Abstract
The gastrointestinal tract comprises a complex ecosystem with extensive opportunities for functional interactions between neoplastic epithelial cells and stromal, immune, neuronal, glial, and other cell types, as well as microorganisms and metabolites within the gut lumen. In this Review, we focus on interactions between gastrointestinal cancers and elements of the central and enteric nervous systems. This previously understudied but rapidly emerging area of investigation has blossomed in recent years, particularly with respect to improved understanding of neural contributions to the development and progression of esophageal, gastric, pancreatic, and colon neoplasia. Cancer neuroscience offers great promise to advance our understanding of how neural-cancer interactions promote alimentary tract neoplasia. The resulting mechanistic insights can be leveraged to identify diagnostic and prognostic biomarkers, and to develop novel therapeutic interventions.
Introduction
Advanced cancers of the gastrointestinal (GI) tract are highly resistant to chemotherapy, radiotherapy, and biologicals (1). Even new immunotherapies benefit only a subset of patients with colorectal cancer (CRC) (2). Thus, CRC and gastric cancer remain, respectively, the second and third leading causes of global cancer mortality (3, 4). Although CRC incidence and mortality decreased substantially in the United States over the past 30 years, increasing incidence and mortality in persons younger than 50 years are concerning (5, 6). Moreover, the COVID-19 pandemic adversely impacted cancer screening, thereby upstaging newly diagnosed lesions (7–9). In the United States, the incidence of esophageal and pancreatic ductal adenocarcinomas, both commonly diagnosed at advanced stages, is increasing; esophageal adenocarcinoma and pancreatic ductal adenocarcinoma (PDAC), respectively, cause more than 16,000 and 47,000 deaths yearly (5, 10). Five-year survival rates for advanced esophageal, gastric, pancreatic, and colorectal cancers are all less than 20% (5, 11). Clearly, developing more effective ways to detect and manage these cancers is a high priority — gut-brain interactions impacting GI cancer development and progression provide a largely untapped reservoir of novel diagnostic, prognostic, and therapeutic opportunities.
Abundant evidence implicates the central nervous system (CNS) in GI cancer progression. Chronic behavioral stress is linked to increased cancer risk by mechanisms involving neuroendocrine signaling (12). In preclinical models, stress-induced adrenergic signaling promotes PDAC progression (13, 14), in part by inducing matrix metalloproteinases (MMPs). MMPs degrade extracellular matrix, facilitating tumor expansion and metastasis (15); these effects are attenuated by β-adrenergic blockade (16). In murine and human studies, pharmacological inhibition of β-adrenergic signaling and chemical denervation of the pancreas improve chemotherapeutic efficacy (17).
Recent attention focused on cancer cell heterogeneity and the role of the tumor microenvironment in modulating tumor growth, invasion, and dissemination. Nonetheless, the spotlight has shone primarily on cancer, stromal, and immune cells (18), with less attention paid to neurons and glial cells (19). While the CNS can modulate disease, the GI tract possesses a unique intrinsic nervous system, the enteric nervous system (ENS), sometimes called the “second brain” or “little brain,” which, alone or in coordination with the CNS, modulates the diverse functions of the gut in health and disease (20). ENS neurons and glial cells are anatomically poised to transmit information multidirectionally to normal and neoplastic GI mucosal cells, stromal cells, immunocytes, and the brain. These complex interactions are further complicated by input from enteroendocrine cells sprinkled throughout the GI tract and by the gut microbiome. GI cancers profit from a landscape uniquely combining neuronal postsynaptic, endocrine, and paracrine signaling with diverse cell-cell contacts and access to key metabolites (Figure 1 and ref. 21).
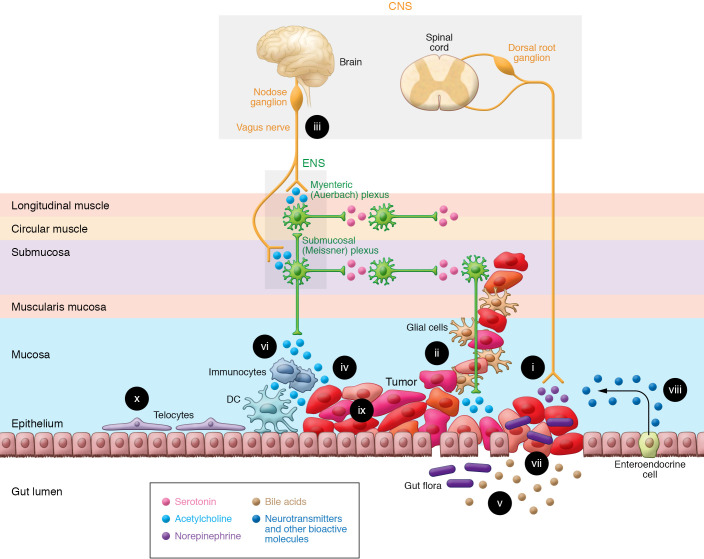
Figure 1. The GI neuron-cancer interface.
The ability of the gut-brain axis to modulate GI cancer progression is enhanced by the proximity and multidirectional crosstalk between numerous elements; these complex interactions provide opportunities for therapeutic intervention. (i) Cancer cells release nerve growth factors that promote neuronal tropism toward the tumor, enhancing access to neurotransmitters, metabolites, and the neural scaffold. Advanced cancer stages correlate with increased neural density. (ii) Perineural invasion, associated with worse outcomes, provides a path for tumor spread, access to neurotransmitters, and shielding from immune attack. (iii) Vagal innervation stimulates cancer progression by muscarinic mechanisms and modulates immune function. (iv) Neurotransmitters, like acetylcholine, produced and released by neurons, cancer cells, immunocytes, and possibly gut bacteria stimulate tumor growth, invasion, and dissemination. (v) Fecal bile acids, modified by gut bacteria, modulate immune and cancer cell function by several mechanisms, including activation of cancer cell muscarinic receptors. (vi) Immunocyte function is modulated by neurotransmitters released from the ENS, and cancer, immune, and enteroendocrine cells. (vii) Disruption of the intestinal barrier in the cancer field permits translocation of microorganisms that modulate immune and neural function. (viii) In response to bacterial and neural input, enteroendocrine cells, sprinkled throughout the mucosa, release neurotransmitters and other bioactive molecules. (ix) Cancer cells display intratumor heterogeneity and overexpress receptors for neurotransmitters and bioactive molecules. (x) Subepithelial telocytes are a critical source of pro-proliferative signaling for the intestinal stem cell niche; despite their prominent location, a functional role for telocytes at the neuron-cancer interface remains to be established.
In this Review, we analyze published findings, experimental models, and approaches used to uncover the mechanisms whereby the gut-brain axis modulates GI cancer development and progression. We consider how neoplastic cells advance their survival and progression by hijacking neurotransmitters, growth factors, signaling molecules, and metabolites that normally maintain tissue homeostasis and repair. In so doing, we identify potential therapeutic targets and highlight unresolved questions that can direct future research. Focusing on cancers of the esophagus, stomach, pancreas, and colon, we leave in-depth analysis of the anatomical gut-brain interface and the role of the gut microbiome to other contributors to this Review series.
The GI neuron-cancer interface
There is growing interest in understanding the role neurons play in the genesis and growth of non-CNS cancers, particularly with respect to cancers of the GI tract (21, 22). Nonetheless, many interactions between cancers and the nervous system are highly context-dependent. Thus, differences in the innervation of the esophagus, stomach, pancreas, and colon influence crosstalk between neuronal, glial, and cancer cells. The unique proximity of intestinal cancers to the gut microbiome and other fecal contents, and the specialized gut immune system, add layers of complexity. Appreciating the prominence of this tumor microenvironment generated interest in “ecological therapy,” wherein cells that nourish cancer cells are targeted to retard cancer growth (23).
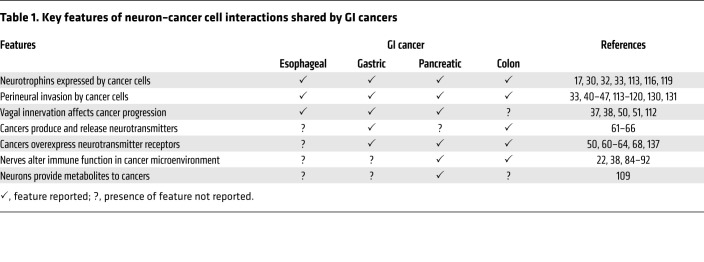
Anatomical features facilitate interactions between neurons, immunocytes, gut microorganisms, and other constituents of this complex ecosystem, acting in concert to modulate GI cancer cell proliferation, survival, and invasion (Figure 1). The ganglia of the ENS are concentrated in myenteric (Auerbach) plexuses spanning the entire length of the GI tract, and submucosal (Meissner) plexuses in the small and large intestines (20, 22). Enteric glial cells are positioned in the muscularis propria and mucosa, especially at the base of normal intestinal crypts (24, 25). Beyond providing support for neurons, enteric glial cells, which outnumber neurons, participate actively in a variety of ENS functions, including those vital for neuron maintenance and survival (24). Like neurons, enteric glia express neurotransmitter receptors and transporters, and respond to neurotransmitters, largely by changes in intracellular calcium that modulate cell function (26). Enteric neurons and glia are classified by their roles in regulating cellular architecture, neurotransmitter release, receptor activation, electrophysiological activity, and other functional characteristics; single-cell sequencing may modify classification based on molecular or genetic features (27). Figure 1 illustrates the broad framework of neural–GI cancer interactions in the context of the colon cancer microenvironment — features shared by cancers of the esophagus, stomach, and pancreas (Table 1). Figure 2 zooms in on key interactions between GI cancer cells and the gut neural/glial cell network.
Table 1. Key features of neuron–cancer cell interactions shared by GI cancers.
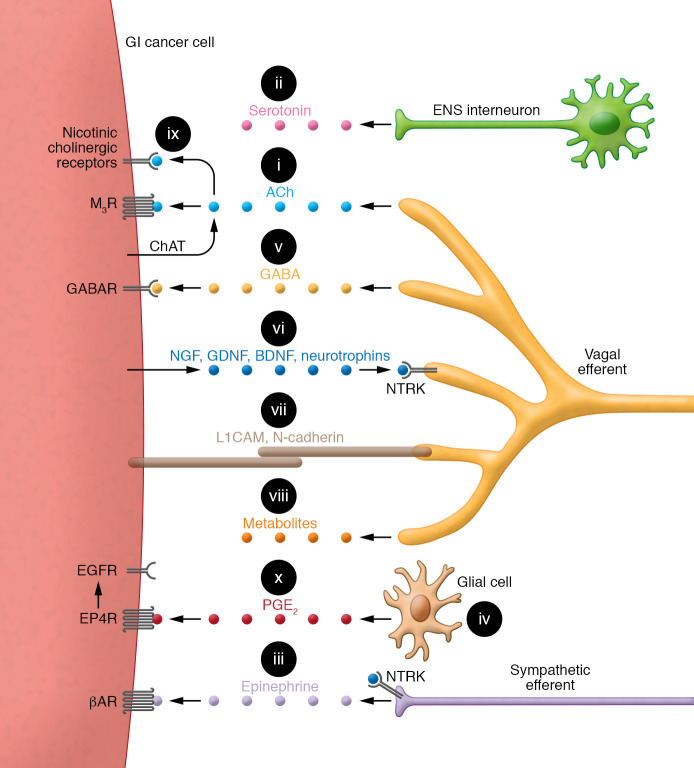
Figure 2. Key interactions between a generic GI cancer cell and the neural/glial cell network in the tumor microenvironment.
