Abstract
Rationale.
Substantial variability in response to asthma treatment with inhaled corticosteroids (ICS) has been described among individuals and populations, suggesting the contribution of genetic factors. Nonetheless, only a few genes have been identified to date. We aimed to identify genetic variants associated with asthma exacerbations despite ICS use in European children and young adults and to validate the findings in non-Europeans. Moreover, we explored whether a gene-set enrichment analysis could suggest potential novel asthma therapies.
Methods.
A genome-wide association study (GWAS) of asthma exacerbations was tested in 2,681 European-descent children treated with ICS from eight studies. Suggestive association signals were followed up for replication in 538 European asthma patients. Further evaluation was performed in 1,773 non-Europeans. Variants revealed by published GWAS were assessed for replication. Additionally, gene-set enrichment analysis focused on drugs was performed.
Results.
Ten independent variants were associated with asthma exacerbations despite ICS treatment in the discovery phase (p≤5×10−6). Of those, one variant at the CACNA2D3-WNT5A locus was nominally replicated in Europeans (rs67026078, p = 0.010), but this was not validated in non-European populations. Five other genes associated with ICS response in previous studies were replicated. Additionally, an enrichment of associations in genes regulated by trichostatin A treatment was found.
Conclusions.
The intergenic region of CACNA2D3 and WNT5A was revealed as a novel locus for asthma exacerbations despite ICS treatment in European populations. Genes associated were related to trichostatin A, suggesting that this drug could regulate the molecular mechanisms involved in treatment response.
Keywords: Childhood asthma, Europeans, Exacerbations, Pharmacogenomics, treatment, trichostatin A
INTRODUCTION
Asthma is the most common chronic condition in children and young adults [1]. Inhaled corticosteroids (ICS) are the first-line treatment recommended by current international guidelines to control and prevent asthma symptoms [1]. Although ICS are the most effective medication for improving symptoms and preventing severe exacerbations [2], high interindividual variability in ICS response has been described [3]. Studies have shown that 30 to 40% of the asthmatic children treated with ICS do not show an improvement of their symptoms and that 10 to 15% of them may even experience worsening of asthma exacerbations despite the regular use of this medication [3]. Moreover, marked variation in ICS response has been described among populations [4].
The contribution of genetic factors in asthma-related traits has been widely suggested [5]. Specifically, the variation in ICS response has been suggested to be the result of the interaction of several factors such as the specific asthma endotype, comorbidities, ancestry, the environment, and the individual’s genetic composition [6]. Approximately 40–60% of the total variation in ICS response may be explained by genetic factors [7]. Pharmacogenetic studies of ICS response have focused mostly on a few genes with known biological implications in the mechanisms of action of ICS [5]. More recently, genome-wide association studies (GWAS), have explored the role of genetic variation in the ICS response [8–10]. Overall, these GWAS have identified 13 genes associated with different definitions of ICS response, most of which were not previously associated with asthma-related phenotypes, except for PDE10A [11]. However, it is expected that more genes are involved in the response to this asthma treatment. Moreover, the genetic architecture of clinical markers of disease severity, such as asthma exacerbations or lung function measurements, is not completely disentangled [12,13]. The studies performed to date have been limited by the relatively small number of study participants. Therefore, there is a need for studies including a large number of individuals to increase the power to detect significant associations with asthma severity and ICS response [5]. Increasing the knowledge about the genetic markers involved in asthma progression and therapeutic response would be of special importance in clinical practice since current international guidelines for the management of asthma propose pharmacological stepwise approaches based on the occurrence and persistence of clinical outcomes as indicators of disease severity [1].
In the present study, we aimed to replicate suggested associations in a candidate gene approach and to identify novel genetic variants involved in the occurrence of asthma exacerbations despite ICS treatment by performing a large GWAS in Europeans and to examine whether this genetic variation is shared with other populations. We also explored whether a gene-set enrichment analysis of the GWAS results could suggest alternative treatments that could be potential therapeutic alternatives in patients who do not respond to ICS therapy.
METHODS
Ethics statement
All studies included were approved by their local institutional review boards and written informed consent was provided by participants or their parents/caregivers. All methods were carried out following guidelines and regulations for human subject research under the principles of the Declaration of Helsinki.
Study Populations
A total of fourteen independent studies participating in the Pharmacogenomics in Childhood of Asthma (PiCA) consortium [14] were included in this study. Eight available studies in populations of European descent at the time of data collecting were included in the discovery phase, whereas replication of association results was evaluated in three additional independent European studies. Further validation was performed in three non-Europeans studies from Hispanic/Latino, African American, and Asian populations.
Discovery phase
Asthma patients from eight independent European studies were analysed in the discovery phase: the Pharmacogenetics of Asthma Medication in Children: Medication with Anti-inflammatory effects (PACMAN); the Paediatric Asthma Gene-Environment Study (PAGES); BREATHE; the Genetics of the Scottish Health Research Register (GoSHARE); the Pharmacogenetics of Adrenal Suppression study (PASS); SLOVENIA; the follow-up stage of the Multicenter Asthma Genetics in Childhood Study (followMAGICS); Effectiveness and Safety of Treatment with Asthma Therapy in Children (ESTATe). All these studies included children and young adults aged 2 to 25 years recruited in five different European countries. Among the participants, only individuals with reported use of ICS, information about asthma exacerbations, and genome-wide genotyping data were included. ICS use was based on declared use of any type of ICS and/or combination with long-acting β2 agonists at least once in the previous 12 months based on self-reports, pharmacy, or medical records [15]. A period of the last 6 months was considered for those studies without data available related to the previous year. A detailed description of each study is provided in the Supplementary Material.
The presence or absence of at least one asthma exacerbation episode during the 6 or 12 months preceding the study enrolment was assessed. Severe asthma exacerbations were defined by a need for emergency care, hospitalizations, or administration of systemic corticosteroids because of asthma for PACMAN, GoSHARE, PASS, SLOVENIA, and ESTATe (Table 1) [16]. Definitions of moderate asthma exacerbations were used in BREATHE-PAGES, BREATHE, and followMAGICS (Table 1), since no information was available for any of the previous variables [16]. Therefore, data related to unscheduled general practitioner or respiratory system specialist visits and school absence were also considered in the definition of asthma exacerbations for BREATHE-PAGES, BREATHE, and followMAGICS (Table 1), as described elsewhere [15].
Table 1.
Clinical and demographic characteristics of the European populations included in the discovery phase.
| PACMAN |
BREATHE-PAGES |
GoSHARE |
PASS |
SLOVENIA |
BREATHE |
followMAGICS |
ESTATe |
|
|---|---|---|---|---|---|---|---|---|
| Sample size | 654 | 540 | 472 | 402 | 182 | 182 | 147 | 102 |
| Gender (% male) | 61.6 | 60.4 | 24.8 | 55.0 | 57.1 | 59.3 | 59.9 | 58.8 |
| Mean age ± SD (years) | 8.7 ± 2.3 | 10.2 ± 3.5 | 11.3 ± 5.7 | 12.0 ± 2.0 | 10.8 ± 3.4 | 8.9 ± 4.0 | 17.2 ± 3.0 | 10.6 ± 4.2 |
| Recruitment country | Netherlands | United Kingdom | United Kingdom | United Kingdom | Slovenia | United Kingdom | Germany/Austria | Netherlands |
| Asthma exacerbations in the last 12 months (%) | 11.0 | 54.1a | 13.8 | 51.7a | 34.1 | 52.7a | 53.1 | 48.0 |
| Definition | ER visits/OCS use | hospitalizations/OCS use/school absences | hospitalizations/OCS use | OCS use | ER visits/hospitalizations/OCS use | OCS use/hospitalizations/school absences | ER visits/hospitalizations/GP visits/specialist visits | ER visits/hospitalizations/OCS use |
| ER visits (%) b | 6.1 | NA | NA | NA | 28.0 | NA | 7.5 | NA |
| OCS use (%) c | 6.7 | 35.0 | 13.8 | 51.7 | 12.6 | 48.4 | NA | 35.3 |
| Hospitalizations (%)d | NA | 13.5 | 0.21 | NA | 9.9 | 46.7 | 3.4 | 12.7 h |
| GP visits (%)e | NA | NA | NA | NA | NA | NA | 49.0 | NA |
| Specialist visits (%)f | NA | NA | NA | NA | NA | NA | 21.8 | NA |
| School absences (%) g | NA | 43.1 | NA | NA | NA | 47.2 | NA | NA |
| Treatment steps i | ||||||||
| Step 2 (%) j | 70.6 | 37.6 | 97.3 | 7.5 | NA | 61.0 | 29.3 | 63.7 |
| Step 3 (%) k | 20.8 | 32.6 m,n | 2.5 m,n | 32.1 n | NA | 29.1 m,n | 59.8 n | 33.3 n |
| Step 4 (%) l | 5.4 | 29.8 ñ | 0.2 ñ | 57.2 | NA | 9.9 ñ | 10.9 | 2.0 |
| No classification | 3.2 | NA | NA | 3.2 | NA | NA | NA | 1.0 |
| Genotyping platform | Illumina Infinium CoreExome-24 BeadChip (Illumina) | Axiom Precision Medicine Research Array (Affymetrix) | Axiom Precision Medicine Research Array (Affymetrix) | Illumina Omni Express 8v1 (Illumina) | Illumina Global Screening Array-24 v1.0 BeadChip | Illumina Infinium CoreExome-24 BeadChip (Illumina) | Illumina Sentrix HumanHap300 BeadChip (Illumina) | Illumina Infinium CoreExome-24 BeadChip (Illumina) |
Asthma exacerbations-related data was available for the 6 precedent months of the study enrolment;
Proportion of patients with any exacerbations who sought emergency care due to asthma;
Proportion of patients with any exacerbations who needed the use oral corticosteroids because of asthma;
Proportion of patients with any exacerbations who needed to be hospitalized because of asthma;
Proportion of patients with any exacerbations who needed any unscheduled general practitioner visits because of asthma;
Proportion of patients with any exacerbations who needed any respiratory system specialist visits because of asthma;
Proportion of patients with any exacerbations who were absent from school because of asthma;
ER visits and hospitalizations were considered as a single variable;
Adapted from British Thoracic Society/Scottish Intercollegiate Guidelines Network guidelines;
As-needed SABA plus regular ICS;
As-needed SABA plus regular ICS and LABA;
As-needed SABA plus regular ICS, LABA and LTRA;
As-needed SABA plus combinations of ICS and LABA; as-needed SABA plus ICS and combinations of ICS and LABA;
As-needed SABA plus ICS and LTRA was also considered;
As-needed SABA plus LABA, combinations of ICS and LABA, and LTRA; as-needed SABA plus ICS, combinations of ICS and LABA, and LTRA; or as-needed SABA plus combinations of ICS and LABA, and LTRA was also considered.
