The structure of an infectious immature flavivirus redefines viral architecture and maturation.
Abstract
Flaviviruses are the cause of severe human diseases transmitted by mosquitoes and ticks. These viruses use a potent fusion machinery to enter target cells that needs to be restrained during viral assembly and egress. A molecular chaperone, premembrane (prM) maintains the virus particles in an immature, fusion-incompetent state until they exit the cell. Taking advantage of an insect virus that produces particles that are both immature and infectious, we determined the structure of the first immature flavivirus with a complete spike by cryo–electron microscopy. Unexpectedly, the prM chaperone forms a supporting pillar that maintains the immature spike in an asymmetric and upright state, primed for large rearrangements upon acidification. The collapse of the spike along a path defined by the prM chaperone is required, and its inhibition by a multivalent immunoglobulin M blocks infection. The revised architecture and collapse model are likely to be conserved across flaviviruses.
INTRODUCTION
Flaviviruses are commonly known for encephalitic and hemorrhagic diseases caused by arthropod-borne viruses such as Zika virus (ZIKV), dengue virus (DENV), and tick-borne encephalitis virus (TBEV). Collectively, these viruses have a large disease burden (1). Less known flaviviruses regularly emerge as global pathogens, while others remain localized or restricted to their arthropod host (2). Some of the latter are insect-specific flaviviruses (ISFs), which are transmitted exclusively between arthropods with no involvement of a vertebrate host (3). Lineage II ISFs are particularly interesting, because they cluster phylogenetically with the vertebrate-infecting flaviviruses (VIFs) but exhibit an insect-specific phenotype with no growth in vertebrates in vitro or in vivo (3, 4). Their study informs us on the evolution of pathogenic flaviviruses and the nature of host restriction factors. Binjari virus (BinJV), a lineage II ISF, has been used for the generation of chimeric viruses, which involves exchanging the premembrane (prM) and envelope (E) genes for the corresponding genes of pathogenic flaviviruses, such as ZIKV, West Nile virus (WNV), and DENV (5). These chimeras produce structurally authentic virions that are antigenically indistinguishable from the donor pathogenic virus but are unable to replicate in a range of vertebrate cell lines. The observed tolerance for exchange of BinJV structural proteins and sequence similarity between ISF lineage II and VIFs suggests a common viral architecture, but until now, this had not been confirmed at the atomic level.
Single-particle cryo–electron microscopy (cryo-EM) analysis has revealed the molecular architecture of the infectious VIF virion to be a near-spherical icosahedral particle composed of 180 copies of E and membrane (M) proteins arranged in a distinctive herringbone-like pattern of 90 E dimers (6, 7). The infectious virion is derived from an immature particle that assembles in the endoplasmic reticulum and consists of 60 trimeric spikes of the E and prM complex (8). During morphogenesis and maturation, the prM protein acts as a molecular chaperone and prevents premature triggering of the pH-activated E fusion machinery during transit through the acidified secretory vesicles of the host cell (9). Release of the pr domain is considered a prerequisite for viral infectivity. For release to occur, a site within prM is cleaved by furin in the acidic trans-Golgi compartment (10). The pr domain dissociates upon viral release, priming the E- and M-mediated fusion mechanism required for virus cell entry (11).
Maturation efficiency is variable between flaviviruses (12). In the case of DENV, a notable proportion of the released virions retain prM and stimulate the production of nonneutralizing antibodies. This contributes to the antibody-dependent enhancement (ADE) of disease, whereby prM antibodies can facilitate uptake of otherwise noninfectious, immature DENV particles into host immune cells (13). Previous studies on ISFs have found a higher ratio of prM to E compared with traditional flaviviruses, suggesting inefficient cleavage (4, 14, 15). The immature virions of VIFs are heterogeneous, which has limited our understanding of their structure to main-chain models.
In this work, we show that the immature form predominates the viral preparations of BinJV and, unlike for VIFs, is infectious without the requirement of additional antibody-mediated uptake. Antibody-mediated inhibition of maturation neutralizes BinJV but is isotype dependent. The homogeneity of the BinJV particles allowed for the first structure determination of an immature flavivirus with an unambiguous topology that is likely to be conserved across all flaviviruses. The unexpected architecture supports a model of maturation where a centrally placed prM activates and guides a collapse of the spike.
RESULTS
BinJV is secreted as a fully immature flavivirus particle
To investigate the structure of an ISF, we purified BinJV from infected C6/36 cells for cryo-EM imaging. SDS–polyacrylamide gel electrophoresis (PAGE) analysis of the preparation revealed that it was mostly immature, with a prominent band for prM and only traces of mature M (Fig. 1A). Similar levels of prM were observed over multiple infection and purification rounds and were also observed for two other ISFs, Hidden Valley virus (HVV) and Parramatta River virus (PaRV), suggesting a conserved ISF feature (Fig. 1A). For comparison, a preparation of the VIF WNV kunjin subtype (WNVKUN) is also shown (Fig. 1A). Supernatant titers of BinJV are similar to WNVKUN, suggesting that viral release is likely efficient and not a major contributing factor to explain the predominance of uncleaved prM in BinJV preparations.
Fig. 1. Cryo-EM reconstruction of BinJV.
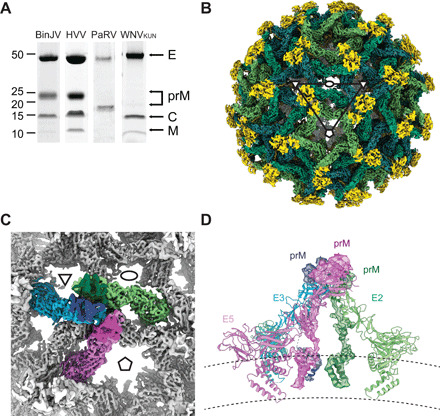
(A) SDS-PAGE analysis of gradient purified BinJV, HVV, PaRV, and WNVKUN virions. Virus preparations are nonreduced and stained with SYPRO Ruby stain. E, prM, capsid (C), and M proteins indicated by arrows. (B) Surface cryo-EM reconstruction of BinJV. Density attributed to the pr proteins within the immature form is shown in yellow, and the three E proteins that make up the asymmetric unit were shown in shades of green. Ellipses, triangles, and pentagons represent the two-, three-, and fivefold icosahedral axes, respectively. (C) The asymmetric unit of BinJV is composed of three copies of E (green, pink, and cyan) and prM (dark green, magenta, and blue). (D) Side view of the asymmetric unit displayed as a cartoon and colored by symmetry position. Density is shown for prM. Lines illustrate the extent of the lipid bilayer.
When imaged by cryo-EM, most of the BinJV particles had a spiky appearance that resembles the immature structure of DENV and ZIKV and were mostly homogenous with no structural evidence of partial maturation. Some smooth particles were noted but, consistent with gel profiles, were in the minority (fig. S1A). The cryo-EM reconstruction of the immature virion was resolved to an average resolution of 4.4 Å (fig. S1, B to F, and table S1). Sixty asymmetrical trimers project from the surface, creating the spiky appearance (Fig. 1B). The asymmetric unit of the immature BinJV particle consists of a spike formed by three prM/E protein complexes (Fig. 1, C and D). Both E and prM have the typical fold of flavivirus surface proteins. E consists of an elongated three-domain ectodomain (ED-I, ED-II, and ED-III), a membrane-proximal stem region and two C-terminal transmembrane (TMs) helices (Fig. 2, A and B). BinJV E sequence has no N-glycosylation sequons, and peptide N-glycosidase F (PNGase) treatment together with our structural data indicate that, unlike VIFs, BinJV E is not glycosylated. The pr domain is a compact β-sandwich with two glycans at position N13 and N71 (fig. S2). This globular domain is linked to the M domain of prM by an extended linker containing the furin cleavage site and helix M-H0. M is composed of a membrane-proximal helix and two TM helices (Fig. 2, A and B).
Fig. 2. The structure of the prM-E heterodimer reveals a furin site buried within the spike and a central pillar supporting the spike rather than the proposed drawstring.
