Abstract
In recent years, children in Thailand have been infected with zoonotic Brugia pahangi. However, the local environment of rubber or oil palm plantations, which would increase their exposure to risk factors of the infection due to mosquito transmission, is unclear. The present study first sought to determine the extent to which variations in the local landscape, such as the elevated versus low-lying ecotope of rubber or oil palm plantations, in a 2-km radius of the geographically defined landscape in a rural area of Suratthani, Southern Thailand could influence the abundance of Armigeres subalbatus and its susceptibility to zoonotic filarial parasite infections compared to Mansonia, Aedes, and Culex, and Coquillettidia. Thereafter, the filarial larvae found in the infected mosquitoes were molecularly investigated. Ar. subalbatus plantation ecotype was not only found to outnumber the local mosquitoes, but was identified as the predominant species that adapted well to the elevated ecotopes of the rubber or oil palm plantations, which existed at altitudes of 60–80 m. The overall rate of zoonotic filarial parasite infections (L1, L2, or L3 larvae) of Ar. subalbatus was 2.5% (95% CI, −0.2 to 4.1), with an average L3 load of 2.3 larvae per infected Ar. subalbatus (95% CI, −0.6 to 13.0); this is because the infections were found to be concentrated in the elevated ecotopes alone. Based on filarial orthologous β-tubulin gene-specific touchup-nested PCR and sequence analysis using 30 L3 larva clones as representatives of 9 Ar. subalbatus infectious pools, Ar. subalbatus either carried B. pahangi or Dirofilaria immitis, or both species. Such findings suggest that Ar. subalbatus might have played an imperative role in the transmission of B. pahangi in the plantation areas infested with Ar. subalbatus.
Keywords: Armigeres subalbatus, Brugia pahangi, Filarial β-tubulin genes, Local landscape variation, Plantation ecotype, Touchup-nested PCR
Graphical abstract

Highlights
-
•
Ar. subalbatus is the natural vector for zoonotic Brugia pahangi.
-
•
Ar. subalbatus plantation ecotype adapted to rubber or oil palm plantations.
-
•
Local landscape variation influencing its abundance and susceptibility to zoonotic filarial infection.
-
•
Touchup-nested PCRs specific for amplifying filarial beta-tubulin genes of L3 larvae present in Ar. subalbatus.
-
•
Implying that Ar. subalbatus carried either B. pahangi or D. immitis, or both.
1. Introduction
Vector-borne parasitic zoonosis is geographically distributed on a global scale [1,2]. As a result, it is a public health problem that has a disproportionate burden due to its occurrence among persons that live below the poverty level [[3], [4], [5]]. Re-emerging and emerging zoonotic Brugia malayi and Brugia pahangi lymphatic filarial parasites can be transmitted by domestic animal reservoirs, such as cats and dogs, to humans [[6], [7], [8], [9]]. Of note, emerging zoonotic infections with B. pahangi parasites have recently been reported to be sporadic in Southeast Asia (SEA) [7,8]. Owing to expansion in geographical locations, host or vector range, and the dynamics of transmission patterns that involve biological, ecological, and social factors in different complex eco-epidemiological settings [5,10], the prevalence of lingering zoonotic infections with B. pahangi parasites is unknown.
In recent years, there has been an emergence of zoonotic B. pahangi infections in children younger than 2 years of age in Thailand (see the supplementary Table S1). Thus, addressing the risk at the interface of human, animal, and the environment, and understanding the vulnerability of children and how they acquire these infections locally through mosquito transmission, are urgently required. Children might be vulnerable to infection with B. pahangi owing to poor social or environmental conditions [3]; the infestation of local mosquitoes from five genera, namely Armigeres, Anopheles, Mansonia, Aedes, and Culex [11]; and the presence of cats or dogs infected with B. pahangi [12] within a 2-km radius of their houses. Of the five mosquito genera mentioned above, Mansonia mosquitoes, such as Ma. uniformis, Ma. indiana, Ma. annulifera, and Ma. bonneae, are the main vectors for B. malayi in Thailand [13] and other countries in SEA [14]. Armigeres subalbatus is the natural vector for zoonotic B. pahangi [15,16] and may also transmit B. pahangi to humans in SEA [8]. There are two ecotypes of Ar. subalbatus that share common characteristics: the forest and plantation ecotypes. The forest ecotype is indigenous to the forest and forest fringe and can breed in natural containers with organic substances. In contrast, the plantation ecotype is well adapted to plantation areas where breeding can occur in either artificial or natural containers with organic substances. The present study attempted to clarify some ecologic and epidemiologic questions regarding the occurrence of zoonotic B. pahangi infections in children by determining the fitness of Ar. subalbatus plantation ecotype (whether this species adapted to the local environment of rubber or oil palm plantations) and its potential to transmit epizootic B. pahangi parasites.
We mimicked local environmental conditions of cases infected with B. pahangi in the plantation areas of Suratthani, Southern Thailand (case no. 1), and Rayong, Eastern Thailand (case no. 2) (Table S1) by employing a 2-km radius of geographically defined local landscape as the study area. This study area covered four different ecotopes, which served as the study sites in Suratthani. Thereafter, we sought to empirically determine the extent to which variations in the local landscape could influence the infestation of Ar. subalbatus plantation ecotypes and zoonotic filarial infections (with L1/L2/L3 juveniles) of Ar. subalbatus compared to other local mosquitoes; and to investigate the representative L3 larva clones of B. pahangi, originally derived from any infectious mosquito pool using a newly developed touchup-nested PCR specific for the amplification of filarial orthologous β-tubulin genes and sequence analysis [[17], [18], [19]].
