Abstract
Bacteriophages encode diverse anti-CRISPR (Acr) proteins that inhibit CRISPR-Cas immunity during infection of their bacterial hosts. Although detailed mechanisms have been characterized for multiple Acr proteins, an understanding of their role in phage infection biology is just emerging. Here, we review recent work in this area and propose a framework of “phage autonomy” to evaluate CRISPR-immune evasion strategies. During phage infection, Acr proteins are deployed by a tightly regulated “fast on-fast off” transcriptional burst, which is necessary, but insufficient, for CRISPR-Cas inactivation. Instead of a single phage shutting down CRISPR-Cas immunity, a community of acr-carrying phages cooperate to suppress bacterial immunity, displaying low phage autonomy. Enzymatic Acr proteins with novel mechanisms have been recently revealed and are predicted to enhance phage autonomy, while phage DNA protective measures offer the highest phage autonomy observed. These varied Acr mechanisms and strengths also have unexpected impacts on the bacterial populations and competing phages.
In brief:
While CRISPR-Cas immune systems target invasive bacteriophages (phages), co-evolved anti-CRISPR mechanisms can disable CRISPR-Cas activity. However, phage anti-CRISPR mechanisms vary in potency, leading to a spectrum of interesting ecological and evolutionary outcomes. Here, we review recent studies and discuss potential future directions on this frontier.
Introduction:
Bacterial viruses, also known as bacteriophages (phages), are the most abundant biological entities on the planet and outnumber their bacteria host by tenfold (Brüssow and Hendrix, 2002). Bacteria and phage have been co-evolving for over three billion years. The incessant arms race between pathogens and their host is a major force that drives the evolution of all living forms on earth (Forterre and Prangishvili, 2009). It has been estimated that ~1025 phage infections happen every second and cause up to 80% of bacterial mortality (Rohwer et al., 2009). These extreme selection pressures led bacteria to evolve an arsenal of anti-phage immune mechanisms. Some of the most well-studied bacterial immunity pathways include restriction-modification, toxin-antitoxin systems, abortive infection systems, and CRISPR-Cas (clustered regularly interspaced short palindromic repeats; CRISPR-associated) systems. Many new immune processes, such as prokaryotic argonautes and cyclic nucleotide based systems have recently been uncovered (Bernheim and Sorek, 2020).
CRISPR-Cas systems are the only adaptive immune system observed to date in the prokaryotic world. During phage infection, a Cas protein complex (often Cas1-Cas2) recognizes short (30–40 nucleotides) DNA fragments, called “protospacers”, from the invading viral genome, followed by integration of spacers into the bacterial CRISPR array and synthesis of a new repeat (Barrangou et al., 2007). Spacer sequences, flanked by direct repeats, comprise the CRISPR array and are inherited by progeny (Fig. 1A, Adaptation). The CRISPR array thus serves as a memory reservoir of past infections and the DNA template for the generation of CRISPR RNAs (crRNAs) (Fig. 1A, Expression) (Brouns et al., 2008; Carte et al., 2008; Haurwitz et al., 2010). Upon phage re-infection, crRNAs guide Cas proteins to recognize and destroy viral DNA via base-pairing (Fig. 1B, Interference) (Barrangou et al., 2007; Brouns et al., 2008; Garneau et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012; Marraffini and Sontheimer, 2008; Sapranauskas et al., 2011), often relying on a 2–6 nucleotide (nt) protospacer adjacent motif (PAM) (Deveau et al., 2008; Gasiunas et al., 2012; Marraffini and Sontheimer, 2010; Mojica et al., 2009). Despite these general mechanistic commonalities, CRISPR-Cas effector complexes are very diverse, divided into multi- (Class 1) and single-effector (Class 2) systems, and further broken into six types (I-VI), each with multiple subtypes (Koonin and Makarova, 2019). In addition, CRISPR-Cas systems are a prevalent defense mechanism in prokaryotes and are estimated to exist in 50% bacteria and 90% archaea (Makarova et al., 2015).
Figure 1: CRISPR-Cas immunity.

A) During phage infection, adaptation occurs when Cas proteins (Cas1&Cas2) acquire spacers from phage genome (i.e. the boxed region) and insert them into CRISPR array. During expression, pre-crRNA is transcribed from the CRISPR array and further processed into crRNA. B) Upon subsequent phage infection, base-pairing between the crRNA-guided surveillance complex (Cascade is shown) and phage protospacer guides cleavage of phage DNA, a process called “interference”. C) By contrast, partial base-pairing between crRNA and phage protospacer leads to priming adaptation where crRNA-guided Cas proteins (Cascade complex, Cas1, Cas2, and Cas3 are shown) rapidly acquire spacers surrounding the partially matched phage genomic region, for instance, the boxed region. The white space between B) and C) indicates two broad types of Acr proteins that 1. inhibit crRNA-guided DNA-binding and priming adaptation and that 2. block the cleavage of phage DNA. Created with BioRender.com
As a prevalent and effective defense mechanism, CRISPR-Cas undoubtedly plays a major role in the ecology and evolution of bacteria and their corresponding phages. On one hand, CRISPR-Cas systems benefit bacteria by protecting them from phage invasion. On the other hand, CRISPR-Cas systems may hinder the acquisition of beneficial genes and generate autoimmunity risk. First, CRISPR-Cas systems cannot distinguish between self and non-self DNA during spacer acquisition and thus, occasionally acquire spacers targeting its own genome, resulting in immunopathology and bacterial death (Levy et al., 2015; Vercoe et al., 2013; Wei et al., 2015). Second, CRISPR-Cas immunity inhibits bacterial cells from obtaining external DNA, which often contribute to the development of novel functions (i.e. pathogenicity and antibiotic resistance) in bacteria (Horvath and Barrangou, 2010; Husnik and McCutcheon, 2018). The detrimental effects caused by CRISPR-Cas systems, however, can be diminished if CRISPR-Cas expression is under tight regulation. Indeed, studies have found that multiple cues or pathways are involved in the regulation of CRISPR-Cas activity. For instance, by coupling with the quorum sensing (QS) pathway and the KinB-AlgB two-component system, CRISPR-Cas expression is not turned on until cell density is high and the bacterial population is more vulnerable to phage invasion (Borges et al., 2020; Høyland-Kroghsbo et al., 2017; Patterson et al., 2016). As a result, CRISPR-Cas may facilitate the re-wiring of gene regulation in bacteria.
Furthermore, the CRISPR-Cas mechanism has been shown to benefit phages unexpectedly by increasing phage mutation rate and accelerating phage evolution (Tao et al., 2018).
