Abstract
Gastric cancer remains one of the most dangerous cancers, bringing suffering and economic burden to people worldwide. Long noncoding RNAs (lncRNAs) exhibit great potentials for targeted therapy of various cancers. In this investigation, we tested mechanisms by which LINC01021 may regulate gastric cancer progression. We collected gastric cancer tissues and procured cell lines to explore the potential factors by which LINC01021 had effects on angiogenesis, invasion, and migration, by quantitative reverse-transcription polymerase chain reaction (qRT-PCR), Transwell assay, and western blot analysis. Relationships among LINC01021, Caudal-type homeobox 2 (CDX2), and KISS1 were validated by dual-luciferase gene reporter, RNA pull-down, and RNA immunoprecipitation assays. Additionally, a murine model was developed to further explore the impact of LINC01021 on tumors in vivo. LINC01021 was upregulated in gastric cancer tissues and cells. LINC01021 regulated KISS1 through CDK2, which promoted phosphorylation and nuclear export in CDX2. Inhibition of LINC01021 suppressed the tumorigenesis of gastric cancer. Further, silencing LINC01021 exerted an inhibitory effect on cancer cell migration, invasion, and angiogenesis by promoting the binding between CDX2 and KISS1, while inhibiting that between CDK2 and CDX2. Taken altogether, high LINC01021 expression in gastric cancer promotes malignant cell migration and angiogenesis by downregulation of KISS1 through CDK2-mediated CDX2 phosphorylation.
Keywords: LINC01021, CDX2, CDK2, gastric cancer, KISS1, phosphorylation
Graphical Abstract
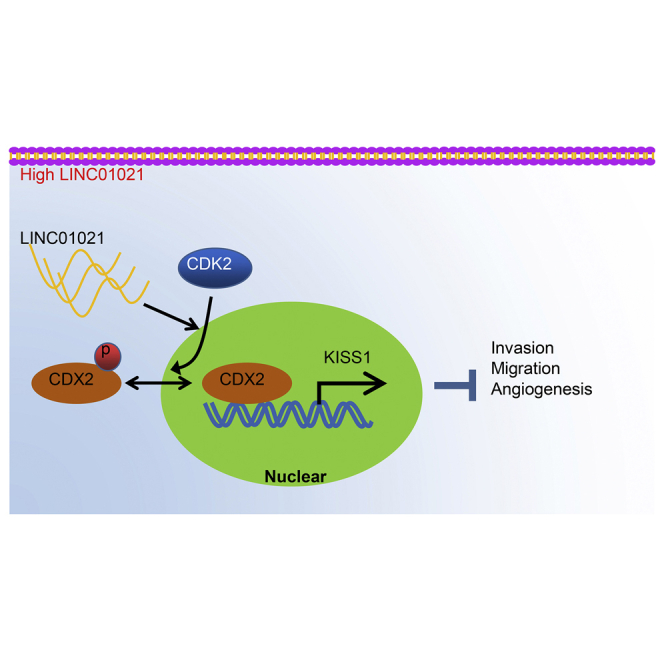
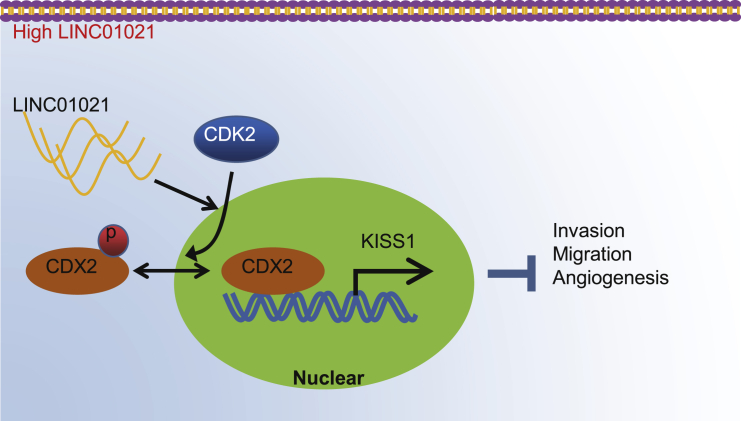
In this investigation, the mechanisms by which LINC01021 regulates gastric cancer progression were investigated by in vivo and in vitro experiments. It was found that high LINC01021 expression in gastric cancer promoted malignant cell migration and angiogenesis by downregulation of KISS1 through CDK2-mediated CDX2 phosphorylation.
Introduction
In spite of great improvements in treatment, gastric cancer remains the fifth most frequently diagnosed cancer, with more than 1,000,000 new cases in 2018 and 783,000 deaths reported annually.1 Because angiogenesis is correlated with gastric cancer development, treatments with angiogenesis inhibitors present a significant aspect of treatments for advanced disease.2,3 Although surgical resection brings a significantly improved survival and quality of life of early-stage patients, the prognosis for patients with advanced gastric cancer remains unsatisfactory.4 Therefore, it is necessary to explore and develop effective new biomarkers and therapeutic targets for altering the status quo.
Thus far, considerable gastric cancer research has focused on the deregulation of protein-coding genes to identify oncogenes and tumor suppressors. Long noncoding RNAs (lncRNAs), which are a family of transcripts exceeding 200 nt in length, are considered as chromatin-restricted regulators in many cancers, such as liver cancer and gastric cancer.5,6 It has been underlined that lncRNAs play key roles in gene regulation and thus contribute to cellular homeostasis, including proliferation, survival, migration, or genomic stability.7 For example, in gastric cancer, lncRNA HOXC-AS3 modulates gastric cancer cell proliferation and migrations, while correlating with poor diagnosis and worse tumorigenesis.8 As such, lncRNA GClnc1 is mechanistically, functionally, and clinically involved in gastric cancer growth with significant effects on prognosis, all due to its property of binding to the key component histone methyltransferase.9 Among lncRNAs, LINC01021/p53 upregulated regulator of p53 levels (PURPL) suppresses basal p53 levels and induces cell-cycle arrest, apoptosis, and tumor suppression in colorectal cancer.10 In addition, a recent study highlighted that the PURPL was associated with tumor size and cancer progression in gastric cancer.11 However, the mechanism by which LINC01021 has effects on cancer progression and growth requires further studies.
Emerging evidence suggests that lncRNAs may recruit a regulatory factor by occupying its site of transcription or by RNA-protein interactions, and may also inhibit the binding of a transcriptional regulatory factor, either directly or indirectly.12 Caudal-type homeobox 2 (CDX2) is a transcription factor involved in intestinal differentiation, which may act to suppress progression and carcinogenesis of gastric carcinoma.13 KISS1, the gene encoding kisspeptin, is known to be critical for the development of the reproductive system and in certain cancers, such that KISS1 signaling could suppress metastases and maintain dormancy of disseminated malignant cells and modulate their glucose and lipid metabolism.14,15 KISS1 has been suggested to be a suppressor of metastasis in various types of malignancy, including gastric, esophageal carcinoma, pancreatic, ovarian, bladder, and prostate cancers.16 Notably, KISS1 expression has been indicated to decline during the process of carcinogenesis in gastric mucosa in association with increased aggressiveness of gastric cancer cells.17,18 After our preliminary examination of an online dataset and subsequent assays, we tested the hypothesis that LINC01021 was upregulated in gastric cancer tissues and cells and modulated KISS1 expression through transcription factor CDX2, with consequences for gastric cancer cell migration, invasion, and angiogenesis.
Results
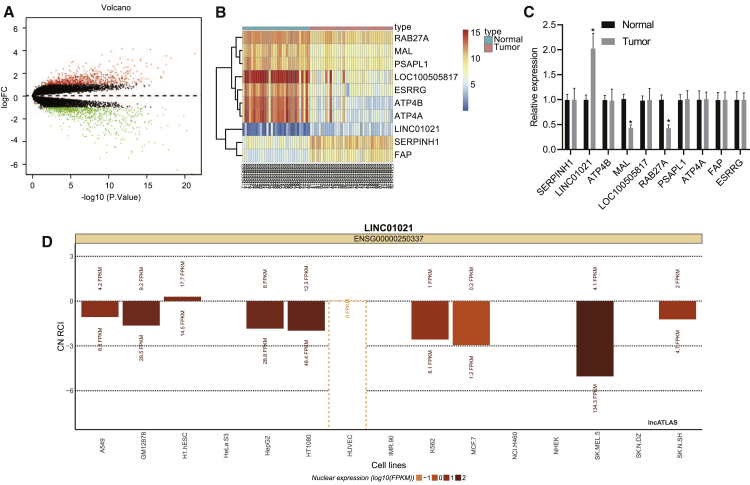
Bioinformatics analysis concerning differently expressed genes in gastric cancer-related dataset
Through differential analysis on the gastric cancer-related dataset Gene Expression Omnibus (GEO): GSE13911, we obtained 1,489 significantly differently expressed genes in gastric cancer (Figure 1A). The expression pattern of the top 10 differentially expressed genes (SERPINH1, LINC01021, ATP4B, MAL, LOC100505817, RAB27A, PSAPL1, ATP4A, FAP, and ESRRG) in GEO: GSE13911 was displayed in Figure 1B, among which LINC01021 expression was significantly increased in gastric cancer when compared with normal controls (Figure 1C), but rare literature reported its regulatory function in gastric cancer. Further prediction on the LINC01021 expression site revealed that LINC01021 was mainly expressed in the nucleus (Figure 1D), suggesting that LINC01021 probably plays a regulatory role by modulating transcription factors.
