Abstract
BACKGROUND
Current clinical management of patients with pulmonary nodules involves either repeated low-dose CT (LDCT)/CT scans or invasive procedures, yet causes significant patient misclassification. An accurate noninvasive test is needed to identify malignant nodules and reduce unnecessary invasive tests.
METHOD
We developed a diagnostic model based on targeted DNA methylation sequencing of 389 pulmonary nodule patients’ plasma samples and then validation in 140 plasma samples independently. We tested the model in different stages and subtypes of pulmonary nodules.
RESULTS
A 100-feature model was developed and validated for pulmonary nodule diagnosis; the model achieved a receiver operating characteristic curve–AUC (ROC-AUC) of 0.843 on 140 independent validation samples, with an accuracy of 0.800. The performance was well maintained in (a) a 6 to 20 mm size subgroup (n = 100), with a sensitivity of 1.000 and adjusted negative predictive value (NPV) of 1.000 at 10% prevalence; (b) stage I malignancy (n = 90), with a sensitivity of 0.971; (c) different nodule types: solid nodules (n = 78) with a sensitivity of 1.000 and adjusted NPV of 1.000, part-solid nodules (n = 75) with a sensitivity of 0.947 and adjusted NPV of 0.983, and ground-glass nodules (n = 67) with a sensitivity of 0.964 and adjusted NPV of 0.989 at 10% prevalence. This methylation test, called PulmoSeek, outperformed PET-CT and 2 clinical prediction models (Mayo Clinic and Veterans Affairs) in discriminating malignant pulmonary nodules from benign ones.
CONCLUSION
This study suggests that the blood-based DNA methylation model may provide a better test for classifying pulmonary nodules, which could help facilitate the accurate diagnosis of early stage lung cancer from pulmonary nodule patients and guide clinical decisions.
FUNDING
The National Key Research and Development Program of China; Science and Technology Planning Project of Guangdong Province; The National Natural Science Foundation of China National.
Keywords: Genetics, Oncology
Keywords: Diagnostics, Lung cancer
Introduction
Lung cancer is the leading cause of cancer-related mortality globally (1). It has been shown that the prognosis of lung cancer is highly correlated with the stage of the disease at diagnosis, with a 5-year overall survival rate decreasing dramatically from 85% for stage IA to 6% for stage IV disease (2). This makes lung cancer screening a highly favorable strategy for saving lives and reducing related medical costs.
The National Lung Screening Trial (NLST) has demonstrated that lung cancer screening by low-dose CT (LDCT) reduces mortality by 20% among current and former smokers of high lung cancer risk (>55 years old, >30 packs per year), which has led to a quick adoption of LDCT screening worldwide (3). Although LDCT does identify small nodules more effectively than conventional x-rays, this advantage comes with the challenge of distinguishing the small percentage of malignant nodules (~10%–20%) from the majority of the detected nodules that are deemed benign (4). Clinical nodule assessment tools, such as Mayo Clinic and Veterans Affairs (VA) models, based on imaging parameters as well as other risk factors, are widely used (5). However, the sensitivity of these tools is largely affected by nodule size and location. Suspected lung cancer lesions identified by LDCT can be further diagnosed via invasive approaches (e.g., bronchoscopy, transthoracic needle aspiration [TTNA], and surgery); however, complications may emerge, including hemorrhage, infection, pneumothorax, and even death. To avoid high false-positive rates, the new Lung Imaging Reporting And Data System (lung-RADS) classification and guidelines set the detection of nodules of 6 mm as the threshold for positivity. Nevertheless, positive CT scans can still be indecisive clinically, particularly for the class of intermediate-risk nodules (usually ranging from 6 to 20 mm in size, with a 5%–65% probability of malignancy as calculated by the clinical assessment tools, ref. 6).
Liquid biopsy has been considered as an easier and safer, more cost-effective, and less invasive method for cancer diagnosis and monitoring. Most noninvasive early detection approaches depend on identification of tumor-derived nucleic acids or proteins present in blood. For example, a blood test of proteomic biomarkers — Pulmonary Nodule Plasma Proteomic Classifier (PANOPTIC) — has been developed (7). Circulating tumor DNA (ctDNA) is exquisitely specific for an individual’s tumor; therefore, it can bypass the issue of false positivity encountered with other circulating biomarkers. Advancement in digital PCR and next-generation sequencing–based (NGS-based) technologies have drastically improved accuracy and sensitivity of ctDNA analysis in the detection of early stage cancers (8). This fast-developing field has drawn attention from international societies, such as the International Association for the Study of Lung Cancer (IASLC), which advocates using liquid biopsy in the management of non–small cell lung cancer (NSCLC).
In this study, we conducted ctDNA methylation profiling instead of somatic mutation detection to develop and validate a blood-based pulmonary nodule diagnosis test. When combined with standard care, it provides a more accurate clinical measurement for pulmonary nodule management.
Results
Clinical cohort.
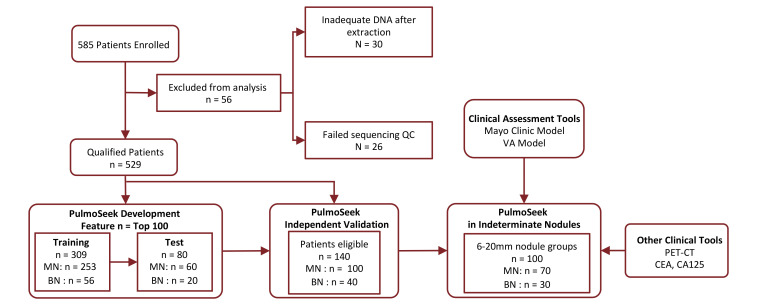
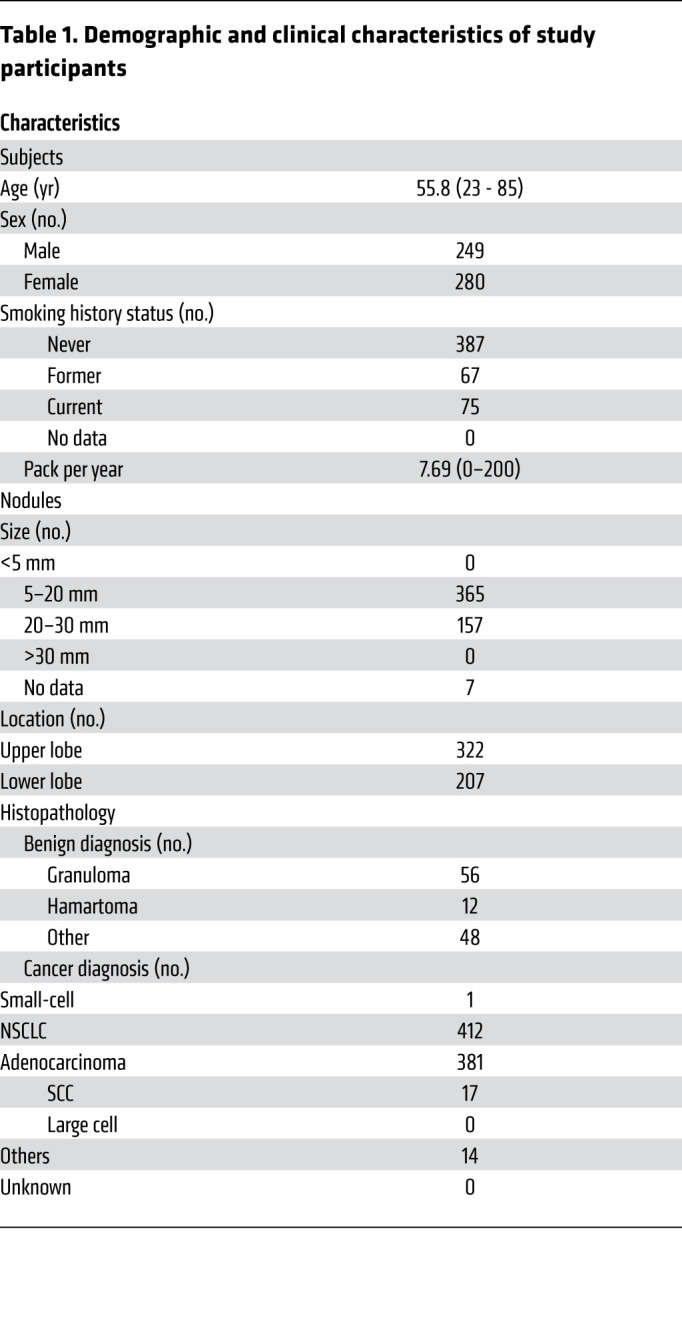
A total of 585 LDCT-positive patients were enrolled from thoracic surgery departments of 14 clinical sites across 8 different provinces in China. The percentage of malignancy based on pathological diagnosis from each province ranged from 75% to 88% (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI145973DS1). Fifty-six samples were excluded from analysis due to failed experimental quality control (QC), e.g., an inadequate circulating free (cfDNA) amount extracted from plasmas. The remaining 529 patients’ plasma samples (116 benign and 413 malignant) were used for DNA methylation profiling, model development, and validation. An overview of the study design is shown in Figure 1, and the demographic characteristics for the 529 patients are shown in Table 1.
