Abstract
The conserved protein Hfq is a key factor in the RNA-mediated control of gene expression in most known bacteria. The transient intermediates Hfq forms with RNA support intricate and robust regulatory networks. In Pseudomonas, Hfq recognizes repeats of adenine–purine–any nucleotide (ARN) in target mRNAs via its distal binding side, and together with the catabolite repression control (Crc) protein, assembles into a translation–repression complex. Earlier experiments yielded static, ensemble-averaged structures of the complex, but details of its interface dynamics and assembly pathway remained elusive. Using explicit solvent atomistic molecular dynamics simulations, we modeled the extensive dynamics of the Hfq–RNA interface and found implications for the assembly of the complex. We predict that syn/anti flips of the adenine nucleotides in each ARN repeat contribute to a dynamic recognition mechanism between the Hfq distal side and mRNA targets. We identify a previously unknown binding pocket that can accept any nucleotide and propose that it may serve as a ‘status quo’ staging point, providing nonspecific binding affinity, until Crc engages the Hfq–RNA binary complex. The dynamical components of the Hfq–RNA recognition can speed up screening of the pool of the surrounding RNAs, participate in rapid accommodation of the RNA on the protein surface, and facilitate competition among different RNAs. The register of Crc in the ternary assembly could be defined by the recognition of a guanine-specific base–phosphate interaction between the first and last ARN repeats of the bound RNA. This dynamic substrate recognition provides structural rationale for the stepwise assembly of multicomponent ribonucleoprotein complexes nucleated by Hfq–RNA binding.
Keywords: Hfq protein, molecular dynamics, RNA-binding protein, RNA metabolism, protein–nucleic acid interaction, Crc protein, ARN repeats, dynamic recognition
Abbreviations: 4BPh, type-4 base–phosphate; ARN, adenine–purine–any nucleotide; Crc, catabolite repression control; PDB, Protein Data Bank; sRNA, small regulatory noncoding RNA molecule; vdW, van der Waals
Hfq is a conserved RNA-binding protein and a pleiotropic regulator of translation and RNA stability in diverse bacteria. Some of its best studied roles are to suppress translation of target mRNAs by annealing them with small regulatory noncoding RNA molecules (sRNAs) (1, 2, 3, 4) or by directly binding an A-rich sequence in the 5′-untranslated region of mRNAs (5, 6). Six Hfq protomers assemble to form a hexameric, ring-like chaperone (Fig. 1A). The hexamer can bind RNAs via three surfaces (7, 8), commonly termed as proximal, distal, and rim faces or sides (9, 10). Furthermore, the intrinsically disordered C-terminal regions can also interact with RNA and autoregulate the activity of Hfq (11, 12). The homo-oligomeric nature of Hfq favors recognition of nucleotide repeats in target RNAs, such as the ARN-triplet repeat motif (where A is an adenine and R and N are a purine and any nucleotide, respectively), which binds on the distal side (Fig. 1B) (13).
Figure 1.
Hfq architecture and assembly into higher order ribonucleoprotein particles.A, the structure of the Pseudomonas aeruginosa Hfq hexamer with differently colored monomeric units and its RNA-binding distal side highlighted in pink. B, a tilted view into the distal side with the amiE6ARN mRNA engaged. The A, R, and N nucleotides are colored in yellow, red, and orange, respectively. The RNA sequence is specified. The Hfq protein is colored in dark gray, and the RNA backbone is in pink. C, the quaternary complex formed by Hfq, Crc, and amiE6ARN. From left to right, the quaternary complex can contain two, three, or four Crc units (light gray), respectively. Crc, catabolite repression control.
In the gram-negative bacterium Pseudomonas aeruginosa, Hfq was identified as a versatile contributor to metabolic regulation (14) whose influences on different pathways are facilitated via interactions with other proteins (15). One such partner is the catabolite repression control (Crc) protein (16). In Pseudomonas, Crc is responsible for directing the metabolic pathways toward preferring succinate over other potential carbon sources (17). The mechanism of Hfq and Crc cooperation involves binding of both proteins to ARN repeats in the 5′-untranslated region of target mRNAs, thus repressing expression of enzymes involved in alternative metabolic pathways (14). When succinate is depleted, the sRNA CrcZ is expressed (18) and proceeds to sequester Hfq and Crc from their mRNA targets, allowing the mRNAs of alternative metabolic genes, such as the amiE, to be translated (6).
Crc has no intrinsic RNA-binding or Hfq-binding capabilities of its own (16), yet it can bind to Hfq–RNA complexes and strengthen Hfq interactions with target RNAs (14). The structural basis of this cooperative action has been unraveled by cryo-EM, which showed Hfq, Crc, and a short segment of amiE mRNA forming a quaternary complex (19). The structure of this complex encompasses two Hfq hexamers, each complexed with an RNA octadecamer containing six ARN repeats, the amiE6ARN fragment. At a minimum, there is a homodimer of two Crc proteins positioned between the two Hfq–RNA complexes. We henceforth refer to this structure as the quaternary complex (Fig. 1C). Depending on availability, up to two additional Crc proteins can be recruited into the quaternary complex (19).
The cryo-EM structures of the quaternary complex reveal a recognition motif seen in earlier X-ray structures of Hfq complexed with polyadenine RNAs (8, 20). In the crystal structures, the A and R nucleotides of ARN repeats are specifically bound by the Hfq, whereas the N nucleotides are bulged away from Hfq and interact with the neighboring crystallographic cells. The same RNA recognition pattern is present in the quaternary complex except that the N nucleotides are instead engaged in nonspecific interactions with the Crc proteins (19).
In this study, we used atomistic molecular dynamics (MD) simulations to explore the conformational variation of the Hfq–RNA binary complex and the higher order quaternary complexes formed with Crc. Our MD simulations utilize a set of carefully calibrated molecular mechanics models (21, 22) that have been applied in many studies of protein–RNA complexes with predictive power (21, 23, 24, 25, 26, 27, 28). MD allows the study of atomic movements at spatiotemporal resolutions inaccessible to any currently available experimental method and can help rationalize experimental observations (21). Although simulation timescales are generally short, well-executed simulations can provide insights into biomolecular dynamics that are not apparent from static models obtained by structural experiments, for which the data are typically time- and ensemble-averaged (23, 29).
Biomolecular dynamics can be invaluable for understanding the nature of complex intramolecular interfaces, such as those of protein–RNA complexes, where it underpins binding affinities, specificities, and formation rates. The interface dynamics between biomolecules can involve competing local conformational substates rather than a fixed geometry (30), resulting in dynamic recognition. The substates associated with the dynamical ensemble may be important for the detailed mechanisms of the process of binding and unbinding (31). Dynamic recognition can be biologically significant as it could facilitate highly specific recognition of RNAs by a protein and would provide a mechanism by which a large pool of cellular RNAs can be interrogated with speed, specificity, and high affinity for target sequences. Different interaction intermediates can be preferred by different binding partners in quaternary complexes. We propose that Hfq must utilize a form of dynamic recognition because its in vivo RNA cycling at both proximal and distal sides was shown to be disproportionally fast relative to its low-nanomolar RNA-binding affinity, as measured by in vitro experiments (32, 33).
Our results suggest that extensive equilibrium local dynamics indeed occur at the distal side interface of the Hfq–RNA complex and offer a concrete example of a conformational switch that can influence rates of translocation along a length of RNA. Namely, the first nucleotide in each ARN repeat (i.e., the adenines) can undergo frequent syn/anti flips, before Crc binding. The anti and syn conformations are supported by Hfq via unique adenine-specific interactions in both positions. The frequency of the flips is lowered upon formation of the quaternary complex, that is, after Crc is bound, and the syn conformation becomes less favored. We also identify a previously unknown binding pocket at the distal side of Hfq which can weakly bind the N nucleotides of the ARN repeats in the absence of Crc. Finally, we suggest a potential assembly pathway in which the Crc initially recognizes an intramolecular RNA interaction in the amiE6ARN.
Results
Design, stability, and reproducibility of the MD simulations
The simulations of Hfq–RNA and quaternary complexes showed no loss of structural compactness and integrity, which indicates good performance of the force field (21) and sufficient quality of the experimental structures (8, 19) used as the starting states for the simulations. For such large systems, we did not expect to achieve a full thermodynamic convergence within affordable computational time (34). In fact, such convergence is not fully achieved even in longer MD simulations of much smaller systems such as RNA tetraloops and tetranucleotides (35). Nevertheless, even without achieving full quantitative convergence, the MD simulations can provide a wealth of information about the RNA systems (21) inaccessible to experimental methods. The simulations presented here can be considered qualitatively converged in a sense that the same simulation trends were observed in multiple independent parallel trajectories of the individual systems (Table 1). In addition, we extended selected simulations up to 5, 10, or 15 μs (Table 1), observing the same trends even on these longer timescales. The analyses presented in the main text are based mainly on these extended trajectories. For the remaining systems, the analyses were performed on a combined simulation ensemble and are described in Supporting information.
