Summary
CRISPR interference is an increasingly popular method for perturbing gene expression. Guided by single-guide RNAs (sgRNAs), nuclease-deficient Cas9 proteins bind to specific DNA sequences and hinder transcription. Specificity is achieved through complementarity of the sgRNAs to the DNA. Changing complementarity by introducing single-nucleotide mismatches can be exploited to tune knockdown. Here, we present a computational pipeline to identify sgRNAs targeting specific genes in a bacterial genome, filter them, and titrate their activity by introducing mismatches.
For complete details on the use and execution of this protocol, please refer to Hawkins et al. (2020).
Subject areas: Bioinformatics, High Throughput Screening, Gene Expression, CRISPR
Graphical abstract

Highlights
-
•
Identify all CRISPR interference targets for a bacterial gene or genome of interest
-
•
Filter associated sgRNAs based on few simple metrics such as potential off-target hits
-
•
Predict knockdown efficacies of mismatch sgRNAs for introducing graded knockdowns
CRISPR interference is an increasingly popular method for perturbing gene expression. Guided by single-guide RNAs (sgRNAs), nuclease-deficient Cas9 proteins bind to specific DNA sequences and hinder transcription. Specificity is achieved through complementarity of the sgRNAs to the DNA. Changing complementarity by introducing single-nucleotide mismatches can be exploited to tune knockdown. Here, we present a computational pipeline to identify sgRNAs targeting specific genes in a bacterial genome, filter them, and titrate their activity by introducing mismatches.
Before you begin
Timing: 10 min
Our computational pipeline generates single-guide RNAs (sgRNAs) for the widely employed Type II-A CRISPRi system from Streptococcus pyogenes (Liu et al., 2017; Peters et al., 2016; Qi et al., 2013), as used for the mismatch-CRISPRi by Hawkins et al. (2020). For the purpose of CRISPR interference, a catalytically deactivated nuclease effector protein, dCas9, is typically co-expressed with an sgRNA, which together form a dCas9-sgRNA complex. This complex recognizes a short DNA sequence (‘NGG’), called protospacer adjacent motif (PAM), and unwinds the DNA to probe complementarity to the sgRNA (Figure 1). When the spacer sequence of the sgRNA complements the DNA sequence adjacent to the PAM, the DNA progressively unwinds from the PAM proximal to the PAM distal site (Boyle et al., 2017), until the dCas9-sgRNA complex fully associates with the DNA. The bound complex is stable for hours (Sternberg et al., 2014). When bound to a gene or promoter, dCas9 hinders RNA polymerase from either binding or elongating and thereby lowers gene expression (Figure 1). Mismatches between the spacer sequence of the sgRNA and DNA can affect the association between dCas9 and the DNA (Gilbert et al., 2014; Hawkins et al., 2020; Jost et al., 2020; Qi et al., 2013), thereby decreasing knockdown efficacy. Since mismatches have a predictable effect on the expression knockdown (Hawkins et al., 2020; Jost et al., 2020; Mathis et al., 2021), mismatch-CRISPRi makes it possible to simultaneously study many expression knockdowns and thereby quantify how expression affects functionality.
Figure. 1.
CRISPR interference (CRISPRi) and mismatch CRISPRi
The dCas9 protein forms a complex with the single-guide RNA (sgRNA). When a PAM region is present, the dCas9-sgRNA complex binds to the DNA and further unwinds the strands when the 20nt spacer sequence of the sgRNA complements the DNA. The strength of transcriptional interference is highest with (A) perfect complementarity between the sgRNA spacer sequence and the non-template strand and lower (B) in the presence of mismatches (red arrow) between the spacer sequence of the sgRNA and DNA sequence.
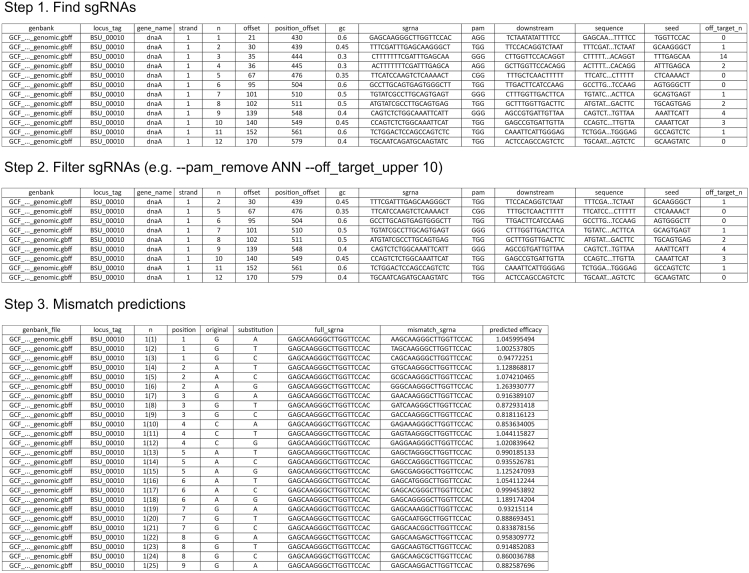
Here, we present a three-step computational pipeline that (1) lists all sgRNAs that target the gene(s) of interest, (2) filters those sgRNAs based on several simple criteria and (3) generates all potential single-nucleotide mismatch sgRNAs and predicts their knockdown efficacies. The pipeline can be used for any bacterial species. As we will detail below, at each step, a CSV file is generated with the information of interest (Figure 2). The computational pipeline can be run step-by-step or in one go, and relies on a single Python script. Before focusing on the computational pipeline, the following preparations are required:
Figure. 2.