Figure 1. Study flow of participants in the study.
Total 585 enrolled; 30 excluded due to limited cfDNA extracted (<5 ng) and 26 excluded due to failing sequencing QC. The model was developed, tested on 389 samples, and validated independently on 140 samples. The model was further validated on 100 indeterminate nodules (6–20 mm) in the validation set.
Table 1. Demographic and clinical characteristics of study participants.
The 529 plasma samples were first split into a model development set and an independent validation set at a 3:1 ratio. Furthermore, the model development set was divided into a training set (56 benign + 253 malignant) and a test set (20 benign + 60 malignant), so that the distribution of malignancy, age, and sex of the test set matched that of the training set, as shown in Figure 1. The percentages of malignancy were 82% and 75% in the training and test sets, respectively. The samples used for model development were primarily from early stage NSCLC. Specifically, stage I and II cancers comprised 94% or 98% of the total cancer patients in the training set and the test set, respectively. Benign and malignant samples were matched with respect to sex and smoking status (P > 0.05). The average size of the nodules in the benign group was 15.8 mm (9.6–22.0 mm), which is statistically smaller (P < 0.05) than that of the malignant group, which was 16.4 mm (9.9–22.9 mm). A summary of nodule types and American Joint Committee on Cancer (AJCC) stage information is shown in Supplemental Tables 2 and 3.
Development and validation of the diagnosis model PulmoSeek for pulmonary nodule diagnosis.
Methylation profiles of 309 plasma samples (Supplemental Table 1, training set) were analyzed using AnchorDx’s proprietary targeted methylation sequencing platform with a panel of 12,899 preselected lung cancer–specific methylation regions, corresponding to 105,844 CpG sites (9). A specific methylation signature was selected based on its performance of differentiating malignant from benign nodules.
The derived classification model, comprising 500 methylation target regions (features) achieved a receiver operating characteristic curve–AUC (ROC-AUC) of 0.823 (0.771–0.884) in the test set. Compared with the 500-feature model, the top 10 features within the model showed AUC values between 0.561 and 0.754 in the training set and 0.525 and 0.720 in the test set, demonstrating the necessity for building a multiple feature–based model (Supplemental Figure 2). For further downstream analysis, we annotated the selected 500 CpG features and performed a gene enrichment analysis. A total of 89 Gene Ontology (GO) categories were significantly enriched (Supplemental Table 4). The enriched categories include tissue proliferation and differentiation, such as embryonic morphogenesis (q value = 10−9.3), cell-fate commitment (q value = 10−4.7), stem cell proliferation (q value = 10−4.6), and epithelial tube morphogenesis (q value = 10−3.0). In addition, transcriptional factor activities, such as RNA polymerase II–specific DNA-binding transcription activator activity, were also significantly enriched (q value = 10−7.5). This result suggested that specific epigenetic signaling responsible for cell differentiation/reprogramming might be essential for pulmonary nodule development.
The performance of the model remained stable during a recursive feature elimination process: the smallest number of features that maintained an AUC within 1% of the 0.829 was 20, with an AUC of 0.810 (0.783–0.850) in the test set (Supplemental Figure 3). This indicates that a robust signature is maintained across different numbers of features selected.
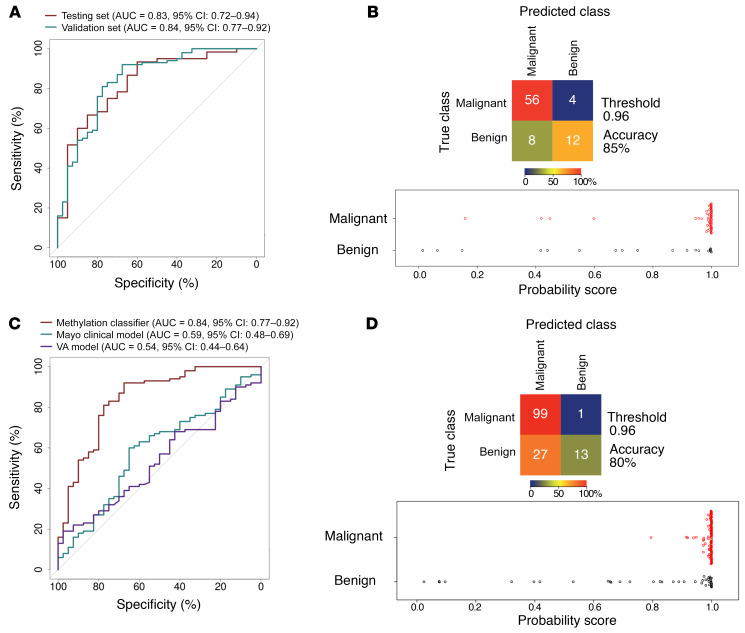
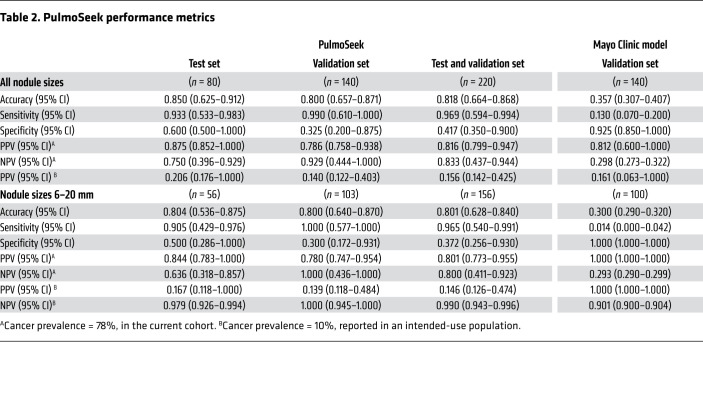
We then chose the 100-feature model, PulmoSeek, for the follow-up analysis. PulmoSeek achieved an overall AUC of 0.829 (0.719–0.942), with a high sensitivity of 0.933 (0.533–0.983) at a specificity of 0.600 (0.500–1.000) in the test set, corresponding to an accuracy of 0.850 (0.625–0.912) (Figure 2, A and C, and Table 2). The detailed information for each methylation feature of PulmoSeek is listed in Supplemental Table 5. Given excessive false positives and overdiagnosis in LDCT screening, unnecessary invasive procedures should be avoided under conditions of high-screening sensitivity in patients with benign nodules; that is, one should not sacrifice sensitivity (misclassify true positives) to pursue a reduction of unnecessary invasive procedures. This argues for a test with high sensitivity and high NPV, instead of a test with high specificity and high PPV. We assessed PulmoSeek’s performance with regard to its negative predictive value (NPV) and positive predictive value (PPV). In the current study cohort of 78% prevalence, the NPV was 0.750 (0.396–0.929) and the PPV was 0.875 (0.852–1.000) in the test set (Table 2). The sensitivities of the top 20–, 50–, and 500-feature models were 0.800 (0.675-0.912), 0.800 (0.713–0.912), and 0.900 (0.517–0.967), respectively, as shown in Supplemental Table 6.
