Abstract
Objective
MicroRNAs (miRNA) are known to regulate the expression of genes involved in several physiological processes including metabolism, mitochondrial biogenesis, proliferation, differentiation, and cell death.
Methods
Using “in silico” analyses, we identified 219 unique miRNAs that potentially bind to the 3′UTR region of a critical mitochondrial regulator, the peroxisome proliferator-activated receptor gamma coactivator (PGC) 1 alpha (Pgc1α). Of the 219 candidate miRNAs, miR-696 had one of the highest interactions at the 3′UTR of Pgc1α, suggesting that miR-696 may be involved in the regulation of Pgc1α.
Results
Consistent with this hypothesis, we found that miR-696 was highly expressed in the skeletal muscle of STZ-induced diabetic mice and chronic high-fat-fed mice. C2C12 muscle cells exposed to palmitic acid also exhibited a higher expression of miR-696. This increased expression corresponded with a reduced expression of oxidative metabolism genes and reduced mitochondrial respiration. Importantly, reducing miR-696 reversed decreases in mitochondrial activity in response to palmitic acid. Using C2C12 cells treated with the AMP-activated protein kinase (AMPK) activator AICAR and skeletal muscle from AMPKα2 dominant-negative (DN) mice, we found that the signaling mechanism regulating miR-696 did not involve AMPK. In contrast, overexpression of SNF1-AMPK-related kinase (SNARK) in C2C12 cells increased miR-696 transcription while knockdown of SNARK significantly decreased miR-696. Moreover, muscle-specific transgenic mice overexpressing SNARK exhibited a lower expression of Pgc1α, elevated levels of miR-696, and reduced amounts of spontaneous activity.
Conclusions
Our findings demonstrate that metabolic stress increases miR-696 expression in skeletal muscle cells, which in turn inhibits Pgc1α, reducing mitochondrial function. SNARK plays a role in this process as a metabolic stress signaling molecule inducing the expression of miR-696.
Keywords: miR-696, Mitochondrial function, Skeletal muscle, SNARK, Pgc1α
Highlights
-
•
MicroRNA miR-696 is expressed under metabolic stress.
-
•
miR-696 downregulates Pgc1α in skeletal muscle.
-
•
Downregulation of miR-696 increases mitochondrial function.
-
•
AMPK-related kinase SNARK, but not AMPK, regulates miR-696 expression in skeletal muscle.
-
•
SNARK plays a novel role in mediating mitochondrial function.
1. Introduction
Metabolic syndrome is a systemic disorder associated with several pathological conditions including type 2 diabetes, obesity, hypertension, glucose intolerance, insulin resistance, and dyslipidemia. There is evidence that mitochondrial dysfunction and alterations in the expression of metabolic genes correlate with these diseases [1,2]. Increased inflammatory processes and defects in fatty acid or glucose metabolism have also been linked to diet-induced obesity and the development of insulin resistance in several tissues including skeletal muscle [3], liver [4], and adipose tissue [5].
It is widely accepted that the peroxisome proliferator-activated receptor gamma coactivator-1α (Pgc1α) regulates mitochondrial biogenesis in skeletal muscle through coactivation of the nuclear receptors PPARβ, ERRα, and thyroid hormone receptor and nuclear respiratory factors NRF1 and NRF2 [[6], [7], [8]]. For example, NRF1 and 2 induce transcription factor A, mitochondrial (TFAM), leading to the transcription of several mitochondrial DNA genes [9]. Therefore, Pgc1α is a critical regulator of mitochondrial biogenesis, oxidative metabolism, fiber-type switching, fatty acid oxidation, and angiogenesis [[10], [11], [12]]. Pgc1α transcription is regulated by an alternative splicing mechanism, resulting in four unique isoforms: Pgc1α-1, α-2, α-3, and α-4 [13]. Pgc1α is also regulated by post-translational modifications involving 5′ AMP-activated protein kinase (AMPK) [14] and sirtuin 1 (SIRT1) [15].
Another level of Pgc1α regulation may be through microRNAs, small non-coding RNA molecules that disrupt post-transcriptional processes by impairing protein translation [16]. As examples, microRNA miR-23a was shown to downregulate Pgc1α and was ascribed as a major cause of mitochondrial dysfunction in estrogen-deficient mice [17], whereas miR-761 was reported to be involved in the biogenesis of mitochondria through the downregulation of Pgc1α [18]. miR-696 was first reported to be expressed in prenatal development (E9.5) of mouse embryos at an intergenic loci on chromosome 3 (mm10, 30852687-30852704) [19]. miR-696 was shown to be decreased by exercise training and increased by hindlimb immobilization in skeletal muscles of wild-type mice [20]. Moreover, C2C12 muscle cells transfected with a miR-696 mimic oligonucleotide markedly reduced mitochondrial DNA content and the ATP/ADP ratio, while miR-696 antisense oligonucleotide (ASO) treatment decreased intracellular triacylglycerol content [21]. The potential roles of miR-696 in regulating muscle metabolism under conditions of metabolic dysregulation such as insulin resistance and obesity have not been investigated.
AMPK activity increases in response to increase in AMP and ADP concentrations [22]. AMPK has been implicated in the regulation of a plethora of cellular effects including glucose uptake, fatty acid oxidation, and mitochondrial gene expression [23]. Decreased AMPK activity correlates with reduced Pgc1α expression and thus metabolic consequences including mitochondrial dysfunction [14,24]. In addition to AMPK, there is a family of AMPK-related kinases with different cellular functions that share the same upstream kinase in LKB1 [25] such as sucrose non-fermenting-related kinase (SNRK), a protein involved in the metabolic properties of cardiac and adipose tissues [[26], [27], [28]] or SNF1-AMPK-related kinase (SNARK), whose expression has been reported to be increased under a variety of altered metabolic conditions including C2C12 cells exposed to palmitic acid and tumor necrosis factor alpha (TNFα), skeletal muscle of chronic high-fat-fed mice, and skeletal muscle from obese and older subjects [29,30]. Given that all of these stimuli provoke cellular stress, it is interesting that SNARK has been shown to function as a nuclear stress-response transcription modulator [31]. Herein, we used cell and in vivo mouse models to test the hypothesis that increased miR-696 reduces mitochondrial function in skeletal muscle through the inhibition of Pgc1α. Furthermore, we determined the role of AMPK and the AMPK-related kinase SNARK in this signaling axis.
2. Methods
2.1. Streptozotocin-treated (STZ) mice
C57BL/6J male mice (10–16 weeks old) were maintained on a 12:12-h light–dark cycle with ad libitum access to water and chow. To induce insulin-deficient diabetes, the mice were injected with streptozotocin (STZ, 70 mg kg−1) through the jugular vein under isoflurane anesthesia. STZ was dissolved in 0.01 M of citrate buffer (pH 4.5). Control mice received only citrate buffer (vehicle) under the same conditions. The mice were sacrificed three days after STZ injection and blood glucose levels were determined by a glucometer. The tibialis anterior (TA) muscle was dissected from both groups, immediately frozen in liquid nitrogen, and stored at −80 °C. Biochemical blood parameters and body weights are available in the Supplementary material Table 1.
2.2. High-fat diet (HFD) mice
Six-week-old C57BL/6J mice were maintained on a 12:12-h light–dark cycle with ad libitum access to water and diet. HFD mice were fed a diet composed of 20% protein, 20% carbohydrate, and 60% fat (Research Diets; product data D12492) for 10 weeks to induce obesity. Control mice were fed a diet composed of 20% protein, 70% carbohydrate, and 10% fat (Research Diets; product data D12450B). The mice were sacrificed at 16 weeks of age and their TA and extensor digitorum longus (EDL) muscles were dissected for experimental analyses. Biochemical blood parameters and body weights are available in the Supplementary material Table 2.
2.3. Generation of AMPK and SNARK transgenic mice
Transgenic mice expressing AMPK with an inactivating D157A mutation in the α2 subunit (AMPKα2 DN) and transgenic mice expressing AMPK with an activating R70Q mutation in the γ1 subunit (AMPKγ1 R70Q) were generated on an FVB background as previously described [32,33]. Dominant-negative SNARK (SNARK DN) and overexpressing wild-type SNARK (SNARK WT) mice were generated on a C57BL/6 background as previously described [29]. Wild-type (WT) littermates were used as controls. The mice were sacrificed at 10–18 weeks of age and their TA muscles were dissected for experimental analyses.
2.4. Exercise capacity tes
Mice (5–7 males) were placed in treadmill lanes to perform maximal exercise capacity. Shocking grids with the frequency set at 75 per minute and intensity at 45% (3.4 mA) were placed at end of the treadmill to force the mice to run at their maximum. The protocol started with speed at 6 m/min followed by incremental adjustments of 2 m/min every 2 min until fatigue was reached. Fatigue was defined as the mouse being stationary on the shocking grid with no attempts to climb off for 20 s. The maximum speed, time, and laps were then recorded and used to calculate the total time, total distance, and work.
2.5. Indirect calorimetry
Mice were individually housed at 22 °C and subjected to indirect calorimetry (Comprehensive Laboratory Animal Monitoring System, CLAMS; Columbus Instruments) for a period of 3 consecutive days under a 12-h light–dark cycle. During this period, we measured their food and water intake, spontaneous activity, and volume of oxygen and carbon dioxide consumed. These data allowed us to estimate the total energy expenditure (TEE) and respiratory exchange ratio (RER). We recorded SDN and SWT mice in three phases: 1) Acclimation, first 24 h of measurement. 2) Fed, 24 h after the end of phase 1, the mice were fed ad libitum. 3) Fasted, 24 h after the end of phase 2, the mice were fasted during the period. Spontaneous activity was extrapolated from the real-time analysis of total mouse activity with a 0.25 cm calculated centroid (how much each mouse moved in the cage). The software analysis package provided a graphical representation of their position within the cage, calculated the total distance traveled, displayed rearing information, correlated activity and distance traveled, and stored the raw position vs time data for other traceable and quantifiable analyses of activity levels.