Figure 1.
Bioinformatics analysis predicted lncRNA with differential expression in gastric cancer
(A) GEO: GSE13911 volcano plot for differential gene expression. The abscissa represents p value of log10, and the ordinate represents the log(fold change), with upregulated genes (red) and downregulated genes (green) in tumor samples. (B) Heatmap displaying expression profile of top 10 differentially expressed genes in microarray dataset GEO: GSE13911. (C) Expression of top 10 differentially expressed genes in microarray dataset GEO: GSE13911 determined by qRT-PCR. (D) Predicted sites of LINC01021 expression in the nucleus (below abscissa) and cytoplasm (above abscissa).
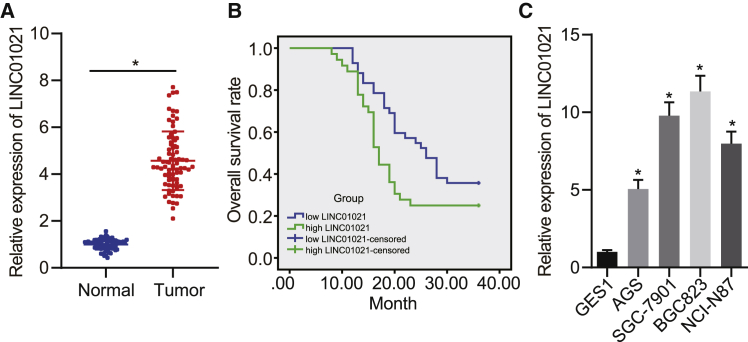
LINC01021 is upregulated in gastric cancer tissues and cells
To validate our database results, we detected LINC01021 expression in gastric cancer tissues and adjacent normal tissues from 78 patients using quantitative reverse-transcription polymerase chain reaction (qRT-PCR). Compared with adjacent normal tissues, LINC01021 was highly upregulated in cancer tissues (p < 0.05) (Figure 2A). Taking the median relative expression of LINC01021 as the cutoff point, we stratified the 78 patients into the LINC01021-low and LINC01021-high groups. We statistically analyzed the relation between the LINC01021 expression and clinical indicators, and discovered significant correlations between LINC01021 and pathological stage, distant metastasis, differentiation degree, and tumor size, but not with age or gender (Table 1). We then confirmed using the Kaplan-Meier method that LINC01021 expression was negatively associated with overall survival time (log rank p < 0.05) (Figure 2B). Furthermore, qRT-PCR analysis of LINC01021 in four gastric cancer cell lines and human normal gastric mucosal cell lines showed that, compared with human normal gastric mucosal cell GES1, there was higher expression of LINC01021 in human gastric cancer cell lines AGS, SGC-7901, BGC823, and NCI-N87. We selected BGC823 and SGC-7901 cells with the much higher LINC01021 expression for subsequent experiments (p < 0.05) (Figure 2C). Thus, LINC01021 was highly expressed in both gastric cancer tissues and cells.
Figure 2.
Highly expressed LINC01021 is found in gastric cancer tissues and cells
(A) qRT-PCR analysis of LINC01021 in cancer tissues and adjacent normal tissues (n = 78). (B) Kaplan-Meier curve for LINC01021 and the overall survival time of patients with gastric cancer. (C) qRT-PCR analysis of LINC01021 expression in four gastric cancer cell lines and normal gastric mucosal cell lines. ∗p < 0.05 versus GES1 cells. Measurement data were expressed as mean ± standard deviation. Comparisons among multiple groups were conducted by one-way ANOVA with Tukey’s post hoc test. The data between two groups were analyzed by paired t test. Survival of patients was calculated using Kaplan-Meier method, and the survival difference was analyzed by log rank test.
Table 1.
Correlation between LINC01021 and clinical indicators
| Variable | Number | Percentage (%) | High LINC01021.n | Low LINC01021.n | p value | |
|---|---|---|---|---|---|---|
| Age (year) | ≤60 | 27 | 34.62 | 16 | 11 | >0.05 |
| >60 | 51 | 65.38 | 20 | 31 | ||
| Gender | Male | 40 | 51.28 | 15 | 25 | >0.05 |
| Female | 38 | 48.72 | 21 | 17 | ||
| Size (cm) | ≤5 | 42 | 53.85 | 15 | 27 | 0.046 |
| >5 | 36 | 46.15 | 21 | 15 | ||
| Stage | I/II | 51 | 65.38 | 18 | 33 | 0.008 |
| III/IV | 27 | 34.62 | 18 | 9 | ||
| Differentiation | Low | 19 | 24.36 | 13 | 6 | 0.019 |
| Moderate | 25 | 32.05 | 13 | 12 | ||
| High | 34 | 43.59 | 10 | 24 | ||
The measurement data were presented as %, detected by chi-square test.
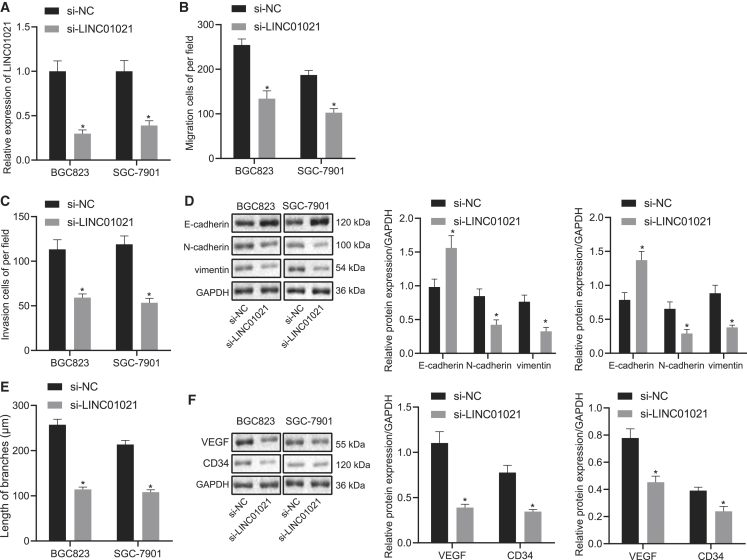
LINC01021 promotes invasion and migration of gastric cancer cells and angiogenesis of endothelial cells
After BGC823 and SGC-7901 cells were transfected, expression of LINC01021 was detected using qRT-PCR, which demonstrated that LINC01021 was significantly reduced after small interfering RNA against LINC01021 (si-LINC01021) treatment (p < 0.05) (Figure 3A). To assess the effect of decreased LINC01021 on the migration and invasion, we conducted Transwell assay and detected expression of E-cadherin, N-cadherin, and vimentin by western blot analysis. The results revealed that downregulation of LINC01021 inhibited cell migration and invasion and decreased expression of N-cadherin and vimentin while increasing E-cadherin expression (Figures 3B–3D). Tube formation assay and western blot analysis showed that si-LINC01021 inhibited tube formation and vascular endothelial growth factor (VEGF) and CD34 expression (p < 0.05) (Figures 3E and 3F). Taken altogether, downregulation of LINC01021 could inhibit the migration and invasion of gastric cancer cells and vascularization of endothelial cells.
Figure 3.
LINC01021 is involved in cell migration, invasion, and angiogenesis in gastric cancer cells
(A) qRT-PCR analysis of LINC01021 expression in BGC823 and SGC-7901 cells treated with si-NC and si-LINC01021. (B) Transwell assay of cell migration after treatment with si-NC and si-LINC01021. (C) Transwell assay of cell invasion after treatment with si-NC and si-LINC01021. (D) Western blot analysis of E-cadherin, N-cadherin, and vimentin in cells after treatment with si-NC and si-LINC01021. (E) Tube formation assay for number of tubes after treatment with si-NC and si-LINC01021. (F) Western blot analysis of VEGF and CD34 protein expression in cells after treatment with si-NC and si-LINC01021. ∗p < 0.05 versus si-NC treatment. Measurement data were expressed as mean ± standard deviation. Comparisons among multiple groups were conducted by one-way ANOVA. The data between two groups were analyzed by paired t test.