LABA: long-acting β2 agonists; LTRA: leukotriene receptor antagonists; SABA: short-acting β2 agonists; SD: standard deviation; ER: emergency room; OCS: systemic corticosteroids; GP: general practitioner; NA: not available.
Replication phase
Validation of the results found in the discovery phase was carried out in three independent European studies: the Avon Longitudinal Study of Parents and Children (ALSPAC); the Childhood Asthma Management Program (CAMP) and, the Children Allergy Milieu Stockholm an Epidemiological Study (BAMSE). Definitions of ICS use and asthma exacerbations were based on retrospective information about the 12 months prior to study enrolment adopting the same criteria applied in the discovery phase, except for prospective data from CAMP. Further details about these studies are described in the Supplementary Material.
Assessment of ICS associations in non-European populations
Association signals with evidence of replication (p≤0.05) among Europeans were evaluated in Latinos/Hispanics from the Genes-Environment and Admixture in Latino Americans (GALA II) study, African Americans included from the Study of African Americans, Asthma, Genes and Environments (SAGE), and Asians from The Singapore Cross-Sectional Genetic Epidemiology Study (SCSGES). Information about the presence or absence of asthma exacerbations despite ICS use in the 12 previous months to study enrolment was considered. The details on these studies are described in Supplementary Material.
Genotyping, genetic ancestry estimation, and imputation
Samples from the studies included in the discovery phase were genotyped using different platforms for previous studies (Table 1) [15], except for PAGES, GoSHARE, and part of the samples from BREATHE. These studies were genotyped with the Axiom™ Precision Medicine Research Array (Affymetrix Inc.) by Centro Nacional de Genotipado (CeGen; www.cegen.org). The same QC procedures described in Hernandez-Pacheco et al. were applied to all the studies [15]. Further details are available in the Supplementary Material.
Details about the genotyping of the replication samples are provided in the Supplementary Material and summarized in Table S1. Similarly, the genotyping methods used for the non-European studies (Table S2) are described in the Supplementary Material.
Assessment of the genetic ancestry was carried out through Principal Component (PC) analyses or by model-based assessments of the proportions of genetic ancestry (GALA II and SAGE) [15]. For SCSGES, estimation of ancestry was not performed since genome-wide genotyping was not available. The second release of the Haplotype Reference Consortium (r1.1 2016) was used as reference panel for imputation [17], except for CAMP and ALSPAC, where phase 3 of the 1000 Genomes Project (1KGP) was used [18].
Association analysis in the discovery phase
GWAS analyses were carried out separately for each study, except for PAGES and a subset of individuals from BREATHE that were genotyped together with PAGES. These two studies were analysed together since the similarities of the study design, type of biological samples, demographic and clinical characteristics, and genotyping platform used, and are denoted as BREATHE-PAGES. Association between genetic variants and the binary variable of asthma exacerbations was tested employing the binary Wald logistic regression model implemented in EPACTS 3.2.6 [19]. Regression models included as covariates age, gender, and the PCs needed to control for population stratification within each study.
Results for single nucleotide polymorphisms (SNPs) with a minor allele frequency (MAF) ≥1% and imputation quality (Rsq) ≥0.3 obtained for each study included in the discovery phase were meta-analysed. Fixed-effects or random-effects models were applied using METASOFT [20], depending on the significance of the Cochran Q-test evidencing heterogeneity among the studies analysed. Association with asthma exacerbations despite the use of ICS treatment was considered at suggestive significance level (p-value ≤5×10−6), which was arbitrarily set based on the criteria commonly adopted in GWAS studies [15].
Independent association signals were detected from these results through conditional and joint multiple-SNP analyses (COJO), as implemented in GCTA 1.92.0 [21]. Stepwise model selection was carried out to select independently associated SNPs within each genomic region with a suggestive association signal through a linkage disequilibrium (LD) correlation matrix obtained with the data from PACMAN, the largest study included in the discovery phase. Independent SNPs associated (p-value≤5×10−6) with asthma exacerbations were followed up for replication.
Association analysis in the replication phase
Association analyses were performed in three different PiCA studies of European descent. The definition of asthma exacerbations used for each replication population is described in Table S1. Association testing in BAMSE was performed following the same methodology as in the discovery phase. Logistic regressions were carried out in CAMP and ALSPAC using PLINK 1.9 [22] and SNPTEST 2.5.2 [23], respectively. Association results obtained from the European replication studies for variants associated with asthma exacerbations despite ICS use at nominal level (p-value≤0.05), and with the same direction of the effects as in the discovery phase were meta-analysed following the same methodology as described above.
Association analysis in non-European populations
The association of the variant with evidence of replication was further assessed in GALA II and SAGE using the same statistical methodology applied for the studies included in the discovery phase. In SCSGES (Table S2), association with asthma exacerbations was evaluated using logistic regressions adjusted by age and gender using PLINK 1.9 [22].
Evidence of validation was considered if the variant assessed showed a p-value ≤0.05 and the same direction of the effect as the one found in European populations.
Association analysis accounting for ICS dosage and asthma severity
Several sensitivity analyses were performed to ascertain whether the effect of the associations found in different populations was driven by potential confounders of the response to asthma medication or disease severity. Specifically, association analyses with asthma exacerbations were performed for the variant with evidence of replication. First, logistic regressions were carried out evaluating the association with the presence/absence of asthma exacerbations accounting for the daily ICS dosage in PACMAN, the only study with available information for this variable, as described in the Supplementary Material. Additionally, association analyses were carried out accounting for asthma severity based on the classification into treatment steps based on a modification of the guidelines established by the British Thoracic Society and the Scottish Intercollegiate Guidelines Network (BTS/SIGN) [24]. Only those individuals with available information about the use of the medications included in the classification into treatment steps were selected and they were classified as described in the Supplementary Material.
In silico functional evaluation of variants associated with asthma exacerbations despite ICS use
Functional evaluation of the variant with evidence of replication was carried out using publicly available databases. Evaluation of functional evidence described in the Encyclopedia of DNA Elements (ENCODE) was used to assess the role as expression quantitative trait loci (eQTL), DNase hypersensitivity sites, and histone marks using HaploReg v4.1 [25], and the Portal for the Genotype-Tissue Expression (GTEx) was also queried [26]. Previous significant evidence as protein quantitative trait loci (pQTL) or methylation quantitative trait loci (meQTL) was also explored using publicly available information by means of the PhenoScanner v2 tool [27,28].
Validation of previously reported ICS genes in European populations
Previous studies identified a total of 26 SNPs located near or within 15 genes associated with ICS response in different populations (Table S3). These variants were analysed in the present dataset using the meta-analysis results of the discovery phase of the current GWAS.
Validation of previous associations was performed at the SNP level, searching for consistent association at the nominal level (p≤0.05). Additionally, replication was also assessed as genomic regions, analysing variants located within 100 kilobases (kb) upstream and downstream from the gene limits. A Bonferroni-corrected significance threshold was estimated for each genomic region as α = 0.05/number of independent variants analysed, using the same methodology as described elsewhere [15].
Enrichment analysis of drug targets
A gene-set enrichment analysis focused on drugs was performed using the summary association results from the discovery phase of this GWAS. An overlap between the genes associated with asthma exacerbations in the discovery phase and gene sets with previous evidence of expression inhibition or induction after exposure to drugs or small molecules was inspected. For that, variants were first assigned to the nearest gene using the UCSC Table Browser tool [29]. Not only SNPs associated (p≤5×10−6) with asthma exacerbations despite ICS treatment in the discovery phase were included, but also those significant at p≤1×10−4 were analysed to increase the statistical power to detect genes previously identified to show drug-induced changes in expression levels. This threshold was arbitrarily set as it is commonly carried out in gene-set enrichment approaches [30,31]. For this, the information available at the Drug SIGnatures DataBase and DrugMatrix was used utilizing the Enrichr tool [32]. Evidence of significant enrichment at drugs was considered for those genes with significant drug-related expression changes after accounting for the multiple comparisons tested (false discovery rate (FDR) ≤0.05).
RESULTS
Characteristics of the study populations
A total of 2,681 children and young adults with asthma from eight studies were analysed in the discovery phase (Table 1), whereas 538 patients from different populations were included in the replication stage of this GWAS in Europeans (Table S1). Individuals from the studies analysed in the discovery phase showed a similar mean age, except for followMAGICS, which included individuals with older ages (17.2 ± 3.0 years) (Table 1). Although different definitions of asthma exacerbations were used, similar proportions of exacerbations were found across European populations included in the discovery phase, except for PACMAN and GoSHARE, which showed the lowest asthma exacerbations rates (11.0% and 13.8%, respectively) (Table 1). Among the non-European samples, Latinos/Hispanics from GALA II had the highest proportion of asthma exacerbations occurrence despite the treatment with ICS (66.4%) (Table S2).
Association results in European populations
Association results for a total of 8.1 million common SNPs (MAF≥1%) with Rsq≥0.3 and shared among the eight European populations included in the discovery phase were meta-analysed. No major evidence of genomic inflation due to population stratification was found when each study was individually analysed (Figure S1A–S1H), neither after combining them in a meta-analysis (λGC = 1.04, Figure S1I). Although no associations were detected at the genome-wide significance level (p-value≤5×10−8), a total of 19 variants near or within 10 loci showed p-value≤5×10−6 in European children and young adults (Table S4, Figure 1). Among those polymorphisms, one independent variant per locus was found after performing pairwise regressions conditioned on the most significant variant for each locus with more than one association signal. Thus, a total of ten independent signals were detected (Table 2), which were followed up for replication.
Figure 1. Manhattan plot of association results of asthma exacerbations in ICS users included in the discovery phase.
