Abstract
Leptospirosis is a zoonosis caused by pathogenic Leptospira spp. Cats have been reported to be infected with Leptospira spp. and shed the bacteria in the urine. However, the importance of cats as an infection source for humans remains unclear. In this study, Leptospira infection in cats in Okinawa Prefecture, Japan, where leptospirosis is endemic, was investigated by leptospiral antibody and DNA detection using microscopic agglutination test and nested PCR, respectively. Moreover, multilocus sequence typing (MLST) and whole genome sequencing (WGS) were conducted on the Leptospira borgpetersenii serogroup Javanica isolated from cats, black rats, a mongoose, and humans. Anti-Leptospira antibodies were detected in 16.6% (40/241) of the cats tested, and the predominant reactive serogroup was Javanica. The leptospiral flaB gene was detected in 7.1% (3/42) of cat urine samples, and their sequences were identical and identified as L. borgpetersenii. MLST and WGS revealed the genetic relatedness of L. borgpetersenii serogroup Javanica isolates. This study indicated that most seropositive cats had antibodies against the serogroup Javanica and that cats excreted L. borgpetersenii in the urine after infection. Further, genetic relatedness between cat and human isolates suggests that cats may be a maintenance host for L. borgpetersenii serogroup Javanica and a source for human infection.
Subject terms: Bacterial infection, Infectious-disease epidemiology
Introduction
Leptospirosis is a zoonotic disease caused by infection with pathogenic spirochetes of the genus Leptospira, composed of 64 species divided into 24 serogroups and more than 300 serovars1–4. Leptospira spp. colonize the proximal renal tubules of maintenance hosts, including wild animals such as rats and boars, livestock such as cattle and pigs, and companion animals such as dogs, and are shed in their urine2,5,6. Humans are infected percutaneously or permucosally with Leptospira spp. by direct contact with the urine of maintenance hosts or by indirect contact with soil or water contaminated with infected urine3,6,7.
Cats have not been considered an important source of Leptospira infection for humans. Besides antibody detection, however, several recent studies from various regions reported that Leptospira spp. were isolated or leptospiral DNAs were detected from cat urine or kidney samples8–21; carriage rate was related to some factors such as rearing style (cat external behavior), climate, and living environment (urban or rural)15,20. Moreover, leptospiral DNA was continuously detected in the urine of naturally infected cats for 8 months20. Although symptomatic cases with polyuria, polydipsia, hematuria, ascites, and diarrhea have been reported, the clinical presentation of leptospirosis in cats is rare, usually mild, or subclinical, and symptoms in feline leptospirosis remain undefined9,13,14,17,20,22. These studies suggest that cats can carry and shed pathogenic Leptospira in urine for a long period after infection. Asymptomatic or uncertain symptoms of infected cats make the diagnosis and appropriate treatment difficult and prevent infected cats from becoming a chronic carrier, resulting in a potential infection source for humans.
Cats are the most common companion animal in Japan. According to a survey conducted by the Japan Pet Food Association, the number of cats has been increasing year by year, and the number of cats raised in Japan is estimated to be 9,778,000 in 2019, higher than that of dogs23. Canine leptospirosis and its causative Leptospira spp. have been recently reported24,25, but the current status of Leptospira infection in cats in Japan remains limited. In the southern Kyushu District, the prevalence of anti-Leptospira antibodies in domestic cats was reported to be 7.7%26. In Okinawa Prefecture, the southernmost part of Japan, where leptospirosis is endemic, it was reported 30 years ago that the seroprevalence and isolation rate of Leptospira spp. among cats ranged from 4.8 to 9.1% and 1.0% to 3.1%, respectively27,28. Although Leptospira spp. were isolated, molecular characterization of the isolates was not performed in these studies.
Understanding the Leptospira genotype-host association in maintenance hosts is important for elucidating and controlling the source for human infection. Currently, molecular typing methods, such as multilocus sequence typing (MLST) and multilocus variable-number tandem repeat analysis (MLVA), are the main methods for characterizing Leptospira isolates1. As a genotyping method having higher resolution, whole genome sequencing (WGS) has been rapidly developed in recent years. These methods have enabled the understanding of host specificity of certain Leptospira genotypes as well as the geographic structuring of genetic diversity and host switching event in Leptospira spp29–32.
In this study, to clarify Leptospira infection and carriage in cats in Okinawa Island (Okinawa Main Island), anti-leptospiral antibodies in cats were investigated by the microscopic agglutination test (MAT) using 13 reference strains. Leptospiral DNA was detected from cat urine samples by nested PCR. Moreover, MLST and WGS were performed to gain further insights into the genetic relatedness of Leptospira isolates from cats, mongooses, rats, and humans.