To evade bacterial CRISPR-Cas systems, phages have evolved counter-defense mechanisms by encoding anti-CRISPR (Acr) proteins. Acr proteins inhibit CRISPR-Cas activity through a diversity of mechanisms, which enables phage to successfully reproduce via the replicative lytic cycle or through the lysogenic cycle, where phages insert their DNA into the bacterial genome. Acr proteins are typically small (~80–150aa) proteins that directly bind to target Cas proteins and inactivate them. Due to this attribute, Acr proteins have been adapted as an “off-switch” to inactivate CRISPR-Cas functions in various heterologous hosts, enabling the development of more controllable and precise CRISPR-Cas tools (Marino et al., 2020). For a detailed discussion of Acr discovery, biochemical mechanisms, and structures that have been elucidated, we direct the reader to reviews on the subject (Borges et al., 2017; Davidson et al., 2020; Pawluk et al., 2018; Stanley and Maxwell, 2018; Wiegand et al., 2020). By comparison, how Acr proteins affect bacteria-phage interactions and their ecology and evolution has drawn less attention and remains elusive. There is a long road ahead of us to understand the role of Acr proteins in driving the massive diversity of bacteria and phage species on our planet. Nonetheless, several studies have started to address Acr ecology and evolution by well-controlled experiments in laboratory conditions. In this review, we focus on the impact of CRISPR-Acr interactions on phage infection biology, the role of Acr proteins in the ecology and evolution of bacteria and phages, and lastly, how novel classes of anti-CRISPR mechanisms influence “phage autonomy.”
Anti-CRISPR proteins inhibit CRISPR-Cas immunity
With a fast multiplication rate and a large population size, phages can readily accumulate mutations in the cognate target sequences (protospacers and PAMs) to evade CRISPR-Cas targeting (Barrangou et al., 2007; Deveau et al., 2008; Semenova et al., 2011). Surprisingly, this approach is not as effective as one may expect due to at least three reasons:
First, CRISPR-Cas “priming” adaptation allows a fast restoration of CRISPR-Cas interference towards mutated phage escapers (Fig. 1C) (Datsenko et al., 2012; Vorontsova et al., 2015). Best characterized in the Type I multi-effector Cascade systems, the CRISPR-Cas spacer acquisition machinery can acquire spacers from proximal genomic regions after binding to a mismatched target (for instance, mutant phage DNA) (Fig. 1C) (Datsenko et al., 2012), a process named priming adaptation. By contrast to CRISPR-Cas cleavage, which demands strict complementarity between protospacer and crRNA, CRISPR-Cas priming adaptation is extremely lenient and tolerates up to 13 mismatches (Fineran et al., 2014). This flexibility enables CRISPR-Cas to quickly adapt to divergent viral genomes that bypass CRISPR-Cas defense. Additionally, interference-driven spacer acquisition, which is analogous to the priming adaptation, allows additional spacer acquisition from phage genomes during CRISPR-Cas interference (Staals et al., 2016). The interference-driven spacer acquisition increases the number of spacers targeting the same phage and decreases the possibility of phage escaping the CRISPR-Cas immunity via protospacer mutations (Staals et al., 2016). Second, phage escapers are precluded by the distributed CRISPR-Cas immunity in a diverse bacterial community, where cells carry different spacers. To overcome the distributed immunity, phages have to acquire mutations in all cognate target sequences simultaneously, which is highly unlikely (Childs et al., 2014; van Houte et al., 2016). Therefore, the synergy within a well-mixed bacterial population, which happens often in natural environments (van Houte et al., 2016; Paez-Espino et al., 2013), increases the chance of purging out invaders in a timely manner and decreases the spread of invaders. Third, it has been demonstrated that compared to wild-type phage, mutated phage escapers often suffer from fitness costs (Chabas et al., 2019). This fitness cost limits the growth and spread of these escapers. Together, these three reasons limit the efficacy of escape mutations, suggesting the need for alternative and potentially more sophisticated “anti-CRISPR” mechanisms.
Anti-CRISPR mechanisms were first uncovered in a 2013 study, which discovered that several prophages encoded 1–2 distinct anti-CRISPR proteins (AcrIF1–5) that inactivated the Pseudomonas aeruginosa Type I-F CRISPR-Cas system (Bondy-Denomy et al., 2013). Subsequently, genes in the same locus (AcrIE1–4) were shown to inhibit Type I-E CRISPR-Cas systems, another highly abundant CRISPR-Cas system in P. aeruginosa (Pawluk et al., 2014). While mature crRNA levels were not impacted by the AcrIF proteins (Bondy-Denomy et al., 2013), ruling out biogenesis inhibition, in vitro binding assays revealed that AcrIF1–4 each bind directly to different CRISPR-Cas components, preventing Cas proteins from binding or cleaving phage DNA (Fig. 1B&1C) (Bondy-Denomy et al., 2015). More recently discovered anti-CRISPR proteins also often interact tightly with Cas protein targets, which enables Acr proteins to disable the immune system, independent of the spacer sequence or the origin of the target (i.e. phage Acr proteins inactivate plasmid targeting and plasmid Acr proteins block phage targeting). Since the initial identification of Acr proteins, 54 distinct families of Acr proteins, inhibiting CRISPR-Cas type I, II, III, V, and VI, have been identified. Acr proteins often interact tightly with target Cas proteins and block amino acid residues that are highly conserved and essential for bacterial immune function (Davidson et al., 2020; Wiegand et al., 2020).
Anti-CRISPR deployment during infection
While acr genes are highly variable and non-homologous, conserved genes that encode predicted helix-turn-helix (HTH) DNA-binding domains are often found next to acr genes, which led to their naming as “anti-CRISPR associated” genes (aca). Despite their frequent observation, the functional importance of aca genes was unaddressed until recently.
The model P. aeruginosa phage JBD30, which contains an acrIF1-aca1 locus, rapidly expresses acrIF1 at the onset of infection. This expression is subsequently attenuated by accumulated Aca1 (Stanley et al., 2019). Deletion of aca1 from the phage genome results in toxic over-expression of the acr locus (Fig. 2 left panel). The uncontrolled transcription from the strong acr promoter dramatically decreases the transcription of downstream essential genes and results in phage lethality (Stanley et al., 2019). A similar role has been observed for Pectobacterium carotovorum aca2 (Birkholz et al., 2019; Stanley et al., 2019) and Neisseria aca3 (Stanley et al., 2019). Moreover, a recent study found that AcrIIA1 in Listeria monocytogenes temperate phages has dual functions as both an Acr protein that binds Cas9 and an acr autorepressor (Osuna et al., 2020a). Again, this autorepression activity was shown to be important for phage fitness. The dual function of AcrIIA1 is realized by a two-domain architecture with its C-terminal domain (CTD) serving as an Acr protein and its N-terminal domain (NTD) as a transcriptional repressor (Fig. 2 right panel). When cas9 expression is high, AcrIIA1 binds and inhibits cas9 and de-represses its own promoter. Such a bi-functional protein thus enables phages to sense the host immunity and adjust its anti-CRISPR activity accordingly. Interestingly, the NTD repressor domain was also identified in some bacterial genomes, where it was suggested to act as an anti-CRISPR repressor or “anti-anti-CRISPR” (Osuna et al., 2020a). Other HTH-Acr fusions with autorepression capabilities were also recently reported for AcrIIA13-AcrIIA15 found in several Staphylococcus genomes (Watters et al., 2020), although their importance for phage replication was not assessed. Additionally, AcrIIA6 and AcrVA4 possess HTH domains, with unknown functions (Hynes et al., 2018; Zhang et al., 2019). These studies together demonstrate the general repressive role of Aca proteins and HTH-Acr fusions in both Gram negative and positive bacteria and their importance as insulators from surrounding genetic elements.
Figure 2: Regulation of acr expression.