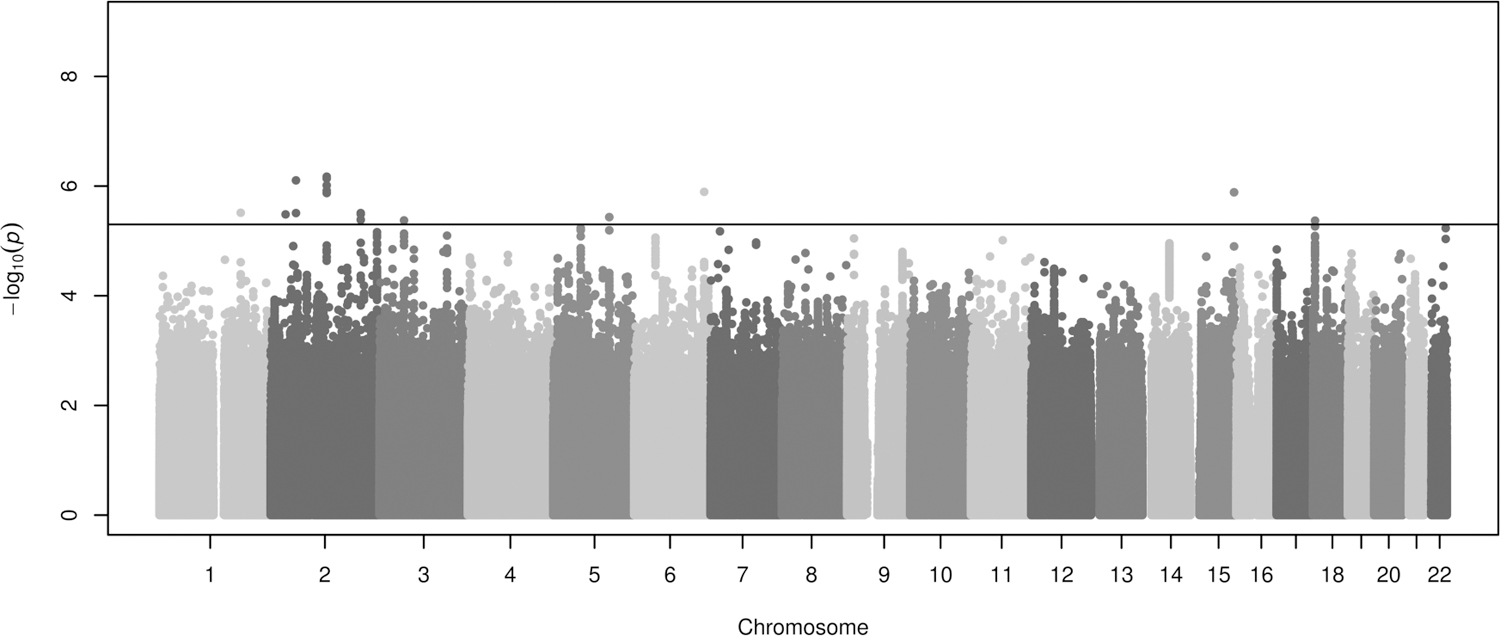
Association results are represented as -log10 p-value on the y-axis along the chromosomes (x-axis). The horizontal black line shows the suggestive significance threshold for replication (p≤5×10−6).
Table 2.
Summary of the conditional regression models for each locus suggestively associated with asthma exacerbations in patients treated with ICS in the discovery phase.
| Meta-analysis (n=2,681) |
Conditional regression model |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nearest gene(s) | SNP | Chr. a | Position b | E/NE | Freq. c | OR (95% CI) d | p-value | Conditioned on | p-value |
| ZNF648-GLUL | rs71632139 | 1 | 182326506 | C/G | 0.109 | 1.60 (1.31–1.94) | 3.07 × 10−6 | NA | NA |
| LTBP1 | rs11681246 | 2 | 33466620 | G/A | 0.436 | 0.72 (0.63–0.83) | 3.28 × 10−6 | NA | NA |
| CCDC85A-VRK2 | rs113364932 | 2 | 56668971 | A/G | 0.063 | 2.20 (1.61–3.01) | 7.86 × 10−7 | rs113364932 | NA |
| rs72805125 | 2 | 56684554 | T/C | 0.063 | 2.09 (1.53–2.85) | 3.11 × 10−6 | 0.888 | ||
| CNTNAP5 | rs76496334 | 2 | 125427606 | T/C | 0.022 | 2.29 (1.64–3.19) | 9.69 × 10−7 | rs144289311 | 0.491 |
| rs146921813 | 2 | 125432412 | C/G | 0.022 | 2.26 (1.63–3.16) | 1.34 × 10−6 | 0.534 | ||
| rs141194780 | 2 | 125432413 | A/G | 0.022 | 2.26 (1.63–3.16) | 1.34 × 10−6 | 0.534 | ||
| rs144289311 | 2 | 125432440 | A/G | 0.022 | 2.33 (1.67–3.25) | 6.73 × 10−7 | NA | ||
| rs145694710 | 2 | 125434780 | T/C | 0.022 | 2.28 (1.63–3.17) | 1.21 × 10−6 | 0.515 | ||
| rs17011852 | 2 | 125440426 | G/A | 0.022 | 2.32 (1.66–3.24) | 7.27 × 10−7 | NA | ||
| AOX1 | rs2465662 | 2 | 201501145 | C/T | 0.283 | 1.13 (0.77–1.66) | 4.08 × 10−6 e | rs7587871 | 0.847 |
| rs7587871 | 2 | 201505269 | A/C | 0.318 | 1.09 (0.75–1.58) | 3.10 × 10−6 e | NA | ||
| rs7420798 | 2 | 201506713 | G/A | 0.318 | 1.09 (0.75–1.58) | 3.24 × 10−6 e | NA | ||
| rs12988162 | 2 | 201507154 | A/T | 0.318 | 1.08 (0.75–1.57) | 4.14 × 10−6 e | NA | ||
| CACNA2D3-WNT5A | rs67026078 | 3 | 55162698 | C/T | 0.085 | 1.50 (0.93–2.43) | 4.22 × 10−6 e | NA | NA |
| ZNF608-GRAMD3 | rs444610 | 5 | 125315286 | A/T | 0.398 | 1.36 (1.09–1.69) | 3.68 × 10−6 e | NA | NA |
| NOX3-ARID1B | rs2493700 | 6 | 156826363 | G/C | 0.677 | 0.71 (0.62–0.82) | 1.28 × 10−6 | NA | NA |
| SPATA8-ARRDC4 | rs72759231 | 15 | 97550165 | G/A | 0.058 | 1.97 (1.50–2.59) | 1.30 × 10−6 | NA | NA |
| DLGAP1-ZBTB14 | rs28761328 | 18 | 4746271 | A/T | 0.148 | 1.56 (1.29–1.89) | 4.26 × 10−6 | NA | NA |
Chromosome;
Positions based on GRCh37/hg19 build;
Frequency of the effect allele in European populations from the 1,000 Genomes Project phase 3;
Odds ratio for the effect alleles (additive model);
Random-effect model was applied since heterogeneity was found between European studies.
CI: Confidence Interval; E: Effect allele; NA: not available; NE: Non-effect allele; SNP: single-nucleotide polymorphism.
Independent SNPs of each gene region are in boldface.
Of the 10 variants associated with asthma exacerbations despite ICS treatment in the discovery phase (p-value≤5×10−6) only the SNP rs67026078, located within the intergenic region of CACNA2D3 and WNT5A (Figure 2), showed nominal replication after meta-analysing the European studies included in the replication (odds ratio (OR) for C allele: 1.83, 95% Confidence Interval (CI): 1.16–2.90, p = 0.010) (Table 3). The association had a consistent effect as in the discovery phase (OR for C allele: 1.50, 95%CI: 0.93–2.43, p = 4.22×10−6) (Table 3). Suggestive genome-wide association was found for this SNP after performing a meta-analysis across the European studies analysed in both phases (OR for C allele: 1.58, 95%CI: 1.11–2.26, p = 4.34×10−7) (Figure 3). Nonetheless, the association effect of this variant was mostly driven by the studies with information about the occurrence of asthma exacerbations available for a 12- month period. This could be explained by the fact that a wider timeframe makes exacerbation events likely to occur, but also by the larger sample size analysed compared to the studies with information based on the previous 6 months (n=1,557 vs. n=1,124).
Figure 2. Regional plot of association results for the CACNA2D3-WNT5A locus for the European populations included in the discovery phase.
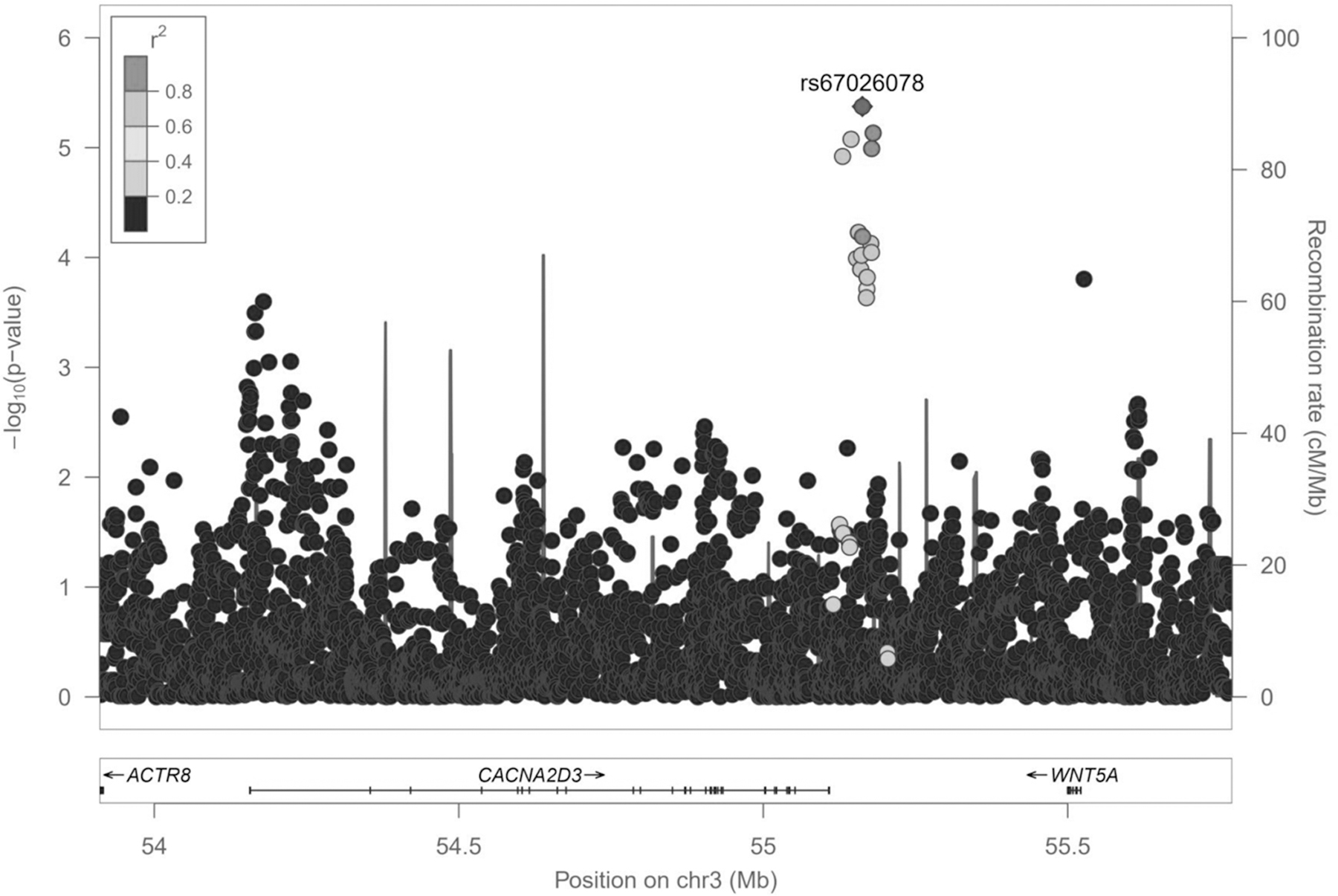
Logarithmic transformation of the association results (-log10 p-value) is represented in the y-axis by chromosome position (x-axis) for each SNP as a dot. The SNP rs67026078 with evidence of replication in the European populations included in the replication phase is represented by a diamond. The remaining variants are grey color-coded based on pairwise r2 values with that SNP for European populations from 1KGP.
