum miRNA analysis (see the Supplementary material). Final results were calculated relative to the mean of all samples in the control group.
As expected, we found no difference in miR-375-3p levels between the teratoma and control groups (Fig. 1A). At a relative expression threshold of 0.6, as selected by Youden’s J statistic, serum miR-375-3p demonstrated 86% sensitivity and 32% specificity to detect residual teratoma. Eighteen of 31 patients with miR-375-3p serum levels above threshold actually harbored teratoma on final pathology (58% positive predictive value). Nine patients had negative miR-375-3p tests, of whom six had benign pathology (67% negative predictive value). A receiver operating characteristic analysis confirmed that serum miR-375-3p lacked discriminatory capacity, with an area under the receiver operating characteristic curve (AUC) of 0.506 (95% confidence interval [CI]: 0.32–0.69, p = 0.95; Fig. 1B). We next examined serum miR-375-5p in a subset of the patient pool (20 patients total, ten each in the teratoma and control groups). Although miRNA passenger strands are generally presumed to be rapidly degraded, previous reports indicated detectable levels of miR-375-5p in the serum [6]. Serum miR-375-5p also lacked any predictive capacity for teratoma, with an AUC of 0.556 (95% CI: 0.30–0.81, p = 0.68; Fig. 1C and 1D). At a relative expression threshold of 1.16, sensitivity and specificity were calculated as 55% and 67%, respectively. To ensure that the probes were both sensitive and specific to their targets, we ran the assay with a dilution series of known concentrations of miR-375-3p and miR-375-5p. The relative level of a consistent exogenous spike-in, cel-miR-39-3p, remained unchanged across samples. The probes targeting each strand of miR-375 were found to specifically detect their target strand (Supplementary Fig. 1). A univariate analysis revealed that the only significant predictor of teratoma on final PC-RPLND pathology was the presence of teratoma at original orchiectomy specimen (p = 0.02; Supplementary Table 2). We therefore conclude that neither miR-375-3p nor miR-375-5p is a suitable serum marker of teratoma.
Fig. 1.
Serum miR-375 does not predict teratoma at PC-RPLND. Boxplots depicting serum (A) miR-375-3p and (C) miR-375-5p expression in the control and teratoma groups. Expression is displayed relative to the control group. Receiver operating characteristic curve depicting discriminatory performance of (B) miR-375-3p and (D) miR-375-5p. Area under the curve is displayed on each graph. AUC = area under the receiver operating characteristic curve; PC-RPLND = postchemotherapy retroperitoneal lymph node dissection.
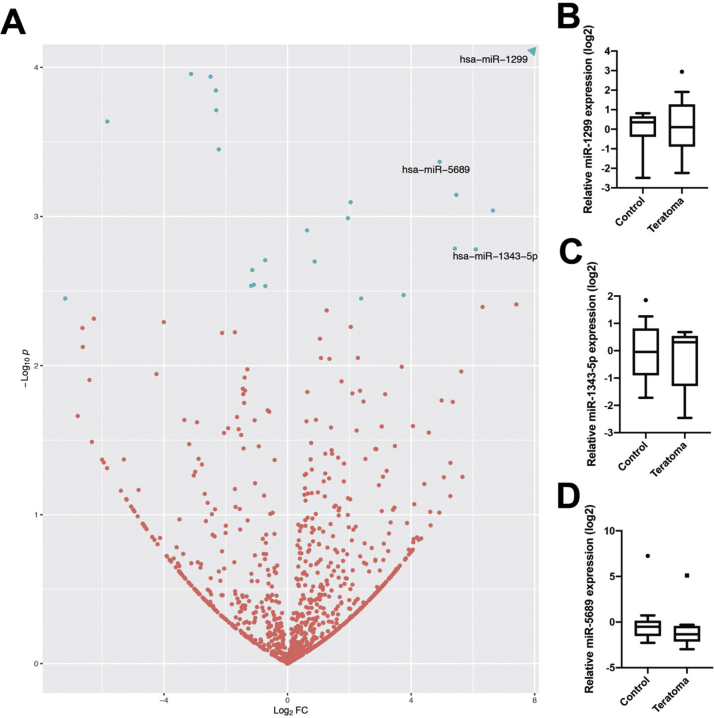
We next searched for other serum miRNAs predictive of pure mature teratoma using small RNA sequencing. Out of 779 detected miRNAs, we identified 28 miRNAs as differentially expressed (adjusted p < 0.1; Fig. 2A). Fourteen of these were higher expressed in teratoma versus controls. We selected three of these targets to validate by qPCR based on expression levels, namely, miR-1299, miR-5689, and miR-1343-5p. None of these targets were found to be significantly different between the teratoma and control groups by qPCR (Fig. 2B–D).