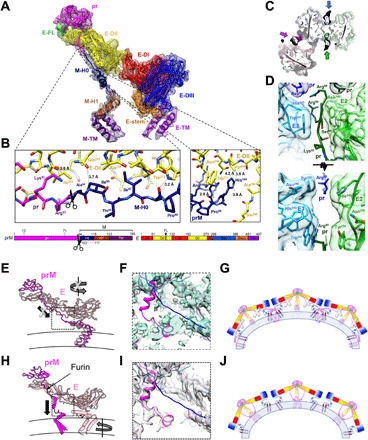
(A) The electron density and structure of the fivefold prM-E heterodimer. The color scheme matches the domain diagram (bottom), which also indicates glycans (cyan), furin cleavage site (scissors), and unmodeled region (dashed line). (B) Zoom of the prM linker. (C) Section of the trimeric spike near the furin site viewed from virion exterior. Molecular surface and stick representation are colored according to the prM-E proximity to the twofold (green/lime), threefold (blue/cyan), and fivefold (magenta/pink) symmetry axes. (D) Zooms of the prM/E interfaces as in (C) (bottom) or in orthogonal orientation (top). (E and F) Structure of BinJV (E) compared to a main-chain model of immature DENV-1 [(H), PDB: 4B03]. E and prM is in pink and magenta, respectively. Arrows indicate that movements proposed in the drawstring model (E) are incompatible with the BinJV topology, supporting a prM-guided collapse (H). (F and I) Linker region electron density for BinJV (F) and DENV-1 [(I) EMD-2141]. Density is only visible for the BinJV model (magenta). None of the reconstructions show density for the proposed DENV-1 prM linker (blue; PDB: 4B03). (G and J) Schematics of the immature virion colored as (A) with membrane in blue. Instead of the previously proposed domain swap (G), prM forms a supporting pillar at the center of each BinJV spike (J).
The spike resembles a splayed tripod with the fusion loop-containing ED-IIs forming the tip of the spike, capped by three pr domains (Fig. 1D). In available main-chain models (16–18), prM and E were modeled parallel to each other along the whole length of E. The TMs of prM and E were thus proposed to be bundled together at the periphery (Fig. 2, E to G). Our structure reveals a different topology where the prM linker, membrane-proximal stem, and TMs are situated in proximity to the center of the spike, away from the feet of the tripod formed by E. This connectivity markedly changes the spike architecture and its network of interactions within the particles (Fig. 2, H to J).
Fully resolved prM linker reveals a buried furin recognition site
In contrast to available structures of immature VIFs we were able to model the complete linker of prM based on readily interpretable electron density (Fig. 2A and fig. S3). This region is well defined in prM subunits next to the twofold (prM2) and fivefold axes (prM5) but missing H0 for the other subunit (prM3). The first section of the linker is in an extended conformation tucked onto the side of E. This region includes the furin recognition site, followed by a hydrophobic stretch that zips the linker to a hydrophobic patch in E-DII (Fig. 2, A and B). A sharp kink orients the rest of prM toward the center of the spike, away from the main body of E. The last interacting residue between prM and E is residue H100, which is conserved in both ISFs and VIFs (Fig. 2B and fig. S3A). The interactions between H100 and E are maintained in mature flavivirus structures, providing an anchor point that remains stable throughout maturation. The M moiety of prM adopts a conformation that is similar to M in mature flaviviruses.
The furin recognition site is buried at the top of the spike and unavailable for cleavage without substantial structural rearrangements. The linker is clamped between the E proteins at the tip of the spike (Fig. 2, C and D). Specifically, residues R88 and R90 of prM2 and prM3 are engaged in salt bridges with D64 in the pr domain of the neighboring subunit, locking the furin site accessibility. D64 and R90 are strictly conserved across flaviviruses.
BinJV virions can be matured by exogenous furin in vitro
Despite variations upstream from the furin cleavage site, the R88-D64 interaction between the furin site and the neighboring prM is also present in other lineage II ISFs, DENV-4, and possibly other DENV serotypes that have a histidine residue at the equivalent of BinJV position 88. This additional salt bridge could potentially explain the predominance of uncleaved prM in purified BinJV and other viruses sharing this feature. At the sequence level, the cleavage site of BinJV (R-S-R-S-K-R) appears compatible with the recognition site of the proprotein convertase, furin (R-X-R/K-R), suggesting that cleavage should occur if the site is accessible. To empirically assess the capacity for processing, we subjected purified BinJV particles to in vitro furin cleavage at a range of pH conditions (pH 5.0 to 8.0). An immature WNVKUN particle preparation produced using the addition of NH4Cl was included as a VIF control. As determined by SDS-PAGE, cleavage occurred between pH 5.5 and 6.5 for BinJV and pH 5.0 and 6.0 for WNVKUN (Fig. 3A). This is in agreement with previous studies, which have demonstrated that cleavage is dependent on the low pH of the secretory pathway (18–20) and suggest that, like VIFs, BinJV immature virions undergo acid-triggered conformational rearrangements that facilitate furin-like protease processing of prM.
Fig. 3. Maturation of BinJV and WNVKUN particles by in vitro furin cleavage.
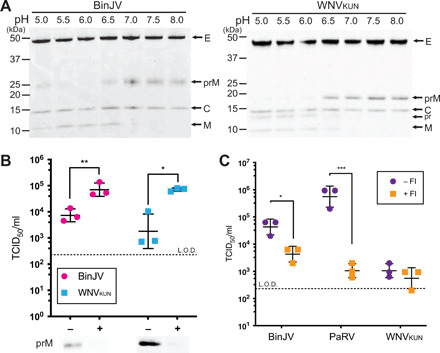
(A) Purified BinJV (left) and immature WNVKUN (right) were incubated with furin in buffer ranging from pH 5 to 8 and analyzed by nonreducing SDS-PAGE and visualized by SYPRO Ruby stain. Viral proteins indicated by arrows. (B) Infectivity of BinJV and immature WNVKUN virions in C6/36 cells with or without furin treatment. Cleavage was analyzed by SDS-PAGE, and virus titers were determined by TCID50. n = 3 biological replicates, and statistical analysis was performed by two-stage multiple t tests against nontreated (furin negative). *P ≤ 0.05 and **P ≤ 0.01. Means ± SD. Limit of detection (L.O.D.) for TCID50 is 2.3 log10 TCID50 ml−1. (C) Infectivity of BinJV, PaRV, and WNVKUN in C6/36 cells pretreated with or without 25 μM furin inhibitor (FI). Virus titers were determined by TCID50; n = 3 biological replicates, and statistical analysis was performed by two-stage multiple t tests against nontreated (FI negative). *P ≤ 0.05 and ***P ≤ 0.001. Means ± SD. Limit of detection for TCID50 is 2.3 log10 TCID50 ml−1.
Earlier studies have shown that in vitro maturation by furin of immature VIFs significantly increases infectivity (9, 20, 21). We tested the impact of cleavage on the infectivity of BinJV particles by infecting C6/36 cells and assessing the output by median tissue culture infective dose (TCID50). We observed that both BinJV and WNVKUN had an approximately 10-fold increase in infectivity when the virus was matured by furin (Fig. 3B). These results confirm that, despite its predominantly immature phenotype, the cleavage of prM enhances BinJV infectivity.
Furin-like activity upon entry is required for ISF replication
Processing of prM is essential for membrane fusion in VIFs (9, 19, 21, 22). Although the BinJV virion does not appear to be readily processed during egress, cleavage may still occur upon cell entry, which would enable the formation of the fusogenic conformation of M and E. A similar maturation pathway has been observed for immature DENV, although this is dependent on antibody-mediated immature virus uptake (13, 23). To test the role of proprotein convertase activity during infectious entry for BinJV, we treated the C6/36 cells before and during infection with decanoyl-RVKR-CMK, a broad spectrum inhibitor of furin-like proprotein convertases. The inhibitor has a half-life of 4 to 8 hours in aqueous solution and is expected not to interfere with the production of initial viral particles (13, 23, 24). Inhibition of furin-like activity caused a significant decrease in BinJV infectivity, while WNVKUN infectivity was unchanged (Fig. 3C). The effect of inhibiting furin was even greater with PaRV, which is a lineage I ISF and also contains high amounts of prM (Fig. 3A) These results suggest that BinJV and PaRV rely on furin-like activity upon entry, indicative of a feature shared across both ISF lineages.