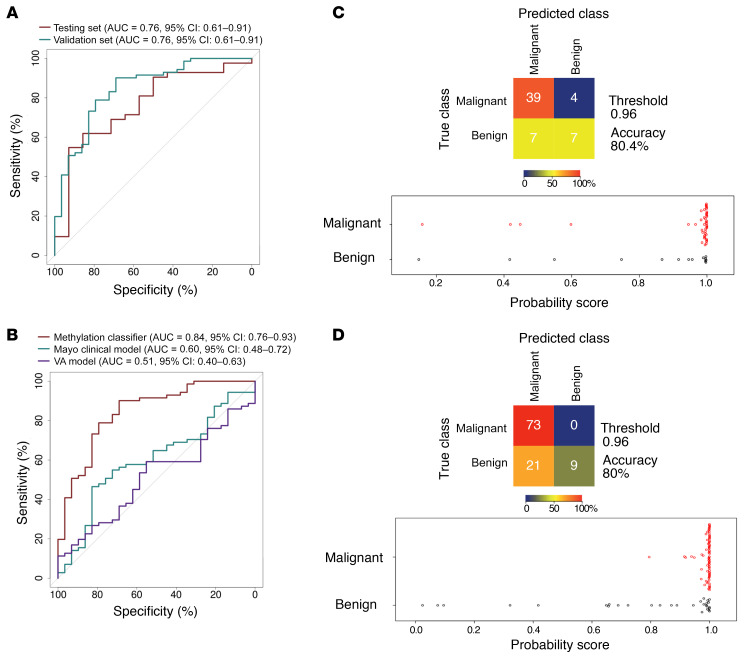
Figure 2. PulmoSeek performance compared with Mayo Clinic/VA model in all nodule sizes.
A representative ROC displays the classification performance of PulmoSeek. (A) In the test set, the AUC was 0.83 (0.72–0.94). In the validation set, the AUC was 0.84 (0.77–0.92). (B) In the validation set, the AUC of the Mayo Clinic classifier was 0.59 (0.48–0.69), and the AUC of the VA classifier was 0.54 (0.44–0.64). (C) Confusion matrices for PulmoSeek comparing the true class with the predicted class for benign (n = 20) and malignant (n = 60) nodule samples and distribution of PulmoSeek scores (range, 0 to 1) in the test set. (D) Confusion matrices for PulmoSeek comparing the true class with the predicted class for benign (n = 40) and malignant (n = 100) nodule samples and distribution of PulmoSeek scores (range, 0 to 1) in the validation set.
Table 2. PulmoSeek performance metrics.
We then used an independent cohort of 140 patient plasma samples (40 benign and 100 malignant; Supplemental Table 2, validation set) to further evaluate the performance of PulmoSeek. PulmoSeek achieved an AUC of 0.843 (0.769–0.918; Figure 2, A and D) with sensitivity of 0.990 (0.610–1.000) at specificity of 0.325 (0.200–0.875) and an overall accuracy of 0.800 (0.657–0.871). The NPV was 0.929 (0.444–1.000), and the PPV was 0.786 (0.758–0.938). In an intended-use population with a prevalence of malignant nodule at 10% (10), the NPV was calculated as 0.997 (0.947–1.000; Table 2). We further split the validation cohort into 3 subcohorts from high to low prevalences. We found that the NPV increased from 0.790 (0.370–1.000) to 1.000 (1.000–1.000) when the subcohort prevalence decreased from 79% to 23% (Supplemental Table 7).
The performance of PulmoSeek in patients with nodules of different histological types was further explored. Robust sensitivity for different subtypes, including minimally invasive adenocarcinoma (MIA) (95.2%), invasive adenocarcinoma (IA) (98.2%), and squamous cell carcinoma (SCC) (90.0%) were observed (Supplemental Table 8).
We also compared the performance of PulmoSeek to 2 clinical assessment models — the Mayo Clinic and VA models, which are based on clinical information and radiological characteristics, including nodule size and location, among others. In the validation set, PulmoSeek outperformed both of the clinical models, with an AUC of 0.843 (0.769–0.918) versus AUC of 0.591 (0.482–0.688) for the Mayo Clinic model and 0.544 (0.442–0.640) for the VA model (Figure 2B)
Classification accuracy of the model in very early stage lung cancers.
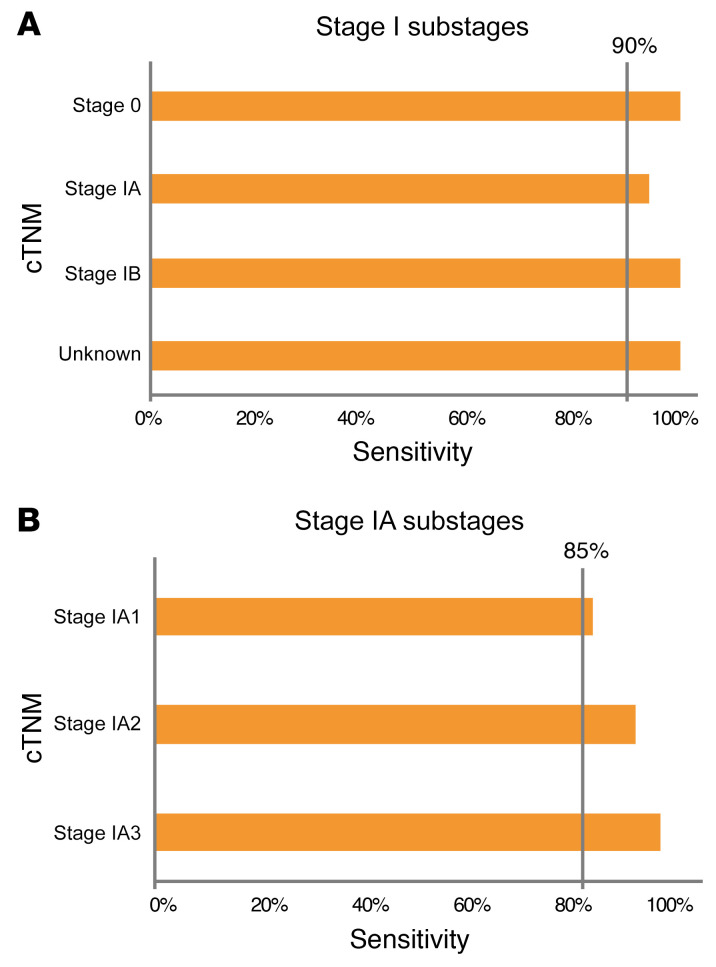
Very early stage cancer (tumor, node, metastasis [TNM] stage I) poses the greatest challenge for cancer diagnosis using a liquid biopsy (11). We tested PulmoSeek in different stage I substages in the validation cohort: it achieved sensitivities of 0.941 and 1.00 in stage IA (n = 85) and stage IB (n = 5), more specifically, 0.864, 0.950, and 1.000 in stage IA1 (n = 22), IA2 (n = 40), and IA3 (n = 23), respectively (Figure 3, A and B). In the combined test and validation set, PulmoSeek detected malignancies with sensitivity of 0.971 (0.942–0.993) for stage 0–I and 0.875 (0.625–1.000) for later stage cancers (Supplemental Figure 4A). The decreased sensitivity in late-stage cancers could be due to the limited number of late-stage samples (n = 8), which was not statistically significant (P = 0.248). Besides, the differences in performance for PulmoSeek in different groups were also calculated, and we observed no significant differences between groups (Supplemental Figure 4, B and C). Taken together, these results validated the accuracy of PulmoSeek, especially in detecting very early stage lung cancers.