2.6. Plasmids and constructs
Full-length Pgc1α plasmid, GFP, and RFP reporter plasmids were purchased from Addgene (cat. #4, #35625, and #35626). Approximately 165 bp of mouse Pgc1α 3′UTR was synthesized from brown adipose tissue cDNA through PCR using AccuPrime Taq DNA polymerase and cloned into Pgc1α (Pgc1α-3′UTR), GFP (GFP-PGC1-3′UTR), and GFP (GFP-PGC1-3′UTRmt (mutation of two nucleotides in positions 4 and 5 at the seed region)) reporter plasmids with T4 ligase enzyme according to the manufacturer's instructions (New England Biolabs). Pgc1α 3′UTR was synthesized with a mutated site for miR-696 interaction using AccuPrime Taq DNA polymerase and cloned into GFP (GFP-PGC1-3′UTRmt) reporter plasmid. The 3′ and 5′ miR-696 oligonucleotide sequences (Eurofins) with BspQ1 overhangs were annealed for 5 min at 95 °C, followed by 10 min at 4 °C. Then the annealing product was cloned into an RFP reporter (RFP-miR696) plasmid using a BspQ1 enzyme (New England Biolabs). SNARK plasmids were generated as previously described [29]. Constructs were sequenced by the DNA Sequencing Core Facility at Brigham and Women's Hospital.
2.7. Cell culture
To isolate and culture primary rat myocytes, cells were seeded in dishes covered with Matrigel (0.1%) until they reached maturity (5–6 days) at a density of 2.5 × 105 cells/well as previously described [30]. For C2C12 cells, approximately 2 × 105 myoblast cells were seeded and cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin at 37 °C with 5% CO2 for 2 days until they reached 90% of confluence. Differentiation was induced by incubating the cells in DMEM containing horse serum (2%) and penicillin/streptomycin (1%) for 4 days, and the medium was changed every 2 days. On day 5 of differentiation, the cells were incubated with AICAR (250 μM), palmitic acid (500 or 700 μM containing fatty acid-free BSA 1%), or vehicle (ethanol containing fatty acid-free BSA 1%) for 24 h. For HEK293 cells, 2 × 105 human embryonic kidney 293 (HEK293) cells were seeded and maintained in DMEM containing FBS (10%) and penicillin/streptomycin (1%) at 37 °C with 5% CO2.
2.8. Plasmid, siRNA, miR, and anti-miR transfection
For rat myotube cell cultures, full-length Pgc1α plasmid was mixed with Lipofectamine 2000 in Opti-MEM medium according to the manufacturer's instructions (Life Technologies) and then added to the cell cultures on day 3 of differentiation (∼60% confluence) and maintained at 37 °C with 5% CO2. The cells were washed with DPBS, and then primary growth medium (PGM) containing FBS (10%), HS (10%), and penicillin/streptomycin (1%) was added 12 h after transfection. Experiments were performed on day 5 of differentiation. Control cells were transfected with pcDNA3 empty vector or GFP. Myoblasts from C2C12 cells were exposed to the reverse transfection protocol using full-length human SNARK plasmid conjugated to Viromer Red reagent in buffer E medium as recommended by the manufacturer (Lipocalyx). Twelve hours after transfection, the cells were washed with PBS and incubated with DMEM medium containing horse serum (2%). Experiments were performed 48 h after reverse transfection. Control cells were transfected with either pcDNA3 empty vector or GFP. For RNA transfection, C2C12 cells were transfected on day 3 of differentiation using Lipofectamine RNAiMAX reagent (Invitrogen-Life Technologies). siRNA against SNARK or scrambled sequences were obtained from Origene (SR30004, SR418439B). The miR-696 mimic, miR-696 ASO, and scrambled sequences were synthesized by miRIDIAN (Dharmacon-GE).
2.9. Reporter assay
HEK293 cells were transfected with GFP-3′UTR, GFP-3′UTRmt, RFP-miR696, or RFP-miR30 using Viromer Red reagent in buffer E medium (Lipocalyx). The medium was changed 24 h after transfection with 1 μg/mL of doxycycline in DMEM containing FBS (10%). After transfection for 72 h, the cells were fixed with paraformaldehyde (4%) and stained with 1 μg/mL of DAPI (Cat. No. D9564; Sigma-Aldrich). Images were obtained under 40× magnification on a fluorescence microscope (Zeiss) and quantified using ImageJ software.
2.10. Western blotting analysis
Protein samples isolated from C2C12 cells (30 μg) and rat primary myocyte-cultured cell lysates (100 μg) were separated by SDS-PAGE on an acrylamide gel (10%). Non-specific binding was blocked with fat-free milk (10%) for 60 min. The membranes were incubated overnight at 4 °C with primary antibodies: PGC-1alpha (Calbiochem, ST1202), p-Akt Ser473 (Cell Signaling, #9271), total Akt (Cell Signaling, #9272), total SNARK (anti-NUAK2 Abcam, ab107287), p-SNARK Thr208 (Cell Signaling), FLAG (Sigma-Aldrich, F7425), and β-actin (Santa Cruz, sc-47778). The membranes were incubated with HRP-conjugated secondary antibodies for 60 min at room temperature. Antibody binding was detected using Amersham ECL reagent (GE) as described by the manufacturer. Blots were scanned using a ChemiDoc imaging system (Bio Rad), and protein band densitometry was determined using ImageLab software (Bio-Rad).
2.11. Real-time quantitative PCR (qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen). One microgram of total RNA was used for complementary DNA synthesis performed using a high-capacity reverse-transcription kit (Applied Biosystems). For real-time PCR analysis, RNA was reverse transcribed using Power SYBR Green Master Mix (Applied Biosystems). The relative expression of mRNAs was determined after normalization using the ΔΔCt method. Quantitative PCR was performed using a StepOne Real-Time PCR System (Applied Biosystems). The sequence of all primers can be found in the Supplementary material. β-actin, GAPDH, or RPL39 were used as reference genes.
2.12. TaqMan MicroRNA qRT-PCR
Using a TaqMan miRNA reverse-transcription kit (Applied Biosystems), 7.5 μL of cDNA reaction products were produced from 5 ng of total RNA with specific stem-loop primers for mmu-miR-696 and mmu-SnoR-202. The cDNA was then diluted (15×) and amplified using the TaqMan miRNA assay together with TaqMan Universal PCR Master Mix (Applied Biosystems) on an ABI Prism 7500 system (Applied Biosystems). The mmu-miR-696 Ct range was between 29 and 34 and mmu-SnoR-202 between 25 and 27. The mmu-SnoR-202 was used as a reference. The miRNA expression was quantified by the ΔΔCt method.
2.13. Oxygen consumption in myotubes
C2C12 cells at a density of 1 × 106 were seeded in 100 mm culture plates. After treatment, the cells were trypsinized, centrifuged (1250 RPM), and incubated with 2 mL of air-saturated respiration medium in an Oxygraph-2k (O2k, OROBOROS Instruments). During measurements of oxygen consumption, oligomycin (1 μg/mL), an ATP synthase inhibitor, and carbonyl cyanide m-chlorophenylhydrazone (CCCP, 1 μM), a mitochondrial uncoupling activator, were used for oxidative phosphorylation inhibition and mitochondrial uncoupling, respectively.
2.14. Oxygen consumption in myoblasts
Cells at a density of 2 × 104/well were seeded in XF 24-well cell culture microplates in 350 μL of FBS (10%) medium and incubated at 37 °C with 5% CO2. After 48 h, experiments were initiated in 675 μL of DMEM medium without phenol containing 1.85 g/L of NaCl, 2 mM of GlutaMax-1, 1 mM of sodium pyruvate, 15 mM of d-glucose, and 20 mM of HEPES at a pH of 7.4. The cells were incubated at 37 °C without CO2 for 60 min before the first measurement. During oxygen consumption measurements, oligomycin (1 μg/mL), an ATP synthase inhibitor, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP, 1 μM), a mitochondrial uncoupling activator, and rotenone (0.1 μM), a mitochondrial complex I inhibitor, were added at various intervals. The Δ oxygen consumption rate (ΔOCR) was measured according to the following calculations: basal respiration = basal − rotenone; uncoupled respiration = oligomycin − rotenone; maximal respiration = FCCP − rotenone. Experiments were performed using an XF 24-well Seahorse (Seahorse Biosciences).
2.15. Determining ROS production
The cells were maintained at 37 °C in PBS containing glucose (5.6 mM), fluorescent Amplex UltraRed (50 μM), and horseradish peroxidase (HRP) (0.1 U/mL) for 60 min. The medium was collected and fluorescence intensity determined at 568 nm excitation and 581 nm emission. The cells were transfected with scramble oligo, miR-696 mimic or anti-miR-696 with palmitic acid (700 μM), or palmitic acid alone (700 μM). The antioxidants glutathione (GSH) and glutathione peroxidase were added as a negative control.
2.16. Tricarboxylic acid cycle intermediates (ICAT)
A total of 2 × 105 C2C12 cells were seeded in a 6-well plate. After treatment, the cells were collected with the addition of perchloric acid (0.5 M) containing EDTA (1 mM) and neutralized with KHCO3 (2.2 M), then centrifuged at 7000 rpm for 10 min. The metabolites citrate, malate, oxaloacetate, and α-ketoglutarate were detected as previously described [64].
2.17. ATP content
ATP was measured in C2C12 cells exposed to either AICAR (250 μM) or palmitic acid (500 μM and 700 μM) using an ATP assay kit (Sigma-Aldrich, Cat. Number FLAA) according to the manufacturer's protocol.
2.18. Determining glucose uptake
Cells were incubated at 37 °C in DMEM with 2 mM of glucose without serum for 2–3 h. The media was changed to DPBS (1 mL) containing glucose (5.6 mM), insulin (100 nM), and 2-deoxy-[1,2-3H]-glucose (0.1 μCi/mL). After 30 min, the cells were washed in DPBS and lysed in NaOH (0.1 N). Radioactivity was determined by liquid scintillation counting and 2-deoxyglucose uptake was calculated based on the DPM results divided per sample protein concentration.