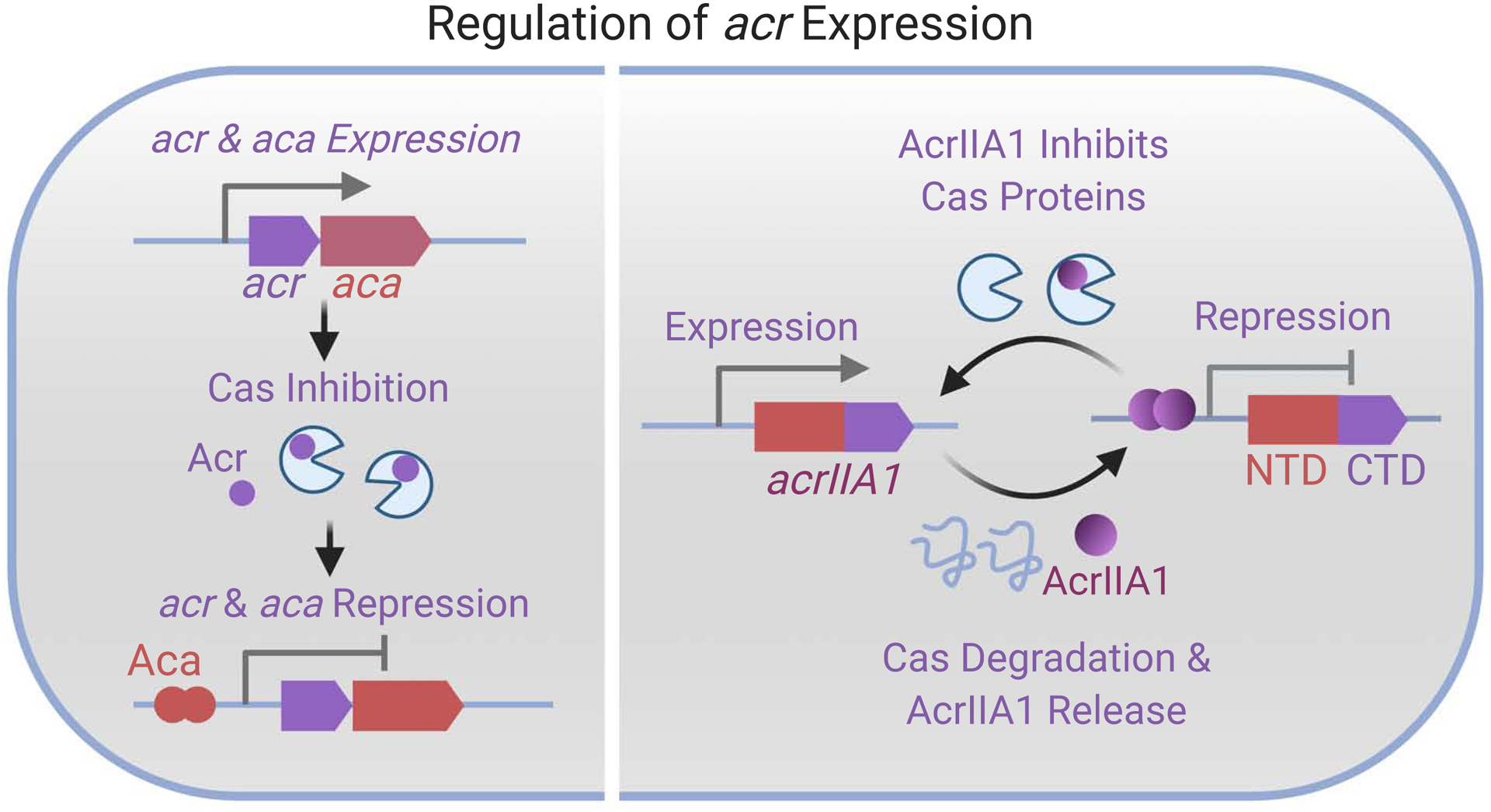
At the onset of phage infection, the acr-aca operon is quickly transcribed and translated to inhibit Cas proteins. Eventually, the dimeric Aca proteins bind to the promoter of the acr-aca locus and repress the expression of acr and aca. Right panel shows the self-regulation of AcrIIA1 proteins (purple-red). AcrIIA1 has dual functions with the N-terminal domain (NTD, red color) as self-repressor and the C-terminal domain (CTD, purple) as anti-CRISPR. Created with BioRender.com
The role of Acr proteins in phage ecology and evolution
As a strong anti-immunity mechanism, Acr proteins undoubtedly play a significant role in shaping the outcome of phage-bacteria interactions. There are many interesting and outstanding questions about how Acr proteins affect phage ecology and evolution. For instance, do phages lacking acr genes benefit from the presence of Acr-encoding phages? Are all Acr proteins equally efficient in deactivating CRISPR-Cas function in spite of different mechanisms? Is one acr gene sufficient for phages to overcome CRISPR-Cas systems in a timely manner? Several critical aspects have been investigated in recent studies.
Time is the key when it comes to the battle between CRISPR-Cas and Acr proteins. While it has been observed that acr expression increases up to 100-fold in the first 20 minutes of infection (Stanley et al., 2019), CRISPR-Cas systems are able to cleave a previously encountered phage genome in as little as two minutes (Garneau et al., 2010). Indeed, the presumed difficulty of inactivating pre-existing CRISPR-Cas complexes, despite prompt acr expression, was experimentally supported: low concentrations of P. aeruginosa phages encoding Type I-F Acr proteins were unable to replicate when faced with CRISPR-Cas targeting (Borges et al., 2018; Landsberger et al., 2018). Interestingly, the tested Acr proteins vary in “strength”: when being produced in an isogenic phage background, certain Acr proteins enable a higher efficiency of phage plaquing in the presence of CRISPR-Cas targeting than others and are considered strong Acr proteins. More importantly, the authors observed that a critical phage multiplicity of infection (MOI) threshold was required for each acr-containing phage, which was inversely proportional to Acr protein strength and directly proportional to the number of targeting spacers. Below this threshold MOI, acr-carrying phages were driven to extinction (comparably to control phages lacking acr genes), and above this threshold, phage replication proceeded as if there was no CRISPR-Cas immunity. A similar observation was also made with an engineered Cas9 system and AcrIIA4 (Borges et al., 2018). Through a series of modeling and plasmid challenge experiments (Landsberger et al., 2018) and the use of replication defective anti-CRISPR “donor” phages and prophage capture experiments (Borges et al., 2018), phage cooperation was implicated as the mechanism for the observed critical MOI thresholds. Cooperation is presumed to manifest when a targeted phage genome rapidly expresses acr transcripts prior to genome destruction (Fig. 3A), which enables gradual cellular immunosuppression (Fig. 3B&C), benefiting subsequent phage infections. Note that the immunosuppression effect varies among different Acr proteins with strong Acrs (Fig. 3B) inactivating CRISPR-Cas activity more efficiently than weak Acrs (Fig. 3C) after the same number of failed infection(s). Therefore, while inhibiting CRISPR-Cas is challenging, this family of temperate phages achieves it by working together in an altruistic manner, with low phage autonomy (Fig. 3E&G). However, when MOI is below the critical threshold, the probability of phage reinfection is low. As a result, the immunosuppressed host generated from previous failed infection events will be spared due to the lack of consecutive phage infection (Fig. 3F). Notably, this demonstrates a clear benefit to maintain CRISPR-Cas immunity even when bacteria face phages bearing Acr proteins.