Figure 3. PulmoSeek performance in early stage lung cancer.
In the independent validation set (A), PulmoSeek performance in early stage cancer: sensitivity was 100% in stage 0 (n = 2), 94.1% in stage IA (n = 85), and 100% in stage 1B (n = 5). (B) PulmoSeek performance in stage IA substages: sensitivity was 86.4% in stage IA1 (n = 22), 95.0% in stage IA2 (n = 40), and 100% in stage 1A3 (n = 23).
PulmoSeek outperformed clinical prediction models and conventional cancer biomarker tests in indeterminate nodules.
Diagnosis of indeterminate pulmonary nodules (IPN) (nodules ranging between 6 and 20 mm in size) is challenging for clinicians due to the lack of well-specified optimal action strategies (12). The 6 to 20 mm size nodules made up about 70% of the test set (56 of 80) and the independent validation set (100 of 140) in this study (Supplemental Table 9). PulmoSeek achieved an AUC of 0.762 (0.610–0.913), sensitivity of 0.905 (0.429–0.976), and specificity of 0.500 (0.286–1.000) in the test set (Figure 4, A and B, and Table 2). In the independent validation set, PulmoSeek achieved an AUC of 0.844 (0.759–0.932), sensitivity of 1.000 (0.577–1.000), and specificity of 0.300 (0.172–0.931; Figure 4, A and D, Table 2, and Supplemental Table 10). For nodules above 20 mm (n = 59), PulmoSeek had an AUC of 0.860 (0.740–0.964) with sensitivity of 0.977 (0.628–1.000) and specificity of 0.562 (0.375–0.938; Supplemental Figure 5).
Figure 4. PulmoSeek performance compared with Mayo Clinic/VA model in 6–20 mm nodule sizes.
A representative ROC displays the classification performance of PulmoSeek. (A) In the test set, the AUC was 0.76 (0.61–0.91). In the validation set, the AUC was 0.84 (0.76–0.93). (B) In the validation set, the AUC of the Mayo Clinic classifier was 0.60 (0.48–0.72) and the AUC of the VA classifier was 0.51 (0.40–0.63). (C) Confusion matrices for PulmoSeek comparing the true class with the predicted class for benign (n = 14) and malignant (n = 43) nodule samples and distribution of PulmoSeek scores (range, 0 to 1) in the test set. (D) Confusion matrices for PulmoSeek comparing the true class with the predicted class for benign (n = 30) and malignant (n = 73) nodule samples and distribution of PulmoSeek scores (range, 0 to 1) in the validation set.
When compared with the Mayo Clinic and VA models, PulmoSeek outperformed both clinical models in the validation set in which an AUC of 0.602 (0.482–0.719) was obtained with the Mayo Clinic model and an AUC of 0.512 (0.402–0.633) was obtained with the VA model (Figure 4C).
Consistent with previous studies, conventional cancer biomarkers such as carcinoembryonic antigen (CEA), cancer antigen 125 (CA-125), and cancer antigen 135 (CA-135) alone failed to effectively identify malignant nodules in our cohort (13). The corresponding sensitivity of CEA, CA-125, and CA-135 was only 0.010, 0.030, and 0.030, respectively, as compared with sensitivity of 0.950 by using PulmoSeek (Supplemental Figure 6).
PulmoSeek outperformed PET-CT in different nodule types, including ground-glass nodule.
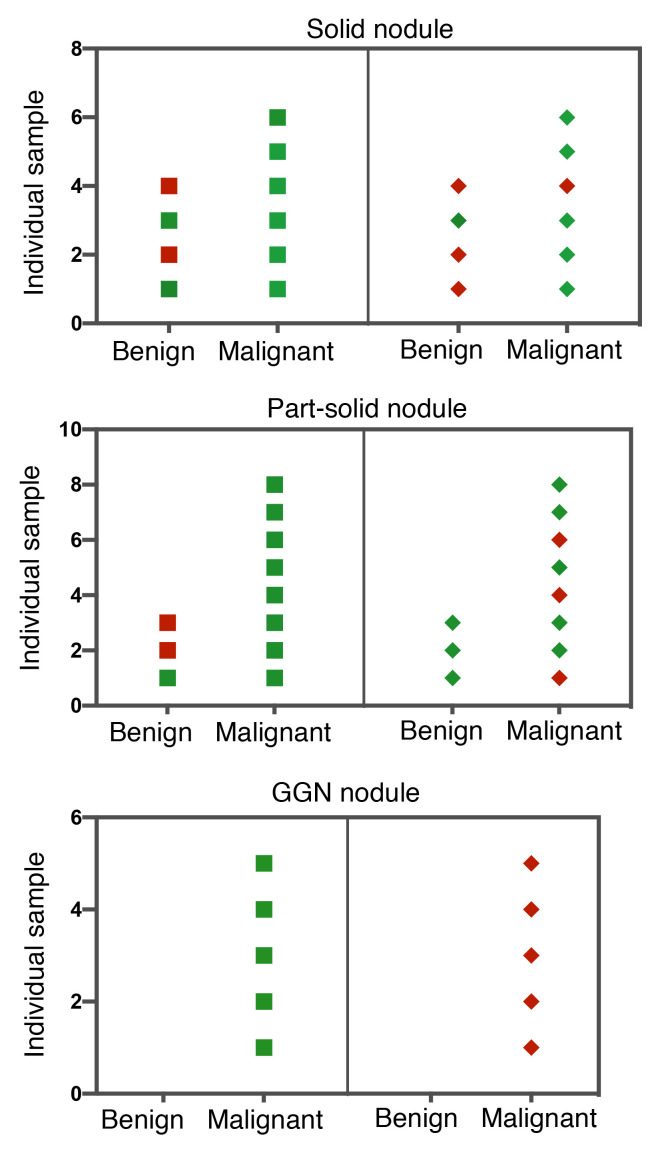
PET-CT is known to be more accurate than CT alone for characterizing solid-type pulmonary nodules, resulting in fewer equivocal findings (14). Thus, low- to intermediate-risk nodules are usually recommended to be further evaluated by PET-CT. However, PET-CT performance drops substantially for subsolid nodules (part-solid and ground-glass nodule [GGN]). We assessed the performance of PulmoSeek in comparison with PET-CT on the participants with established PET-CT records in our independent validation set. The accuracy of PulmoSeek was significantly higher than that of PET-CT: it correctly classified 8 out of 10 patients in the solid nodule (SN) subgroup, 9 out of 11 in the part-solid nodule subgroup, and 5 out of 5 in the GGN subgroup, while PET-CT correctly classified 6 out of 10 patients in the SN subgroup, 7 out of 11 in the part-solid nodule subgroup, and 0 out of 5 in the GGN subgroup (Figure 5). This performance was maintained across all nodule types in the combined test and independent validation sets: the model demonstrated a sensitivity of 1.000 (0.702–1.000) in the solid subgroup (n = 78), 0.947 (0.509–1.000) in the part-solid subgroup (n = 75), and 0.964 (0.518–1.000) in the GGN subgroup (n = 67; Supplemental Figure 7).