2.19. In silico analysis
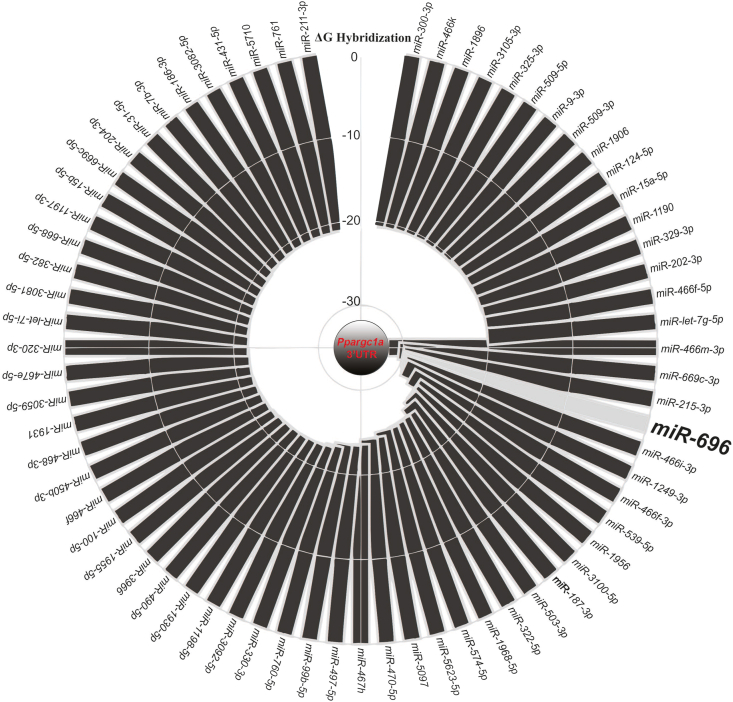
The raw data were collected through Sfold 2.2 online software (http://sfold.wadsworth.org/cgi-bin/index.pl). To acquire the raw data, we used the STarMirDB database prediction model training data (species: HITS-CLIP, mouse) to show the potential Ppargc1a binding microRNA candidates. Hybridization free energy (hΔG) was selected to rank the microRNAs listed. The data were plotted in Plotly online software (https://plotly.com/) to create the graph in Figure 1. A cut-off inferior to −20 kcal/mol was applied for illustrative purposes.
Figure 1.
Database interaction analysis of Pgc1α gene and microRNAs. “In silico” bioinformatics analysis using Sfold 2.2 (http://sfold.wadsworth.org/cgi-bin/index.pl) was conducted to determine all reported Mus musculus microRNAs that bind to Pgc1α 3′UTR. Hybridization free energy (ΔG) indicates interactions between all listed microRNAs and Pgc1α 3′UTR. A cut-off inferior to −20 kcal/mol was applied for illustrative purposes.
2.20. Study approval
SNARK experiments were performed in accordance with a protocol approved by the Joslin Diabetes Center Institutional Animal Care and Use Committee and were in accordance with the Guide for the Care and Use of Laboratory Animals (eighth edition, National Academies Press, 2011). Procedures for inducing diabetes by STZ and isolating rat cells were approved by the Institutional Ethical Committee for the use of laboratory animals from the University of Sao Paulo, Ribeirao Preto Campus (Approval Nos. 092/2010 and 130/2012).
2.21. Statistics
Results were expressed as a mean ± standard error of the mean (S.E.M) and analyzed by t tests or one-way ANOVA followed by Tukey's test for multiple comparisons. p ≤ 0.05 was taken as the criterion for significance.
3. Results
3.1. Database interaction analysis of Pgc1α gene and microRNAs
Reduced mitochondrial function is associated with the downregulation of a large number of metabolic genes. Of these, Pgc1α has been identified as the major regulator of mitochondrial biogenesis, and its low expression is highly correlated with obesity and insulin resistance [2,[34], [35], [36]]. Therefore, our aim was to determine whether there was a microRNA that strongly correlates with Pgc1α based on the hybridization free energy (hΔG) and could be intimately linked to mitochondrial function and metabolic disorders in skeletal muscle. An in silico analysis was performed through the Sfold2.2 online tool to determine which Mus musculus microRNAs bind to Pgc1α 3′UTR [37]. From 219 candidates (Supplementary material), miR-696 had one of the highest hΔG values (−29.8 kcal/mol) in relation to Pgc1α 3′UTR (Figure 1). The miR-696 sequence was paired to extra nucleotide bases in the Pgc1α 3′UTR that were not considered by the program as the “seed” region. These observations suggested that miR-696 has a higher hΔG than the one predicted by the software analysis and may interact with Pgc1α mRNA, possibly impairing its expression.
3.2. Pgc1α was targeted by miR-696
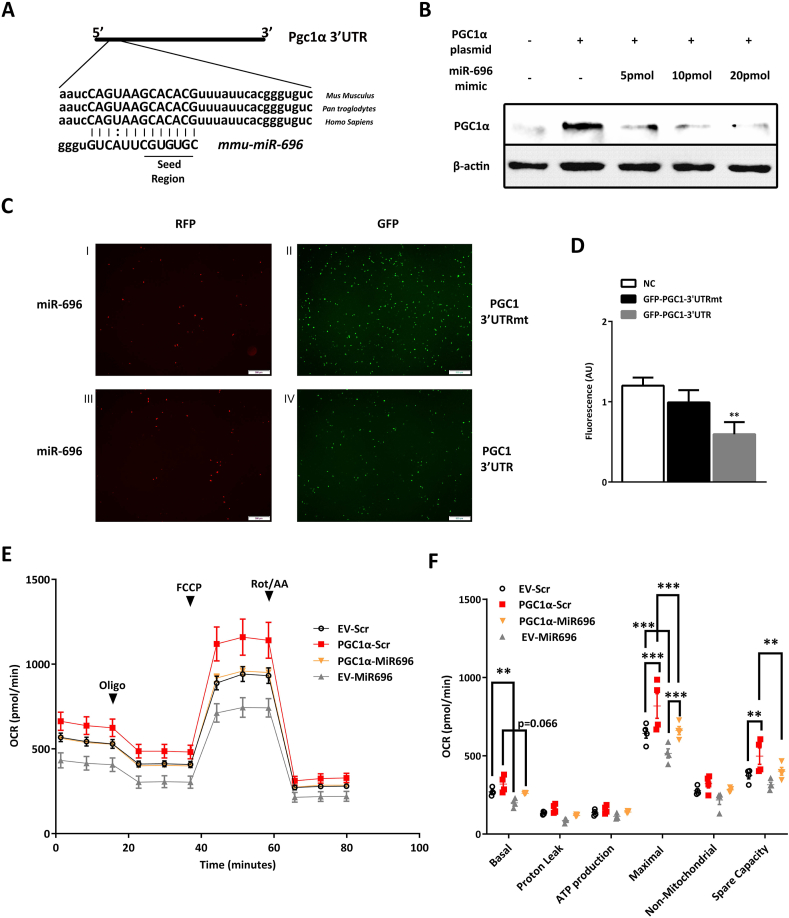
The gene sequence analysis demonstrated a highly conserved site in the Pgc1α mRNA 3′UTR region matching the nucleotide seed sequence of miR-696. The conservation of this sequence suggested that Pgc1α expression may be regulated by miR-696 (Figure 2A). To test this hypothesis, we determined the PGC1α protein expression after co-transfection of primary rat myocytes with full-length Pgc1α and an oligonucleotide with the same sequence of mature miR-696 (miR-696 mimic). The presence of the miR-696 mimic markedly reduced PGC1α protein expression, indicating that miR-696 is a strong repressor of Pgc1α (Figure 2B). To investigate the physical interactions between miR-696 and Pgc1α 3′UTR, we cloned 245–400 bp of Pgc1α-3′UTR mRNA after the GFP sequence and performed a reporter assay overexpressing RFP-miR696 and either GFP-PGC1-3′UTR or a mutant version of the promoter that cannot bind miR696 (GFP-PGC1-3′UTRmt) into HEK 293 cells. Mutation of the PGC1-3′UTR seed region in GFP-PGC1-3′UTRmt plasmid increased the GFP reporter expression, indicating that incomplete binding of miR696 to the 3′UTR region did not block GFP translation (Figure 2C and D). Moreover, given the well-known effect of PGC1α on mitochondrial biogenesis and function [9], we performed an experiment to characterize the impact of miR-696 on the oxygen consumption rate (OCR) of C2C12 myoblasts. Cells overexpressing Pgc1α and transfected with a scrambled sequence of miR-696 (PGC1α-Scr) increased both the maximal and spare capacity OCR compared to cells co-transfected with an empty vector and a scrambled sequence of miR-696 (Ev-Scr). PGC1α-Scr showed a tendency of higher basal OCR than cells transfected with both Pgc1α and miR-696 mimic (PGC1α-MiR696), while the maximal and spare capacity significantly increased. Increasing miR-696 with a miR-696 mimic (Ev-MiR696) reduced the basal and maximal OCR (Figure 2E and F).
Figure 2.
Pgc1α was targeted by miR-696. (A) The schematic interaction between miR-696 and Pgc1α 3′UTR. (B) Primary rat-cultured cells were transfected with full-length Pgc1α plasmid on day 3 of differentiation and then transfected with miR-696 mimic on day 4 of differentiation. After 24 h, protein was collected and Western blotting was performed to detect Pgc1α content (100 μg of protein was loaded). β-actin was used as internal control (n = 2). (C) C2C12 cells were co-transfected either with RFP-miR-696 and GFP-Pgc1α-3′UTR or GFP-Pgc1α-3′UTRmt. GFP fluorescence signals were measured to quantify the interactions between miR-696 and Pgc1α 3′UTR (n = 3 experiments). (D) Quantitative analysis of Figure 4C. Scramble oligonucleotide plus GFP (NC) was used as a negative control (white bar), RFP-miR-696 plus GFP-Pgc1α-3′UTRmt (black bar), and RFP-miR-696 plus GFP-Pgc1α-3′UTR (gray bar). (E–F) Cells were transfected with an empty vector and a scrambled oligonucleotide (EV-Scr), Pgc1α vector and a scrambled (Pgc1α-Scr), Pgc1α vector and a miR-696 mimetic (Pgc1α-MiR696), an empty vector, and a miR-696 mimetic (EV-MiR696) (n = 4). (E) The oxygen consumption rate at a basal state and in the presence of oligomycin, FCCP, and rotenone. (F) Bars indicate the variations in the oxygen consumption rate (ΔOCR, basal = basal − rotenone; uncoupled = oligomycin − rotenone; maximal = FCCP − rotenone) from kinetics depicted in Figure 2E. ∗∗p < 0.01 and ∗∗∗p < 0.001 vs black bar (one-way ANOVA and Tukey's post-test). Each bar indicates mean ± SEM.
