Abstract
Extensive pesticides use is negatively disturbing the environment and humans. Pesticide bioremediation with eco-friendly techniques bears prime importance. This study evaluates the bioremediation of chlorpyrifos in soil using indigenous Bacillus cereus Ct3, isolated from cotton growing soils. Strains were identified through ribotyping (16s rRNA) by Macrogen (Macrogen Inc. Geumchen-gu, South Korea). Bacillus cereus Ct3 was resistant up to 125 mg L−1 of chlorpyrifos and successfully degraded 88% of chlorpyfifos in 8 days at pH 8. Bacillus cereus Ct3 tolerated about 30–40 °C of temperature, this is a good sign for in situ bioremediation. Green compost, farmyard manure and rice husk were tested, where ANOVA (P < 0.05) and Plackett–Burman design, results indicated that the farm yard manure has significant impact on degradation. It reduced the lag phase and brought maximum degradation up to 88%. Inoculum size is a statistically significant (P < 0.05) factor and below 106 (CFU g−1) show lag phase of 4–6 days. Michaelis–Menten model results were as follows; R2 = 0.9919, Vmax = 18.8, Ks = 121.4 and Vmax/Ks = 0.1546. GC–MS study revealed that chlorpyrifos first converted into diethylthiophosphoric acid and 3,5,6-trichloro-2-pyridinol (TCP). Later, TCP ring was broken and it was completely mineralized without any toxic byproduct. Plackett–Burman design was employed to investigate the effect of five factors. The correlation coefficient (R2) between experimental and predicted value is 0.94. Central composite design (CBD) was employed with design matrix of thirty one predicted and experimental values of chlorpyrifos degradation, having “lack of fit P value” of “0.00”. The regression coefficient obtained was R2 = 0.93 which indicate that the experimental vales and the predicted values are closely fitted. The most significant factors highlighted in CBD/ANOVA and surface response plots were chlorpyrifor concentration and inoculum size. Bacillus cereus Ct3 effectively degraded chlorpyrifos and can successfully be used for bioremediation of chlorpyrifos contaminated soils.
Subject terms: Environmental sciences, Environmental microbiology
Introduction
Pesticide dispersal and persistence depends on soil properties, nature of active compound and abiotic/biotic conditions1. In soil, pesticide fate is quite complex, they may degrade, volatilize, hydrolyze or mineralize. Soil has the tendency to restrict the movement of pesticides. Excessive use of pesticides disturbs soil microflora/fauna and as a result decreases soil fertility. Leaching of pesticide results in the ground water contamination. Studies showed that very less amount (0.1%) of applied chlorpyrifos enter the target pest, where as 99.9% disperse in environment2. Chlorpyrifos (CP) is “moderately toxic” (class II) broad range organophosphate insecticide with half life of 60–120 days and is used on almonds, cotton, oranges, apples and corn. It inhibit cholinesterase (AChE) in peripheral and central nervous system. In children/fetus small quantity of pesticide imparts neurotoxic effects and huge quantities exhibit severe toxicity. It may lead to lethal damage to heart and pancreas. The hydroxyl radicals volatilize in atmosphere and become more toxic in the form of chlorpyrifos-oxon. This remains in air over more than 4 h3. The search of new methods is in high demand to restore contaminated soils4,5. Bioremediation is gaining popularity due to more efficiency, eco-friendly, selective destruction and low cost6,7. Bioremediation has successfully decontaminated agricultural soils, wetlands, sludge and ground water8. Pesticides contaminated soil can be restored by injecting specific microbes9–11. Pesticide polluted soils are the most appropriate site to find out the resistant microbes. Native microbes develop resistance over period of time in polluted sites or soils8,12.
Hamzah et al.10 efficiently degraded CP by Pseudomonas aeruginosa in 5 days. Singh et al.13 recommended the injection of Pseudomonas sp. in Australian soils, this causes the fast degradation of CP. The experiment lasted for 90 days and the maximum degradation was observed in basic soils. Enterobacter sp. B-14 degraded 35 mg kg−1 of CP in soils14. Mixed culture proved less effective as compared to the mono-culture. For the bioremediation of diazinon Pseudomonas sp. was examined15. Pseudomonas showed 80–92% degradation of 100 mg K−1 in sterile soil in presence of glucose. The rate of degradation varies from 0.085 to 0.032 mg day−1 and half life varies from 25 to 11 days. Moreover, in 7 days only 2% degradation of diazinon was achieved. In statistical analysis the microbial community and biochemical processes shows positive correlation. Samuel et al.16 reported Pseudomonas putida, which has the ability to detoxify aflatoxin into non-toxic products.
Fungi are also the potential candidate for biodegradation studies in soil. Verticillium sp. exhibited the successful degradation of CP in different mediums. The degradation process becomes speedy by using this strain and it also competed with native microbes17. Similarly, 3.59 times fast degradation was reported by Fang et al.18. Enzymes are effective biocatalyst which can be successfully employed for environmental bioremediation19. Other than native strains and their enzymes, the use of genetically engineered microbes is also gaining popularity for biodegradation20. Current study is designed to investigate the potential of Bacillus cereus Ct3 to degrade CP in soil. Bacillus cereus Ct3 was isolated from cotton fields. Optimization of biodegradation conditions like; kinetic analysis, Plackett–Burman Design, Central Composite Design, response surface plots, ribotyping and biodegradation pathway was studied.
Materials and methods
Microbial strain selection, identification and Inoculum preparation
Bacillus strain Ct3 was selected for this study, this stain was isolated from cotton growing agricultural soils. Soil (30 g), sterile MSM (minimal salt medium), 15 mL chlorpyrifos were mixed and shaked for 1 week. After 1 week, 10 mL of this solution was transferred to freshly sterile MSM. Chlorpyrifos concentration was increased gradually up to 150 mL L−1. Culture was then transferred to sterile nutrient agar plates having 150 mL L−1 of chlorpyrifos21. Only resistant colonies which show growth were selected for further experimentation. Strain Ct3 showed the most prominent growth in presence of chlorpyrifos. The strain Ct3 was also tested for its resistance against other pesticides. Strains were identified through ribotyping (16s rRNA) by using reverse primer (5ʹCCGTCAATTCMTTTRAGTTT3ʹ) and universal forward (5ʹGGATTAGATACCCTGGTA3ʹ). Testing facility was provided by Macrogen (Macrogen Inc. Geumchen-gu, South Korea). The bacterial species sequences were deposited in GenBank, which showed more than 98% similarity and thus identified as Bacillus sp. Inoculums size was quantified by haemocytometer.
Bio-degradation in laboratory soil and pot
Impact of concentration on bioremediation was investigated by adding sterile soil (0.1 kg), chlorpyrifos (measured quantity) and inoculum (quantified) in a sterile glass beaker. The set up was kept at desired temperature. Control was without inoculums and whole experiment was setup with three replicates. After regular intervals soil samples were tested for metabolites and residual pesticide. Effect of temperature, pH and inoculum sizes was testing using different concentration ranges. Green compost, farmyard manure and rice husk were tested as source of carbon. Measured quantity of carbon source, sterile soil, chlorpyrifos and inoculum was aseptically mixed. Similar arrangement without added carbon source was setup as control. The optimum inoculation size and chlorpyrifos concentration with other standardized conditions were tested.
Kinetics study
Kinetic studies were carried out using Michaelis–Menten model22
| 1 |
here; ‘S’ is the ‘substrate concentration’, ‘Vmax’ is the ‘maximum biodegradation rate’ and ‘Ks’ is the ‘half saturation constant’. Rearrangement of Eq. (1) generates “Hanes plot”, which shows straight line between [S] and [S]/v. As the experiment used intact cell instead of isolated enzymes, so we replaced the “Ks” with “Km”22.
Chlorpyrifos extraction and analysis
CP and its metabolites were identified by protocol of Xu et al.23. Petroleum ether was used to get cell-free supernatant. Supernatant was dried, redisssolved in methanol and filtered (0.45 mm pore size). Metabolites were identified by GC–MS system (Agilent, USA).