Table 1.
List of simulations
| Simulation name | Source PDBa | #Hfq | #RNA | #Crc | Number of simulations × length (μs) |
|---|---|---|---|---|---|
| Quaternary complex simulations | |||||
| 2Hfq_2Crc_2RNA | 6o1k | 2 | 2 | 2 | 1 × 5, 2 × 1 |
| 2Hfq_3Crc_2RNA | 6o1l | 2 | 2 | 3 | 2 × 1 |
| 2Hfq_4Crc_2RNA | 6o1m | 2 | 2 | 4 | 1 × 5, 1 × 1 |
| 2Hfq_4Crc_2RNA_AA-synb | 6o1m | 2 | 2 | 4 | 2 × 5, 1 × 1 |
| Hfq_Crc1_RNAc | 6o1k | 1 | 1 | 1 | 2 × 2 |
| Hfq_Crc2_RNAc | 6o1k | 1 | 1 | 1 | 2 × 2 |
| Hfq_2Crc_RNA | 6o1k | 1 | 1 | 2 | 2 × 1 |
| 2Hfq_4Crc_2RNA_noHBfixd | 6o1m | 2 | 2 | 4 | 1 × 1 |
| Quaternary complex simulations–modified systems | |||||
| 2Hfq_2Crc_2RNA_G18Ce | 6o1k | 2 | 2 | 2 | 2 × 1 |
| 2Hfq_2Crc_2RNA_circe | 6o1k | 2 | 2 | 2 | 2 × 1 |
| 2Hfq_2Crc_2RNA_exte | 6o1k | 2 | 2 | 2 | 2 × 1 |
| Other simulations | |||||
| Hfq_RNA | 6o1k | 1 | 1 | 0 | 1 × 15, 2 × 1 |
| Hfq_RNA_allGRf | 6o1k | 1 | 1 | 0 | 1 × 10, 2 × 2 |
| Hfq_RNA_I30Ae | 6o1k | 1 | 1 | 0 | 2 × 1 |
| Hfq_RNA_pol-Ag | 3gib | 1 | 1 | 0 | 1 × 2 |
| 2Crc | 6o1k | 0 | 0 | 2 | 2 × 1 |
| 2Crc_4jg3 | 4jg3 | 0 | 0 | 2 | 2 × 1 |
| 4Crc | 6o1m | 0 | 0 | 4 | 1 × 2 |
| Hfq_RNA_noHBfixd | 6o1k | 1 | 1 | 0 | 2 × 2 |
| Hfq_RNA_2×HBfixh | 6o1k | 1 | 1 | 0 | 1 × 2 |
Crc, catabolite repression control; PDB, Protein Data Bank.
PDB ID of the experimental structure which was utilized as the initial structure. In some simulations, only selected parts of the experimental structure were used (see Selection of initial structures and Fig. S1).
All bases of the “A” nucleotides within the ARN repeats were modified to be in syn conformation before the simulation start.
Crc1 and Crc2 refer to the Crc proteins that bind near and away from G18, respectively (Fig. S1).
Simulations were done without the HBfix (see System building and simulation protocol and Supporting information).
In “G18C”, “circ”, “ext”, or “I30A” simulations, the G18 nucleotide of the RNA was replaced with C18, the RNA chain was circularized by covalently connecting the 5′- and 3′-nucleotides via a newly modeled phosphate, the RNA was extended from its 3′-end by adding nucleotides U19 and G20, or I30 was mutated into alanine, respectively.
All the “R” nucleotides within the ARN repeats were modified to be guanosines.
A circular polyadenine RNA octadecamer was bound to Hfq.
HBfix with 2 kcal/mol penalty was utilized (see System building and simulation protocol and Supporting information).
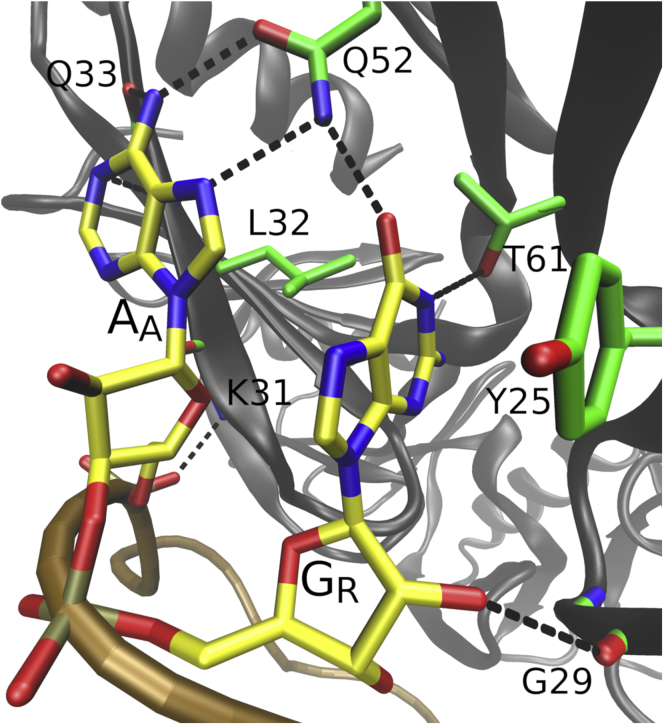
The bound RNA in the quaternary complex consists of six ARN trinucleotide repeats while the Hfq itself is a hexamer (Fig. 1B). All ARN motif nucleotides in the positions “A” (includes A1, A4, A7, A10, A13, and A16) and “R” (includes A2, A5, A8, A11, A14, and G17), respectively, are bound in identically organized binding pockets, and their positions are clearly defined by H-bonds, base stacking, and van der Waals (vdW) interactions (Fig. 2A). The pockets exhibited highly similar behavior in simulations, and their protein–RNA interactions were all maintained with only reversible fluctuations (Tables S1 and S2). To simplify descriptions, we will collectively refer to the first and second nucleotides of the repeats as either AA or AR/GR, with the lower index signifying the nucleotide’s position within the ARN repeat. For example, notation Q33(N)–AA(N7) describes backbone amide nitrogen of residues Q33 in the individual Hfq chains forming H-bonds with N7 of RNA nucleotides A1, A4, A7, A10, A13, or A16. Wherever the behavior significantly differed among the repeats, we refer directly to the specific nucleotides by their residue numbering (Fig. 1). To help distinguish Crc amino acids, they are labeled with an accent (e.g., R162′).
Figure 2.
Reversible syn/anti flip on the distal face of Hfq. Interactions observed for the AA/AR binding pocket with the AA base in (A) anti, (B) a transition intermediate, and (C) syn. The interacting nucleotides and amino acids are labeled on the left panel. Carbon atoms are colored yellow and green and the backbones traced in brown and dark gray, in RNA and Hfq, respectively. The black dashed lines indicate H-bonds. Unless specified otherwise, we use this labeling and color coding in rest of the text. See below for description of the AA/GR binding pocket.
Simulations of isolated Hfq–RNA reveal syn/anti flips of the AA nucleotides
The most significant conformational change observed in the simulations of Hfq–RNA complexes, in absence of Crc, was the dynamic equilibrium between anti and syn conformations of the AA nucleobases. Most strikingly, the anti-AA and syn-AA nucleotides established interactions with the same amino acids. Structurally, this was possible by the N7 atom replacing the N1 atom as the H-bond acceptor and vice versa, whereas the second hydrogen of the N6 amino group was utilized as a H-bond donor (Fig. 2). There is also a single water bridge between AA and AR, which is formed solely with AA in anti (Fig. 3B). This water bridge is present in the X-ray structure of poly-A RNA bound to Hfq (8) and was regularly observed in all our MD simulations as long as AA was in anti while being abolished in syn. Finally, there is a close presumable repulsive atomic contact between O4′ atom of AA ribose and the backbone carbonyl of K31 associated with the anti conformation of AA (Fig. 3B). In MD simulations, the K31(O)-AA(O4′) distance increased when AA flipped into syn, thus relieving the repulsion. The average simulation time between the flips differed significantly among the individual simulations and AA nucleotides, ranging from tens to hundreds of nanoseconds (Fig. 3; Fig. S2). The transition intermediate of these flips (Fig. 2B) was stabilized by a temporary formation of an intranucleotide H-bond, and the transition time of the flips was in the range of tens of picoseconds. In each simulation, several back-and-forth syn/anti flips were observed for every AA nucleotide with few exceptions (Table 2; Table S3). We note the relatively large variability of syn/anti populations among individual AA nucleotides. After comparing multiple simulation trajectories (Table S3), we conclude that it reflects randomness of simulation sampling and the fact that the syn/anti dynamics are not synchronized across the six ARN repeats.