(i) Acetylcholine (ACh) production, mediated by choline acetyltransferase (ChAT), stimulates tumor growth, invasion, and dissemination. Relative quantities of ACh production by cancer cells versus neurons are uncertain. (ii) Serotonin released from ENS interneurons may stimulate colon cancer growth by currently obscure mechanisms. (iii) Epinephrine released from sympathetic neurons stimulates the progression of GI cancer cells overexpressing α- and β-adrenergic receptors (βAR). (iv) In response to neurotransmitters, glial cells play a major role in modulating and supporting the neuron–cancer cell interface. Glial cells also produce and release tumor growth factors into the tumor microenvironment. (v) GABA stimulates cancer cell proliferation via overexpressed GABA receptors (GABAR). (vi) Nerve growth factors (NGF, GDNF, BDNF, neurotrophins) released from cancer cells interact with neuronal receptors (e.g., NTRK) to promote axonal growth and tropism toward the tumor. (vii) Cancer cells express surface molecules, L1 cell adhesion molecule (L1CAM) and N-cadherin, facilitating adherence and migration along enteric neurons. Homophilic interactions allow L1CAM on cancer cells to adhere to L1CAM expressed on neurons. (viii) Neurons release key metabolites (e.g., serine) into the tumor microenvironment or reprogram cancer cell metabolic pathways. (ix) ACh interaction with nicotinic cholinergic receptors expressed on PDAC stimulates tumor progression. (x) Bidirectional interactions between glial and GI cancer cells involve cancer cell–derived interleukins that stimulate prostaglandin E2 (PGE2) biosynthesis and paracrine release by enteric glia. PGE2 stimulates tumor expansion via EP4 receptor–mediated (EP4R-mediated) transactivation of EGFR.
Nerve growth factors.
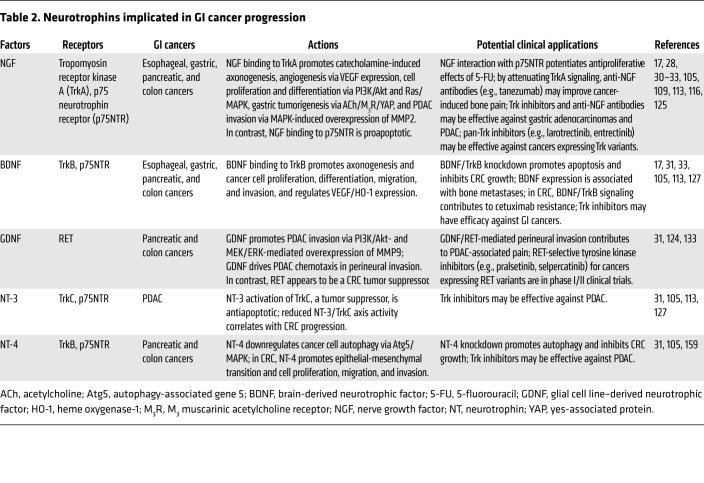
Nerve growth factors, or neurotrophins, comprise a highly homologous family of precursor proteins cleaved to active peptides including nerve growth factor (NGF) (28), brain-derived neurotrophic factor (BDNF), glial cell line–derived neurotrophic factor (GDNF), and neurotrophin-3 and neurotrophin-4 (NT-3 and NT-4) (Table 2 and refs. 29, 30). These proteins stimulate nerve development and survival through diverse signaling mechanisms; for example, binding of NGF to tropomyosin tyrosine receptor kinase fusion proteins stimulates receptor homodimerization, autophosphorylation of the tyrosine kinase domain, and activation of PI3K, Ras, phospholipase C (PLC), and other downstream effectors (31).
Table 2. Neurotrophins implicated in GI cancer progression.
Early-stage cancers release neurotrophins that stimulate local neuronal growth and increased nerve density, features correlated with more aggressive cancers (30, 32). These effects are bidirectional; cancers release neurotrophins that encourage neurogenesis, axonogenesis, and neural migration, while neurons and glial cells release neurotransmitters that stimulate tumor growth and invasion (Figure 2). This was studied extensively in the genesis of PDAC, where release of neurotrophic growth factors (e.g., NGF) by neoplastic cells and expression of their receptors on neurons correlates with nerve density and tumor aggressiveness (33), a mechanism replicated by NGF overexpression in murine PDAC models (17). In a transgenic mouse PDAC model, surgical denervation of celiac and superior mesenteric ganglia enhanced chemotherapeutic efficacy, supporting the importance of CNS input (17). The complexity and specificity of nerve-cancer interactions is highlighted by murine PDAC models wherein chemical denervation of the pancreas attenuates pancreatic intraepithelial neoplasia and progression (34–36), but surgical vagus nerve transection has opposite actions (37, 38).
Perineural invasion and neural scaffold.
Although definitions vary, perineural invasion (PNI) is commonly defined as cancer invasion into any of the three layers of the nerve sheath, or cancer surrounding at least 33% of the neural circumference (39, 40); PNI impacts tumor growth, progression, and responses to therapy. PNI advances cancer progression by facilitating biochemical and physical interactions between neural, glial, and neoplastic cells that promote neural and cancer cell proliferation and stimulate cancers to spread along neural planes. Although PNI is associated with worse clinical outcomes for any GI cancer studied (41–44), regional factors such as neural density may selectively augment the importance of PNI for some cancers versus others (40).
The precise molecular mechanisms underlying PNI are uncertain (40), although the release of nerve growth factors and cytokines from cancer and immune cells into the tumor microenvironment is important (45). As illustrated in Figure 1, neurons in the cancer microenvironment can provide a physical scaffold for GI cancer invasion and metastasis (40, 46); cancer cell nests are reported near the myenteric plexus (47). Adherence to and migration along enteric neurons are facilitated by cancer cell expression of key surface molecules (e.g., L1CAM and N-cadherin) (Figure 2 and ref. 46). Notably, retrograde traffic along ENS neurons may include gut bacteria (48), reflecting a perineural cancer scaffold that provides a hub for crosstalk between cancer cells, neurons, glial cells, immunocytes, and translocated microorganisms. Few, if any, experimental models reflect the multitude of interactions between cell types in this complex tumor microenvironment. No treatments currently target PNI.
Vagal innervation.
Although highly context-dependent, vagal innervation is the most prominent way the CNS regulates GI neoplasia. Surgical interruption of the vagus (vagotomy) appears to reduce gastric cancer risk (49, 50); gastric neuronal density and cancer stage are correlated (50). In mice, surgical or pharmacological hemivagotomy attenuates proneoplastic Wnt signaling and reduces gastric tumor formation in the denervated stomach (50, 51). Notably, opposite effects are observed in murine PDAC models wherein vagotomy promotes neoplasia (38). As discussed below, this conundrum may be explained by differential expression of M1 and M3 muscarinic receptors (M1R and M3R) with conflicting actions on cancer progression. Lastly, a liver-brain-gut neural arc identified by retrograde tracing of hepatic vagal branches may modulate immune responses to CRC (52).
Neurotransmitters and their receptors on cancer cells.
For decades, traditional neurotransmitters, like acetylcholine (ACh), were considered to derive primarily if not uniquely from neurons. Over the past 15 years, growing interest has focused on non-neuronal neurotransmitter production and release from cancer and immune cells in the tumor microenvironment (53–55) and bacteria in the gut microbiome (56–59). In this regard, most work pertains to effects of ACh on muscarinic receptors expressed by GI cancers (Figure 3 and refs. 60, 61). Human gastric (62, 63) and colon (64) cancer cells express choline acetyltransferase (ChAT) and synthesize and release ACh. Normal pancreatic stellate cells produce ACh (65) and pancreatic cancer cells express choline transporters (66), a surrogate marker of ACh production. Yet, to our knowledge, ACh production by PDAC has not been proved. Cancer cell types that release ACh commonly overexpress M3R (64); M3R expression correlates with gastric cancer stage and metastasis (62–64). This, and the relatively low concentrations of ACh released by cancer cells, suggest that non-neuronal release of neurotransmitters by cancer, tuft (67), immune, and other cells in the tumor microenvironment modulates cell function by autocrine and paracrine actions. Consistent with these observations, M3R deficiency in murine CRC models attenuates neoplasia (68, 69).
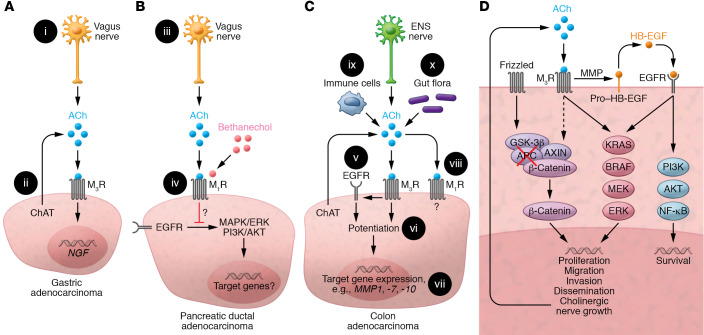
Figure 3. Muscarinic receptor activation in GI cancer.
(A) Gastric adenocarcinoma. (i) ACh release from vagal efferents activates M3 muscarinic receptors (M3R); vagotomy attenuates neoplasia. (ii) Cancer cells express ChAT, key for non-neuronal ACh synthesis; resulting ACh levels and their autocrine and paracrine impact on tumor progression remain uncertain. M3R activation induces nerve growth factor (NGF) expression. (B) PDAC. (iii) Treating mice with bethanechol, a non–subtype-selective muscarinic receptor agonist, activates muscarinic receptors. (iv) M1R activation attenuates PDAC progression by undefined mechanisms involving repressed EGFR signaling. (C) CRC. (v) M3R signaling transactivates EGFR; this is mediated by MMP7-mediated release of HB-EGF, an EGFR ligand. (vi) Concurrent activation of M3R and EGFR potentiates target gene expression. (vii) M3R activation selectively induces MMP1, MMP7, and MMP10 expression. MMP1 and MMP7 facilitate cell invasion. MMP7 also catalyzes the release of EGFR ligands (e.g., HB-EGF). (viii) M1R expression and activation attenuate colon cancer progression by unknown mechanisms. (ix) Immunocytes and (x) gut flora provide additional sources of non-neuronal ACh. (D) Post-M3R signaling alters gene expression and cancer cell function by impacting various signaling pathways. APC and/or β-catenin gene mutations free β-catenin from proteasomal destruction, promoting transcription of β-catenin target genes. M3R activation transactivates EGFR and augments β-catenin signaling. Resulting changes in downstream gene transcription stimulate cancer cell proliferation, survival, migration, invasion, and dissemination. Notably, induction of neurotrophin expression can promote neural growth and tropism, a feedback loop providing additional access to ACh and other neurotransmitters.