2. Materials and methods
2.1. Study area and site
Prior to the study carried out in 2013, the local environmental conditions of the receptive plantation areas (Table S1) were considered ideal for the establishment of the selection criterion (i.e., the selection of the local landscape as the study area where domestic animal reservoirs carrying B. pahangi and human blood-seeking mosquitoes inhabit). The selection criteria were based on field data obtained using field surveys. A topographical survey was conducted to gather spatial information and to locate both natural and man-made land surface features using land use maps with elevation contours and geographical positioning system (GPS). This provided the correct information of the scaled and detailed boundary of the selected study area and the GPS coordinates of the ecotopes, which were the selected study sites, based on spatial considerations such as latitude, longitude, altitude, rubber or oil palm plantation polygons, roads, water bodies, building, built-up land, and potential larval breeding sites [20,21]. The ecotope can be defined as a small-scale landscape of the plantation area that is geographically associated with the infestation of locally adapted mosquitoes (e.g., Mansonia, Armigeres, Aedes, Culex, Coquillettidia, or Anopheles). Parasitological survey was carried out to examine zoonotic filarial infections in domestic cats or dogs within the boundary of the selected study area. Blood samples collected from cats or dogs at night were used for this survey. Larval survey was carried out to assess the availability of larval habitats of local mosquitoes within, or proximal to, the selected ecotopes. Household survey was conducted to assess the physical environmental conditions inside and outside houses with domestic animals.
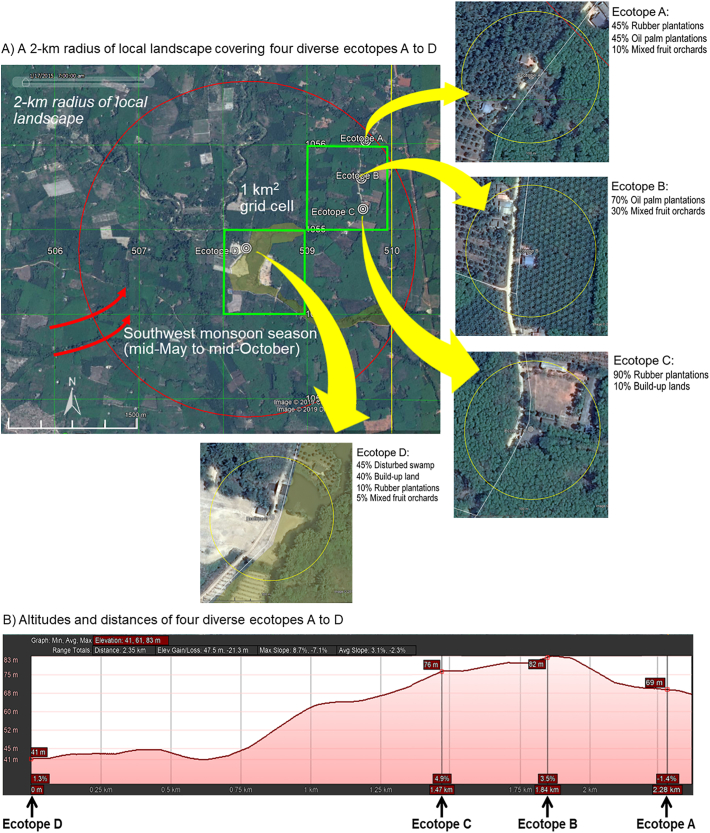
The study area that met the selection criteria covered a 2-km radius of geographically defined local landscape. The study area was also comprised of four diverse ecotopes, which served as the study sites using a 1-km2 universal transverse mercator (UTM) grid cell (Fig. 1A). Of note, the study area was geographically associated with the infestation of local mosquitoes (i.e., Mansonia, Armigeres, Aedes, Culex, and Coquillettidia). The elevated ecotopes (i.e., A to C) that were mainly covered with either rubber or oil palm plantations were located approximately 70 m above sea level (MASL), whereas the low-lying ecotope (i.e., D) with only 10% rubber plantation was located at 40 MASL in a disturbed swamp, which is the breeding habitat of Mansonia mosquitoes (Fig. 1B). The locations of ecotopes A to D were used as the sites of sampling for local mosquitoes as shown in Table S2.
Fig. 1.
Local landscape of the study area and study sites. A) Google Earth maps illustrating a 2-km radius with variations in the geographically defined local landscape and four diverse ecotopes (A to D). A 100-m radius of land use is shown. B) Location of the four ecotopes with different altitudes and distances. Google Earth maps illustrating diverse filariasis ecotopes (A to D). Ecotopes A to C (A) represent land use and land cover distinguishable from ecotope D and (B) geographically associated with the infestation of Mansonia vectors.
2.2. Mosquito collection and dissection
Periodic assessments of species compositions and abundances of the local mosquito populations were carried out between May 2014 and May 2015. At each timepoint of mosquito collection from each ecotope, the human landing catch (HLC) collection method was used to detect the species and numbers of mosquitoes, both indoors and outdoors, between 18:00 h and 21:00 h for 3 consecutive nights (i.e., 3 pools of identified mosquitoes were obtained). The taxonomic identification of individual adult female mosquitoes was blindly performed by two expert entomologists. The abundance of local mosquitoes was expressed as man landing rate (MLR), which infers the number of human blood-seeking mosquitoes per night per person for each ecotope.