CSV files generated in computational pipeline
In step 1, the computational pipeline lists all sgRNAs targeting the non-template strand of a gene of interest as well as its accompanying information (e.g., offset, GC content, off-target hits). This list is shown in the “sgRNA.csv” file. In step 2, this list is filtered according to a few simple criteria specified by the researcher. In this example, sgRNAs are filtered based on the associated PAM sequence and number of off-target hits, including only sgRNAs for which the PAM sequence does not match “ANN” (--pam_remove ANN) and for which there are fewer than 10 off-target hits (--off_target_upper 10). The filtered sgRNAs are in the “sgRNA_filtered.csv” file. In step 3, for each of the filtered sgRNAs a complete list of all mismatches is generated, for which the knockdown efficacy is determined. This information is listed in the “sgRNA_mismatched.csv”.
Download the GenBank file
-
1.
Search for the full genome sequence of your organism of interest or a close relative through https://www.ncbi.nlm.nih.gov/genome. Make sure to download the GenBank file that matches your specific species or strain. For example, when searching for the genome of Bacillus subtilis, NCBI automatically refers to the reference genome sequence of Bacillus subtilis 168 (NCBI Reference Sequence: NC_000964.3). There are however more than 400 Bacillus subtilis genomes available, so one could specifically download the GenBank file corresponding to your strain.
-
2.
Download the GenBank file (common extensions: .gb, .gbk, and .gbff) with both genome features and full genome sequence (or genome contigs). When compressed, unzip the GenBank file. For an example, see GenBank file of Bacillus subtilis 168 (Kunst et al., 1997) included in our GitHub repository (https://github.com/jordivangestel/sgRNAs-for-mismatch-CRISPRi), which can be opened using any standard text editor (e.g., Notepad, TextEdit).
Install Python and packages
-
3.Download Python from https://www.python.org/downloads/ and confirm that Python version 3.6.0 or greater is installed using the following command in the terminal prompt:
- > python --version
-
4.The computational pipeline relies on two Python modules: (1) pandas and (2) Biopython. Determine if those packages are installed:
- > python -c "import pandas; import Bio"
-
5.If an error message appears, install the missing package(s) using your python package manager. With Python’s standard package management system, use the following commands in the terminal prompt:
- > pip install pandas
- > pip install biopython
Download Python script
-
6.
Download the “generate_sgrnas.py” Python script and the “model_param.csv” file available from our GitHub repository (https://github.com/jordivangestel/sgRNAs-for-mismatch-CRISPRi) and store them in the same folder, or clone the repository directly using Git (https://git-scm.com/docs/git-clone).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| Python version 3.6.0 or higher | Python Software Foundation https://www.python.org/downloads/ |
N/A |
| generate_sgrnas.py | This paper https://github.com/jordivangestel/sgRNAs-for-mismatch-CRISPRi |
N/A |
| model_param.csv | This paper https://github.com/jordivangestel/sgRNAs-for-mismatch-CRISPRi |
N/A |
Step-by-step method details
Step 1: Find sgRNAs
Timing: 5 min
Although CRISPRi can target both coding sequences and promoters (Gilbert et al., 2013; Cui et al., 2018), in prokaryotes, transcriptional interference is most effective when blocking RNA polymerase elongation (Rishi et al., 2020). Targeting within the protein coding sequence of a gene guarantees an elongating RNAP and does not require knowledge of promoters. For dCas9, it has furthermore been shown that knockdown efficacy is higher when targeting the non-template strand of a gene (Figures 3A and 3B). The computational pipeline therefore screens for sgRNAs by identifying PAM sequences (NGG) on the template strand of genes. The 20 nucleotides upstream of a PAM sequence corresponds to the spacer sequence of an sgRNA that would target the complementary non-template strand, causing effective expression knockdown (Figure 1).
-
1.To identify all sgRNAs targeting the non-template strand of the protein coding sequence of the gene(s) of interest, one can run the first step of the computational pipeline in the following way:
- >python <path to “generate_sgrnas.py” script>
- --genbank <path to genbank file>
- --locus_tag <locus_tag>
- --step find
-
a.Here, the --genbank and --locus_tag are the only mandatory arguments and specify the path to the GenBank file and the locus tag(s) (not gene names) of the gene(s) of interest, respectively.
-
b.The --step argument is used to only run the first step of the computational pipeline (i.e., find). Without specifying this argument, the script will automatically go through all steps. For a complete list of arguments and the default settings see Table 1.Note: one can also run the help-command:
- >python <path to “generate_sgrnas.py” script> -h
-
2.Step 1 will generate a CSV file (“sgRNA.csv”) in the same folder as the python script, which lists all sgRNAs targeting the gene(s) of interest, as well as their properties (Figure 2 and Table 2). This includes:
-
a.Index of sgRNA (n)
-
b.Offset between the sgRNA and the start codon
-
c.GC content of sgRNA spacer
-
d.sgRNA spacer sequence (20nt upstream of the PAM)
-
e.PAM sequence
-
f.Sequence downstream of the PAM
-
g.Full sequence: sgRNA spacer + PAM + downstream sequence
-
h.Seed sequence from sgRNA spacer used to identify potential off-target hits
-
i.Number of potential off-target hits based on seed sequence
-
a.