α2A-Adrenergic receptor activation in normal gut epithelial cells may stimulate EGFR transactivation and downstream MEK/ERK signaling, which enhances cell migration and wound healing (70, 71). Although adrenergic receptor activation was implicated in PDAC progression, compared with muscarinic neurotransmitters, the role of adrenergic receptor agonists (e.g., epinephrine) in modulating GI cancer growth and progression remains relatively unexplored (72). Gauging the importance of neurotransmitter release from neurons and cancer, immune, enteroendocrine, and other cells in the tumor microenvironment is limited by the challenges of accurate spatial and temporal measurement of very low neurotransmitter concentrations. Moreover, when evaluating neurotransmitter effects in vitro, it is crucial to discriminate physiological from pharmacological (i.e., supraphysiological) neurotransmitter concentrations that may lack disease relevance.
The GI cancer microenvironment
Bile acids.
Bile acids (BAs), produced in the liver, excreted into the intestinal lumen, and modified by bacteria in the gut microbiome, are recycled via enterohepatic circulation (73). BAs modulate the function of normal (74) and neoplastic (75) GI epithelial cells by interacting with Takeda G-coupled receptor 5 (TGR5; GPBAR1) and M3 muscarinic GPCRs (Figure 3 and refs. 75, 76). Long associated with CRC risk, BAs have pleiotropic effects including gut immune modulation (77) and functional interactions with muscarinic receptors (78) overexpressed in CRC (79). These functional interactions mimic those of cholinergic neurotransmitters (e.g., ACh) and, among other actions, result in transactivation of EGFR and signal transduction that stimulates cancer cell proliferation, survival, and invasion (80, 81). Interestingly, chenodeoxycholic acid also inhibits the pro-oncogenic effects of Bacteroides fragilis toxin (82).
Gut immunocytes.
By modulating lymphatic traffic, egress from lymph nodes, and T cell activation, CNS adrenergic nerve fibers suppress immune activity in highly innervated GI organs like the stomach and pancreas (83–85). This may limit immune surveillance and checkpoint inhibitor efficacy (86, 87); surgical or chemical denervation may improve the efficacy of immunotherapy (83, 87). Neurons in the ENS also regulate the activity of enteric immunocytes (88–90) that synthesize and release non-neuronal ACh in the cancer microenvironment (Figure 3 and refs. 91, 92). Macrophages in the endoneurium release cytokines that facilitate PNI by attracting tumor cells to neurons (45). A recently identified neural arc connecting the brain and gut via the liver may modulate immune responses to GI cancers by a mechanism involving ACh neurotransmission (52).
Gut microbiome.
Intestinal barrier disruption encourages transmural infiltration of bacteria and fungi comprising the gut microbiome (93). Cancers arising from GI epithelial cells at the host–gut microbiome interface break the single-layer barrier formed by tight junctions between normal epithelial cells (Figure 1 and refs. 94, 95). Dysbiosis resulting from “barrier-breaking” effects of cancer can activate multiple signaling systems (96, 97). For example, NF-ĸB and STAT3 pathways regulate the function of regional immune cells and neurons (98). Tumors hijack these developmental, wound healing, and antiinflammatory signaling programs to foster their own progression. Some bacterial metabolites, e.g., ACh and BAs, are GPCR agonists that can alter both neuron and cancer cell function (Figure 3 and refs. 99, 100). Mucosal microbial biofilms from humans with CRC are carcinogenic in murine models (101), and some bacterial products (e.g., B. fragilis toxin) contribute to barrier-breaking effects of cancers (102).
Enteroendocrine cells.
Sprinkled throughout the epithelial lining of the GI tract (Figure 1), enteroendocrine cells under neuronal, hormonal, and paracrine control synthesize and release a variety of neurotransmitters. Paracrine signaling by enteroendocrine cell–derived serotonin modulates the activity of neurons, immunocytes, and cancer cells (103, 104). Whether serotonin release from enteroendocrine cells or neurons in the ENS advances or retards GI cancer progression remains uncertain; this is likely context- and concentration-dependent (Figure 2 and ref. 105). Similar to pulmonary epithelial neuroendocrine cells that produce and release ACh, which stimulates small cell lung cancer progression (106), neuroendocrine tumors such as pheochromocytomas may release neurotransmitters and growth factors that enhance GI cancer progression (107, 108).
Metabolites.
Cancer cells have high metabolic requirements and limited blood supply. Intriguing work suggests neuronal axons can supply scarce amino acids and nutrients to GI cancer cells in this nutrient-depleted tumor microenvironment (109, 110). Banh and colleagues found that serine deprivation impaired PDAC growth but permitted the selective translation and secretion of NGF to increase neural density and provide PDAC cells with access to axon-derived serine (109). Rabben et al. found that gastric cancers are glutamine-dependent; vagotomy induced a metabolic switch from glutaminolysis to oxidative phosphorylation and glycolysis (Warburg effect) (110). These studies uncover the provision of neuron-derived metabolites and neuron-induced metabolic reprogramming of GI cancer cells as potential therapeutic targets (Figure 2).
Interactions of specific GI cancers with gut neural networks
Organ-specific features of GI cancer–nervous system interactions are reported for the esophagus, stomach, pancreas, and colon. In most cases, distinguishing between generic and truly organ-specific features awaits further clarification.
Esophageal cancer.
Esophageal cancers are composed of squamous cell carcinomas and adenocarcinomas; the impact of the gut-brain axis on esophageal cancer is evidenced by the surge in adenocarcinomas, once uncommon and now the most frequent form of esophageal cancer in developed nations (111). Gastroesophageal reflux, due primarily to poorly understood defects in CNS (vagal parasympathetic and spinal sympathetic) and ENS control of lower esophageal sphincter pressure and esophageal motility, predisposes to preneoplastic Barrett’s epithelium and esophageal adenocarcinoma (112). Thus, although the impact of neurons and glia on esophageal cancer progression is less clear than their impact on progression of other GI cancers (Table 1), there is compelling evidence that the nervous system plays a central role in the development of esophageal adenocarcinomas.
Among other mechanisms, neural innervation promotes esophageal tumor progression via neurotrophins and their receptors (Table 2) (45, 113–115). Nerve bundles and neuropeptide-immunoreactive nerve fibers expressing neurotrophic receptor kinase 1 (NTRK1, also called TrkA), an NGF-binding receptor, are commonly observed in esophageal cancers that overexpress NGF (113, 116); an esophageal cancer subtype expresses high levels of Trk-T1 neurotrophin receptor mRNA (114). Low-affinity p75 neurotrophin receptors (p75NTR), expressed in the stem cell population of normal esophageal epithelial cells, were detected in approximately half of 187 esophageal squamous cell carcinomas (113). RNAi knockdown of p75NTR expression in esophageal squamous cancer cells inhibited proliferation and induced apoptosis (117). Notably, NTRK gene fusions involving NTRK1, NTRK2, or NTRK3 detected in a subset of esophageal, pancreatic, and colon cancers are targets for two FDA-approved TRK inhibitors, entrectinib and larotrectinib (118).
PNI in approximately half of esophageal squamous carcinomas identifies a clinical subset with a worse prognosis (43) and reduced survival (119). Meta-analysis identified PNI as a biomarker for advanced esophageal and esophagogastric junction cancers (120). PNI correlates with advanced TNM stage, poor cell differentiation (120), shorter disease-free survival, and increased rates of local recurrence (121), factors associated with overexpression of NGF (116).
Gastric cancer.
The proximal two-thirds of the stomach is endowed with extensive vagal innervation (122) that regulates secretion of gastric acid and pepsinogen by cholinergic mechanisms (123). Epidemiological observations suggest a link between cholinergic innervation and gastric neoplasia; neuronal density correlates with more advanced stages of gastric cancer (50), and vagotomy for peptic ulcer disease may reduce long-term cancer risk (49). Robust evidence for this association was provided by elegant murine studies showing reduced gastric neoplasia following surgical or pharmacological denervation along with improved responses to chemotherapy and prolonged survival (50), findings confirmed by others (51). In these murine models, vagotomy attenuated nuclear translocation of β-catenin and expression of several Wnt/β-catenin target genes, including Ccnd1, Axin2, Myc, Lgr5, and Cd44 (50), providing a plausible mechanism underlying the benefits of denervation.
As for other GI cancers (68, 69), M3R deficiency or inhibition suggests a prominent role for this receptor subtype (50, 51). Gastric cancer cells express ChAT, synthesize and release ACh, and overexpress M3R (Figure 2); M3R expression correlates with gastric cancer stage and lymph node metastasis (62, 63). M3R activation by autocrine release of ACh stimulates cell proliferation by an M3R/EGFR/ERK-dependent mechanism (63); M3R knockdown suppressed growth and promoted apoptosis of human gastric cancer cell xenografts (62). In murine gastric epithelial cells, ACh release from Dclk1-positive tuft cells and neurons induced NGF expression by a YAP-mediated mechanism (ACh/NGF/M3R/YAP axis), which promotes neuron proliferation and cancer progression (Figure 3A and ref. 32).
Pancreatic ductal adenocarcinoma.
Neurotrophins are overexpressed by PDACs and intrapancreatic cancer neurons; adding neurotrophins and coculture of PDAC and neural cells stimulates PDAC cell proliferation (Table 2 and ref. 124). Exogenous NGF dose-dependently increases MMP2 expression and enhances pancreatic cancer cell invasion by activating ERK signaling (125). Pancreatic cancers overexpress GDNF, which may have chemokinetic effects on tumor cells and upregulates MMP9 expression and activity. MMP9, a gelatinase (type IV collagenase), facilitates cancer cell invasion and metastasis (15, 126). BDNF and NT-3 also stimulate PDAC invasion into the basement membrane (127, 128).
Following retroperitoneal nerve dissections that revealed neural involvement in PDAC (129), strong evidence accumulated linking neural input to cancer progression. Neural invasion, almost uniformly present in PDAC, shortens survival (44, 130–133). Neuronal support and mutations in axon guidance genes are also implicated in PDAC progression (134). Rare RET mutations in PDAC are associated with GDNF-dependent tumor invasion (Table 2 and ref. 135).
Neurotransmitters like ACh, adrenergic agonists, γ-aminobutyric acid (GABA), and glutamate, released from neuron and glial cell networks infiltrating PDAC, play important roles in tumor growth and dissemination (136). Cholinergic signaling via muscarinic receptors directly and indirectly suppresses pancreatic tumorigenesis and cancer stemness (37). In genetically engineered mice, subdiaphragmatic vagotomy accelerated, and a muscarinic agonist, bethanechol, suppressed, PDAC development; bethanechol, which improved survival by an M1R-dependent mechanism (Figure 3B and ref. 37), is in early clinical trials. Likewise, in a murine PDAC model, subdiaphragmatic vagotomy promoted tumor growth and reduced survival, but not in mice deficient in TNF-α (38). In line with non-neuronal ACh production (54), ACh produced by human and rat pancreatic stellate cells may modulate pancreatic exocrine secretion and neoplasia (65). The specific downstream target genes for muscarinic receptor signaling in PDAC remain uncertain (Figure 3B).