Figure 3.
Time development of the anti (green)/syn (blue) of the AAnucleotides in selected simulations. For the graphs of the remaining simulations and of the second RNA molecules of systems containing two RNAs, see Fig. S2. See Table 1 for details of the individual systems.
Table 2.
Transitions between the anti and syn conformations of individual AA nucleotides in selected MD simulationsa
| Simulation name and length | I: number of transitions/syn populationb II: time until first transition (ns)c III: Average lifetimes of syn/anti (ns)d |
||||||
|---|---|---|---|---|---|---|---|
| A1 | A4 | A7 | A10 | A13 | A16 | ||
| Hfq_RNA – 15 μs | I | 14/0.95 | 39/0.63 | 15/0.30 | 22/0.62 | 7/0.24 | 11/0.54 |
| II | 17 | 800 | 140 | 905 | 2210 | 212 | |
| III | 752/25 | 235/131 | 240/522 | 385/214 | 272/819 | 1148/871 | |
| Hfq_RNA_allGR – 10 μs | I | 1/0.96 | 5/0.91 | 3/0.97 | 5/0.47 | 27/0.85 | 3/0.82 |
| II | 426 | 487 | 57 | 120 | 70 | 373 | |
| III | – | 3036/298 | 4868/132 | 938/1062 | 356/59 | 4095/905 | |
| 2Hfq_2Crc_2RNA – 5 μs | I | 0/0.00 | 2/0.57 | 0/0.00 | 0/0.00 | 0/0.00 | 4/0.44 |
| 0/0.00 | 0/0.00 | 0/0.00 | 0/0.00 | 3/0.31 | 3/0.91 | ||
| II | – | 1122 | – | – | – | 38 | |
| – | – | – | – | 483 | 431 | ||
| III | – | 2844/1077 | – | – | – | 544/564 | |
| – | – | – | – | 777/1148 | 2284/215 | ||
| 2Hfq_4Crc_2RNA – 5 μs | I | 0/0.00 | 0/0.00 | 0/0.00 | 0/0.00 | 0/0.00 | 2/0.19 |
| 0/0.00 | 0/0.00 | 0/0.00 | 0/0.00 | 0/0.00 | 2/0.10 | ||
| II | – | – | – | – | – | 2873 | |
| – | – | – | – | – | 174 | ||
| III | – | – | – | – | – | 342/994 | |
| – | – | – | – | – | 292/1031 | ||
| 2Hfq_4Crc_2RNA_AA-syn_1 – 5 μs | I | 0/1.00 | 11/0.59 | 4/0.99 | 3/0.84 | 0/1.00 | 2/0.99 |
| 0/1.00 | 1/0.99 | 6/0.56 | 2/0.98 | 0/1.00 | 0/1.00 | ||
| II | – | 66 | 3440 | 111 | – | 608 | |
| – | 4954 | 785 | 654 | – | – | ||
| III | – | 267/262 | 1243/9 | 1392/275 | – | 2497/6 | |
| – | – | 463/318 | 2439/123 | – | – | ||
| 2Hfq_4Crc_2RNA_AA-syn_2 – 5 μs | I | 0/1.00 | 4/0.65 | 1/0.04 | 3/0.03 | 0/1.00 | 6/0.51 |
| 0/1.00 | 3/0.89 | 1/0.84 | 3/0.25 | 0/1.00 | 0/1.00 | ||
| II | – | 858 | 195 | 66 | – | 123 | |
| – | 636 | 4224 | 84 | – | – | ||
| III | – | 1091/864 | – | 95/2405 | – | 637/817 | |
| – | 2218/189 | – | 423/933 | – | – | ||
The two lines in the “2RNA” simulations each describe one of the two RNA molecules contained in these systems. Crc’s sterically obstruct flips of A1 in 2Crc and of A1 and A13 in 4Crc systems, respectively.
Number of syn/anti transitions (in any direction) and the populations of the two states with one and zero corresponding to all-syn and all-anti, respectively. The AA nucleotide was considered to be in syn and anti when its χ dihedral angle was −30° to 150° and 150° to 330°, respectively. We disregarded transitions lasting less than 300 ps.
Simulation time in which the first syn/anti transition (in either direction) occurred. The “–” symbol indicates that no transition occurred.
Average simulation time that AA nucleotide remained in syn and anti, respectively, before flipping. The lifetimes are not stated when there was only a single or no transition observed.
Crc attenuates syn/anti flips of the AA nucleotides
The syn/anti flips were strongly reduced when Crc is bound to the Hfq–RNA intermediate (Table 2 and Fig. 3) for the unmodified experimental structure (19) where all AAs are in anti. For nucleotides A1 and A13, this can be explained by the Crc proteins forming vdW contacts and H-bonds that sterically block the flips and stabilize the anti conformation, respectively (Fig. 4A) (19). For the rest of the AA positions, Crc forms extensive nonspecific contacts with the phosphate groups immediately downstream (Fig. S3). The syn conformation of AA is associated with dihedral angle transitions of this backbone suite (36) but not the anti conformation (Fig. S4). This indirectly promotes the anti conformation as the non-specific contacts with Crc restrict the available conformational space for such dihedral angle transitions. The overall atomic fluctuations of this backbone suite are also generally lower with Crc (Fig. S5). For both syn and anti conformations, the K31(O)-AA(O4′) distance was shorter in the presence of Crc than in isolated Hfq–RNA (Fig. 4C), that is, Crc pushes the AA nucleotide deeper into the A pocket. This could affect the syn/anti balance as a tighter geometry is associated with the anti conformation. Flipping of the AA base into syn to relieve the K31(O)-AA(O4′) repulsion then becomes less favorable when Crc is bound. On the other hand, the penalty for the loss of the water bridge (Fig. 4B) upon flipping into syn remains the same in both systems. In conclusion, rather than simply blocking the flips, the Crc could also promote the anti conformation by subtly shifting the free-energy balance among multiple interactions within and around the AA binding pocket.
Figure 4.
Recruitment of Crc impedes conformational switching on the Hfq distal face.A, binding of Crc proteins sterically obstructs syn/anti flips, stabilizes the anti conformation of A13 nucleobase, and blocks formation of the transition structure in the simulations. The A1 nucleobase was similarly immobilized by another Crc protein. B, the anti conformation of every AA is associated with a water bridge toward AR and a repulsive interaction between the O4′ atom of ribose and the backbone carbonyl of K31. Flip of AA into syn relieves the repulsion but disrupts the water bridge. C, normalized histograms of the average K31(O)–AA(O4′) distances observed in selected simulations for AA nucleotides in either syn or anti conformations. The presence of Crc produces shorter distances. See Table 1 for details of the individual systems. Crc, catabolite repression control.
Prior flipping of AA nucleotides into syn alters protein–RNA interface with Crc
The experimental structures used in this study have all the AA nucleotides in the anti conformation. In a quaternary complex where we flipped all the AA bases into syn before the simulation start (2Hfq_4Crc_2RNA_AA-syn simulations; see Table 1), we observed some AA bases returning to anti on the simulation timescale (Table 2 and Fig. 3). This strongly contrasts with the simulations where the AA bases were in anti from the start (Crc attenuates syn/anti flips of the AA nucleotides) and subsequently showed no signs of flipping into syn. However, often the AA bases in the 2Hfq_4Crc_2RNA_AA-syn simulations which flipped into anti would once again flip into syn in time, seemingly contradicting the idea that Crc promotes anti or suppresses the flips. For many of the more short-lived anti states (Fig. 3), this was because not all of the interactions with Hfq (Fig. 2) had properly formed after AA flipped into anti. This never occurred in simulations without Crc and suggests Hfq’s ability to seamlessly accommodate the spontaneous flips (Fig. 2) is limited by Crc. We also observed lower stability and alterations of the Crc–RNA interactions in the 2Hfq_4Crc_2RNA_AA-syn simulations (Table S4). Even for repeats where these interactions were not lost, the flips back into syn were also often preceded by temporary disruption of the local Crc–RNA interactions (Fig. S6). This suggests that a priori flipping AA into syn perturbs the interface with Crc and this disturbance is not fully relaxed in the course of our simulations. This allows subsequent syn/anti flips in both directions similar to those observed in simulations without Crc. We also suspect there could be some degree of synchronization between the flips of individual AA nucleotides originating from general destabilization of the Crc–RNA interface although this could not be decisively established from our simulations.