Figure 3.
Impact of targeting strand and offset on expression knockdowns and relative fitness
(A and B) Expression knockdown of sgRNAs targeting the template (T) or non-template (NT) strands (box = interquartile range, horizontal line = median, wishers = data within 1.5 times interquartile range above or below box, dots = outliers). (A) Relative GFP expression in strains expressing gfp-targeting sgRNAs from a constitutive promoter in B. subtilis (red) and E. coli (blue) (Hawkins et al., 2020). (B) Relative fitness of sgRNAs targeting essential genes using the template or non-template strand in two different E. coli strains with high (green) or low (purple) dCas9 expression (Cui et al., 2018).
(C and D) Impact of sgRNA offset – distance to start codon (nt) – on expression knockdown. (C) Expression knockdown of sgRNAs targeting the non-template strand of gfp in B. subtilis (red) and E. coli (blue). (D) Relative fitness of sgRNA targeting non-template strand of essential genes in E. coli with high (green, upper regression: p < 0.01, R2=0.003) and low (purple, lower regression: P < 0.001, R2=0.005) dCas9 expression.
Table 1.
Parameter settings
| Parameter | Description | Default |
|---|---|---|
| Required parameters | ||
| --genbank | Genbank file name or path to genbank file name | N/A |
| --locus_tag | Locus tag or list of locus tags for which to generate sgRNAs. Lists should be separated by commas, not spaces | N/A |
| Optional parameters | ||
| --model_param | [Optional] Name of csv file with model parameters | model_param.csv |
| --step | [Optional] Steps to conduct. When conducting multiple steps list them separated by a comma | find,filter,mismatch |
| --downstream | [Optional] Number of nucleotides downstream of the PAM sequence to be included in the analysis | 15 |
| --off_target_seed | [Optional] Number of nucleotides of the seed sequence of the sgRNA proximal to the PAM to be included for off-target analysis | 9 |
| --file_find | [Optional] Output file name of step 1 (find) | sgRNA.csv |
| --file_filter | [Optional] Output file name of step 2 (filter) | sgRNA_filter.csv |
| --file_mismatch | [Optional] Output file name of step 3 (mismatch) | sgRNA_mistmatch.csv |
| Filter parameters | ||
| --sgrna_remove | [Optional] Remove all sgRNAs that match a given sequence, e.g., NNNNNNNNNNNNNNNNNNGG, removes all sgRNAs ending with ‘GG’ | None |
| --pam_remove | [Optional] Remove all sgRNAs that are associated with a PAM that matches a given sequence. | None |
| --downstream_remove | [Optional] Remove all sgRNAs that are associated with an upstream sequence that matches a given sequence, e.g., GNNNNNNNNNNNNNN | None |
| --gc_lower | [Optional] Minimal GC content of sgRNA | 0 |
| --gc_upper | [Optional] Maximum GC content of sgRNA | 1 |
| --offset_upper | [Optional] Maximum distance (nt) from start codon (i.e., offset) | 1000 |
| --off_target_upper | [Optional] Maximum number of allowed off-target hits of seed sequence of sgRNA | 10 |
Table 2.
Output data
| Column name | Description |
|---|---|
| genbank | Genbank file name or path to genbank file name |
| locus_tag | Locus tag |
| gene_name | Gene name associated with locus tag in GenBank file |
| strand | Coding strand (1= positive, −1 = negative) |
| n | Index of identified sgRNA |
| offset | Distance of sgRNA to beginning of start codon |
| position_offset | Genome position of sgRNA nucleotide closest to start codon |
| gc | GC content (fraction) |
| sgrna | 20 nucleotide sgRNA spacer sequence |
| pam | PAM sequence |
| downstream | Nucleotide sequence downstream of PAM |
| sequence | Complete sequence: sgRNA spacer + PAM + downstream sequence |
| seed | Seed sequence of sgRNA that is used to identify potential off-target hits |
| off_target_n | Number of potential off-target hits based on seed sequence of sgRNA |
These properties will be used in step 2 to filter against sgRNAs with unwanted properties. Below we give some examples on what input arguments could be used for running the first step of the pipeline.