β-Adrenergic signaling mediates the accelerated PDAC growth and invasion observed with chronic stress (16). In an orthotopic mouse model of PDAC, in vivo optical imaging revealed that stress-induced neural activation increased tumor growth and metastasis. These effects were reproduced by pharmacological activation of β-adrenergic signaling and reversed by β-blockade, which also extended animal survival (13). Compared with controls, PDAC-bearing mice exposed to chronic stress had larger tumors and shortened lifespans, effects attenuated by a β-blocker (13). β-Blockade in PDAC is being evaluated in clinical trials.
Other neurotransmitters and receptors are implicated in PDAC progression. For example, GABA, which primarily inhibits CNS neuronal excitability, unexpectedly stimulates PDAC cell proliferation (105). These actions are most likely mediated via overexpressed GABA receptor π subunits that signal by elevating intracellular calcium and activating MAPK/ERK signaling (137). In accord with increased PDAC risk in tobacco users, nicotine-derived nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a potent mutagen, carcinogen, and nicotinic ACh receptor agonist, induced PDAC in hamsters (138). Aggressive pancreatic cancers overexpress NMDA glutamate receptors that stimulate pro-growth signaling pathways when activated by glutamatergic nerves (139, 140).
Colorectal cancer.
Neural influences are evident early in CRC development — colon cancer stem cells (CCSCs) proliferate in an environment with a denser enteric glial cell network than in normal colon, particularly at the tumor invasive front (25). In precancerous lesions and CRC, enteric glial networks exhibit structural abnormalities (141, 142); S-100β and glial fibrillary acidic protein immunostaining reveal denser and more branched networks and glial cells intimately associated with both CRC cells and adjacent neurons (Figures 1 and 2 and ref. 25). Bidirectional interactions between glial and CRC cells involve CRC-derived IL-1α/β stimulation of prostaglandin E2 (PGE2) biosynthesis and paracrine release by enteric glia (Figure 2). PGE2 stimulates CCSC growth and expansion via EP4 receptor–mediated transactivation of EGFR (25, 143–146). Activation of enteric glia by IL-1 may promote tumorigenesis by effects on immunocytes similar to those in inflammatory bowel diseases that increase CRC risk (147). In human CRC cell lines, immunodeficient mice, and primary human CRC cells, enteric glia stimulate an increase in the number and size of CCSC-derived tumors (25), identifying enteric glia as potential therapeutic targets.
Neurons also facilitate tumor development by serving as physical scaffolds for CRC cell migration and metastasis (Figure 1 and refs. 40, 46). ENS neurons are uniquely unmyelinated; the lack of perineurium and endoneurium sheath layers presents an unimpeded interface with tumors (46, 148) and glial cells, which, through paracrine signaling, enable CCSC activation (25). Enteric neurons and EpCAM-positive CRC cells are closely associated, particularly at the tumor invasive front, facilitating physical interactions between the two cell types (Figure 2 and ref. 46).
Synergistic interactions between bacterial species are associated with CRC initiation and progression (149); in mouse models, microorganisms (e.g., B. fragilis, E. coli, and Fusobacterium nucleatum; refs. 150–152) release factors, including B. fragilis–derived BFT toxin (101, 102), F. nucleatum–derived FadA and Fap2 adhesins, and NF-κB, that enhance CRC progression (153–155). It is likely that these factors modulate ENS activity; NF-κB, for example, plays a key role in regulating CNS inflammation (156). These multidirectional interactions between the gut microbiome, CRC cells, and enteric neurons are likely to promote cancer progression; stronger evidence awaits better experimental models (153). Because differences in immune, epithelial, and neural cell functions and microbial diversity in one region of the colon may impact cancer development and progression at other sites, purely reductionist approaches to explore links between the microbiome, ENS, and neoplasia may be misleading (157, 158).
The ability of CRC cells to adhere to enteric neurons and migrate to new anatomic locations is facilitated by cell surface molecules (e.g., L1CAM and N-cadherin) (Figure 2 and ref. 46) and can be modulated by neurotrophins (e.g., NT-4) (Table 2 and ref. 159). When cocultured with primary enteric neurons, enteric glia, and mesenchymal cells, CRC cells from established lines and primary CRC cells colocalize with enteric neurons (46). Moreover, in contrast to nontransformed intestinal epithelial cells and mesenchymal cells, CRC cells migrate across longer distances to reach enteric neurons and adhere to them with greater force (46). Successful invasion of CRC cells through the high-resistance neural sheath layers of collagen and basement membrane identifies hardier cells and confers survival and proliferative advantages (40). Besides the prognostic value of PNI, single-cell gene profiling may identify expression patterns predictive of PNI and cancer, neuronal, and glial genes that provide therapeutic opportunities (40–42, 160, 161).
As in gastric cancer and PDAC, cholinergic muscarinic receptors in CRC are the most prominent neurotransmitter targets. Treatment with a non–subtype-selective muscarinic agonist, bethanechol, promotes murine colon neoplasia (162). Of five muscarinic receptor subtypes, M1R and M3R activity most prominently modulates colon cancer progression (Figure 3C). M3R overexpression in primary CRC predicts metastases, and in murine models of sporadic and genetic CRC, global M3R deficiency robustly attenuates intestinal neoplasia (68, 69, 79). M3R activation selectively induces the expression of MMP1, MMP7, and MMP10, which facilitates CRC invasion and spread (163). Blocking expression and activation of MMP1 in vitro abolishes ACh-induced colon cancer cell invasion into endothelial cell monolayers (164). Selective BAs (e.g., deoxycholyltaurine) can activate M3R (78, 80), providing a mechanism whereby increased fecal BA levels augment murine colon neoplasia (165, 166). The mechanisms underlying the actions of M3R in GI cancer are summarized in Figure 3D.
In contrast to M3R, the role of M1R in GI cancer remains obscure. As in animal models of PDAC (37), in azoxymethane-treated mice, M1R deficiency modestly augmented colon neoplasia and, notably, negated the beneficial effects of M3R deficiency (167). A therapeutic strategy directed at muscarinic receptor signaling will likely require targeting of M1R and M3R simultaneously.
Impediments to studying gut-brain interactions in GI cancer
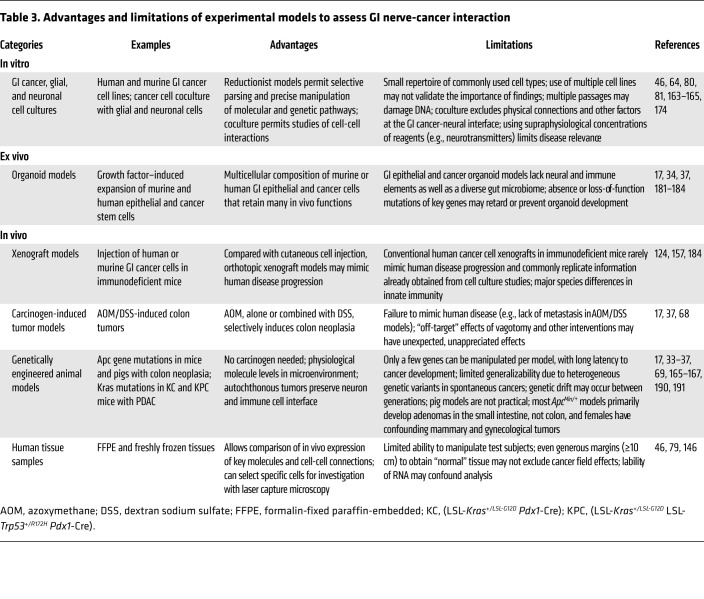
Capturing the intricacies of gut-brain interactions experimentally is challenging. While reductionist experimental systems such as cell coculture are valuable approaches to parse cross-directional cell signaling, they fail to capture the complex milieu and interactions between cells in living organisms. These models may not accurately reflect relevant concentrations of neurotransmitters and other biologically active molecules, diffusion limits in the extracellular space, and other parameters important to distinguish physiological from pharmacological effects (Table 3). The human GI tract features a particularly complex and dynamic ecosystem that may not be reproduced even by in vivo mouse and other animal models, which are also confounded by species differences (Figure 1).
Table 3. Advantages and limitations of experimental models to assess GI nerve-cancer interaction.
Technical limitations and insufficient attention to quality control — e.g., confirming the specificity of antibodies, particularly those directed at GPCRs (168); authenticating cell lines and transgenic mice; optimizing tissue fixation, preservation, and autofluorescence; and ensuring high-quality mRNA measurement (169) — further impact data quality, interpretation, and translational value. An additional challenge is replicating in vitro, ex vivo, and in vivo the physical forces cancer cells exert for PNI and migration along a neural scaffold (Figure 1 and refs. 170, 171). Collectively, these limitations contribute to the poor track record of experimental models in predicting therapeutic success of novel interventions in clinical trials, and to the paucity of treatments directed at the gut-brain axis (172, 173).
In vitro models.
The majority of information regarding the effects of neurotransmitters on oncogenic cell signaling and function derives from in vitro cell models. These use a relatively small repertoire of human cancer cell lines, many established decades ago. Extensive passaging is likely to have altered their genetic makeup and key biological features (174). Use of primary GI cancer cells may address these concerns but is limited by the continuous need to replenish tissue samples and the innate heterogeneity of cancers (174–176). Coculturing primary GI cancer and ENS cells provides useful information but does not fully capture complex in vivo cell interactions (46). Biomedical journals commonly require investigators to replicate findings in multiple cancer cell lines, but adherence to this guidance is not uniform; as recently as 2018, an otherwise exemplary study employed only one established human colon cancer cell line without providing a rationale for cell line selection, although key findings were replicated in primary human colon cancer cells (46). Even using multiple cell lines does not assure scientific validity or reproducibility in more complex systems, particularly given the lack of cross-directional input from the neural and glial components of the tumor microenvironment and substantial intratumoral and neural network diversity (177). To some extent, the use of single-cell RNA sequencing may address the latter limitation (178), but changes in gene expression, which must be confirmed by quantitative PCR, are not necessarily mirrored by commensurate changes in protein expression.
Ex vivo models.
Conventional organoid models developed from GI cancer stem cells can provide useful information regarding the factors promoting growth, invasion, and metastasis, but, among other limitations, organoids lack neural and immune elements (179–182). Even novel 3D organoids-on-a-chip, which permit the growth of mini-intestines on scaffolds that mimic basement membranes, fail to incorporate neural elements (179, 183). These limitations may be overcome by tissue engineering to develop scaffold-guided organoid morphogenesis from tissue stem cells that more faithfully mimic in vivo biology (183). In addition to furthering investigation into the role of neurons in GI neoplasia, development of increasingly accurate patient-derived organoid models may pave the way for advances in precision medicine by predicting the efficacy of novel therapies directed at the gut-brain axis (183).
In vivo models.
Xenografts developed from human cancer cells injected into the skin of immunodeficient mice are common “in vivo” models. More cynically, these models represent only a change in culture medium from in vitro solutions to live organisms; xenograft experiments almost uniformly mirror in vitro findings without offering novel mechanistic insights, providing only an incremental advance and limited validation of in vitro findings. Orthotopic xenografts may more faithfully replicate human cancer progression and metastasis, e.g., human colon cancer cells implanted in the mouse sigmoid colon (157, 184). Patient-derived xenografts (PDXs) can provide real-time information to develop cancer-specific treatment (176).