Plackett–Burman design (PBD) for screening significant factors
In this study activity of five variables (both nutritional and environmental) were selected for statistical analysis. These variables and their higher (+ 1) and lower (− 1) values are presented in Table S5. Plackett–Burman design was designed using Minitab 16.2.2 software24 and the comprehensive matrix (standard 12 run) is presented in Table 1. The impact of each variable on biodegradation of chlorpyrifos was calculated through following equation:
| 2 |
where Y represents the chlorpyrifos biodegradation (response), Ao represents the intercept, Bi represents the linear factor coefficient and Xi is the level of each variable. Probability value (P value < 0.05) in regression analysis is used to identify significant factors for chlorpyrifos biodegradation. ANOVA for PBD was generated using Minitab software. Factors were further optimized by Response surface methodology.
Table 1.
Plackett–Burman design (PBD) of factors (in coded levels) with biodegradation of chlorpyrifos (%) as response.
| Run order | Concentration | Temperature | pH | Carbon | Inoculum | Response | Predicted |
|---|---|---|---|---|---|---|---|
| 1 | − 1 | − 1 | + 1 | + 1 | + 1 | 82.3 | 81.3 |
| 2 | − 1 | + 1 | + 1 | + 1 | − 1 | 50.3 | 53.7 |
| 3 | + 1 | + 1 | − 1 | + 1 | + 1 | 72.4 | 62.5 |
| 4 | + 1 | + 1 | + 1 | − 1 | + 1 | 31.8 | 31.2 |
| 5 | − 1 | − 1 | − 1 | + 1 | + 1 | 79.3 | 76.5 |
| 6 | + 1 | + 1 | − 1 | + 1 | − 1 | 30.3 | 29.3 |
| 7 | − 1 | − 1 | − 1 | − 1 | − 1 | 14.2 | 7.2 |
| 8 | + 1 | − 1 | − 1 | − 1 | + 1 | 7.2 | 20.8 |
| 9 | − 1 | + 1 | + 1 | − 1 | + 1 | 50.7 | 50.7 |
| 10 | + 1 | − 1 | + 1 | − 1 | − 1 | 6.3 | 2.3 |
| 11 | − 1 | + 1 | − 1 | − 1 | − 1 | 6.7 | 12.7 |
| 12 | + 1 | − 1 | + 1 | + 1 | − 1 | 18.4 | 28.7 |
Central composite design (CCD) of experiments and response surface methodology (RSM)
Four significant factors viz concentration of chlorpyrifos, temperature, carbon source and inoculum size were established as critical determinants for biodegradation of chlorpyrifos as the outcome of PBD. Full factorial central composite design (CCD) (24) was adopted with 31 experimental runs, seven center points and eight axial points25. The significance of model, prediction equation, regression coefficient and case statics were calculated through analysis of variance (ANOVA). A second-order polynomial equation was employed to fit results and analyze interaction among factors, as shown by Eq. (3):
| 3 |
where ‘Y’ represents the chlorpyrifos degradation, ‘X’ represents the factors that influence degradation, ‘Ao'’ represents the intercept coefficient, ‘Bi’ represents the linear coefficient, ‘Bii’ represents the quadratic coefficient and ‘Bij’ represents the interaction coefficient. Response surface plots were obtained using Minitab (statistical software, version 16.2.2) to identify effect of factors. ANOVA for CCD was generated using Minitab Software.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent is not-applicable in this study.
Results and discussion
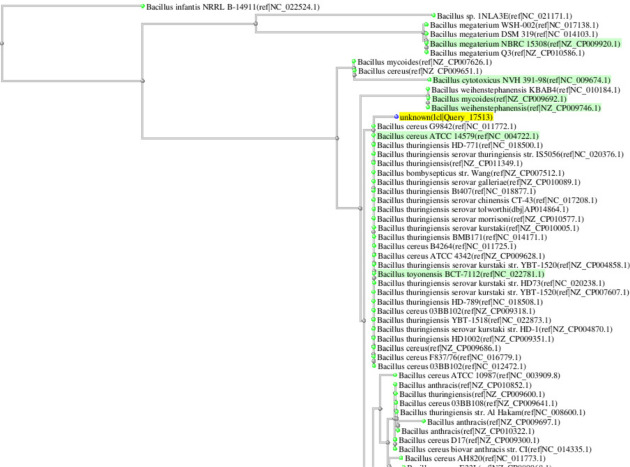
The demand of eco-friendly solution and the use of indigenous species to restore pesticide contaminated soils is growing globally26. Biodegradation is considered best option for in situ restoration operations6. Soils contaminated with polycyclic aromatic hydrocarbon was successfully restored by Nocardia farcinica, Nocardia asteroids and Nocardia cashijiensis27. Chlorpyrifos is widely used on cotton and vegetables to control pests. Excessive use of the pesticide and its accumulation in soil changes the physiochemical properties of soil. Which lead to change in soil micro-flora and fertility. Abigail et al.7 reported that loss of soil fertility is due to toxicity and mutagenicity in nostoc muscorum and anabaena doliolum. To restore chlorpyrifos contaminated soil we isolated resistant strain Ct3, it was resistant up to 150 mg of chlorpyrifos. The resistance detail in different concentrations of chlorpyrifos is given in Table S1. This strain is not much resistant against other pesticides (Table S2). Its phylogenetic trees identified it as Bacillus cereus (Fig. 1). The use of indigenous species is preferred as they do not pose any negative impact on micro-flora. The degradation rate becomes slow at higher CP concentration, whereas at low concentration with apparently no lag phase the rate of biodegradation becomes fast (Fig. 2a). To maintain minimum number of bacteria the more time is required that becomes the reason of longer lag phase28. Results showed that the CP bioremediation is concentration dependent. ANOVA reveled that concentration is a significant (P < 0.05) factor (Tables 1, 2). 4.7% bioremediation was noted with the initial concentration of 75 mg kg−1 and when the concentration was increased up to 125 mg kg−1 the bioremediation decreased to 2% (in 2 days). After 4 days the rapid degradation started when initial concentration of 75 mg kg−1, whereas after 8 days of incubation, rapid degradation started when concentrations was 100 and 125 mg kg−1. At 16th day more degradation (66%) was noted with 75 mg kg−1 of CP and less degradation (43) was noted with 125 mg kg−1 of CP. The 2 main reasons which impact the CP degradation are as follows; firstly, as the parent pollutant (CP) concentration decreases the attractive forces between soil particles and CP molecules increases11. Secondly, the growth of bacterial population was hindered due to the accumulation of intermediate metabolites29. According to these results it is revealed that broad range of CP concentration was tolerated by the Bacillus sp. Hua et al.30 successfully degraded 12 mg kg−1 up to 79.5% in soil (35 days). Ahmad et al.31 reported resistant strain, this strain showed 97% degradation of 50 mg kg−1 in 45 days. This strain was identified as Bacillus pumilus C2A1.
Figure 1.
Phylogenetic tree of strain Ct3.
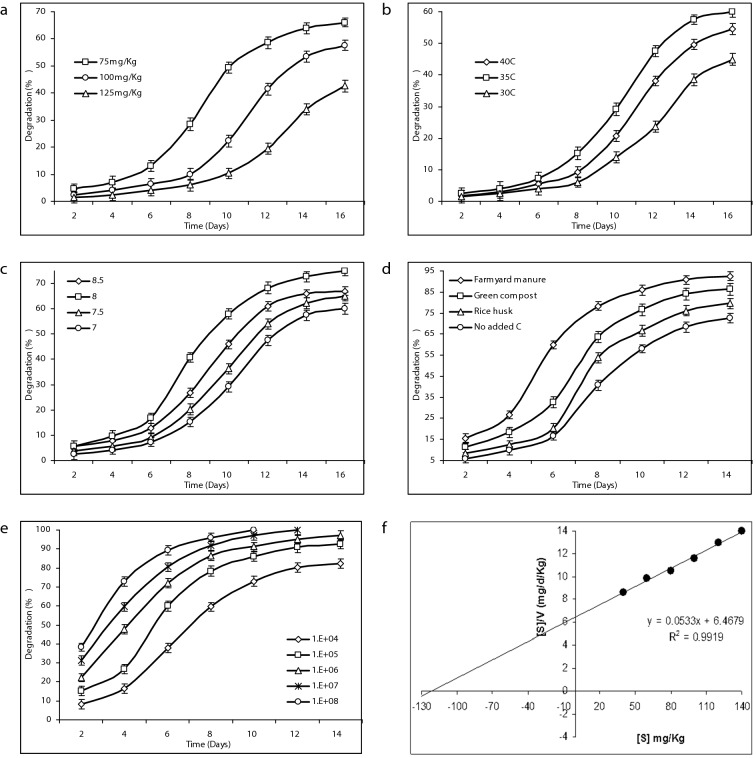
Figure 2.