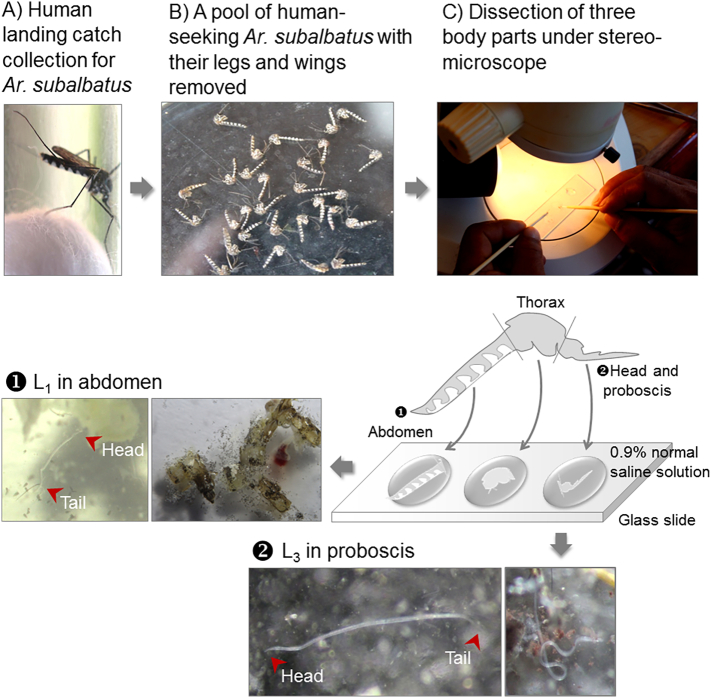
All identified adult female mosquitoes were individually examined for the presence of L1, L2, or L3 larval stages by dissection under a stereomicroscope (Fig. 2). None of the mosquito pools of Culex, Aedes, Mansonia, and Coquillettidia collected from the studied ecotopes was positive, except for the infectious pools of Ar. subalbatus (Fig. 2). The number of larvae recovered from the abdomen (L1), thorax (L2), and head and proboscis (L3) was tallied [18], and the average number and range of L3 per infected mosquito were presented. The infectious mosquito pool must have had at least one adult female mosquito infected with any of the L1, L2, or L3 larval stages. For example, the abundance of the Ar. subalbatus infectious pool was expressed as infectious MLR, which infers the infectious number of human blood-seeking Ar. subalbatus per night per person for each ecotope. The infection rate (%) for each ecotope was expressed as the number of Ar. subalbatus adult female mosquitoes infected with any larval stages (L1, L2, or L3) in the total number of Ar. subalbatus adult female mosquitoes collected by HLC, multiplied by 100. The 95% confidence interval (CI) was used to estimate the infection rate for Ar. subalbatus in all ecotopes and the L3 load for the Ar. subalbatus infectious pools.
Fig. 2.
Ar. subalbatus infectious mosquito pool. Based on HLC collection (A), a pool no. of BAP1, including 29 Ar. subalbatus adult female mosquitoes obtained from ecotope B, was used to represent the Ar. subalbatus infectious mosquito pool that was individually dissected into three body parts (B—C). Compared to L1 obtained from the abdomen ①, the isolation of single L3 larva clone obtained from the proboscis ② is shown.
2.3. Genomic DNA preparation of L3 and positive controls
Immediately after mosquito dissection (Fig. 2), single L3 larva clones originally obtained from the 9 Ar. subalbatus infectious pools of ecotopes A to C were isolated under carefully controlled conditions in the field. Of the 56 L3 larva clones obtained, 30 were used as representatives of the L3 gDNA templates: AAP1 (L01 to L12 clones), AAP2 (L13 to L15 clones), AAP3 (L16 to L18 clones), BAP1 (L19 to L22 clones), BAP2 (L23 clone), BAP3 (L24 to L25 clones), CAP1 (L26 clone), CAP2 (L27 clone), and CAP3 (L28 to L30 clones). These clones were separately prepared for gDNA extraction using the QIAamp® Blood Mini Kit (QIAGEN, Germany), as described elsewhere [19], with modifications. Finally, the eluted L3 gDNA solution (approximate 100–200 μl), which had a 260/280 absorbance ratio of 1.8–2.0, contained the average gDNA content per L3 clone of 994.6 ng, or 7.4 to 12.5 ng/μl.
The positive controls included purified gDNA templates of microfilaremic (Mf) bloods harboring a wide range of microfilarial densities (Mf/ml): Wuchereria bancrofti MMO7 (1246), MDA1 (252), and MMO6 (13) patient isolates [19]; Brugia malayi NT01 (673), NT02 (166), and NT08 (367) patient isolates; B. pahangi DA08 (167) dog isolate and CA12 (300) cat isolate; and Dirofilaria immitis Di106 (>5000) and Di101 (2633) dog isolates. All purified Mf gDNA samples were prepared using the QIAamp® Blood Mini Kit (QIAGEN, Germany) according to the methods described elsewhere [17,19]. Negative controls, such as human, cat, dog, or mosquito gDNAs and nuclease-free deionized water, were used throughout the study.
2.4. Amplification of orthologous β-tubulin genes of L3
2.4.1. Primer design
The nucleotide sequences of filarial orthologous β-tubulin genes were retrieved from the genome databases, GenBank, EMBL, DDBJ, and DFCI. Their homology was analyzed using multiple sequence alignment (Fig. 3) according to the methods described elsewhere [17,19]. Based on the conserved sequences retained in the filarial β-tubulin orthologs, the primer sets designed for the touchup-nested PCR formats (Fig. 3 and Table 1) to yield authentically amplified L3 DNA fragments were tested for specificity and their physical properties, according to the methods described elsewhere [17,19].