Figure 3: Acr-mediated phage interactions.

A) When phage first infect bacteria with a targeting crRNA (pre-immunized bacteria), CRISPR-Cas can target the phage rapidly despite of its acr locus. B&C) Transcription of the acr gene from the failed infection leaves behind Acr proteins, which accumulate in the bacterial cell. B) Strong Acr proteins turn off bacterial CRISPR-Cas activity and result in fully immunosuppressed (FI) bacteria. C) weak Acr proteins, accumulated by the same number of failed infections, lead to partially immunosuppressed (PI) bacterial cells. D) Phages lacking acr gene can exploit and replicate in the fully immunosuppressed bacterial cells generated by the failed infection of strong Acr+ phages. E) Above the critical MOI threshold, Acr+ phages replicate in immunosuppressed cells as there is no CRISPR-Cas system. F) Below the MOI threshold, consecutive phage infection is unlikely. Bacteria propagate as there is no phage infection. G) Above the MOI threshold, weak Acr+ phage re-infect the partially immunosuppressed cell, further turn down the CRISPR-Cas activity and eventually replicate. H) Acr− phages are unable to replicate in partially immunosuppressed cells generated by previous infection of weak Acr+ phages. Created with BioRender.com
The altruistic behavior of Acr-encoding pioneers has a clear benefit for the survival of their kin phages by generating immunosuppressed bacteria cells (Fig. 3E&G). However, this altruistic behavior also comes with a potential shortcoming because the immunosuppressed host could be exploited by phages lacking acr genes (Acr− phages). When bacteria that harbor a targeting spacer are co-infected by phages encoding a strong Acr protein (i.e. AcrIF1 or AcrIIA4) and isogenic Acr− phages, a replicative benefit is also reaped by the Acr− phages, likely due to some “immunosuppression cheating”, however, not encoding their own Acr proteins severely limits the maximum replicative capacity of Acr− phages (Fig. 3D) (Borges et al., 2018). This same protective effect was not seen for the weaker AcrIF4 (Fig. 3H) (Chevallereau et al., 2020), which binds more weakly to the Csy complex (crRNA-guided surveillance complex) than AcrIF1 (Borges et al., 2018). Thus, AcrIF1 is more efficient in shutting down CRISPR interference and requires a lower phage MOI threshold (Borges et al., 2018; Landsberger et al., 2018). Paradoxically, due to the strong protection for Acr− cheaters, AcrIF1+ phages have a lower relative fitness than AcrIF4+ phages when in competition with Acr-phages (Chevallereau et al., 2020). Additionally, both AcrIF1+ and AcrIF4+ phages benefit otherwise isogenic Acr− phages when infecting a naïve bacterial host which does not harbor a phage targeting spacer by generating immunosuppressed host cells and diminishing host immune adaptation, including spacer acquisition and priming adaptation (Chevallereau et al., 2020). The latter demonstrates the ecological importance of previously observed inhibition of primed spacer acquisition by Acr proteins (Vorontsova et al., 2015). However, the importance of this observation when the Acr− phages are non-isogenic (i.e. acquired spacers would not offer cross-protection) remains to be seen. Similar “cheating” by Acr− phages was also observed when Acr proteins were first discovered, where stable prophages produced Acr proteins, inactivating CRISPR-Cas and licensing superinfection by phages that would otherwise be targeted (Bondy-Denomy et al., 2013). This demonstrates a clear cost to constitutive acr expression but is necessary to prevent self-targeting. Moreover, many of the acr-carrying prophages in P. aeruginosa also mediate superinfection exclusion at the cell surface (Bondy-Denomy et al., 2016), potentially compensating for the lost intracellular defense mechanism.
Phages and archaeal viruses often carry more than one acr gene (Bondy-Denomy et al., 2013; Dong et al., 2019; He et al., 2018; Marino et al., 2018; Osuna et al., 2020b; Rauch et al., 2017). In an extreme case, 12 acrID1 paralogues were found in an archaeal virus -- the lytic rudivirus SIRV2 (He et al., 2018). It is unclear whether these Acrs function independently, cooperatively or even antagonistically. The multiple copies of acr genes may benefit viruses by producing Acr proteins at a higher dose and thus inhibiting CRISPR-Cas systems in a timelier manner. Additionally, the existence of multiple acr genes in the same phage or archaeal virus might be a result of the constant arms race between bacteria and phage: bacteria acquire novel CRISPR-Cas systems to combat Acr-encoding phages; in return, phages gain additional acr genes to overcome these novel CRISPR-Cas types. The fact that many bacteria harbor multiple CRISPR-Cas systems is consistent with this hypothesis. Beyond this, a recent study suggests that Acr proteins encoded by the same phage synergize while targeting the same CRISPR-Cas9 system, with one Acr protein acting during the lysogenic state and another during lytic infection (Osuna et al., 2020b). This further leads us to contemplate potential interactions among Acr proteins encoded by different phages. In one work, it has been shown that mechanistically distinct Acr proteins AcrVA1 that cleaves Cas12a-bound guide RNA and AcrVA5 that inhibits Cas12a from recognizing dsDNA are encoded by two different prophages in the same bacterial genome (Dong et al., 2019) and biochemically compete with each other in disabling guide RNA (gRNA) bound Cas12a complex (Knott et al., 2019). However, the consequence of such a competition is unknown. Future studies to address whether the benefit of acrVA1 and acrVA5 is specific to their respective prophage and whether their co-existence enhances or reduces CRISPR-Cas activity in comparison to the presence of each acr alone will shed light on the ecology among acr genes from different phages. Conversely, acrVA4 and acrVA5 have been reported to coexist in the same prophage acr locus (Watters et al., 2018) and interact with each other physically (Zhang et al., 2019), suggesting potential synergy. Such a synergy can be realized, for instance, by AcrVA4 directing the acetyltransferase activity of AcrVA5 (Dong et al., 2019) or via regulation between AcrVA4 and AcrVA5. The biological importance of Acr competition, Acr synergy, and Acr-Acr interactions needs to be further addressed.
How do acr genes affect phage evolutionary trajectories? Even though Acr proteins increase the survival of phages, in order to overcome CRISPR-Cas immunity, cooperation among Acr-encoding phages is important, suggesting a lack of autonomy in these phages. In order to avoid Acr-cheaters, the evolution of strong acr genes which enables one phage particle to turn off the CRIPSR-Cas activity will be favored. Later in this review, we will discuss novel types of Acr mechanisms which may have a higher potency and provide a higher level of phage autonomy. Additionally, we postulate that phages that encapsulate Acr proteins, if they are able to inject these Acr proteins into bacteria upon infection, might quickly turn off CRISPR-Cas activity and increase phage autonomy. Lastly, synergy among Acr proteins encoded by the same phage may reduce a phage’s reliance on its kin and improve phage autonomy.
The role of acr in bacteria ecology and evolution: blessing or curse?
Acr proteins are able to inhibit CRISPR-Cas immunity, which allows phage replication and can result in bacterial death. However, is anti-CRISPR always a problem for bacteria? Surprisingly, Acr-assisted phage infections have been suggested to indirectly benefit bacteria in numerous ways.