Figure 5. PulmoSeek performance in different nodule types and comparison with PET-CT.
In the independent validation set samples with PET-CT records, the diagnosis result for each patient using PulmoSeek (squares) and PET-CT (diamonds) is shown. Green indicates the sample was diagnosed correctly, and the red incorrectly. PulmoSeek correctly identified 8 out 10 patients in the SN subgroup, 9 out of 11 in the part-solid nodule subgroup, and 5 out of 5 in the GGN subgroup. The PET-CT correctly identified 6 out 10 patients in the SN subgroup, 7 out of 11 in the part-solid nodule subgroup, and 0 out of 5 in the GGN subgroup.
A strategy of integrating liquid biopsy–based ctDNA and protein marker analysis followed by PET-CT imaging for cancer screening has been proposed (15). We tried to assess this strategy in our cohort by testing the performance of PET-CT on the malignant nodules identified by our methylation model. In both solid and part-solid nodule groups, integration of PET-CT did not reduce false-positive rates. Rather, it introduced a significant number of false negatives. In SNs, PulmoSeek had a false-positive rate of 14.2% (2 out of 14 misclassified), while integration of PET-CT resulted in a false-positive rate of 16.2% (2 out of 12) and a false-negative rate of 100% (2 out of 2). In all nodules, PulmoSeek had a false-positive rate of 14.8% (4 out of 27 misclassified), while integration of PET-CT had a false-positive rate of 11.7% (2 out of 17) and a false-negative rate of 80% (8 out of 10) (Supplemental Table 11).
Discussion
In this study, we analyzed ctDNA methylation profiles in 529 pulmonary nodule patients from 14 hospitals in China and developed and validated a model called PulmoSeek for pulmonary nodule diagnosis. Notably, PulmoSeek demonstrated high sensitivity and NPV at a moderate specificity across different lesion locations, nodule types, and stages of lung cancer. To the best of our knowledge, this is the largest retrospective study so far to validate a blood-based methylation model for lung nodule diagnosis.
Most recently, a prospective, interventional study of more than 10,000 women using a multiomics blood test coupled with PET-CT imaging demonstrated its clinical potential for early cancer screening (15). However, this strategy may not apply to lung cancer screening efficiently for the following reasons: (a) using a sequencing-based method for initial screening can be costly and throughput limited; and (b) a ctDNA somatic mutation assay may yield a high number of false positives that would need a PET-CT to be filtered out; however, it is reported that PET-CT performs suboptimally in characterizing subsolid nodules.
Our study demonstrates a potential new diagnosis workup of LDCT, followed by a blood-based test as a less invasive and cost-effective strategy for identifying early stage lung cancer. The cost-effectiveness advantage is noteworthy: chest LDCT costs less than $50 (in China), and the process only takes a few minutes (16). This is clinically critical because in the United States, there is a well-documented high rate of pulmonary nodules (17). Similarly, in China, it is estimated that over 100 million people live with lung nodules and the number is growing quickly each year (18). LDCT screening has already been widely adopted as a major tool for lung cancer screening. We believe that a strategy of coupling LDCT with PulmoSeek is more practical and suited for population-based lung cancer screening.
In the NLST study, all enrolled patients had pulmonary nodules of 4 mm or larger in diameter, and the false-positive rate was over 96.4% after 3 rounds of LDCT screening (6). This is particularly impactful for nodules between 6 and 20 mm, i.e., indeterminate nodules (pCA 5%–65%), which account for the majority of the nodules identified by LDCT (50%–76%) and for which the risk of malignancy is hard to determine with current clinical risk assessment models (19). Current guidelines suggest further evaluation with PET-CT scan, endobronchial ultrasound–guided transbronchial forceps biopsy (EBUS-TBB), or TTNA. Integrated PET-CT imaging shows good sensitivity (~88%) and specificity (~75%). However, this performance is limited to SNs, and there are still possibilities of false positives (e.g., granulomatous disease) and false negatives (e.g., carcinoid). It has been reported that the sensitivity of PET-CT dropped to 50% in part-solid nodules and even lower than 20% in GGN nodules (20); the performance of EBUS-TBB on peripheral pulmonary nodules is largely dependent on nodule size: the diagnostic sensitivity is significantly higher for nodules larger than 20 mm than for those of 20 mm or smaller (~50% sensitivity), with only 35% sensitivity for nodules between 5 and 10 mm in diameter (21). The EBUS-TBB procedure is also expertise dependent. TTNA has a 1% risk of hemorrhage and less patient compliance (22). Despite the high medical costs associated with those 3 approaches, clinicians can still be left uncertain in management decisions, leading to potential overdiagnosis and/or overtreatment. To fulfill unmet clinical and economical needs, an alternative/complementary, noninvasive approach for nodule management is needed in order, on the one hand, to provide prompt necessary treatment when the nodule is in the early stages of lung cancer and, on the other, to minimize testing when the nodule is deemed benign.
PulmoSeek provides a potential solution for meeting all of the above needs: this blood-based assay was developed on a group of pathology-confirmed nodules mostly at early stages (stage I and II, 92%) from thoracic departments and with a high prevalence of lung cancer (78%). In the current study, PulmoSeek achieved an AUC of 0.843, high sensitivity of 0.990, and NPV of 0.929 in the independent validation set. It outperformed current clinical assessment models (23). Ultimately, a rule-out test is likely to be most clinically beneficial in the group of a lower prevalence of lung cancer: doctors would have a reliable test to rule out the “true negatives” and effectively reduce the “uncertain cases” so as to avoid overtreatment. When adjusted to an average prevalence of 10%, the model had a very high NPV of 0.997, with specificity over 40%. This suggests that PulmoSeek alone could reduce more than 40% of unnecessary invasive procedures on benign nodules with less than a 0.3% false-negative rate. The superior NPV and sensitivity of PulmoSeek compare favorably to other published rule-out models to date, for which the NPV range is between 85% and 98% (depending on the prevalence) and sensitivity is between 85% and 97% (24–27). Those models are usually used in combination with clinical parameters, such as age, smoking status, nodule size/location, and classic cancer biomarkers (e.g., CEA).
PulmoSeek demonstrated a robust performance in very early stages of lung cancer (stages 0 and I). The slightly lower sensitivity observed in the later stage cancers (87.5%) was due to misclassification of 1 out of the 8 late-stage samples. This misclassified sample was a part-solid nodule of smaller nodule size (11 mm); the subgroup PulmoSeek showed a relatively lower performance in the current study. We further tested with another independent cohort (n = 12) of late-stage nodule samples, and PulmoSeek correctly identified all of them (our unpublished observations). Nevertheless, a larger cohort is required to further validate performance of PulmoSeek in late-stage cancers.