Table 3.
Association results for the independent suggestive associations followed up for replication in populations of European descent.
| SNP | Chr.a | Positionb | Nearest gene(s) | E/NE | Discovery phase |
Replication phase |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Meta-analysis (n=2,681) | ALSPAC (n=258) | CAMP (n=175) | BAMSE (n=105) | Meta-analysis (n=538) | ||||||||||
| OR (95% CI)d | p-value | OR (95% CI)d | p-value | OR (95% CI)d | p-value | OR (95% CI)d | p-value | OR (95% CI)d | p-value | |||||
| rs71632139 | 1 | 182326506 | ZNF648-GLUL | C/G | 1.60 (1.31–1.94) |
3.07 × 10−6 | 1.36 (0.76–2.43) |
0.315 | 1.16 (0.60–2.22) |
0.665 | 1.31 (0.48–3.47) |
0.604 | 1.27 (0.85–1.89) |
0.243 |
| rs11681246 | 2 | 33466620 | LTBP1 | G/A | 0.72 (0.63–0.83) |
3.28 × 10−6 | 1.50 (1.00–2.27) |
0.051 | 1.12 (0.69–1.80) |
0.649 | 1.12 (0.59–2.08) |
0.738 | 1.28 (0.97–1.70) |
0.082 |
| rs113364932 | 2 | 56668971 | CCDC85A-VRK2 | A/G | 2.20 (1.61–3.01) |
7.86 × 10−7 | 0.58 (0.14–2.39) |
0.424 | 1.87 (0.69–5.08) |
0.222 | 0.47 (0.11–1.92) |
0.305 | 1.00 (0.49–2.03) |
0.991 |
| rs144289311 | 2 | 125432440 | CNTNAP5 | A/G | 2.33 (1.67–3.25) |
6.73 × 10−7 | 1.71 (0.72–4.07) |
0.234 | 0.51 (0.14–1.88) |
0.314 | 0.88 (0.13–5.41) |
0.892 | 1.14 (0.58–2.23) |
0.703 |
| rs7587871 | 2 | 201505269 | AOX1 | A/C | 1.09 (0.75–1.58) |
3.10 × 10−6 e | 0.70 (0.44–1.10) |
0.117 | 1.40 (0.89–2.19) |
0.146 | 0.98 (0.53–1.78) |
0.949 | 0.99 (0.75–1.32) |
0.942 |
| rs67026078 | 3 | 55162698 | CACNA2D3-WNT5A | C/T | 1.50 (0.93–2.43) |
4.22 × 10−6 e | 2.06 (1.07–3.97) |
0.032 | 1.68 (0.77–3.65) |
0.193 | 1.53 (0.48–4.71) |
0.471 | 1.83 (1.16–2.90) |
0.010 |
| rs444610 | 5 | 125315286 | ZNF608-GRAMD3 | A/T | 1.36 (1.09–1.69) |
3.68 × 10−6 e | 0.76 (0.51–1.15) |
0.189 | 0.83 (0.53–1.29) |
0.409 | 0.79 (0.43–1.43) |
0.455 | 0.79 (0.61–1.04) |
0.091 |
| rs2493700 | 6 | 156826363 | NOX3-ARID1B | G/C | 0.71 (0.62–0.82) |
1.28 × 10−6 | 1.10 (0.68–1.76) |
0.697 | 1.05 (0.67–1.64) |
0.827 | 0.65 (0.35–1.17) |
0.162 | 0.96 (0.72–1.28) |
0.573 |
| rs72759231 | 15 | 97550165 | SPATA8-ARRDC4 | G/A | 1.97 (1.50–2.59) |
1.30 × 10−6 | 0.91 (0.39–2.14) |
0.829 | 0.60 (0.28–1.27) |
0.180 | 0.83 (0.22–3.05) |
0.785 | 0.74 (0.44–1.24) |
0.247 |
| rs28761328 | 18 | 4746271 | DLGAP1-ZBTB14 | A/T | 1.56 (1.29–1.89) |
4.26 × 10−6 | 0.40 (0.19–0.82) |
0.007 | 1.23 (0.65–2.36) |
0.524 | 0.70 (0.28–1.68) |
0.433 | 0.74 (0.48–1.13) |
0.164 |
Chromosome;
Positions based on GRCh37/hg19 build;
Frequency of the effect allele in European populations from the 1,000 Genomes Project phase 3;
Odds ratio for the effect alleles (additive model);
Random-effect model was applied since heterogeneity was found between European studies.
CI: Confidence Interval; E: Effect allele; NA: not available; NE: Non-effect allele; SNP: single-nucleotide polymorphism.
SNPs with evidence of replication in independent European populations are in boldface.
Figure 3. Forest plot of association effect of rs67026078 across European studies included in the GWAS of asthma exacerbations despite ICS treatment.
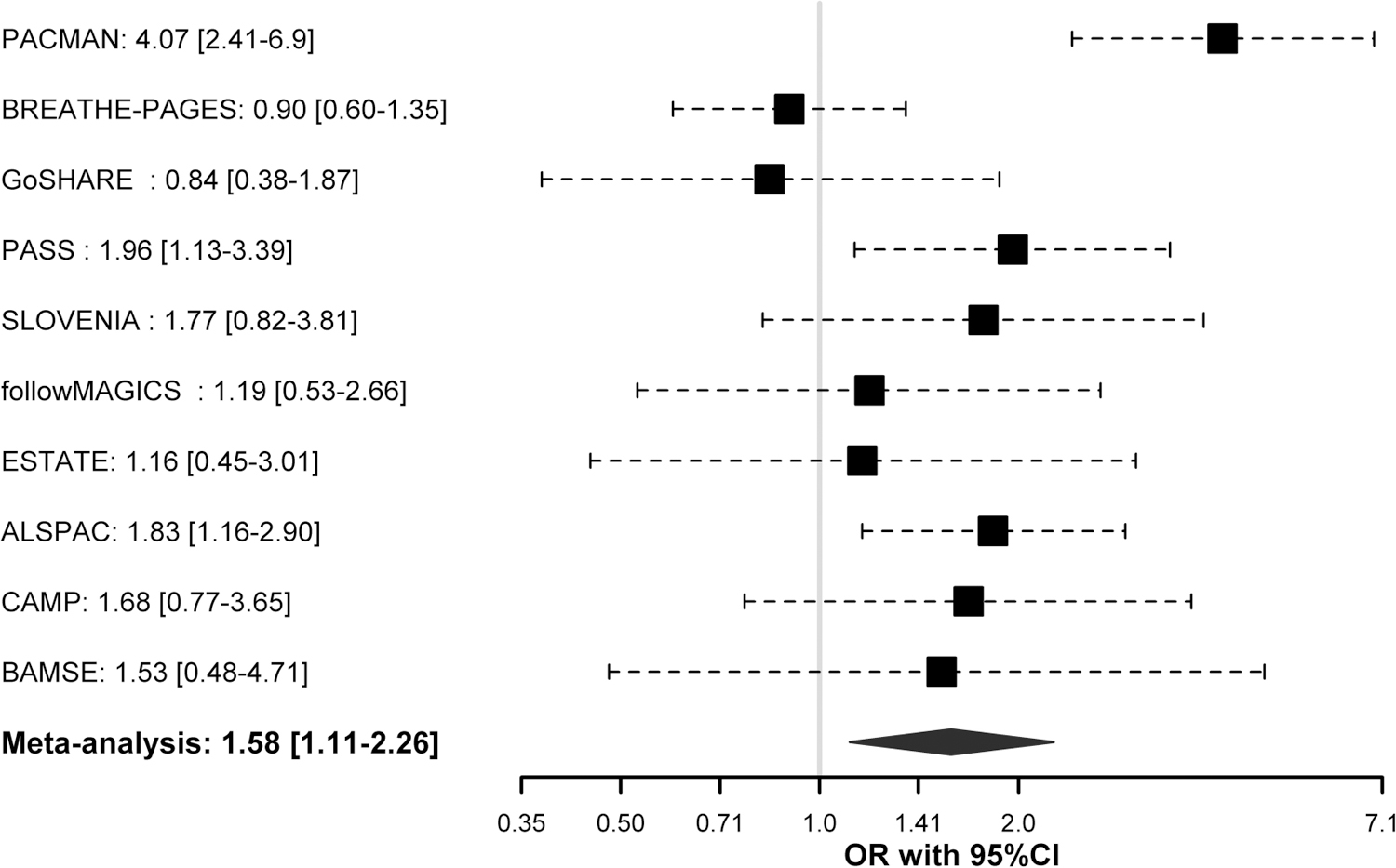
Association effects are shown in terms of odds ratio (OR) for the effect allele (C) for each study and after meta-analysing the results from both phases by black boxes and a blue diamond, respectively. Black dash lines indicate the corresponding 95% Confidence Intervals (95% CI) for each study. Effect of association results is not given for BREATHE since rs67026078 did not pass quality control checks.