Biodegradation rate of CP at different factors in laboratory soil. (a) pH: 7, inoculum density: 105 (CFU mL−1), incubation temperature: 37 °C and C-source: nil. (b) CP conc: 100 mg kg−1, pH: 7, inoculum density: 105 (CFU g−1) and C-source: nil. (c) CP Conc: 100 mg kg−1, inoculum density: 105 (CFU g−1), incubation temperature: 35 °C and C-source: nil. (d) CP Conc: 100 mg kg−1, inoculum density: 105 (CFU g−1), incubation temperature: 35 °C and pH: 8. (e) CP Conc: 100 mg kg−1, pH: 8, incubation temperature: 35 °C and C-source: farmyard manure. (f) Hanes plot for calculating chlorpyrifos biodegradation kinetics of Bacillus sp. in soil.
Table 2.
ANOVA for PBD model.
| Source | DF | Seq SS | Adj SS | Adj MS | F | P |
|---|---|---|---|---|---|---|
| Regression | 5 | 192.247 | 192.247 | 38.4493 | 69.049 | 0.000 |
| Concentration | 1 | 21.960 | 21.960 | 21.9597 | 39.436 | 0.000 |
| Temperature | 1 | 6.579 | 6.579 | 6.5789 | 11.815 | 0.0017437 |
| pH | 1 | 3.613 | 3.613 | 3.6127 | 6.488 | 0.0662283 |
| Carbon | 1 | 92.683 | 92.683 | 92.6833 | 166.444 | 0.0000000 |
| Inoculum | 1 | 67.412 | 67.412 | 67.4120 | 121.061 | 0.0000000 |
| Error | 30 | 16.705 | 16.705 | 0.5568 | ||
| Lack-of-fit | 6 | 16.607 | 16.607 | 2.7678 | 672.568 | 0.0000000 |
| Pure error | 24 | 0.099 | 0.099 | 0.0041 | ||
| Total | 35 | 208.952 |
The failure and success of biodegradation is due to temperature variation32. Degradation capacity of Bacillus Ct3 was significantly affected temperature (P < 0.05) (Tables 1, 2). After 6 days rapid degradation was started (at 35 °C), whereas, lag phase continued up to 8 days at 30 °C and 40 °C. Excessive degradation of 45%, 60% and 54% was observed at 30 °C, 35 °C and 40 °C, respectively (Fig. 2b). According to the results, the isolated Bacillus sp tolerated wide range of temperature; this is a good sign for in situ bioremediation. Liu et al.9 stated that at 30 °C Pseudomonas putida worked most effectively and achieved 78%.
In bioremediation process, soil pH plays one of the most effective roles as abiotic factors. The CP bioremediation rate changes with small fluctuation in pH. The maximum degradation (75%) of CP was observed at 8 pH (Fig. 2c). After 6 days, the CP degradation increase sharply and later at day 14 it entered into stationary phase. Maximum degradation rate of 67% was observed at pH 8.5 and the minimum (60%) was observed at pH 7. All the applied treatments of pH showed significant difference from each other at the 16th day. Whereas, non-significant results were observed at 2nd and 4th day. Impact of pH change was non-significant (P < 0.05) on degradation (Table 2). Results also showed that with wide range of pH the Bacillus can effectively degrade CP (Fig. 2c). Other research projects also highlighted the significance of pH in bioremediation33. In the rapid changing environment, the strains which had the ability to work at variable pH are more likely to succeed in degradation and are more acceptable. Enterobacter sp. was isolated by Singh et al.14, this strain showed optimum degradation at higher pH, whereas low pH had negative effect on this strain. Pseudomonas kilonensis SRK1 was isolated from wastewater which degraded initial concentration of 150 mg L−1 having CFU (3.6 × 106), pH (8) and glucose as additional carbon source34. Samual et al.35 reported optimum biodegradation of p-nitrophenol at pH 7 using Pseudomonas putida. Gong et al.36 stated that optimum pH is one of the critical factor in biodegradation and successive cycling of pollutants in forest.
In bioremediation process the organic amendments significantly impact the rate and course of degradation. In this study, rice husk, farmyard manure and green compost was added in the soil as carbon source. In presence of organic amendments Bacillus sp. showed changed behavior towards bioremediation (Fig. 2d). Farmyard manure shows more promising results towards bioremediation process in comparison to non-carbon treatment. ANOVA (P < 0.05) and PBD indicated that the carbon source has significant impact on degradation (Table 2). It reduced the lag phase and brought maximum degradation up to 88%. During whole experiment the degradation percentage varies significantly. The increase in degradation rate from 1st to 14th day was gradual, but degradation rate becomes slow after day 14th (Fig. 2d). With addition of green compost the degradation was also facilitated and reached to the level of 83% without lag phase. However, addition of rice husk showed different pattern. Initially lag phase was observed, later degradation increased at 6th day and reached at stationary phase at 14th day. Rice husk is 13% less effective than farmyard manure. This result shows that Bacillus sp. can effectively use different organic material to speed up the bioremediation process. Many studies have recommended the effectiveness of amendments especially organic one, common examples includes public green compost, farmyard manure, municipal waste, cow manure37, mushroom spent, nut shells38 and coconut fiber39.
In CP bioremediation the inoculum size/density also play a vital role. At different inoculum densities this strain depicts almost the similar pattern (Fig. 2e). With 108 CFU g−1 the CP degradation was up to 100% (in 10 days). Inoculums density of 107 CFU g−1 also led to 100% degradation. However, medium inoculums densities (104 and 105 CFU g m−1) reported slightly less degradation rate initially but later fast degradation starts. Inoculums density of 107 (CFU g−1) also attained 100% degradation. Cupriavidus sp. DT-1 also degraded 100% of CP and 94% of TCP. Inoculum is a statistically significant (P < 0.05) factor in chlorpyrifos degradation (Table 2). The whole experiment lasted for 30 days with 106 cells g−1 40. The use of sewage sludge for improving bioaugmentation was tested and found effective41. Jariyal et al.42 has reported that instead of single strain, consortia can be used for better degradation. Use of consortium is also very beneficial in synergistic biodegradation of aromatic-aliphatic copolyester plastic. This consortium was isolated from marine ecosystem and have the tendency to use pollutant as sole carbon source in 15 days43.
The Hanes plot’s straightness is signified by the R2 (0.9919) and Vmax (18.7627) (Fig. 2f). Effectiveness of the isolates is usually predicted by the ratio of Vmax/Ks, and in this study this ratio is 0.1546. Maya et al.22 investigated the degradation of CP and TCP with the addition of Pseudomonas, Bacillus and Agrobacterium sp. Km varies from 97 to 142.3 mg L−1 for CP and for TCP Km range was 103.09–148.8 mg L−1. The range of Vmax of CP was from 7.4 to 12.1 mg L−1. Whereas, Vmax of TCP range from 14.90 to 21.20 mg L−1. Fang et al.18 used Verticillium sp. for bioremediation of CP and calculated Vmax = 12.2 and R2 = 0.98. Geed et al.29 calculated bioremediation kinetics of Malathion (pesticide) under inhibitory and non-inhibitory conditions. The kinetic constants were calculated by using Monod models and the values were as follows, Ks = 126.30 mg L−1 and μmax = 0.27 day−1. Samuel et al.44 reported that pseudo-secound order model to be best fit for Pb(II) removal. The kinetic data was as follows, R2 = 0.987, qe = 24.64, K2 (min−1) = 0.0342 and qmax (mg g−1) = 112.35. In GC–MS chromatogram, 2 peaks appeared at retention time (RT) of 9.14 and 15.13 min (Fig. 3). These peaks were designated “A” and “B”. Peak “B” with retention time 15.13 min match with the chlorpyrifos. With the passage of time this peak decreased gradually. Peak “A” with the RT of 9.14 min match with TCP (Fig. 3). Characteristic fragment ion peak and molecular ion (m/z) also supported the identification of TCP. Peak “A” was most prominent around 7–15 days, later this peak started decreasing. The CP first converted into diethylthiophosphoric acid (DETP, m/z = 172) and TCP (m/z = 197) by hydrolysis (Table S3). Figure 4 presents the proposed degradation pathway of chlorpyrifos. The TCP ring was broken and it was completely mineralized without any accumulation of byproduct. The degradation products were non-toxic as they did not hinder the growth of Bacillus cereus Ct3. Very good growth was noted till 14th day of biodegradation experiment (Table S4). Similar pathway and metabolites were reported by other studies22,45. Hamzah et al.10 reported 3,5,6-trichloro-2-pyridinol and CPF-oxon as metabolite of CP degradation by Pseudomonas aeruginosa. The metabolites identified in this study were diethyl phosphate and 3,5,6-trichloro-2-pyridinol, these 2 metabolites were also reported by Liu et al.9. Samuel et al.46 calculated qmax = 71.9 and R2 = 0.99 for biosorption of chromium by Aspergillus niger. Table 3 reports the chlorpyrifos degradation efficiencies of different strains in different conditions.