The architecture of the spike is incompatible with a drawstring role for prM
Having shown that, like VIFs, BinJV requires maturation for infectivity, we considered the implications of the revised spike architecture for the maturation process. The full-length prM defines a topology where it is centrally positioned in the trimeric prM-E spike (Figs. 1D and 2, E to J). Homology modeling and sequence comparison indicate that this topology is likely to be conserved across the Flavivirus genus (fig. S3). The peripheral location of prM in previous models resulted from a proposed linkage of pr domains to M domains that belong to the neighboring spikes in our structure (Fig. 2, E and H, and fig. S4). This arrangement suggested a drawstring mechanism where a pulling force exerted by prM would drive a large rotation of the E ectodomains away from the center of the spike (Fig. 2J) (7). In our structure, the positioning of prM is incompatible with a drawstring mechanism. Instead, prM forms a central pillar that holds the spike in an upright position. This is achieved in three ways: (i) The pr domains clamp the top of the spike by tight interactions with the tip of E and extended dimeric interfaces between pr domains of prM2/prM3 (Figs. 1C and 4, A to C); (ii) the furin site seals the gap between prM-E subunits (Fig. 2C); and (iii) the linkers buttress the inside of the spike (Figs. 1D and 2, E and H).
A collapse model of flavivirus maturation
Most of the intraspike interactions involve prM with only one direct contact interface between E molecules. In the absence of these pr-mediated interactions, the spike is not predicted to be stable with only one remaining interface burying just 146 Å2 (fig. S5). Thus, a collapse of the spike does not require a pull force. The role of prM during maturation may instead guide the collapse toward the underlying M, which undergoes little structural change (Fig. 4D and movie S1).
Fig. 4. Proposed model of maturation.
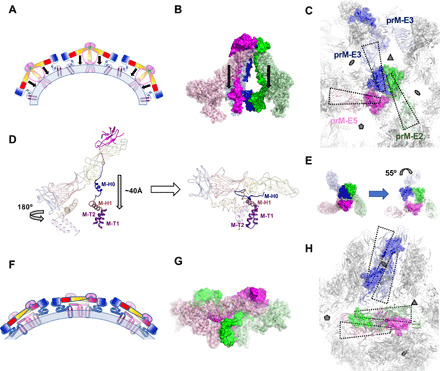
(A and F) A cartoon schematic showing several copies of prM-E arranged on a membrane (light blue), illustrating the collapse maturation model for the immature (A) and mature structure (F). Domains are colored as in Fig. 2. (B) The immature trimer of BinJV colored according to symmetry position with prM and E represented as a solid and semitransparent surfaces, respectively. The collapse model involves a refolding of the pr linker, which guides the E ectodomain toward M (see movie S1). (C and H) The transition from immature (C) to mature (H) structure involves minimal displacement of M and a simple collapse of E (see movie S2). Molecules are allowed to migrate from on icosahedral symmetry position to another (see movie S3).Views from the exterior of the virion. Four prM-E are outlined in wedged boxes and displayed as in (B). For the mature structure, subunits are colored according to their original icosahedral symmetry position as defined in (B) and (C). (D) Comparison of a single prM-E subunit between the immature form (left) to the mature form (right). The translation of pr and the ectodomain of E and the rotation of the E stem/TM are indicated. Domains are colored as in Fig. 2. (E) The pr and E components of the trimeric spike are asymmetric in the immature virion (left). By comparison, the spike would follow a pseudo-threefold symmetry if superposed to the M components of the immature spike (right). The arrow represents the twist of the pr and E component of prM-E3 in the actual immature spike. (G) Side view of a dimer of M-E in the mature structure. Mature structures are modeled from DENV-1 in (D), (G), and (H).
A strictly conserved histidine residue, H243 in ED-II of the E protein, is in close proximity to the prM-prM salt bridges between R88/R90 and D64 (Fig. 2D). Protonation of this residue during egress introduces a buried positive charge that may destabilize the prM-prM dimeric interface and release the furin site, acting as a pH switch to facilitate the maturation process.
How the collapse results in a transition from 60 trimeric spikes to 90 head-to-tail dimers (Fig. 4D and movie S1) may be explained by key elements in the BinJV structure. First, we observe that, as in immature particles, the spike is present as an asymmetric trimer. Departure from a low-energy, symmetrical trimer results from a ~55° twist of the pr/E components of prM-E3 (Fig. 4E). This arrangement is locked in place by a large dimeric interface between the pr domains of prM2 and prM3 at the top of the spike (Fig. 4C). By contrast, the membrane-anchored M domains follow a quasi-threefold symmetry.
At the level of the prM-E subunits, maturation would thus result from dissociation of the pr dimeric interface, release of the furin site, and collapse of the prM linker (movie S2). In this model, refolding of M-H0 region of the linker guides the rest of the prM-E subunit onto the underlying M (Fig. 4D). At the level of the spike, formation of dimers requires prM-E3 to move out of the way during collapse. This is compatible with a release of the 55° twist in prM-E3 (Fig. 4E) to allow the formation of a dimer of prM-E5 and prM-E2 (Fig. 5, F to H, and movie S2). The released prM-E3 can associate with any of the two other prM-E3 located around the icosahedral threefold symmetry axis implying different movements for icosahedrally equivalent molecules. We modeled these transitions for the three spikes as shown in fig. S6 and movie S3. By contrast with previous models of flavivirus maturation, the rearrangements do not require a ~180° rotation of the projecting E ectodomains, and M subunits associate with nearby counterparts. The collapse described above for prM-E is also compatible with rearrangements at the particle level so that all molecules may undergo similar conformation changes, driving the transition from a projecting spike to a membrane lining dimer. However, the newly formed dimeric subunits undergo translational displacements that differ between prM-E subunits, resulting in their redistribution compared to the icosahedral symmetry axes and a temporary break of the icosahedral symmetry. Two of the prM-E molecules closest to a threefold icosahedral symmetry axis move to form a dimer around a twofold icosahedral symmetry axis (blue prM-E3 molecules in Fig. 4C versus Fig. 4H)), while prM-E2 remains close to a threefold symmetry axis. A 180° rotation of the TM and stem region of E allows the formation of a tight bundle with the TMs of M (Figs. 2E and 4D; fig. S6, D to F; and movie S3).
Fig. 5. The pr-specific antibody 2A7 inhibits BinJV in an isotype-dependent manner.
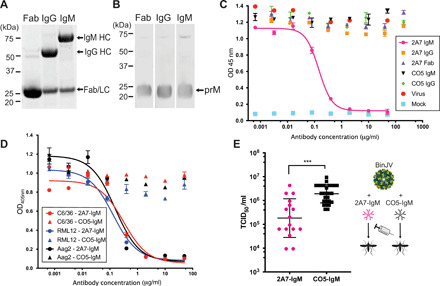
(A) Purified 2A7 in Fab, IgG, and IgM form analyzed by reduced 4 to 12% SDS-PAGE. Heavy (HC) and light chains (LC) indicated by arrows. (B) Western blot probing with recombinant 2A7-Fab (before HRV3C digestion), IgG, and IgM. Purified BinJV from infected C6/36 cells was resolved by nonreducing 4 to 12% SDS-PAGE. (C) Neutralization of BinJV by 2A7-Fab, IgG, and IgM antibodies or anti-influenza CO5 antibody as isotype controls. Neutralization curves were determined by TCID50 on C6/36 cells. Means ± range. OD, optical density. (D) Neutralization by 2A7-IgM or control CO5-IgM of BinJV produced in C6/36 (A. albopictus), RML12 (A. albopictus), and Aag2 (A. aegypti) mosquito cells with 2A7-IgM or control CO5-IgM. Neutralization was determined by TCID50 on C6/36 cells. Means ± range. (E) Viral titers in A. albopictus mosquitoes injected intrathoracically with BinJV:2A7-IgM or BinJV:CO5-IgM. BinJV supernatant was incubated with either antibody (2A7-IgM or CO5-IgM) at 28°C for 1 hour before injection. Each point represents a single infected mosquito; means ± SD. Statistical analysis performed by unpaired t test with Welch’s correction. ***P ≤ 0.001.