Assessment of ICS associations in non-European populations
The SNP rs67026078 with evidence of replication in independent European populations was not associated with asthma exacerbations in patients treated with ICS from Hispanic/Latino nor African American populations (Table S5). In Asians, this variant was not consistently associated with asthma exacerbations in SCSGES neither (Table S5). Differences in the effect allele frequency of this variant were found among the populations evaluated, being higher in the studies of European ancestry included in the discovery (6.1–9.3%) and replication phases (5.7–9.4%), compared to the non-European populations. Specifically, this variant had a frequency of 4.7%, 4.9%, and 1.4% in Hispanics/Latinos, African Americans, and Asians, respectively.
Association analysis accounting for ICS dosage and asthma severity
Sensitivity analyses of asthma exacerbations despite ICS use including daily medication dosages as a covariate in 521 asthma patients of European descent from the PACMAN study revealed that the association effect of rs67026078 adjusted by the ICS did not account for the association with the occurrence of asthma exacerbations (OR for C allele: 1.24, 95% CI: 1.14–1.34, p = 2.30×10−7). These results are equivalent in terms of significance to those obtained applying the original association model for the same individuals with complete data, but the effect sizes are smaller (OR for C allele: 4.30, 95% CI: 2.33–7.92, p = 2.98×10−6 in the model not adjusted by ICS dose). Similar results were found adjusting by a categorical variable related to ICS dose based on age groups: OR for C allele: 1.23, 95% CI: 1.14–1.34, p = 2.02×10−7) (Table S6).
Association analyses adjusted by asthma severity based on treatment steps classification were performed in 2,282 asthma patients from the discovery phase with available data related to the medication use (Table 1). The SNP rs67026078 was suggestively associated with asthma exacerbations after accounting for disease severity (OR for C allele: 1.43, 95% CI: 0.88–2.33, p = 1.05×10−5). These results are equivalent to those obtained applying the original association models to the individuals with available classification into treatment steps (OR for C allele: 1.45, 95% CI: 0.91–2.33, p = 1.03×10−5).
Functional evaluation of the variant associated with asthma exacerbations despite ICS use
According to the ENCODE project, the SNP with evidence of replication among Europeans, rs67026078, is located within a histone H3 lysine 4 mono-methylation (H3K4me1) mark in several tissues, including foetal lung fibroblasts and other foetal pulmonary cells. Its suggestive role in regulating gene expression is also shown by the fact that this is a DNAse hypersensitivity site in lung fibroblast primary cells [33]. However, no evidence of significant eQTL was found for this SNP. Nonetheless, the SNP rs67026078 had been previously significantly identified (p≤0.01) as pQTL and meQTL. Specifically, Sun et al. found this variant to be associated with protein expression levels for 16 different proteins in plasma [27,28,34] (Table S7). Some of these have been related to molecular and cellular processes related to asthma pathophysiology (ADAMTS5) and involved directly or indirectly in the Wnt pathway (PSMA2, ADAMTS5, ATAD2, CHST3, TEAD3) [35]. Moreover, rs67026078 was found to regulate the methylation patterns of a CpG site (cg16278514) at the intergenic region of CACNA2D3 and WNT5A in whole blood by Bonder et al. [27,28,36]. Interestingly, both CACNA2D3 and WNT5A are expressed in pulmonary tissues [26].
Validation of genes previously associated with ICS response
Among the 26 SNPs associated in previous GWAS of ICS response, one variant intergenic to UMAD1 and GLCCI1 (rs37972) showed evidence of replication in European populations included in the PiCA consortium (OR for C allele: 1.20, 95%CI: 1.05–1.37, p = 6.58×10−3) (Table S8). Considering the genomic regions where these genes reside, 33,096 variants located within 100 kb upstream and downstream from the 15 genes of ICS response previously described were evaluated. Accounting for the number of independent association signals within each genomic region, evidence of replication was found for 40 SNPs near five genomic regions: PDE10A-T (SNP with min p-value: rs57042153, OR for T allele: 1.43, 95%CI: 1.20–1.70, p = 5.97 × 10−5), UMAD1-GLCCI1 (rs13235500, OR for G allele: 0.71, 95%CI: 0.60–0.85, p = 2.44 × 10−4), SHB-ALDH1B1 (SNP with min p-value: rs341488, OR for A allele: 2.24, 95%CI: 1.48–3.40, p = 1.44 × 10−4), ZNF432-ZNF841 (SNP with min p-value: rs67834224, OR for A allele: 0.65, 95%CI: 0.52–0.82, p = 2.86 × 10−4), ELMO2-ZNF334 (SNP with min p-value: rs11087003, OR for C allele: 0.77, 95%CI: 0.66–0.89, p = 5.84 × 10−4) (Table S9). However, none of these associations were significant after correction for the total number of SNPs tested across all genomic regions (1,799 independent SNPs: Bonferroni-like correction significance threshold of p≤2.78×10−5).
Enrichment analysis in European asthmatic children and young adults treated with ICS
Enrichment analysis of associations from the GWAS results focused on drugs was carried out, including 782 SNPs associated with asthma exacerbations despite ICS treatment (p≤1×10−4) in the discovery phase. A total of 49 different drugs and small molecules that had been found to regulate expression levels of the genes associated with asthma exacerbations in the GWAS were revealed (Table S10). Of those, trichostatin A (TSA) remained statistically significant after adjusting for multiple comparisons (FDR = 0.035) (Table S10). Specifically, a total of 30 of the 83 genes associated at p≤1×10−4 in our GWAS had been previously proposed as targets of TSA, since changes in expression levels were found to be triggered by the exposure to this drug (Table S11). These genes included several loci previously associated with asthma-related traits and allergic diseases (e.g., RERE, NEGR1, ROBO2, LAMA2, SLC11A2, JMJD1C) or involved in drug metabolism (e.g., AOX1) (Table S12) [35,37].
DISCUSSION
To our knowledge, this study describes the results of the largest GWAS of asthma exacerbations in children and young adults treated with ICS to date. After combining eight different studies of European ancestry, ten independent variants were found to be suggestively associated with asthma exacerbations despite ICS treatment in children and young adults with asthma. One SNP within the intergenic region of CACNA2D3 and WNT5A showed evidence of replication at nominal level in three independent European populations. However, this was not validated in Latinos/Hispanics, African Americans, or Asians, which could be due to ancestry-specific effects. Additionally, we found evidence of replication for five different genes associated with ICS response by previous GWAS studies at SNP or genomic-region level. Furthermore, an enrichment analysis of association signals with asthma exacerbations revealed TSA, which could regulate molecular mechanisms involved in asthma pathogenesis.
CACNA2D3 encodes a member of the alpha-2/delta subunit family, which are voltage-dependent calcium channels consisting of a complex of alpha-1, alpha-2/delta, beta, and gamma subunits. Specifically, CACNA2D3 modulates the calcium current density through the regulation of the influx of calcium ions into the cell upon membrane polarization [38]. CACNA2D3 has important functions given the fact that calcium is a secondary messenger involved in multiple cellular processes such as cell proliferation, apoptosis, adhesion, and migration [39]. This gene could have a role in respiratory diseases since variants located near to CACNA2D3 have been recently associated with different lung function measurements, which are important predictors of asthma severity and progression [40,41]. Specifically, these associations include forced expiratory volume in one second (FEV1), forced vital capacity (FVC), and the ratio FEV1/FVC, in chronic obstructive pulmonary disease (COPD) patients from the large cohort of European descent UKBiobank [42,43], and the change in lung function after administration of bronchodilators in smokers [44]. It is well known that pulmonary function is an important predictor of asthma severity and progression [40,41]. Additionally, an intronic CACNA2D3 variant (rs1820616) has also been associated with the fractional concentration of nitric oxide (FeNO) in exhaled air [45], which is a good indicator of inflammatory patterns in the airways and potentially an alternative approach to support asthma diagnosis in children [46,47] and to monitor the adherence and response to medications [48]. These findings suggest that CACNA2D3 could be involved in asthma progression, including the risk of asthma exacerbations, even in patients under ICS therapy.
WNT5A encodes for the WNT family member 5A, a lipid-modified glycoprotein that activates diverse signalling pathways [49]. This protein has been evidenced to play a crucial role in development during embryogenesis, oncogenesis, and regulation of inflammatory processes in infectious disorders [50]. Moreover, other genes encoding for ligands involved in the WNT signalling pathway are associated with impaired lung function in asthmatic children [51]. This suggests that WNT5A could be also involved in the pulmonary capacity in asthma. Interestingly, genes associated with asthma susceptibility have been linked to WNT signalling through a gene-set enrichment analysis [30]. Specifically, this biological process seems to play regulatory and suppressive roles through the modulation of inflammation and structural changes in airways. WNT ligands have been proposed to act on the major players implicated on inflammatory processes such as dendritic and T-helper type 2 (Th2) cells and macrophages [52]. Indeed, WNT molecules regulate the homeostasis of these cells, avoiding dysregulated immune responses, which could trigger several diseases, including allergic asthma [52].
Specifically, expression of WNT5A has been positively associated with Th2-mediated airway inflammation in asthmatic patients [53]. Additionally, eosinophils derived from asthma patients have been found to enhance expression levels of this gene in airway smooth muscle (ASM) cells, triggering cell proliferation, inflammatory processes, and airway remodelling [54]. It is well known that eosinophilia at blood and tissue levels is one of the most important phenotypes in asthma patients [55], triggered by high levels of chemokines and cytokines. Specifically, eosinophils migrate from lymph nodes to the airway in asthma, where they adhere to the ASM, releasing transforming growth factor β1 (TGF-β1) molecules [56]. Increased levels of TGF-β1 have been related to the overexpression of WNT5A in ASM cells at gene and protein levels compared to healthy individuals. Therefore, production of extracellular matrix proteins is induced, increasing ASM mass and contractility and hence, airway remodelling by means of hypertrophy and hyperplasia [54]. These findings suggest the important role of the WNT5A and the WNT signalling pathway in asthma pathogenesis, making it a promising therapeutic target in asthma [57], throughout inhibition of WNT ligands biogenesis, secretion and blocking their ligand-receptor interactions through small pharmacological molecules [50]. Nonetheless, further research is needed to explore the potential side effects of drugs targeting this pathway, since tumorigenesis-related functions have been also widely attributed to WNT molecules [58].