Figure 3.
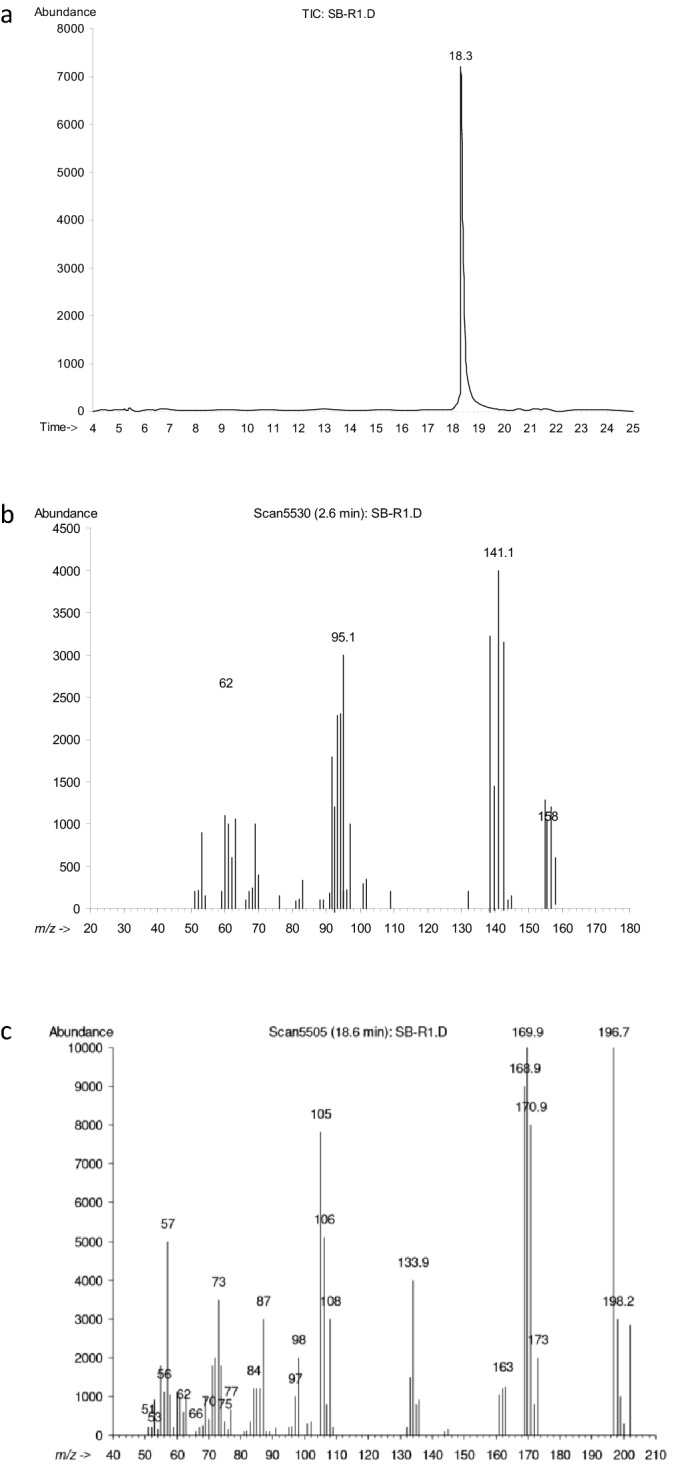
HPLC chromatograms indicating metabolites of chlorpyrifos (a), (b) & (c).
Figure 4.
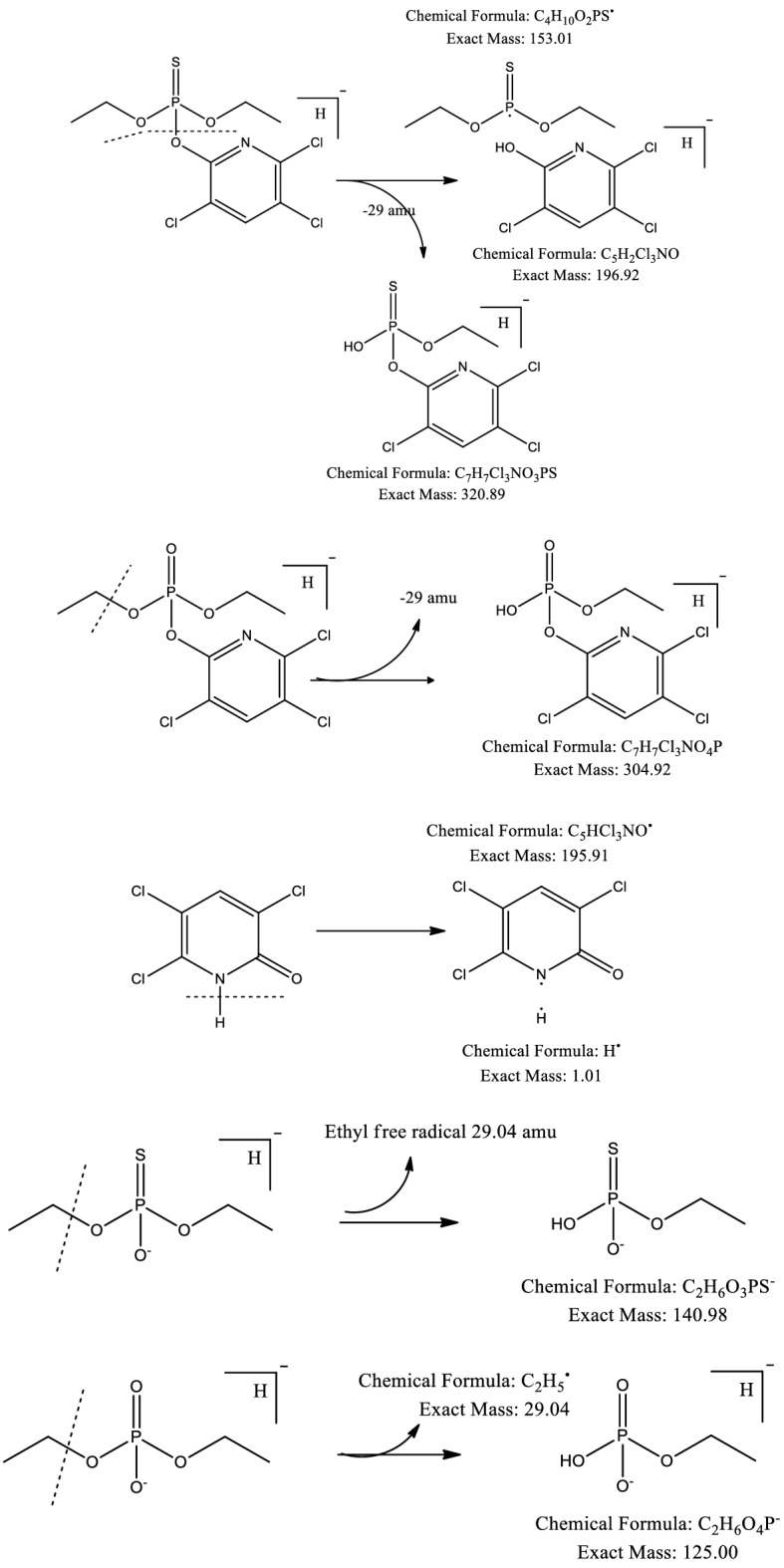
Purposed degradation pathway of chlorpyrifos.
Table 3.