Fig. 3.
Multiple sequence alignment of partially homologous β-tubulin genes of human and non-human filariids, which are involved in benzimidazole susceptibility. The retrieved nucleotide sequences (accession no. and positions): Brugia malayi (Bm) (BRQD553TR, 3–789), Brugia pahangi (Bp) (M36380, 2267–3054), Wuchereria bancrofti (Wb) (AY705383, 109–916), Onchocerca volvulus (Ov) (AF019886, 1582–2400), and Dirofilaria immitis (Di) (HM596854, 1462–2244) are shown as coding (upper case) and non-coding (lower case) sequences. The gap (insertion/deletion) was generated to maximize the homology representing conserved (•) and degenerate nucleotide residues. The deduced amino acid sequences (Thr107 to Leu234) of conserved β-tubulin homologs of all taxa are shown along with the amino acid substitutions (blue color-dashed boxes). The primers that specifically annealed to the regions of target L3 gDNAs of B. malayi (Bmtubb), B. pahangi (Bptubb), D. immitis (Ditubb), and W. bancrofti (Wbtubb), and their direction of amplification (arrows) are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Primers used in touchup-nested PCRs specific for the orthologous β-tubulin genes using L3 gDNAs.
| Primer name | Direction/Sequence (5′ to 3′)d | Target DNA: amplicons with expected size (bp) | Reference |
|---|---|---|---|
| Heterologous primersa | |||
| BT7 | Forward/GCTGAIGGATGCGAITGTCTTCAG | Wbtubb (726), | This study |
| BT10 | Reverse/ACTCCIGACATTGTIACAGA | Ditubb (701), | |
| Bmtubb (705), | |||
| Bptubb (706) | |||
| Species-specific primersb | |||
| BT91 | Forward/GGATCCGGATTTCAACTAACG | Wbtubb (493) | [16] |
| BT122 | Reverse/GAATTCCAAGTGGTTGAGGTCG | ||
| BT93 | Forward/GGATCCGGATTCCAACTGACT | Ditubb (477) | This study |
| BT26 | Reverse/GAATTCCAAGTGATTGAGATCG | ||
| Brugia-specific primersc | |||
| BT91 | Forward/GGATCCGGATTTCAACTAACG | Bmtubb (468), | This study |
| BT123 | Reverse/GAATTCCAAATGGTTGAGGTCA | Bptubb (468) | |
Primer sets used in touchup-nested PCRs: afirst-round amplification of orthologous β-tubulin genes and b,csecond-round amplification of Brugia (Bmtubb or Bptubb), D. immitis (Ditubb), and W. bancrofti (Wbtubb) in separate reactions.
dUnderlined sequences of primers represent the 5′ modification sites with additional recognition sequences, BamH I (GGATCC) and EcoR I (GAATTC), while athe degenerated inosine (I) can substitute any base changed (A, G, T, or C).
2.4.2. Touchup-nested PCR
Initially, the optimization of the touchup-nested PCR (TUPCR) program was carried out empirically using a bracket of specific primer-template annealing temperatures in a 25-μl reaction volume, as described elsewhere [19], except that 5 μl (approximately 30–60 ng) of purified L3 gDNA template was used. In primary TUPCR, the heterologous primers, BT7 and BT10, that could specifically amplify 701 to 726 bp amplicons were originally derived from orthologous β-tubulin genes of human or non-human filarial parasites. The reaction was performed with an initial denaturation at 95 °C for 4 min, followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 57 °C for 1 min and polymerization at 72 °C for 1 min. The final extension was performed at 72 °C for 4 min.
For the secondary TUPCRs containing species-specific or Brugia-specific primer sets, the PCR ingredients were similar to those of the first reaction, except that 2 μl of the primary TUPCR products was used as a template. A touchup program (or TU5660) was performed with initial denaturation at 95 °C for 1 min, followed by 5 cycles with successive annealing temperature increments of 1 °C in every cycle. For the first 5 cycles, the reaction mixture was heated at 95 °C for 1 min, followed by annealing at 56 °C → 60 °C for 1 min and polymerization at 72 °C for 1 min. The subsequent 30 cycles of amplification were similar, except that the annealing temperature was 60 °C for 1 min. Lastly, the extension was performed at 72 °C for 4 min. TUPCRs containing both positive controls (approximately 20–30 ng of Mf gDNA templates) and negative controls were performed in the same manner as those containing the L3 gDNA templates.
2.5. Post-PCR analysis and sequencing
Electrophoresis of the PCR products with expected sizes and the determination of the homology of all sequenced amplicons at the DNA and protein levels were accomplished using the methods described elsewhere [17,19]. The nucleotide sequences from this study were deposited in the GenBank database (accession numbers): W. bancrofti (MT674270-MT674272), B. malayi (MT674273- MT674275), B. pahangi (MT674276- MT674279), and D. immitis (MT674280- MT674288).