An anti-prM IgM inhibits BinJV infection in cell culture and Aedes albopictus mosquitoes
To clarify the relationship between the maturation state and infectivity, we investigated the effect of monoclonal antibodies (mAbs) against BinJV structural proteins on these two viral processes. Notably, an immunoglobulin M (IgM) mAb named 2A7 that recognized prM was neutralizing in vitro (table S2). To determine whether the neutralization activity was isotype dependent, we cloned the variable domains into a human IgM and IgG backbone (Fig. 5A). All three recombinant formats bind to prM (Fig. 5B). Neutralization assays using the panel of 2A7 isotype variants revealed that only the IgM form was capable of inhibiting BinJV (Fig. 5C). These results were confirmed for BinJV produced from three different insect cell lines (C6/36, RML12, and Aag2) (Fig. 5D).
To assess the role of immature BinJV particles in vivo, we investigated the potential for the pr-specific 2A7-IgM to inhibit infection of A. albopictus mosquitoes. C6/36 cell–produced BinJV was incubated with either 2A7-IgM or a control IgM before intrathoracic injection. Mosquitoes were harvested at 3 days post infection (dpi), and virus levels were determined via TCID50 on C6/36 cells. This revealed that 2A7-IgM had a significant inhibitory effect on BinJV replication compared to the control antibody (Fig. 5E). Overall, this supports that BinJV is producing an infectious immature particle and that 2A7-IgM has the ability to neutralize this infectious virion.
The anti-prM 2A7-IgM inhibits maturation
To elucidate the neutralization mechanism of 2A7, the cryo-EM structure of BinJV in complex with a recombinant Fab was determined to a resolution of 9.7 Å (Fig. 6, A and B, and fig. S7). The viral architecture did not appear to be altered by the Fab binding. The 2A7 epitope is conformational, spanning the a, b, and c strands on the outer face of BinJV pr (Fig. 6, C and D). These regions are not conserved across linage I ISFs or VIFs, accounting for the lack of cross-reactivity observed for 2A7 (table S2). The 2A7-Fab engaged all 180 pr molecules in the cryo-EM reconstruction. Since the Fab and IgG forms of 2A7 do not have neutralization activity (Fig. 5C), the mechanism is unlikely to involve a block of a hypothetical receptor binding site in pr.
Fig. 6. Cryo-EM reconstruction of BinJV:2A7.
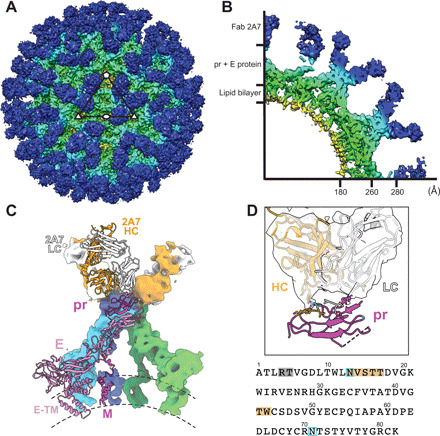
(A) Surface and (B) wedge cross-section of the cryo-EM reconstruction of BinJV:2A7-Fab. Map was colored according to radius (red, 0 to 30 Å; yellow, 31 to 140 Å; green, 141 to 180 Å; cyan, 181 to 280 Å; and blue, >280 Å). (C) Fit of a homology model of the 2A7-Fab and the structure of BinJV within the reconstruction of the complex. Dimers of prM-E are colored according to symmetry position: twofold with green/lime for prM/E, threefold with blue/cyan for prM/E, and fivefold with magenta/pink for prM/E. The HC and LC of the 2A7-Fab is represented in orange and white, respectively. Surfaces for one member of the asymmetric unit are omitted to show a cartoon representation. (D) Zoom of the pr:2A7 interface with the sequence of the N-terminal domain of pr displayed below. Residues of pr that are within 7 Å of the Fab are indicated by dark orange (HC) and gray (LC). N-linked glycan sites of pr are highlighted in light blue.
We next examined the role of 2A7 on prM cleavage. Purified BinJV was incubated with equimolar concentrations of antibody before incubating with or without furin. The addition of 2A7-IgM inhibited furin cleavage, while the IgG and Fab forms of 2A7 did not affect the furin processing (Fig. 7, A to C). Since binding is distal from the furin site and not predicted to prevent access to the 50-kDa furin protein, the maturation block induced by 2A7 is allosteric. We propose that it functions by cross-linking spikes, preventing conformational changes triggered by acidification (Fig. 7D).
Fig. 7. 2A7 blocks in vitro maturation in an isotype-dependent manner.
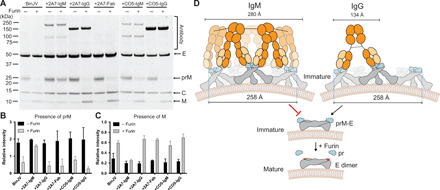
(A) Purified BinJV was incubated with anti-prM 2A7 antibodies (IgM, IgG, and Fab) or the control antibody CO5 (IgM and IgG) for 1 hour before incubating with furin and analyzing by SYPRO Ruby–stained SDS-PAGE. Virus and antibodies were mixed at equimolar concentrations. Viral proteins are indicated by arrows, and antibody fragments are in brackets. The relative intensity of the prM (B) and M (C) bands following incubation with and without furin was quantified. n = 2 independent experiments; means ± SD. (D) Model of neutralization of immature BinJV by the 2A7-IgM. Schematic representation of five prM-E trimers that are presented in gray (E) and blue (pr) surrounding the fivefold axis. Multimeric binding of IgM antibody are proposed to cross-link the prM-E spikes inhibiting low pH–mediated release of the furin site during endocytosis. Distance between antigen binding sites of IgM (51) and IgG (52) antibody isotypes and the span across the fivefold axis is annotated.
DISCUSSION
Here, we show that the predominant form of the ISF BinJV is immature and, through the use of a pr-specific antibody, we demonstrate that this form is infectious in vitro and in vivo. Our findings suggest that cleavage is required for infection, but this likely takes place during cell entry (fig. S8). Maturation during cellular uptake has been previously observed for DENV, but in contrast to BinJV, immature DENV requires antibody-mediated uptake in Fc-γ receptor–bearing cells to facilitate infectious cell entry. In the case of BinJV, prM processing occurring during entry rather than egress may be explained by receptor binding–mediated conformational changes, which have been shown to increase furin-like processing for other viruses (25). Thus, the cellular receptor(s) of BinJV either recognizes pr directly or, contrary to VIFs, it is able to bind the receptor-binding domain of E in the immature-like structure. It is also possible that the subcellular location of insect furin-like proteases may contribute to entry-dependent prM processing. However, given that VIF maturation is efficient in insect cells and we observed equivalent in vitro furin processing for both BinJV and WNVKUN, other factors likely contribute to the high levels of immature particles observed for BinJV and ISFs more generally.