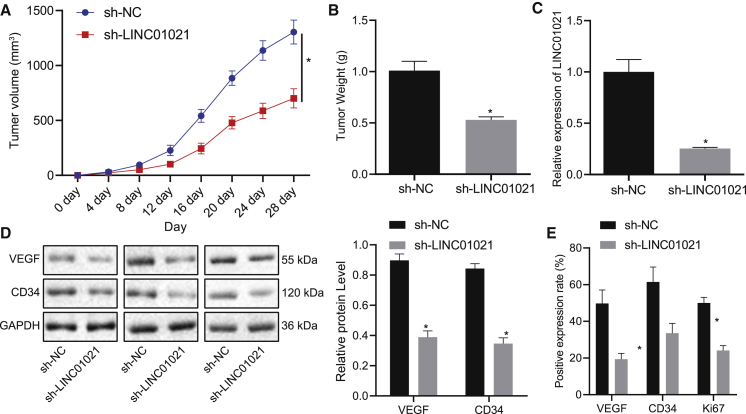
LINC01021 serves as an oncogenic lncRNA in gastric cancer in vivo
We tested the xenograft tumors in nude mice bearing cells infected with negative control (NC) for short hairpin RNA (sh-NC) or sh-LINC01021. Results demonstrated that sh-LINC01021 engendered decreased average volume and weight of transplanted tumors, as well as lower LINC01021 expression (p < 0.05) (Figures 4A–4C), which confirmed the above findings that low LINC01021 inhibited gastric cancer development in vivo. Additionally, western blot analysis depicted lower expression of VEGF and CD34 proteins in xenograft tumors under sh-LINC01021 treatment compared with sh-NC treatment (p < 0.05) (Figure 4D), while immunohistochemistry (IHC) staining gave similar results along with reduced positive expression of Ki67 protein (p < 0.05) (Figure 4E). Therefore, we concluded that downregulation of LINC01021 could inhibit the tumorigenic capacity of gastric cancer cells in vivo.
Figure 4.
LINC01021 promotes tumor growth in gastric cancer in vivo
(A) Average tumor volume of mice bearing treated cells. (B) Average tumor weight of mice bearing treated cells. (C) qRT-PCR of LINC01021 expression in tumors. (D) Western blot analysis of VEGF and CD34 protein expression. (E) IHC of positive expression of VEGF, Ki67, and CD34 in tumors. ∗p < 0.05 versus sh-NC treatment. Measurement data were expressed as mean ± standard deviation. Comparisons among multiple groups were conducted by one-way ANOVA. The data between two groups were analyzed by paired t test. n = 10.
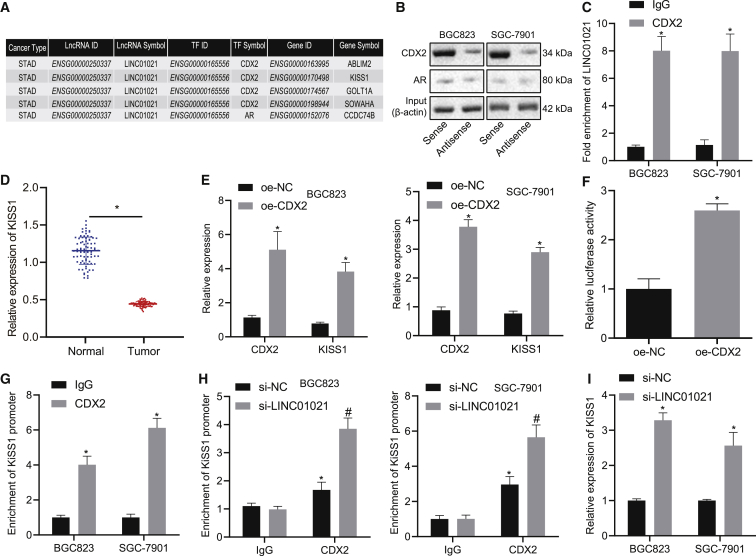
LINC01021 regulates the expression of KISS1 through the transcription factor CDX2
To further investigate the mechanism by which LINC01021 functions in gastric cancer, we used LncMAP to predict the downstream regulatory transcription factors and genes of LINC01021 (Figure 5A), which revealed a possible relation between LINC01021 and CDX2, which was further tested by RNA pull-down assay. Results showed that LINC01021 bound strongly to CDX2 (Figure 5B). In addition, RNA immunoprecipitation (RIP) was applied to test the binding between LINC01021 and CDX2, showing that the CDX2 protein could specifically bind more to LINC01021 than to the control, immunoglobulin G (IgG) (Figure 5C). The LncMAP prediction of target genes of LINC01021 via CDX2 identified KISS1 as a putative gene (Figure 5A). KISS1 has been reported to inhibit tumor progression in many tumors,19,20 but its implication in gastric cancer is rarely reported. Therefore, we inferred that LINC01021 might affect gastric cancer progression via the CDX2/KISS1 axis.
Figure 5.
LINC01021 inhibits KISS1 expression through CDX2
(A) LncMAP prediction of the target genes of CDX2 modulated by LINC01021. (B) RNA pull-down assay of binding between LINC01021 and CDX2 in BGC823 and SGC-7901 cells. (C) RIP assay of binding between LINC01021 and CDX2; ∗p < 0.05 versus IgG. (D) qRT-PCR analysis of KISS1 in gastric cancer tissues and adjacent normal tissues (n = 78). (E) qRT-PCR analysis of CDX2 and KISS1 expression in cells treated with oe-CDX2. ∗p < 0.05 versus oe-NC treatment. (F) Dual-luciferase reporter gene assay of binding between CDX2 and KISS1. ∗p < 0.05 versus oe-NC treatment. (G) ChIP assay of binding between CDX2 and KISS1. ∗p < 0.05 versus IgG. (H) ChIP assay of binding between CDX2 and KISS1 in BGC823 and SGC-7901 cells at the presence of si-LINC01021 or si-NC. ∗p < 0.05 versus IgG; #p < 0.05 versus si-NC treatment. (I) qRT-PCR of KISS1 expression in BGC823 and SGC-7901 cells after silencing LINC01021. ∗p < 0.05 versus si-NC treatment; #p < 0.05 versus si-LINC01021 + si-NC. Measurement data were expressed as mean ± standard deviation. Comparisons among multiple groups were conducted by one-way ANOVA. The data between two groups were analyzed by paired t test.
To investigate the potential role of LINC01021 in gastric cancer, we conducted qRT-PCR assay to determine the expression of target genes ABLIM, GOLT1A, SOWAHA (ANKRD43), and KISS1 regulated by CDX2 in response to LINC01021 silencing. The results indicated no obvious change of the expression of ABLIM, GOLT1A, and SOWAHA but upregulated expression of KISS1 in response to LINC01021 interference (Figure S1). Hence we selected KISS1 for further experiments.
Subsequently, we detected KISS1 expression in gastric cancer using qRT-PCR and showed that KISS1 was significantly downregulated in cancer tissues, compared with adjacent normal tissues (p < 0.05) (Figure 5D). To verify whether CDX2 targets KISS1, after overexpressing CDX2, qRT-PCR was applied to detect mRNA expression of KISS1 and displayed that overexpression (oe)-CDX2 led to increased KISS1 and CDX2 expression (Figure 5E), suggesting overexpressed upregulated KISS1 expression. Furthermore, dual-luciferase reporter gene assay revealed that oe-CDX2 also elevated the luciferase activity of KISS1 (p < 0.05) (Figure 5F), indicating that CDX2 could increase KISS1 expression. Chromatin immunoprecipitation (ChIP) experiments confirmed that CDX2 could specifically bind to the KISS1 promoter (Figure 5G), while silencing LINC01021 could promote binding between CDX2 and KISS1 in BGC823 and SGC-7901 cells (Figure 5H), and KISS1 expression increased (Figure 5I).
Taken altogether, we could conclude that CDX2 could bind to the KISS1 gene, and that silencing LINC01021 increased the binding of CDX2 to KISS1, thereby promoting the expression of KISS1.