Results
Antibodies against Leptospira spp. were detected in 40 of 241 cats (16.6%) included in the analysis. Thirty-seven cats and one cat had antibodies against serogroups Javanica and Hebdomadis, respectively, and two samples were positive for multiple serogroups (Table 1). The reciprocal antibody titers for serogroup Javanica ranged from 160 to 2560 (Table 1). The antibody-positive samples in the northern, central, and southern regions of Okinawa Island were 15.0% (26/173), 22.0% (9/41), and 18.5% (5/27), respectively. Of the 40 anti-leptospiral antibody-positive cats, 23 were male (19.3%) and 17 were female (13.9%). There were no significant differences between seropositivity and capture area nor between seropositivity and sex. In contrast, the seropositivity varied among the age groups based on weight: 3% (1/33) in kittens, 12.1% (7/58) in juveniles, and 21.3% (32/150) in adults. There was a statistically significant difference in the seropositivity among the age groups (p = 0.021), and a trend toward an increase in seropositivity with age was observed (p = 0.006).
Table 1.
Frequency of anti-Leptospira antibodies in cats.
| Reciprocal antibody titer | 160 | 320 | 640 | 1280 | 2560 | No. of positives |
|---|---|---|---|---|---|---|
| Serogroup | ||||||
| Javanica | 13 | 7 | 7 | 6 | 4 | 37 |
| Hebdomadis | 0 | 0 | 1 | 0 | 0 | 1 |
| Multiple | (2†,‡) | (1†) | (1‡) | 2 | ||
†The cat FS13001 exhibited the reciprocal MAT titer 160 and 640 for serogroups Ballum and Javanica, respectively.
‡The cat FS18013 exhibited the reciprocal MAT titer 160 and 1280 for serogroups Autumnalis and Hebdomadis, respectively.
MAT, microscopic agglutination test.
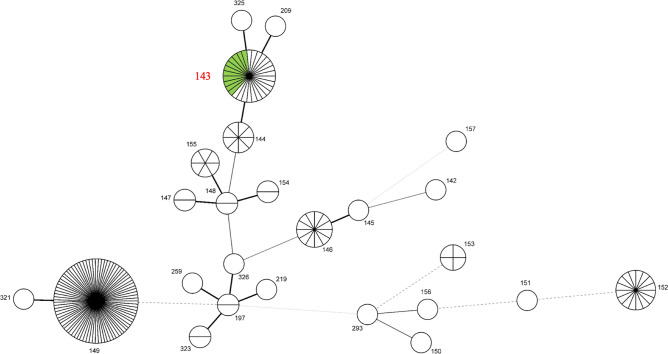
Leptospiral flaB was detected in 3 of 42 urine samples (7.1%, see Supplementary Fig. S1 online) included in the analysis. All three PCR-positive cats were also positive for antibody against serogroup Javanica with the titers of 320 (2 animals) and 640 (1 animal). All three flaB sequences were identical and identified as L. borgpetersenii (DDBJ accession numbers LC596932–LC596934). Amplification of all seven housekeeping genes for MLST was succeeded in two of the three flaB-positive urine samples (FU18017 and FU18027) and nine strains of L. borgpetersenii serogroup Javanica (Table 2). Their sequence types (STs) were all assigned as ST143 (Fig. 1). There were no gross abnormalities in the autopsy findings in the three cats.
Table 2.
L. borgpetersenii serogroup Javanica strains used for MLST and/or WGS.
| Strain | Animal from which Leptospira strain was isolated | Year of isolation | Analytical method |
|---|---|---|---|
| FK-118 | Cat | 1989 | MLST/WGS |
| F-208 | Cat | 1990 | MLST/WGS |
| OHJ2008-88U | Small Indian mongoose | 2008 | MLST/WGS |
| OR2010-56 | Black rat | 2010 | MLST |
| OR2010-61 | Black rat | 2010 | MLST |
| OR2011-1 | Black rat | 2011 | MLST |
| OP104 | Human | 2000 | MLST/WGS |
| 058031 | Human | 2005 | MLST/WGS |
| 078065 | Human | 2007 | MLST/WGS |
MLST, multilocus sequence typing; WGS, whole genome sequencing.
Figure 1.
MST of L. borgpetersenii detected from cat urine samples and L. borgpetersenii serogroup Javanica strains in this study and 182 L. borgpetersenii strains based on the allelic profiles of the MLST seven housekeeping genes. Each circle represents an individual ST, and the numbers represent ST numbers. Circle sizes correspond to the numbers of strains in each ST. The thickness and the dotting of lines indicate the distance between the circles: a thicker line indicates a closer distance than a thin line, and a thin line denotes a closer distance than a dotted line. The green-colored circle and the ST number in red represent L. borgpetersenii serogroup Javanica strains and DNAs determined in this study.
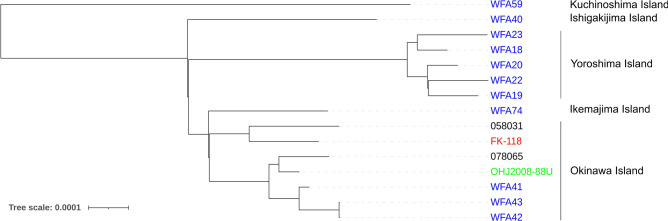
Of the six strains subjected to WGS, enough data for comparison were obtained from four strains, OHJ2008-88U, FK-118, 058031, and 078065. Strains isolated from a cat, a mongoose, and humans were clustered with L. borgpetersenii serogroup Javanica strains isolated from black rats in Okinawa Island sequenced in a previous study30 (Fig. 2).