Dynamics of the AA/GR binding pocket differs compared with the AA/AR consensus
The second nucleotide of the ARN repeats can be either adenosine or guanosine, collectively referred to as AR or GR. The GR nucleotides form interactions with the same Hfq residues as the AR nucleotides (Fig. 2), both in the experimental structure and in simulations. However, the simulations reveal a striking difference in dynamics of the binding pocket and the associated syn/anti flips of the preceding AA nucleotide when GR is present instead of the AR. Namely, in simulations of the Hfq–RNA complex where we replaced all the AR nucleotides with GR (Table 1), the syn population of the AA nucleotides was significantly increased (Table 2 and Fig. 3). We suggest the reason for this is formation of an interaction with the Q52 side chain that can simultaneously interact with AA(N6), AA(N7), and GR(O6) atoms only when AA is in syn (Fig. 5).
Figure 5.
AAnucleotide in syn with GRas the succeeding nucleotide. Q52 side chain could form simultaneous interaction with both nucleotides in simulations, significantly stabilizing the syn conformation of AA when guanosine was the succeeding nucleotide.
The simulations predict a third binding pocket for the N nucleotides on the distal side of Hfq
In the cryo-EM structures of the quaternary complex (19), the N nucleotides of all ARN repeats, except G18, have their bases turned away from the distal side of the Hfq to interact with Crc. The N nucleotides are positioned similarly in the X-ray structure of isolated Hfq bound to poly-A RNA, where they participate in crystal packing (8). In contrast, all N nucleotides bend toward Hfq in our simulations of isolated Hfq–RNA complexes. The nucleotides formed vdW interactions with I30, as well as H-bonding or ion-bridge interactions with the N28(O) atoms of the individual Hfq subunits (Fig. 6 and Table S5). This consistently occurred in all simulations, either with the amiE6ARN mRNA or poly-A RNA sequence bound (Table 1) and was universally observed for all N nucleotides except G18. The simulations thus predict a third binding pocket at the distal side of the Hfq for nondiscriminatory binding of N nucleotides of the ARN repeats (Fig. 6). We henceforth refer to it as the N pocket, in analogy to the previously described A pockets and R pockets, which bind A and R nucleotides, respectively (8). The existence of N pocket binding would be in agreement with the previous report that Hfq mutants lacking I30 have a reduced affinity for RNA sequences that are bound to the distal side (9). Indeed, in simulations of a system where we replaced I30 in every Hfq chain with alanine, the N-pocket binding was either reduced or abolished (Table S6).
Figure 6.
A distal side pocket for the N position of the ARN motif.A, the bases of the N nucleotides of the ARN repeats in the amiE6ARN mRNA are bulged away from the distal side of Hfq in the quaternary complexes (left) (19). In simulations of the isolated Hfq–RNA systems without any Crc, they all bend toward Hfq (right). B, all the bases (i–iv) formed vdW interaction with I30 and either H-bond or ion-bridge interaction with the backbone of N28. This arrangement is a putative binding pocket for N nucleotides at the distal side of Hfq, the N-pocket. The K+ are shown as lilac spheres. The black dashed lines indicate H-bonds or ion bridges. ARN, adenine–purine–any nucleotide; Crc, catabolite repression control; vdW, van der Waals.
In the cryo-EM structures of quaternary complexes (19), as well as in our simulations, the N nucleotides form nonspecific interactions with the Crc partners. These interactions are described in detail in the Supporting information, except for the 3′-terminal G18, which is described in The G18/A3 4BPh base–phosphate interaction is stabilized by Crc. In summary, the novel, putative N-pockets could serve as nondiscriminatory, transient ‘status quo’ binding pocket for the N nucleotides, until a partner molecule, such as Crc, engages the Hfq–RNA intermediate.
The G18–A3 4BPh base–phosphate interaction is stabilized by Crc
The 3′-terminal G18 is different from the other N nucleotides in the quaternary complex. First, the G18 is not flipped away from the distal side of Hfq in the cryo-EM structure (19). Instead, it forms a vdW interaction with the I30 side chain in a manner similar to the N-pocket binding predicted for the other N nucleotides in isolated Hfq–RNA systems (Fig. 6). Second, the G18 is forming a guanine-specific type-4 base–phosphate (4BPh) interaction (37) with the phosphate of A3. Third, in the experimental structure, the G18 is very close to potentially interact with Crc, namely with the K135′, R138′, and K139′ side chains. These interactions were subsequently formed in MD simulations (Fig. 7 and Table S7). Notably, the K139′ interaction was sequence-specific and seemed to stabilize the intramolecular 4BPh interaction by compensating for the repulsion between the guanine’s O6 atom and the A2 phosphate (Fig. 7). This is supported by simulations of the isolated Hfq–RNA system (Table 1), in which the G18–A3 4BPh interaction was visibly fluctuating and then lost early in all simulations, along with the G18 base vdW interaction toward I30 (Table S7). For more details, see Supporting information.
Figure 7.
Registering Crc on the Hfq distal face. The G18 nucleotide as seen in the structure of the quaternary complex (left) (19). The black and purple dashed lines indicate H-bonds and putative H-bonds, respectively. The putative H-bonds between Crc and G18 were fully realized in all MD simulations (right). The K139′ side chain formed a base-specific interaction with the G18 and screened repulsion between the base and the RNA backbone. Crc, catabolite repression control.
amiE6ARN G18 as a putative anchor point for Crc binding to the Hfq–RNA complex
Crc does not dimerize spontaneously (19), which is in agreement with our MD simulations (Supporting information). Yet, a minimum of two Crc proteins is required for full assembly of a quaternary complex with Hfq–RNA (Fig. 1) (19). Therefore, a transiently formed structure with one Crc protein bound to a single Hfq–RNA intermediate could potentially exist at early stages of the quaternary complex formation. We thus simulated two Hfq–RNA–Crc systems that contained either the Crc1 (bound near the G18) or the Crc2 protein (Table 1; Fig. S1). We observed reduced fluctuations in relation to the Hfq–RNA for Crc1 compared with Crc2 (Fig. S7). Interestingly, binding of Crc1 alone was sufficient to stabilize the 4BPh base–phosphate interaction and to establish the base-specific interaction with the K139′ formed by the G18 nucleobase (Fig. 7 and Table S7).
Next, we explored the dependency of Crc binding on the sequence identity and position of the 3′-terminal nucleotide within the RNA chain. G18 is the only N nucleotide recognized specifically by Crc (Fig. 7) and could potentially act as a register-defining marker for its binding. To explore this, we first prepared a system with circularized amiE6ARN mRNA (Table 1). Building the covalent bond between the two RNA termini necessarily involved disruption of the 4BPh interaction as the G18 had to be shifted to make the bond. The G18 4BPh interaction was never reformed in subsequent simulations while no specific interactions between Crc and the G18 occurred. The K139′ side chain formed nonspecific interactions with the newly modeled phosphate (Fig. S8). Therefore, the discontinuity in the bound RNA may be essential for proper assembly of the quaternary complex. Next, we prepared systems where we replaced G18 with C18 (Table 1). There, the C18 neither formed base–phosphate interactions nor made any other specific interactions with Crc. It was consequently unstable in its initial position and interacted with the solvent or flipped over to stack with G15. Finally, we tested whether the 4BPh interaction could be stable without the G18 being a 3′-terminal nucleotide, but still the last nucleotide bound by Hfq, as it would be in a full-length amiE mRNA. Thus, we prepared systems where we extended the amiE6ARN mRNA by two nucleotides, introducing U19 and G20. The newly modeled nucleotides were positioned in a way to avoid any clashes with the rest of the system. The G18 4BPh interaction remained entirely stable in these simulations.
Discussion
Hfq is an RNA chaperone involved in numerous regulatory networks in many bacteria. Its diverse functional roles are underpinned by multiple RNA-binding surfaces, each preferring different sequences (9, 10). Here, we examined the structure and dynamics of RNA binding at the distal side of Hfq, which prefers ARN repeats. In 90 μs of MD simulations, we observed significant equilibrium dynamics occurring at the A-pocket of the Hfq–RNA interface, which is attenuated by Crc binding (Fig. 1). In addition, a putative new binding pocket on the Hfq distal side was discovered. Finally, a possible folding pathway of the quaternary complex involving sequence-specific recognition of a 3′-terminal bound guanosine by the Crc is proposed.
Syn/anti flipping in A pockets may contribute to Hfq’s binding strategy
Our simulations revealed syn/anti transitions of the A-pocket adenosines (Fig. 2) on a submicrosecond timescale. These transitions were seen for the amiE6ARN mRNA and for circular poly-A RNA. The adenosines interacted with the same A-pocket amino acids in both anti and syn conformations (Fig. 2). This strongly suggests that there may be a dynamic equilibrium of syn and anti A-pocket–bound adenosines existing within the solution structure of the Hfq–RNA complex. There have been previous reports of proteins utilizing syn/anti conformational differences to recognize multiple bases (38) or specifically promoting one of the conformations over the other upon binding (39). However, to our knowledge, there are no studies of protein–RNA complexes where a protein could recognize syn/anti conformations of a single base equally well via a fixed set of amino acids. Importantly, the syn/anti flips do not violate the experimentally known specificity of A pockets for adenosines (8) because adenine-specific interactions are formed in both conformations.