3. Results
3.1. Entomological findings from different ecotopes
A total of 1393 local mosquitoes, Ar. subalbtus (953), Cx. vishnui (133), Cx. gelidus (105), Ae. albopictus (80), Ma. uniformis (58), Ae. aegypti (23), Ma. indiana (15), Ma. bonneae (10), Cq. crassipes (6), Cx. quinquefasciatus (5), Cx. sp. (4), and Cx. nigropunctatus (1) were collected using the HLC method (Table 2). Based on the relative ratio (pi), Ar. subalbatus was the predominant species sessile to the elevated ecotopes, A (pi = 0.807), B (pi = 0.829), and C (pi = 0.543), but not the low-lying ecotope, D (pi = 0.103) when compared to other locally adapted mosquitoes such as Cx. vishnui, Cx. gelidus, Ae. albopictus, and Ma. uniformis (Table 2). The more abundant Ar. subalbatus mosquito population, which was locally adapted to the elevated ecotopes, A (29.67), B (31.83), and C (16.90), tended to exhibit an average MLR of 19.85, which was 4-fold to 20-fold greater than that of other mosquito species: Culex spp. (5.17), Aedes spp. (2.15), and Mansonia spp. (1.73) (Table 3).
Table 2.
Species and numbers (pi)a of human blood-seeking adult female mosquitoesb in four ecotopes based HLC collection method, ordered by predominant species.
| Species | Ecotope A (N = 441) |
Ecotope B (N = 461) |
Ecotope C (N = 374) |
Ecotope D (N = 117) |
All (N = 1393) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | pi | n | pi | n | pi | n | pi | n | pi | |
| Ar. subalbatus | 356 | 0.807 | 382 | 0.829 | 203 | 0.543 | 12 | 0.103 | 953 | 0.684 |
| Cx. vishnui | 52 | 0.118 | 20 | 0.043 | 48 | 0.128 | 13 | 0.111 | 133 | 0.095 |
| Cx. gelidus | 11 | 0.025 | 13 | 0.028 | 69 | 0.184 | 12 | 0.102 | 105 | 0.075 |
| Ae. albopictus | 11 | 0.025 | 40 | 0.087 | 26 | 0.069 | 3 | 0.026 | 80 | 0.057 |
| Ma. uniformis | 1 | 0.002 | 2 | 0.004 | 3 | 0.008 | 52 | 0.444 | 58 | 0.042 |
| Ae. aegytpi | 5 | 0.011 | 3 | 0.006 | 10 | 0.027 | 5 | 0.043 | 23 | 0.016 |
| Ma. indiana | 2 | 0.005 | 0 | 0 | 10 | 0.027 | 3 | 0.026 | 15 | 0.011 |
| Ma. bonneae | 0 | 0 | 0 | 0 | 1 | 0.003 | 9 | 0.077 | 10 | 0.007 |
| Cq. crassipes | 3 | 0.007 | 0 | 0 | 3 | 0.008 | 0 | 0 | 6 | 0.004 |
| Cx. quinquefasciatus | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0.043 | 5 | 0.003 |
| Cx. sp. | 0 | 0 | 1 | 0.002 | 1 | 0.003 | 2 | 0.017 | 4 | 0.003 |
| Cx. nigropunctatus | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.008 | 1 | 0.001 |
The predominant species was derived based on the proportion of a mosquito species relative to the total number of all identified mosquito species (pi) present in each ecotope.
The data were obtained from a repeated cross-sectional entomological survey between May 2014 and May 2015.
Table 3.
The abundance of local mosquitoes in the four ecotopes, ordered by predominant local mosquitoes.a
| Local mosquitoes | MLRd |
||||
|---|---|---|---|---|---|
| Ecotope A | Ecotope B | Ecotope C | Ecotope D | Average | |
| Ar. subalbatus | 29.67 | 31.83 | 16.90 | 1.00 | 19.85 |
| Culex spp. a | 5.25 | 2.83 | 9.83 | 2.75 | 5.17 |
| Aedes spp.b | 1.33 | 3.58 | 3.0 | 0.67 | 2.15 |
| Mansonia spp.c | 0.25 | 0.17 | 1.17 | 5.33 | 1.73 |
| Cq. crassipes | 0.25 | 0 | 0.25 | 0 | 0.12 |
Including Cx. vishnui, Cx. gelidus, Cx. quinquefasciatus, Cx. nigropunctatus, and Cx. sp.
Including Ae. albopictus and Ae. aegypti.
Including Ma. bonneae, Ma. uniformis, and Ma. indiana.
MLR expressed as the number of human blood-seeking mosquitoes per night per person.
All 440 HLC-collected adult female mosquitoes belonging to the genera Culex (n = 248), Aedes (n = 103), Mansonia (n = 83), and Coquillettidia (n = 6) (Table 2) were negative based on mosquito dissection. In contrast, of the 941 Ar. subalbatus mosquitoes obtained from the 9 Ar. subalbatus mosquito pools of the elevated ecotopes (A to C) alone, there were 24 infected mosquitoes (2.5% overall infection rate) of varying filarial parasite infections in ecotope C (2.96%), B (2.62%), and A (2.25%) (Table 4). However, Ar. subalbatus tended to exhibit an average infectious MLR of 0.5, with infectious MLRs of 0.83 (ecotope B), 0.67 (ecotope A), and 0.50 (ecotope C) (Table 4). All 9 Ar. subalbatus infectious pools had an average filarial larva number of 5 (110 larvae/24 infected mosquitoes). Further, Ar. subalbatus tended to harbor 56 infective L3, which was found to be three-fold greater than the amount of L1 (29) and L2 (25) harbored (Table S3).
Table 4.