Comparison of chlorpyrifos biodegradation efficiencies.
| Study | Isolate used | Degradation (%) | Initial concentration (mg L−1) | Time to degrade (days) |
|---|---|---|---|---|
| Present study | Bacillus cereus Ct3 | 87 | 125 | 8 |
| Chishti et al.47 | Enterobacter sp. SWLC2 | 87 | 100 | 18 |
| Soares et al.48 | Aspergillus sydowii CBMAI 935 | 32 | 192 | 10 |
| Jin et al.49 | Trametes versicolor | 90 | 150 | 8 |
| Gangola et al.50 | Bacillus subtilis | 95 | 200 | 15 |
| Gao et al.51 | Cladosporium cladosporioides Hu-01 | >90 | 50 | 5 |
| Kulshrestha and Kumari52 | Acremonium sp. (GFRC-1) | 84 | 300 | 20 |
| Zhu et al.53 | Bacillus licheniformis ZHU-1 | 100 | 100 | 14 |
| Anwar et al.54 | Bacillus pumilus C2A1 | 89 | 1000 | 15 |
| Korade and Fulekar55 | Pseudomonas nitroreducens PS-2 | 100 | 100 | 28 |
| Fang et al.56 | Verticillium sp. DSP | 100 | 100 | 7 |
| Li et al.57 | Sphingomonas sp. | 98 | 30 | 10 |
Plackett–Burman design was employed to investigate the effect of five factors. Table 1 represents the experimental design with the actual chlorpyrifos degradation values and the predicted chlorpyrifos degradation values. The correlation coefficient (R2) between experimental and predicted value is 0.94, which is in reasonable agreement. Table 2 represents the ANOVA results for PBD, where P value less than 0.05 represents the significance of the factor for chlorpyrifos biodegradation. The polynomial equation for independent and dependent factors in this study could be written as:
| 4 |
Base on the results of PBD following four significant factors were identified; concentration, temperature, carbon, inoculum size. Central composite design (CBD) was employed and its design matrix with thirty one predicted and experimental values of chlorpyrifos degradation is represented in Table 4. The second-degree polynomial equation obtained was as follows:
| 5 |
Table 4.
Experimental design and results of central composite design (CCD).
| Run order | Concentration | Temperature | Carbon | Inoculum | Response | |
|---|---|---|---|---|---|---|
| Experimental | Predicted | |||||
| 1 | 100 | 35 | 1.5 | 50005000 | 71.1 | 71.2 |
| 2 | 100 | 45 | 1.5 | 50005000 | 30.2 | 38.1 |
| 3 | 75 | 30 | 3 | 100000000 | 80.1 | 73.6 |
| 4 | 125 | 40 | 3 | 10000 | 22.4 | 18.6 |
| 5 | 50 | 35 | 1.5 | 50005000 | 91.2 | 87.6 |
| 6 | 75 | 30 | 3 | 10000 | 22.3 | 30.6 |
| 7 | 125 | 40 | 0 | 10000 | 15.4 | 14.5 |
| 8 | 100 | 35 | − 1.5 | 50005000 | 11.2 | 17.3 |
| 9 | 125 | 40 | 0 | 100000000 | 35.4 | 28.5 |
| 10 | 100 | 35 | 1.5 | 50005000 | 71.5 | 71.2 |
| 11 | 75 | 40 | 0 | 10000 | 37.7 | 43.6 |
| 12 | 100 | 35 | 1.5 | 50005000 | 71.9 | 71.2 |
| 13 | 125 | 30 | 3 | 10000 | 11.1 | 12.0 |
| 14 | 125 | 30 | 3 | 100000000 | 65.7 | 55.5 |
| 15 | 125 | 40 | 3 | 100000000 | 69.2 | 57.1 |
| 16 | 100 | 35 | 1.5 | 50005000 | 69.8 | 71.2 |
| 17 | 75 | 40 | 0 | 100000000 | 65.1 | 57.1 |
| 18 | 125 | 30 | 0 | 100000000 | 33.4 | 23.8 |
| 19 | 100 | 35 | 1.5 | 50005000 | 71.6 | 71.2 |
| 20 | 75 | 40 | 3 | 100000000 | 90.2 | 74.3 |
| 21 | 100 | 25 | 1.5 | 50005000 | 25.4 | 27.8 |
| 22 | 100 | 35 | 1.5 | − 49985000 | 6.2 | 3.4 |
| 23 | 75 | 40 | 3 | 10000 | 34.1 | 36.3 |
| 24 | 150 | 35 | 1.5 | 50005000 | 30.1 | 40.3 |
| 25 | 100 | 35 | 1.5 | 149995000 | 45.2 | 56.9 |
| 26 | 125 | 30 | 0 | 10000 | 3.4 | 4.8 |
| 27 | 100 | 35 | 1.5 | 50005000 | 71.5 | 71.2 |
| 28 | 75 | 30 | 0 | 100000000 | 60.2 | 53.5 |
| 29 | 75 | 30 | 0 | 10000 | 30.2 | 35.0 |
| 30 | 100 | 35 | 4.5 | 50005000 | 30.4 | 41.6 |
| 31 | 100 | 35 | 1.5 | 50005000 | 71.3 | 71.2 |
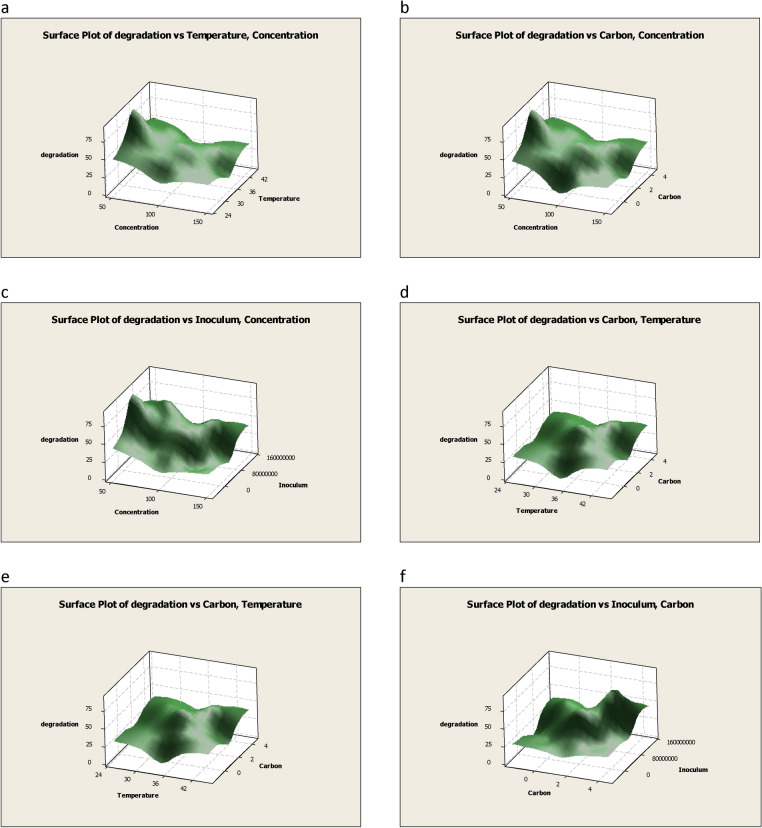
The ANOVA result for CBD is presented in Table 5. It represents the individual main effects and the interaction among different factors. The “F-value” of the model was 1050.7 which suggest that the model was significant. For the present study the “lack of fit P value” was 0.00. Figure 5 represents the experimental values and the predicted values for the biodegradation of chlorpyrifos. The regression coefficient obtained was R2 = 0.93 which indicate that the experimental vales and the predicted values are closely fitted. Figure 6a–f represents the combined effect of different factors for biodegradation of chlorpyrifos. The degradation increase with the increase in concentration, temperature, carbon source and inoculum but at certain point it reaches the maximum. Further increase in any factor beyond this point will decrease the degradation of chlorpyrifos. The most significant factors highlighted in ANOVA and surface response plots were concentration and inoculum size. Changes in concentration and inoculum size impacts more on degradation then other factors. This is in argument with the work reported by Zhou et al.58 and Ungureanu et al.59, wherein the inoculum size was the important factor for phenol degradation. Samuel et al.60 concluded (using response surface methodology) that biosorbent by Certocystis paradoxa MSR2 depends on temperature, pH and initial concentration. Similarly, Alice et al.61 optimized biosorption of chromium based on response surface method. Variables optimized were contact time, initial concentration and biosorbent dose.