Examples
-
•To generate the list of all possible sgRNAs for the essential gene, dnaA (locus tag = BSU_00010), in Bacillus subtilis (GenBank = GCF_000009045.1_ASM904v1_genomic.gbff). One can run the following command:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step find
-
•To examine multiple genes, provide a list of locus tags. For example, to identify the sgRNAs of both DnaA and DnaN one can run the following command:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010,BSU_00020
- --step find
-
•One can examine all genes using the following command:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag all
- --step find
-
•One can also specify the path and file name of the CSV file generated in step 1. By default the CSV file is saved in the same folder as your python script and is named “sgRNA.csv”. In order to change the default settings run the following command:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag all
- --step find
- --file_find new_name.csv
-
•The CSV file stores information for each of the sgRNAs (Figure 2). For example, the computational pipeline determines the number of potential off-target hits in the genome based on sgRNA seed sequence. By default, the seed sequence equals the 9 PAM-proximal nucleotides of an sgRNA spacer. The length of the seed sequence is based on a study showing that DNA complementarity of 9 nucleotides proximal to the PAM is sufficient to cause off-target knockdown effects (Cui et al., 2018). Within our pipeline, one could however adjust the size of the seed sequence using --off_target_seed. For example, the following command line limits the seed sequences to 8 nucleotides only and thus finds all off-target hits for this shorter sequence:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step find
- --off_target_seed 8
-
•Finally, one could specify the number of nucleotides downstream from the PAM that are included for further analyses (--downstream). As we will detail below, the downstream sequence of the PAM can affect the efficacy of CRISPRi-mediated expression knockdown, and therefore plays an important role in filtering out potentially weak sgRNAs. Based on a previous analysis (Calvo-Villamañán et al., 2020), we include a default downstream sequence of 15 nucleotides, but one could adjust this to, for example, 20 downstream nucleotides using the following command:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step find
- --downstream 20
Step 2: Filter sgRNAs
Timing: 1 min
Several factors have been suggested to affect the sgRNA-mediated knockdown efficacy, although in most cases we have little mechanistic understanding as to how these effects come about. Qi et al. (2013) reported a relatively strong decline in knockdown efficiency with the distance of a sgRNA from the start codon, but this effect has not been reproduced in more recent studies. For instance, in Hawkins et al. (2020), the offset between the sgRNAs and start codon had no effect on the knockdown of gfp in both E. coli and B. subtilis (Figure 3C). Similarly, when re-analyzing the relative fitness of sgRNA targeting essential genes in Cui et al. (2018), we only find a weak dependency of the relative fitness on the offset (R2<0.01; Figure 3D). Reanalyzing the same data, Calvo-Villamañán et al. (2020) also did not note an effect of offset. Besides offset, the nucleotide sequence has been suggested to affect knockdown efficacy. Using the knockdown library of Cui et al. (2018), Calvo-Villamañán et al. (2020) showed that a small number of nucleotide positions in the sgRNA, PAM and downstream region affect transcriptional interference by perfectly matched sgRNAs. Inspired by this work, we re-analyzed the E. coli and B. subtilis datasets of Hawkins et al. (2020) as well as the previously analyzed data from Cui et al. (2018). For each dataset, we fitted a linear model on one-hot encoded primary sequence data to predict knockdown efficacy. For the primary sequence, we used the sgRNA spacer sequence (20 nucleotides), the PAM sequence (3 nucleotides, of which only one variable) and 15 nucleotides downstream of the PAM. Following Calvo-Villamañán et al. (2020), we prevent overfitting of the highly parameterized linear models, by penalizing absolute coefficient values using L1 regularization (based on the penalty term with the lowest mean squared error in a 10-fold cross validation; Figure 4).
Figure 4.
Impact of individual nucleotides on knockdown efficacy
Inspired by a recent study (Calvo-Villamañán et al., 2020), the impact of sgRNAs targeting the non-template strand of essential genes in both the datasets of Cui et al. (2018) (top panel) and Hawkins et al. (2020) (bottom panels) using an L1 linear model is shown.
(A) Sequence logo (Wagih, 2017) of coefficients associated with nucleotides in the sgRNA, PAM and downstream sequences. Positive coefficients correspond to improved knockdown efficacy and negative with reduced efficacy.
(B) Relation between penalty term, , in the L1 linear model and the mean squared error of a 10-fold cross validation. Red dots show mean squared error and error bars indicate standard deviation. Vertical dotted line shows penalty term associated with lowest mean squared error. Lower penalties result in overfitting of the data and higher penalties in underfitting.
Figure 4 shows how each nucleotide in the sgRNA-PAM-downstream region affects knockdown efficacy. Positive coefficients indicate stronger knockdowns. As expected, for the dataset of Cui et al. (2018) we retrieve nearly the same results as Calvo-Villamañán et al. (2020): a few nucleotides upstream (19, 20) and downstream from the PAM (+1,+2), together with the first nucleotide of the PAM, largely determine the knockdown efficacy. Similar data for E. coli from Hawkins et al. (2020) shows a much weaker pattern, due to substantially more noise, which likely results from the high-copy number plasmid used in these experiments. Nonetheless, key residues are consistent with the data from Cui et al. (2018). For example, an adenine in the first nucleotide of the PAM strongly reduces knockdown efficiency. The dataset from B. subtilis (which used a chromosomally integrated system) showed a very similar noise levels as that from Cui et al. (2018) and also a similar dependency on nucleotides close to the PAM. However, in addition, we observe a strong negative effect on knockdown efficacy from thymine in the first three positions of the sgRNA (1,2,3), corresponding to the transcription start site (TSS) at the 5’-end of the sgRNA (PAM-distal). This is likely due to inefficient transcriptional initiation from these nucleotides or enhanced degradation. We also observe that a cytosine or guanine in the +8 or +9 position downstream from the PAM negatively impacts knockdown efficiency. It is yet unclear if these effects are specific to B. subtilis or are present in other gram-positive bacteria as well.