First, Acr proteins help bacterial hosts by alleviating CRISPR-Cas auto-immunity. Albeit valuable in destroying lytic phages, CRISPR-Cas immunity can be detrimental to the bacterial genome when it comes to temperate phages and other integrative mobile elements. Upon infection, temperate phages can enter either the lytic cycle or lysogenic cycle. The former results in bacterial lysis, while the later results in the integration of phage DNA into the bacterial genome. During lysogeny, CRISPR-Cas targeting of the prophage genome is deadly for the cell (Edgar and Qimron, 2010) and, this “self-targeting” can be neutralized by Acr proteins (Marino et al., 2018; Rauch et al., 2017; Watters et al., 2018). Moreover, even in naïve bacteria, CRISPR-Cas is at the risk of damaging bacterial/prophage DNA via priming adaptation, which is triggered by partial complementarity between prophage DNA and CRISPR-Cas spacer (Rollie et al., 2020). Priming adaptation facilitates spacer acquisition from the partially matched prophage genome, which subsequently leads to self-targeting and a fitness cost (Rollie et al., 2020). Consistent with the role of priming adaptation in bacteria immunopathology, bacterial lysogen without a mismatched spacer or without the ability to acquire spacers, do not show a fitness cost. Additionally, by constructing phage mutants that encode corresponding Acr proteins, priming adaptation related damage can be prevented by prophage Acr proteins (Rollie et al., 2020). Without Acr proteins, the immunopathology is eventually resolved by disabling the CRISPR-Cas function through large genomic deletions encompassing the CRISPR-Cas locus. Therefore, in this context, Acr proteins benefit not only the invading phage, but also the bacterial host by preventing lethal self-targeting generated via spacer acquisition. One exception is the Type III CRISPR-Cas system, which provides immunity only when the target is transcribed and thus tolerates prophages or other mobile elements as long as they stay silent (Goldberg et al., 2014). However, the conditionally tolerant Type III system is imperfect and can still incur fitness costs due to leaky transcription of prophage targets within otherwise stably lysogenic cells (Goldberg et al., 2018). Broadly, spacers primed against prophages are widespread in natural bacteria populations. It has been found that these potential self-targeting bacterial genomes are more likely to carry acr genes, further suggesting the importance of Acr proteins in diminishing damages caused by CRISPR-Cas autoimmunity (Rollie et al., 2020).
Second, by disabling canonical CRISPR-Cas functions, Acr proteins can promote the acquisition of new functions by allowing the transfer of conjugative elements (Mahendra et al., 2020) and the integration of prophages (Borges et al., 2018), leading to lysogenic conversion. These gene transfer events have contributed to numerous bacterial functions such as antibiotic resistance, virulence, toxin, and even new CRISPR-systems (Horvath and Barrangou, 2010; Husnik and McCutcheon, 2018). Lastly, it has been proposed that Acr proteins promote the evolution of novel functions in bacteria (Wimmer and Beisel, 2020). After being disabled by endogenous Acr proteins, CRISPR-Cas proteins no long participate in bacterial defense and may be re-purposed for alternative functions. For instance, if Cas nucleases are blocked by Acr proteins, the CRISPR-Cas binding machinery can be co-opted for transcription regulation by binding to specific DNA sequences. Interestingly, it has been observed that in one instance, CRISPR-Cas DNA binding machinery functions in bacterial gene regulation even without Acr inhibition (Ratner et al., 2019). Therefore, we conjecture that Acr proteins may promote degeneration of CRISPR-Cas function and that the seemingly non-functional CRISPR-Cas remnants might instead serve novel functions.
Last, by inhibiting specific types of CRIPSR-Cas systems, Acr proteins are likely to drive the evolution of alternate bacterial defense mechanisms. This will help explain the various CRISPR-Cas types and subtypes and many other defense mechanisms harbored by bacteria (i.e. restriction-modification, abortive infection etc.). However, long-term experiments that co-evolve CRISPR-Cas carrying bacteria and acr-bearing phages are required to characterize the effect of Acr proteins on the evolution of bacterial defense mechanisms. At a high level, this is consistent with the recently proposed “pan-immunity” hypothesis, which posits that no single bacterial immune system is dominant in a species, but instead distributed diversity is favored (Bernheim and Sorek, 2020).
Emerging classes of Anti-CRISPR mechanisms
As indicated above, the majority of Acr proteins share several generic properties, including a small protein size, close genomic association with conserved Acr repressors, and direct interactions with CRISPR-Cas proteins. However, new anti-CRISPR mechanisms that do not share all of these properties, have started to be revealed. Although not yet thoroughly experimentally investigated, these new mechanisms likely have a strong impact on the infection biology, ecology and evolution of phages. In this section, we will describe these emerging anti-CRISPR mechanisms, and speculate on how they will impact phage autonomy.
Enzymatic Proteins as anti-CRISPR
Three enzymatic anti-CRISPR proteins have been discovered in the past two years (Fig. 4A). AcrVA1 and AcrVA5 that inhibit Type V CRISPR-Cas immunity were identified on Moraxella prophages (Marino et al., 2018; Watters et al., 2018) and both possess catalytic activity. AcrVA1 cleaves the crRNA in the Cas12a-crRNA complex and thus prevents Cas12a from detecting phage genome (Knott et al., 2019), while AcrVA5, an acetyltransferase, inhibits Cas12a by acetylating one of its critical PAM recognition sites (Dong et al., 2019). This particular site is observed to vary in some closely related Cas12a orthologues, enabling the evasion from AcrVA5 acetylation. AcrIII-1 is a ring nuclease that can rapidly degrade a CRISPR-associated cyclic signaling molecule, cyclic tetra-adenylate (cA4). Upon detecting viral RNA, the Type III CRISPR-Cas protein Cas10 not only cleaves the transcriptionally active DNA target, but also synthesizes the second messenger cA4, which activates non-specific ribonucleases Csm6/Csx1 and leads to an abortive infection-like antiviral response (Kazlauskiene et al., 2017; Niewoehner et al., 2017). By degrading cA4, AcrIII-1 inhibits cA4-elicited CRISPR-Cas defense and allows phage infection (Athukoralage et al., 2020). However, AcrIII-1 cannot inhibit Type III CRISPR-Cas systems that use cA6 for signal relay. Presumably, phages encoding anti-CRISPR proteins that possess catalytic turnover may lower the MOI threshold required for successful CRISPR-Cas inactivation, thus being a more “phage autonomous” anti-CRISPR mechanism compared to stoichiometric Acr proteins. This, however, has yet to be tested. Additionally, it remains to be assessed how phage-phage competition and the emergence of cheaters will be impacted by hyper potent Acr proteins.
Figure 4: Novel types of Acr proteins.