Infectious man landing rates (MLR) for Armigeres subalbatus in the four ecotopes.
| Ecotope | Mosquito pool | Adult female no. (%) | Infectious adult female no. (% infection rate) | Infectious MLRa |
|---|---|---|---|---|
| A | AAP1 | 154 | 4 | |
| AAP2 | 79 | 3 | ||
| AAP3 | 123 | 1 | ||
| Subtotal | 356 (37.36) | 8 (2.25) | 0.67 | |
| B | BAP1 | 155 | 3 | |
| BAP2 | 108 | 4 | ||
| BAP3 | 119 | 3 | ||
| Subtotal | 382 (40.08) | 10 (2.62) | 0.83 | |
| C | CAP1 | 66 | 1 | |
| CAP2 | 45 | 2 | ||
| CAP3 | 92 | 3 | ||
| Subtotal | 203 (21.30) | 6 (2.96) | 0.50 | |
| D | DAP1 | 8 | 0 | |
| DAP2 | 2 | 0 | ||
| DAP3 | 2 | 0 | ||
| Subtotal | 12 (1.26) | 0 | 0 | |
| All | Total | 953 | 24 (2.52)b | 0.50 |
Infectious MLR expressed as the infectious number of human blood-seeking Ar. subalbatus per night per person for each ecotope.
The overall infection rate with 95% CI (−0.17 to 4.08) for all ecotopes.
3.2. Molecular findings
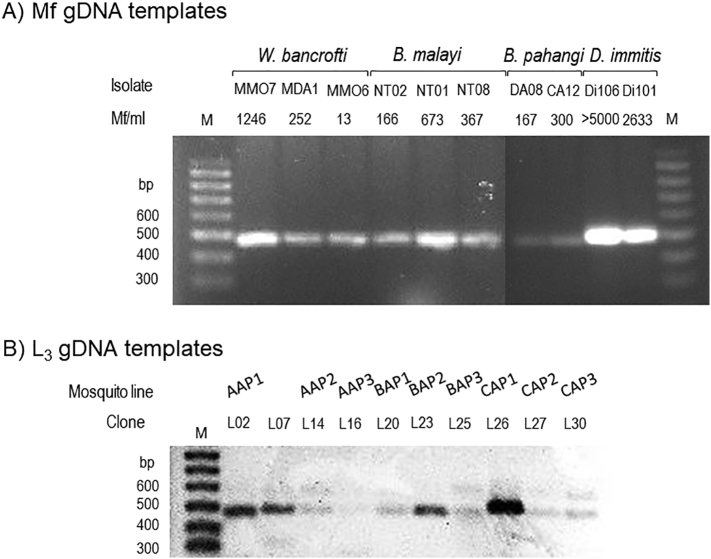
Based on the orthologous β-tubulin gene-specific TUPCR amplification carried out using gDNA templates of 30 representative L3 larva clones, the Ar. subalbatus infectious pool, AAP1, corresponded to infections with both B. pahangi (468-bp amplicons of L02 clone) and D. immitis (477-bp amplicons of the L07 clone) (Fig. 4). The CAP3 infectious pool corresponded with the single infection of B. pahangi, showing 468-bp amplicons of the L28-L30 clones. The other infectious pools, AAP2, AAP3, BAP1, BAP2, BAP3, CAP1, and CAP2 might have carried the single infection of D. immitis, whose reactions putatively yielded 477-bp amplicons. None of the L3 gDNA was positive with the species-specific primers for W. bancrofti and B. malayi. Moreover, the representative L3 larva clones whose sequenced amplicons had 100% homology to the target filarial β-tubulin genes of B. pahangi were L02 and L30, while those of D. immitis were L07, L14, L20, L23, L25, L26, and L27.
Fig. 4.
Touchup-nested PCR (TNPCR) amplification specific for orthologous β-tubulin genes of human and non-human filarial parasites using representative gDNA templates that were used to originally derive the Mf isolates (A) and L3 larva clones (B) as mentioned in the text. A) Amplification patterns of secondary TNPCR containing Mf gDNAs are displayed for specific amplicons with expected sizes (bp) authentically derived from the β-tubulin genes of W. bancrofti (493 bp), B. malayi (468 bp), B. pahangi (468 bp), and D. immitis (477 bp). B) Amplification patterns of secondary TNPCR containing L3 gDNAs are displayed for specific amplicons with expected sizes (bp): 468-bp B. pahangi amplicons (L02 and L30) and 477-bp D. immitis amplicons (L07, L14, L16, L20, L23, L25, L26, and L27).
4. Discussion
Armigeres subalbatus, the natural vector for B. pahangi and D. immitis, is commonly found in urban and rural areas in SEA [8,14,22,23] and South Asia [24,25]. In this study, Ar. subalbatus plantation ecotype was found to be the predominant species carrying zoonotic B. pahangi and D. immitis, and may have played an imperative role in circulating these parasites in the local landscape assessed in the rural area of Suratthani. Based on the results obtained herein, variation in the local landscape and environment influenced the infestation of Ar. subalbatus, enabling them to adapt well to the elevated ecotopes of rubber or oil palm plantations at 60–80 MASL. Rubber or oil palm plantations and not disturbed swamp, which was found in ecotope D, might be the ideal places for the breeding of Ar. subalabatus in both artificial and natural containers with organic matters. Its adaptability markedly contributed to its outnumbering of its counterpart species, such as Mansonia, Aedes, and Culex, and foraging blood meals in human dwellings with domestic cats or dogs. Further, there were high abundances of Ar. subalbatus in the elevated ecotopes as shown in Table 3 and Table S2.