Off-target effects are also a concern when designing sgRNAs. Where possible, sgRNAs with high specificity – few off-target hits – should be chosen. Analyses of mismatch sgRNAs suggest that higher binding affinity resulting from high GC percentage results in lower sensitivity to imperfect complementarity between sgRNA and DNA, which could imply that these sgRNAs are more likely to have off-target knockdown effects as well (Hawkins et al., 2020; Jost et al., 2020; Mathis et al., 2021). Cui et al. (2018) furthermore showed that off-target knockdown effects can occur when only 9 nucleotides in the seed sequence of a sgRNA complement a gene sequence.
Finally, Cui et al. (2018) identified the presence of toxic seed sequences that are consistently associated with negative fitness effects in E. coli. These seed sequences comprise five nucleotides at the 3’ end of the sgRNA, proximal to the PAM. Although dCas9 expression levels were shown to affect this bad-seed effect, the mechanistic basis of this toxicity has not been elucidated (Cui et al., 2018). The top 10 bad-seed sequences are: AGGAA, TGACT, ACCCA, AAAGG, GAGGC, CGGAA, ATATG, AACTA, TGGAA, CACTC (Vigouroux and Bikard, 2020).
-
3.In step 2 of the computational pipeline, the list of sgRNAs is filtered based on a number of pre-specified criteria, thereby removing sgRNAs with potentially low efficacy, off-target hits or toxic side-effects. By default, the pipeline uses lenient filter criteria, only removing sgRNAs with more than 10 potential off-target hits and with an offset (distance to the start codon) of more than 1000 nucleotides. One can however use a number of additional filter criteria, including the sgRNA spacer sequence, PAM sequence, downstream sequence and GC content. To run the second step of the computational pipeline, use the following command line:
- >python <path to “generate_sgrnas.py” script>
- --genbank <path to genbank file>
- --locus_tag <locus_tag>
- --step filter
- --sgrna_remove <string of sgRNA sequence(s) to remove>
- --pam_remove <string of PAM sequence(s) to remove>
- --downstream_remove <string of downstream sequence(s) to remove>
- --gc_lower <minimal GC-content sgRNA spacer>
- --gc_upper <maximum GC-content sgRNA spacer>
- --offset_upper <maximum distance to the start codon>
- --off_target_upper <maximum number of off-target hits>
-
a.All filter criteria are optional. When no filter criteria are specified, only the default filter settings will apply. For the filter step, the Python script will load the CSV file generated in the first step of the pipeline (e.g., “sgRNA.csv”). Importantly, filtering only works when this CSV file is present. When an alternative file name is used in the first step (using the --file_find argument), this should also be specified for the filtering step.
-
4.
Like in step 1, a CSV file is also generated in the second step (“sgRNA_filtered.csv”). It contains the same information as the first file, but includes only the sgRNAs that pass the filtering criteria (Figure 2). The first two steps of the computational pipeline can also be used to design sgRNAs that target the same gene(s) across different strains or species. To this end, one should first generate and filter sgRNAs for each individual strain or species, targeting the same orthologous gene(s), and subsequently compare their “sgRNA_filered.csv” output files. By selecting sgRNAs shared across strains/species, one can identify common CRISPRi targets. Note that these targets might not necessarily yield the same knockdown efficacies across strains/species, for example because of differences in the region upstream of the PAM.
Examples
-
•Based on the sequence analysis in Figure 4, in B. subtilis, we expect higher knockdown efficiencies when filtering against: (1) sgRNAs starting with a T, (2) PAM sequences starting with an A, (3) and downstream sequences associated with either a C or G at position +8 or +9. The following command would filter the list of sgRNAs accordingly:
- >python Generate_sgRNAs.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step filter
- --sgRNA_remove TNNNNNNNNNNNNNNNNNNN
- --PAM_remove ANN
- --off_target_upper 5
- --downstream_remove NNNNNNNGNNNNNNN,NNNNNNNNGNNNNNN,NNNNNNNNCNNNNNN,NNNNNNNCNNNNNNN
All sgRNAs that match the sequence of sgrna_remove will be removed; all sgRNAs associated with a PAM sequence matching pam_remove will be removed; and all sgRNAs associated with a downstream sequence matching downstream_remove will be removed.