Fig. 2.
Small RNA sequencing examination of serum miRNAs to detect teratoma. (A) Volcano plot of detected miRNAs in teratoma and control groups; hsa-miR-1299 fell outside the range of plot and is depicted as an arrow (log2FC = 21.92, –log10p = 13.2). Fold change is calculated as expression in the teratoma group over that in the control group. Blue dots are significantly differentially expressed miRNAs. (Quantitative PCR-based validation of (B) miR-1299, (C) miR-1343-5p, and (D) miR-5689. PCR = polymerase chain reaction.
These results confirm previous findings that serum miR-375-3p does not predict teratoma and demonstrate that serum miR-375-5p does not predict teratoma at PC-RPLND. Based on The Cancer Genome Atlas molecular profiling of TGCT, miR-375 has been considered an attractive candidate for a teratoma marker [7]. This conflicted with earlier reports indicating that miR-375 expression was enriched in yolk sac, but not teratoma, tissue [8], [9]. We first examined this discrepancy and reported the inability of serum miR-375-3p to detect teratoma at primary RPLND, but the small sample size required follow-up studies to confirm the result [4]. Additional reports with more substantial sample sizes examining both pre- and postchemotherapy patients have since confirmed these results [5]. Lobo et al [5] identified other serum miRNAs in their dataset, including miR-885-5p and miR-448, with discriminatory capacity for teratoma. Neither of these miRNAs was found to be differentially expressed in our small RNA sequencing study. Nappi et al [10] have very recently reported positive results regarding the ability of plasma miR-375 in combination with miR-371a-3p to predict teratoma. Although our results initially appear to conflict with those of Nappi et al [10], their study found limited utility of circulating miR-375 alone in detecting teratoma in the validation cohort, in agreement with the findings presented here and those referenced previously. The ultimately negative results from our small RNA sequencing data suggest that although serum miRNAs have performed admirably in the detection of residual viable TGCT, perhaps another avenue of investigation will be more fruitful in the case of teratoma. However, caution must be taken when interpreting these results, as the current study is limited by a small sample size and the relatively low concentration of RNA present in serum. Despite this, the lack of a reliable circulating marker of teratoma remains a critical unmet clinical need.
Author contributions: Aditya Bagrodia had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Lafin, Kapur, Speir, Chesnut, Frazier, Coleman, Murray, Amatruda, Bagrodia.
Acquisition of data: Lafin, Kenigsberg, Meng, Abe, Savelyeva, Singla, Woldu, Lotan, Mauck, Lewis, Margulis, Wong, Jia, Kapur.
Analysis and interpretation of data: Lafin, Kenigsberg, Meng, Abe, Singla, Wong, Jia, Kapur, Xu, Strand, Murray, Amatruda, Bagrodia.
Drafting of the manuscript: Lafin, Kenigsberg, Wong.
Critical revision of the manuscript for important intellectual content: Lafin, Kenigsberg, Meng, Abe, Savelyeva, Singla, Woldu, Lotan, Mauck, Lewis, Margulis, Wong, Jia, Kapur, Xu, Speir, Chesnut, Frazier, Strand, Coleman, Murray, Amatruda, Bagrodia.
Statistical analysis: Lafin, Kenigsberg, Wong, Xu.