(purple). A) Three enzymatic Acr proteins that acetylate Cas12 PAM recognition site (acetyltransferase), cleave Cas12 crRNA (nuclease), and degrade cyclic tetra-adenylate (cA4) (ring nuclease), respectively. B) Transcriptional repressor of cas genes act as Acr proteins by repressing the expression of cas genes. C) Some jumbo phages assemble a nucleus-like shell structure to prevent DNA-targeting Cas proteins from accessing phage DNA. The shell structure hosts phage genome, DNA transcription and replication. Phage mRNA molecules are transported and translated outside of the shell. Created with BioRender.com
CRISPR-Cas Repressors as anti-CRISPR proteins
While CRISPR-Cas immunity is potent in defending against phage infection, this multicomponent immune system is costly to maintain (Vale et al., 2015; Westra et al., 2015). Additionally, as CRISPR systems can accidentally acquire spacers from the host genome, keeping CRISPR-Cas constantly on poses a risk of autoimmunity (Vercoe et al., 2013; Wei et al., 2015). To minimize these side effects, some CRISPR-Cas systems are not turned on until the detection of phage infection (Agari et al., 2010; Quax et al., 2013; Young et al., 2012) and others are regulated by certain environmental cues, such as sugar availability (Patterson et al., 2015), envelop stress (Perez-Rodriguez et al., 2011), cell density via the QS pathway (Høyland-Kroghsbo et al., 2017; Patterson et al., 2016) and others. In one work, it has been shown that multiple positive regulators of alginate production, a polysaccharide component in Pseudomonas aeruginosa biofilms, also act as negative regulators of Type I-F cas genes (Borges et al., 2020). Homologs of one repressor, named AmrZ, are present in multiple mobile genetic elements (MGEs), including plasmids, lytic phages and prophages, and some of these proteins also repress Type I-F CRISPR-Cas expression, demonstrating their role as bona fide anti-CRISPR proteins. AmrZ+ phages were used for phage infection experiments revealing that while ArmZ homologs are able to reduce CRISPR-Cas expression during lysogeny, they are ineffective during lytic infection (Borges et al., 2020). Thus, it is likely that ArmZ homologs and other MGE-encoded repressors will not be strong enough to fully inhibit CRISPR-Cas targeting during lytic or lysogenic growth, but likely require co-encoded anti-CRISPRs that are yet to be discovered. It remains to be seen whether phages that encode both a traditional strong stoichiometric inhibitor and a biogenesis inhibitor together, would be a more autonomous phage, due to a prolonged immunosuppressed state and an inability to create more CRISPR-Cas complexes.
Nucleus-like structure for CRISPR evasion
Despite the fact that most anti-CRISPR proteins have been discovered from prophages and temperate phages, recent work has shifted attention to understanding how obligately lytic phages interact with CRISPR-Cas (Hynes et al., 2018). Two recent studies have identified a remarkable new mechanism of CRISPR-Cas evasion possessed by lytic “jumbo” phages (genome size > 200 kb) infecting Pseudomonas and Serratia. As opposed to inhibition, jumbo phages evade CRISPR-Cas targeting via genome protection (Malone et al., 2020; Mendoza et al., 2020). Upon infection, these phages assemble a proteinaceous shell structure that encloses the phage DNA (Fig. 4C) (Chaikeeratisak et al., 2017). This shell structure is reminiscent to the eukaryotic nucleus, with phage DNA replication and transcription happening inside of the shell and protein translation happening outside of the shell. The shell structure serves as a physical barrier blocking DNA-targeting immune proteins from accessing the phage DNA. However, both phages remain vulnerable to RNA-targeting immune enzymes (Type III-A and VI-A CRISPR-Cas). This mechanism was shown to be highly effective against over-expressed CRISPR-Cas and restriction enzymes, even at vanishingly low infectious doses of phage (i.e. MOI = 10−7) (Mendoza et al., 2020), suggesting that protection in cis as opposed to inhibition in trans generates a high degree of phage autonomy. It remains to be addressed whether this mechanism can be cheated upon by any parasitic phage and whether any CRISPR or immune variants can circumvent the shell exclusion mechanism.
Hyper-potent AcrVIA1 enables phage autonomy
To fight against viral infection, Type VI CRISPR-Cas system recognizes complementary viral transcripts, which activates Cas13 nuclease to cleave both host and viral RNA indiscriminately (Abudayyeh et al., 2016; East-Seletsky et al., 2016; Meeske et al., 2019). The non-specific RNA degradation arrests host growth and prevents phage spread (Meeske et al., 2019). A recent study discovered an anti-CRISPR protein AcrVIA1, encoded by a DNA phage (Meeske et al., 2020). AcrVIA1 binds to Cas13 to prevent it from recognizing viral transcripts and being activated. Surprisingly, unlike Acr proteins from Type I or Type II systems which requires multiple phage infections to inhibit CRISPR-Cas activity, AcrVIA1 enables phage autonomy -- a single viral infection (a MOI as low as 10−6) is sufficient to turn off CRIPSR-Cas immunity. The hyper-potency of AcrVIA1 may originate from the fact that Type VI-A CRISPR-Cas system is unable to cleave viral DNA. Thus, even though viral RNA is targeted, the intact viral DNA can still be transcribed at a low level, which eventually leads to Acr accumulation and the inhibition of CRISPR-Cas immunity in the bacterial cell.
Conclusions and outlook
Many anti-CRISPR discovery and characterization efforts have focused mostly on model systems that assess function outside of the context of the endogenous elements. Thus, much anti-CRISPR phage/virus biology likely remains to be uncovered. For example, MGEs with low reproductive rates, such as phages with small burst sizes (i.e. jumbo phages) and large plasmids, may be unable to rely on Acr proteins that require MGE cooperation to turn down the CRISPR-Cas activity and thus may encode hyper-potent anti-CRISPR proteins that await discovery. Future work should explore the autonomy that hyper-potent Acrs could provide to MGEs, and diverse MGEs should continue to be mined for new Acr proteins as they will likely yield new and exciting mechanisms of CRISPR-Cas neutralization. In the same vein, the degree to which “cheater” MGEs could benefit from these hyper-potent CRISPR inhibitors is worthy of investigation. In that vein, the degree of phage autonomy may not be easily predicted from Acr mechanism alone. For example, while it was established that high affinity stoichiometric inhibitors like AcrIF1 and AcrIF2 require phage cooperation to neutralize CRISPR-Cas immunity (Borges et al., 2018; Landsberger et al., 2018), recently discovered stoichiometric inhibitor AcrVIA1Lse was shown to function when deployed from its natural phage host at very low MOIs, suggesting no such need for cooperation (Meeske et al., 2020). The authors speculated that this is due to RNA-targeting mechanism that underlies Type VI CRISPR-Cas systems, which may leave the phage genome intact until inhibition can occur. In addition to the recently characterized jumbo phage CRISPR-Cas evasion mechanism via a nucleus-like structure, these are the two most autonomous anti-CRISPR mechanisms described to date.
CRISPR-Cas system and Acr proteins are under strong selection pressures and provide a powerful study system to understand the dynamics and the molecular basis of co-evolution. Understanding the factors that dictate the host range, strength and mechanism of Acr proteins, and the rise and fall of CRISPR-Cas or Acr systems would help us learn what makes a “successful” defense or anti-defense strategy, and eventually predict evolutionary outcomes of a mixed bacterial and phage community. Such knowledge is critical in the rational design of phage therapy to overcome current “superbug” crisis and in developing secure practices of CRISPR-Cas genetic editing. More broadly, it is of great interest to determine the role of the conflict between CRISPR-Cas and Acr proteins in their respective phylogenetic distribution and in maintaining the diversity of bacteria and phage species.
As a result, in the future, it is important to study how CRISPR and Acr antagonism shapes the dynamics of phage and bacteria populations and to dissect the evolutionary outcomes and the genetic basis of bacteria-phage interactions in both short and long timescales using, for instance, experimental evolution or comparative genomics approaches. Furthermore, even though host-pathogen interactions often happen in diverse communities (multi-species interactions) in natural settings, our current knowledge is largely restricted to the interaction of one pathogen and one host species at a given time. Thus, it is appealing to expand our current methodology into multi-species systems to understand host-pathogen interactions in a higher dimension.