In vivo models commonly fail to account for the impact of human immune and neural cells on GI cancer progression, even when PDX models employ “humanized” mice. Variability in gut microbiota can also confound outcomes; causal inferences based solely on murine studies should be avoided (185, 186). Investigators using human surgical tissues commonly use adjacent uninvolved tissue as control, but even the use of broad margins, 10 cm or more from the cancer, may be confounded by macroscopically indistinguishable cancer “field effects” affecting “control” cells (187). Innervation maps forming the basis for understanding nerve-tumor interactions derive largely from studies of noncancerous tissue, whereas GI cancers may restructure and rewire neuronal networks (30). Off-target effects of surgical manipulations, e.g., vagotomy, may alter the GI cancer-neuron interface in unexpected ways (38, 50, 51).
Genetically engineered mice and pigs (188–190) are limited by fundamental species differences in physiology and pathophysiology (191). For example, the mouse and human immune systems have very different major histocompatibility genes. Humanized genetically engineered mouse models have long latency periods and fail to recapitulate late-stage human disease, the most difficult clinical management problem. Even combining advanced techniques and models fails to mimic faithfully the complexity of the human GI tumor microenvironment.
Conclusions and perspectives
Despite impressive progress, therapeutic interventions directed at the GI cancer gut-brain axis are currently limited to targeting neurotrophin, muscarinic, and β-adrenergic receptors. To advance the field, a more comprehensive understanding of the GI neuronal-glial-cancer cell interface is needed, akin to that for the gut microbiome (154, 192) and immune system (18, 193). Specific areas ripe for exploration include (a) determining how precancerous changes in the GI tumor secretome alter enteric glial networks and facilitate tumorigenesis; (b) using single-cell RNA-Seq and spatial transcriptomics to develop a more complete inventory of the cells, genes, and proteins comprising the tumor-neuron adhesion complex and molecular guidance factors, and better understanding how their expression alters cancer progression; (c) using similar methods to learn how immune cells, e.g., tumor-associated macrophages, mediate interactions between the ENS and GI cancer cells; (d) elucidating how GI cancers attract neurons and other constituents of the ENS and vice versa; (e) exploring whether molecules like L1CAM, whose expression correlates with PNI in PDAC (194), are viable therapeutic targets; (f) investigating whether PNI provides a mechanism for GI cancer cells to evade immune detection and treatment; (g) cataloging axon-derived metabolites that enhance cancer cell survival and growth (109, 195); and (h) improving experimental models to more faithfully capture the extraordinary complexity of the GI tumor microenvironment and the integration of neural and glial networks (196).
Exciting research opportunities in GI cancer neuroscience will result from leveraging advances in tissue preparation, clearing, and higher-resolution optical imaging to resolve CNS and ENS circuitry; computational biology for single-cell mRNA sequencing and metabolomics; optogenetics using light to monitor and control the activity of individual neurons and biochemical pathways modified by gene editing; 3D electrophysiological recording; and artificial intelligence (18, 197). Integrating newly discovered cell types and signaling pathways will yield novel mechanistic insights and therapeutic targets. For example, subepithelial telocytes, which provide pro-proliferative signals to stem cells throughout the small intestine and colon (198, 199), may contribute to crosstalk between colon cancer stem cells and components of the tumor microenvironment (Figure 1 and ref. 200). Filling key gaps in knowledge has great potential to advance our understanding of the role the gut-brain axis plays in GI cancer progression and empower us to leverage this information to improve therapeutic outcomes. Because of shared pathways and mechanisms (Figure 2 and Figure 3D), novel therapeutics targeting the gut-brain connection for one GI cancer will likely be applicable to others.
Author contributions
AS, GX, and JPR conceived, wrote, reviewed, edited, and approved the submitted manuscript.
Acknowledgments
This work was supported by VA Merit Awards BX002777 (to GX), BX002129 (to JPR), and BX004890 (to JPR) from the US Department of Veterans Affairs Biomedical Laboratory Research and Development Program. The contents do not represent the views of the US Department of Veterans Affairs or the US government.
Version 1. 05/17/2021
Electronic publication
Footnotes
Conflict of interest: Aspects of treating cancer with anticholinergic agents are the subject of a patent (“Hybrid cholinergic agents and compositions, methods of making, and methods of using to treat a cholinergic disorder,” US 6,624,155) issued on September 23, 2003, to the University of Arkansas; JPR is an inventor on this patent. JPR owns equities in Agile Therapeutics, Gilead Sciences, and Procter & Gamble.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(10):e143776.https://doi.org/10.1172/JCI143776.
Contributor Information
Alyssa Schledwitz, Email: alyssa.schledwitz@som.umaryland.edu.
Guofeng Xie, Email: gxie@som.umaryland.edu.
Jean-Pierre Raufman, Email: jraufman@som.umaryland.edu.
References
- 1.Ganesh K, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–375. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre T, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med. 2020;383(23):2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, et al. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19(8):1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, et al. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 6.Siegel RL, et al. Global patterns and trends in colorectal cancer incidence in young adults. Gut. 2019;68(12):2179–2185. doi: 10.1136/gutjnl-2019-319511. [DOI] [PubMed] [Google Scholar]
- 7.Sharpless NE. COVID-19 and cancer. Science. 2020;368(6497):1290. doi: 10.1126/science.abd3377. [DOI] [PubMed] [Google Scholar]
- 8.Corley DA, et al. Association between time to colonoscopy after a positive fecal test result and risk of colorectal cancer and cancer stage at diagnosis. JAMA. 2017;317(16):1631–1641. doi: 10.1001/jama.2017.3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patt D, et al. Impact of COVID-19 on cancer care: how the pandemic is delaying cancer diagnosis and treatment for american seniors. JCO Clin Cancer Inform. 2020;4:1059–1071. doi: 10.1200/CCI.20.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rawla P, et al. Epidemiology of pancreatic cancer: global trends, etiology and risk factors. World J Oncol. 2019;10(1):10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol. 2020;18(3):534–542. doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai S, et al. Chronic stress promotes cancer development. Front Oncol. 2020;10:1492. doi: 10.3389/fonc.2020.01492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Partecke LI, et al. Chronic stress increases experimental pancreatic cancer growth, reduces survival and can be antagonised by beta-adrenergic receptor blockade. Pancreatology. 2016;16(3):423–433. doi: 10.1016/j.pan.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Schuller HM, et al. Regulation of pancreatic cancer by neuropsychological stress responses: a novel target for intervention. Carcinogenesis. 2012;33(1):191–196. doi: 10.1093/carcin/bgr251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kessenbrock K, et al. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim-Fuchs C, et al. Chronic stress accelerates pancreatic cancer growth and invasion: a critical role for beta-adrenergic signaling in the pancreatic microenvironment. Brain Behav Immun. 2014;40:40–47. doi: 10.1016/j.bbi.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renz BW, et al. β2 adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell. 2018;33(1):75–90. doi: 10.1016/j.ccell.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schurch CM, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020;182(5):1341–1359. doi: 10.1016/j.cell.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faulkner S, et al. Tumor neurobiology and the war of nerves in cancer. Cancer Discov. 2019;9(6):702–710. doi: 10.1158/2159-8290.CD-18-1398. [DOI] [PubMed] [Google Scholar]
- 20.Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol. 2012;9(5):286–294. doi: 10.1038/nrgastro.2012.32. [DOI] [PubMed] [Google Scholar]
- 21.Monje M, et al. Roadmap for the emerging field of cancer neuroscience. Cell. 2020;181(2):219–222. doi: 10.1016/j.cell.2020.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rademakers G, et al. The role of enteric neurons in the development and progression of colorectal cancer. Biochim Biophys Acta Rev Cancer. 2017;1868(2):420–434. doi: 10.1016/j.bbcan.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Kareva I. What can ecology teach us about cancer? Transl Oncol. 2011;4(5):266–270. doi: 10.1593/tlo.11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharkey KA. Emerging roles for enteric glia in gastrointestinal disorders. J Clin Invest. 2015;125(3):918–925. doi: 10.1172/JCI76303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vales S, et al. Tumor cells hijack enteric glia to activate colon cancer stem cells and stimulate tumorigenesis. EBioMedicine. 2019;49:172–188. doi: 10.1016/j.ebiom.2019.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boesmans W, et al. Neurotransmitters involved in fast excitatory neurotransmission directly activate enteric glial cells. Neurogastroenterol Motil. 2013;25(2):e151–e160. doi: 10.1111/nmo.12065. [DOI] [PubMed] [Google Scholar]
- 27.Schneider S, et al. Unexpected roles for the second brain: enteric nervous system as master regulator of bowel function. Annu Rev Physiol. 2019;81:235–259. doi: 10.1146/annurev-physiol-021317-121515. [DOI] [PubMed] [Google Scholar]
- 28.Levi-Montalcini R. The nerve growth factor and the neuroscience chess board. Prog Brain Res. 2004;146:525–527. [PubMed] [Google Scholar]
- 29.Jones KR, Reichardt LF. Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc Natl Acad Sci U S A. 1990;87(20):8060–8064. doi: 10.1073/pnas.87.20.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zahalka AH, Frenette PS. Nerves in cancer. Nat Rev Cancer. 2020;20(3):143–157. doi: 10.1038/s41568-019-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farina AR, et al. The oncogenic neurotrophin receptor tropomyosin-related kinase variant, TrkAIII. J Exp Clin Cancer Res. 2018;37(1):119. doi: 10.1186/s13046-018-0786-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayakawa Y, et al. Nerve growth factor promotes gastric tumorigenesis through aberrant cholinergic signaling. Cancer Cell. 2017;31(1):21–34. doi: 10.1016/j.ccell.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stopczynski RE, et al. Neuroplastic changes occur early in the development of pancreatic ductal adenocarcinoma. Cancer Res. 2014;74(6):1718–1727. doi: 10.1158/0008-5472.CAN-13-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha S, et al. PanIN neuroendocrine cells promote tumorigenesis via neuronal cross-talk. Cancer Res. 2017;77(8):1868–1879. doi: 10.1158/0008-5472.CAN-16-0899-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saloman JL, et al. Ablation of sensory neurons in a genetic model of pancreatic ductal adenocarcinoma slows initiation and progression of cancer. Proc Natl Acad Sci U S A. 2016;113(11):3078–3083. doi: 10.1073/pnas.1512603113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bai H, et al. Inhibition of chronic pancreatitis and pancreatic intraepithelial neoplasia (PanIN) by capsaicin in LSL-KrasG12D/Pdx1-Cre mice. Carcinogenesis. 2011;32(11):1689–1696. doi: 10.1093/carcin/bgr191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renz BW, et al. Cholinergic signaling via muscarinic receptors directly and indirectly suppresses pancreatic tumorigenesis and cancer stemness. Cancer Discov. 2018;8(11):1458–1473. doi: 10.1158/2159-8290.CD-18-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Partecke LI, et al. Subdiaphragmatic vagotomy promotes tumor growth and reduces survival via TNFα in a murine pancreatic cancer model. Oncotarget. 2017;8(14):22501–22512. doi: 10.18632/oncotarget.15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liebig C, et al. Perineural invasion is an independent predictor of outcome in colorectal cancer. J Clin Oncol. 2009;27(31):5131–5137. doi: 10.1200/JCO.2009.22.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liebig C, et al. Perineural invasion in cancer: a review of the literature. Cancer. 2009;115(15):3379–3391. doi: 10.1002/cncr.24396. [DOI] [PubMed] [Google Scholar]