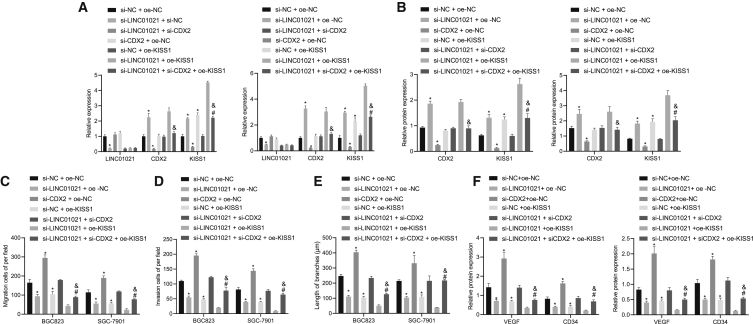
LINC01021/CDX2/KISS1 regulates the invasion, migration, and tube formation of gastric cancer cells
To measure the expression of KISS1 in BGC823 and SGC-7901 cells affected by LINC01021/CDX2/KISS1, we performed qRT-PCR for quantification on expression of LINC01021, CDX2, and KISS1 following treatment with si-NC + oe-NC, si-LINC01021 + oe-NC, si-CDX2 + oe-NC, si-NC + oe-KISS1, si-LINC01021 + si-CDX2, si-LINC01021 + oe-KISS1, and si-LINC01021 + si-CDX2 + oe-KISS1. Results showed that si-LINC01021 alone led to an increase of CDX2 and KISS1 expression, along with a reduction in LINC01021 expression, while si-CDX2 alone evoked decreased KISS1 and CDX2 expression, but no significant difference of LINC01021. In the presence of oe-KISS1 alone, LINC01021 and CDX2 expression did not differ significantly, whereas KISS1 was upregulated. In comparison with treatment with si-LINC01021 + si-CDX2, the si-LINC01021 + si-CDX2 + oe-KISS1 treatment induced no significant difference of LINC01021 and CDX2 expression, along with upregulation of KISS1. When compared with transfection of si-LINC01021 + oe-KISS1, further addition of si-CDX2 + oe-KISS1 resulted in no significant difference of LINC01021 expression, as well as downregulated CDX2 and KISS1 (Figure 6A) (p < 0.05), consistent with the results of western blot analysis (Figure 6B).
Figure 6.
LINC01021 downregulates the expression of KISS1 through CDX2, thereby promoting cell migration, invasion, and angiogenesis in gastric cancer
(A) qRT-PCR of LINC01021, CDX2, and KISS1 expression. (B) Western blot analysis of CDX2 and KISS1 expression. (C) Transwell assay to detect cell migration. (D) Transwell assay to detect cell invasion. (E) Tube formation assay to measure blood vessels. (F) Western blot analysis of VEGF and CD34 protein expression. ∗p < 0.05 versus si-NC + oe-NC treatment; #p < 0.05 versus si-LINC01021 + si-CDX2 treatment; &p < 0.05 versus si-LINC01021 + oe-KISS1 treatment. Measurement data were expressed as mean ± standard deviation. Comparisons among multiple groups were conducted by one-way ANOVA. The data between two groups were analyzed by paired t test.
In addition, Transwell assay was employed to detect the influence of LINC01021/CDX2/KISS on BGC823 and SGC-7901 cell invasion and migration. Results showed that si-LINC01021 or oe-KISS1 led to reduced invasion and migration ability, while si-CDX2 had opposite effects. Additional oe-KISS1 in the presence of si-LINC01021 + si-CDX2 led to suppressed migration and invasion, while additional si-CDX2 in the presence of si-LINC01021 + oe-KISS1 led to opposite results (p < 0.05) (Figures 6C and 6D).
We co-cultured cancer cells and human umbilical vein endothelial cells (HUVECs) for conducting tube formation assays, which showed that si-LINC01021 or oe-KISS1 led to impaired tube formation ability, whereas si-CDX2 increased tube numbers (p < 0.05) (Figure 6E). Moreover, oe-KISS1 reversed the promoted tube formation ability induced by si-LINC01021 + si-CDX2, while si-CDX2 counteracted the inhibitory action of si-LINC01021 + oe-KISS1. In addition, western blot analysis also identified that si-LINC01021 or oe-KISS1 led to a decline of VEGF and CD34 protein expression in HUVECs, while si-CDX2 increased the expression of VEGF and CD34 protein (p < 0.05) (Figure 6F). When LINC01021 and CDX2 were both silenced, overexpression of KISS1 induced downregulation of VEGF and CD34. Moreover, silencing CDX2 increased protein expression of VEGF and CD34 in HUVECs with silenced LINC01021 and overexpressed KISS1. The above results indicated that LINC01021 downregulated the expression of KISS1 through CDX2, thereby promoting cell migration, invasion, and angiogenesis in gastric cancer cells.
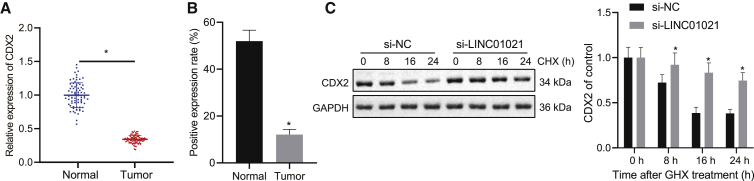
LINC01021 regulates the expression of CDX2 and affects the stability of CDX2
We detected the expression of CDX2 in gastric cancer tissues and adjacent normal tissues using qRT-PCR, and we noticed that CDX2 was significantly downregulated in cancer tissues (Figure 7A), consistent with IHC results (Figure 7B). After silencing LINC01021, western blot analysis unraveled upregulated CDX2 expression in BGC823 and SGC-7901 cells (Figure 6B). In addition, as BGC823 cells were treated with the protein synthesis inhibitor cycloheximide (CHX) for 8 h, si-LINC01021 significantly improved the protein stability of CDX2 (Figure 7C). Therefore, LINC01021 could regulate the expression of CDX2 and affect the stability of CDX2.
Figure 7.
Silencing LINC01021 promotes CDX2 expression and stability
(A) qRT-PCR analysis of CDX2 expression in cancer tissues and adjacent normal tissues (n = 78); ∗p < 0.05. (B) IHC of CDX2 expression in cancer tissues and adjacent normal tissues. (C) Western blot analysis of CDX2 protein expression in BGC823 cell lines after CHX treatment. CDX2 expression was quantified relative to GAPDH expression at indicated times and normalized to the 0-hour time point (before CHX treatment); ∗p < 0.05, ∗∗p < 0.01 versus si-NC treatment. Measurement data were expressed as mean ± standard deviation. The data between two groups were analyzed by paired t test.
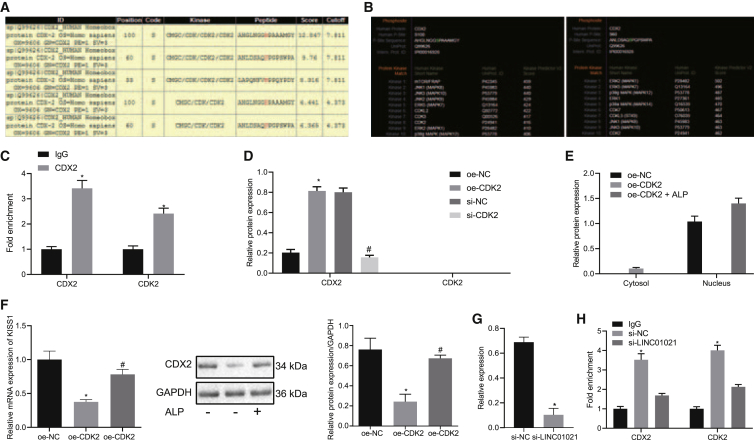
LINC01021 regulates CDX2 phosphorylation, CDX2 nuclear export, and transcriptional activity of downstream target genes through CDK2
It is highlighted that CDX2 phosphorylation can block CDX2 multiubiquitination and stable proteins.21 CDK2 can phosphorylate CDX2 in vitro and in vivo when activating phosphorylation of serine 60 residues.22 In this work, we predicted the sites where CDK2 promoted CDX2 phosphorylation through the database (http://gps.biocuckoo.org/links.php#l1) and obtained two sites, S100 and S60, with highest scores (Figure 8A). The website (http://www.phosphonet.ca/) predicted that the phosphorylation enzymes at S100 and S60 sites of CDX2 and CDK2 were included in the enzymes with high score (Figure 8B). Accordingly, it is reasonable to speculate that CDK2 regulates the CDX2 phosphorylation.
Figure 8.