Figure 2.
Core genome SNP-based maximum likelihood tree of L. borgpetersenii serogroup Javanica strains isolated in Japan. The strain Piyasena, belonging to serogroup Javanica serovar Ceylonica, was used as the reference, omitted from the tree. WGS of strains 058031, 078065, FK-118, and OHJ2008-88U was performed in this study, and strains named WFA have been analyzed in a previous study30. Black, human isolate; blue, black rat isolate; green, mongoose isolate; red, cat isolate.
Discussion
Identification of prevalent serovar(s) and their maintenance host(s) and seroprevalence in the host(s) is important for understanding the epidemiology of leptospirosis in a particular geographic region33. In the Okinawa Island, which has a subtropical climate, Leptospira spp. have been isolated or detected from various animals such as rat, mouse, shrew, mongoose, wild boar, and dog24,34–36. The animals excrete the bacteria into the environment where Leptospira spp. can survive and remain infective for several weeks7. Humans can get infected with Leptospira spp. through contact with the contaminated environment during recreational activities in rivers and agricultural activities37. Therefore, the approach of “One Health” recognizing the interconnection between humans, animals, and their shared environment is important to elucidate the epidemiology of leptospirosis in this island. In this study, we demonstrated that free-roaming stray cats in Okinawa Island were predominantly infected with L. borgpetersenii serogroup Javanica and shed the bacterium in urine, suggesting a potential role of cats in transmitting leptospirosis to humans.
The prevalence of anti-Leptospira antibodies and the detection of leptospiral DNA among cats worldwide range from 4 to 33% and 0% to 67.8%, respectively9–16,18–21. In this study, the seropositivity was 16.6%, and more than 90% of the cat serum samples reacted with serogroup Javanica (Table 1). Pathogenic Leptospira DNA was detected in three cat urine samples (7.1%, 3/42). The seropositivity in this study was slightly higher than that in previous studies done in Okinawa Prefecture (mainly in the Okinawa Island) 30 years ago (4.8% and 9.1%), but reactive serogroups were much diverse in the previous studies27,28. Previous studies reported that Leptospira spp. were isolated from 1.0% and 3.1% of the cats tested, lower than the DNA detection in this study, but success in isolation is generally lower than that in DNA detection. The positive rate of anti-leptospiral antibodies and leptospiral DNA detection in cats could be affected by various factors, such as age, rearing styles, season, geographical region, presence of maintenance hosts, panel of serovars, and/or cutoff value used for MAT, sensitivity, and specificity of the primer (and probe) set used for PCR9,15,16,38, which may be true for this study, and primer sets for other genes may be able to detect leptospiral DNA in more samples. This study employed reciprocal MAT titer 160 as the cutoff value because MAT titer 100 is generally accepted to indicate a previous infection3,5. Under this definition, there was no significant association between seropositivity and capture area in Okinawa Island. A previous report indicated that the prevalence of anti-leptospiral antibodies in cats was higher in rural areas than in urban areas9. The southern part of Okinawa Island is an urban area, and it changes to rural areas as it moves northward. In addition, human leptospirosis is often reported in the northern area due to recreational activities in rivers37. These facts suggest no regional difference in the risk of leptospiral infection in cats in Okinawa Island. The previous study mentioned above included only domestic cats9, whereas this study included free-roaming, stray cats. Stray cats have more risk of contact with maintenance hosts than pet cats, even in urban areas, resulting in discrepant results between the two studies. In addition, as more than 90% of seropositive cats had antibodies against serogroup Javanica, the transmission of L. borgpetersenii serogroup Javanica may easily occur between cats. A significant association between seropositivity and cat age groups based on weight was shown in this study (p = 0.021), and seropositivity tended to increase with age (p = 0.006). This is consistent with the previous report describing that older cats had higher seroprevalence due to an increased opportunity for exposure to the source of infection16.
Each serovar tends to be maintained in specific animal species: host-maintained infections of global importance are Icterohaemorrhagiae in the brown rat, Hardjo in cattle and sheep, Canicola in dogs, and Bratislava in pigs5. Infections with several serovars/serogroups have been identified in cats, such as Australis, Autumnalis, Ballum, Bataviae, Bratislava, Canicola, Copenhageni, Cynopteri, Grippotyphosa, Hardjo, Icterohaemorrhagiae, Javanica, Panama, Pomona, Pyrogenes, Rachmati, or Shermani9,12,14–16,18–20. A serological survey revealed that cats were predominantly infected with serogroup Javanica in Okinawa Island (Table 1). Serogroup Javanica has also been isolated from black rats and mongooses in the northern part of Okinawa Island34. MLST revealed that L. borgpetersenii detected in urine samples and isolated from cats, black rats, a mongoose, and humans all belonged to ST143. Although ST143 has been isolated from mongooses, L. interrogans serogroup Hebdomadis is the predominant strain isolated from mongooses34. Since cats are carnivorous and avoid water, they are more likely to be infected by rat predation than by waterborne infections39. These facts suggest an infection cycle between cats and rats in Okinawa Island. Monitoring acute leptospirosis in dogs as sentinels is suggested to aid in estimating the risk to humans in specific areas40. Cats may also act as sentinels, but their clinical manifestations seem to be less apparent than those of dogs9,13,14,22 and their implications may need further verification.