Taking advantage of the homogeneity of BinJV virions, we determined a structure of an immature flavivirus that reveals a complete spike including the prM linker that was disordered in all available VIF structures, including the recent high-resolution locally reconstructed immature Spondweni virus by Renner et al. (26). Unexpectedly, we find that the prM-E spike in the BinJV virion structure has a different connectivity from the current model of immature flaviviruses (16, 17, 19). Homology modeling and conservation of key residues suggest that this topology is shared among the entire flavivirus family. The structure reveals three features important for the role of the immature spike in forming a particle that is both sufficiently stable and primed for maturation. First, extensive interactions between the globular pr domains of two prM proteins maintain the spike in an asymmetric conformation, priming it for maturation. Second, the prM region including the furin site not only is buried in interactions with the tips of the E proteins and pr but also contributes to the stability of the spike. This organization provides a coupling between a dissociation of the spike and exposure of the furin cleavage site. Third, the prM stem provides structural support at the center of the spike rather than pulling away from it. Notably, this architecture is incompatible with the proposed drawstring maturation model (7), which involves large movements of prM and a 180° rotation of the E ectodomain. In a model based on our observed topology of the spike, the prM function is analogous to a ripcord. Dissociation of the clamp at the top of the spikes releases the furin site and initiates a 40-Å collapse of E onto the membrane guided by the prM linker without the requirement for large migration of M or a 180° rotation of E (movies S1 and S2). The collapse rearrangement of the spike is likely to be similar for all molecules in the virion. However, topological constraints to prevent clashes and tangles of prM molecules mean that some of the prM-E complexes migrate away from their original symmetry positions. Initiation of the transition at the fivefold axis minimizes clashes and displacements in our model (fig. S8 and movie S3), and it will be interesting to see whether this is supported by experimental evidence. Away from this site, larger sliding motions are required, and symmetry mismatches must be temporarily generated.
The valency-dependent inhibition observed for the pr-specific 2A7 mAb conforms with two aspects of a ripcord maturation mechanism: the requirement for conformational changes in the pr region of the spike to allow exposure of the furin site and a potential initiation of maturation at the fivefold axis, which is propagated through the rest of the particle by migration of prM-E subunits away from their original symmetry position. All forms of the antibody are able to recognize prM and do not occlude directly the furin site. Since the binding site of 2A7 is distal to the cleavage site in the linker region, inhibition must occur through allosteric mechanisms. The splayed angle of Fab approach suggests that 2A7-IgG bivalent interactions may reach across two prM-E trimers but not prM within the same trimer. Irrespective of whether it is mono- or divalent, this interaction is insufficient to effectively inhibit furin cleavage. Only the larger pentameric IgM is able to cross-link more than two adjacent spikes. This is likely to lock the trimer in the up position and prevent both the initial collapse at the IgM bound sites and its propagation through the particle if initiated elsewhere (Fig. 7D).
While these findings involve an ISF, they have potential implications for pathogenic flaviviruses where immature and partially mature virions have been implicated in disease severity. Previous studies have used IgG forms of pr-specific antibodies to assess the neutralization and ADE (13). Our findings show that pr-specific IgM antibodies can block furin processing on entry. They may therefore offer protection against ADE in the case of DENV infection, where a significant fraction of the virions are immature. As IgM antibodies predominate the initial response in naïve individuals, pr-specific responses may offer some level of disease protection in primary infection. While in secondary infection, early class switching to IgG would remove this protective effect and contribute to enhanced disease.
MATERIALS AND METHODS
Cell and virus culture
C6/36 cells (A. albopictus; American Type Culture Collection CRL1660) were maintained in RPMI 1640 supplemented with 5% fetal bovine serum (FBS) at 28°C. RML-12 cells (A. albopictus) were cultured in Leibovitz’s L-15 medium supplemented with 10% FBS and 10% tryptose phosphate buffer at 28°C. Aag2 cells (Aedes aegypti) were maintained in medium with a 1:2 mixture of Mitsuhashi-Maramorosch and Schneider’s insect media (Invitrogen) supplemented with 10% FBS at 28°C. All media contained streptomycin (50 μg ml−1), penicillin (50 U ml−1), and l-glutamine (2 mM). The viruses used in this study include BinJV (GenBank MG587038), WNVKUN (GenBank JN887352), PaRV (GenBank KT192549) (15), and HVV (GenBank MN954647) (27). All viruses were propagated on C6/36 cell monolayers at a multiplicity of infection (MOI) of 0.1 and harvested 5 dpi. Virus titers were determined by TCID50 and assessed by fixed-cell enzyme-linked immunosorbent assay (ELISA) using the anti-NS1 antibody 4G4 (28), as previously described (5).
Virus purification
C6/36 cells were infected with BinJV, WNVKUN, PaRV, and HVV for 1 hour at 28°C at an MOI of 0.1. BinJV, HVV, and WNVKUN were harvested 4 and 7 dpi. PaRV was harvested 5, 7, and 9 dpi. Immature particles of WNVKUN were generated by replacing the media 1 dpi with medium containing 20 mM NH4Cl and harvested 5 dpi. Each virus was precipitated by polyethylene glycol (8% PEG 8000) by gently mixing overnight before centrifugation for 1 hour at 4°C and 8000 revolutions per minute (rpm) using an Avanti J-26 JLA10.5 rotor. Virus pellet was resuspended in NTE [10 mM tris (pH 8), 120 mM NaCl, and 1 mM EDTA (pH 8)] buffer before ultracentrifugation through a 20% sucrose cushion and left in NTE (600 μl) overnight. The precipitated virus pellet was resuspended in NTE and then clarified at 5000g for 10 min, with the resulting supernatant put through 25 to 40% potassium tartrate gradient. The virus bands resolved by the gradient were harvested and buffer exchanged into NTE. The purity and concentration of purified virions were determined by resolving proteins using SDS-PAGE (4 to 12% bis-tris; Life Technologies). Proteins were stained using SYPRO Ruby stain (Invitrogen) following the manufacturers’ guidelines. Serial dilutions of bovine serum albumin standards (Thermo Fisher Scientific) were used to determine the concentration of E protein.
PNGase F digestion
For treatment with PNGase F [New England Biolabs (NEB)], purified virions were separated in glycoprotein denaturing buffer (1×) and H2O to make a total volume of 20 μl. The reaction was then heated at 100°C for 10 min and chilled before adding GlycoBuffer 2 (1×), 10% NP-40, and H2O with or without 500 U of PNGase F. For digestion to occur, the reaction was incubated at 37°C for 1 hour. The digestion was analyzed by 4 to 12% SDS-PAGE and SYPRO Ruby stain.
Single-particle cryo-EM of BinJV
An aliquot of 3.5 μl of purified viral particles was applied to a glow-discharged R1.2/1.3 holey carbon grid (Quantifoil Micro Tools GmbH, Germany). The grid was blotted (2.5-s blot time, −4 blot force, and 1-s drain) at 100% relative humidity (RH) and plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI/Thermo Fisher Scientific). Grids were transferred under liquid nitrogen to a Titan Krios transmission EM (FEI/Thermo Fisher Scientific) operated at 300 kV and set for parallel illumination. Movies were recorded using EPU 2 (FEI) on a K2 Summit direct electron detector (Gatan Inc., USA) in super-resolution mode with energy filtering. The data collection parameters are summarized in table S1.
The movies were binned two times by Fourier cropping before motion correction and integrated with MotionCor2 (v1.1.0) (29), giving a final pixel size of 1.34 Å. The contrast transfer function (CTF) parameters of each image were determined using Gctf (v. 1.06) (30), and images with significant astigmatism or drift were removed. The remaining micrographs were used for particle picking and three-dimensional (3D) reconstruction.
Using RELION (v. 3.1) (31), particles were manually selected from a subset of micrographs and extracted for 2D classification to generate templates for autopicking of the entire dataset. Reference-free 2D classification was sufficient to remove the disordered particles for 3D refinement. A reconstruction of immature DENV1 (EMD-2141) (16) was low-pass–filtered to 20 to 30 Å as a reference for the first 3D refinement. A soft mask excluding the solvent, internal capsid, and genome was used to improve alignment. Before subsequent rounds of 3D refinement, the defocus and motion of each particle were refined as well as the astigmatism and anisotropic magnification of the micrograph. The final map was filtered according to its local resolution using a B-factor estimated from the post-processed masked map.