In the local landscape examined herein, the zoonotic filarial infection rate was approximately 2.5% or up to 4% in the natural population of Ar. subalbatus. However, an accurate estimate of the B. pahangi infection in Ar. subalbatus was not established. When filarial orthologous beta-tubulin gene-specific TUPCR was carried out using representative L3 larva clones, any Ar. subalbatus infectious pool might have carried either B. pahangi or D. immitis, or both B. pahangi and D. immitis. This finding strongly suggests that Ar. subalbatus is susceptible to these zoonotic filarial parasites [8,[14], [15], [16]] with a wide range of infective L3 loads. Ar. subalbatus might have a disproportionate L3 load of B. pahangi relative to D. immitis, despite its infection rate being estimated to be less than 1% in the natural population. Such finding might be explained by the feeding behaviors of Ar. subalbatus that more likely carried out a vicious attack on dogs in the outdoor settings than cats when seeking animal blood meals during the peak hour of 18:30–19:30 h. Moreover, Ar. subalbatus might have a flight range longer than 100 m, even up to 1000 m. However, this might not relate B. pahangi infected cats to B. pahangi infections in Ar. subalbatus in the elevated ecotopes of A to C owing to its animal host range [23]. Similar to that observed in Malaysia, the local transmission of zoonotic B. pahangi might be due to the fitness of the Ar. subalbatus vectors, their ability to produce large numbers, and the source of infections in domestic cats [7,8,12]; a further study is required to verify the above findings.
Collectively, our findings are general considerations of the epidemiological catchment area comprised of designated sites scalable for vector surveillance and case investigation of B. pahangi infections in children. However, several factors, including the variation in the local landscape and environment, availability of Ar. subalbatus breeding sites proximal to the dwellings of domestic animals, the abundance of Ar. subalbatus, and the prevalence of B. pahangi infection in domestic animals, might be important.
5. Conclusion
Variations in the local landscape and environment, such as elevated instead of a low-lying ecotope of rubber plantations or oil palm plantations, could influence higher abundances of Ar. subalbatus plantation ecotypes and higher rates of zoonotic filarial parasite infections of B. pahangi and D. immitis in Ar. subalbatus than other local mosquitoes belonging to the genera Mansonia, Aedes, Culex, and Coquillettidia. Based on the molecular investigation of the L3 larva clones, which were representatives of the Ar. subalbatus infectious pools, Ar. subalbatus could either carry B. pahangi or D. immitis, or both species. Such findings imply the potential role of Ar. subalbatus in the natural transmission of not only zoonotic B. pahangi infection, but also D. immitis in plantation areas in Thailand, regardless of altitude.
Ethics approval
The authors assert that all procedures contributing to this work complied with the ethical standards and were approved by the Institutional Review Board at the Faculty of Veterinary Medicine, Mahanakorn University of Technology (Approval No. ACUC-MUT-2014/001).
Funding resources
The research was supported by The Thailand Research Fund; Office of the Higher Education Commission; and Mahanakorn University of Technology, Bangkok, Thailand (grant number MRG5680087, 2013).
Authors' contributions
Apiradee Intarapuk: Conceptualiation; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Validation; Writing - original draft.
Adisak Bhumiratana: Conceptualiation; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Validation; Visualization; Writing - original draft; Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Suntorn Pimnon and Supit Yodmek for identifying the mosquito species in the study. The authors also thank the staff of the Vector Borne Disease Control Unit 11.3.5 Chaiya for supporting the field works.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2021.100261.
Appendix A. Supplementary data
The Supplementary Table S1
The Supplementary Table S2
The Supplementary Table S3
Amplicon sequences of L3 clones
References
- 1.World Health Organization . 2020. Zoonoses.https://www.who.int/news-room/fact-sheets/detail/zoonoses [Google Scholar]
- 2.World Health Organization, Food and Agriculture Organization of the United Nations, World Organisation for Animal Health . World Health Organization; 2019. Taking a multisectoral, one health approach: a tripartite guide to addressing zoonotic diseases in countries.https://apps.who.int/iris/handle/10665/325620 [Google Scholar]
- 3.World Health Organization, United Kingdom. Dept. for International Development, Animal Health Programme, Food and Agriculture Organization of the United Nations & World Organisation for Animal Health . WHO Headquarters, Geneva, with the Participation of FAO and OIE. World Health Organization; 2006. The control of neglected zoonotic diseases: a route to poverty alleviation: report of a joint WHO/DFID-AHP meeting, 20 and 21 September 2005.https://apps.who.int/iris/handle/10665/43485 [Google Scholar]
- 4.World Health Organization, Regional Office for South-East Asia . 2005. Combating emerging infectious diseases in the South-East Asia Region, WHO Regional Office for South-East Asia.https://apps.who.int/iris/handle/10665/204878 [Google Scholar]