Conversely, all urine PCR-positive cats showed high antibody titer against serogroup Javanica, indicating that they shed leptospires in urine for some period after infection. Therefore, infected stray cats contaminate the environment and can be a source for infection to humans. In this study, more than 90% of seropositive cats had antibodies against serogroup Javanica (Table 1), although a variety of Leptospira serogroups/serovars exist on the island24,34–37. In cases of leptospirosis in humans on this island during 2007–2016, Hebdomadis was the most frequently detected serogroup (40.1%, 57/142), whereas Javanica was rare (2.8%, 4/142)37. Urine PCR-positive DNA samples showed the presence of L. borgpetersenii ST143, which was the same ST with cat isolates on this island; furthermore, the serogroup of these isolates was Javanica (Table 2). It has been reported that naturally infected cats subclinically shed leptospires (leptospiral DNAs) for 8 months after infection20. These results suggest that this genotype of L. borgpetersenii serogroup Javanica can be easily transmitted among cats and that cats may act as their maintenance host. Antibodies against serogroup Javanica were also detected from cats in Taiwan12. Although no genetic information on serogroup Javanica strain from cats was obtained, the same L. borgpetersenii serogroup Javanica genotype has been isolated from rats in Taiwan and in other Asian countries, such as China, Indonesia, Laos, Thailand, and Sri Lanka30 (PubMLST; https://pubmlst.org/organisms/leptospira-spp). These facts suggest that cats may carry L. borgpetersenii serogroup Javanica in other Asian countries.
In addition to MLST, WGS revealed that L. borgpetersenii serogroup Javanica isolated from a cat, a mongoose, black rats, and humans in Okinawa Island belonged to the same cluster, supporting their genetic relatedness and the geographic structuring of genetic diversity of Leptospira species as with the previous studies30,31 (Fig. 2). The previous report indicated that this genotype of L. borgpetersenii serogroup Javanica could infect various rodent species, suggesting that they are a generalist pathogen30. Moreover, this study supports this L. borgpetersenii serogroup Javanica as a generalist, as they can colonize the kidney tissues of cats and mongooses. In this study, only one cat and mongoose succeeded in WGS. WGS of more cat and mongoose isolates could identify animal species-specific characteristics, which may gain new insights into the mechanism of renal colonization and evolution of Leptospira spp. in different animals. It could also identify the precise animal source for human infection.
In conclusion, this study reports that cats are commonly infected with and excrete L. borgpetersenii serogroup Javanica that are genetically closely related to those isolated from black rats, mongooses, and humans in Okinawa Island, Japan. Although genetic relatedness suggests that black rats and mongooses are the source of infection for cats, a high proportion of serogroup Javanica infection and urinary excretion of L. borgpetersenii after infection also suggest that cats may be a maintenance host of L. borgpetersenii serogroup Javanica and the source for human infection.
Methods
Sample collection
There were 241 serum samples and 42 urine samples collected in Okinawa Island, the main island of Okinawa Prefecture, Japan. Of these, 121 serum samples and 42 urine samples were collected from free-roaming, stray cats captured/accommodated at the Okinawa Prefectural Animal Protection and Control Center from June 2012 to November 2018, based on the Act on Welfare and Management of Animals. Cats were euthanized by carbon dioxide gas inhalation under the Act, not for this study, and all methods were performed in accordance with the American Veterinary Medical Association guidelines. Autopsy findings, body weight, sex, and capture area were recorded. Blood was collected in a serum separation tube by cardiocentesis and centrifuged at 1710 × g for 15 min to separate the serum. Urine was aseptically collected directly from the bladder using a syringe. Blood and urine collection from euthanized cats was conducted with permission from the Okinawa Prefectural Animal Protection and Control Center.
The other 120 serum samples were derived from the residual blood collected from free-roaming, stray cats during the free-roaming neutering program in the northern part of Okinawa Island from 2016 to 2018 carried out by a nonprofit organization. Serum was separated as described above, and the body weight, sex, and capture area of cats were recorded.
The study was carried out in compliance with the ARRIVE guidelines (https://arriveguidelines.org/).