In addition, PulmoSeek is accurate in diagnosing 6 to 20 mm IPNs and subsolid nodules (part-solid and GGN nodules), unlike other tests that are limited to SNs. We are currently combining PulmoSeek with LDCT image artificial intelligence (AI) to further augment the overall diagnostic performance (our unpublished observations).
To gauge the potential clinical utility, a trade-off value was calculated as follows: specificity/(1 – sensitivity) ≥ (prevalence/1 – prevalence) × harm/benefit, where the harm/benefit ratio is defined as the net harm of a false-negative test to the net benefit of a true-negative test (28); PulmoSeek produced a harm/benefit value of 292.5 (e.g., 292.5 true-negative results accompanied with one false-negative result) in the intended-use population (10% prevalence). These results suggest that the trade-off is acceptable and warrant a future clinical utility study. As a matter of fact, a large prospective clinical validation study — The Thunder Project — was started in 2018 aiming to enroll more than 10,000 patients across 23 top hospitals in China (ClinicalTrials.gov NCT03651986; ref. 29). As of January 2021, over 9500 patients have already been enrolled (our unpublished observations).
In summary, we have developed and validated a ctDNA methylation assay for diagnosis of malignant and benign pulmonary nodules. It showed superior performance as compared with existing clinical procedures. Coupled with LDCT, it could become a robust tool for pulmonary nodule management and lung cancer screening.
Methods
Study design and participants.
We performed a multicenter, retrospective diagnostic study using plasma samples collected from 14 hospitals’ thoracic departments in China. From May 2017 to February 2019, 585 patients with malignant and benign pulmonary nodules were enrolled. The participating hospitals were the First Affiliated Hospital of Guangzhou Medical University, Second Xiangya Hospital of Central South University, The First Affiliated Hospital of Sun Yat-sen University, Shenzhen People’s Hospital, Nanfang Hospital of Southern Medical University, Jiangsu Province Hospital, West China Hospital of Sichuan University, Xuanwu Hospital of Capital Medical University, Beijing Cancer Hospital, Qilu Hospital of Shandong University, The Second Affiliated Hospital of Nanhua University of Hunan Province, Anhui Chest Hospital, Xiangya Hospital of Central South University, and the Fourth Affiliated Hospital of Harbin Medical University.
Adult patients 18 years old or older were included with the following criteria: either sex; single pulmonary nodules detected by standard or LDCT screening with nodule size between 5 and 30 mm; and nodule types of SNs, part-solid nodules (mixed GGNs [mGGN]), and pure GGNs (pGGN). Exclusion criteria included pregnant or lactating females, patients with 2 or more nodules with lesion size of 5 mm or more, patients with metastasis symptoms, such as pleural effusion or mediastinal lymph node’s shorter diameter larger than 10 mm, patients without confirmed pathological diagnosis after surgery, or patients with cancer confirmed pathologically within 2 years prior to enrollment (except for nonmelanoma skin cancer). All patients underwent pathological examination, and the detailed deidentified clinical information, including demographics, LDCT imaging reports, and pathology reports, were transferred to the investigators.
Procedures.
All blood samples were collected in Streck cell-free DNA BCT tubes (Streck, catalog 218962) according to the manufacturer’s instructions and shipped to AnchorDx’s certified molecular diagnosis laboratory. Plasma was separated immediately from the whole blood samples upon receipt using a standard protocol described previously (9) and stored at –80°C until use. Repeated freezing and thawing of plasma was avoided to prevent cfDNA degradation and genomic DNA contamination from WBCs. cfDNA was isolated by the Thermo MagMAX Cell‑Free DNA Kit (Thermo Fisher Scientific, catalog A29319) according to the manufacturer’s protocol. The concentration of cfDNA was measured by Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, catalog Q32854), and the quality was examined using the Agilent High Sensitivity DNA Kit (catalog 5067–4626).
Full details of sample preparation and targeted cfDNA methylation sequencing were described previously (9). In brief, bisulfite conversion was performed using the EZ DNA Methylation-Lightning Kit (catalog D5031, Zymo Research) according to the manufacturer’s protocol. Targeted genome methylation analysis was conducted using a proprietary AnchorIRIS technology on 10 ng input cfDNA. AnchorIRIS prelibrary construction was carried out using the AnchorDx EpiVisio Methylation Library Prep Kit (AnchorDx, catalog A0UX00019) and the AnchorDx EpiVisio Indexing PCR Kit (AnchorDx, catalog A2DX00025). The amplified prehybridization libraries were subsequently purified using IPB1 Magnetic Beads, and concentration was determined using the Qubit dsDNA HS Assay Kit. Prehybridization libraries containing more than 400 ng DNA were considered qualified for target enrichment. Next, target enrichment was performed using the AnchorDx EpiVisio Target Enrichment Kit (AnchorDx, catalog A0UX00031). A custom-made lung cancer methylation panel (see below), which consisted of 12,899 preselected regions enriched for lung cancer specific methylations, was used for this study.
After probe hybridization, specific portions of the DNA libraries bound with biotinylated probes were pulled down using Dynabeads M270 streptavidin beads (Thermo Fisher Scientific, catalog 65306). These enriched libraries were further amplified with P5 and P7 primers using KAPA HiFi HotStart Ready Mix (KAPA Biosystems, catalog KK2602), and PCR product was then purified with Agencourt AMPure XP Magnetic Beads (Beckman Coulter, catalog A63882). The resulting libraries were sequenced on the NovaSeq 6000 System (Illumina Inc).
Lung cancer–specific methylation panel development.
Early stage lung cancer methylation profiles were generated by targeted bisulfite sequencing. DNA extracted from a total of 232 tissue samples, including 133 benign pulmonary nodule samples (inflammation, granulomas, tuberculosis, fungal infection, hamartomas, and sclerosing hemangioma) and 99 malignant pulmonary nodules (IA, MIA, adenocarcinoma in situ, SCC), were analyzed by the TruSeq Methyl Capture EPIC Library Kit (Illumina, catalog FC-151-1002). Differentially methylated CpG sites were discovered using R package DSS, version 2.14.0 (30). By using a filtering criteria of P < 0.001 and Δ (i.e., group difference) > 0.02, hypermethylated and hypomethylated sites were identified. Using this information together with the lung cancer–specific DNA methylation markers discovered from The Cancer Genome Atlas (TCGA) database, we developed a targeted methylation panel consisting of 12,899 lung cancer–associated informative methylation regions covering 105,844 CpG sites.
Sequencing data analysis.
Sequencing data were processed as previously reported (9). Briefly, the sequencing quality was evaluated by the Illumina Sequencing Analysis Viewer and FastQC software (Babraham Bioinformatics). Sequencing adapters and 3′ low quality bases were trimmed from raw sequencing reads using a custom algorithm and then aligned to the C→T in silico converted hg19 reference genome, using Bismark version 0.17.0 (Bowtie2 as the default aligner behind Bismark). Aligned reads were then evaluated by Picard, version 2.5.0, for metrics that measured the performance of target-capture based bisulfite sequencing assays (http://broadinstitute.github.io/picard).The biases of specific motifs or GC-enriched regions were excluded. After the preliminary analysis, we calculated the average coverage as well as the missing rate for each CpG site. The CpG sites with coverage less than 30× and/or with missing rate greater than 0.20 were filtered out.
Differential methylation signature analysis.