The two available X-ray structures of P. aeruginosa Hfq (PDB ID: 3gib, 5new) (8, 40) with poly-A RNA bound at its distal side, have the A-pocket–bound adenosines solely in the anti conformation. In the structure of S. aureus Hfq bound to an RNA A tract via its distal side (PDB ID: 3qsu), some of the adenines are in the syn conformation, and the study also revealed evidence of syn/anti flips (41). However, Hfq’s distal sides in S. aureus and P. aeruginosa are quite different. It should be noted that the assignment of syn/anti nucleobase conformers in X-ray structures is often challenging and mistakes can occur even in high-quality structures because of phase errors, resolution limitations, and the time-averaged and ensemble-averaged nature of the data collection (42, 43, 44). The crystal lattice or the cryo temperatures could also enforce the anti conformation exclusively (45). Our visual inspection of the electron density maps (8, 40) is consistent with the assigned anti conformation, although this does not exclude possibility of phase error.
Populations of the syn/anti states predicted by MD simulations may be influenced by potential mild force field imbalances, and thus, the results may not reach quantitative accuracy. However, we suggest that the simulations unambiguously predict that the Hfq A pocket can readily host A in syn conformation. The syn conformation, even if overpopulated by MD, could represent a transient (higher energy) binding pattern (46) involved in the process of prebinding of RNA to Hfq or in substrate cycling. Such a binding pattern would be undetectable in ground-state experimental structures because of its low population. The syn conformation of A-pocket adenosines was significantly more populated when the succeeding R-pocket nucleotide was guanosine instead of adenosine (Fig. 5), suggesting a degree of structural communication between the two pockets. There might also be Hfq cofactors that profit from accessibility of the syn orientation or are able to capture the transition conformation (Fig. 2B) for binding.
Crc prefers anti conformation of A-pocket nucleotides
Specific protein–RNA interactions between AA nucleobases and Crc directly promoting the anti conformation are formed only for the A1 and A13 (Fig. 4). Hence, at first sight, the Crc should be able to tolerate syn in the other A-pocket nucleotides (i.e., A4, A7, A10, and A16). Despite this, our simulations of the quaternary complex revealed very few flips toward syn for these nucleotides when starting from the experimental structure where they all possess the anti conformation (Table 2 and Fig. 3). When we manually flipped all A-pocket adenosines into syn before starting the simulations, the AA nucleobases subsequently had the tendency to flip back into anti (Table 2 and Fig. 3), despite all the Hfq–RNA syn interactions having been established, suggesting that the syn conformation is not supported in the presence of Crc. Furthermore, some of the Crc–RNA interactions are destabilized by the syn conformation (Table S4). Some flips back into syn were observed in case the Hfq interactions did not properly form after flipping into anti or during disruptions of some local Crc–RNA interactions (Fig. S6). In other words, the simulation timescale may be insufficient to fully relax the Crc–RNA interface after the initial introduction of the syn conformation. We suggest that Crc may promote the anti conformation via nonspecific interactions it forms with the phosphates downstream of the AA nucleotide (Fig. S3) and by sterically restricting the ability of the A pocket to accommodate the flips that shifts the balance in favor of anti (Fig. 4). This observation illustrates the delicate balance of interactions at the Hfq–RNA interface and its potential utilization by various cofactors which may prefer different conformation of AA nucleobases. We acknowledge that the proposed mechanism affecting the syn/anti balance could include additional components which were not sampled on the timescale of our simulations, such as the partial or complete unbinding of Crc from the quaternary complex.
The dynamic recognition of RNA could be important for substrate cycling by Hfq
The avid binding affinity of Hfq for various RNA targets appears incompatible with its fast biological binding turnover and cellular response (32, 33, 47). The KDs measured in vitro are in the subnanomolar range, which would indicate binding half-lives of well over 1 h. In contrast, the cellular responses facilitated by Hfq are on a timescale of 1 to 2 min, suggesting that nascent RNAs are rapidly cycled through the cellular Hfq pool. To reconcile these two observations, it has been suggested that RNA bound by Hfq can be displaced by competitors from the cellular pool in a stepwise process, which was termed as active cycling (32, 33). We note that the extensive dynamics observed in our simulations for RNA bound to the distal side of Hfq could provide an entry point for its displacement by a competing RNA. Importantly, displacement could occur one ARN repeat at a time or even a single nucleotide at a time, as envisaged by the active cycling model (32, 33). Crc and possibly other Hfq-binding proteins could have coevolved to actively suppress the intrinsic dynamics of the Hfq–RNA interface to slow down RNA cycling. However, as our study involved only RNA bound to the distal side of Hfq, we do not claim this interaction fully accounts for the affinity-response discrepancy (32, 33). Moreover, it is likely that we capture only part of the dynamics immediately pertinent to the dominantly bound state because of the 1 to 15 μs timescales of the simulations.
Hfq provides a weak binding pocket for the N nucleotides
In the quaternary complex (19), the N nucleotides are flipped away from Hfq (Fig. 6) and form many nonspecific interactions with Crc (Supporting information). The exception is G18 which forms an intramolecular 4BPh interaction with the A3 phosphate (Fig. 7). The N nucleotides are also flipped away in the two X-ray structures of the isolated Hfq–RNA complexes (8, 40) where they form extensive crystal packing interactions.
In our simulations of the isolated Hfq–RNA complexes, all the N nucleotides quickly flipped toward Hfq and formed vdW interactions with I30 side chain and H-bonding or ion bridging with the protein backbone of N28 (Fig. 6). Based on our simulations, we suggest that the I30 and N28 residues might constitute a third binding pocket (N-pocket) at the distal side of Hfq. The N-pocket likely offers weaker contribution to the overall binding affinity than the known A and R pockets (8), given the smaller number of intermolecular interactions that define it. Indeed, the bound nucleotides were very dynamical in our simulations and we regularly observed a drift of the base along the molecular interface formed by vdW interaction with the I30 side chain. Such movement would become especially pronounced during temporary disruptions of the interaction toward the N28. We suggest this type of dynamical binding is realistic as it was reported to be in agreement with NMR data in other protein–RNA systems (48). In addition, we suggest that vdW interactions between nucleobase aromatic faces and other molecules are generally very well described by the utilized AMBER force fields with electrostatic potential–derived charges. This is true even for stacking interactions among nucleobases and with aromatic amino acids. Although these interactions are sometimes described as “π-π” in the literature, rigorous quantum chemical studies (49, 50, 51, 52) showed that AMBER type of force fields provides good description of these interactions. There are no substantial “π-π” orbital effects neglected by the force fields. Rather, these interactions primarily involve electrostatic interaction, London dispersion attraction, and short-range repulsion, all of which can be approximated by the current molecular mechanics model. This does not rule out overstabilization or understabilization of vdW interactions of nucleobases in MD simulations. However, such imbalance would rather involve the solvation and not inaccuracy in description of the direct (intrinsic) vdW contact (21).
The flexible behavior and weak binding explain why the N-pocket RNA recognition is supplanted by crystal packing interactions in the two X-ray structures of the isolated Hfq–RNA complexes (8, 40). However, such disordered-like binding could be useful for dynamic recognition of molecules interacting with the Hfq–RNA surface and could serve as a ‘status quo’ state of the Hfq–RNA intermediate. For example, Crc is recognizing the N nucleotides in a nonspecific manner, whereas other proteins might be able to directly read out their sequence. Owing to their dynamical binding to Hfq, the N-pocket nucleotides are readily available to rapidly establish contacts with other partners. The N-pocket binding observed in our simulations is similar to what is seen in the X-ray structure of E. coli Hfq bound to A-rich linker from OxyS sRNA (PDB ID: 4qvc) (53). However, this structure also shows deviations from the ARN repeat consensus and has multiple repulsive and crystal packing interactions affecting its protein–RNA interface. In addition, only three nucleotides could be resolved. Therefore, we opted not to use it for MD simulations.
In summary, we suggest that the “disordered” N-pockets weakly contribute to binding affinity of RNAs to the Hfq in a sequence-independent manner. The dynamical N-nucleotides are at the same time able to quickly establish interactions with other partners, which can already be sequence dependent.
Crc might sample the ARN repeats during quaternary complex formation, searching for 3′-terminal guanosine
The cryo-EM structure of the quaternary complex (19) shows extensive but base nonspecific interactions between the N nucleotides and Crc, suggesting that the Crc may interact equally well with any of the ARN repeats. It is therefore puzzling how the Crc selects its binding register during the quaternary complex formation or indeed if such selection occurs. The fact that it was possible to resolve a high-resolution structure of the quaternary complex by cryo-EM (19), which relies on averaging of many individual images of the complex, suggests that a specific binding register has been selected.