- 41.van Wyk HC, et al. The role of perineural invasion in predicting survival in patients with primary operable colorectal cancer: a systematic review. Crit Rev Oncol Hematol. 2017;112:11–20. doi: 10.1016/j.critrevonc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Knijn N, et al. Perineural invasion is a strong prognostic factor in colorectal cancer: a systematic review. Am J Surg Pathol. 2016;40(1):103–112. doi: 10.1097/PAS.0000000000000518. [DOI] [PubMed] [Google Scholar]
- 43.Chen JW, et al. The prognostic effect of perineural invasion in esophageal squamous cell carcinoma. BMC Cancer. 2014;14:313. doi: 10.1186/1471-2407-14-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee D, et al. Perineural and intraneural invasion in posttherapy pancreaticoduodenectomy specimens predicts poor prognosis in patients with pancreatic ductal adenocarcinoma. Am J Surg Pathol. 2012;36(3):409–417. doi: 10.1097/PAS.0b013e31824104c5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amit M, et al. Mechanisms of cancer dissemination along nerves. Nat Rev Cancer. 2016;16(6):399–408. doi: 10.1038/nrc.2016.38. [DOI] [PubMed] [Google Scholar]
- 46.Duchalais E, et al. Colorectal cancer cells adhere to and migrate along the neurons of the enteric nervous system. Cell Mol Gastroenterol Hepatol. 2018;5(1):31–49. doi: 10.1016/j.jcmgh.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueno H, et al. Histogenesis and prognostic value of myenteric spread in colorectal cancer: a Japanese multi-institutional study. J Gastroenterol. 2014;49(3):400–407. doi: 10.1007/s00535-013-0822-1. [DOI] [PubMed] [Google Scholar]
- 48.Rao M, Gershon MD. The bowel and beyond: the enteric nervous system in neurological disorders. Nat Rev Gastroenterol Hepatol. 2016;13(9):517–528. doi: 10.1038/nrgastro.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bahmanyar S, et al. Long-term risk of gastric cancer by subsite in operated and unoperated patients hospitalized for peptic ulcer. Am J Gastroenterol. 2007;102(6):1185–1191. doi: 10.1111/j.1572-0241.2007.01161.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhao CM, et al. Denervation suppresses gastric tumorigenesis. Sci Transl Med. 2014;6(250):250ra115. doi: 10.1126/scitranslmed.3009569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, et al. Muscarinic acetylcholine receptor 3 mediates vagus nerve-induced gastric cancer. Oncogenesis. 2018;7(11):88. doi: 10.1038/s41389-018-0099-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teratani T, et al. The liver-brain-gut neural arc maintains the Treg cell niche in the gut. Nature. 2020;585(7826):591–596. doi: 10.1038/s41586-020-2425-3. [DOI] [PubMed] [Google Scholar]
- 53.Beckmann J, Lips KS. The non-neuronal cholinergic system in health and disease. Pharmacology. 2013;92(5-6):286–302. doi: 10.1159/000355835. [DOI] [PubMed] [Google Scholar]
- 54.Wessler I, Kirkpatrick CJ. Acetylcholine beyond neurons: the non-neuronal cholinergic system in humans. Br J Pharmacol. 2008;154(8):1558–1571. doi: 10.1038/bjp.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawashima K, Fujii T. Basic and clinical aspects of non-neuronal acetylcholine: overview of non-neuronal cholinergic systems and their biological significance. J Pharmacol Sci. 2008;106(2):167–173. doi: 10.1254/jphs.FM0070073. [DOI] [PubMed] [Google Scholar]
- 56.Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693(pt B):128–133. doi: 10.1016/j.brainres.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asano Y, et al. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am J Physiol Gastrointest Liver Physiol. 2012;303(11):G1288–G1295. doi: 10.1152/ajpgi.00341.2012. [DOI] [PubMed] [Google Scholar]
- 58.Barrett E, et al. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol. 2012;113(2):411–417. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- 59.Stanaszek PM, et al. Isolation, extraction, and measurement of acetylcholine from Lactobacillus plantarum. Appl Environ Microbiol. 1977;34(2):237–239. doi: 10.1128/AEM.34.2.237-239.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Konishi M, et al. Role of muscarinic acetylcholine signaling in gastrointestinal cancers. Biomedicines. 2019;7(3):E58. doi: 10.3390/biomedicines7030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shah N, et al. Muscarinic receptors and ligands in cancer. Am J Physiol Cell Physiol. 2009;296(2):C221–C232. doi: 10.1152/ajpcell.00514.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang L, et al. Muscarinic receptor M3 mediates cell proliferation induced by acetylcholine and contributes to apoptosis in gastric cancer. Tumour Biol. 2016;37(2):2105–2117. doi: 10.1007/s13277-015-4011-0. [DOI] [PubMed] [Google Scholar]
- 63.Yu H, et al. Acetylcholine acts through M3 muscarinic receptor to activate the EGFR signaling and promotes gastric cancer cell proliferation. Sci Rep. 2017;7:40802. doi: 10.1038/srep40802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng K, et al. Acetylcholine release by human colon cancer cells mediates autocrine stimulation of cell proliferation. Am J Physiol Gastrointest Liver Physiol. 2008;295(3):G591–G597. doi: 10.1152/ajpgi.00055.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phillips PA, et al. Pancreatic stellate cells produce acetylcholine and may play a role in pancreatic exocrine secretion. Proc Natl Acad Sci U S A. 2010;107(40):17397–17402. doi: 10.1073/pnas.1000359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirai K, et al. Molecular and functional analysis of choline transporters and antitumor effects of choline transporter-like protein 1 inhibitors in human pancreatic cancer cells. Int J Mol Sci. 2020;21(15):E5190. doi: 10.3390/ijms21155190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider C, et al. Regulation of immune responses by tuft cells. Nat Rev Immunol. 2019;19(9):584–593. doi: 10.1038/s41577-019-0176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raufman JP, et al. Genetic ablation of M3 muscarinic receptors attenuates murine colon epithelial cell proliferation and neoplasia. Cancer Res. 2008;68(10):3573–3578. doi: 10.1158/0008-5472.CAN-07-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raufman JP, et al. Muscarinic receptor subtype-3 gene ablation and scopolamine butylbromide treatment attenuate small intestinal neoplasia in Apcmin/+ mice. Carcinogenesis. 2011;32(9):1396–1402. doi: 10.1093/carcin/bgr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buffin-Meyer B, et al. EGF receptor transactivation and PI3-kinase mediate stimulation of ERK by alpha(2A)-adrenoreceptor in intestinal epithelial cells: a role in wound healing. Eur J Pharmacol. 2007;574(2-3):85–93. doi: 10.1016/j.ejphar.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 71.Schaak S, et al. Alpha(2) adrenoceptors regulate proliferation of human intestinal epithelial cells. Gut. 2000;47(2):242–250. doi: 10.1136/gut.47.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perrone MG, et al. Upregulation of beta3-adrenergic receptor mRNA in human colon cancer: a preliminary study. Oncology. 2008;75(3-4):224–229. doi: 10.1159/000163851. [DOI] [PubMed] [Google Scholar]
- 73.Wahlstrom A, et al. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24(1):41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 74.Sorrentino G, et al. Bile acids signal via TGR5 to Activate intestinal stem cells and epithelial regeneration. Gastroenterology. 2020;159(3):956–968. doi: 10.1053/j.gastro.2020.05.067. [DOI] [PubMed] [Google Scholar]
- 75.Raufman JP, et al. Selective interaction of bile acids with muscarinic receptors: a case of molecular mimicry. Eur J Pharmacol. 2002;457(2-3):77–84. doi: 10.1016/S0014-2999(02)02690-0. [DOI] [PubMed] [Google Scholar]
- 76.Yang F, et al. Structural basis of GPBAR activation and bile acid recognition. Nature. 2020;587(7834):499–504. doi: 10.1038/s41586-020-2569-1. [DOI] [PubMed] [Google Scholar]
- 77.Winkler ES, et al. The intestinal microbiome restricts alphavirus infection and dissemination through a bile acid-type I IFN signaling axis. Cell. 2020;182(4):901–918. doi: 10.1016/j.cell.2020.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng K, et al. Functional interaction of lithocholic acid conjugates with M3 muscarinic receptors on a human colon cancer cell line. Biochim Biophys Acta. 2002;1588(1):48–55. doi: 10.1016/s0925-4439(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 79.Cheng K, et al. Differential expression of M3 muscarinic receptors in progressive colon neoplasia and metastasis. Oncotarget. 2017;8(13):21106–21114. doi: 10.18632/oncotarget.15500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng K, Raufman JP. Bile acid-induced proliferation of a human colon cancer cell line is mediated by transactivation of epidermal growth factor receptors. Biochem Pharmacol. 2005;70(7):1035–1047. doi: 10.1016/j.bcp.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 81.Shant J, et al. Akt-dependent NF-kappaB activation is required for bile acids to rescue colon cancer cells from stress-induced apoptosis. Exp Cell Res. 2009;315(3):432–450. doi: 10.1016/j.yexcr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Metz P, et al. Drug discovery and repurposing inhibits a major gut pathogen-derived oncogenic toxin. Front Cell Infect Microbiol. 2019;9:364. doi: 10.3389/fcimb.2019.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salmon H, et al. Host tissue determinants of tumour immunity. Nat Rev Cancer. 2019;19(4):215–227. doi: 10.1038/s41568-019-0125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakai A, et al. Control of lymphocyte egress from lymph nodes through β2-adrenergic receptors. J Exp Med. 2014;211(13):2583–2598. doi: 10.1084/jem.20141132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qiao G, et al. β-Adrenergic signaling blocks murine CD8+ T-cell metabolic reprogramming during activation: a mechanism for immunosuppression by adrenergic stress. Cancer Immunol Immunother. 2019;68(1):11–22. doi: 10.1007/s00262-018-2243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mohammadpour H, et al. β2 adrenergic receptor-mediated signaling regulates the immunosuppressive potential of myeloid-derived suppressor cells. J Clin Invest. 2019;129(12):5537–5552. doi: 10.1172/JCI129502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bucsek MJ, et al. β-adrenergic signaling in mice housed at standard temperatures suppresses an effector phenotype in cd8+ t cells and undermines checkpoint inhibitor therapy. Cancer Res. 2017;77(20):5639–5651. doi: 10.1158/0008-5472.CAN-17-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Godinho-Silva C, et al. Neuro-immune cell units: a new paradigm in physiology. Annu Rev Immunol. 2019;37:19–46. doi: 10.1146/annurev-immunol-042718-041812. [DOI] [PubMed] [Google Scholar]