-
•The following command removes sgRNAs that have a spacer sequence starting with three consecutive Ts or ending with an A:
- >python Generate_sgRNAs.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step filter
- --sgRNA_remove TTTNNNNNNNNNNNNNNNNN,NNNNNNNNNNNNNNNNNNNA
-
•One can also run the find and filter steps in one go:
- >python Generate_sgRNAs.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step find,filter
-
•When you would like to filter sgRNAs based on a downstream nucleotide sequence of only 10 nucleotides (default is 15), one should specify the length of the downstream sequence before specifying the filter criteria:
- >python Generate_sgRNAs.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step find,filter
- --downstream 10
- --downstream_remove NNNNNNNGNN,NNNNNNNNGN,NNNNNNNNCN,NNNNNNNCNN
-
•One can also remove sgRNAs with either a low or high GC content. The following command filters out all sgRNAs with a GC content below 0.4 and above 0.6:
- >python Generate_sgRNAs.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step find,filter
- --gc_lower 0.4
- --gc_upper 0.6
-
•You can also adjust the default filter criteria, for example the following command removes all sgRNAs with more than 5 off-target hits or an offset larger than 200nt:
- >python Generate_sgRNAs.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step find
- --off_target_upper 5
- --offset_upper 200
-
•When needed, it is possible to perform multiple consecutive rounds of filtering by both specifying the input and output files. For example:
-
(1)First round: remove sgRNA starting with T:
- >python Generate_sgRNAs.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step find,filter
- --sgRNA_remove TNNNNNNNNNNNNNNNNNNN
-
(2)Second round: remove sgRNA of PAM sequences that start with A:
- >python Generate_sgRNAs.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step filter
- --PAM_remove ANN
- --file_find sgRNA_filter.csv
- --file_filter sgRNA_filter_new.csv
-
(1)
In the second round of filtering, the filtered sgRNAs from the first step (“sgRNA_filter.csv”) is used as input file (--file_find sgRNA_filter.csv) to generate a new CSV file (“sgRNA_filter_new.csv”; --file_filter sgRNA_filter_new.csv). If the new file is not specified, the old one (“sgRNA_filter.csv”) will be overwritten. Consecutive rounds of filtering can be used to determine how each additional filter criterion prunes the list of sgRNAs.
Step 3: Generate mismatch sgRNAs
Timing: 5 min
In the third and final part of the computational pipeline, mismatch sgRNAs are generated for all sgRNAs that passed the filtering criteria. As detailed by Hawkins et al. (2020), imperfect complementarity between the sgRNA spacer and target DNA sequence can be used to predictably tune sgRNA activity. By examining over 1500 single-nucleotide mismatch sgRNAs in both E. coli and B. subtilis, corresponding to 33 sgRNA spacers targeting gfp, Hawkins et al. (2020) trained a species-independent linear model that predicts knockdown efficacy, based on the location and type of nucleotide mismatch and the GC percentage of the parent sgRNA (E. coli: p<10−16, R2=0.48 and B. subtilis: p<10−16, R2=0.57). PAM proximal mismatches more strongly affect sgRNA activity, while PAM distal mismatches have little effect (Figure 5A). These results corroborate previous findings that demonstrate the importance of the sgRNA seed sequence for binding of the dCas9-sgRNA complex to the DNA (Cui et al., 2018; Hsu et al., 2013; Jost et al., 2020; Mathis et al., 2021; Qi et al., 2013). Besides predicting knockdown efficacy for single mismatched sgRNAs, the predictive model was also successfully applied to predict knockdown efficacies of double mismatched sgRNAs using a multiplicative model, as suggested in Qi et al., 2013. As a next step, Hawkins et al. (2020) generated a library of mismatched sgRNAs targeting the essential genes in E. coli and B. subtilis to examine how graded knockdowns affect fitness. As expected, reduced expression knockdowns, associated with PAM proximal mismatches, were associated with higher relative fitness (Figure 5B).
-
5.The coefficients of the species-independent linear model are stored in the file “model_param.csv” and can be manually updated based on new experimental data. When running the final step of the computational pipeline, both the “sgRNA_filtered.csv” and “model_param.csv” files are loaded to determine the predicted expression knockdowns of all 60 possible sgRNA spacer mismatches per sgRNA. This final step of the Python script does not rely on any additional arguments and can be run using the following command:
- >python <path to “generate_sgrnas.py” script>
- --genbank <path to genbank file>
- --locus_tag <locus_tag>
- --step mismatch
-
6.Like in the previous two steps of the pipeline (Figure 2), a CSV file will be generated, “sgRNA_mismatched.csv”, which shows the complete list of mismatched sgRNAs with their predicted knockdown efficacy. This file contains the following information:
-
a.Index of sgRNA, as shown in sgRNA.csv and sgRNA_filtered.csv, with the index of the associated mismatch in between parenthesis
-
b.Nucleotide position in sgRNA spacer that is substituted
-
c.Original base
-
d.Substituted base
-
e.Sequence of parental (fully complementary) sgRNA spacer
-
f.Sequence of mismatch sgRNA spacer
-
g.Predicted knockdown efficacy: 1 or higher indicating full knockdown (as seen in fully complementary sgRNA) and 0 no knockdown.
-
a.
Figure 5.
Efficacy of mismatch CRISPRi
Effect of single-nucleotide mismatches on (A) knockdown of GFP expression and (B) relative fitness of essential genes. Left panels show how mismatches in different positions of sgRNA affect GFP knockdown and relative fitness in datasets of E. coli and B. subtilis. Middle panels show relation between predicted knockdowns and (A) observed knockdowns in E. coli (p<10−16, R2=0.48) and B. subtilis (p<10−16, R2=0.57) and the (B) relative fitness in E. coli and B. subtilis. Right panels show distribution of observed GFP knockdowns and relative fitness.