Modeling and refinement of the BinJV structure
An initial model was produced by alignment of the reference and target sequences using Clustal Omega (32) and mapping of the sequence alignment onto an existing structure using Sculptor (33) from the Phenix suite (v. 1.16) (34). Different reference models were used for each component: YFV [E and pr; Protein Data Bank (PDB): 6EPK], DENV-2 (pr; PDB: 36CE), and TBEV (M; 5O6V). Modeling was performed in Coot (v. 0.8.9.1) (35), and the model was refined in real space with the phenix.real_space_refine program (PHENIX v. 1.16) (34) using secondary structure and noncrystallographic symmetry restraints. The geometry and quality of the model were evaluated using a combination of MolProbity (36) and Phenix. PyMOL (the PyMOL Molecular Graphics System, version 2.3.0, Schrödinger LLC) and University of California, San Francisco (UCSF) Chimera (v. 1.14) (37) were used to render images of the structure. The buried surfaces were calculated by using the Proteins, Interfaces, Structures, and Assemblies server (38). To visualize the location of side chains in DENV-1 prM, a homology model was generated using the Phyre2 one-to-one threading server (39) with the prM structure as a reference. Modeling of the transition between the immature and mature forms of the virion was performed by morphing in PyMOL. Morphing was performed on a single spike representing the asymmetric unit of the virion, providing only the initial and final structures consisting of our BinJV immature structure and a model of a low-pH DENV-1 virion with the pr domain attached, respectively. To account for a temporary break of symmetry in the intact virion, we modeled the transition for three spikes around a threefold axis and expanded the trajectories to other molecules in third of the virion by applying icosahedral symmetries. In addition to the initial and final structures, an intermediate structure manually generated in PyMOL was provided to guide the morphing and limit clashes. Movie clips were generated in PyMOL and QuickTime and merged into a single movie in iMovie.
Analysis of furin cleavage
Approximately 12 μg of purified immature BinJV and WNVKUN virions was adjusted to the desired pH (pH 5.0 to 8.0) using 1 M MES in NTE. Furin (NEB) was added according to the concentration of E (0.2 U per μg) along with 3 mM CaCl2 and incubated at 30°C for 16 hours. Cleavage was observed by resolving the protein by nonreducing SDS-PAGE using SYPRO Ruby stain as described above. To assess infectivity of the virions after furin cleavage, each reaction was prepared in either pH 6 or 8 conditions, with one-fifth of the reaction (~2 μg of whole protein) assessed by SDS-PAGE and the remaining reaction (~10 μg of whole protein) used to infect C6/36 cells. The cells were seeded at a density of 1 × 105 in a 24-well plate 24 hours before infection. Virus was diluted in 2% FBS RPMI 1640, and monolayers were inoculated for 1 hour at 28°C. Following infection, the cells were washed in triplicate with sterile phosphate-buffered serum (PBS) and replaced with 2% FBS RPMI 1640 growth medium. The supernatant was harvested 2 dpi, and the virus titers were established using TCID50 as described above. To assess cleavage when the virus is bound to Fab, IgG, and IgM antibodies, the virus was initially incubated with molar equivalent ratios of each antibody at 4°C for 2 hours. The virus and antibody complex were then prepared as described above.
Furin inhibition
C6/36 cells were seeded 1 × 105 in a 24-well plate 24 hours before infection. One hour before infection, the cells were treated with 25 μM furin-specific inhibitor (decanoyl-RVKR-CMK; Merck). Each virus had a nontreated control. Cells were infected at an MOI of 1 for 1 hour at 28°C. The cell monolayer was washed in triplicate with sterile PBS, and 2% FBS RPMI 1640 growth medium was added and incubated at 28°C. Virus supernatant was harvested at 24 hours, and the titers were assessed by TCID50 as described above. To probe for each virus, anti-NS1 antibody 4G4 (28) was used for BinJV, and WNVKUN and anti-E antibody 7D11 were used for PaRV (40).
mAb production
To generate antibodies that recognized the prM protein of BinJV, BALB/c mouse (Animal Resources Centre) were immunized with two doses of purified BinJV and inulin-based adjuvant Advax (Vaxine Ltd.) by subcutaneous injection. The mouse was administered a booster dose intravenously 4 days before harvesting the spleen. All animal procedures were approved by the University of Queensland Animal Ethics Committee. The spleen cells were fused with NS0 myeloma cells (European Collection of Cell Cultures) as previously described (27). BinJV-reactive antibodies were selected on the basis of positive reactivity in fixed-cell ELISA. Hybridomas were maintained at 37°C with 5% CO2 in hybridoma serum-free media supplemented with streptomycin (50 μg ml−1), penicillin (50 U ml−1), and 2 mM l-glutamine with 20% FBS initially and then weaned to serum-free.
Recombinant antibody and Fab production
The cellular RNA from BinJV 2A7 hybridoma cells was obtained using a FavorPrep Tissue Total RNA kit. The variable domains were amplified using a set of primers (41) with the successful products cloned into pUC19-infusion vector using infusion cloning (Clontech) and checked for the complete open reading frame (note S1). The putative variable domains were then recloned into human constant domain expression scaffolds for transient expression in Chinese hamster ovary (CHO) cells (42–44). For 2A7-Fab, a human rhinovirus 3C protease (HRV3C) site was inserted at the heavy chain (HC) hinge region (45). The equimolar ratio of HC and light chain (LC) plasmids were transfected with polyethylenimine (4 mg of PEI per 1 μg of plasmid DNA) for 4 hours before replacing the media with CHO growth media supplemented with 8 mM GlutaMAX (Invitrogen) and Feeds A and B at 7.5% (v/v). The CHO cells were incubated for 7 days, shaking at 120 rpm, 37°C with 7.5% CO2. After 7 days, the supernatant was clarified and filtered before protein A purification (HiTrap Protein A, GE healthcare). IgM was purified using a 1 ml HiTrap N-hydroxysuccinimide–activated high-performance column coupled with anti-IgM antibody, DA4 (46). The antibodies were buffer exchanged into PBS. 2A7-Fab was digested using the GST-HRV3C protease (47) for 48 hours at 37°C before removing the Fc fragment and protease using protein A–sepharose 4B (Life Technologies) and glutathione S-transferase-agarose (Pierce). All antibodies were assessed for purity using reducing SDS-PAGE and binding ability by ELISA and Western blot.
Single-particle cryo-EM of the BinJV-2A7 complex
Purified BinJV was complexed with 2A7-Fab at a molar ration of 2:1 Fab:E protein and incubated at 4°C for 2 hours. An aliquot of 4 μl of complex was applied to a glow-discharged 400-mesh holey carbon–coated grid (ProSciTech). The grid was blotted (7-s blot time) at 100% RH and plunge-frozen in liquid ethane using a Vitrobot MKII (FEI/Thermo Fisher Scientific). Grids were transferred under liquid nitrogen to a Tecnai F30 G2 transmission EM (FEI/Thermo Fisher Scientific) operated at 300 kV and set for parallel illumination. Movies were recorded using SerialEM (48) on a K2 Summit direct electron detector (Gatan Inc., USA) in counting mode. The data collection parameters are summarized in table S1.
The movies were motion corrected and integrated with MotionCor2 (v1.1.0) (29), and the CTF parameters of each image were determined using CTFFIND (v. 4.1) (49). Using RELION (v. 2.1) (50), particles were manually selected from the micrographs and extracted for 2D classification with a box size of 384 pixels. A reconstruction of an immature flavivirus virion (EMD-20439) was low-pass–filtered to 60 Å and used as a reference for 3D classification of the best 2D classes. The most ordered class from the 3D classification was further refined using the RELION 3D autorefine procedure. The half-maps were masked with a spherical mask that excluded the capsid density, and the Fourier Shell Correlation (FSC) was calculated using the post-processing procedure in RELION with the gold-standard FSC cutoff of 0.143.
To visualize the interface of 2A7 and BinJV, a homology model of 2A7 was generated using the Phyre2 one-to-one threading server (39). A Fab crystal structure (PDB: 5F3B) was used as a reference model (with 92.3 or 76.9% sequence identity to the heavy or light chains, respectively). The homology model was docked into the map with the BinJV structure using sequential rigid-body fitting in UCSF Chimera.
Microneutralization assays
Antibodies were titrated by serial 10-fold dilutions for hybridoma supernatant (starting undiluted) or fivefold dilutions for recombinant antibodies (starting 50 or 200 μg ml−1) in 50 μl of 2% FBS RPMI 1640 media. BinJV was added at 100 TCID50 U per well, and incubated for 1 hour at 28°C. C6/36 wells were added at 1 × 104 per well and incubated for 48 hour at 28°C. The presence of virus was detected in ELISA using murine anti-NS1 mAb, 4G4 (28).