Antibody detection from cats
To detect anti-Leptospira antibodies in serum samples, MAT was performed using 13 reference strains of serogroups: Australis (serovar Australis), Autumnalis (Autumnalis and Rachmati), Ballum (Castellonis), Bataviae (Bataviae), Canicola (Canicola), Grippotyphosa (Grippotyphosa), Hebdomadis (Hebdomadis), Icterohaemorrhagiae (Icterohaemorrhagiae), Javanica (Javanica), Pomona (Pomona), Pyrogenes (Pyrogenes), and Sejroe (Hardjo). These reference strains were cultivated in Ellinghausen-McCullough-Johnson-Harris medium at 30°C7. Twenty-five microliters of twofold serially diluted serum samples [1:80–1:5120 by phosphate-buffered saline] were incubated with the same volume of leptospiral cultures for 3 h at 30 °C. The endpoint was determined by ≥ 50% decrease of free, unagglutinated leptospires compared with the control suspension3. Reciprocal MAT titer 160 was used for the cutoff antibody titer.
DNA detection from cat urine samples
DNA was extracted from 200 µL urine using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) and subjected to nested PCR targeting flaB for the pathogenic Leptospira spp. First, 5 µl of extracted DNA were used for the first PCR using the primer set L-flaB-F1 5′-CTCACCGTTCTCTAAAGTTCAAC-3′ and L-flaB-R1 5′-TGAATTCGGTTTCATATTTGCC-3′ in a 50 µl reaction volume. Then, 1 µl of the first PCR product was added to 19 µl of the second PCR mixture with the primer set L-flaB-F2 5′-TGTGGACAAGACGATGAAAGC-3′ and L-flaB-R2 5′-AACATTGCCGTACCACTCTG-3′. The positive first PCR samples (FU18017, FU18027, and FU18028) were subjected to DNA sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA).
MLST
MLST was performed for the flaB-positive DNA samples (FU18017, FU18027, and FU18028) and DNA samples extracted from nine strains of Leptospira borgpetersenii serogroup Javanica isolated from cats, a mongoose, black rats, and humans using the QIAamp DNA Blood Mini Kit, which were stored at − 80 °C at the Okinawa Prefectural Institute of Health and Environment27,28 (Table 2). MLST using seven housekeeping genes (glmU, pntA, sucA, tpiA, pfkB, mreA, and caiB) for the isolates was performed as previously described40. MLST for the flaB-positive DNAs, FU18017 and FU18028, was performed via nested PCR as previously described42. For FU18027, since five of the seven genes were not amplified using the original primer sets41,42, new primer sets were designed based on L. borgpetersenii sequences (accession numbers CP000350, CP012029, CP015044, CP015046, CP015048, CP015050, CP015052, CP015814, CP021412, CP026671, and CP033440) as described in Table 3. glmU, sucA, tpiA, pfkB, and caiB were amplified by nested PCR while pntA and mreA were amplified by the first PCR alone. Nucleotide sequences of the amplicons were determined using the BigDye Terminator v3.1 Cycle Sequencing Kit. The concatenated sequences were aligned in MEGA10, and STs were assigned through the MLST database (https://pubmlst.org/organisms/leptospira-spp). A minimum spanning tree (MST) based on the allelic profiles determined in this study and those of 182 L. borgpetersenii strains registered in the MLST database was created using BioNumerics Software (version 7.6; Applied-Maths, Sint Maartens-Latem, Belgium) with default settings (MST for categorical data).
Table 3.
MLST primer sets for Leptospira borgpetersenii clone FU18027.
| Locus | PCR | Primer name | Sequence (5′–3′) | References |
|---|---|---|---|---|
| glmU | 1st | glmU-FM | AGGATAAGGTCGCTGTGGTA | 41 |
| 1st | glmU-RM | AGTTTTTTTCCGGAGTTTCT | 41 | |
| 2nd | 1-glmU-2F_M13 | TGTAAAACGACGGCCAGTCGYATGAAAACGGATCAG | 42 | |
| 2nd | 1-glmU-2R_M13 | CAGGAAACAGCTATGACCGGAAGRTARTATTCDCCCTG | 42 | |
| pntA | 1st | pntA-F-borg | GCCGGAGCAAATCTTGTATC | This study |
| 1st | pntA-R-borg | TGACCGATTACCGTTACCCC | This study | |
| sucA | 1st | sucA-FM | TCATTCCACTTYTAGATACGAT | 41 |
| 1st | sucA-RM | TCTTTTTTGAATTTTTGACG | 41 | |