Examples
-
•In order to run the mismatch step for our B. subtilis example, use the following command line:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
- --step mismatch
Running the entire pipeline in one go
-
7.Although the computational pipeline can be conducted step-by-step, one can also run the entire pipeline, by leaving out the step argument (default, --step find,filter,mismatch):
- >python <path to “generate_sgrnas.py” script>
- --genbank <path to genbank file>
- --locus_tag <locus_tag>
Examples
-
•In the case of our B. subtilis example, one can generate all sgRNA targeting DnaA (step 1), filter these sgRNA based on default filter criteria (step 2) and generate all mismatches (step 3) using the following command:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag BSU_00010
-
•To generate sgRNA for all genes in the genome, simply add --locus_tag all (e.g., this takes ∼10 minutes on a regular laptop for all 4325 genes of B. subtilis):
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag all
-
•One could also specify filter criteria when running the entire pipeline. As explained above, these filter criteria are applied in the second step of the pipeline:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag all
- --sgrna_remove TNNNNNNNNNNNNNNNNNNN
-
•When you would like to include additional filter criteria, after running the entire pipeline, you can simply redo the last two steps of the pipeline and specify the new output files:
- >python generate_sgrnas.py
- --genbank GCF_000009045.1_ASM904v1_genomic.gbff
- --locus_tag all
- --step filter,mismatch
- --pam_remove ANN
- --offset_upper 200
- --file_filter sgRNA_filtered_new.csv
- --file_mismatch sgRNA_mismatched_new.csv
Expected outcomes
The pipeline should result in three CSV output files: “sgRNA.csv”, containing the full list of sgRNAs; “sgRNA_filtered.csv” containing the filtered list of sgRNAs; “sgRNA_mismatched.csv”, containing all single-nucleotide mismatches and the predicted knockdown efficacies (Figure 2 and Table 2). Depending on the GC content, the number of available PAM sequences and thus the number of sgRNA targets will differ between organisms. For example, B. subtilis (43.5% GC content; Kunst et al., 1997) has a median of 25 sgRNA targets per gene (median±MAD=25±20.8). 0.5% of the genes (23/4325) have no sgRNA target. For comparison, in Escherichia coli K-12 MG1655 (50.8% GC content; Blattner et al., 1997), there is a median of 41 sgRNA targets per gene (median±MAD=41±31.1) and also 0.5% of the genes cannot be targeted (23/4357). Although dCas9-mediated CRISPRi has successfully been applied to a wide range of species (Todor et al., 2021), for organisms with low GC content the number of available PAM sequences could be limiting. For those organisms, employing CRISPR-Cas systems that target alternative PAM sequences might be more appropriate (Kleinstiver et al., 2015; Kim et al., 2017).
Limitations
There are several general limitations to mismatch CRISRPi. First, since genes are often part of larger operons, transcriptional interference often leads to polar effects, knocking down other genes in the operon as well (Qi et al., 2013; Peters et al., 2015). Second, knockdown efficacies of mismatch sgRNAs can only be predicted with limited accuracy (Figure 5; Hawkins et al., 2020; Jost et al., 2020; Mathis et al., 2021). Similarly, although we can filter against potentially ineffective or bad sgRNAs, we cannot entirely exclude them (Cui et al., 2018). We therefore advise designing multiple fully complementary and mismatch sgRNAs per gene, which can serve as internal controls. Third, the predicted knockdown efficacies may in part depend on our genetic implementation of the CRISPRi system (Hawkins et al., 2020). In our case, sgRNAs are expressed from a constitutive promoter, while dCas9 is expressed using a xylose-inducible promoter. For alternative genetic constructs, predicted efficacies might require additional verification. Finally, dCas9 is often costly to express and, especially when targeting essential genes, suppressors might rapidly appear, either lowering dCas9 expression or compensating for essential gene knockdowns (Mathis et al., 2021).
Troubleshooting
Various error messages can appear while running the computational pipeline. Here we provide a brief list of errors and explanations on how to overcome them:
-
1.python: can't open file 'script.py': [Errno 2] No such file or directory
- (step 1 error) The path to the Python script is incorrectly specified, making it impossible for python to run the code.
-
2.FileNotFoundError: [Errno 2] No such file or directory: <path to GenBank file>
- (step 1 error) The path to the GenBank file is incorrectly specified, making it impossible to load the GenBank file in the computational pipeline.
-
3.ValueError: args.pam_remove value 0 was NNN which filtered out all sgRNAs.
- (step 2 error) Wrong filter setting for the PAM sequence (--pam_remove NNN), resulting in filtering out all sgRNAs.
-
4.ValueError: args.pam_remove value 0 was NGN which contained a 'G' in 2nd or 3rd position.
- (step 2 error) Wrong filter setting for the PAM sequence (--pam_remove NGN), resulting in filtering out all sgRNAs.
-
5.ValueError: args.downstream_remove included G which is not 15 nt long.
- (step 2 error) Wrong filter setting for downstream sequence (--downstream_remove G), which is too short given the default setting of --downstream 15.
-
6.generate_sgrnas.py: error: unrecognized arguments: ANNNNNNNNNNNNNNNNNNN
- (step 2 error) When specifying multiple sequences for the --sgrna_remove filter, make sure to separate them by a comma without spaces. When having a space (--sgrna_remove GNNNNNNNNNNNNNNNNNNN, ANNNNNNNNNNNNNNNNNNN) the above error appears.