The C allele of the SNP rs67026078, which is located 54.1 kb from the 3’ UTR of CACNA2D3, was found to be associated with an increased risk of asthma exacerbations despite the ICS treatment across the European studies analysed in the discovery and replication phases. Sensitivity analyses accounting for baseline asthma severity suggested that the effect of this association is related to the response to asthma medications or to the biologic drivers of asthma exacerbations. Nonetheless, this did not show to be significantly associated with asthma exacerbations in patients treated with ICS from Hispanic/Latino, African American, or Asian populations. This result could be explained by ancestry-driven effects evidenced by the lower frequency of the effect allele of this variant in non-European populations. This polymorphism had not been previously associated with asthma treatment response, although functional evidence suggests that this variant could be actively involved in the regulation of gene expression in cells from lung tissue [33].
We also performed a gene-set enrichment analysis focusing on drugs, finding evidence of enrichment of TSA, which had been proposed to target several genes previously associated with asthma-related traits and drug metabolism, suggesting that TSA could be involved in the molecular mechanisms underlying the occurrence of asthma exacerbations despite ICS treatment. These findings demonstrate that GWAS approaches in combination with gene-set enrichment analyses seem to be a powerful strategy to explore potential novel therapeutic interventions, even in the absence of genome-wide associations [59,60].
TSA is a hydroxamic acid extracted from the bacterial genus Streptomyces with a wide range of histone deacetylase (HDAC) inhibitor activities in mammalian cells [61]. Specifically, TSA belongs to a family of compounds acting on metal-dependent HDACs, inhibiting histone deacetylation, and causing hyperacetylation of core histones, which is one of the major regulators of the chromatin structure [62]. Nonetheless, HDAC inhibitors have been also demonstrated to act on diverse non-histone substrates involved in several functions such as cell signalling, chromatin structure, and DNA repair, among others [63].
Interestingly, the potential clinical utility of HDAC inhibitors in asthma has been investigated [63]. Several studies in animal models [63–65] have suggested that the inhibition of HDACs by TSA could play an important role in the reduction of asthma development by decreasing airway inflammation and hyperresponsiveness [66]. These findings, together with evidence that HDACs regulate sensitivity to glucocorticosteroids [63], suggest that histone acetylation may play a key role in asthma development [67], and seems to be a promising target for alternatives to the standard medications currently used in the management of asthma. Specifically, in vivo experiments in allergen-challenged mice have demonstrated that treatment with TSA decreases eosinophils and lymphocytes levels in bronchial alveolar lavage. Reduced expression levels of inflammatory mediators such as Th2 cytokines were also detected [67]. Moreover, it has been found that TSA shows additive effects in combination with glucocorticosteroids, suggesting that it might target the main pathological processes in asthma through mechanisms of action different from the classical asthma anti-inflammatory medications [64]. Additionally, Banerjee et al. also found that TSA could have important functions in the inhibition of bronchoconstriction by inducing remodelling changes [64]. It has been demonstrated that TSA treatment might inhibit the release of intracellular calcium, reducing ASM contraction in human lung slices and ASM cells in vitro expose to contractile agonists [64].
Although the effects of TSA on chromatin structure and regulation of gene expression in pulmonary tissues are still unclear [64], these findings suggest that TSA could potentially play an important role in asthma through epigenetic modifications and regulate the molecular mechanisms involved in response to ICS. Nonetheless, to the best of our knowledge, the effect of TSA on asthma patients has not been tested in clinical trials yet and little is known about the potential side effects of this drug. For this reason, there is still a long way for the potential introduction of TSA as controller therapy in clinical practice.
The current study has some limitations that need to be acknowledged. First, the genome-wide significance level was reached neither in the discovery phase nor after combining the results with independent European studies. Although to our knowledge our study includes the largest sample size analysed in any GWAS of exacerbations despite ICS use performed in children and young adults with asthma to date, the lack of genome-wide associations could be explained by reduced statistical power given by differences in patient recruitment and definition of asthma exacerbations tested in association in both discovery and replication phases. Additionally, no covariates related to the aetiology of asthma exacerbations and exposure to potential environmental triggers were considered in the association analyses. Second, retrospective information about the occurrence or absence of asthma exacerbations partly based on self-reports was used, which could not be fully informative of the real ICS response. Moreover, a period of 6 or 12 months preceding the study enrolment was considered, which could have introduced substantial heterogeneity in the interpretation of treatment response, since more exacerbations are possible in additional 6 months and non-response might be more likely to occur in 12 months. Third, although the standard definition of severe asthma exacerbations established by the European Respiratory Society (ERS) and the American Thoracic Society (ATS) considering them as the need for unscheduled medical care because of asthma [16] was used, this information was incomplete for some of the European studies included in the discovery or replication phases. Therefore, data regarding unscheduled visits to general practitioners or respiratory disease specialists and school absences due to asthma were considered instead, which captures moderate asthma exacerbations. Additionally, no variables indicating whether ICS therapy had been initiated before or after exacerbations episodes were available. Altogether, this heterogeneity in data availability could represent a potential interpretation bias in terms of response to asthma treatment. Fourth, specific ICS dose and type or any index of treatment adherence were not included as covariates in the association analyses, since information related to these variables was not available for most of the studies included in this GWAS. Fifth, although in silico evaluation of the functional implication of CACNA2D3 and WNT5A on asthma exacerbations was carried out, in vitro experiments, pharmacogenomic research of pre-existing randomized controlled trials, and longitudinal asthma studies are needed to confirm their role in asthma treatment response.
In summary, our GWAS of asthma exacerbations in children and young adults treated with ICS revealed a novel association in Europeans. We also found evidence of replication of variants previously associated with different definitions of ICS response in asthma patients of European descent and suggested TSA as a potential novel therapy that could be implicated in mechanisms controlling asthma symptoms and moderate-to-severe exacerbations in ICS non-responders. These findings suggest that the integration of different analytical methods could be a powerful strategy providing new insights into the molecular mechanisms underlying ICS response and suggesting alternative asthma therapies.
Supplementary Material
TAKE-HOME MESSAGE.
A genome-wide association study of asthma exacerbations despite inhaled corticosteroids treatment in childhood asthma revealed a novel association at the CACNA2D3-WNT5A locus and suggested trichostatin A as a potential asthma therapy.
ACKNOWLEDGEMENTS
The authors acknowledge the patients, families, recruiters, health care providers and community clinics for their participation in all the studies included in the PiCA consortium (http://pica-consortium.org). The authors thank the contribution of Teide High-Performance Computing facilities (http://teidehpc.iter.es) provided by the Instituto Tecnológico y de Energías Renovables (ITER, S.A.) to the results of this research and also the Centro Nacional de Genotipado-Plataforma de Recursos Biomoleculares-Instituto de Salud Carlos III (CeGen-PRB3-ISCIII; www.cegen.org) for the genotyping services provided.
We acknowledge all the families who took part in this ALSPAC, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses.
The GALA II study collaborators include Shannon Thyne, UCSF; Harold J. Farber, Texas Children’s Hospital; Denise Serebrisky, Jacobi Medical Center; Rajesh Kumar, Lurie Children’s Hospital of Chicago; Emerita Brigino-Buenaventura, Kaiser Permanente; Michael A. LeNoir, Bay Area Pediatrics; Kelley Meade, UCSF Benioff Children’s Hospital, Oakland; William Rodriguez-Cintron, VA Hospital, Puerto Rico; Pedro C. Avila, Northwestern University; Jose R. Rodriguez-Santana, Centro de Neumologia Pediatrica; Luisa N. Borrell, City University of New York; Adam Davis, UCSF Benioff Children’s Hospital, Oakland; Saunak Sen, University of Tennessee and Fred Lurmann, Sonoma Technologies, Inc.
The authors acknowledge the families and patients for their participation and thank the numerous health care providers and community clinics for their support and participation in GALA II. In particular, the authors thank study coordinator Sandra Salazar; the recruiters who obtained the data: Duanny Alva, MD, Gaby Ayala-Rodriguez, Lisa Caine, Elizabeth Castellanos, Jaime Colon, Denise DeJesus, Blanca Lopez, Brenda Lopez, MD, Louis Martos, Vivian Medina, Juana Olivo, Mario Peralta, Esther Pomares, MD, Jihan Quraishi, Johanna Rodriguez, Shahdad Saeedi, Dean Soto, Ana Taveras; and the lab researcher Celeste Eng who processed the biospecimens.
The SAGE study collaborators include Harold J. Farber, Texas Children’s Hospital; Emerita Brigino-Buenaventura, Kaiser Permanente; Michael A. LeNoir, Bay Area Pediatrics; Kelley Meade, UCSF Benioff Children’s Hospital, Oakland; Luisa N. Borrell, City University of New York; Adam Davis, UCSF Benioff Children’s Hospital, Oakland and Fred Lurmann, Sonoma Technologies, Inc.
The authors acknowledge the families and patients for their participation and thank the numerous health care providers and community clinics for their support and participation in SAGE. In particular, the authors thank study coordinator Sandra Salazar; the recruiters who obtained the data: Lisa Caine, Elizabeth Castellanos, Brenda Lopez, MD, Shahdad Saeedi; and the lab researcher Celeste Eng who processed the biospecimens.