Table 5.
ANOVA for central composite design model (CCD).
| Source | DF | Adj SS | Adj MS | F | P |
|---|---|---|---|---|---|
| Regression | 14 | 17,523.8 | 1251.70 | 4.09 | 0.004 |
| Linear | 4 | 9282.5 | 2320.63 | 7.58 | 0.001 |
| Concentration | 1 | 3360.7 | 3360.67 | 10.97 | 0.004 |
| Temperature | 1 | 160.2 | 160.17 | 0.52 | 0.480 |
| Carbon | 1 | 888.2 | 888.17 | 2.90 | 0.108 |
| Inoculum | 1 | 4873.5 | 4873.50 | 15.91 | 0.001 |
| Square | 4 | 7473.6 | 1868.39 | 6.10 | 0.004 |
| Concentration * concentration | 1 | 95.9 | 95.93 | 0.31 | 0.584 |
| Temperature * temperature | 1 | 2625.3 | 2625.26 | 8.57 | 0.010 |
| Carbon * carbon | 1 | 3126.6 | 3126.64 | 10.21 | 0.006 |
| Inoculum * inoculum | 1 | 3277.9 | 3277.93 | 10.70 | 0.005 |
| Interaction | 6 | 767.8 | 127.96 | 0.42 | 0.857 |
| Concentration * temperature | 1 | 1.0 | 1.00 | 0.00 | 0.955 |
| Concentration * carbon | 1 | 132.2 | 132.25 | 0.43 | 0.520 |
| Concentration * inoculum | 1 | 0.2 | 0.25 | 0.00 | 0.978 |
| Temperature * carbon | 1 | 9.0 | 9.00 | 0.03 | 0.866 |
| Temperature * inoculum | 1 | 25.0 | 25.00 | 0.08 | 0.779 |
| Carbon * inoculum | 1 | 600.3 | 600.25 | 1.96 | 0.181 |
| Residual error | 16 | 4901.4 | 306.34 | ||
| Lack-of-fit | 10 | 4898.6 | 489.86 | 1050.7 | 0.000 |
| Pure error | 6 | 2.8 | 0.47 | ||
| Total | 30 | 22,425.2 |
Figure 5.
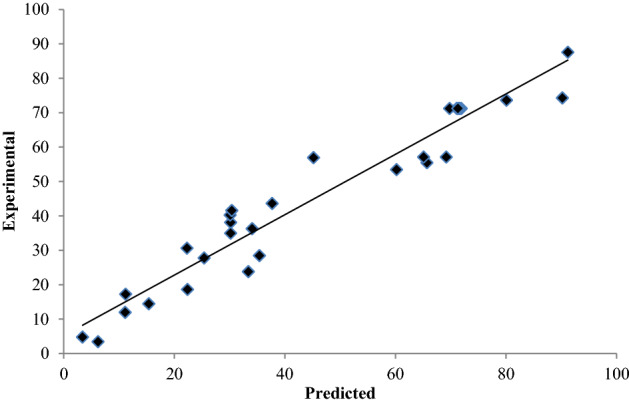
The predicted values plotted against actual values derived from CCD model for chlorpyrifos biodegradation.
Figure 6.
Surface plot showing interaction between different variables. (a) Interaction between degradation, temperature and chlorpyrifos concentration. (b) Interaction between degradation, carbon source and chlorpyrifos concentration. (c) Interaction between degradation, inoculum size and chlorpyrifos concentration. (d) Interaction between degradation, carbon source and temperature. (e) Interaction between degradation, carbon source and temperature. (f) Interaction between degradation, inoculum size and carbon source.
Conclusion
Rapid increase in pesticide is negatively linked with ecological disturbance, decrease in soil fertility and changes in microbial community. Natural and eco-friendly techniques are gaining popularity to protect ecosystems from the side effects of pesticides. To restore chlorpyrifos contaminated soil, we isolated 156 strains from cotton growing Punjab region of Pakistan. Bacillus cereus Ct3 was most resistant up to 175 mg L−1 of chlorpyrifos. The use of indigenous specie is preferred as they do not pose any negative impact on micro-flora and has better survival chance. The process of biodegradation was optimized using Michaelis–Menten model and 88% degradation was achieved in 8 days at pH 8. Biodegradation pathway was proposed, where CP first degraded into diethylthiophosphoric acid and 3,5,6-trichloro-2-pyridinol (by hydrolysis, TCP further mineralized. Plackett–Burman design, Central composite design and surface response methods was employed to investigate the effect of five factors. Base on the results significant factors identified were chlorpyrifos concentration and inoculum size. Bacillus cereus Ct3 can be employed to degraded chlorpyrifos without producing toxic metabolite and can successfully be used for bioremediation of chlorpyrifos contaminated soils. Future study will focus on the whole genome sequencing of Bacillus cereus Ct3, identification of genes involved in biodegradation and action of enzymes responsible.
Supplementary information
Acknowledgements
We are thankful to “Irrigation Department, Government of the Punjab, Pakistan” for helping us in sampling and analysis.
Author contributions
M.F., M.A. and A.W. planned this research. M.F., A.K., S.A.R., Z.A. conducted this research. Q.F., H.Q., A.K. and M.F. analyze the data. All authors contributed in research paper writing.
Funding
This project was funded by Government College University, Lahore, under Grant # 92-GCU-PHD-ENV-09.
Competing interests
Dr. Muhammad Farhan has received research grants from Government College University Lahore Pakistan. Dr. Muhammad Farhan has received ‘Professor J.H. Orbison Medal’ from Forman Christian College (A Chartered University) Lahore. Dr. Muhammad Farhan has received ‘Best Teacher Award’ from Forman Christian College (A Chartered University) Lahore. Dr. Abdul Wahid has received ‘Best Teacher Award’ from Higher Education Commission Islamabad Pakistan. Dr. Abdul Wahid has received ‘Out Standing Researcher Award’ from Pakistan Science Foundation.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88264-x.