- 89.Chavan SS, et al. Mechanisms and therapeutic relevance of neuro-immune communication. Immunity. 2017;46(6):927–942. doi: 10.1016/j.immuni.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huh JR, Veiga-Fernandes H. Neuroimmune circuits in inter-organ communication. Nat Rev Immunol. 2020;20(4):217–228. doi: 10.1038/s41577-019-0247-z. [DOI] [PubMed] [Google Scholar]
- 91.Fujii T, et al. Expression and function of the cholinergic system in immune cells. Front Immunol. 2017;8:1085. doi: 10.3389/fimmu.2017.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cox MA, et al. Beyond neurotransmission: acetylcholine in immunity and inflammation. J Intern Med. 2020;287(2):120–133. doi: 10.1111/joim.13006. [DOI] [PubMed] [Google Scholar]
- 93.Ha CWY, et al. Translocation of viable gut microbiota to mesenteric adipose drives formation of creeping fat in humans. Cell. 2020;183(3):666–683. doi: 10.1016/j.cell.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou J, Boutros M. JNK-dependent intestinal barrier failure disrupts host-microbe homeostasis during tumorigenesis. Proc Natl Acad Sci U S A. 2020;117(17):9401–9412. doi: 10.1073/pnas.1913976117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pope JL, et al. Claudin-1 overexpression in intestinal epithelial cells enhances susceptibility to adenamatous polyposis coli-mediated colon tumorigenesis. Mol Cancer. 2014;13:167. doi: 10.1186/1476-4598-13-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375(24):2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 97.Patterson L, et al. Glucosylceramide production maintains colon integrity in response to Bacteroides fragilis toxin-induced colon epithelial cell signaling. FASEB J. 2020;34(12):15922–15945. doi: 10.1096/fj.202001669R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature. 2016;529(7586):307–315. doi: 10.1038/nature17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen H, et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell. 2019;177(5):1217–1231. doi: 10.1016/j.cell.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Colosimo DA, et al. Mapping interactions of microbial metabolites with human G-protein-coupled receptors. Cell Host Microbe. 2019;26(2):273–282. doi: 10.1016/j.chom.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tomkovich S, et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest. 2019;129(4):1699–1712. doi: 10.1172/JCI124196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wick EC, et al. Stat3 activation in murine colitis induced by enterotoxigenic Bacteroides fragilis. Inflamm Bowel Dis. 2014;20(5):821–834. doi: 10.1097/MIB.0000000000000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alcaino C, et al. A population of gut epithelial enterochromaffin cells is mechanosensitive and requires Piezo2 to convert force into serotonin release. Proc Natl Acad Sci U S A. 2018;115(32):E7632–E7641. doi: 10.1073/pnas.1804938115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Worthington JJ, et al. Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 2018;11(1):3–20. doi: 10.1038/mi.2017.73. [DOI] [PubMed] [Google Scholar]
- 105.Di YZ, et al. Role of the brain-gut axis in gastrointestinal cancer. World J Clin Cases. 2019;7(13):1554–1570. doi: 10.12998/wjcc.v7.i13.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song P, et al. M3 muscarinic receptor antagonists inhibit small cell lung carcinoma growth and mitogen-activated protein kinase phosphorylation induced by acetylcholine secretion. Cancer Res. 2007;67(8):3936–3944. doi: 10.1158/0008-5472.CAN-06-2484. [DOI] [PubMed] [Google Scholar]
- 107.von Rosenvinge EC, et al. Bedside to bench: role of muscarinic receptor activation in ultrarapid growth of colorectal cancer in a patient with pheochromocytoma. Mayo Clin Proc. 2013;88(11):1340–1346. doi: 10.1016/j.mayocp.2013.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cives M, et al. The tumor microenvironment in neuroendocrine tumors: biology and therapeutic implications. Neuroendocrinology. 2019;109(2):83–99. doi: 10.1159/000497355. [DOI] [PubMed] [Google Scholar]
- 109.Banh RS, et al. Neurons release serine to support mRNA translation in pancreatic cancer. Cell. 2020;183(5):1202–1218. doi: 10.1016/j.cell.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rabben HL, et al. Neural signaling modulates metabolism of gastric cancer. iScience. 2021;24(2):102091. doi: 10.1016/j.isci.2021.102091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Runge TM, et al. Epidemiology of barrett’s esophagus and esophageal adenocarcinoma. Gastroenterol Clin North Am. 2015;44(2):203–231. doi: 10.1016/j.gtc.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mittal R, Vaezi MF. Esophageal motility disorders and gastroesophageal reflux disease. N Engl J Med. 2020;383(20):1961–1972. doi: 10.1056/NEJMra2000328. [DOI] [PubMed] [Google Scholar]
- 113.Blondy S, et al. Neurotrophins and their involvement in digestive cancers. Cell Death Dis. 2019;10(2):123. doi: 10.1038/s41419-019-1385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou Y, et al. A subtype of oral, laryngeal, esophageal, and lung, squamous cell carcinoma with high levels of TrkB-T1 neurotrophin receptor mRNA. BMC Cancer. 2019;19(1):607. doi: 10.1186/s12885-019-5789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bakst RL, Wong RJ. Mechanisms of perineural invasion. J Neurol Surg B Skull Base. 2016;77(2):96–106. doi: 10.1055/s-0036-1571835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tsunoda S, et al. Significance of nerve growth factor overexpression and its autocrine loop in oesophageal squamous cell carcinoma. Br J Cancer. 2006;95(3):322–330. doi: 10.1038/sj.bjc.6603255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Okumura T, et al. The biological role of the low-affinity p75 neurotrophin receptor in esophageal squamous cell carcinoma. Clin Cancer Res. 2006;12(17):5096–5103. doi: 10.1158/1078-0432.CCR-05-2852. [DOI] [PubMed] [Google Scholar]
- 118.Cocco E, et al. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15(12):731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Griffin N, et al. Clinicopathological significance of nerves in esophageal cancer. Am J Pathol. 2020;190(9):1921–1930. doi: 10.1016/j.ajpath.2020.05.012. [DOI] [PubMed] [Google Scholar]
- 120.Gao A, et al. Prognostic value of perineural invasion in esophageal and esophagogastric junction carcinoma: a meta-analysis. Dis Markers. 2016;2016:7340180. doi: 10.1155/2016/7340180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu G, et al. Prognosis and progression of ESCC Patients with perineural invasion. Sci Rep. 2017;7:43828. doi: 10.1038/srep43828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Goldberg E, Raufman JP. In: Yamada T, ed. Textbook of Gastroenterology. Wiley-Blackwell; 2015;60–72. [Google Scholar]
- 123.Xie G, et al. Cholinergic agonist-induced pepsinogen secretion from murine gastric chief cells is mediated by M1 and M3 muscarinic receptors. Am J Physiol Gastrointest Liver Physiol. 2005;289(3):G521–G529. doi: 10.1152/ajpgi.00105.2004. [DOI] [PubMed] [Google Scholar]
- 124.Ketterer K, et al. Reverse transcription-PCR analysis of laser-captured cells points to potential paracrine and autocrine actions of neurotrophins in pancreatic cancer. Clin Cancer Res. 2003;9(14):5127–5136. [PubMed] [Google Scholar]
- 125.Okada Y, et al. Nerve growth factor stimulates MMP-2 expression and activity and increases invasion by human pancreatic cancer cells. Clin Exp Metastasis. 2004;21(4):285–292. doi: 10.1023/B:CLIN.0000046131.24625.54. [DOI] [PubMed] [Google Scholar]
- 126.Okada Y, et al. Glial cell-derived neurotrophic factor upregulates the expression and activation of matrix metalloproteinase-9 in human pancreatic cancer. Surgery. 2003;134(2):293–299. doi: 10.1067/msy.2003.239. [DOI] [PubMed] [Google Scholar]
- 127.Miknyoczki SJ, et al. Neurotrophins and Trk receptors in human pancreatic ductal adenocarcinoma: expression patterns and effects on in vitro invasive behavior. Int J Cancer. 1999;81(3):417–427. doi: 10.1002/(SICI)1097-0215(19990505)81:3<417::AID-IJC16>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 128.Albini A. Tumor and endothelial cell invasion of basement membranes. The matrigel chemoinvasion assay as a tool for dissecting molecular mechanisms. Pathol Oncol Res. 1998;4(3):230–241. doi: 10.1007/BF02905254. [DOI] [PubMed] [Google Scholar]
- 129.Wang X, et al. The concept and controversy of retroperitoneal nerve dissection in pancreatic head carcinoma (Review) Int J Oncol. 2015;47(6):2017–2027. doi: 10.3892/ijo.2015.3190. [DOI] [PubMed] [Google Scholar]
- 130.Takahashi T, et al. Perineural invasion by ductal adenocarcinoma of the pancreas. J Surg Oncol. 1997;65(3):164–170. doi: 10.1002/(SICI)1096-9098(199707)65:3<164::AID-JSO4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 131.Crippa S, et al. Implications of perineural invasion on disease recurrence and survival after pancreatectomy for pancreatic head ductal adenocarcinoma. Ann Surg. doi: 10.1097/sla.0000000000004464. [published online October 19, 2020]. [DOI] [PubMed] [Google Scholar]
- 132.Ceyhan GO, et al. Pancreatic neuropathy results in “neural remodeling” and altered pancreatic innervation in chronic pancreatitis and pancreatic cancer. Am J Gastroenterol. 2009;104(10):2555–2565. doi: 10.1038/ajg.2009.380. [DOI] [PubMed] [Google Scholar]
- 133.Liebl F, et al. The impact of neural invasion severity in gastrointestinal malignancies: a clinicopathological study. Ann Surg. 2014;260(5):900–907. doi: 10.1097/SLA.0000000000000968. [DOI] [PubMed] [Google Scholar]
- 134.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mulligan LM. GDNF and the RET receptor in cancer: new insights and therapeutic potential. Front Physiol. 2018;9:1873. doi: 10.3389/fphys.2018.01873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tan X, et al. Nerve fibers in the tumor microenvironment in neurotropic cancer-pancreatic cancer and cholangiocarcinoma. Oncogene. 2021;40(5):899–908. doi: 10.1038/s41388-020-01578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Takehara A, et al. Gamma-aminobutyric acid (GABA) stimulates pancreatic cancer growth through overexpressing GABAA receptor pi subunit. Cancer Res. 2007;67(20):9704–9712. doi: 10.1158/0008-5472.CAN-07-2099. [DOI] [PubMed] [Google Scholar]
- 138.Al-Wadei HA, Schuller HM. Nicotinic receptor-associated modulation of stimulatory and inhibitory neurotransmitters in NNK-induced adenocarcinoma of the lungs and pancreas. J Pathol. 2009;218(4):437–445. doi: 10.1002/path.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zeng Q, et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature. 2019;573(7775):526–531. doi: 10.1038/s41586-019-1576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li L, et al. GKAP acts as a genetic modulator of NMDAR signaling to govern invasive tumor growth. Cancer Cell. 2018;33(4):736–751. doi: 10.1016/j.ccell.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Neunlist M, et al. Enteric glia inhibit intestinal epithelial cell proliferation partly through a TGF-beta1-dependent pathway. Am J Physiol Gastrointest Liver Physiol. 2007;292(1):G231–G241. doi: 10.1152/ajpgi.00276.2005. [DOI] [PubMed] [Google Scholar]