LINC01021 plays roles in CDX2 phosphorylation, nuclear export, and transcriptional activity of downstream target genes via CDK2
(A) The website (http://gps.biocuckoo.org/links.php#l1) predicted sites that CDK2 promoted CDX2 phosphorylation while highest score was observed in S100 and S60. (B) The website (http://www.phosphonet.ca/) predicted the phosphorylation enzymes at the S100 and S60 sites of CDX2, including CDK2. (C) coIP showed that CDK2 can bind to CDX2 in BGC823 cells. ∗p < 0.05 versus IgG. (D) Western blot analysis of CDX2 expression in BGC823 after IP with anti-phosphoserine. ∗p < 0.05 versus oe-NC treatment; #,∗p < 0.05 versus si-NC treatment. (E) Western blot of CDX2 expression in cytoplasm and nucleus after overexpressing CDK2 in cytoplasm and nucleus with GAPDH used as internal reference in neoplasm and Lamin A/C in nucleus. (F) qRT-PCR analysis of KISS1 expression and western blot analysis of CDX2 in BGC823 cells treated with oe-CDK2. ∗p < 0.05 versus oe-NC; #p < 0.05 versus oe-CDK2 treatment. (G) Western blot analysis of CDX2 expression in BGC823 cells treated with si-LINC01021 after IP with anti-phosphoserine. ∗p < 0.05 versus IgG. (H) CoIP of binding between CDK2 and CDX2 in BGC823 cells treated with si-LINC01021. ∗p < 0.05 versus IgG.
Furthermore, it was noticed that CDK2 binds to CDX2, according to co-immunoprecipitation (coIP) assay (Figure 8C); IP was performed with anti-phosphoserine, and CDX2 expression was detected by western blot analysis. The results showed that overexpression of CDK2 could significantly promote CDX2 serine phosphorylation and when CDK2 was silenced, CDX2 serine phosphorylation was significantly inhibited. When S60 and S100 were replaced with Ala in CDX2, overexpression of CDK2 did not affect the phosphorylation (Figure 8D), indicating that it is through S100 and S60 that CDK2 regulates CDX2 phosphorylation.
The nucleocytoplasmic separation experiments showed that overexpression of CDK2 promoted the accumulation of CDX2 protein in the cytoplasm, and following alkaline phosphatase treatment for 1 h, CDX2 was mainly expressed in the nucleus, indicating that CDK2 promotes CDX2 phosphorylation and nuclear export (Figure 8E). Additionally, qRT-PCR and western blot demonstrated that overexpressed CDK2 significantly reduced the expression of CDX2 and KISS1. After the addition of ALP, CDX2 and KISS1 expression was rescued (Figure 8F). The above results indicated that CDK2 allowed CDX2 protein to maintain in the cytoplasm by promoting CDX2 phosphorylation, thus preventing CDX2 from activating transcription of target gene KISS1.
Then, after silencing LINC01021 in BGC823 cells, we performed IP with anti-phosphoserine and western blot analysis and showed reduced CDX2 phosphorylation (Figure 8G). CoIP assay also showed that the binding between CDK2 and CDX2 decreased after silencing LINC01021 (Figure 8H). Taken altogether, CDK2 could promote CDX2 serine phosphorylation, thereby promoting the nuclear export of CDX2, reducing the expression of CDX2, and inhibiting the transcriptional activity of CDX2. Moreover, silencing LINC01021 could inhibit the binding of CDK2 and CDX2 and reduce the level of CDX2 serine phosphorylation.
Discussion
Gastric cancer imposes a huge burden on healthcare around the world, despite surgery and the introduction of targeted therapies for clinical use.23 Interestingly, gastric cancer shows aberrant methylation in assorted relevant gene classes, such as tumor suppressors, DNA repair genes, cell-cycle regulators, and transcription factors. In addition, lncRNAs were functionally implicated in the kinds of biological pathway, such as cell proliferation, apoptosis, migration, invasion, and chemosensitivity.24 In this work, we investigated the role of a newly described lncRNA, LINC01021, in gastric cancer and tested the relationships between LINC01021 and the transcription factors CDX2, KISS1, and CDK2. Finally, we present evidence demonstrating that silencing LINC01021 could repress the binding between CDK2 and CDX2 and could likewise suppress gastric cancer cell invasion, migration, and tube formation through upregulation of KISS1 (Figure 9).
Figure 9.
Molecular schematic diagram concerning LINC01021 in gastric cancer
LINC01021 was upregulated in gastric cancer and could promote phosphorylation and nuclear export of CDX2 by CDK2. Silencing LINC01021 inhibited the binding of CDK2 and CDX2, as well as CDX2 phosphorylation. Additionally, silencing LINC01021 suppressed cell invasion, migration, and angiogenesis by upregulating the expression of KISS1.
Obtaining an in-depth understanding of cellular mechanisms is crucial to promote better therapy development, especially for targeted therapy. Available biomarkers are not absolutely predictive of treatment response, but they may identify groups of patients with a greater likelihood of response, thereby guiding clinical decision-making for treatment sequencing.25 Rapid development of next-generation sequencing and bioinformatics technology has revealed increasing numbers of lncRNAs with aberrant expression that is associated with the progression in various types of cancers.26,27 Mechanistically, lncRNAs may recruit chromatin regulatory complexes to specific targets on genomic DNA to control gene expression, while engendering assemblies of chromatin regulatory complexes and other protein complexes that do not normally form protein-protein interactions.28 Among lncRNA, the newly discovered lncRNA LINC01021 is the focus of the present study. It has recently been reported that anomalous expression of this lncRNA in colorectal cancer reduces instability and protein levels of p53, thus indicating that LINC01021 acts as a pro-survival gene.10 Our present assay results confirmed that LINC01021 was upregulated in gastric cancer tissues and cells, which was in line with a previous study.11 Moreover, we showed that LINC01021 could play a deleterious role in gastric cancer cell migration and angiogenesis, whereas its downregulation inhibited these processes.
Additionally, we note that CDX2 transactivates its own promoter and positively regulates its expression in gastrointestinal human carcinoma cell lines while binding to its promoter in the mouse ileum and in human stomach.29 CDX2 is differentially expressed in normal stomach but progressively reduced in gastric dysplasia and further in the transition to gastric cancer, where its expression is inversely correlated with the expression of gastric mucins.30 In this work, statistical analyses showed that CDX2 was poorly expressed in gastric cancer. Moreover, we also predicted and confirmed that LINC01021 could strongly bind to CDX2 protein. The emerging evidence highlighted that this lncRNA could bind to and modulate a transcription factor, or indeed other protein engaged in gene expression, thus exerting specific effects on the cell.31 For instance, previous work showed that AC093818.1 could upregulate the 3-phosphoinositide-dependent kinase-1 (PDK1) level by transcriptional activation, which occurred by recruiting the transcription factors signal transducer and activator of transcription 3 and SP1 to the PDK1 promotor, thus enhancing metastasis in gastric cancer.32
Migration and metastasis of tumor cells represent a major factor limiting the efficacy of various cancer treatments.33 KISS1 is an established tumor suppressor, which is absent in metastatic cells but present in non-metastatic cells, and KISS1 is associated with poorer prognosis in cancer patients.34 Physiologically, KISS1 signaling inhibits metastases and maintains dormancy of disseminated malignant cells.15 Moreover, KISS-1 has been suggested to inhibit the proliferation and invasion of gastric carcinoma cells in vitro and in vivo through the downregulation of matrix metalloproteinase-9.35 In this study, we confirmed that KISS1 was a target gene of CDX2 and then showed that silencing LINC01021 even affected KISS1 expression. Similarly, as indicted by one recent study, lncRNA lung cancer associated transcript 1 could promote prostate cancer cell migration and invasion through downregulation of KISS1.36
Moreover, we indicated that CDK2 bound to CDX2, and that silencing LINC01021 could stimulate more binding. Meanwhile, LINC01021 regulated CDX2 phosphorylation through CDK2. It was previously reported that CDK2 negatively regulated silent information regulator 5 (SIRT5), a tumor suppressor in gastric cancer cells, and it was also shown that SIRT5 inhibited gastric cancer cell proliferation, aerobic glycolysis, and tumor formation capacity.37 Boulanger et al.38 had shown that aberrant expression of CDK2 led to reduced CDX2 expression, while CDK2 interacted with CDX2 and phosphorylated CDX2.
In conclusion, the present results, along with previous findings, show that LINC01021 could play a significant role in regulating biological process through the interaction between CDX2, CDK2, and KISS1, which highlights the potential of LINC01021 as a biomarker for gastric cancer treatment. Nonetheless, details of the interaction between LINC01021 and CDX2, including the regions responsible for the interaction of LINC01021 and CDX2, the location where the interaction takes place, and how it affects CDX2-CDK2, merit further investigation before translation to clinical trials.
Materials and methods
Ethics statement
The study followed the Declaration of Helsinki and received approval by the Ethics Committee of Xuzhou Cancer Hospital, and written informed consent was obtained from each patient. All animals in the study were utilized for medical research and use of Laboratory Animal, and the procedures were approved by the Animal Ethics Committee of Xuzhou Cancer Hospital.