Our simulations indicated that the terminal G18 nucleotide and its 4BPh interaction toward the A3 phosphate might be the marker which, at least in case of the amiE6ARN mRNA, provided a specific binding site for Crc. The guanine-specific 4BPh interaction is the strongest base–phosphate interaction occurring in folded RNAs (37). The simulations additionally predict a guanine-specific protein–RNA interaction between G18 and the K139′ side chain (Fig. 7). To establish both of these interactions, the guanosine does not need to be the 3′-terminal nucleotide, but merely the last nucleotide bound by the Hfq after which the RNA chain exits the Hfq’s distal side. This would be the case in, for example, a full-length amiE mRNA, or during binding of multiple shorter RNA segments rich in ARN repeats, such as the CrcZ (Fig. 8) (14). In either case, recognition of these 3′-terminal guanine-specific interactions by Crc would be enough to overcome binding register degeneracy in the quaternary complex, as the binding sites of the next Crc would then be predefined by the first. It is however unclear whether preventing the degenerate binding of Crc could have effects on Hfq/RNA/Crc recognition in vivo or if it is just a system-specific coincidence which may have helped the resolution of the cryo-EM structures (19). We do not expect that the free-energy gain associated with the 3′-terminal guanosine recognition is large enough to completely abolish other Crc-binding patterns. However, even small increases in affinity could play a role in balancing the overall complex kinetic and thermodynamic networks of interactions in which Hfq is involved. In addition, it is possible that binding of some other proteins could be weakened by the same marker.
Figure 8.
A model for scanning and locking register in the Hfq–Crc–RNA assembly.A, cartoon of the ARN repeat–rich RNA binding to Hfq. The Hfq monomeric units are shown as pink ovals. The individual ARN repeats are differently colored. B, when the N nucleotide from the last ARN repeat bound by Hfq is guanosine, it can potentially form a base–phosphate interaction (red dotted double line) with the first repeat (top). This could happen when binding a single RNA sequence with six ARN repeats but also with multiple RNA sequences bound at the same time (bottom). C, the base–phosphate interaction formed by the guanosine could serve as mark for initial Crc binding (top). Without this mark, a degenerate multiple-register binding would be the likely result (bottom). ARN, adenine–purine–any nucleotide; Crc, catabolite repression control.
Concluding remarks
In this study, we explore the dynamics of RNA recognition by a conserved and pleotropic RNA chaperone, Hfq. We used state-of-the art atomistic MD simulations to obtain high-resolution insight into local dynamics and substates associated with RNA binding to the Hfq distal site. We show that Hfq partly utilizes dynamic recognition of RNA substrates, a type of molecular recognition that is difficult to fully resolve in structural experiments. However, we suggest that dynamic recognition is likely an important contribution to structural mechanism by which Hfq engages RNA targets with adequate specificity while maintaining high RNA turnover rates in the cell. Upon presentation of a target RNA to effector molecules, such as Crc, this turnover is slowed down significantly, allowing for a downstream cellular response.
Experimental procedures
Selection of initial structures
We have used cryo-EM structures of Hfq–Crc–RNA quaternary complexes with molecular composition ratios of 2:2:2, 2:3:2, and 2:4:2 (Protein Data Bank [PDB] IDs: 6o1k, 6o1l, and 6o1m) (19) as starting structures for MD simulations. Models of isolated Hfq–RNA, partially assembled quaternary complexes, and the Crc dimer and tetramer were prepared by removal of subunits from the experimental 2:2:2 structure. The simulations of the Crc dimer were also started based on X-ray structure of the isolated Crc protein (PDB ID: 4jg3) (16) with its dimer structure obtained via crystallographic symmetry. We have also used the X-ray structure of Escherichia coli Hfq bound to RNA poly-A octadecamer for simulations of the isolated Hfq–RNA system (8). Starting structures for simulations with modified, extended, or circularized RNA sequences or modified nucleobase conformations were prepared by molecular modeling of the experimental structures. A complete list of simulated systems is presented in Table 1 and visualized in Fig. S1.
System building and simulation protocol
The starting files for MD simulations were prepared in tLeap module of AMBER 18 (54). We have used bsc0χOL3 (i.e., OL3) (55) and ff12SB (56) force fields for description of RNA and protein, respectively; the preference for ff12SB rather than ff14SB in protein–RNA simulations is explained elsewhere (57).
A mild 1 kcal/mol stabilizing HBfix potential (58) was applied to the native H-bond interactions K31(N)–AA(OP2) and G29(O)–AR(O2′). Both of these H-bonds are present in all available experimental structures of P. aeruginosa Hfq bound to RNA-containing ARN repeats (8, 19, 40). The HBfix was applied to reduce likelihood of potential random spurious departures of the trajectories from the experimental geometry in longer simulations due to some force-field imperfectness. It allows us to more efficiently examine, within the affordable simulation time, the dynamics corresponding to the Hfq–RNA bound state as indicated by the experiments. We emphasize that unlike the restrained or targeted explicit solvent MD (or even Gō potentials in coarse-grained modeling (59)), HBfix introduces force potential between two atoms only within the narrow region corresponding to the H-bonding distance. Therefore, although somewhat bolstering these H-bonds by increasing their lifetime, it still allows the system to explore other geometries and should not bias the results derived in our study. Nevertheless, to verify this, we performed control simulations without any HBfix and simulations where we used increased 2 kcal/mol potential. These simulations confirmed that the results presented below do not depend on the use of the HBfix. Further details of the HBfix and of the control simulations are extensively described in Supporting information, together with explanation why the HBfix does not bias the results of the study.
In all simulations, the biomolecular systems were surrounded in a truncated octahedral box of SPC/E water molecules (60) with a minimal distance of 13 Å from the box border. The systems were neutralized and a salt concentration of 0.15 M was established by addition of K+ and Cl− atoms (61).
The systems built in tLeap were minimized and equilibrated according to the protocol extensively described in Ref. (62) utilizing the pmemd.MPI module (54). Afterward, the production simulations were carried out with the pmemd.cuda module (63). The typical simulation timescale was 1 μs with selected simulations further extended afterward. We have used the SHAKE protocol along with HMR (hydrogen mass repartitioning) to allow a 4-fs integration step (64, 65). Particle mesh Ewald (66) and periodic boundary conditions were used to handle long-range electrostatics and to prevent the box-border bias. The cut-off distance for Lennard–Jones interactions was set to 9 Å. The Langevin thermostat and Monte Carlo barostat (54) were used to keep the systems at temperature and pressure of 300 K and 1 bar, respectively.
Analyses
The cpptraj (67) was used to perform analyses of all simulation trajectories, and visual molecular dynamics (68) was used for their visual inspection. Raster3D (69) and POV-Ray were used for preparation of figures. LibreOffice and Inkscape were used to prepare graphs and schemes, respectively. The presence of H-bonds was evaluated based on the donor–acceptor distance and donor–hydrogen–acceptor angle, with 3.5 Å and 120° cutoffs, respectively. For selected simulations, principal component analysis (67) was used to evaluate RNA backbone dynamics and global interdomain movements between the Hfq and Crc. Every fifth frame of the trajectories was used to calculate the coordinate covariance matrix which was then diagonalized and used to obtain the first ten eigenvectors (principal components). The principal components of motion were visualized by projecting them along the utilized simulation frames.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its Supporting information. The raw MD simulation trajectories can be obtained from the corresponding author (Miroslav Krepl) upon reasonable request.
Supporting information
This article contains supporting information (6, 21, 35, 36, 57, 58, 70, 71, 72, 73, 74, 75, 76, 77) (Table S8).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge the use of CESNET storage facilities [Grant number CESNET LM2018140] for this project.
Author contributions
M. K., T. D., B. F. L., and J. S. conceptualization; M. K. data curation; M. K. formal analysis; M. K., T. D., B. F. L., and J. S. investigation; M. K. and T. D. methodology; M. K., B. F. L., and J. S. project administration; M. K. validation; M. K. visualization; M. K. writing - original draft; T. D., B. F. L., and J. S. writing - review & editing; J. S. funding acquisition; J. S. resources.
Funding and additional information
This work was supported by the Czech Science Foundation [Grant number 20-16554S to M. K. and J. S.]; Praemium Academiae to J. S.; AstraZeneca studentship to T. D.; and Wellcome Trust Investigator award [Grant number 200873/Z/16/Z to B. F. L.].