3.3. miR-696 and Pgc1α had inverse expression profiles
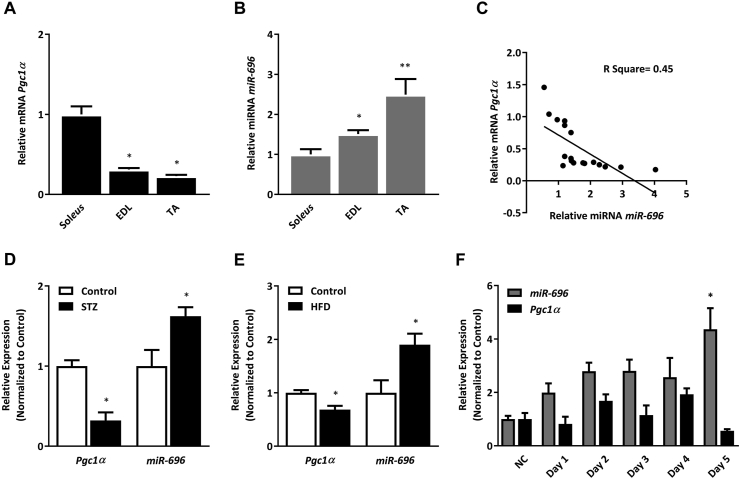
Numerous physiologic conditions are known to regulate Pgc1α expression. Basal levels of Pgc1α mRNA are higher in slow-twitch oxidative muscles than in more fast-twitch glycolytic muscles [38,39]. Muscle-specific overexpression of Pgc1α in mice results in a higher number of slow-twitch fibers in skeletal muscle [39], whereas ablation of Pgc1α induces a significant shift from oxidative toward glycolytic muscle fibers [40]. To determine whether skeletal muscle fiber-type composition is associated with miR-696 and Pgc1α expression, we examined gene expression in the soleus, a primarily oxidative muscle, and the extensor digitorum longus (EDL) and tibialis anterior (TA), primarily glycolytic muscles. As expected, Pgc1α expression was more abundant in the soleus than in the EDL and TA muscles (Figure 3A). Consistent with our hypothesis, miR-696 expression was significantly higher in the EDL and TA (Figure 3B), and there was a negative correlation between Pgc1α and miR-696 (Figure 3C).
Figure 3.
miR-696 and Pgc1α had inverse expression profiles. (A) Transcriptional levels of Pgc1α expression in the soleus, EDL, and TA muscles from the C57BL/6 mice (n = 6 per group), where comparisons were made between the soleus (oxidative) and EDL and TA (glycolytic) (β-actin was used as a reference gene). (B) Transcriptional levels of miR-696 expression in the soleus, EDL, and TA muscles from the C57BL/6 mice (n = 6 per group) (SnoR-202 was used as a reference gene). (C) Correlation between Pgc1α and miR-696 expression through linear regression. (D) Transcriptional levels of Pgc1α and miR-696 in TA muscle from the C57BL/6 mice treated with intravenous injections of streptozotocin (STZ, 70 mg kg−1 in overnight fasted mice) or DMSO/saline (control) (n = 5 per group) (β-actin and SnoR-202 were used as reference genes, respectively). (E) Transcriptional levels of Pgc1α and miR-696 measured in TA muscle from the C57BL/6 mice treated with a normal diet (control) or high-fat diet (HFD) for 10 weeks (n = 5 per group) (β-actin and SnoR-202 were used as reference genes, respectively). (F) Pgc1α and miR-696 expression measured in C2C12 cells without serum (NC) or with horse serum on days 1, 2, 3, 4, and 5 of differentiation (n = 4) (β-actin and SnoR-202 were used as reference genes, respectively). ∗∗p < 0.01 and ∗p < 0.05 (Student's t test, one-way ANOVA, or Tukey's post-test). Each bar indicates mean ± SEM.
Another potent regulator of Pgc1α expression is metabolic status. We found that Pgc1α expression significantly decreased in two animal models of diabetes: STZ-induced diabetic mice and high-fat diet (HFD)-fed mice (Figure 3D and 3E, respectively). These effects were concomitant with an increase in miR-696 expression (Figure 3D and E). Using C2C12 cells, we found that by day 5 of differentiation, miR-696 had significantly increased, accompanying a significant decrease in Pgc1α compared with their respective controls (Figure 3F). Taken together, these findings demonstrated that there is a negative correlation between miR-696 and Pgc1α expression in mouse skeletal muscle.
3.4. Reduction of miR-696 protected C2C12 cells from the metabolic effects of palmitic acid overload
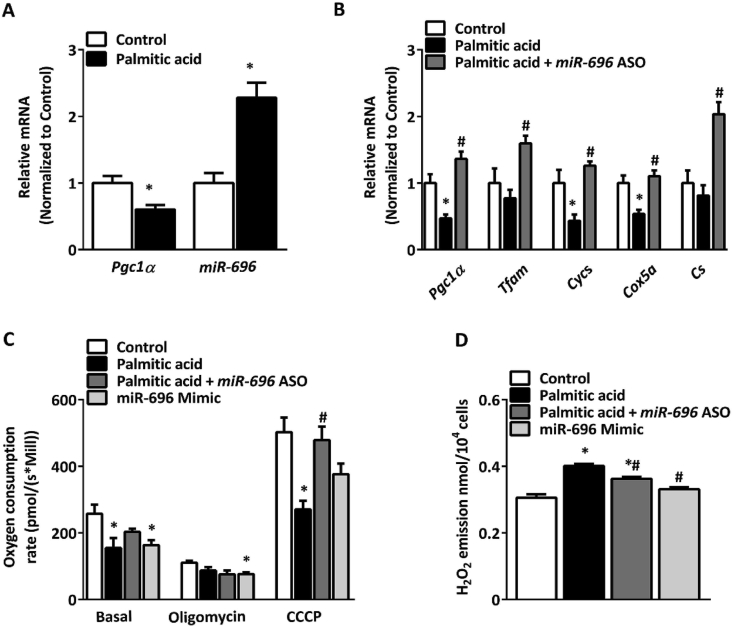
Palmitic acid overload is a model commonly used to downregulate mitochondrial function in skeletal muscle cells [30,41]. C2C12 cells exposed to palmitic acid for 24 h exhibited decreased expression of Pgc1α and elevated expression of miR-696 (Figure 4A). We also measured the expression of key mitochondrial genes and found that palmitic acid significantly decreased the expression of cytochrome c (Cycs) and the mitochondrial respiratory chain complex IV subunit Cox5a. Reduction of miR-696 using its antisense oligonucleotide (ASO) reversed the effects of palmitic acid, increasing the expression of these two genes, as well as Pgc1α, Tfam, and Cs (Figure 4B). To study mitochondrial function, we evaluated OCR in C2C12 cells and found that palmitic acid reduced both the basal and CCCP-induced OCR, with no effect on the OCR after ATP-synthase inhibition with oligomycin. Transfection of C2C12 cells with miR-696 ASO before palmitic acid treatment was sufficient to reverse the palmitic acid-induced reduction of OCR (Figure 4C). Increasing miR-696 with a miR-696 mimic mirrored the effects of palmitic acid treatment by reducing the basal and oligomycin-induced OCR, tending to decrease the CCCP-induced OCR (Figure 4C).
Figure 4.
Reduction of miR-696 protected C2C12 cells from the metabolic effects of palmitic acid overload. (A) Pgc1α and miR-696 expression were measured in C2C12 myotubes treated with either ethanol (control) or palmitic acid (700 μM) for 24 h; both groups were treated with vehicle (ethanol in 1% BSA) (n = 3) (β-actin and SnoR-202 were used as reference genes, respectively). (B) Expression of genes associated with mitochondrial function and biogenesis. Cells were either transfected with scrambled oligonucleotide (control), scrambled oligonucleotide and treated with 700 μM of palmitic acid (palmitic acid), or miR-696 antisense oligonucleotide and treated with 700 μM of palmitic acid (palmitic acid + miR-696 ASO). All the groups were treated with vehicle (ethanol in 1% BSA) (n = 5) (β-actin was used as a reference gene). (C and D) Cells were transfected with scrambled oligonucleotide (control), scrambled oligonucleotide and treated with 700 μM of palmitic acid (palmitic acid), miR-696 antisense oligonucleotide and treated with 700 μM of palmitic acid (palmitic acid + miR-696 ASO), and miR-696 full-length oligonucleotide sequence (miR-696 mimic). All the groups were treated with vehicle (ethanol in 1% BSA) (n = 5). (C) Mitochondrial respiration rate in C2C12 cells. Oligomycin was used as an ATP-synthase inhibitor and CCCP was used for mitochondrial uncoupling (n = 5). (D) Hydrogen peroxide emission as determined by an Amplex UltraRed fluorescent probe after 24 h of palmitic acid treatment (n = 6). ∗p < 0.05 vs control and #p < 0.05 vs palmitic acid (Student's t test). Each bar indicates mean ± SEM.
We next determined whether miR-696 alters reactive oxygen species (ROS) production in C2C12 cells, since this indicates mitochondrial dysfunction and metabolic stress [30,41]. The addition of palmitic acid increased the rate of H2O2 emission in the C2C12 cells, an effect that was reduced by decreasing miR-696 through transfection of miR-696 ASO (Figure 4D). There was no effect of miR-696 mimic on H2O2 emission in the C2C12 cells relative to the control group (Figure 4D).
Together, these data suggested that metabolic stress can increase miR-696, which in turn can downregulate mitochondrial respiration in skeletal muscle cells. Consistent with this finding, we demonstrated that preventing miR-696 upregulation under metabolically stressful situations can prevent reduction of mitochondrial function in muscle.