| 2nd | 3-sucA-2F_M13 | TGTAAAACGACGGCCAGTGCSGGTRATCATCWBATGG | 42 | |
| 2nd | 3-sucA-2R_M13 | CAGGAAACAGCTATGACCGRAAWCCYTTYGCAAGATC | 42 | |
| tpiA | 1st | tpiA-F-borg | AAATCGCATGCGCAAAACGG | This study |
| 1st | tpiA-R-borg | GAGCGCTTGTATGTTATCCG | This study | |
| 2nd | tpiA-F2-borg | CGCCGGAAACTGGAAAATGA | This study | |
| 2nd | tpiA-R2-borg | TTCCGCAATTTCTTTCGCGC | This study | |
| pfkB | 1st | pfkB-F-borg | CCGGGAAGGTTTCCAAAGAC | This study |
| 1st | pfkB-R-borg | TAAAACCGTGGGTCAGTCCG | This study | |
| 2nd | pfkB-F2-borg | GGAAAACGCCGGAATCCTTT | This study | |
| 2nd | pfkB-RM | AGAACACCCGCCGCAAAACAAT | 41 | |
| mreA | 1st | mreA-F-borg | GGTGGAAAGATATGGCTCGC | This study |
| 1st | mreA-R-borg | TTCCTATCGCGGTCATGGAC | This study | |
| caiB | 1st | caiB-F | CAACTTGCGGAYATAGGAGGAG | 41 |
| 1st | caiB-R-borg | TCCGAGAGATCGGTAAATCG | This study | |
| 2nd | 7-caiB-2F_M13 | TGTAAAACGACGGCCAGTCTTKCTTCRATYTTGGCG | 42 | |
| 2nd | 7-caiB-2R_M13 | CAGGAAACAGCTATGACCAMCGATATGTWAYMGGRGTT | 42 | |
| glmU | 1st | glmU-FM | AGGATAAGGTCGCTGTGGTA | 41 |
| 1st | glmU-RM | AGTTTTTTTCCGGAGTTTCT | 41 | |
| 2nd | 1-glmU-2F_M13 | TGTAAAACGACGGCCAGTCGYATGAAAACGGATCAG | 42 | |
| 2nd | 1-glmU-2R_M13 | CAGGAAACAGCTATGACCGGAAGRTARTATTCDCCCTG | 42 |
MLST, multilocus sequence typing.
WGS
Genomic DNA from the six strains in Table 2 was prepared as described above. Genomic DNA libraries were prepared using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions and sequenced on MiSeq (Illumina) with 300 bp paired-end reads. Core genome single nucleotide variants (SNVs) were extracted using BactSNP v.1.1.03743 with the genome of L. borgpetersenii serogroup Javanica serovar Ceylonica strain Piyasena as the reference (GenBank accession no. CP026671.1 and CP026672.1). For phylogenetic analysis, SNVs in the recombinogenic regions detected using Gubbins version 2.3.444 and those in the repetitive regions of the Piyasena genome identified using MUMmer v.3.225945 were excluded. Phylogenetic relationships were determined by reconstructing a phylogenetic tree via the maximum likelihood method using IQ-TREE46 with 1000 ultrafast bootstrap replicates. The data have been deposited with links to BioProject accession number PRJDB10861 in the DDBJ BioProject database.
Statistical methods
To define the capture areas, the island was divided into northern, central, and southern areas. Cats were categorized according to three age groups based on weight as previously described 47: male: kitten, < 1.0 kg; juvenile, 1–2.4 kg; and adult, ≥ 2.5 kg and female: kitten, < 1.0 kg; juvenile, 1–1.9 kg; and adult, ≥ 2.0 kg. Associations of seropositivity with capture area, sex, and age based on weight were analyzed using χ2 test or 2 × 3 Fisher’s exact test and χ2 test for trend.
Ethics declarations
No ethical approval was required as the samples were collected from cats sacrificed under an act or derived from residual blood from health examinations.
Supplementary Information
Acknowledgements
We thank Okinawa Prefectural Animal Protection and Control Center and Okinawa Wildlife Federation for collecting the samples. We are grateful to Kanako Oba for her technical assistance. This work was supported by the Research Program on Emerging and Re-emerging Infectious Diseases (JP21fk0108139) from the Japan Agency for Medical Research and Development (AMED) (N.K.).
Author contributions
T.K. and N.K. conceived and designed the study. T.K., Y.K., S.O., M.M., and N.K. conducted the experiments. T.K., M.M., and N.K. analyzed the data. H.K. supervised the study. T.K., M.M., and N.K. wrote the manuscript. All authors read and approved the final manuscript.
Data availability
The flaB sequences have been deposited in a public database (DDBJ accession numbers LC596932–LC596934). The WGS data have been deposited with links to BioProject accession number PRJDB10861 in the DDBJ BioProject database.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89872-3.