Bioinformatics analysis
Based on the GEO database (https://www.ncbi.nlm.nih.gov/geo/), we obtained gastric cancer-related dataset GEO: GSE13911, including 31 normal samples and 38 gastric cancer samples, and next conducted differential analysis with |logFoldChange| > 1 and adjusted (adj.) p < 0.05 using limma package of R language. The location of LINC01021 was predicted using the database lncATLAS (http://lncatlas.crg.eu), and its downstream transcription factors and regulators were detected using the database LncMAP (http://bio-bigdata.hrbmu.edu.cn/LncMAP/). In addition, the binding sequence between CDX2 and KISS1 was predicted via the database JASPAR (http://jaspar.genereg.net), while CDK2 and CDX2 phosphorylation were predicted by the database GPS3.0 (http://gps.biocuckoo.org/links.php#l1) and by PhosphoNET (http://www.phosphonet.ca/).
Study subjects
A total of 78 gastric cancer tissues and their corresponding adjacent noncancerous tissues (>5 cm away from the tumor) were collected from patients with advanced gastric cancer who underwent surgical resection in Xuzhou Cancer Hospital between October 2015 and October 2016. No patients had undergone chemoradiotherapy before surgery, and their tissues were pathologically confirmed. The collected tissues were immediately placed in liquid nitrogen for temporary preservation and stored in a refrigerator (−80°C) for long-term storage.
Cell culture and transfection
Human gastric cancer cell lines SGC-7901 (ZQ0062), NCI-N87 (ZQ0060), BGC-823 (ZQ0055), and AGS (ZQ0240) and human gastric mucosal cell line GES1 (ZQ0905) and mediums were purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology (Shanghai, China). SGC-7901, NCI-N87, and BGC-823 cells were cultured with 90% Roswell Park Memorial Institute-1640 medium (ZQ-201), AGS with F-12K medium (ZQ-501), and GES1 with medium (ZQ-101) containing Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), and penicillin – streptomycin in a cell incubator at 37°C with 5% CO2. When grown to 90% confluence, the cells were passaged. As for transfection, cells were transfected with si-CDX2, oe-CDK2, si-LINC01021, si-LINC01021 + si-CDX2, si-LINC01021 + oe-KISS1, and si-LINC01021 + siCDX2 + oe-KISS1, as well as their corresponding controls. The BGC823 and SGC-7901 cells in logarithmic growth phase were seeded into a 24-well plate and transfected according to the instructions of the Lipofectamine 3000 Reagent kit (Thermo Fisher Scientific, Waltham, MA, USA) with medium renewed 12 h after transfection.
qRT-PCR
Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and reversely transcribed into cDNA with PrimeScript RT kit (Takara, Japan). qRT-PCR was conducted using the SYBR Premix EX Taq kit (Takara) in a fluorescent quantitative real-time PCR instrument (ABI7500; ABI, Foster City, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal reference. The fold changes between the experiment group and the control group were calculated using relative quantification 2−ΔΔCt. The used primers are shown in Table 2.
Table 2.
Primer sequence of qRT-PCR
| Gene | Sequences |
|---|---|
| LINC01021 | F: 5′-ACGGGAAGGCTTTGCTCAAT-3′ |
| R: 5′-CACCTGCATGCTGCATCAAC-3′ | |
| CDX2 | F: 5′-CGCCGCAGAACTTCGTCAG-3′ |
| R: 5′-CGTAGCCATTCCAGTCCTCCC-3′ | |
| KISS1 | F: 5′-GAACCCAAGGAGTGTGACCC-3′ |
| R: 5′-CATCATCTGCTTACCGCACC-3′ | |
| ABLIM2 | F: 5′-AGCTGACTATCACGCCAAGT-3′ |
| R: 5′-GGAAGGGTGGTAGTGCTTCT-3′ | |
| GOLT1A | F: 5′-GGGCCTGTCCCTCATCATT-3′ |
| R: 5′-TTTGTGCCGTTGGAAGAAGAA-3′ | |
| SOWAHA | F: 5′-GGTTTAGTGCCCAGTCTGCC-3′ |
| R: 5′-CTTAGACAGCGTGCCACAGG-3′ | |
| GAPDH | F: 5′-GTCAGTGGTGGACCTGACCT-3′ |
| R: 5′-TGACCTTGACAAAGTGGTCG-3′ |
CDX2, Caudal-type homeobox 2; F, forward; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; R, reverse.
RNA pull-down assay
Biotin-labeled RNAs were transcribed by the Biotin RNA Labeling Mix (Roche, Indianapolis, IN, USA) and T7 RNA polymerase (Promega, Madison, WI, USA), followed by treatment of RNase-free DNase I (Promega) and purification with RNeasy Mini Kit (QIAGEN, Dusseldorf, German). Biotinylated RNA (5 g) was heated at 95°C for 5 min, placed on ice for 5 min, and then placed at room temperature for 20 min to form a secondary structure. The folded RNAs were then mixed with the cell extracts for 2 h. The remaining lysate was incubated with 50 μL of streptavidin beads (Invitrogen) for 1.5 h. After the beads were washed, treated with ribonuclease, and dissolved in sodium dodecyl sulfate (SDS) buffer, the total protein was extracted and detected by western blot analysis.
RIP assay
The RIP kit (Millipore, Bedford, MA, USA) was adopted to examine the binding of LINC01021 and CDX2. The cells were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer and centrifuged at 14,000 rpm for 10 min at 4°C. A part of the cell extract was taken out as an input, and the remaining part was co-precipitated by incubation with the antibody. Next, 50 μL magnetic beads was resuspended in 100 μL RIP wash buffer and incubated with 5 μg antibody. The magnetic bead-antibody complex was resuspended in 900 μL RIP wash buffer and incubated with 100 μL cell extracting solution at 4°C overnight. The sample was placed on the magnetic base, and the magnetic bead-protein complex was collected. The sample and input were detached with Proteinase K to extract RNA for subsequent PCR detection of LINC01021. The antibodies recruited for RIP included the anti-CDX2 (ab70458; Abcam, Shanghai, China) and the IgG (ab109489; Abcam) as a NC.
ChIP assay
Cells were fixed with 1% formaldehyde at room temperature for 10 min to cross-link the DNA and protein. Afterward, the ultrasonic breaker was set to 10 s per ultrasonic cycle with 10-s intervals with 15 cycles to break the chromatin, and the cells were centrifuged at 12,000 rpm at 4°C for 10 min. The supernatants were collected and divided into two tubes, then incubated overnight at 4°C with anti-CDX2 (ab70458; Abcam) and the IgG (ab109489; Abcam). The endogenous DNA-protein complex was precipitated by protein agarose/Sepharose. After centrifugation, with supernatant removed and the non-specific complex washed, the cross-linking was reversed overnight by incubation at 65°C. The DNA fragments were extracted, purified, and recovered using phenol/chloroform with the KISS1 promoter-specific primers for qRT-PCR (forward: 5′-TTCTCCCCAGCTCCCTGATCACATCC-3′ and reverse: 5′-CTGCCTCCAGTCACAGAGC-3′).
Dual-luciferase reporter gene assay
KISS1 promoter (forward: 5′-TTCTCCCCAGCTCCCTGATCACATCC-3′ and reverse: 5′-CTGCCTCCAGT-CACAGAGC-3′)39 was constructed into pGL3-Basic vector (Promega) to form the recombinant vector through restriction sites KpnI and XbaI, which was co-transfected with oe-CDX2 and oe-NC into HEK293T cells with Renilla luciferase as internal interference. After 48 h of transfection, the relative light unit (RLU) was measured by a Renilla luciferase assay kit (K801-200; Biovision, CA, USA) and dual-luciferase reporter gene assay (Promega). With Renilla luciferase as the internal reference, the dual-luciferase reporter gene assay system (Promega) was employed for analysis. The activity of the target reporter gene was considered as the ratio of RLUs of firefly luciferase to that of Renilla luciferase.
Immunoprecipitation and western blot analysis
Mouse monoclonal anti-phosphoserine (ab6639, 1:2,000) was used for immunoprecipitation. The complex was subjected to SDS polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis. The protein was separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membrane, which was then blocked by 10% bovine serum albumin (BSA) at room temperature for 2 h. The PVDF membrane was incubated with primary antibodies at 4°C overnight: anti-rabbit CDX2 (ab88129, 1:1,000), anti-rabbit KISS1 (ab19028, 1:1,000), rabbit monoclonal VEGF (ab52917, 1:10,000), rabbit monoclonal CD34 (ab81289, 1:10,000), mouse monoclonal E-cadherin (ab76055, 1:1,000), rabbit polyclonal N-cadherin (ab18203, 1:1,000), rabbit polyclonal Vimentin (ab137321, 1:1,000), rabbit monoclonal CDK2 (ab32147, 1:1,000), rabbit monoclonal GAPDH (ab128915, 1:10,000), rabbit monoclonal Lamin A/C (ab108922, 1:1,000), and mouse monoclonal β-actin (ab8226, 1:1,000). After that, the membrane was re-probed with horseradish peroxidase-labeled secondary goat anti-rabbit IgG (ab205718, 1:2,000) or goat anti-mouse IgG (ab205719, 1:2,000) at room temperature for 2 h. Image-Pro Plus 6.0 software was applied to quantify band intensity. All the above-mentioned antibodies were provided by Abcam.