3.5. miR-696 reduced glucose uptake in C2C12 cells
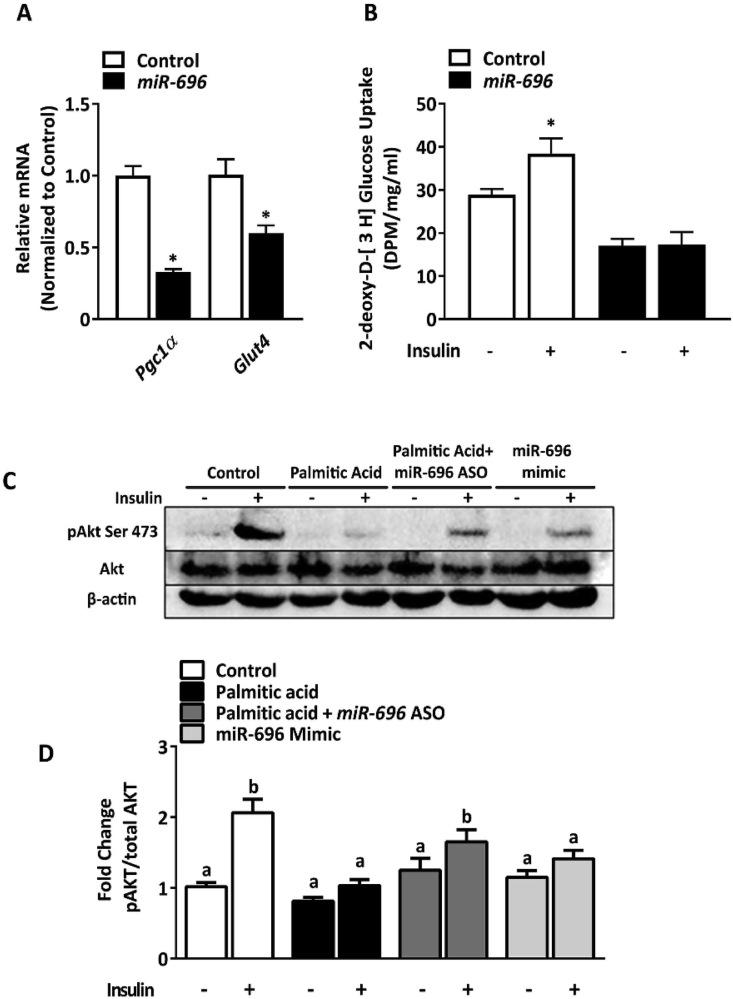
GLUT4 is a major transporter involved in glucose uptake in skeletal muscle cells [42]. Because Pgc1α can regulate Glut4 transcription [43] and glucose uptake [14,44], we investigated whether miR-696 has an effect on Glut4 transcription. Plasmid overexpression containing the full-length sequence of miR-696 (miR-696) in C2C12 cells significantly reduced Pgc1α and Glut4 expression (Figure 5A). Consistent with decreased Glut4, both basal and insulin-stimulated glucose uptake were significantly reduced in C2C12 cells overexpressing miR-696 (Figure 5B). We next investigated the effects of miR-696 on the key insulin signaling intermediary Akt by measuring Akt phosphorylation on the activating site Ser473. Palmitic acid treatment and miR-696 mimic transfection decreased insulin-stimulated Akt Ser473 phosphorylation. Reducing miR-696 with miR-696 ASO partially reversed palmitic acid-induced decreases in Akt phosphorylation (Figure 5C and D). Therefore, our data demonstrated that miR-696 downregulates glucose uptake and Akt signaling in C2C12 cells.
Figure 5.
miR-696 reduced glucose uptake in C2C12 cells. (A–B) C2C12 cells were transfected with RFP (control) or RFP-miR-696 (miR-696) (n = 6). (A) Gene expression of Pgc1α and Glut4, in both the presence and absence of miR-696 (β-actin was used as a reference gene). (B) Glucose uptake was measured with or without insulin (100 nM) in the control and miR-696-transfected groups. (C) C2C12 cells were transfected with scrambled oligonucleotide (control), scrambled oligonucleotide and treated with 700 μM of palmitic acid (palmitic acid), miR-696 antisense oligonucleotide and treated with 700 μM of palmitic acid (palmitic acid + miR-696 ASO), and miR-696 full-length oligonucleotide sequence (miR-696 mimic). All the groups were treated with vehicle (ethanol in 1% BSA) (n = 5). Western blotting of total Akt (Akt) and phosphorylated Akt on serine 473 (pAkt Ser 473) with or without insulin (100 nM) for 10 min. β-actin (30 μg) was used as an internal control. (D) The bars are a quantification of the Western blotting for the treatment groups represented in panel (C) (n = 3). ∗p < 0.05 vs control (Student's t test). Each bar indicates mean ± SEM.
3.6. Effect of AMPK on miR-696 expression
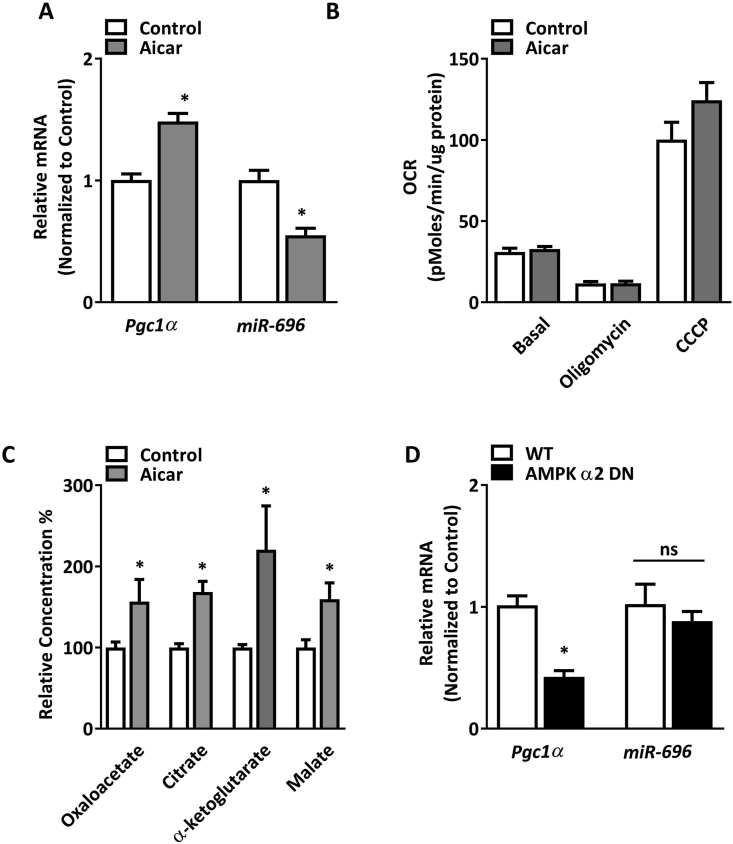
AMPK is a key regulator of Pgc1α expression and mitochondrial function in skeletal muscle [14,24,45]. In addition, AMPK can regulate GLUT4 transcription and glucose uptake in skeletal muscle, both of which were downregulated by miR-696 (Figure 5). Therefore, we tested the hypothesis that AMPK may be a negative regulator of miR-696. C2C12 cells treated with the AMPK activator AICAR significantly increased Pgc1α expression, concomitant with a significant decrease in miR-696 expression (Figure 6A). We next explored whether mitochondrial function was altered through AICAR treatment in skeletal muscle cells using the oxygen consumption rate (OCR) as our metric. AICAR did not significantly alter basal, oligomycin, or CCCP-stimulated OCR (Figure 6B), but there were significant increases in the concentrations of four tricarboxylic acid (TCA) cycle metabolites (Figure 6C). Because AICAR did not exclusively alter AMPK activity in cells, we determined the specific effects of decreased activity of skeletal muscle AMPKα2, the major isoform expressed in skeletal muscle. Interestingly, AMPKα2 dominant-negative mice (AMPKα2 DN) [46] had reduced Pgc1α but no alteration in miR-696 gene expression (Figure 6D). This suggested that AICAR works through an AMPKα2-independent mechanism to mediate miR-696 expression and that AMPKα2 may not be a major regulator of miR-696 in skeletal muscle.
Figure 6.
Effect of AMPK on miR-696 expression. (A) Pgc1α and miR-696 expression in C2C12 cells either under control conditions or treated with AICAR (250 μM) (n = 5) (β-actin and SnoR-202 were used as reference genes, respectively). (B) Mitochondrial respiration rate in C2C12 cells as measured by the oxygen consumption rate (OCR). Oligomycin and CCCP were used for oxidative phosphorylation inhibition and mitochondrial uncoupling, respectively. (C) Tricarboxylic acid (TCA) cycle intermediate concentrations in C2C12 cells treated with AICAR (250 μM) compared to untreated controls (n = 5). (D) Pgc1α and miR-696 expression in the TA muscle from transgenic AMPKα2 DN mice (n = 5). ∗p < 0.05 vs control (Student's t test). Each bar indicates mean ± SEM.