Western blot
Lysates of virus-infected C6/36 cells were prepared by incubating cells monolayers (3.5 ml per 107 cells) with NP-40 lysis buffer [1% NP-40, 50 mM tris-Cl, 150 mM NaCl, and 2 mM EDTA (pH 7.5); 1:1000; Sigma-Aldrich protease inhibitor] at 4°C for 30 min. Lysate was clarified by centrifugation at 14,000g for 10 min at 4°C and stored at −80°C. The lysate or purified virus was resolved by electrophoresis on a 4 to 12% bis-tris SDS-PAGE gel (Life Technologies) before transferring onto a nitrocellulose membrane. The membrane was blocked in TENTC blocking buffer [0.05 M Tris-HCl (pH 8.0), 1 mM EDTA, 0.15 M NaCl, 0.05% (v/v) Tween-20, and 0.2% (w/v) casein] before probing with the appropriate antibody for 1 hour. After washing with TBST (tris-buffered saline and 0.05% Tween 20) in triplicate, the membrane was incubated for 1 hour with a 1:8000 dilution of goat anti-mouse or goat anti-human IgG (DyLight 800 4× PEG conjugate; Thermo Fisher Scientific). The membrane was once again washed in triplicate with TBST with a final wash with H2O and imaged with the Odyssey Imaging System (LI-COR).
Maintenance of mosquitoes
A. albopictus mosquitoes were obtained from a colony established in the QIMR Berghofer Medical Research Insectary from collections made from Hammond Island, Torres Strait, in 2014 and 2015. The colony was maintained at 27°C, 70% RH, and 12-hour light/12-hour dark cycle with 30-min crepuscular periods. For these experiments, eggs were hatched in rainwater, and larvae were transferred to plastic trays (48 cm by 40 cm by 7 cm) at a density of 300 larvae per tray in 3 liters of rainwater. The larvae were fed ground TetraMin Tropical Flakes fish food (Tetra, Melle, Germany) ad libitum. Pupae were transferred to containers of rainwater inside 30 cm by 30 cm by 30 cm cages (BugDorm, MegaView Science Education Services Co., Taichung, Taiwan). Adult mosquitoes were provided with cotton balls soaked with 10% sugar solution.
Intrathoracic inoculation of A. albopictus mosquitoes
Before injection, BinJV at 105.8 TCID50 ml−1 was incubated with 2A7-IgM or control IgM (0.31 mg ml−1) at 28°C for 1 hour in 2% FBS RPMI 1640 cell culture medium. Female A. albopictus mosquitoes (3 to 5 days old) were aspirated from cages, anesthetized with CO2, and placed on a petri dish on ice. The mosquitoes were intrathoracically injected with 100 nl of BinJV:IgM (approximately 60 TCID50 U per mosquito) using a programmable nanoliter injector (Nanoject III, Drummond Scientific, Broomall, PA, USA). The injected mosquitoes were transferred to 750-ml plastic screw-top containers with gauze lids and provided with 10% sucrose solution on cotton balls. The containers of mosquitoes were maintained in an environmental chamber (HiPoint 740HLED, HiPoint Corporation, Taiwan) set at 28°C, 75% RH, and 12-hour light/12-hour dark cycling with 30-min crepuscular periods. The mosquitoes were harvested at 3 dpi from the containers, anesthetized with CO2, and placed in a petri dish on ice. Single whole mosquitoes were transferred to 2-ml screw cap vials together with three to four 2.3-mm zirconium silica beads and 500 μl of 2% FBS RPMI 1640 with 0.1% amphotericin B (Fungizone, Life Technologies Inc., Grand Island, NY). The mosquitoes were homogenized using a Mini-BeadBeater 96 (BioSpec Products, Bartlesville, OK, USA). The samples were then clarified by centrifuging at 15,000g, 4°C for 10 min, and 400 μl of supernatant was transferred to sterile tubes to be stored at −80°C. To determine the titer of BinJV from the mosquito homogenates, an additional 600 μl of 2% FBS RPMI 1640 with 0.1% amphotericin B was added and filtered through a 0.2-μm syringe filter (Pall Acrodisc). The supernatant was titrated 10-fold, and the titer was calculated by TCID50.
Acknowledgments
We acknowledge the facilities and the scientific and technical assistance of the Ramaciotti Centre for Cryo-Electron Microscopy at Monash University, the MASSIVE HPC facility (www.massive.org.au) and the Australian Microscopy and Microanalysis Research Facility at the Centre for Microscopy and Microanalysis, University of Queensland. We acknowledge the technical assistance provided by H. Bielefeldt-Ohmann for the generation BinJV-specific antibodies. Funding: This work was supported by NHMRC project grants APP1164216 and APP1138611 and NHMRC ideas grant APP2004582. Author contributions: N.D.N., J.M.H., R.A.H., J.H.-P., F.C., and D.W. conceived and designed the study. N.D.N. prepared and purified the virus samples. N.D.N. and A.A.A. performed the functional in vitro assays. N.D.N. and L.E.H. performed the in vivo experiments. N.D.N., J.J.H., and J.H.-P. isolated the monoclonal antibodies. N.D.N., N.M., S.B., R.J.T., and D.W. produced the recombinant antibodies. J.M.H., N.D.N., and D.W. screened the samples for cryo-EM. J.M.H., N.D.N., D.W., and H.V. collected the cryo-EM data. J.M.H. and N.D.N. performed the cryo-EM data processing, refinement, and model building with assistance from F.C. and D.W. F.C. performed modeling and visualization of the pH-induced virion maturation. N.D.N., J.M.H., F.C., and D.W. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Materials relating to the chimeric viral system can be provided by the University of Queensland pending scientific review and a completed material transfer agreement. Requests for chimeric viruses or related materials should be submitted to D.W. (d.watterson@uq.edu.au), J.H.-P. (j.peters2@.uq.edu.au), or R.A.H. (roy.hall@uq.edu.au).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/20/eabe4507/DC1
REFERENCES AND NOTES
- 1.Pierson T. C., Diamond M. S., The continued threat of emerging flaviviruses. Nat. Microbiol. 5, 796–812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver S. C., Barrett A. D. T., Transmission cycles, host range, evolution and emergence of arboviral disease. Nat. Rev. Microbiol. 2, 789–801 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blitvich B. J., Firth A. E., Insect-specific flaviviruses: A systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses 7, 1927–1959 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cammisa-Parks H., Cisar L. A., Kane A., Stollar V., The complete nucleotide sequence of cell fusing agent (CFA): Homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology 189, 511–524 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Hobson-Peters J., Harrison J. J., Watterson D., Hazlewood J. E., Vet L. J., Newton N. D., Warrilow D., Colmant A. M. G., Taylor C., Huang B., Piyasena T. B. H., Chow W. K., Setoh Y. X., Tang B., Nakayama E., Yan K., Amarilla A. A., Wheatley S., Moore P. R., Finger M., Kurucz N., Modhiran N., Young P. R., Khromykh A. A., Bielefeldt-Ohmann H., Suhrbier A., Hall R. A., A recombinant platform for flavivirus vaccines and diagnostics using chimeras of a new insect-specific virus. Sci. Transl. Med. 11, eaax7888 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Kuhn R. J., Zhang W., Rossmann M. G., Pletnev S. V., Corver J., Lenches E., Jones C. T., Mukhopadhyay S., Chipman P. R., Strauss E. G., Baker T. S., Strauss J. H., Structure of dengue virus: Implications for flavivirus organization, maturation. Cell 108, 717–725 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X., Ge P., Yu X., Brannan J. M., Bi G., Zhang Q., Schein S., Zhou Z. H., CryoEM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 20, 105–110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plevka P., Battisti A. J., Junjhon J., Winkler D. C., Holdaway H. A., Keelapang P., Sittisombut N., Kuhn R. J., Steven A. C., Rossmann M. G., Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 12, 602–606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elshuber S., Allison S. L., Heinz F. X., Mandl C. W., Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus FN1. J. Gen. Virol. 84, 183–191 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Stadler K., Allison S. L., Schalich J., Heinz F. X., Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71, 8475–8481 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modis Y., Ogata S., Clements D., Harrison S. C., Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Pierson T. C., Diamond M. S., Degrees of maturity: The complex structure and biology of flaviviruses. Curr. Opin. Virol. 2, 168–175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodenhuis-Zybert I. A., van der Schaar H. M., da Silva Voorham J. M., van der Ende-Metselaar H., Lei H.-Y., Wilschut J., Smit J. M., Immature dengue virus: A veiled pathogen? PLOS Pathog. 6, e1000718 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crabtree M. B., Sang R. C., Stollar V., Dunster L. M., Miller B. R., Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Arch. Virol. 148, 1095–1118 (2003). [DOI] [PubMed] [Google Scholar]