Differential methylation (DM) analysis was performed on the training cohort of lung cancer patients and controls using R package DSS, version 2.14.0 (30). Differentially methylated CpG sites were identified by comparing malignant to benign samples (P < 0.001, Δ > 0.02) and further assembled into differentially methylated regions (DMRs). Targeted regions of the capture panel covered by DMRs (at least 50% bases of a target region covered) were selected as candidate features to build classification models of malignant/benign states.
Deep learning–based benign-malignant prediction modeling.
Methylation features were selected by calculating the comethylated reads (reads having at least 3 methylated CpGs within a sliding window of 5 CpGs or at least 2 methylated CpGs within a sliding window of 3 CpGs) ratios within the DMRs (9). Then in light of the heuristic nature of various methylation metrics, such as comethylation and epiallele (31), an autoencoder (AE) neural network (32) was applied to further construct the representative methylation features. The AE is a type of unsupervised neural network with wide applications, particularly in image processing. A general AE architecture was shown in Supplemental Figure 8. We took advantage of it when analyzing methylation sequencing data to convert intractable high-dimensional sequencing reads into lower dimensional numerical representative features (31).
In our model, the input matrix X represented each DMR and the hidden vector h was the low-dimensional representative feature of the DMR methylation status after training. The encoder was implemented by a ResNet model–based convolutional neural network model (33). For the decoder, to reconstruct the region from h, deconvolutional layers, composed of the reverse operations of the convolutional layers in the encoder, were implemented (34). The whole model was further optimized by the Adam algorithm (35).
We then built a gradient boosted trees-based classifier with Scikit-Learn LightGBM using the AE-based methylation features. During the training process, we tuned the number of trees, maximum tree depth, and the number of leaves used by the lightGBM model, as these were major parameters to overcoming the overfitting problem. The learning rate and other parameters were kept at its default values. The number of leaves was set up between 3 and 20 and the depth of trees between 3 and 10 and up to 1000 for each model.
Comparison with Mayo Clinic and VA models.
The Mayo Clinic model for malignancy in pulmonary nodules calculated the malignancy probability as a function of 3 clinical and 3 radiographic variables (36): probability of malignancy = ex/(1 + ex), where x = –6.8272 + (0.0391 × age) + (0.7917 × smoking) + (1.3388 × cancer) + (0.1274 × nodule diameter) + (1.0407 × spiculation) + (0.7838 × upper lobe), where e is Euler’s number, a mathematical constant approximately equal to 2.71828.
The VA model for malignancy in pulmonary nodules calculated the malignancy probability as a function of 3 clinical and 1 radiographic variables (37): probability of malignancy = 100 × [e(logx)/ 1 + e(logx)], where x = −8.404 + 2.061 × smoke + 0.779 × age/10 + 0.112 × diameter + 0.567 × yearsquit/10, where smoke is 1 if a current or former smoker (otherwise 0), age/10 is age in years divided by 10, diameter is the largest diameter of the nodule in millimeters, yearsquit/10 is the number of years since quitting smoking divided by 10, and e is Euler’s number.
Gene set enrichment analysis.
We performed gene set enrichment analysis using the R-package Metascape (38).
Statistics.
Statistical analysis was performed as described in each figure legend, and sample sizes are given in each figure legend. Categorical variables, including sex, nodule subtypes, etc., were compared using Fisher’s exact test. Sensitivities of different AJCC stages for malignant nodules were also compared with Fisher’s exact test. Continuous variables such as age were compared using Student’s t test, and 95% CIs were calculated based on 2000 bootstrap resamplings of the classification results. The sensitivity, specificity, accuracy, PPV, and NPV of PulmoSeek and other models in detecting malignant nodules were obtained by comparison with pathological outcomes. ROCs were obtained using the pROC R package (version 1.15.3). Positive and negative classifications of PulmoSeek were determined by the cutoff value (0.960) using Youden’s index, while positive and negative values for CEA, CA-125, and CA-135 were determined by the clinical report. Unless otherwise specified, all statistical tests were 2 sided. FDR (Benjamini-Hochberg method) correction was used for multiple test correction. All statistical analysis was performed with R software, version 3.32.
Study approval.
This study was approved by the IRBs at the hospitals involved. Written consent was obtained from each participant.
Author contributions
JH and JBF conceived the study. WL, ZC, JBF, and JH designed the experiments. HL, DZ, and ZC performed the experiments. HJ, ZJ, XC, JL, and XT performed the data modeling. CL, JL, WY, HC, CC, FY, CZ, LL, HT, KC, Xiang Liu, ZW, NX, QD, LC, YY, XZ, ZJ, LM, JW, and all other authors contributed to data acquisition. JL, ZC, and WL contributed to data visualization. Xin Liu, WL, ZC, JBF, and JH contributed substantially to the development of this manuscript. All authors reviewed and approved the manuscript.
Supplementary Material
Acknowledgments
We thank all the participants for their generosity. This study was supported by the China National Science Foundation (nos. 82022048 and 81871893), the Key Project of Guangzhou Scientific Research Project (no. 201804020030), the National Key Research and Development Project (nos. 2017YFC0907903, 2017YFC1309002, and 2017YFC0112704), the Scheme of Guangzhou Economic and Technological Development District for Leading Talents in Innovation and Entrepreneurship (no. 2017-L152), the Scheme of Guangzhou for Leading Talents in Innovation and Entrepreneurship (no. 2016007), the Scheme of Guangzhou for Leading Team in Innovation (no. 201909010010), the Science and Technology Planning Project of Guangdong Province, China (no. 2017B020226005), the High-Level University Construction Project of Guangzhou Medical University (no. 20182737, 201721007, 201715907, 2017160107), and the Guangdong High Level Hospital Construction “Reaching Peak” Plan.
Version 1. 04/01/2021
In-Press Preview
Version 2. 05/17/2021
Electronic publication
Footnotes
Conflict of interest: JBF, ZC, JT, Xin Liu, DZ, HL, XT, and ZJ are current employees of AnchorDx Medical Co. or AnchorDx Inc.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(9):e145973.https://doi.org/10.1172/JCI145973.
Contributor Information
Wenhua Liang, Email: liangwh1987@163.com.
Zhiwei Chen, Email: zhiwei_chen@anchordx.com.
Caichen Li, Email: yzg_lcc@163.com.
Jun Liu, Email: liujun9707@sina.com.
Jinsheng Tao, Email: jinsheng_tao@anchordx.com.
Xin Liu, Email: xin_liu@anchordx.com.
Dezhi Zhao, Email: dezhi_zhao@anchordx.com.
Weiqiang Yin, Email: yinwq88@21cn.com.
Hanzhang Chen, Email: hanzhangchen@163.com.
Chao Cheng, Email: drchengchao@163.com.
Fenglei Yu, Email: yufenglei@csu.edu.cn.
Chunfang Zhang, Email: zhcf3801@csu.edu.cn.
Hui Tian, Email: tianhuiql@126.com.
Kaican Cai, Email: doc_cai@163.com.
Xiang Liu, Email: crisis163@163.com.
Zheng Wang, Email: shwnzn@163.com.
Ning Xu, Email: xuning200901@aliyun.com.
Qing Dong, Email: dongqing100859@163.com.
Liang Chen, Email: 2434794740@qq.com.
Yue Yang, Email: zlyangyue@bjmu.edu.cn.
Xiuyi Zhi, Email: xiuyizhi2015@163.com.
Hui Li, Email: hui_li@anchordx.com.
Xixiang Tu, Email: xixiang_tu@anchordx.com.
Xiangrui Cai, Email: caixiangrui@dbis.nankai.edu.cn.