Protein stability analysis
BGC823 and SGC-7901 cells were transfected with the siRNAs or infected with overexpressed lentivirus and their corresponding controls. After 48 h, cells were exposed to CHX (#2112, 50 μg/mL; Cell Signaling Technology, Danvers, MA, USA) to inhibit protein synthesis. Afterward, cells were collected for western blot analysis at 0, 8, 16, and 24 h.
CoIP
Transfected cells were lysed in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 10% glycerol, 1 mM ethylene diamine tetraacetic acid [EDTA], 0.5% Nonidet P-40, and protease inhibitors cocktail) and centrifuged to move cell debris. Cleared cell lysates were incubated with 1 μg anti-CDX2 (ab70458; Abcam) and 15 μL protein A/G beads (Santa Cruz Biotechnology, Dallas, TX, USA) for 2 h. After extensive washing, beads were boiled at 100°C for 5 min. Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes (Millipore), followed by immunoblotting.
Nucleocytoplasmic separation experiments
Cells were resuspended with protease inhibitor, RNase inhibitor (N8080119; Thermo Fisher Scientific), and Hypotonic buffer A (10 mM HEPES [pH 7.5], 0.5 mM DTT, 10 mM KCl, 1.5 mM MgCl2). After incubation for 10 min on ice and centrifugation for 10 min at 1,000 × g and 4°C, the cytoplasm was obtained and the precipitate was washed twice with hypotonic buffer and resuspended in hypotonic buffer B (10 mM HEPES [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.5% Nonidet P-40), followed by centrifugation at 6,000 × g and 4°C. After that, the precipitate was washed with hypotonic buffer and resuspended in RIPA buffer (50 mM Tris HCl [pH 7.5], 1,500 mM KCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA [pH 8.0]). Finally, after further incubation and centrifugation at 15,000 × g, the obtained supernatant was the nuclear RNA.
Plasmid construction
According to the manufacturer’s instructions, recombinant plasmids expressing the pcDNA3.1 vector and CDX2-wild-type (WT) or CDX2-S60, 100A (S60, 100 replaced by A) were generated and transfected into BGC823 cells, which were then selected with 750 μg/mL G418 (Calbiochem, Darmstadt, Germany). Mutated CDX2 was constructed by the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Foster City, CA, USA). Moreover, plasmids containing pcDNA3.1-Flag-RhoA-WT and pcDNA3.1-Flag-CDX2-S60, 100A (S60, 100 replaced by A) were constructed.
Alkaline phosphatase assay
The cell lysate (5–20 g) was interacted with 3–6 U alkaline phosphatase (Roche, Indianapolis, IN, USA) in the buffer at 37°C for at least 1 h. In the NC, the lysate alkaline was treated with buffer (25 mM Tris [pH 7.6], 1 mM MgCl2, 0.1 mM ZnCl2, 50% glycerol w/v) instead of alkaline phosphatase. The reaction was paused with Laemmli’s sample buffer. Migration of the corresponding proteins on SDS-PAGE was examined by immunoblotting, as described above.
Transwell migration and Matrigel invasion assays
For migration assay, 2 × 105 cells were seeded in the apical chamber with cell suspension and the lower chamber with DMEM (700 μL) containing 10% FBS. For invasion assay, 0.2 mL (2.5 × 105 cells/mL) of cells was seeded into the apical chamber with extracellular matrix gel. After 24 h, the migrated or invaded cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Then pictures were taken of the migratory or invading cells under a microscope with five randomly selected fields and counted.
Matrigel tube formation assay
Matrigel that thawed overnight at 4°C was placed on a pre-cooled 96-well plate (75 μL/well) and placed at 37°C for 60 min. HUVEC suspension was placed to the plate at a density of 2.5 × 104 cells/well. After the cells adhered, the suspension was replaced with BGC823 and SGC-7901 cell supernatants for incubation. After 4–6 h, the cells were observed from randomly chosen fields under a microscope.
Mouse xenograft model
Twenty nude BALB/c mice (4–6 weeks, 18–25 g, irrespective of gender) were purchased from Shanghai SLAC Laboratory Animal (Shanghai, China) and maintained under specific pathogen-free condition. The gastric cancer BGC823 cells that were infected with sh-NC and sh-LINC01021 (Shanghai Gene Pharma, Shanghai, China) (n = 10) were detached with 0.25% trypsin. The collected cells were mixed with serum-free matrix. A total of 100 μL of cell suspension maintaining 1 × 106 cells was injected into the neck of mice and maintained under the same condition. Tumor formation was observed every 4 days to calculate tumor volumes (TVs) according to the following formula: TV = (a × b2)/2, where a is maximum diameter and b is the minimum diameter. On day 28, mice were euthanized via excess anesthesia, and tumors were excised to make a paraffin section before recording weight of tumor.
IHC
The paraffin-embedded sections were dewaxed with xylene and rehydrated with gradient alcohol. The sections were next immersed in 3% H2O2 for 10 min, washed with phosphate-buffered saline (PBS) twice for 5 min, and subjected to high-pressure antigen retrieval buffer (Biyuntian Biotechnology, Shanghai, China) for 90 s. After that, the sections were incubated with BSA blocking solution at 37°C for 30 min, with primary antibody rabbit anti-mouse monoclonal VEGF (ab52917, 1:250; Abcam), rabbit anti-mouse monoclonal CD34 (ab81289, 1:250; Abcam), and rabbit anti-mouse monoclonal Ki67 (ab16667, 1:1,000; Abcam) at 4°C overnight and with 50 μL biotinylated mouse anti-goat IgG (ab6789; Abcam) at 37°C for 30 min.
After that, the samples were developed with 3,3-diaminobenzidine tetrahydrochloride (Sigma-Aldrich, St. Louis, MO, USA) and counterstained with hematoxylin (Sigma-Aldrich) for 5 min. Finally, the sections were subjected to washing, dehydration, clearing, sealing, and microscopic examination. Meanwhile, in NC, primary antibody was replaced with PBS buffer. Positive expression referred to positive cells accounting for over 10%, and the coloration (yellow) was mainly located in the cytoplasm or cell membrane and vascular endothelium. Positive expression ratio of VEGF and CD34 was observed and calculated from five randomly chosen fields under the microscope.
Statistical analysis
The data were processed using SPSS 21.0 statistical software (IBM SPSS Statistics, Chicago, IL, USA). Measurement data were expressed as mean ± standard deviation. When the data conformed to normality and homogeneity of variance, the paired t test was applied for comparison within groups, and unpaired t test was employed for comparison between groups. Analysis between multiple groups was conducted by one-way analysis of variance (ANOVA). Data at different time points among groups were compared by repeated measures ANOVA, followed by Bonferroni post hoc test. The survival rate was calculated by Kaplan-Meier method, and the survival difference was analyzed by log rank test. Enumeration data were expressed as rate (%), and difference between groups was assessed by chi-square test. p <0.05 was considered statistically significant. All experiments were repeated at least three times independently.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article.
Acknowledgments
We acknowledge and appreciate our colleagues for their valuable suggestions and technical assistance for this study.
Author contributions
Y.W. and Q.W. conducted the experiments. R.J. and H.Z. designed the experiments. Y.W. and H.Z. collected the data. Y.L. and Z.S. wrote the paper. All authors approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2021.01.025.