Edited by Karin Musier-Forsyth
Supporting information
References
- 1.Vogel J., Luisi B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoekzema M., Romilly C., Holmqvist E., Wagner E.G.H. Hfq-dependent mRNA unfolding promotes sRNA-based inhibition of translation. EMBO J. 2019;38 doi: 10.15252/embj.2018101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azam M.S., Vanderpool C.K. Translational regulation by bacterial small RNAs via an unusual Hfq-dependent mechanism. Nucleic Acids Res. 2017;46:2585–2599. doi: 10.1093/nar/gkx1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwiatkowska J., Wroblewska Z., Johnson K.A., Olejniczak M. The binding of class II sRNA MgrR to two different sites on matchmaker protein Hfq enables efficient competition for Hfq and annealing to regulated mRNAs. RNA. 2018;24:1761–1784. doi: 10.1261/rna.067777.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vecerek B., Moll I., BLASI U. Translational autocontrol of the Escherichia coli Hfq RNA chaperone gene. RNA. 2005;11:976–984. doi: 10.1261/rna.2360205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonnleitner E., Wulf A., Campagne S., Pei X.-Y., Wolfinger M.T., Forlani G., Prindl K., Abdou L., Resch A., Allain F.H.-T., Luisi B.F., Urlaub H., Bläsi U. Interplay between the catabolite repression control protein Crc, Hfq and RNA in Hfq-dependent translational regulation in Pseudomonas aeruginosa. Nucleic Acids Res. 2018;46:1470–1485. doi: 10.1093/nar/gkx1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacher M.A., Pearson R.F., Møller T., Valentin-Hansen P., Brennan R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq–RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Link T.M., Valentin-Hansen P., Brennan R.G. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikulecky P.J., Kaw M.K., Brescia C.C., Takach J.C., Sledjeski D.D., Feig A.L. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan R.G., Link T.M. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Santiago-Frangos A., Kavita K., Schu D.J., Gottesman S., Woodson S.A. C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E6089–E6096. doi: 10.1073/pnas.1613053113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen B., Wang W., Zhang J., Gong Q., Shi Y., Wu J., Zhang Z. Structural and dynamic properties of the C-terminal region of the Escherichia coli RNA chaperone Hfq: Integrative experimental and computational studies. Phys. Chem. Chem. Phys. 2017;19:21152–21164. doi: 10.1039/c7cp01044c. [DOI] [PubMed] [Google Scholar]
- 13.Robinson K.E., Orans J., Kovach A.R., Link T.M., Brennan R.G. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 2014;42:2736–2749. doi: 10.1093/nar/gkt1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonnleitner E., Bläsi U. Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van den Bossche A., Ceyssens P.-J., De Smet J., Hendrix H., Bellon H., Leimer N., Wagemans J., Delattre A.-S., Cenens W., Aertsen A., Landuyt B., Minakhin L., Severinov K., Noben J.-P., Lavigne R. Systematic identification of hypothetical bacteriophage proteins targeting key protein complexes of Pseudomonas aeruginosa. J. Proteome Res. 2014;13:4446–4456. doi: 10.1021/pr500796n. [DOI] [PubMed] [Google Scholar]
- 16.Milojevic T., Grishkovskaya I., Sonnleitner E., Djinovic-Carugo K., Bläsi U. The Pseudomonas aeruginosa catabolite repression control protein Crc is devoid of RNA binding activity. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolff J.A., MacGregor C.H., Eisenberg R.C., Phibbs P.V. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J. Bacteriol. 1991;173:4700–4706. doi: 10.1128/jb.173.15.4700-4706.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnleitner E., Abdou L., Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21866–21871. doi: 10.1073/pnas.pnas.0910308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pei X.Y., Dendooven T., Sonnleitner E., Chen S., Bläsi U., Luisi B.F. Architectural principles for Hfq/Crc-mediated regulation of gene expression. Elife. 2019;8 doi: 10.7554/eLife.43158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikulin A., Stolboushkina E., Perederina A., Vassilieva I., Blaesi U., Moll I., Kachalova G., Yokoyama S., Vassylyev D., Garber M., Nikonov S. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr. D Biol. Crystallogr. 2005;61:141–146. doi: 10.1107/S0907444904030008. [DOI] [PubMed] [Google Scholar]
- 21.Šponer J., Bussi G., Krepl M., Banáš P., Bottaro S., Cunha R.A., Gil-Ley A., Pinamonti G., Poblete S., Jurečka P., Walter N.G., Otyepka M. RNA structural dynamics as captured by molecular simulations: A comprehensive overview. Chem. Rev. 2018;118:4177–4338. doi: 10.1021/acs.chemrev.7b00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nerenberg P.S., Head-Gordon T. New developments in force fields for biomolecular simulations. Curr. Opin. Struct. Biol. 2018;49:129–138. doi: 10.1016/j.sbi.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Campagne S., Krepl M., Sponer J., Allain F.H.T. Chapter fourteen - combining NMR spectroscopy and molecular dynamic simulations to solve and analyze the structure of protein–RNA complexes. In: Wand A.J., editor. Methods Enzymol. Academic Press; Cambridge, MA: 2019. pp. 393–422. [DOI] [PubMed] [Google Scholar]
- 24.Borišek J., Saltalamacchia A., Gallì A., Palermo G., Molteni E., Malcovati L., Magistrato A. Disclosing the impact of carcinogenic SF3b mutations on pre-mRNA recognition via all-atom simulations. Biomolecules. 2019;9 doi: 10.3390/biom9100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma M., Sharma S., Alawada A. Understanding the binding specificities of mRNA targets by the mammalian quaking protein. Nucleic Acids Res. 2019;47:10564–10579. doi: 10.1093/nar/gkz877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casalino L., Palermo G., Spinello A., Rothlisberger U., Magistrato A. All-atom simulations disentangle the functional dynamics underlying gene maturation in the intron lariat spliceosome. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6584–6589. doi: 10.1073/pnas.1802963115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palermo G., Casalino L., Magistrato A., Andrew McCammon J. Understanding the mechanistic basis of non-coding RNA through molecular dynamics simulations. J. Struct. Biol. 2019;206:267–279. doi: 10.1016/j.jsb.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma M., Anirudh C.R. Mechanism of mRNA-STAR domain interaction: Molecular dynamics simulations of mammalian quaking STAR protein. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-12930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Górecka K.M., Krepl M., Szlachcic A., Poznański J., Šponer J., Nowotny M. RuvC uses dynamic probing of the holliday junction to achieve sequence specificity and efficient resolution. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-11900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ripin N., Boudet J., Duszczyk M.M., Hinniger A., Faller M., Krepl M., Gadi A., Schneider R.J., Šponer J., Meisner-Kober N.C., Allain F.H.-T. Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2935–2944. doi: 10.1073/pnas.1808696116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borkar A.N., Bardaro M.F., Camilloni C., Aprile F.A., Varani G., Vendruscolo M. Structure of a low-population binding intermediate in protein-RNA recognition. Proc. Natl. Acad. Sci. U. S. A. 2016;113:7171–7176. doi: 10.1073/pnas.1521349113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fender A., Elf J., Hampel K., Zimmermann B., Wagner E.G.H. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner E.G.H. Cycling of RNAs on Hfq. RNA Biol. 2013;10:619–626. doi: 10.4161/rna.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krepl M., Havrila M., Stadlbauer P., Banas P., Otyepka M., Pasulka J., Stefl R., Sponer J. Can we execute stable microsecond-scale atomistic simulations of protein-RNA complexes? J. Chem. Theor. Comput. 2015;11:1220–1243. doi: 10.1021/ct5008108. [DOI] [PubMed] [Google Scholar]