Zeyu Jiang, Email: zeyu_jiang@anchordx.com.
Hua Ji, Email: mike_ji@anchordx.com.
Lili Mo, Email: 542856111@qq.com.
Jiaxuan Wang, Email: 1174527917@qq.com.
Jian-Bing Fan, Email: jianbingfan1115@smu.edu.cn.
Jianxing He, Email: jianxing@gird.cn.
References
- 1.Didkowska J, et al. Lung cancer epidemiology: contemporary and future challenges worldwide. Ann Transl Med. 2016;4(8):150. doi: 10.21037/atm.2016.03.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Torre LA, et al. Lung cancer and personalized medicine. In: Ahmad A, Gadgeel S, eds. Advances in Experimental Medicine and Biology. Springer; 2016:1–19. [PubMed] [Google Scholar]
- 3.Kramer BS, et al. Lung cancer screening with low-dose helical CT: results from the National Lung Screening Trial (NLST) J Med Screen. 2011;18(3):109–111. doi: 10.1258/jms.2011.011055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McWilliams A, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med. 2013;369(10):910–919. doi: 10.1056/NEJMoa1214726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair VS, et al. Accuracy of models to identify lung nodule cancer risk in the National Lung Screening Trial. Am J Respir Crit Care Med. 2018;197(9):1220–1223. doi: 10.1164/rccm.201708-1632LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinsky PF, et al. Performance of lung-RADS in the National Lung Screening Trial: a retrospective assessment. Ann Intern Med. 2015;162(7):485–491. doi: 10.7326/M14-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silvestri GA, et al. Assessment of plasma proteomics biomarker’s ability to distinguish benign from malignant lung nodules: results of the PANOPTIC (pulmonary nodule plasma proteomic classifier) trial. Chest. 2018;154(3):491–500. doi: 10.1016/j.chest.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Postel M, et al. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Rev Mol Diagn. 2018;18(1):7–17. doi: 10.1080/14737159.2018.1400384. [DOI] [PubMed] [Google Scholar]
- 9.Liang W, et al. Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA) Theranostics. 2019;9(7):2056–2070. doi: 10.7150/thno.28119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacMahon H, et al. Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology. 2017;284(1):228–243. doi: 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- 11.Castro-Giner F, et al. Cancer diagnosis using a liquid biopsy: challenges and expectations. Diagnostics (Basel) 2018;8(2):31. doi: 10.3390/diagnostics8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Team NLSTR. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassanein M, et al. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res (Phila) 2012;5(8):992–1006. doi: 10.1158/1940-6207.CAPR-11-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groheux D, et al. FDG PET-CT for solitary pulmonary nodule and lung cancer: literature review. Diagn Interv Imaging. 2016;97(10):1003–1017. doi: 10.1016/j.diii.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Lennon AM, et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369(6499):eabb9601. doi: 10.1126/science.abb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaine R, et al. Cost-effectiveness of a low-dose computed tomography screening programme for lung cancer in New Zealand. Lung Cancer. 2018;124:233–240. doi: 10.1016/j.lungcan.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Gould MK, et al. Recent trends in the identification of incidental pulmonary nodules. Am J Respir Crit Care Med. 2015;192(10):1208–1214. doi: 10.1164/rccm.201505-0990OC. [DOI] [PubMed] [Google Scholar]
- 18.Yaguang F, et al. China national guideline of classification, diagnosis and treatment for lung nodules (2016 version) Zhongguo Fei Ai Za Zhi. 2016;19(12):793–798. doi: 10.3779/j.issn.1009-3419.2016.12.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peikert T, et al. Radiomics-based management of indeterminate lung nodules? Are we there yet? Am J Respir Crit Care Med. 2020;202(2):165–167. doi: 10.1164/rccm.202004-1279ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Velloso MJ, et al. Assessment of indeterminate pulmonary nodules detected in lung cancer screening: diagnostic accuracy of FDG PET/CT. Lung Cancer. 2016;97:81–86. doi: 10.1016/j.lungcan.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 21.Schuhmann M, et al. Endobronchial ultrasound for peripheral lesions: a review. Endosc Ultrasound. 2013;2(1):3–6. doi: 10.4103/2303-9027.117710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chockalingam A, Hong K. Transthoracic needle aspiration: the past, present and future. J Thorac Dis. 2015;7(Suppl 4):S292–S299. doi: 10.3978/j.issn.2072-1439.2015.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi HK, et al. Models to estimate the probability of malignancy in patients with pulmonary nodules. Ann Am Thorac Soc. 2018;15(10):1117–1126. doi: 10.1513/AnnalsATS.201803-173CME. [DOI] [PubMed] [Google Scholar]
- 24.Li X-j, et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci Transl Med. 2013;5(207):207ra142. doi: 10.1126/scitranslmed.3007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trivedi NN, et al. Analytical validation of a novel multi-analyte plasma test for lung nodule characterization. Biomed Res Rev. 2018;2(3):1–10. doi: 10.15761/brr.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren S, et al. Early detection of lung cancer by using an autoantibody panel in Chinese population. Oncoimmunology. 2018;7(2):e1384108. doi: 10.1080/2162402X.2017.1384108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, et al. Ultrasensitive DNA hypermethylation detection using plasma for early detection of NSCLC: a study in Chinese patients with very small nodules. Clin Epigenetics. 2020;12(1):39. doi: 10.1186/s13148-020-00828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazzone PJ, et al. Evaluating molecular biomarkers for the early detection of lung cancer: when is a biomarker ready for clinical use? An official American Thoracic Society policy statement. Am J Respir Crit Care Med. 2017;196(7):e15–e29. doi: 10.1164/rccm.201708-1678ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang W, et al. Evaluating the diagnostic accuracy of a ctDNA methylation classifier for incidental lung nodules: protocol for a prospective, observational, and multicenter clinical trial of 10,560 cases. Transl Lung Cancer Res. 2020;9(5):2016–2026. doi: 10.21037/tlcr-20-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng H, et al. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014;42(8):e69. doi: 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett JE, et al. Quantification of tumour evolution and heterogeneity via Bayesian epiallele detection. BMC Bioinformatics. 2017;18(1):1–10. doi: 10.1186/s12859-016-1414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rumelhardt DE, et al. Learning internal representations by error propagation. In: Rumelhart DE, et al, eds. Parallel Distributed Processing: Explorations In The Microstructure Of Cognition, 1: Foundations. MIT Press; 1986:318–362. [Google Scholar]
- 33. doi: 10.1109/CVPR.2016.90. He K, et al. Deep residual learning for image recognition. Presented at: 29th IEEE Conference On Computer Vision And Pattern Recognition; June 26–July 1, 2016; Las Vegas, Nevada, USA. Accessed March 22, 2021. [DOI]
- 34. Zeiler MD, et al. Deconvolutional networks. Presnted at: 2010 IEEE Computer Society Conference On Computer Vision And Pattern Recognition; June 13–18, 2010; San Francisco, California, USA. https://doi.ieeecomputersociety.org/10.1109/CVPR.2010.5539957 Accessed March 22, 2021.
- 35. Kingma DP, Ba J. Adam: a method for stochastic optimization [preprint]. https://arxiv.org/abs/1412.6980v9 Posted on arXiv December 22, 2014.
- 36.Swensen SJ, et al. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med. 1997;157(8):849–855. [PubMed] [Google Scholar]
- 37.Gould MK, et al. A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules. Chest. 2007;131(2):383–388. doi: 10.1378/chest.06-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.