3.7. SNARK regulated miR-696 expression
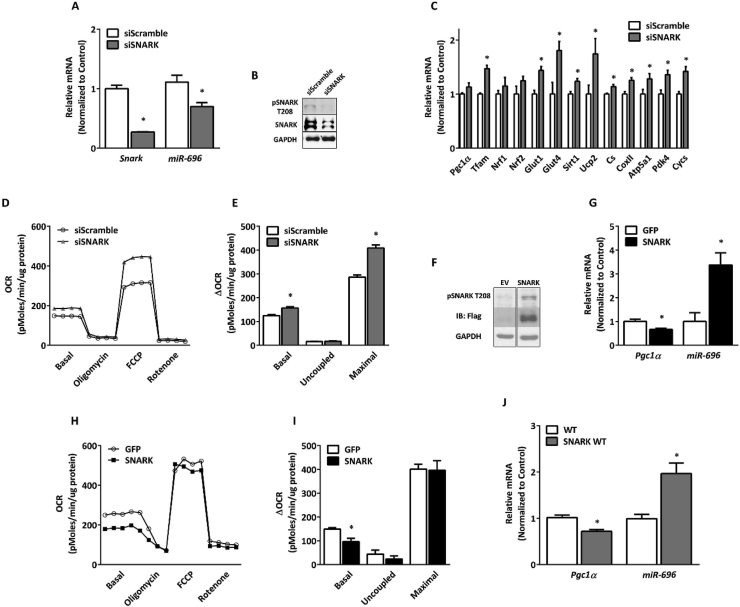
Similar to our data demonstrating an increase in miR-696 by metabolic stress, expression of the AMPK-related kinase SNARK was found to be increased in skeletal muscle under several metabolic stress conditions including humans with obesity [47], high-fat diet-fed mice (HFD), and C2C12 cells treated with palmitic acid [29]. Furthermore, SNARK activity is regulated by AICAR [31], thus identifying it as a potential mediator of AICAR-induced miR-696 expression. Therefore, we hypothesized that SNARK may play a regulatory role in miR-696 expression and consequently Pgc1α expression. Genetic knockdown of Snark in C2C12 cells by small interfering RNA (siSNARK) significantly reduced Snark mRNA (75%) and protein concentrations (60%) (Figure 7A and B, respectively). Snark knockdown also resulted in a 38% reduction in miR-696 expression (Figure 7A) and interestingly resulted in the upregulation of several Pgc1α-regulated genes (Figure 7C). Consistent with the upregulation of mitochondrial genes, Snark knockdown in C2C12 cells resulted in increased basal and maximal OCR, indicating enhanced mitochondrial function (Figure 7D and E). We also found that the overexpression human full-length SNARK in C2C12 cells (Figure 7F) significantly decreased Pgc1α expression, while miR-696 significantly increased (Figure 7G). SNARK overexpression reduced the basal OCR without affecting either the oligomycin or FCCP-induced OCR (Figure 7H and I). These findings suggested that SNARK is a negative regulator of mitochondrial function through the upregulation of miR-696.
Figure 7.
SNARK regulated miR-696 expression. (A–E) C2C12 cells were transfected with scrambled oligonucleotide (control) or siSNARK oligonucleotide (siSNARK) for 48 h (n = 3). (A) Comparison of Snark and miR-696 gene expression with and without oligonucleotide transfection (Gapdh and SnoR-202 were used as reference genes, respectively). (B) Total and phosphorylated SNARK protein content. GAPDH was used as a loading control; 30 μg of protein was loaded. (C) Gene expression analysis of mitochondrial-related genes in skeletal muscle: Pgc1α, Tfam, Nrf1, Nrf2, Sirt1, Ucp2, Cs CoxII, Atp5a1, Pdk4, and Cycs and glucose metabolism-related genes Glut1 and Glut4 in skeletal muscle. (D) The oxygen consumption rate at a basal state and in the presence of oligomycin, FCCP, and rotenone. (E) Bars indicate the variations in the oxygen consumption rate (ΔOCR, basal = basal − rotenone; uncoupled = oligomycin − rotenone; maximal = FCCP − rotenone) from kinetics depicted in (D). (F–I) C2C12 cells were transfected with GFP or empty vector (EV) or SNARK vector (SNARK) for 48 h (n = 3). (F) Protein expression of Flag and phosphorylated SNARK protein content. GAPDH (30 μg) was used as a loading control. (G) Pgc1α and miR-696 gene expression (Gapdh and SnoR-202 were used as reference genes, respectively). (H) Oxygen consumption rate at a basal state and in the presence of oligomycin, FCCP, and rotenone. (I) Bars indicate the difference in the oxygen consumption rate between the treatment and control groups under different metabolic conditions (basal = basal − rotenone; uncoupled = oligomycin − rotenone; maximal = FCCP − rotenone). Bar graph of the kinetics depicted in Figure 7H. (J) Pgc1α and miR-696 gene expression in TA muscle from overexpressing SNARK wild-type (SNARK WT) compared to wild-type control mice (WT) (n = 5) (Gapdh and SnoR-202 were used as reference genes, respectively). ∗p < 0.05 (Student's t test). Each bar indicates mean ± SEM.
To investigate SNARK's effects on miR-696 and Pgc1α transcription in vivo, we used a muscle-specific SNARK transgenic mouse line that overexpresses the wild-type human full-length Snark sequence (SNARK WT) [29]. TA muscle from SNARK WT had a significant reduction in Pgc1α mRNA and a significant increase in miR-696 compared to control mice (Figure 7J). Thus, in cells and muscles in vivo, alterations in SNARK regulated miR-696 and Pgc1α.
3.8. Modulation of SNARK expression changed spontaneous activity
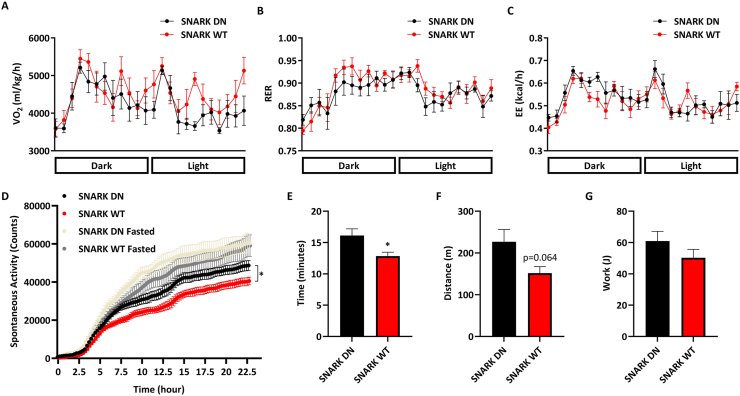
Functional analysis from metabolic chambers comparing the SNARK WT to the dominant-negative SNARK transgenic mouse (SNARK DN) showed that, regarding overall metabolism, neither the volume of oxygen consumed (VO2), the respiratory exchange ratio (RER), or the total body energy expenditure (EE) changed between the two strains (Figure 8A–C). However, when we analyzed the total amounts of spontaneous locomotor activity in these mice, we observed a significant reduction in the SNARK WT mouse activity (Figure 8D). Moreover, by performing a maximal endurance capacity test, time spent on the treadmill was lower for the SNARK WT mice (Figure 8E), while the distance tended to be lower (Figure 8F, p = 0.064). No changes in total mice work performed were observed given the reduction of total body weight in the SNARK WT group (Figure 8G). These data revealed the in vivo relevance of the SNARK-miR696-Pgc1α pathway by demonstrating that the SNARK transgenic mice had altered locomotor activity.
Figure 8.
Modulation of SNARK expression changed spontaneous activity. (A–C) ANCOVA analysis of SNARK DN and SNARK WT mice housed under 22 °C in metabolic cages. (A) Volume of oxygen (VO2) consumed over time. (B) Respiratory exchange ratio (RER) over time. (C) Energy expenditure (EE) over time. (D) ANCOVA analysis of cumulative activity over time of fed and fasted SNARK DN and SNARK WT mice housed under 22 °C in metabolic cages. (E) Time, (F) distance, (G) and work achieved by SNARK DN and SNARK WT mice on a maximal endurance performance test (running on a treadmill until exhaustion). ∗p < 0.05 (Student's t test or 2-way ANOVA). Graphs are mean ± SEM.
4. Discussion
Increased oxidative stress and impaired mitochondrial function are primary contributors to the pathology of type 2 diabetes. Given the large number of people who are affected by metabolic diseases, determining mechanisms that affect mitochondrial function is crucial to developing appropriate prevention and potential therapeutic interventions. Using in silico analysis, we identified miR-696 as a potentially novel regulator of Pgc1α, a PPARγ co-activator involved in mitochondrial function and biogenesis [48]. Confirming these findings, we observed direct regulation of Pgc1α by miR-696 using a reporter assay (Figure 2C). A similar result was previously validated in primary hepatocytes through an experiment using a luciferase reporter assay [49]. In addition, Pgc1α content was markedly reduced in skeletal muscle cells exposed to increasing concentrations of miR-696 mimic (Figure 2B). Thus, our data established miR-696 as a novel suppressor of Pgc1α expression.
As Pgc1α is a critical regulator of mitochondrial function, we next sought to determine the physiological implications of changes in miR-696 expression in skeletal muscle using in vitro and in vivo models. Pgc1α expression has been shown to be higher in skeletal muscles predominantly composed of oxidative (type I) fibers compared to skeletal muscle tissues mainly composed of glycolytic (type II) fibers [[50], [51], [52]]. In the current study, we found a negative correlation between miR-696 and Pgc1α expression in muscle, with oxidative muscle having lower miR-696 and higher Pgc1a levels than glycolytic muscle. Thus, our data identified miR-696 as a potential mechanism for differential Pgc1α expression and mitochondrial content in skeletal muscles with different metabolic characteristics (Figure 3C).
Our data demonstrated that the expression of miR-696 increased in the skeletal muscle of C57BL/6 mice exposed to streptozotocin or a high-fat diet (Figure 3D and E), both of which are well-known models for inducing metabolic stress and reducing mitochondrial function [34,45,53]. We also observed increased expression of mir-696 throughout the differentiation process of C2C12 cells (Figure 3F). Interestingly, inhibition of miR-696 was shown to accelerate differentiation in 3D-engineered human skeletal muscle bundles [54]. In addition, we showed that C2C12 cells exposed to a high dose of palmitic acid had elevated miR-696, and the transfection of C2C12 cells with a miR-696 oligonucleotide mimic reduced both Pgc1α expression and basal oxygen consumption. These effects were associated with increased reactive oxygen species production and low levels of several mitochondrial biogenesis-related genes, indicating a close relationship between miR-696 expression and mitochondrial function. In contrast, miR-696 knockdown partially reversed the effects of fatty acid overload on mitochondrial function in muscle cells (Figure 4). Thus, our data identified miR-696 as a novel regulator of mitochondrial function in response to metabolic stress in muscle.
AMPK, an intracellular energetic sensor, has been described as a positive mediator of Pgc1α expression by direct phosphorylation of the Pgc1a promoter in myotubes [14]. Herein, we demonstrated that AICAR, a well-known activator of AMPK, can upregulate Pgc1α and downregulate miR-696 expression, suggesting AMPK as a potential mediator of our newly discovered miR-696/Pgc1α axis. However, we observed no effect of AMPK on miR-696 expression in skeletal muscle of mice overexpressing AMPK with a dominant-negative mutation at the α2 subunit (Figure 6D), suggesting that a target other than AMPK may be responsible for AICAR-induced regulation of miR-696/Pgc1α.