- 35.Bergonzo C., Henriksen N.M., Roe D.R., Cheatham T.E. Highly sampled tetranucleotide and tetraloop motifs enable evaluation of common RNA force fields. RNA. 2015;21:1578–1590. doi: 10.1261/rna.051102.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson J.S., Schneider B., Murray L.W., Kapral G.J., Immormino R.M., Headd J.J., Richardson D.C., Ham D., Hershkovits E., Williams L.D., Keating K.S., Pyle A.M., Micallef D., Westbrook J., Berman H.M. RNA backbone: Consensus all-angle conformers and modular string nomenclature (an RNA Ontology Consortium contribution) RNA. 2008;14:465–481. doi: 10.1261/rna.657708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zirbel C.L., Sponer J.E., Sponer J., Stombaugh J., Leontis N.B. Classification and energetics of the base-phosphate interactions in RNA. Nucleic Acids Res. 2009;37:4898–4918. doi: 10.1093/nar/gkp468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daubner G.M., Cléry A., Jayne S., Stevenin J., Allain F.H.-T. A syn–anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31:162–174. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kligun E., Mandel-Gutfreund Y. The role of RNA conformation in RNA-protein recognition. RNA Biol. 2015;12:720–727. doi: 10.1080/15476286.2015.1040977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulz E.C., Seiler M., Zuliani C., Voigt F., Rybin V., Pogenberg V., Mücke N., Wilmanns M., Gibson T.J., Barabas O. Intermolecular base stacking mediates RNA-RNA interaction in a crystal structure of the RNA chaperone Hfq. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-10085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horstmann N., Orans J., Valentin-Hansen P., Shelburne S.A., III, Brennan R.G. Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res. 2012;40:11023–11035. doi: 10.1093/nar/gks809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wlodawer A., Minor W., Dauter Z., Jaskolski M. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 2008;275:1–21. doi: 10.1111/j.1742-4658.2007.06178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chou F.-C., Sripakdeevong P., Dibrov S.M., Hermann T., Das R. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat. Methods. 2012;10:74. doi: 10.1038/nmeth.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krepl M., Blatter M., Cléry A., Damberger F.F., Allain F.H.T., Sponer J. Structural study of the fox-1 RRM protein hydration reveals a role for key water molecules in RRM-RNA recognition. Nucleic Acids Res. 2017;45:8046–8063. doi: 10.1093/nar/gkx418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atakisi H., Moreau D.W., Thorne R.E. Effects of protein-crystal hydration and temperature on side-chain conformational heterogeneity in monoclinic lysozyme crystals. Acta Crystallogr. D Struct. Biol. 2018;74:264–278. doi: 10.1107/S2059798318000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganser L.R., Kelly M.L., Herschlag D., Al-Hashimi H.M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019;20:474–489. doi: 10.1038/s41580-019-0136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santiago-Frangos A., Woodson S.A. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA. 2018;9 doi: 10.1002/wrna.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krepl M., Cléry A., Blatter M., Allain F.H.T., Sponer J. Synergy between NMR measurements and MD simulations of protein/RNA complexes: Application to the RRMs, the most common RNA recognition motifs. Nucleic Acids Res. 2016;44:6452–6470. doi: 10.1093/nar/gkw438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Šponer J., Leszczyński J., Hobza P. Nature of nucleic acid−base stacking: Nonempirical ab Initio and empirical potential characterization of 10 stacked base dimers. Comparison of stacked and H-bonded base pairs. J. Phys. Chem. 1996;100:5590–5596. [Google Scholar]
- 50.Šponer J., Jurečka P., Marchan I., Luque F.J., Orozco M., Hobza P. Nature of base stacking: Reference quantum-chemical stacking energies in ten unique B-DNA base-pair steps. Chemistry. 2006;12:2854–2865. doi: 10.1002/chem.200501239. [DOI] [PubMed] [Google Scholar]
- 51.Šponer J., Riley K.E., Hobza P. Nature and magnitude of aromatic stacking of nucleic acid bases. Phys. Chem. Chem. Phys. 2008;10:2595–2610. doi: 10.1039/b719370j. [DOI] [PubMed] [Google Scholar]
- 52.Sponer J., Sponer J.E., Mladek A., Jurecka P., Banas P., Otyepka M. Nature and magnitude of aromatic base stacking in DNA and RNA: Quantum chemistry, molecular mechanics, and experiment. Biopolymers. 2013;99:978–988. doi: 10.1002/bip.22322. [DOI] [PubMed] [Google Scholar]
- 53.Wang L., Wang W., Li F., Zhang J., Wu J., Gong Q., Shi Y. Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq. Nucleic Acids Res. 2015;43:2400–2411. doi: 10.1093/nar/gkv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Case IYB-SDA, Brozell S.R., Cerutti D.S., Cheatham T.E., III, Cruzeiro V.W.D., Darden T.A., Duke R.E., Ghoreishi D., Gilson M.K., Gohlke H., Goetz A.W., Greene D., Harris R., Homeyer N., Izadi S. University of California; San Francisco, San Francisco, CA: 2018. AMBER 18. [Google Scholar]
- 55.Zgarbova M., Otyepka M., Sponer J., Mladek A., Banas P., Cheatham T.E., Jurecka P. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theor. Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K., Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Šponer J., Krepl M., Banáš P., Kührová P., Zgarbová M., Jurečka P., Havrila M., Otyepka M. How to understand atomistic molecular dynamics simulations of RNA and protein–RNA complexes? Wiley Interdiscip. Rev. RNA. 2017;8 doi: 10.1002/wrna.1405. [DOI] [PubMed] [Google Scholar]
- 58.Kuhrova P., Best R., Bottaro S., Bussi G., Sponer J., Otyepka M., Banas P. Computer folding of RNA tetraloops: Identification of key force field deficiencies. J. Chem. Theor. Comput. 2016;12:4534–4548. doi: 10.1021/acs.jctc.6b00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Noel J.K., Levi M., Raghunathan M., Lammert H., Hayes R.L., Onuchic J.N., Whitford P.C. SMOG 2: A versatile software package for generating structure-based models. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berendsen H.J.C., Grigera J.R., Straatsma T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- 61.Joung I.S., Cheatham T.E. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krepl M., Vögele J., Kruse H., Duchardt-Ferner E., Wöhnert J., Sponer J. An intricate balance of hydrogen bonding, ion atmosphere and dynamics facilitates a seamless uracil to cytosine substitution in the U-turn of the neomycin-sensing riboswitch. Nucleic Acids Res. 2018;46:6528–6543. doi: 10.1093/nar/gky490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le Grand S., Götz A.W., Walker R.C. SPFP: Speed without compromise—a mixed precision model for GPU accelerated molecular dynamics simulations. Comput. Phys. Commun. 2013;184:374–380. [Google Scholar]
- 64.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical-integration of cartesian equations of motion of a system with constraints - molecular-dynamics of N-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 65.Hopkins C.W., Le Grand S., Walker R.C., Roitberg A.E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theor. Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 66.Darden T., York D., Pedersen L. Particle mesh Ewald - an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98 [Google Scholar]
- 67.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 68.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 69.Merritt E.A., Bacon D.J. Raster3D: Photorealistic molecular graphics. In: Carter C.W., Sweet R.M., editors. Macromolecular Crystallography, Pt B. Elsevier Academic Press Inc; San Diego, CA: 1997. pp. 505–524. [Google Scholar]
- 70.Zhao J., Kennedy S.D., Berger K.D., Turner D.H. Nuclear magnetic resonance of single-stranded RNAs and DNAs of CAAU and UCAAUC as benchmarks for molecular dynamics simulations. J. Chem. Theor. Comput. 2020;16:1968–1984. doi: 10.1021/acs.jctc.9b00912. [DOI] [PubMed] [Google Scholar]
- 71.Mlýnský V., Kührová P., Kühr T., Otyepka M., Bussi G., Banáš P., Šponer J. Fine-tuning of the AMBER RNA force field with a new term adjusting interactions of terminal nucleotides. J. Chem. Theor. Comput. 2020;16:3936–3946. doi: 10.1021/acs.jctc.0c00228. [DOI] [PubMed] [Google Scholar]
- 72.Kuhrova P., Mlynsky V., Zgarbova M., Krepl M., Bussi G., Best R.B., Otyepka M., Sponer J., Banas P. Improving the performance of the RNA amber force field by tuning the hydrogen-bonding interactions. J. Chem. Theor. Comput. 2019;15:3288–3305. doi: 10.1021/acs.jctc.8b00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bottaro S., Bussi G., Kennedy S.D., Turner D.H., Lindorff-Larsen K. Conformational ensembles of RNA oligonucleotides from integrating NMR and molecular simulations. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aar8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Szabla R., Havrila M., Kruse H., Šponer J. Comparative assessment of different RNA tetranucleotides from the DFT-D3 and force field perspective. J. Phys. Chem. B. 2016;120:10635–10648. doi: 10.1021/acs.jpcb.6b07551. [DOI] [PubMed] [Google Scholar]
- 75.Bergonzo C., Henriksen N.M., Roe D.R., Swails J.M., Roitberg A.E., Cheatham T.E. Multidimensional replica exchange molecular dynamics yields a converged ensemble of an RNA tetranucleotide. J. Chem. Theor. Comput. 2014;10:492–499. doi: 10.1021/ct400862k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Condon D.E., Kennedy S.D., Mort B.C., Kierzek R., Yildirim I., Turner D.H. Stacking in RNA: NMR of four tetramers benchmark molecular dynamics. J. Chem. Theor. Comput. 2015;11:2729–2742. doi: 10.1021/ct501025q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banáš P., Mládek A., Otyepka M., Zgarbová M., Jurečka P., Svozil D., Lankaš F., Šponer J. Can we accurately describe the structure of adenine tracts in B-DNA? Reference quantum-chemical computations reveal overstabilization of stacking by molecular mechanics. J. Chem. Theor. Comput. 2012;8:2448–2460. doi: 10.1021/ct3001238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and its Supporting information. The raw MD simulation trajectories can be obtained from the corresponding author (Miroslav Krepl) upon reasonable request.