FINANCIAL SUPPORT STATEMENT
This study was supported by the awards (AC15/00015 and AC15/00058) funded by the Instituto de Salud Carlos III (ISCIII) through Strategic Action for Health Research (AES) and European Community (EC) within the Active and Assisted Living (AAL) Programme framework (MP-Y, OS-P), the SysPharmPedia grant from the ERACoSysMed 1st Joint Transnational Call from the European Union under the Horizon 2020, and the grant SAF2017–83417R by the Spanish Ministry of Science, Innovation and Universities. The PACMAN study was funded by a strategic alliance between GlaxoSmithKline and Utrecht Institute for Pharmaceutical Sciences. The SLOVENIA study was financially supported by the Slovenian Research Agency (research core funding No. P3–0067) and from SysPharmPedia grant, co-financed by Ministry of Education, Science and Sport Slovenia (MIZS) (contract number C3330–16-500106). GALA II was supported by the National Heart, Lung, and Blood Institute of the National Institute of Health (NIH) grants R01HL117004 and X01HL134589; study enrolment supported by the Sandler Family Foundation, the American Asthma Foundation, the RWJF Amos Medical Faculty Development Program, Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II and the National Institute of Environmental Health Sciences grant R01ES015794. SAGE was funded by the National Heart, Lung, and Blood Institute of the National Institute of Health (NIH) grants R01HL117004 and X01HL134589; study enrolment supported by the Sandler Family Foundation, the American Asthma Foundation, the RWJF Amos Medical Faculty Development Program, Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II. The SHARE Bioresource (GoSHARE) and SHARE have ongoing funding from NHS Research Scotland and established by funding from The Wellcome Trust Biomedical Resource [Grant No. 099177/Z/12/Z]. Genotyping of samples from BREATHE-PAGES, GoSHARE, and SCSGES was carried out at CeGen-PRB3-ISCIII; supported by ISCIII and European Regional Development Fund (ERDF) (PT17/0019). ALSPAC was supported by the UK Medical Research Council and Wellcome (102215/2/13/2) and the University of Bristol. The Swedish Heart-Lung Foundation, the Swedish Research Council and Region Stockholm (ALF project and database maintenance) funded the BAMSE study. ESTATe was funded by an independent research grant by ZonMw project (113201006). CAMP was supported by grants from NIH (R01HL127332 and R01NR013391). The PASS study was funded by the NHS Chair of Pharmacogenetics via the UK Department of Health. MP is Emeritus NIHR Senior Investigator.
REFERENCES
- 1.Global strategy for asthma management and prevention. Global Initiative for Asthma (GINA) 2020. http://ginasthma.org/.
- 2.Cerasoli F Jr. Developing the ideal inhaled corticosteroid. Chest 2006;130(1 Suppl):54S–64S. [DOI] [PubMed] [Google Scholar]
- 3.Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF, Strunk RC, Zeiger RS, Larsen G, Spahn JD, Bacharier LB, Bloomberg GR, Guilbert TW, Heldt G, Morgan WJ, Moss MH, Sorkness CA, Taussig LM. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol 2005;115:233–242. [DOI] [PubMed] [Google Scholar]
- 4.Mersha TB. Mapping asthma-associated variants in admixed populations. Front Genet 2015;6:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez-Pacheco N, Pino-Yanes M, Flores C. Genomic Predictors of Asthma Phenotypes and Treatment Response. Front Pediatr. 2019;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramadan AA, Gaffin JM, Israel E, Phipatanakul W. Asthma and Corticosteroid Responses in Childhood and Adult Asthma. Clin Chest Med 2019;40:163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park HW, Tantisira KG, Weiss ST. Pharmacogenomics in asthma therapy: where are we and where do we go? Annu Rev Pharmacol Toxicol. 2015;55:129–47. [DOI] [PubMed] [Google Scholar]
- 8.Farzan N, Vijverberg SJ, Arets HG, Raaijmakers JA, Maitland-van der Zee AH. Pharmacogenomics of inhaled corticosteroids and leukotriene modifiers: a systematic review. Clin Exp Allergy 2017;47:271–293. [DOI] [PubMed] [Google Scholar]
- 9.Levin AM, Gui H, Hernandez-Pacheco N, Yang M, Xiao S, Yang JJ, Xiao S, Yang JJ, Hochstadt S, Barczak AJ, Eckalbar WL, Rynkowski D, Samedy LA, Kwok PY, Pino-Yanes M, Erle DJ, Lanfear DE, Burchard EG, Williams LK. Integrative approach identifies corticosteroid response variant in diverse populations with asthma. J Allergy Clin Immunol 2019;143:1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keskin O, Farzan N, Birben E, Akel H, Karaaslan C, Maitland-van der Zee AH, et al. Genetic associations of the response to inhaled corticosteroids in asthma: a systematic review. Clin Transl Allergy 2019;9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tantisira KG, Damask A, Szefler SJ, Schuemann B, Markezich A, Su J, Klanderman B, Sylvia J, Wu R, Martinez F, Boushey HA, Chinchilli VM, Mauger D, Weiss ST, Israel E. Genome-wide association identifies the T gene as a novel asthma pharmacogenetic locus. Am J Respir Crit Care Med 2012;185:1286–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrera-Luis E, Hernandez-Pacheco N, Vijverberg SJ, Flores C, Pino-Yanes M. Role of genomics in asthma exacerbations. Curr Opin Pulm Med 2019;25:101–112. [DOI] [PubMed] [Google Scholar]
- 13.Hall R, Hall IP, Sayers I. Genetic risk factors for the development of pulmonary disease identified by genome-wide association. Respirology 2019;24:204–214. [DOI] [PubMed] [Google Scholar]
- 14.Farzan N, Vijverberg SJ, Andiappan AK, Arianto L, Berce V, Blanca-Lopez N, Bisgaard H, Bonnelykke K, Burchard EG, Campo P, Canino G, Carleton B, Celedon JC, Chew FT, Chiang WC, Cloutier MM, Daley D, Den Dekker HT, Dijk FN, Duijts L, Flores C, Forno E, Hawcutt DB, Hernandez-Pacheco N, de Jongste JC, Kabesch M, Koppelman GH, Manolopoulos VG, Melen E, Mukhopadhyay S, Nilsson S, Palmer CN, Pino-Yanes M, Pirmohamed M, Potocnik U, Raaijmakers JA, Repnik K, Schieck M, Sio YY, Smyth RL, Szalai C, Tantisira KG, Turner S, van der Schee MP, Verhamme KM, Maitland-van der Zee AH. Rationale and design of the multiethnic Pharmacogenomics in Childhood Asthma consortium. Pharmacogenomics 2017;18:931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hernandez-Pacheco N, Farzan N, Francis B, Karimi L, Repnik K, Vijverberg SJ, Soares P, Schieck M, Gorenjak M, Forno E, Eng C, Oh SS, Perez-Mendez L, Berce V, Tavendale R, Samedy LA, Hunstman S, Hu D, Meade K, Farber HJ, Avila PC, Serebrisky D, Thyne SM, Brigino-Buenaventura E, Rodriguez-Cintron W, Sen S, Kumar R, Lenoir M, Rodriguez-Santana JR, Celedon JC, Mukhopadhyay S, Potocnik U, Pirmohamed M, Verhamme KM, Kabesch M, Palmer CAN, Hawcutt DB, Flores C, Maitland-van der Zee AH, Burchard EG, Pino-Yanes M. Genome-wide association study of inhaled corticosteroid response in admixed children with asthma. Clin Exp Allergy 2019;49:789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddel HK, Taylor DR, Bateman ED, Boulet LP, Boushey HA, Busse WW, Casale TB, Chanez P, Enright PL, Gibson PG, de Jongste JC, Kerstjens HA, Lazarus SC, Levy ML, O’Byrne PM, Partridge MR, Pavord ID, Sears MR, Sterk PJ, Stoloff SW, Sullivan SD, Szefler SJ, Thomas MD, Wenzel SE. An official American Thoracic Society/European Respiratory Society statement: asthma control and exacerbations: standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med. 2009;180:59–99. [DOI] [PubMed] [Google Scholar]
- 17.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki AE, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PI, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang HM. EPACTS (Efficient and Parallelizable Association Container Toolbox). http://genome.sph.umich.edu/wiki/EPACTS (2016).