References
- 1.Kumar U, Berliner J, Totan A, Prakash CR, Avro D, Somnath SP, Nitiprasad NJ, Panneerselvam P, Anjani K, Shyamranjan DM. Non-target effect of continuous application of chlorpyrifos on soil microbes, nematodes and its persistence under sub-humid tropical rice cropping system. Ecotoxicol. Environ. Saf. 2017;135:225–235. doi: 10.1016/j.ecoenv.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Tariq MI, Afzal S, Hussain J. Pesticides exposure in Pakistan: a review. Environ. Int. 2007;33:1107–1122. doi: 10.1016/j.envint.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Harnly M, McLaughlin R, Bradman A, Anderson M, Gunier R. Correlating agricultural use of organophosphates with outdoor air concentrations: a particular concern for children. Environ. Health Persp. 2005;113:1184–1189. doi: 10.1289/ehp.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abraham J, Silambarasa S. Biodegradation of chlorpyrifos and its hydrolysis product 3, 5, 6- trichloro-2-pyridinol using a novel bacterium Ochrobactrum sp. JAS2: a proposal of its metabolic pathway. Pest Biochem. Physiol. 2016;126:13–21. doi: 10.1016/j.pestbp.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 5.González-Gaya B, Martínez-Varela A, Vila-Costa M. Biodegradation as an important sink of aromatic hydrocarbons in the oceans. Nat. Geosci. 2019;12:119–125. doi: 10.1038/s41561-018-0285-3. [DOI] [Google Scholar]
- 6.Maria SF, Enzo E, Raimondo A, María JA, Claudia S. Removal of a mixture of pesticides by a Streptomyces consortium: influence of different soil systems. Chemosphere. 2017;173:359–367. doi: 10.1016/j.chemosphere.2017.01.044. [DOI] [PubMed] [Google Scholar]
- 7.Abigail MEA, Samuel SM, Ramalingam C. Addressing the environmental impacts of butachlor and the available remediation strategies: a systematic review. Int. J. Environ. Sci. Technol. 2015;12:4025–4036. doi: 10.1007/s13762-015-0866-2. [DOI] [Google Scholar]
- 8.Sasikala SC, Jiwal S, Rout P, Ramya M. Biodegradation of chlorpyrifos by bacterial consortium isolated from agriculture soil. World J. Microbiol. Biotechnol. 2012;28:1301–1308. doi: 10.1007/s11274-011-0879-z. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Luming T, Jing W, Zhiyong W, Hong N, Lin L. Complete biodegradation of chlorpyrifos by engineered Pseudomonas putida cells expressing surface-immobilized laccases. Chemosphere. 2016;157:200–207. doi: 10.1016/j.chemosphere.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 10.Hamzah AK, Shubo H, Sherrice A, Shirley LC. Metabolism of chlorpyrifos by Pseudomonas aeruginosa increases toxicity in adult zebrafish (Danio rerio) Int. Biodeter. Biodegrad. 2017;121:114–121. doi: 10.1016/j.ibiod.2017.03.024. [DOI] [Google Scholar]
- 11.Wang C, Zhiren Z, Hongdan L, Junjie L, Ying W, Heng X. Application of acclimated sewage sludge as a bio-augmentation/biostimulation strategy for remediating chlorpyrifos contamination in soil with/without cadmium. Sci. Total Environ. 2017;579:657–666. doi: 10.1016/j.scitotenv.2016.11.044. [DOI] [PubMed] [Google Scholar]
- 12.Tang X, Yang Y, Wenda H, Murray B, McBride JG, Ran T, Yunv D. Transformation of chlorpyrifos in integrated re-circulating constructed wetlands (IRCWs) as revealed by compound-specific stable isotope (CSIA) and microbial community structure analysis. Biores. Technol. 2017;233:264–270. doi: 10.1016/j.biortech.2017.02.077. [DOI] [PubMed] [Google Scholar]
- 13.Singh BK, Walker A, Morgan JA, Wright DJ. Effects of soil pH on the biodegradation of chlorpyrifos and isolation of a chlorpyrifos-degrading bacterium. Appl. Environ. Microbiol. 2003;69:5198–5206. doi: 10.1128/AEM.69.9.5198-5206.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh BK, Walker A, Morgan JA, Wright DJ. Biodegradation of chlorpyrifos by Enterobacter strainB-14 and its use in bioremediation of contaminated soils. Appl. Environ. Microbiol. 2004;70:4855–4863. doi: 10.1128/AEM.70.8.4855-4863.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cycon M, Wójcik M, Piotrowska-Seget Z. Biodegradation of the organophosphorus insecticide diazinon by Serratia sp. and Pseudomonas sp. and their use in bioremediation of contaminated soil. Chemosphere. 2009;76:494–501. doi: 10.1016/j.chemosphere.2009.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Samuel MS, Sivaramakrishna A, Mehta A. Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeter. Biodegrad. 2014;86:202–209. doi: 10.1016/j.ibiod.2013.08.026. [DOI] [Google Scholar]
- 17.Yu YL, Fang H, Wang X, Wu XM, Shan M, Yu JQ. Characterization of a fungal strain capable of degrading chlorpyrifos and its use in detoxification of the insecticide on vegetables. Biodegradation. 2006;17:487–494. doi: 10.1007/s10532-005-9020-z. [DOI] [PubMed] [Google Scholar]
- 18.Fang H, Xiang YQ, Hao YJ, Chu XQ, Pan XD, Yu JQ, Yu YL. Fungal degradation of chlorpyrifos by Verticillium sp. DSP in pure cultures and its use in bioremediation of contaminated soil and pakchoi. Int. J. Biodeter. Biodegrad. 2008;61:294–303. doi: 10.1016/j.ibiod.2007.10.001. [DOI] [Google Scholar]
- 19.Saptashwa D, Veena R, Melvin S, Samuel F, Selvarajan E. Immobilization of laccases and applications for the detection and remediation of pollutants: a review. Environ. Chem. Lett. 2020 doi: 10.1007/s10311-020-01081-y. [DOI] [Google Scholar]
- 20.Duraj-Thatte AM, Manjula-Basavanna A, Courchesne NMD, et al. Water-processable, biodegradable and coatable aquaplastic from engineered biofilms. Nat. Chem. Biol. 2021 doi: 10.1038/s41589-021-00773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farhan M, Khan AU, Wahid A, Ahmad M, Ahmad F, Butt ZA. Enhanced biodegradation of chlorpyrifos by agricultural soil isolate. Asian J. Chem. 2014;26:3013–3017. doi: 10.14233/ajchem.2014.16463. [DOI] [Google Scholar]
- 22.Maya K, Singh RS, Upadhyay SN, Dubey SK. Kinetic analysis reveals bacterial efficacy for biodegradation of chlorpyrifos and its hydrolyzing metabolite TCP. Process Biochem. 2011;46:2130–2136. doi: 10.1016/j.procbio.2011.08.012. [DOI] [Google Scholar]
- 23.Xu G, Zheng W, Li Y, Wang S, Zhang J, Yan Y. Biodegradation of chlorpyrifos and 3,5,6-trichloro-2-pyridinol by a newly isolated Paracoccus sp. strain TRP. Int. J. Biodeter. Biodeg. 2008;62:51–56. doi: 10.1016/j.ibiod.2007.12.001. [DOI] [Google Scholar]
- 24.Mohanty SS, Jena HM. Process optimization of butachlor bioremediation by Enterobacter cloacae using Plackett Burman design and response surface methodology. Process Saf. Environ. Prot. 2018;119:198–206. doi: 10.1016/j.psep.2018.08.009. [DOI] [Google Scholar]
- 25.Hassan M, Essam T, Yassin S, Salama A. Optimization of rhamnolipid production by biodegrading bacterial isolates using Plackett–Burman design. Int. J. Biolog. Macromol. 2016;82:573–579. doi: 10.1016/j.ijbiomac.2015.09.057. [DOI] [PubMed] [Google Scholar]
- 26.Akbar S, Sikander S. Soil bacteria showing a potential of chlorpyrifos degradation and plant growth enhancement. Braz. J. Microbiol. 2016;47:563–570. doi: 10.1016/j.bjm.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azadi D, Shojaei H. Biodegradation of polycyclic aromatic hydrocarbons, phenol and sodium sulfate by Nocardia species isolated and characterized from Iranian ecosystems. Sci. Rep. 2020;10:21860. doi: 10.1038/s41598-020-78821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lizano-Fallas V, Masís-Mora M, Espinoza-Villalobos D, Lizano-Brenes M, Rodríguez-Rodríguez CE. Removal of pesticides and eco-toxicological changes during the simultaneous treatment of triazines and chlorpyrifos in biomixtures. Chemosphere. 2017;182:106–113. doi: 10.1016/j.chemosphere.2017.04.147. [DOI] [PubMed] [Google Scholar]
- 29.Geed SR, Kureel MK, Giri BS, Singh RS, Rai BN. Performance evaluation of Malathion biodegradation in batch and continuous packed bed bioreactor (PBBR) Biores. Technol. 2017;227:56–65. doi: 10.1016/j.biortech.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Hua F, Yunlong Y, Xiaoqiang C, Xiuguo W, Xiaoe Y, Jingquan Y. Degradation of chlorpyrifos in laboratory soil and its impact on soil microbial functional diversity. J. Environ. Sci. 2009;21:380–386. doi: 10.1016/S1001-0742(08)62280-9. [DOI] [PubMed] [Google Scholar]