-
7.Empty CSV files
- (step 1 or 2 error) When all CSV files are empty, there can be three possible problems: (i) there might be no sgRNAs targeting the gene(s) of interest, (ii) the wrong locus tag(s) might have been specified (--locus_tag) or (iii) a wrong GenBank file was provided. Please make sure the GenBank file contains both genome features (CDS, gene names, locus tags, etc.) and sequence data (i.e., full nucleotide sequences of all contigs). One can open the GenBank file in any regular text editor to confirm both types of information are included. When only genome features are included, all CSV files will be empty. When only the “sgRNA_filtered.csv” and “sgRNA_mismatched.csv” are empty, it means that the specified filter criteria are too stringent and none of the sgRNAs passes the filter criteria.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Carol A. Gross (cgrossucsf@gmail.com).
Material availability
Not applicable
Data and code availability
Code can be downloaded from https://github.com/jordivangestel/sgRNAs-for-mismatch-CRISPRi
Acknowledgments
J.v.G. is supported by a Swiss National Science Foundation Postdoc.Mobility fellowship (P400PB_186789). C.A.G. is supported by the National Institutes of Health (R35 GM118061) and the Innovative Genomics Institute, UC Berkeley.
Author contributions
J.v.G. and J.S.H. wrote the Python script. J.v.G. made figures and wrote the first version of the draft. H.T., J.S.H., and C.A.G. edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Jordi van Gestel, Email: jordivangestel@gmail.com.
Horia Todor, Email: horia.todor@gmail.com.
Carol A. Gross, Email: cgrossucsf@gmail.com.
References
- Blattner F.R., Plunkett G., Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Boyle E.A., Andreasson J.O.L., Chircus L.M., Sternberg S.H., Wu M.J., Guegler C.K., Doudna J.A., Greenleaf W.J. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. PNAS. 2017;114:5461–5466. doi: 10.1073/pnas.1700557114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Villamañán A., Ng J.W., Planel R., Ménager H., Chen A., Cui L., Bikard D. On-target activity predictions enable improved CRISPR–dCas9 screens in bacteria. Nucleic Acids Res. 2020;48:e64. doi: 10.1093/nar/gkaa294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Vigouroux A., Rousset F., Varet H., Khanna V., Bikard D. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 2018;9:1912. doi: 10.1038/s41467-018-04209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J.S., Silvis M.R., Koo B.-M., Peters J.M., Osadnik H., Jost M., Hearne C.C., Weissman J.S., Todor H., Gross C.A. Mismatch-CRISPRi reveals the co-varying expression-fitness relationships of essential genes in Escherichia coli and Bacillus subtilis. Cell Syst. 2020;11:523–535. doi: 10.1016/j.cels.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost M., Santos D.A., Saunders R.A., Horlbeck M.A., Hawkins J.S., Scaria S.M., Norman T.M., Hussmann J.A., Liem C.R., Gross C.A. Titrating gene expression using libraries of systematically attenuated CRISPR guide RNAs. Nat. Biotechnol. 2020;38:355–364. doi: 10.1038/s41587-019-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Kim H., Ahn W.C., Park K.H., Woo E.J., Lee D.H., Lee S.G. Efficient transcriptional gene repression by Type V-A CRISPR-Cpf1 from Eubacterium eligens. ACS Synth. Biol. 2017;6:1273–1282. doi: 10.1021/acssynbio.6b00368. [DOI] [PubMed] [Google Scholar]
- Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.R.J. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Ogasawara N., Moszer I., Albertini A.M., Alloni G., Azevedo V., Bertero M., Bessieres P., Bolotin A., Borchert S. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Liu X., Gallay C., Kjos M., Domenech A., Slager J., van Kessel S.P., Knoops K., Sorg R.A., Zhang J., Veening J. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol. Syst. Biol. 2017;13:931. doi: 10.15252/msb.20167449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A.D., Otto R.M., Reynolds K.A. A simplified strategy for titrating gene expression reveals new relationships between genotype, environment, and bacterial growth. Nucleic Acids Res. 2021;49:e6. doi: 10.1093/nar/gkaa1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M., Silvis M.R., Zhao D., Hawkins J.S., Gross C.A., Qi L.S. Bacterial CRISPR: accomplishments and prospects. Curr. Opin. Microbiol. 2015;27:121–126. doi: 10.1016/j.mib.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M., Colavin A., Shi H., Czarny T.L., Larson M.H., Wong S., Hawkins J.S., Lu C.H.S., Koo B.M., Marta E. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishi H.S., Toro E., Liu H., Wang X., Qi L.S., Arkin A.P. Systematic genome-wide querying of coding and non-coding functional elements in E. coli using CRISPRi. bioRxiv. 2020 doi: 10.1101/2020.03.04.975888. [DOI] [Google Scholar]
- Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todor H., Silvis M.R., Osadnik H., Gross C.A. Bacterial CRISPR screens for gene function. Curr. Opin. Microbiol. 2021;59:102–109. doi: 10.1016/j.mib.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux A., Bikard D. CRISPR tools to control gene expression in bacteria. Microbiol. Mol. Biol. Rev. 2020;84 doi: 10.1128/MMBR.00077-19. e00077–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics. 2017;33:3645–3647. doi: 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Code can be downloaded from https://github.com/jordivangestel/sgRNAs-for-mismatch-CRISPRi