- 20.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet 2011;88:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, Frayling TM, McCarthy MI, Hirschhorn JN, Goddard ME, Visscher PM. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369–375, S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet 2010;11:499–511. [DOI] [PubMed] [Google Scholar]
- 24.British Thoracic Society and the Scottish Intercollegiate Guidelines Network. British guideline on the management of asthma. Thorax 2014;69 Suppl 1:1–192. [PubMed] [Google Scholar]
- 25.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 2016;44:D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, Young R, Butterworth AS. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 2016;32:3207–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, Butterworth AS, Staley JR. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019;35:4851–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 2004;32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barreto-Luis A, Corrales A, Acosta-Herrera M, Gonzalez-Colino C, Cumplido J, MartinezTadeo J, Carracedo A, Villar J, Carrillo T, Pino-Yanes M, Flores C. A pathway-based association study reveals variants from Wnt signalling genes contributing to asthma susceptibility. Clin Exp Allergy 2017;47:618–626. [DOI] [PubMed] [Google Scholar]
- 31.Srikanth K, Lee SH, Chung KY, Park JE, Jang GW, Park MR, Kim NY, Kim TH, Chai HH, Park WC, Lim D. A Gene-Set Enrichment and Protein-Protein Interaction Network-Based GWAS with Regulatory SNPs Identifies Candidate Genes and Pathways Associated with Carcass Traits in Hanwoo Cattle. Genes (Basel) 2020;11:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A.Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Genomic atlas of the human plasma proteome. Nature 2018;558:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics 2016;54:1301–1313. [DOI] [PubMed] [Google Scholar]
- 36.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, van Iterson M, van Dijk F, van Galen M, Bot J, Slieker RC, Jhamai PM, Verbiest M, Suchiman HE, Verkerk M, van der Breggen R, van Rooij J, Lakenberg N, Arindrarto W, Kielbasa SM, Jonkers I, van ‘t Hof P, Nooren I, Beekman M, Deelen J, van Heemst D, Zhernakova A, Tigchelaar EF, Swertz MA, Hofman A, Uitterlinden AG, Pool R, van Dongen J, Hottenga JJ, Stehouwer CD, van der Kallen CJ, Schalkwijk CG, van den Berg LH, van Zwet EW, Mei H, Li Y, Lemire M, Hudson TJ, Slagboom PE, Wijmenga C, Veldink JH, van Greevenbroek MM, van Duijn CM, Boomsma DI, Isaacs A, Jansen R, van Meurs JB, t Hoen PA, Franke L, Heijmans BT. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet 2017;49:131–138. [DOI] [PubMed] [Google Scholar]
- 37.Carvalho-Silva D, Pierleoni A, Pignatelli M, Ong C, Fumis L, Karamanis N, Carmona M, Faulconbridge A, Hercules A, McAuley E, Miranda A, Peat G, Spitzer M, Barrett J, Hulcoop DG, Papa E, Koscielny G, Dunham I et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res. 2019;47:D1056–D1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies A, Hendrich J, Van Minh AT, Wratten J, Douglas L, Dolphin AC. Functional biology of the alpha(2)delta subunits of voltage-gated calcium channels. Trends Pharmacol Sci 2007;28:220–228. [DOI] [PubMed] [Google Scholar]
- 39.P Parkash J, Asotra K. Calcium wave signaling in cancer cells. Life Sci 2010;87:587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grossman NL, Ortega VE, King TS, Bleecker ER, Ampleford EA, Bacharier LB, Cabana MD, Cardet JC, Carr TF, Castro M, Denlinger LC, Denson JL, Fandino N, Fitzpatrick AM, Hawkins GA, Holguin F, Krishnan JA, Lazarus SC, Nyenhuis SM, Phipatanakul W, Ramratnam SK, Wenzel S, Peters SP, Meyers DA, Wechsler ME, Israel E. Exacerbation-prone asthma in the context of race and ancestry in Asthma Clinical Research Network trials. J Allergy Clin Immunol 2019;144:1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grossman NL, Doros GD, Fandino N, Fuhlbrigge AL, Pace WD, Wechsler ME, Yawn BP, Israel E. Susceptibility to exacerbations in Black adults with asthma. J Asthma 2019;56:704–710. [DOI] [PubMed] [Google Scholar]
- 42.Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, Schoech A, Pasaniuc B, Price AL. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am J Hum Genet 2019;104:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shrine N, Guyatt AL, Erzurumluoglu AM, Jackson VE, Hobbs BD, Melbourne CA, Batini C, Fawcett KA, Song K, Sakornsakolpat P, Li X, Boxall R, Reeve NF, Obeidat M, Zhao JH, Wielscher M, Weiss S, Kentistou KA, Cook JP, Sun BB, Zhou J, Hui J, Karrasch S, Imboden M, Harris SE, Marten J, Enroth S, Kerr SM, Surakka I, Vitart V, Lehtimaki T, Allen RJ, Bakke PS, Beaty TH, Bleecker ER, Bosse Y, Brandsma CA, Chen Z, Crapo JD, Danesh J, DeMeo DL, Dudbridge F, Ewert R, Gieger C, Gulsvik A, Hansell AL, Hao K, Hoffman JD, Hokanson JE, Homuth G, Joshi PK, Joubert P, Langenberg C, Li L, Lin K, Lind L, Locantore N, Luan J, Mahajan A, Maranville JC, Murray A, Nickle DC, Packer R, Parker MM, Paynton ML, Porteous DJ, Prokopenko D, Qiao D, Rawal R, Runz H, Sayers I, Sin DD, Smith BH, Soler Artigas M, Sparrow D, Tal-Singer R, Timmers Prhj, Van den Berge M, Whittaker JC, Woodruff PG, Yerges-Armstrong LM, Troyanskaya OG, Raitakari OT, Kahonen M, Polasek O, Gyllensten U, Rudan I, Deary IJ, Probst-Hensch NM, Schulz H, James AL, Wilson JF, Stubbe B, Zeggini E, Jarvelin MR, Wareham N, Silverman EK, Hayward C, Morris AP, Butterworth AS, Scott RA, Walters RG, Meyers DA, Cho MH, Strachan DP, Hall IP, Tobin MD, Wain LV. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet 2019;51:481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lutz SM, Cho MH, Young K, Hersh CP, Castaldi PJ, McDonald ML, Regan E, Mattheisen M, DeMeo DL, Parker M, Foreman M, Make BJ, Jensen RL, Casaburi R, Lomas DA, Bhatt SP, Bakke P, Gulsvik A, Crapo JD, Beaty TH, Laird NM, Lange C, Hokanson JE, Silverman EK. A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genet 2015;16:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Valk RJ, Duijts L, Timpson NJ, Salam MT, Standl M, Curtin JA, Genuneit J, Kerhof M, Kreiner-Moller E, Caceres A, Gref A, Liang LL, Taal HR, Bouzigon E, Demenais F, Nadif R, Ober C, Thompson EE, Estrada K, Hofman A, Uitterlinden AG, van Duijn C, Rivadeneira F, Li X, Eckel SP, Berhane K, Gauderman WJ, Granell R, Evans DM, St Pourcain B, McArdle W, Kemp JP, Smith GD, Tiesler CM, Flexeder C, Simpson A, Murray CS, Fuchs O, Postma DS, Bonnelykke K, Torrent M, Andersson M, Sleiman P, Hakonarson H, Cookson WO, Moffatt MF, Paternoster L, Melen E, Sunyer J, Bisgaard H, Koppelman GH, Ege M, Custovic A, Heinrich J, Gilliland FD, Henderson AJ, Jaddoe VW, de Jongste JC. Fraction of exhaled nitric oxide values in childhood are associated with 17q11.2-q12 and 17q12-q21 variants. J Allergy Clin Immunol 2014;134:46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan A, Hardjojo A, Yu S, Price D. Asthma Across Age: Insights From Primary Care. Front Pediatr 2019;7:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pijnenburg MW. The Role of FeNO in Predicting Asthma. Front Pediatr 2019;7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petsky HL, Cates CJ, Kew KM, Chang AB. Tailoring asthma treatment on eosinophilic markers (exhaled nitric oxide or sputum eosinophils): a systematic review and meta-analysis. Thorax 2018;73: 1110–1119. [DOI] [PubMed] [Google Scholar]
- 49.van Amerongen R, Mikels A, Nusse R. Alternative wnt signaling is initiated by distinct receptors. Sci Signal 2008;1:re9. [DOI] [PubMed] [Google Scholar]
- 50.Baarsma HA, Konigshoff M. ‘WNT-er is coming’: WNT signalling in chronic lung diseases. Thorax 2017;72:746–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma S, Tantisira K, Carey V, Murphy AJ, Lasky-Su J, Celedon JC, Lazarus R, Klanderman B, Rogers A, Soto-Quiros M, Avila L, Mariani T, Gaedigk R, Leeder S, Torday J, Warburton D, Raby B, Weiss ST. A role for Wnt signaling genes in the pathogenesis of impaired lung function in asthma. Am J Respir Crit Care Med 2010;181:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reuter S, Beckert H, Taube C. Take the Wnt out of the inflammatory sails: modulatory effects of Wnt in airway diseases. Lab Invest 2016;96:177–185. [DOI] [PubMed] [Google Scholar]
- 53.Choy DF, Modrek B, Abbas AR, Kummerfeld S, Clark HF, Wu LC, Fedorowicz G, Modrusan Z, Fahy JV, Woodruff PG, Arron JR. Gene expression patterns of Th2 inflammation and intercellular communication in asthmatic airways. J Immunol 2011;186:1861–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Januskevicius A, Vaitkiene S, Gosens R, Janulaityte I, Hoppenot D, Sakalauskas R, Malakauskas K. Eosinophils enhance WNT-5a and TGF-beta1 genes expression in airway smooth muscle cells and promote their proliferation by increased extracellular matrix proteins production in asthma. BMC Pulm Med 2016;16:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bousquet J, Chanez P, Lacoste JY, Barneon G, Ghavanian N, Enander I, Venge P, Ahlstedt S, Simony-Lafontaine J, Godard P, Michel FB. Eosinophilic inflammation in asthma. N Engl J Med 1990;323:1033–1039. [DOI] [PubMed] [Google Scholar]
- 56.Kay AB, Phipps S, Robinson DS. A role for eosinophils in airway remodelling in asthma. Trends Immunol 2004;25:477–482. [DOI] [PubMed] [Google Scholar]
- 57.Skronska-Wasek W, Gosens R, Konigshoff M, Baarsma HA. WNT receptor signalling in lung physiology and pathology. Pharmacol Ther 2018;187:150–166. [DOI] [PubMed] [Google Scholar]
- 58.El-Sahli S, Xie Y, Wang L, Liu S. Wnt Signaling in Cancer Metabolism and Immunity. Cancers (Basel) 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Z, Lee PH, Chaffin MD, Chung W, Loh PR, Lu Q, Christiani DC, Liang L. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet 2018;50:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun R, Hui S, Bader GD, Lin X, Kraft P. Powerful gene set analysis in GWAS with the Generalized Berk-Jones statistic. PLoS Genet 2019;15:e1007530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mogal A, Abdulkadir SA. Effects of Histone Deacetylase Inhibitor (HDACi); Trichostatin-A (TSA) on the expression of housekeeping genes. Mol Cell Probes 2006;20:81–86. [DOI] [PubMed] [Google Scholar]
- 62.Ma J, Li N, Zhao J, Lu J, Ma Y, Zhu Q, Dong Z, Liu K, Ming L. Histone deacetylase inhibitor trichostatin A enhances the antitumor effect of the oncolytic adenovirus H101 on esophageal squamous cell carcinoma in vitro and in vivo. Oncol Lett 2017;13:4868–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Royce SG, Karagiannis TC. Histone deacetylases and their role in asthma. J Asthma 2012;49:121–128. [DOI] [PubMed] [Google Scholar]
- 64.Banerjee A, Trivedi CM, Damera G, Jiang M, Jester W, Hoshi T, Epstein JA, Panettieri RA. Trichostatin A abrogates airway constriction, but not inflammation, in murine and human asthma models. Am J Respir Cell Mol Biol 2012;46:132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toki S, Goleniewska K, Reiss S, Zhou W, Newcomb DC, Bloodworth MH, Stier MT, Boyd KL, Polosukhin VV, Subramaniam S, Peebles RS. The histone deacetylase inhibitor trichostatin A suppresses murine innate allergic inflammation by blocking group 2 innate lymphoid cell (ILC2) activation. Thorax 2016;71:633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adcock IM, Tsaprouni L, Bhavsar P, Ito K. Epigenetic regulation of airway inflammation. Curr Opin Immunol. 2007;19:694–700. [DOI] [PubMed] [Google Scholar]
- 67.Choi JH, Oh SW, Kang MS, Kwon HJ, Oh GT, Kim DY. Trichostatin A attenuates airway inflammation in mouse asthma model. Clin Exp Allergy 2005;35:89–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