Supplemental information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Yu J., Zhang Y., Leung L.H., Liu L., Yang F., Yao X. Efficacy and safety of angiogenesis inhibitors in advanced gastric cancer: a systematic review and meta-analysis. J. Hematol. Oncol. 2016;9:111. doi: 10.1186/s13045-016-0340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sammarco G., Varricchi G., Ferraro V., Ammendola M., De Fazio M., Altomare D.F., Luposella M., Maltese L., Currò G., Marone G. Mast Cells, Angiogenesis and Lymphangiogenesis in Human Gastric Cancer. Int. J. Mol. Sci. 2019;20:2106. doi: 10.3390/ijms20092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thrumurthy S.G., Chaudry M.A., Chau I., Allum W. Does surgery have a role in managing incurable gastric cancer? Nat. Rev. Clin. Oncol. 2015;12:676–682. doi: 10.1038/nrclinonc.2015.132. [DOI] [PubMed] [Google Scholar]
- 5.Carlevaro-Fita J., Johnson R. Global Positioning System: Understanding Long Noncoding RNAs through Subcellular Localization. Mol. Cell. 2019;73:869–883. doi: 10.1016/j.molcel.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Martens-Uzunova E.S., Böttcher R., Croce C.M., Jenster G., Visakorpi T., Calin G.A. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 8.Zhang E., He X., Zhang C., Su J., Lu X., Si X., Chen J., Yin D., Han L., De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154. doi: 10.1186/s13059-018-1523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun T.T., He J., Liang Q., Ren L.L., Yan T.T., Yu T.C., Tang J.Y., Bao Y.J., Hu Y., Lin Y. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 10.Li X.L., Subramanian M., Jones M.F., Chaudhary R., Singh D.K., Zong X., Gryder B., Sindri S., Mo M., Schetter A. Long Noncoding RNA PURPL Suppresses Basal p53 Levels and Promotes Tumorigenicity in Colorectal Cancer. Cell Rep. 2017;20:2408–2423. doi: 10.1016/j.celrep.2017.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moridi H., Karimi J., Tavilani H., Khodadadi I., Emami Razavi A.N. Overexpression of PURPL and downregulation of NONHSAT062994 as potential biomarkers in gastric cancer. Life Sci. 2019;237:116904. doi: 10.1016/j.lfs.2019.116904. [DOI] [PubMed] [Google Scholar]
- 12.Long Y., Wang X., Youmans D.T., Cech T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito M., Okayama H., Saito K., Ando J., Kumamoto K., Nakamura I., Ohki S., Ishi Y., Takenoshita S. CDX2 is involved in microRNA-associated inflammatory carcinogenesis in gastric cancer. Oncol. Lett. 2017;14:6184–6190. doi: 10.3892/ol.2017.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semaan S.J., Kauffman A.S. Emerging concepts on the epigenetic and transcriptional regulation of the Kiss1 gene. Int. J. Dev. Neurosci. 2013;31:452–462. doi: 10.1016/j.ijdevneu.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corno C., Perego P. KiSS1 in regulation of metastasis and response to antitumor drugs. Drug Resist. Updat. 2019;42:12–21. doi: 10.1016/j.drup.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Ji K., Ye L., Mason M.D., Jiang W.G. The Kiss-1/Kiss-1R complex as a negative regulator of cell motility and cancer metastasis (Review) Int. J. Mol. Med. 2013;32:747–754. doi: 10.3892/ijmm.2013.1472. [DOI] [PubMed] [Google Scholar]
- 17.Kostakis I.D., Agrogiannis G., Vaiopoulos A.G., Mylona E., Patsouris E., Kouraklis G., Koutsilieris M. KISS1 and KISS1R expression in gastric cancer. J. BUON. 2018;23:79–84. [PubMed] [Google Scholar]
- 18.Li C., Yuan L., Han S., Xuan M., Liu D., Tian B., Yu W. Reduced Kiss-1 expression is associated with clinical aggressive feature of gastric cancer patients and promotes migration and invasion in gastric cancer cells. Oncol. Rep. 2020;44:1149–1157. doi: 10.3892/or.2020.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell D.C., Abdelrahim M., Weng J., Stafford L.J., Safe S., Bar-Eli M., Liu M. Regulation of KiSS-1 metastasis suppressor gene expression in breast cancer cells by direct interaction of transcription factors activator protein-2alpha and specificity protein-1. J. Biol. Chem. 2006;281:51–58. doi: 10.1074/jbc.M506245200. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y., Berk M., Singh L.S., Tan H., Yin L., Powell C.T., Xu Y. KiSS1 suppresses metastasis in human ovarian cancer via inhibition of protein kinase C alpha. Clin. Exp. Metastasis. 2005;22:369–376. doi: 10.1007/s10585-005-8186-4. [DOI] [PubMed] [Google Scholar]
- 21.Gross I., Lhermitte B., Domon-Dell C., Duluc I., Martin E., Gaiddon C., Kedinger M., Freund J.N. Phosphorylation of the homeotic tumor suppressor Cdx2 mediates its ubiquitin-dependent proteasome degradation. Oncogene. 2005;24:7955–7963. doi: 10.1038/sj.onc.1208945. [DOI] [PubMed] [Google Scholar]
- 22.Rings E.H., Boudreau F., Taylor J.K., Moffett J., Suh E.R., Traber P.G. Phosphorylation of the serine 60 residue within the Cdx2 activation domain mediates its transactivation capacity. Gastroenterology. 2001;121:1437–1450. doi: 10.1053/gast.2001.29618. [DOI] [PubMed] [Google Scholar]
- 23.Van Cutsem E., Sagaert X., Topal B., Haustermans K., Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 24.Padmanabhan N., Ushijima T., Tan P. How to stomach an epigenetic insult: the gastric cancer epigenome. Nat. Rev. Gastroenterol. Hepatol. 2017;14:467–478. doi: 10.1038/nrgastro.2017.53. [DOI] [PubMed] [Google Scholar]
- 25.Topalian S.L., Taube J.M., Anders R.A., Pardoll D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T., Xie J., Shen C., Cheng D., Shi Y., Wu Z., Deng X., Chen H., Shen B., Peng C. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 27.Alvarez-Dominguez J.R., Lodish H.F. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130:1965–1975. doi: 10.1182/blood-2017-06-788695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinodoz S., Guttman M. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24:651–663. doi: 10.1016/j.tcb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barros R., da Costa L.T., Pinto-de-Sousa J., Duluc I., Freund J.N., David L., Almeida R. CDX2 autoregulation in human intestinal metaplasia of the stomach: impact on the stability of the phenotype. Gut. 2011;60:290–298. doi: 10.1136/gut.2010.222323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q., Teh M., Ito K., Shah N., Ito Y., Yeoh K.G. CDX2 expression is progressively decreased in human gastric intestinal metaplasia, dysplasia and cancer. Mod. Pathol. 2007;20:1286–1297. doi: 10.1038/modpathol.3800968. [DOI] [PubMed] [Google Scholar]
- 31.Marchese F.P., Raimondi I., Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ba M.C., Ba Z., Long H., Cui S.Z., Gong Y.F., Yan Z.F., Lin K.P., Wu Y.B., Tu Y.N. LncRNA AC093818.1 accelerates gastric cancer metastasis by epigenetically promoting PDK1 expression. Cell Death Dis. 2020;11:64. doi: 10.1038/s41419-020-2245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuelten C.H., Parent C.A., Montell D.J. Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat. Rev. Cancer. 2018;18:296–312. doi: 10.1038/nrc.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciaramella V., Della Corte C.M., Ciardiello F., Morgillo F. Kisspeptin and Cancer: Molecular Interaction, Biological Functions, and Future Perspectives. Front. Endocrinol. (Lausanne) 2018;9:115. doi: 10.3389/fendo.2018.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li N., Wang H.X., Zhang J., Ye Y.P., He G.Y. KISS-1 inhibits the proliferation and invasion of gastric carcinoma cells. World J. Gastroenterol. 2012;18:1827–1833. doi: 10.3748/wjg.v18.i15.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C., Wang L., Li Y.W., Cui Y.S., Wang Y.Q., Liu S. Long noncoding RNA LUCAT1 promotes migration and invasion of prostate cancer cells by inhibiting KISS1 expression. Eur. Rev. Med. Pharmacol. Sci. 2019;23:3277–3283. doi: 10.26355/eurrev_201904_17689. [DOI] [PubMed] [Google Scholar]
- 37.Tang Z., Li L., Tang Y., Xie D., Wu K., Wei W., Xiao Q. CDK2 positively regulates aerobic glycolysis by suppressing SIRT5 in gastric cancer. Cancer Sci. 2018;109:2590–2598. doi: 10.1111/cas.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulanger J., Vézina A., Mongrain S., Boudreau F., Perreault N., Auclair B.A., Lainé J., Asselin C., Rivard N. Cdk2-dependent phosphorylation of homeobox transcription factor CDX2 regulates its nuclear translocation and proteasome-mediated degradation in human intestinal epithelial cells. J. Biol. Chem. 2005;280:18095–18107. doi: 10.1074/jbc.M502184200. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell D.C., Stafford L.J., Li D., Bar-Eli M., Liu M. Transcriptional regulation of KiSS-1 gene expression in metastatic melanoma by specificity protein-1 and its coactivator DRIP-130. Oncogene. 2007;26:1739–1747. doi: 10.1038/sj.onc.1209963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.