Acknowledgments
We thank Adair Borges and anonymous reviewers for their helpful comments. CRISPR-Cas and phage work in the Bondy-Denomy lab is supported by the UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation, the Searle Fellowship, the Vallee Foundation, the Innovative Genomics Institute, and an NIH Director’s Early Independence Award DP5-OD021344, R01GM127489.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interests
J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics and a scientific advisory board member and co-founder of Acrigen Biosciences. UCSF has filed patent applications relating to anti-CRISPR and CRISPR technology on which J.B.-D. is an inventor.
References:
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, and Shinkai A (2010). Transcription Profile of Thermus thermophilus CRISPR Systems after Phage Infection. J. Mol. Biol 395, 270–281. [DOI] [PubMed] [Google Scholar]
- Athukoralage JS, McMahon SA, Zhang C, Grüschow S, Graham S, Krupovic M, Whitaker RJ, Gloster TM, and White MF (2020). An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577, 572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, and Horvath P (2007). CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Bernheim A, and Sorek R (2020). The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Rev. Microbiol 18, 113–119. [DOI] [PubMed] [Google Scholar]
- Birkholz N, Fagerlund RD, Smith LM, Jackson SA, and Fineran PC (2019). The autoregulator Aca2 mediates anti-CRISPR repression. Nucleic Acids Res. 47, 9658–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, and Davidson AR (2013). Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, and Davidson AR (2015). Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, and Maxwell KL (2016). Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 10, 2854–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges AL, Davidson AR, and Bondy-Denomy J (2017). The Discovery, Mechanisms, and Evolutionary Impact of Anti-CRISPRs. Annu. Rev. Virol 4, 37–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges AL, Zhang JY, Rollins MF, Osuna BA, Wiedenheft B, and Bondy-Denomy J (2018). Bacteriophage Cooperation Suppresses CRISPR-Cas3 and Cas9 Immunity. Cell 174, 917–925.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges AL, Castro B, Govindarajan S, Solvik T, Escalante V, and Bondy-Denomy J (2020). Bacterial alginate regulators and phage homologs repress CRISPR-Cas immunity. Nat. Microbiol 5, 679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, and van der Oost J (2008). Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science 321, 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüssow H, and Hendrix RW (2002). Phage Genomics: Small Is Beautiful. Cell 108, 13–16. [DOI] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, and Terns MP (2008). Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22, 3489–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabas H, Nicot A, Meaden S, Westra ER, Tremblay DM, Pradier L, Lion S, Moineau S, and Gandon S (2019). Variability in the durability of CRISPR-Cas immunity. Philos. Trans. R. Soc. B Biol. Sci 374, 20180097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikeeratisak V, Nguyen K, Khanna K, Brilot AF, Erb ML, Coker JKC, Vavilina A, Newton GL, Buschauer R, Pogliano K, et al. (2017). Assembly of a nucleus-like structure during viral replication in bacteria. Science 355, 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallereau A, Meaden S, Fradet O, Landsberger M, Maestri A, Biswas A, Gandon S, van Houte S, and Westra ER (2020). Exploitation of the Cooperative Behaviors of Anti-CRISPR Phages. Cell Host Microbe 27, 189–198.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs LM, England WE, Young MJ, Weitz JS, and Whitaker RJ (2014). CRISPR-Induced Distributed Immunity in Microbial Populations. PLoS ONE 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, and Semenova E (2012). Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun 3, 945. [DOI] [PubMed] [Google Scholar]
- Davidson AR, Lu W-T, Stanley SY, Wang J, Mejdani M, Trost CN, Hicks BT, Lee J, and Sontheimer EJ (2020). Anti-CRISPRs: Protein Inhibitors of CRISPR-Cas Systems. Annu. Rev. Biochem 89, null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, and Moineau S (2008). Phage Response to CRISPR-Encoded Resistance in Streptococcus thermophilus. J. Bacteriol 190, 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Guan X, Li N, Zhang F, Zhu Y, Ren K, Yu L, Zhou F, Han Z, Gao N, et al. (2019). An anti-CRISPR protein disables type V Cas12a by acetylation. Nat. Struct. Mol. Biol 26, 308–314. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JHD, Tjian R, and Doudna JA (2016). Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, and Qimron U (2010). The Escherichia coli CRISPR System Protects from λ Lysogenization, Lysogens, and Prophage Induction. J. Bacteriol 192, 6291–6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Gerritzen MJH, Suárez-Diez M, Künne T, Boekhorst J, Hijum SAFT van, Staals RHJ, and Brouns SJJ (2014). Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl. Acad. Sci 111, E1629–E1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, and Prangishvili D (2009). The Great Billion-year War between Ribosome- and Capsid-encoding Organisms (Cells and Viruses) as the Major Source of Evolutionary Novelties. Ann. N. Y. Acad. Sci 1178, 65–77. [DOI] [PubMed] [Google Scholar]
- Garneau JE, Dupuis M-È, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, and Moineau S (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, and Siksnys V (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci 109, E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, and Marraffini LA (2014). Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg GW, McMillan EA, Varble A, Modell JW, Samai P, Jiang W, and Marraffini LA (2018). Incomplete prophage tolerance by type III-A CRISPR-Cas systems reduces the fitness of lysogenic hosts. Nat. Commun 9, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, and Doudna JA (2010). Sequence- and Structure-Specific RNA Processing by a CRISPR Endonuclease. Science 329, 1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Bhoobalan-Chitty Y, Van LB, Kjeldsen AL, Dedola M, Makarova KS, Koonin EV, Brodersen DE, and Peng X (2018). Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat. Microbiol 3, 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, and Barrangou R (2010). CRISPR/Cas, the Immune System of Bacteria and Archaea. Science 327, 167–170. [DOI] [PubMed] [Google Scholar]
- van Houte S, Ekroth AKE, Broniewski JM, Chabas H, Ashby B, Bondy-Denomy J, Gandon S, Boots M, Paterson S, Buckling A, et al. (2016). The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J, and Bassler BL (2017). Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc. Natl. Acad. Sci. U. S. A 114, 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F, and McCutcheon JP (2018). Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol 16, 67–79. [DOI] [PubMed] [Google Scholar]
- Hynes AP, Rousseau GM, Agudelo D, Goulet A, Amigues B, Loehr J, Romero DA, Fremaux C, Horvath P, Doyon Y, et al. (2018). Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun 9, 2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G, and Siksnys V (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609. [DOI] [PubMed] [Google Scholar]
- Knott GJ, Thornton BW, Lobba MJ, Liu J-J, Al-Shayeb B, Watters KE, and Doudna JA (2019). Broad-spectrum enzymatic inhibition of CRISPR-Cas12a. Nat. Struct. Mol. Biol 26, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, and Makarova KS (2019). Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. B Biol. Sci 374, 20180087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberger M, Gandon S, Meaden S, Rollie C, Chevallereau A, Chabas H, Buckling A, Westra ER, and van Houte S (2018). Anti-CRISPR Phages Cooperate to Overcome CRISPR-Cas Immunity. Cell 174, 908–916.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, and Sorek R (2015). CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendra C, Christie KA, Osuna BA, Pinilla-Redondo R, Kleinstiver BP, and Bondy-Denomy J (2020). Broad-spectrum anti-CRISPR proteins facilitate horizontal gene transfer. Nat. Microbiol 5, 620–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol 13, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone LM, Warring SL, Jackson SA, Warnecke C, Gardner PP, Gumy LF, and Fineran PC (2020). A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat. Microbiol 5, 48–55. [DOI] [PubMed] [Google Scholar]