Obtaining funding: Lewis, Speir, Chesnut, Frazier, Wong, Coleman, Murray, Amatruda, Bagrodia.
Administrative, technical, or material support: Savelyeva, Singla, Woldu, Lotan, Mauck, Lewis, Margulis, Kapur, Xu, Speir, Chesnut, Frazier, Strand, Coleman, Murray, Amatruda, Bagrodia.
Supervision: Murray, Amatruda, Bagrodia.
Other: None.
Financial disclosures: Aditya Bagrodia certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: This work was supported by the National Cancer Institute of the National Institutes of Health under award number 5 P30 CA142543 09 (Cheryl Lewis), a St. Baldrick’s Consortium award under grant 358099 (A. Lindsay Frazier, Matthew J. Murray, Nicholas Coleman, and James F. Amatruda), grant RP170152 from the Cancer Prevention and Research Institute of Texas (Aditya Bagrodia and James F. Amatruda), a Rally Foundation award (Matthew J. Murray, James F. Amatruda, and A. Lindsay Frazier), and Malignant Germ Cell International Consortium (Aditya Bagrodia, Matthew J. Murray, A. Lindsay Frazier, and James F. Amatruda) and Dedman Family Scholarship in Clinical Care (Aditya Bagrodia).
Acknowledgments: We would like to thank the University of Texas Southwestern Tissue Repository (UTSTR) for acquisition and storage of patient serum samples.
Associate Editor: Guillaume Ploussard
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.euros.2021.02.003.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Leão R., van Agthoven T., Figueiredo A. Serum miRNA predicts viable disease after chemotherapy in patients with testicular nonseminoma germ cell tumor. J Urol. 2018;200:126–135. doi: 10.1016/j.juro.2018.02.068. [DOI] [PubMed] [Google Scholar]
- 2.Miranda Ede P., Abe D.K., Nesrallah A.J. Predicting necrosis in residual mass analysis after retroperitoneal lymph node dissection: a retrospective study. World J Surg Oncol. 2012;10:203. doi: 10.1186/1477-7819-10-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen H., Shih J., Hollern D.P. Integrated molecular characterization of testicular germ cell tumors. Cell Rep. 2018;23:3392–3406. doi: 10.1016/j.celrep.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafin J.T., Singla N., Woldu S.L. Serum microRNA-371a-3p levels predict viable germ cell tumor in chemotherapy-naïve patients undergoing retroperitoneal lymph node dissection. Eur Urol. 2020;77:290–292. doi: 10.1016/j.eururo.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lobo J., Gillis A.J.M., van den Berg A. Identification and validation model for informative liquid biopsy-based microRNA biomarkers: insights from germ cell tumor in vitro, in vivo and patient-derived data. Cells. 2019;8:1637. doi: 10.3390/cells8121637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakhter A.J., Pratt R.E., Moore R.E. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia. 2018;61:1124–1134. doi: 10.1007/s00125-018-4559-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen H., Shih J., Hollern D.P. Integrated molecular characterization of testicular germ cell tumors. Cell Rep. 2018;23:3392–3406. doi: 10.1016/j.celrep.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okpanyi V., Schneider D.T., Zahn S. Analysis of the adenomatous polyposis coli (APC) gene in childhood and adolescent germ cell tumors. Pediatr Blood Cancer. 2011;56:384–391. doi: 10.1002/pbc.22669. [DOI] [PubMed] [Google Scholar]
- 9.Murray M.J., Saini H.K., van Dongen S. The two most common histological subtypes of malignant germ cell tumour are distinguished by global microRNA profiles, associated with differential transcription factor expression. Mol Cancer. 2010;9:290. doi: 10.1186/1476-4598-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nappi L., Thi M., Adra N. Integrated expression of circulating miR375 and miR371 to identify teratoma and active germ cell malignancy components in malignant germ cell tumors. Eur Urol. 2021;79:16–19. doi: 10.1016/j.eururo.2020.10.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.