References
- 1.Cerqueira GM, Picardeau M. A century of Leptospira strain typing. Infect. Genet. Evol. 2009;9:760–768. doi: 10.1016/j.meegid.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Guglielmini J, et al. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019;13:e0007374. doi: 10.1371/journal.pntd.0007374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levett PN. Leptospirosis. Clin. Microbiol. Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vincent AT, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019;13:e0007270. doi: 10.1371/journal.pntd.0007270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis WA. Animal leptospirosis. In: Adler B, editor. Leptospira and Leptospirosis. Berlin: Springer; 2015. pp. 99–137. [Google Scholar]
- 6.Haake DA, Levett PN. Leptospirosis in humans. In: Adler B, editor. Leptospira and Leptospirosis. Berlin: Springer; 2015. pp. 65–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faine S, Adler B, Bolin C, Perolat P. Leptospira and Leptospirosis. 2. Amsterdam: MediSci; 1999. [Google Scholar]
- 8.Alashraf AR, et al. First report of pathogenic Leptospira spp. isolated from urine and kidneys of naturally infected cats. PLoS ONE. 2020;15:e0230048. doi: 10.1371/journal.pone.0230048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azócar-Aedo L, Monti G, Jara R. Leptospira spp. in domestic cats from different environments: prevalence of antibodies and risk factors associated with the seropositivity. Animals. 2014;4:612–626. doi: 10.3390/ani4040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dybing NA, Jacobson C, Irwin P, Algar D, Adams PJ. Leptospira species in feral cats and black rats from Western Australia and Christmas Island. Vector Borne Zoonotic Dis. 2017;17:319–324. doi: 10.1089/vbz.2016.1992. [DOI] [PubMed] [Google Scholar]
- 11.Gomard Y, et al. Feral cats do not play a major role in leptospirosis epidemiology on Reunion Island. Epidemiol. Infect. 2019;147:e97. doi: 10.1017/S0950268819000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan K-W, et al. Serological and PCR detection of feline Leptospira in Southern Taiwan. Vector Borne Zoonotic Dis. 2014;14:118–123. doi: 10.1089/vbz.2013.1324. [DOI] [PubMed] [Google Scholar]
- 13.Lapointe C, Plamondon I, Dunn M. Feline leptospirosis serosurvey from a Quebec referral hospital. Can. Vet. J. 2013;54:497–499. [PMC free article] [PubMed] [Google Scholar]
- 14.Millán J, et al. Leptospirosis in wild and domestic carnivores in natural areas in Andalusia, Spain. Vector Borne Zoonotic Dis. 2009;9:549–554. doi: 10.1089/vbz.2008.0081. [DOI] [PubMed] [Google Scholar]
- 15.Murillo A, et al. Leptospira detection in cats in Spain by serology and molecular techniques. Int. J. Environ. Res. Public Health. 2020;17:1600. doi: 10.3390/ijerph17051600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mylonakis ME, et al. Leptospiral seroepidemiology in a feline hospital population in Greece. Vet. Rec. 2005;156:615–616. doi: 10.1136/vr.156.19.615. [DOI] [PubMed] [Google Scholar]
- 17.Ojeda J, Salgado M, Encina C, Santamaria C, Monti G. Evidence of interspecies transmission of pathogenic Leptospira between livestock and a domestic cat dwelling in a dairy cattle farm. J. Vet. Med. Sci. 2018;80:1305–1308. doi: 10.1292/jvms.16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pratt N, Conan A, Rajeev S. Leptospira seroprevalence in domestic dogs and cats on the Caribbean Island of Saint Kitts. Vet. Med. Int. 2017;2017:5904757. doi: 10.1155/2017/5904757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talebkhan Garoussi M, Mehravaran M, Abdollahpour G, Khoshnegah J. Seroprevalence of leptospiral infection in feline population in urban and dairy cattle herds in Mashhad, Iran. Vet. Res. Forum. 2015;6:301–304. [PMC free article] [PubMed] [Google Scholar]
- 20.Weis S, et al. Detection of Leptospira DNA in urine and presence of specific antibodies in outdoor cats in Germany. J. Feline Med. Surg. 2017;19:470–476. doi: 10.1177/1098612X16634389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaidi S, et al. Urinary shedding of pathogenic Leptospira in stray dogs and cats, Algiers: a prospective study. PLoS ONE. 2018;13:1–11. doi: 10.1371/journal.pone.0197068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murillo A, Goris M, Ahmed A, Cuenca R, Pastor J. Leptospirosis in cats: Current literature review to guide diagnosis and management. J. Feline Med. Surg. 2020;22:216–228. doi: 10.1177/1098612X20903601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Japan Pet Food Association. The dog and cat breeding fact-finding survey in Japan. Retrieved from https://petfood.or.jp/data/chart2019/index.html (2019).