- 31.Ahmad F, Iqbal S, Anwar S, Afzal M, Islam E, Mustafa T, Khan QM. Enhanced remediation of chlorpyrifos from soil using ryegrass (Lollium multiflorum) and chlorpyrifos-degrading bacterium Bacillus pumilus C2A1. J. Hazard Mater. 2012;237:110–115. doi: 10.1016/j.jhazmat.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Chin-Pampillo JS, Masís-Mora M, Ruiz-Hidalgo K, Carazo-Rojas E, Rodríguez-Rodríguez CE. Removal of carbofuran is not affected by co-application of chlorpyrifos in a coconut fiber/compost based biomixture after aging or pre-exposure. J. Environ. Sci. 2016;46:182–189. doi: 10.1016/j.jes.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 33.Xie H, Zhu L, Ma T, Wang J, Wang J, Su J, Shao B. Immobilization of an enzyme from a Fusarium fungus WZ-I for chlorpyrifos degradation. J. Environ. Sci. 2010;22:1930–1935. doi: 10.1016/S1001-0742(09)60341-7. [DOI] [PubMed] [Google Scholar]
- 34.Khalid S, Imran H, Sher JK. Bacterial assisted degradation of chlorpyrifos: the key role of environmental conditions, trace metals and organic solvents. J. Environ. Manag. 2016;168:1–9. doi: 10.1016/j.jenvman.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 35.Samuel MS, Sivaramakrishna A, Mehta A. Bioremediation of p-Nitrophenol by Pseudomonas putida 1274 strain. J. Environ. Health Sci. Eng. 2014;12:53. doi: 10.1186/2052-336X-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong P, Xu H, Wang C. Persistent organic pollutant cycling in forests. Nat. Rev. Earth Environ. 2021;2:182–197. doi: 10.1038/s43017-020-00137-5. [DOI] [Google Scholar]
- 37.Tejada M, Gomez I, Toro M. Use of organic amendments as a bioremediation strategy to reduce the bioavailability of chlorpyrifos insecticide in soils. Effects on soil biology. Eco-toxicol. Environ. Saf. 2011;74:2075–2081. doi: 10.1016/j.ecoenv.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Kadian N, Malik A, Satya S, Dureja P. Effect of organic amendments on microbial activity in chlorpyrifos contaminated soil. J. Environ. Manag. 2012;95:199–202. doi: 10.1016/j.jenvman.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 39.Juan S, Chin P, Mario M, Karla R. Removal of carbofuran is not affected by co-application of chlorpyrifos in a coconut fiber/compost based biomixture after aging or pre-exposure. J. Environ. Sci. 2016;46:182–189. doi: 10.1016/j.jes.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 40.Lu P, Li Q, Liu H, Feng Z, Yan X, Hong Q, Li S. Biodegradation of chlorpyrifos and 3,5,6-trichloro-2-pyridinol by Cupriavidus sp. DT-1. Biores. Technol. 2013;127:337–342. doi: 10.1016/j.biortech.2012.09.116. [DOI] [PubMed] [Google Scholar]
- 41.Can W, Zhiren Z, Hongdan L, Junjie L, Ying W, Heng X. Application of acclimated sewage sludge as a bio-augmentation/biostimulation strategy for remediating chlorpyrifos contamination in soil with/without cadmium. Sci. Total Environ. 2017;579:657–666. doi: 10.1016/j.scitotenv.2016.11.044. [DOI] [PubMed] [Google Scholar]
- 42.Jariyal M, Vikas J, Kousik M, Virash KG, Balwinder S. Bioremediation of organophosphorus pesticide phorate in soil by microbial consortia. Ecotoxicol. Environ. Saf. 2018;159:310–316. doi: 10.1016/j.ecoenv.2018.04.063. [DOI] [PubMed] [Google Scholar]
- 43.Meyer-Cifuentes IE, Werner J, Jehmlich N. Synergistic biodegradation of aromatic-aliphatic copolyester plastic by a marine microbial consortium. Nat. Commun. 2020;11:5790. doi: 10.1038/s41467-020-19583-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samuel MS, Shah S, Bhattacharya J, Subramaniam K, Singh ND. Adsorption of Pb(II) from aqueous solution using a magnetic chitosan/grapheme oxide composite and its toxicity studies. Int. J. Biol. Macromol. 2018;115:1142–1150. doi: 10.1016/j.ijbiomac.2018.04.185. [DOI] [PubMed] [Google Scholar]
- 45.Tiwari MK, Guha S. Kinetics of biotransformation of chlorpyrifos in aqueous and soil slurry environments. Water Res. 2014;51:73–85. doi: 10.1016/j.watres.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 46.Samuel MS, Abigail AE, Chidambaram R. Isotherm modelling, kinetic study and optimization of batch parameters using response surface methodology for effective removal of Cr(VI) using fungal biomass. PLoS ONE. 2015;10(3):e0116884. doi: 10.1371/journal.pone.0116884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chishti Z, Ahmad Z, Zhang X, Jha SK. Optimization of biotic and abiotic factors liable for biodegradation of chlorpyrifos and their modeling using neural network approaches. Appl. Soil Ecol. 2021;166:103990. doi: 10.1016/j.apsoil.2021.103990. [DOI] [Google Scholar]
- 48.Soares PR, Willian GB, Ferreira IM, Porto AM. Biodegradation pathway of the organophosphate pesticides chlorpyrifos, methyl parathion and profenofos by the marine-derived fungus Aspergillus sydowii CBMAI 935 and its potential for methylation reactions of phenolic compounds. Mar. Pollut. Bull. 2021;166:112185. doi: 10.1016/j.marpolbul.2021.112185. [DOI] [PubMed] [Google Scholar]
- 49.Jin X, Yu X, Zhu G. Conditions optimizing and application of Laccase-mediator system (LMS) for the Laccase-catalyzed pesticide degradation. Sci. Rep. 2016;6:35787. doi: 10.1038/srep35787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gangola S, Sharma A, Bhatt P. Presence of esterase and laccase in Bacillus subtilis facilitates biodegradation and detoxification of cypermethrin. Sci. Rep. 2018;8:12755. doi: 10.1038/s41598-018-31082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao Y, Chen S, Hu M, Hu Q, Luo J, Li Y. Purification and Characterization of a novel chlorpyrifos hydrolase from Cladosporium cladosporioides Hu-01. PLoS ONE. 2012;7(6):38137. doi: 10.1371/journal.pone.0038137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulshrestha G, Kumari A. Fungal degradation of chlorpyrifos by Acremonium sp. strain (GFRC-1) isolated from a laboratory-enriched red agricultural. Biol. Fertil. Soils. 2011;47:219–225. doi: 10.1007/s00374-010-0505-5. [DOI] [Google Scholar]
- 53.Zhu J, Zhao Y, Qiu J. Isolation and application of a chlorpyrifos-degrading Bacillus licheniformis ZHU-1. Afr. J. Microbiol. Res. 2010;4:2410–2413. [Google Scholar]
- 54.Anwar S, Liaquat F, Khan QM, Khalid ZM, Iqbal S. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J. Hazard. Mater. 2009;168:400–405. doi: 10.1016/j.jhazmat.2009.02.059. [DOI] [PubMed] [Google Scholar]
- 55.Korade DL, Fulekar MH. Rhizosphere remediation of chlorpyrifos in mycorrhizospheric soil using ryegrass. J. Hazard. Mater. 2009;172:1344–1350. doi: 10.1016/j.jhazmat.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Fang H, Xiang YQ, Hao YJ, Chu XQ, Pan XD, Yu JQ, Yu YL. Fungal degradation of chlorpyrifos by Verticillium sp. DSP in pure cultures and its use in bioremediation of contaminated soil and pakchoi. Int. Biodeter. Biodegr. 2008;61:294–303. doi: 10.1016/j.ibiod.2007.10.001. [DOI] [Google Scholar]
- 57.Li X, He J, Li S. Isolation of chlorpyrifos degrading bacterium, Sphingomonas sp. strain Dsp-2, and cloning of the mpd gene. Res. Microbiol. 2007;158:143–149. doi: 10.1016/j.resmic.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Zhou J, Yu X, Ding C, Wang Z, Zhou Q, Pao H, Cai W. Optimization of phenol degradation by Candida tropicalis Z-04 using Plackett–Burman design and response surface methodology. J. Environ. Sci. 2011;23(1):22–30. doi: 10.1016/S1001-0742(10)60369-5. [DOI] [PubMed] [Google Scholar]
- 59.Ungureanu CP, Favier L, Bahrim G, Amrane A. Response surface optimization of experimental conditions for carbamazepine biodegradation by Streptomyces MIUG 4.89. New Biotechnol. 2015;32(3):347–357. doi: 10.1016/j.nbt.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 60.Samuel MS, Abigail MEA, Chidambaram R. Biosorption of Cr(VI) by Ceratocystis paradoxa MSR2 using isotherm modelling, kinetic study and optimization of batch parameters using response surface methodology. PLoS ONE. 2015;10(3):e0118999. doi: 10.1371/journal.pone.0118999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alice AM, Samuel MS, Chidambaram R. Hexavalent chromium biosorption studies using Penicillium griseofulvum MSR1 a novel isolate from tannery effluent site: Box-Behnken optimization, equilibrium, kinetics and thermodynamic studies. J. Taiwan Inst. Chem. Eng. 2015;49:156–164. doi: 10.1016/j.jtice.2014.11.026. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.