The AMPK-related kinase SNARK has been associated with multiple cellular processes including glucose transport during skeletal muscle contraction [55], metabolic stress [47], and regulation of skeletal muscle mass and cell survival [29]. Interestingly, a gradual increase in SNARK expression during differentiation of C2C12 cells and a gradual reduction of Pgc1α expression in C2C12 cells throughout differentiation [56] were demonstrated. Furthermore, SNARK expression was markedly increased in human cultured myocytes in response to metabolic stressors including palmitate and TNFα, two conditions well known to decrease PGC1α transcription [47]. Given previous evidence that SNARK and Pgc1α are divergently regulated, we hypothesized that SNARK may be a negative regulator of Pgc1a expression via miR-696. In line with this hypothesis, we found that overexpression of SNARK caused a reduction in the Pgc1α expression and mitochondrial oxygen consumption rate in C2C12 cells (Figure 7G and H). Importantly, the impact of SNARK overexpression on Pgc1a expression and mitochondrial function occurred in conjunction with increased miR-696 transcription in C2C12 cells and SNARK transgenic mice (Figure 7G and J). Thus, our data identified a novel role of SNARK in regulating mitochondrial function through the miR-696/Pgc1a axis.
The physiological purpose of this newly discovered SNARK-miR-696-Pgc1α axis is not known. However, we have previously shown that SNARK is necessary to prevent apoptosis in muscle cells exposed to metabolic stressors such as palmitate [29]. Moreover, we have demonstrated that loss of SNARK activity leads to accelerated loss of muscle mass with age. Thus, previous evidence indicated that SNARK plays a protective role in muscle by preventing apoptosis and muscle loss under metabolic and physiological stress conditions. We postulate that the protective effect of SNARK on myocyte survival may be due in part to its ability to reduce Pgc1α expression via miR-696. In line with that assertion, mice overexpressing Pgc1α exhibited severe muscle atrophy, leading to a 25% reduction in body weight by 25 weeks of age [57]. In addition, mice overexpressing Pgc1α in muscle were more prone to fat-induced insulin resistance and had no metabolic improvements in response to exercise or caloric restriction [58,59]. These data indicated that an overabundance of mitochondrial activity had a deleterious effect on cellular function and myocyte survival. In support of this, long-term overexpression of PGC1β, a member of the PGC family, induced mitochondrial biogenesis and activity and apoptosis [60]. Thus, although increased mitochondrial function is often thought to be a positive adaptation, SNARK may play an important role in limiting excess mitochondrial activity during times of metabolic increases, protecting cells from mitochondria-induced apoptosis.
It is important to acknowledge that miR-696 is a mouse-specific microRNA and was studied in the context of metabolic stress. We believe that it is crucial to search for microRNAs that can regulate mitochondrial biogenesis through PGC1Α in human cells and provide biological relevance for humans. For this reason, we performed an “in silico” analysis (TargetScanHuman prediction toll) using the human 3′UTR of the PGC1Α gene and found the human microRNA hsa-miR-218-5p sitting exactly on the same region targeted by the microRNA miR-696 in the mouse 3′UTR of the Pgc1α gene. Interestingly, hsa-mir-218 was found to be increased in urinary exosomes of type 1 diabetic children [61]. In another study, hsa-mir-218-5p was reduced in gastric cancer cells allowing metastatic behavior, which could be associated with an increase in PGC1A expression [62]. Moreover, “in silico” analysis four microRNAs mitochondrial sub-localization indicated that hsa-mir-218-5p is located in the mitochondria [63]. Taken together, this suggests that microRNA hsa-mir-218-5p may regulate PGC1Α in humans and indicates a possible regulatory function of hsa-mir-218-5p over mitochondrial homeostasis.
5. Conclusion
In conclusion, we provided evidence of a mechanism in which high levels of metabolic stress in skeletal muscle cells increased the microRNA miR-696 levels and reduced Pgc1α expression and mitochondrial function. We also identified SNARK as a novel regulator of mitochondrial function via miR-696 and Pgc1α. These findings could lead to novel therapeutic and preventive strategies for metabolic diseases, including type 2 diabetes.
Acknowledgments
This study was supported by the State Science-Funding Agency FAPESP (2013/22733-0), National Institutes of Health grants R01DK099511 (to L.J.G.), and the Joslin Diabetes Center DRC (P30 DK36836). CAPES sponsored the first author via a doctorate scholarship 99999.009038/2014-00. Special thanks to Alison Slack for proofreading this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101226.
Contributor Information
Laurie J. Goodyear, Email: Laurie.Goodyear@joslin.harvard.edu.
Leonardo R. Silveira, Email: leors@unicamp.br.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Perry R.J., Zhang D., Zhang X.-M., Boyer J.L., Shulman G.I. Controlled-release mitochondrial protonophore reverses diabetes and steatohepatitis in rats. Science. 2015;347:1253–1256. doi: 10.1126/science.aaa0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 3.Pal M., Wunderlich C.M., Spohn G., Brönneke H.S., Schmidt-Supprian M., Wunderlich F.T. Alteration of JNK-1 signaling in skeletal muscle fails to affect glucose homeostasis and obesity-associated insulin resistance in mice. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodama K., Toda K., Morinaga S., Yamada S., Butte A.J. Anti-CD44 antibody treatment lowers hyperglycemia and improves insulin resistance, adipose inflammation, and hepatic steatosis in diet-induced obese mice. Diabetes. 2015;64:867–875. doi: 10.2337/db14-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wueest S., Rapold R.A., Schumann D.M., Rytka J.M., Schildknecht A., Nov O. Deletion of Fas in adipocytes relieves adipose tissue inflammation and hepatic manifestations of obesity in mice. Journal of Clinical Investigation. 2010;120:191–202. doi: 10.1172/JCI38388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright D.C., Han D.-H., Garcia-Roves P.M., Geiger P.C., Jones T.E., Holloszy J.O. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1alpha expression. Journal of Biological Chemistry. 2007;282:194–199. doi: 10.1074/jbc.M606116200. [DOI] [PubMed] [Google Scholar]
- 7.Bocco B.M.L.C., Louzada R.A.N., Silvestre D.H.S., Santos M.C.S., Anne-Palmer E., Rangel I.F. Thyroid hormone activation by type 2 deiodinase mediates exercise-induced peroxisome proliferator-activated receptor-γ coactivator-1α expression in skeletal muscle. Journal of Physiology (London) 2016;594:5255–5269. doi: 10.1113/JP272440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho Y., Hazen B.C., Russell A.P., Kralli A. Peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. Journal of Biological Chemistry. 2013;288:25207–25218. doi: 10.1074/jbc.M113.489674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Arany Z. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Current Opinion in Genetics & Development. 2008;18:426–434. doi: 10.1016/j.gde.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safdar A., Little J.P., Stokl A.J., Hettinga B.P., Akhtar M., Tarnopolsky M.A. Exercise increases mitochondrial PGC-1alpha content and promotes nuclear-mitochondrial cross-talk to coordinate mitochondrial biogenesis. Journal of Biological Chemistry. 2011;286:10605–10617. doi: 10.1074/jbc.M110.211466. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Scarpulla R.C. Nuclear control of respiratory gene expression in mammalian cells. Journal of Cellular Biochemistry. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- 13.Ruas J.L., White J.P., Rao R.R., Kleiner S., Brannan K.T., Harrison B.C. A PGC-1α isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151:1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jäger S., Handschin C., St-Pierre J., Spiegelman B.M. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemoto S., Fergusson M.M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} Journal of Biological Chemistry. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 16.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun L.-Y., Wang N., Ban T., Sun Y.-H., Han Y., Sun L.-L. MicroRNA-23a mediates mitochondrial compromise in estrogen deficiency-induced concentric remodeling via targeting PGC-1α. Journal of Molecular and Cellular Cardiology. 2014;75:1–11. doi: 10.1016/j.yjmcc.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y., Zhao C., Sun X., Liu Z., Zhang J. MicroRNA-761 regulates mitochondrial biogenesis in mouse skeletal muscle in response to exercise. Biochemical and Biophysical Research Communications. 2015;467:103–108. doi: 10.1016/j.bbrc.2015.09.113. [DOI] [PubMed] [Google Scholar]
- 19.Mineno J., Okamoto S., Ando T., Sato M., Chono H., Izu H. The expression profile of microRNAs in mouse embryos. Nucleic Acids Research. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoi W., Naito Y., Mizushima K., Takanami Y., Kawai Y., Ichikawa H. The microRNA miR-696 regulates PGC-1{alpha} in mouse skeletal muscle in response to physical activity. American Journal of Physiology. Endocrinology and Metabolism. 2010;298:E799–E806. doi: 10.1152/ajpendo.00448.2009. [DOI] [PubMed] [Google Scholar]
- 21.Wen F., Zhang H., Bao C., Yang M., Wang N., Zhang J. Resistin increases ectopic deposition of lipids through miR-696 in C2C12 cells. Biochemical Genetics. 2015;53:63–71. doi: 10.1007/s10528-015-9672-2. [DOI] [PubMed] [Google Scholar]