- 5.Coker R.J., Hunter B.M., Rudge J.W., Liverani M., Hanvoravongchai P. Emerging infectious diseases in Southeast Asia: regional challenges to control. Lancet. 2011;337:599–609. doi: 10.1016/S0140-6736(10)62004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paniz-Mondolfi A.E., Gárate T., Stavropoulos C., Fan W., González L.M., Eberhard M., Kimmelstiel F., Sordillo E.M. Zoonotic filariasis caused by novel Brugia sp. nematode, United States, 2011. Emerg. Infect. Dis. 2014;20:1248–1250. doi: 10.3201/eid2007.131654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan L.H., Fong M.Y., Mahmud R., Muslim A., Lau Y.L., Kamarulzaman A. Zoonotic Brugia pahangi filariasis in a suburbia of Kuala Lumpur City, Malaysia. Parasitol. Int. 2011;60:111–113. doi: 10.1016/j.parint.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Muslim A., Fong M.Y., Mahmud R., Sivanandam S. Vector and reservoir host of a case of human Brugia pahangi infection in Selangor, peninsular Malaysia. Trop. Biomed. 2013;30 727–230. [PubMed] [Google Scholar]
- 9.Ravindran R., Varghese S., Nair S.N., Balan V.M., Lakshmanan B., Ashruf R.M., Kumar S.S., Kumar A., Gopalan K., Nair A.S., Malayil A., Chandrasekhar L., Juliet S., Kopparambil D., Ramachandran R., Kunjupillai R., Kakada S.A.M. Canine filarial infections in a human Brugia malayi endemic area of India. Biomed. Res. Int. 2014;2014:630160. doi: 10.1155/2014/630160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaffry K.T., Ali S., Rasool A., Raza A., Gill Z.J. Zoonoses. Int. J. Agric. Biol. 2009;11:217–220. [Google Scholar]
- 11.Woodbridge A., Foster A., Walker E.D. Medical and Veterinary Entomology (Third Edition) 2019. Chapter 15 - Mosquitoes (Culicidae) pp. 261–325. [DOI] [Google Scholar]
- 12.Nuchprayoon S., Junpee A., Nithiuthai S., Chungpivat S., Suvannadabba S., Poovorawan Y. Detection of filarial parasites in domestic cats by PCR-RFLP of ITS1. Vet. Parasitol. 2006;140:366–372. doi: 10.1016/j.vetpar.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Zielke E., Hinz E., Sucharit S. Lymphatic filariasis in Thailand: a review on distribution and transmission. Mitt. Österr. Ges.Tropenmed. Parasitol. 1993;15:141–148. [Google Scholar]
- 14.Mulyaningsih B., Umniyati S.R., Hadisusanto S., Edyansyah E. Study on vector mosquito of zoonotic Brugia malayi in Musi Rawas, South Sumatera, Indonesia. Vet. World. 2019;12:1729–1734. doi: 10.14202/vetworld.2019.1729-1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aliota M.T., Fuchs J.F., Rocheleau T.A., Clark A.K., Hillyer J.F., Chen C.C., Christensen B.M. Mosquito transcriptome profiles and filarial worm susceptibility in Armigeres subalbatus. PLoS Negl. Trop. Dis. 2010;4 doi: 10.1371/journal.pntd.0000666. e666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto H., Kobayashi M., Ogura N., Tsuruoka H., Chigusa Y. Studies on filariasis VI: the encapsulation of Brugia malayi and B. pahangi larvae in the mosquito, Armigeres subalbatus. Jpn. J. Sanit. Zool. 1985;36:1–6. doi: 10.7601/mez.36.1_1. [DOI] [Google Scholar]
- 17.Bhumiratana A., Pechgit P., Koyadun S., Siriaut C., Yongyuth P. Imported bancroftian filariasis: Diethylcarbamazine response and benzimidazole susceptibility of Wuchereria bancrofti in dynamic cross-border migrant population targeted by the National Program to eliminate lymphatic Filariasis in South Thailand. Acta Trop. 2010;113:121–128. doi: 10.1016/j.actatropica.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Bhumiratana A., Intarapuk A., Sangthong D., Koyadun S., Pechgit P., Pothikasikorn J. Molecular diagnosis and monitoring of benzimidazole susceptibility of human filariids. In: Rodriguez-Morales A.J., editor. Current Topics in Tropical Medicine. 2012. pp. 397–424. [DOI] [Google Scholar]
- 19.Pechgit P., Intarapuk A., Pinyoowong D., Bhumiratana A. Touchdown-touchup nested PCR for low-copy gene detection of benzimidazole-susceptible Wuchereria bancrofti with a Wolbachia endosymbiont imported by migrant carriers. Exp. Parasitol. 2011;127:559–568. doi: 10.1016/j.exppara.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Kaewwaen W., Bhumiratana A. Landscape ecology and epidemiology of malaria-associated rubber plantations in Thailand: integrated approaches to malaria ecotoping. Interdiscip. Perspect. Infect. Dis. 2015 doi: 10.1155/2015/909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorosjinda-Nunthawarasilp P., Bhumiratana A. Ecotope-based entomological surveillance and molecular xenomonitoring of multidrug resistance malaria in Anopheles vectors. Interdiscip. Perspect. Infect. Dis. 2014 doi: 10.1155/2014/969531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheong W.H., Mak J.W., Naidu S., Mahadevan S. Armigeres subalbatus incriminated as an important vector of the dog heartworm Dirofilaria immitis and the bird Cardiofilaria in urban Kuala Lumpur. Southeast Asian J. Trop. Med. Public Health. 1981;12:611–612. [PubMed] [Google Scholar]
- 23.Boonserm R., Jantorn R., Phumee A., Sor-suwan S., Jariyapan N., Tiawsirisup S. P Siriyasatien, identification of blood meal from field collected filarial vector mosquitoes, Armigeres subalbatus by multiplex PCR. Thai J. Vet. Med. 2019;49:155–160. [Google Scholar]
- 24.Das P., Bhattacharya S., Chakraborty S., Palit A., Das S., Ghosh K.K., Hati A.K. Diurnal man-biting activity of Armigeres subalbatus (Coquillet, 1898) In a village in West Bengal. Indian. J. Med. Res. 1983;78:794–798. [PubMed] [Google Scholar]
- 25.Rajavel A.R. Larval habitat of Armigeres subalbatus (COQ) and its characteristics in Pondicherry. Southeast Asian J. Trop. Med. Public Health. 1992;23:470–473. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary Table S1
The Supplementary Table S2
The Supplementary Table S3
Amplicon sequences of L3 clones