- 142.Liu YA, et al. 3-D imaging, illustration, and quantitation of enteric glial network in transparent human colon mucosa. Neurogastroenterol Motil. 2013;25(5):e324–e338. doi: 10.1111/nmo.12115. [DOI] [PubMed] [Google Scholar]
- 143.Pai R, et al. Prostaglandin E2 transactivates EGF receptor: a novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy. Nat Med. 2002;8(3):289–293. doi: 10.1038/nm0302-289. [DOI] [PubMed] [Google Scholar]
- 144.Buchanan FG, et al. Role of beta-arrestin 1 in the metastatic progression of colorectal cancer. Proc Natl Acad Sci U S A. 2006;103(5):1492–1497. doi: 10.1073/pnas.0510562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang D, et al. Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology. 2015;149(7):1884–1895. doi: 10.1053/j.gastro.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Doherty GA, et al. Proneoplastic effects of PGE2 mediated by EP4 receptor in colorectal cancer. BMC Cancer. 2009;9:207. doi: 10.1186/1471-2407-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pesic M, Greten FR. Inflammation and cancer: tissue regeneration gone awry. Curr Opin Cell Biol. 2016;43:55–61. doi: 10.1016/j.ceb.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 148.Gershon MD, Bursztajn S. Properties of the enteric nervous system: limitation of access of intravascular macromolecules to the myenteric plexus and muscularis externa. J Comp Neurol. 1978;180(3):467–488. doi: 10.1002/cne.901800305. [DOI] [PubMed] [Google Scholar]
- 149.Drewes JL, et al. Transmission and clearance of potential procarcinogenic bacteria during fecal microbiota transplantation for recurrent Clostridioides difficile. JCI Insight. 2019;4(19):130848. doi: 10.1172/jci.insight.130848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nakatsu G, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Feng Q, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 152.Bullman S, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Huybrechts I, et al. The human microbiome in relation to cancer risk: a systematic review of epidemiologic studies. Cancer Epidemiol Biomarkers Prev. 2020;29(10):1856–1868. doi: 10.1158/1055-9965.EPI-20-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kasai C, et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol Rep. 2016;35(1):325–333. doi: 10.3892/or.2015.4398. [DOI] [PubMed] [Google Scholar]
- 155.Rubinstein MR, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu T, et al. NF-kappaB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Felton J, et al. Two sides to colon cancer: mice mimic human anatomical region disparity in colon cancer development and progression. J Cancer Metastasis Treat. 2018;4:51. doi: 10.20517/2394-4722.2018.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bergstrom K, et al. Proximal colon-derived O-glycosylated mucus encapsulates and modulates the microbiota. Science. 2020;370(6515):467–472. doi: 10.1126/science.aay7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang Z, et al. Upregulated NTF4 in colorectal cancer promotes tumor development via regulating autophagy. Int J Oncol. 2020;56(6):1442–1454. doi: 10.3892/ijo.2020.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Murakawa K, et al. Prediction of lymph node metastasis and perineural invasion of biliary tract cancer by selected features from cDNA array data. J Surg Res. 2004;122(2):184–194. doi: 10.1016/j.jss.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 161.Zhang J, et al. Molecular markers associated with perineural invasion in pancreatic ductal adenocarcinoma. Oncol Lett. 2020;20(4):5. doi: 10.3892/ol.2020.11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Peng Z, et al. Cholinergic muscarinic receptor activation augments murine intestinal epithelial cell proliferation and tumorigenesis. BMC Cancer. 2013;13:204. doi: 10.1186/1471-2407-13-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xie G, et al. Acetylcholine-induced activation of M3 muscarinic receptors stimulates robust matrix metalloproteinase gene expression in human colon cancer cells. Am J Physiol Gastrointest Liver Physiol. 2009;296(4):G755–G763. doi: 10.1152/ajpgi.90519.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Raufman JP, et al. Muscarinic receptor agonists stimulate matrix metalloproteinase 1-dependent invasion of human colon cancer cells. Biochem Biophys Res Commun. 2011;415(2):319–324. doi: 10.1016/j.bbrc.2011.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Raufman JP, et al. Slc10a2-null mice uncover colon cancer-promoting actions of endogenous fecal bile acids. Carcinogenesis. 2015;36(10):1193–1200. doi: 10.1093/carcin/bgv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cheng K, et al. Diminished gallbladder filling, increased fecal bile acids, and promotion of colon epithelial cell proliferation and neoplasia in fibroblast growth factor 15-deficient mice. Oncotarget. 2018;9(39):25572–25585. doi: 10.18632/oncotarget.25385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cheng K, et al. Divergent effects of muscarinic receptor subtype gene ablation on murine colon tumorigenesis reveals association of M3R and zinc finger protein 277 expression in colon neoplasia. Mol Cancer. 2014;13:77. doi: 10.1186/1476-4598-13-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Michel MC, et al. How reliable are G-protein-coupled receptor antibodies? Naunyn Schmiedebergs Arch Pharmacol. 2009;379(4):385–388. doi: 10.1007/s00210-009-0395-y. [DOI] [PubMed] [Google Scholar]
- 169.Sriram K, et al. Detection and quantification of GPCR mRNA: an assessment and implications of data from high-content methods. ACS Omega. 2019;4(16):17048–17059. doi: 10.1021/acsomega.9b02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang K, et al. Hidden in the mist no more: physical force in cell biology. Nat Methods. 2016;13(2):124–125. doi: 10.1038/nmeth.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mierke CT. The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells. Rep Prog Phys. 2019;82(6):064602. doi: 10.1088/1361-6633/ab1628. [DOI] [PubMed] [Google Scholar]
- 172.Pound P, Ritskes-Hoitinga M. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. J Transl Med. 2018;16(1):304. doi: 10.1186/s12967-018-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Van Norman GA. Limitations of animal studies for predicting toxicity in clinical trials: is it time to rethink our current approach? JACC Basic Transl Sci. 2019;4(7):845–854. doi: 10.1016/j.jacbts.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gisselsson D, et al. Clonal evolution through genetic bottlenecks and telomere attrition: potential threats to in vitro data reproducibility. Genes Chromosomes Cancer. 2019;58(7):452–461. doi: 10.1002/gcc.22685. [DOI] [PubMed] [Google Scholar]
- 175.Lipinski KA, et al. Cancer evolution and the limits of predictability in precision cancer medicine. Trends Cancer. 2016;2(1):49–63. doi: 10.1016/j.trecan.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Goto T. Patient-derived tumor xenograft models: toward the establishment of precision cancer medicine. J Pers Med. 2020;10(3):64. doi: 10.3390/jpm10030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bedard PL, et al. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kuipers J, et al. Advances in understanding tumour evolution through single-cell sequencing. Biochim Biophys Acta Rev Cancer. 2017;1867(2):127–138. doi: 10.1016/j.bbcan.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ranganathan S, et al. Research in a time of enteroids and organoids: how the human gut model has transformed the study of enteric bacterial pathogens. Gut Microbes. 2020;12(1):1795492. doi: 10.1080/19490976.2020.1795389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Crespo M, et al. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat Med. 2017;23(7):878–884. doi: 10.1038/nm.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dedhia PH, et al. Organoid models of human gastrointestinal development and disease. Gastroenterology. 2016;150(5):1098–1112. doi: 10.1053/j.gastro.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kechele DO, Wells JM. Recent advances in deriving human endodermal tissues from pluripotent stem cells. Curr Opin Cell Biol. 2019;61:92–100. doi: 10.1016/j.ceb.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 183.Nikolaev M, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 2020;585(7826):574–578. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- 184.Roper J, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol. 2017;35(6):569–576. doi: 10.1038/nbt.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Vujkovic-Cvijin I, et al. Host variables confound gut microbiota studies of human disease. Nature. 2020;587(7834):448–454. doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Walter J, et al. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell. 2020;180(2):221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 187.Bernstein C, et al. Field defects in progression to gastrointestinal tract cancers. Cancer Lett. 2008;260(1–2):1–10. doi: 10.1016/j.canlet.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Flisikowska T, et al. A porcine model of familial adenomatous polyposis. Gastroenterology. 2012;143(5):1173–1175. doi: 10.1053/j.gastro.2012.07.110. [DOI] [PubMed] [Google Scholar]
- 189.Callesen MM, et al. A genetically inducible porcine model of intestinal cancer. Mol Oncol. 2017;11(11):1616–1629. doi: 10.1002/1878-0261.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Principe DR, et al. KRAS(G12D) and TP53(R167H) cooperate to induce pancreatic ductal adenocarcinoma in sus scrofa pigs. Sci Rep. 2018;8(1):12548. doi: 10.1038/s41598-018-30916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee JW, et al. Genetically engineered mouse models of pancreatic cancer: the KPC model (LSL-KrasG12D/+;LSL-Trp53R172H/+;Pdx-1-Cre), its variants, and their application in immuno-oncology drug discovery. Curr Protoc Pharmacol. 2016;73:14.39.1–14.39.20. doi: 10.1002/cpph.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Garrett WS. The gut microbiota and colon cancer. Science. 2019;364(6446):1133–1135. doi: 10.1126/science.aaw2367. [DOI] [PubMed] [Google Scholar]
- 193.Abdullah N, et al. Neural control of gut homeostasis. Am J Physiol Gastrointest Liver Physiol. 2020;319(6):G718–G732. doi: 10.1152/ajpgi.00293.2020. [DOI] [PubMed] [Google Scholar]
- 194.Ben QW, et al. Positive expression of L1-CAM is associated with perineural invasion and poor outcome in pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2010;17(8):2213–2221. doi: 10.1245/s10434-010-0955-x. [DOI] [PubMed] [Google Scholar]
- 195.Shen S, et al. Persistent cancer cells: the deadly survivors. Cell. 2020;183(4):860–874. doi: 10.1016/j.cell.2020.10.027. [DOI] [PubMed] [Google Scholar]
- 196.Schonkeren SL, et al. The emerging role of nerves and glia in colorectal cancer. Cancers (Basel) 2021;13(1):E152. doi: 10.3390/cancers13010152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wetzstein G, et al. Inference in artificial intelligence with deep optics and photonics. Nature. 2020;588(7836):39–47. doi: 10.1038/s41586-020-2973-6. [DOI] [PubMed] [Google Scholar]
- 198.Samuelson LC. Debate over the identity of an intestinal niche-cell population settled. Nature. 2018;558(7710):380–381. doi: 10.1038/d41586-018-05281-z. [DOI] [PubMed] [Google Scholar]
- 199.Kaestner KH. The intestinal stem cell niche: a central role for Foxl1-expressing subepithelial telocytes. Cell Mol Gastroenterol Hepatol. 2019;8(1):111–117. doi: 10.1016/j.jcmgh.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Rajagopal C, Harikumar KB. The origin and functions of exosomes in cancer. Front Oncol. 2018;8:66. doi: 10.3389/fonc.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]