- 24.Koizumi N, et al. Molecular and serological investigation of Leptospira and leptospirosis in dogs in Japan. J. Med. Microbiol. 2013;62:630–636. doi: 10.1099/jmm.0.050039-0. [DOI] [PubMed] [Google Scholar]
- 25.Koizumi N, Muto MM, Izumiya H, Suzuki M, Ohnishi M. Multiple-locus variable-number tandem repeat analysis and clinical characterization of Leptospira interrogans canine isolates. J. Med. Microbiol. 2015;64:288–294. doi: 10.1099/jmm.0.000027. [DOI] [PubMed] [Google Scholar]
- 26.Akuzawa M, Maruyama T, Endo Y, Oishi A, Nakamura K. Survey of Leptospira antibodies in domestic cats in the southern Kyushu district. J. Jpn. Vet. Med. Assoc. 2006;59:45–48. doi: 10.12935/jvma1951.59.45. [DOI] [Google Scholar]
- 27.Yonahara Y, Tokumura K, Shingaki Y, Kinjo E, Chiba Y. Investigation on the prevalence of leptospirosis among cats in Okinawa Islands. Ann. Rep. Okinawa Pref. Inst. Public Health. 1990;24:40–45. [Google Scholar]
- 28.Yonahara Y, et al. Investigation on the prevalence of leptospirosis among cats in Okinawa (Continuation) Ann. Rep. Okinawa Pref. Inst. Public Health. 1991;25:33–40. [Google Scholar]
- 29.Dietrich M, et al. Biogeography of Leptospira in wild animal communities inhabiting the insular ecosystem of the western Indian Ocean islands and neighboring Africa. Emergy. Microbes Infect. 2018;7:57. doi: 10.1038/s41426-018-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koizumi N, Wada T, Morita M, Mu J-J, Ohnishi M. Comparative genomic analysis of Leptospira borgpetersenii serogroup Javanica isolated from Rattus species in Southern Japan, Philippines, and Taiwan. Infect. Gen. Evol. 2020;85:104447. doi: 10.1016/j.meegid.2020.104447. [DOI] [PubMed] [Google Scholar]
- 31.Santos LA, et al. Genomic comparison among global isolates of L. interrogans serovars Copenhageni and Icterohaemorrhagiae identified natural genetic variation caused by an indel. Front. Cell. Infect. Microbiol. 2018;8:193. doi: 10.3389/fcimb.2018.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, et al. Genetic characteristics of pathogenic Leptospira in wild small animals and livestock in Jiangxi Province, China, 2002–2015. PLoS Negl. Trop. Dis. 2019;13:e0007513. doi: 10.1371/journal.pntd.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghneim GS, et al. Use of a case-control study and geographic information systems to determine environmental and demographic risk factors for canine leptospirosis. Vet. Res. 2007;38:37–50. doi: 10.1051/vetres:2006043. [DOI] [PubMed] [Google Scholar]
- 34.Ishibashi O, et al. Distribution of Leptospira spp. on the small Asian mongoose and the roof rat inhabiting the northern part of Okinawa Island. Jpn. J. Zoo Wildl. Med. 2006;11:35–41. doi: 10.5686/jjzwm.11.35. [DOI] [Google Scholar]
- 35.Koizumi N, et al. Multiple-locus variable-number tandem repeat analysis of Leptospira interrogans and Leptospira borgpetersenii isolated from small feral and wild mammals in East Asia. Infect. Gen. Evol. 2015;36:434–440. doi: 10.1016/j.meegid.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura M, et al. A survey of leptospiral reservoir animals in Okinawa Prefecture. J. Jpn. Vet. Med. Assoc. 2004;57:321–325. doi: 10.12935/jvma1951.57.321. [DOI] [Google Scholar]
- 37.National Institute of Infectious Diseases and Tuberculosis and Infectious Diseases Control Division, Ministry of Health, Labour and Welfare. Leptospirosis in Japan, January 2007–April 2016. Infect. Agents Surveill. Rep.37, 103–104. https://www0.niid.go.jp/niid/idsc/iasr/37/436.pdf. (2016).
- 38.Rodriguez J, Lapointe C, Arsenault J, Carioto L, Harel J. Serologic and urinary PCR survey of leptospirosis in healthy cats and in cats with kidney disease. J. Vet. Intern. Med. 2014;28:284–293. doi: 10.1111/jvim.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartmann K, et al. Leptospira species infection in cats: ABCD guidelines on prevention and management. J. Feline Med. Surg. 2013;15:576–581. doi: 10.1177/1098612X13489217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martins G, Penna B, Lilenbaum W. The dog in the transmission of human leptospirosis under tropical conditions: victim or villain? Epidemiol. Infect. 2012;140:207–208. doi: 10.1017/S0950268811000276. [DOI] [PubMed] [Google Scholar]
- 41.Boonsilp S, et al. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl. Trop. Dis. 2013;7:e1954. doi: 10.1371/journal.pntd.0001954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss S, et al. An extended multilocus sequence typing (MLST) scheme for rapid direct typing of Leptospira from clinical samples. PLoS Negl. Trop. Dis. 2016;10:1–11. doi: 10.1371/journal.pntd.0004996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshimura D, et al. Evaluation of SNP calling methods for closely related bacterial isolates and a novel high-accuracy pipeline: BactSNP. Microb. Genom. 2019;5:1–8. doi: 10.1099/mgen.0.000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Algar D, Hamilton N, Pink C. Progress in eradicating cats (Felis catus) on Christmas Island to conserve biodiversity. Raffles Bull. Zool. 2014;Suppl. 30:45–53. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The flaB sequences have been deposited in a public database (DDBJ accession numbers LC596932–LC596934). The WGS data have been deposited with links to BioProject accession number PRJDB10861 in the DDBJ BioProject database.