- Marino ND, Zhang JY, Borges AL, Sousa AA, Leon LM, Rauch BJ, Walton RT, Berry JD, Joung JK, Kleinstiver BP, et al. (2018). Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 362, 240–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino ND, Pinilla-Redondo R, Csörgő B, and Bondy-Denomy J (2020). Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods 17, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, and Sontheimer EJ (2008). CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA. Science 322, 1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, and Sontheimer EJ (2010). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Nakandakari-Higa S, and Marraffini LA (2019). Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 570, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Jia N, Cassel AK, Kozlova A, Liao J, Wiedmann M, Patel DJ, and Marraffini LA (2020). A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza SD, Nieweglowska ES, Govindarajan S, Leon LM, Berry JD, Tiwari A, Chaikeeratisak V, Pogliano J, Agard DA, and Bondy-Denomy J (2020). A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature 577, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, and Almendros C (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology, 155, 733–740. [DOI] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostøl JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, and Jinek M (2017). Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548. [DOI] [PubMed] [Google Scholar]
- Osuna BA, Karambelkar S, Mahendra C, Sarbach A, Johnson MC, Kilcher S, and Bondy-Denomy J (2020a). Critical Anti-CRISPR Locus Repression by a Bi-functional Cas9 Inhibitor. Cell Host Microbe 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna BA, Karambelkar S, Mahendra C, Christie KA, Garcia B, Davidson AR, Kleinstiver BP, Kilcher S, and Bondy-Denomy J (2020b). Listeria Phages Induce Cas9 Degradation to Protect Lysogenic Genomes. Cell Host Microbe 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, and Banfield JF (2013). Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat. Commun 4, 1–7. [DOI] [PubMed] [Google Scholar]
- Patterson AG, Chang JT, Taylor C, and Fineran PC (2015). Regulation of the Type I-F CRISPR-Cas system by CRP-cAMP and GalM controls spacer acquisition and interference. Nucleic Acids Res. 43, 6038–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AG, Jackson SA, Taylor C, Evans GB, Salmond GPC, Przybilski R, Staals RHJ, and Fineran PC (2016). Quorum Sensing Controls Adaptive Immunity through the Regulation of Multiple CRISPR-Cas Systems. Mol. Cell 64, 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Bondy-Denomy J, Cheung VHW, Maxwell KL, and Davidson AR (2014). A New Group of Phage Anti-CRISPR Genes Inhibits the Type I-E CRISPR-Cas System of Pseudomonas aeruginosa. MBio 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Davidson AR, and Maxwell KL (2018). Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol 16, 12–17. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, and DeLisa MP (2011). Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol 79, 584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TEF, Voet M, Sismeiro O, Dillies M-A, Jagla B, Coppée J-Y, Sezonov G, Forterre P, van der Oost J, Lavigne R, et al. (2013). Massive Activation of Archaeal Defense Genes during Viral Infection. J. Virol 87, 8419–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner HK, Escalera-Maurer A, Le Rhun A, Jaggavarapu S, Wozniak JE, Crispell EK, Charpentier E, and Weiss DS (2019). Catalytically Active Cas9 Mediates Transcriptional Interference to Facilitate Bacterial Virulence. Mol. Cell 75, 498–510.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, and Bondy-Denomy J (2017). Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell 168, 150–158.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Prangishvili D, and Lindell D (2009). Roles of viruses in the environment. Environ. Microbiol 11, 2771–2774. [DOI] [PubMed] [Google Scholar]
- Rollie C, Chevallereau A, Watson BNJ, Chyou T, Fradet O, McLeod I, Fineran PC, Brown CM, Gandon S, and Westra ER (2020). Targeting of temperate phages drives loss of type I CRISPR-Cas systems. Nature 578, 149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, and Siksnys V (2011). The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJJ, and Severinov K (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U. S. A 108, 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RHJ, Jackson SA, Biswas A, Brouns SJJ, Brown CM, and Fineran PC (2016). Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system. Nat. Commun 7, 12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SY, and Maxwell KL (2018). Phage-Encoded Anti-CRISPR Defenses. Annu. Rev. Genet 52, 445–464. [DOI] [PubMed] [Google Scholar]
- Stanley SY, Borges AL, Chen K-H, Swaney DL, Krogan NJ, Bondy-Denomy J, and Davidson AR (2019). Anti-CRISPR-Associated Proteins Are Crucial Repressors of Anti-CRISPR Transcription. Cell 178, 1452–1464.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao P, Wu X, and Rao V (2018). Unexpected evolutionary benefit to phages imparted by bacterial CRISPR-Cas9. Sci. Adv 4, eaar4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale PF, Lafforgue G, Gatchitch F, Gardan R, Moineau S, and Gandon S (2015). Costs of CRISPR-Cas-mediated resistance in Streptococcus thermophilus. Proc. R. Soc. B Biol. Sci 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, and Fineran PC (2013). Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands. PLoS Genet. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorontsova D, Datsenko KA, Medvedeva S, Bondy-Denomy J, Savitskaya EE, Pougach K, Logacheva M, Wiedenheft B, Davidson AR, Severinov K, et al. (2015). Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery. Nucleic Acids Res. 43, 10848–10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Fellmann C, Bai HB, Ren SM, and Doudna JA (2018). Systematic discovery of natural CRISPR-Cas12a inhibitors. Science 362, 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters KE, Shivram H, Fellmann C, Lew RJ, McMahon B, and Doudna JA (2020). Potent CRISPR-Cas9 inhibitors from Staphylococcus genomes. Proc. Natl. Acad. Sci 117, 6531–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Terns RM, and Terns MP (2015). Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 29, 356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Houte S, Oyesiku-Blakemore S, Makin B, Broniewski JM, Best A, Bondy-Denomy J, Davidson A, Boots M, and Buckling A (2015). Parasite Exposure Drives Selective Evolution of Constitutive versus Inducible Defense. Curr. Biol 25, 1043–1049. [DOI] [PubMed] [Google Scholar]
- Wiegand T, Karambelkar S, Bondy-Denomy J, and Wiedenheft B (2020). Structures and Strategies of Anti-CRISPR-Mediated Immune Suppression. Annu. Rev. Microbiol 74, null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer F, and Beisel CL (2020). CRISPR-Cas Systems and the Paradox of Self-Targeting Spacers. Front. Microbiol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Dill BD, Pan C, Hettich RL, Banfield JF, Shah M, Fremaux C, Horvath P, Barrangou R, and VerBerkmoes NC (2012). Phage-Induced Expression of CRISPR-Associated Proteins Is Revealed by Shotgun Proteomics in Streptococcus thermophilus. PLoS ONE 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li Z, Daczkowski CM, Gabel C, Mesecar AD, and Chang L (2019). Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins. Cell Host Microbe 25, 815–826.e4. [DOI] [PubMed] [Google Scholar]