- 22.Xiao B., Sanders M.J., Underwood E., Heath R., Mayer F.V., Carmena D. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teodoro B.G., Sampaio I.H., Bomfim L.H.M., Queiroz A.L., Silveira L.R., Souza A.O. Long-chain acyl-CoA synthetase 6 regulates lipid synthesis and mitochondrial oxidative capacity in human and rat skeletal muscle. The Journal of Physiology. 2017;595:677–693. doi: 10.1113/JP272962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palacios O.M., Carmona J.J., Michan S., Chen K.Y., Manabe Y., Ward J.L. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging. 2009;1:771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koh H.-J., Arnolds D.E., Fujii N., Tran T.T., Rogers M.J., Jessen N. Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Molecular and Cellular Biology. 2006;26:8217–8227. doi: 10.1128/MCB.00979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thirugnanam K., Cossette S.M., Lu Q., Chowdhury S.R., Harmann L.M., Gupta A. Cardiomyocyte-specific Snrk prevents inflammation in the heart. Journal of the American Heart Association. 2019;8 doi: 10.1161/JAHA.119.012792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rines A.K., Chang H.-C., Wu R., Sato T., Khechaduri A., Kouzu H. Snf1-related kinase improves cardiac mitochondrial efficiency and decreases mitochondrial uncoupling. Nature Communications. 2017;8:14095. doi: 10.1038/ncomms14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., An R., Lai S., Li L., Liu S., Xu H. Dysregulation of PP2A-Akt interaction contributes to sucrose non-fermenting related kinase (SNRK) deficiency induced insulin resistance in adipose tissue. Molecular Metabolism. 2019;28:26–35. doi: 10.1016/j.molmet.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lessard S.J., Rivas D.A., So K., Koh H.-J., Queiroz A.L., Hirshman M.F. The AMPK-related kinase SNARK regulates muscle mass and myocyte survival. Journal of Clinical Investigation. 2016;126:560–570. doi: 10.1172/JCI79197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbosa M.R., Sampaio I.H., Teodoro B.G., Sousa T.A., Zoppi C.C., Queiroz A.L. Hydrogen peroxide production regulates the mitochondrial function in insulin resistant muscle cells: effect of catalase overexpression. Biochimica et Biophysica Acta. 2013;1832:1591–1604. doi: 10.1016/j.bbadis.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 31.Kuga W., Tsuchihara K., Ogura T., Kanehara S., Saito M., Suzuki A. Nuclear localization of SNARK; its impact on gene expression. Biochemical and Biophysical Research Communications. 2008;377:1062–1066. doi: 10.1016/j.bbrc.2008.10.143. [DOI] [PubMed] [Google Scholar]
- 32.Fujii N., Hirshman M.F., Kane E.M., Ho R.C., Peter L.E., Seifert M.M. AMP-activated protein kinase alpha2 activity is not essential for contraction- and hyperosmolarity-induced glucose transport in skeletal muscle. Journal of Biological Chemistry. 2005;280:39033–39041. doi: 10.1074/jbc.M504208200. [DOI] [PubMed] [Google Scholar]
- 33.Barré L., Richardson C., Hirshman M.F., Brozinick J., Fiering S., Kemp B.E. Genetic model for the chronic activation of skeletal muscle AMP-activated protein kinase leads to glycogen accumulation. American Journal of Physiology. Endocrinology and Metabolism. 2007;292:E802–E811. doi: 10.1152/ajpendo.00369.2006. [DOI] [PubMed] [Google Scholar]
- 34.Patti M.E., Butte A.J., Crunkhorn S., Cusi K., Berria R., Kashyap S. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Yang L., Gao Y.-F., Fan Z.-M., Cai X.-Y., Liu M.-Y. MicroRNA-106b induces mitochondrial dysfunction and insulin resistance in C2C12 myotubes by targeting mitofusin-2. Molecular and Cellular Endocrinology. 2013;381:230–240. doi: 10.1016/j.mce.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Greene N.P., Lee D.E., Brown J.L., Rosa M.E., Brown L.A., Perry R.A. Mitochondrial quality control, promoted by PGC-1α, is dysregulated by Western diet-induced obesity and partially restored by moderate physical activity in mice. Physiological Reports. 2015;3 doi: 10.14814/phy2.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Y., Lawrence C.E. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Research. 2001;29:1034–1046. doi: 10.1093/nar/29.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., Jia Y., Rogers H., Suzuki N., Gassmann M., Wang Q. Erythropoietin contributes to slow oxidative muscle fiber specification via PGC-1α and AMPK activation. The International Journal of Biochemistry & Cell Biology. 2013;45:1155–1164. doi: 10.1016/j.biocel.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin J., Wu H., Tarr P.T., Zhang C.-Y., Wu Z., Boss O. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 40.Zechner C., Lai L., Zechner J.F., Geng T., Yan Z., Rumsey J.W. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metabolism. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson E.J., Lustig M.E., Boyle K.E., Woodlief T.L., Kane D.A., Lin C.-T. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. Journal of Clinical Investigation. 2009;119:573–581. doi: 10.1172/JCI37048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuchiya A., Kanno T., Nishizaki T. PI3 kinase directly phosphorylates Akt1/2 at Ser473/474 in the insulin signal transduction pathway. Journal of Endocrinology. 2014;220:49–59. doi: 10.1530/JOE-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michael L.F., Wu Z., Cheatham R.B., Puigserver P., Adelmant G., Lehman J.J. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGee S.L., van Denderen B.J.W., Howlett K.F., Mollica J., Schertzer J.D., Kemp B.E. AMP-activated protein kinase regulates GLUT4 transcription by phosphorylating histone deacetylase 5. Diabetes. 2008;57:860–867. doi: 10.2337/db07-0843. [DOI] [PubMed] [Google Scholar]
- 45.Franko A., von Kleist-Retzow J.C., Böse M., Sanchez-Lasheras C., Brodesser S., Krut O. Complete failure of insulin-transmitted signaling, but not obesity-induced insulin resistance, impairs respiratory chain function in muscle. Journal of Molecular Medicine. 2012;90:1145–1160. doi: 10.1007/s00109-012-0887-y. [DOI] [PubMed] [Google Scholar]
- 46.Musi N., Hirshman M.F., Arad M., Xing Y., Fujii N., Pomerleau J. Functional role of AMP-activated protein kinase in the heart during exercise. FEBS Letters. 2005;579:2045–2050. doi: 10.1016/j.febslet.2005.02.052. [DOI] [PubMed] [Google Scholar]
- 47.Rune A., Osler M.E., Fritz T., Zierath J.R. Regulation of skeletal muscle sucrose, non-fermenting 1/AMP-activated protein kinase-related kinase (SNARK) by metabolic stress and diabetes. Diabetologia. 2009;52:2182–2189. doi: 10.1007/s00125-009-1465-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohas L.M., St-Pierre J., Uldry M., Jäger S., Handschin C., Spiegelman B.M. A fundamental system of cellular energy homeostasis regulated by PGC-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7933–7938. doi: 10.1073/pnas.0702683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang Z., Li P., Jia W., Jiang T., Wang Z., Xiang Y. miR-696 plays a role in hepatic gluconeogenesis in ob/ob mice by targeting PGC-1α. International Journal of Molecular Medicine. 2016;38:845–852. doi: 10.3892/ijmm.2016.2659. [DOI] [PubMed] [Google Scholar]
- 50.Rasbach K.A., Gupta R.K., Ruas J.L., Wu J., Naseri E., Estall J.L. PGC-1alpha regulates a HIF2alpha-dependent switch in skeletal muscle fiber types. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21866–21871. doi: 10.1073/pnas.1016089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Handschin C., Chin S., Li P., Liu F., Maratos-Flier E., Lebrasseur N.K. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. Journal of Biological Chemistry. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 52.Matsuura T., Li Y., Giacobino J.-P., Fu F.H., Huard J. Skeletal muscle fiber type conversion during the repair of mouse soleus: potential implications for muscle healing after injury. Journal of Orthopaedic Research. 2007;25:1534–1540. doi: 10.1002/jor.20451. [DOI] [PubMed] [Google Scholar]
- 53.Rocha-Rodrigues S., Rodríguez A., Gouveia A.M., Gonçalves I.O., Becerril S., Ramírez B. Effects of physical exercise on myokines expression and brown adipose-like phenotype modulation in rats fed a high-fat diet. Life Sciences. 2016;165:100–108. doi: 10.1016/j.lfs.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Cheng C.S., Ran L., Bursac N., Kraus W.E., Truskey G.A. Cell density and Joint microRNA-133a and microRNA-696 inhibition enhance differentiation and contractile function of engineered human skeletal muscle tissues. Tissue Engineering Part A. 2016;22:573–583. doi: 10.1089/ten.tea.2015.0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koh H.-J., Toyoda T., Fujii N., Jung M.M., Rathod A., Middelbeek R.J.-W. Sucrose nonfermenting AMPK-related kinase (SNARK) mediates contraction-stimulated glucose transport in mouse skeletal muscle. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15541–15546. doi: 10.1073/pnas.1008131107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adamovich Y., Shlomai A., Tsvetkov P., Umansky K.B., Reuven N., Estall J.L. The protein level of PGC-1α, a key metabolic regulator, is controlled by NADH-NQO1. Molecular and Cellular Biology. 2013;33:2603–2613. doi: 10.1128/MCB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miura S., Tomitsuka E., Kamei Y., Yamazaki T., Kai Y., Tamura M. Overexpression of peroxisome proliferator-activated receptor gamma co-activator-1alpha leads to muscle atrophy with depletion of ATP. American Journal Of Pathology. 2006;169:1129–1139. doi: 10.2353/ajpath.2006.060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi C.S., Befroy D.E., Codella R., Kim S., Reznick R.M., Hwang Y.-J. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19926–19931. doi: 10.1073/pnas.0810339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong K.E., Mikus C.R., Slentz D.H., Seiler S.E., DeBalsi K.L., Ilkayeva O.R. Muscle-specific overexpression of PGC-1α does not augment metabolic improvements in response to exercise and caloric restriction. Diabetes. 2015;64:1532–1543. doi: 10.2337/db14-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sopariwala D.H., Yadav V., Badin P.-M., Likhite N., Sheth M., Lorca S. Long-term PGC1β overexpression leads to apoptosis, autophagy and muscle wasting. Scientific Reports. 2017;7:10237. doi: 10.1038/s41598-017-10238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kong Q., Guo X., Guo Z., Su T. Urinary exosome miR-424 and miR-218 as biomarkers for type 1 diabetes in children. Clinical Laboratory. 2019;65 doi: 10.7754/Clin.Lab.2018.180921. [DOI] [PubMed] [Google Scholar]
- 62.Lin J., Zhang Y., Zeng X., Xue C., Lin X. Circrna circrims acts as a microrna sponge to promote gastric cancer metastasis. ACS Omega. 2020;5:23237–23246. doi: 10.1021/acsomega.0c02991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan L.-R., Wang A., Lv Z., Yuan Y., Xu Q. Mitochondria-related core genes and TF-miRNA-hub mrDEGs network in breast cancer. Bioscience Reports. 2021:41. doi: 10.1042/BSR20203481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bergmeyer H.U. 2nd ed. Academic Press; New York (NY): 1974. Methods of enzymatic analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.