- 15.McLean B. J., Hobson-Peters J., Webb C. E., Watterson D., Prow N. A., Nguyen H. D., Hall-Mendelin S., Warrilow D., Johansen C. A., Jansen C. C., van den Hurk A. F., Beebe N. W., Schnettler E., Barnard R. T., Hall R. A., A novel insect-specific flavivirus replicates only in Aedes-derived cells and persists at high prevalence in wild Aedes vigilax populations in Sydney, Australia. Virology 486, 272–283 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Kostyuchenko V. A., Zhang Q., Tan J. L., Ng T.-S., Lok S.-M., Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J. Virol. 87, 7700–7707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad V. M., Miller A. S., Klose T., Sirohi D., Buda G., Jiang W., Kuhn R. J., Rossmann M. G., Structure of the immature Zika virus at 9 Å resolution. Nat. Struct. Mol. Biol. 24, 184–186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L., Lok S.-M., Yu I. M., Zhang Y., Kuhn R. J., Chen J., Rossmann M. G., The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science 319, 1830–1834 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Yu I.-M., Zhang W., Holdaway H. A., Li L., Kostyuchenko V. A., Chipman P. R., Kuhn R. J., Rossmann M. G., Chen J., Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319, 1834–1837 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Yu I. M., Holdaway H. A., Chipman P. R., Kuhn R. J., Rossmann M. G., Chen J., Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J. Virol. 83, 12101–12107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zybert I. A., van der Ende-Metselaar H., Wilschut J., Smit J. M., Functional importance of dengue virus maturation: Infectious properties of immature virions. J. Gen. Virol. 89, 3047–3051 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Heinz F. X., Stiasny K., Püschner-Auer G., Holzmann H., Allison S. L., Mandl C. W., Kunz C., Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198, 109–117 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Rodenhuis-Zybert I. A., Moesker B., da Silva Voorham J. M., van der Ende-Metselaar H., Diamond M. S., Wilschut J., Smit J. M., A fusion-loop antibody enhances the infectious properties of immature flavivirus particles. J. Virol. 85, 11800–11808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garten W., Hallenberger S., Ortmann D., Schäfer W., Vey M., Angliker H., Shaw E., Klenk H. D., Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie 76, 217–225 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Walls A. C., Tortorici M. A., Snijder J., Xiong X., Bosch B.-J., Rey F. A., Veesler D., Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 114, 11157–11162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renner M., Dejnirattisai W., Carrique L., Martin I. S., Karia D., Ilca S. L., Ho S. F., Kotecha A., Keown J. R., Mongkolsapaya J., Screaton G. R., Grimes J. M., Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins. Nat. Commun. 12, 1238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison J. J., Hobson-Peters J., Colmant A. M. G., Koh J., Newton N. D., Warrilow D., Bielefeldt-Ohmann H., Piyasena T. B. H., O’Brien C. A., Vet L. J., Paramitha D., Potter J. R., Davis S. S., Johansen C. A., Setoh Y. X., Khromykh A. A., Hall R. A., Antigenic characterization of new lineage II insect-specific flaviviruses in Australian mosquitoes and identification of host restriction factors. mSphere 5, e00095-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark D. C., Lobigs M., Lee E., Howard M. J., Clark K., Blitvich B. J., Hall R. A., In situ reactions of monoclonal antibodies with a viable mutant of Murray Valley encephalitis virus reveal an absence of dimeric NS1 protein. J. Gen. Virol. 88, 1175–1183 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zivanov J., Nakane T., Scheres S. H. W., Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sievers F., Higgins D. G., Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bunkoczi G., Read R. J., Improvement of molecular-replacement models with Sculptor. Acta Crystallogr. D Biol. Crystallogr. 67, 303–312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piyasena T. B. H., Setoh Y. X., Hobson-Peters J., Newton N. D., Bielefeldt-Ohmann H., McLean B. J., Vet L. J., Khromykh A. A., Hall R. A., Infectious DNAs derived from insect-specific flavivirus genomes enable identification of pre- and post-entry host restrictions in vertebrate cells. Sci. Rep. 7, 2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. Toleikis, O. Broders, S. Dübel, Cloning single-chain antibody fragments (scFv) from hybridoma cells, in Molecular Diagnosis of Infectious Diseases, J. Decler, U. Reischl, Eds. (Humana Press, 2004), pp. 447–458. [DOI] [PubMed] [Google Scholar]
- 42.Jones M. L., Seldon T., Smede M., Linville A., Chin D. Y., Barnard R., Mahler S. M., Munster D., Hart D., Gray P. P., Munro T. P., A method for rapid, ligation-independent reformatting of recombinant monoclonal antibodies. J. Immunol. Methods 354, 85–90 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Li J., Watterson D., Chang C.-W., Che X.-Y., Li X.-Q., Ericsson D. J., Qiu L.-W., Cai J.-P., Chen J., Fry S. R., Cheung S. T. M., Cooper M. A., Young P. R., Kobe B., Structural and Functional characterization of a cross-reactive dengue virus neutralizing antibody that recognizes a cryptic epitope. Structure 26, 51–59.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Jaberolansar N., Chappell K. J., Watterson D., Bermingham I. M., Toth I., Young P. R., Skwarczynski M., Induction of high titred, non-neutralising antibodies by self-adjuvanting peptide epitopes derived from the respiratory syncytial virus fusion protein. Sci. Rep. 7, 11130–11130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLellan J. S., Pancera M., Carrico C., Gorman J., Julien J.-P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K., O’Dell S., Patel N., Shahzad-ul-Hussan S., Yang Y., Zhang B., Zhou T., Zhu J., Boyington J. C., Chuang G.-Y., Diwanji D., Georgiev I., Kwon Y. D., Lee D., Louder M. K., Moquin S., Schmidt S. D., Yang Z.-Y., Bonsignori M., Crump J. A., Kapiga S. H., Sam N. E., Haynes B. F., Burton D. R., Koff W. C., Walker L. M., Phogat S., Wyatt R., Orwenyo J., Wang L.-X., Arthos J., Bewley C. A., Mascola J. R., Nabel G. J., Schief W. R., Ward A. B., Wilson I. A., Kwong P. D., Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee T. K., Rollence M. L., Hallberg P. L., Oelkuct M. S., Dodd S. W., Nagle J. W., Filpula D. R., Production of engineered IgM-binding single-chain antibodies in Escherichia coli. J. Ind. Microbiol. 14, 371–376 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Walker P. A., Leong L. E. C., Ng P. W. P., Tan S. H., Waller S., Murphy D., Porter A. G., Efficient and rapid affinity purification of proteins using recombinant fusion proteases. Biotechnology 12, 601–605 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimanius D., Forsberg B. O., Scheres S. H., Lindahl E., Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharp T. H., Boyle A. L., Diebolder C. A., Kros A., Koster A. J., Gros P., Insights into IgM-mediated complement activation based on in situ structures of IgM-C1-C4b. Proc. Natl. Acad. Sci. U.S.A. 116, 11900–11905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sosnick T. R., Benjamin D. C., Novotny J., Seeger P. A., Trewhella J., Distances between the antigen-binding sites of three murine antibody subclasses measured using neutron and x-ray scattering. Biochemistry 31, 1779–1786 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/20/eabe4507/DC1


