Abstract
Introduction
KRAS mutations drive tumorigenesis by altering cell signaling and the tumor immune microenvironment. Recent studies have shown promise for KRAS-G12C covalent inhibitors, which are advancing rapidly through clinical trials. The sequencing and combination of these agents with other therapies including immune checkpoint blockade (ICB) will benefit from strategies that also address the immune microenvironment to improve durability of response.
Areas covered
This paper reviews KRAS signaling and discusses downstream effects on cytokine production and the tumor immune microenvironment. RAS targeted therapy is introduced and perspectives on therapeutic targeting of KRAS-G12C and its immunosuppressive tumor microenvironment are offered.
Expert opinion
The availability of KRAS-G12C covalent inhibitors raises hopes for targeting this pervasive oncogene and designing better therapeutic combinations to promote anti-tumor immunity. A comprehensive mechanistic understanding of KRAS immunosuppression is required in order to prioritize agents for clinical trials.
Keywords: IL-1β, KRAS, KRAS-G12C inhibitor, STING, tumorigenesis, oncogene, cancer, targeted therapies
1. Introduction
1.1. RAS oncogenes
Following its original discovery from a murine sarcoma virus, decades of study of have firmly established the human KRAS oncogene as a key driver of tumorigenesis in non-small cell lung cancer (NSCLC), pancreatic ductal adenocarcinoma (PDAC), colorectal cancer (CRC), and multiple other tumor types (1-3). KRAS has intrinsic GTPase activity and is active when bound to GTP and inactive when bound to GDP, a process that is catalyzed by RAS GTPase activating proteins (GAPs). The three commonly observed missense mutations in KRAS: G12, G13, and Q61, impair GAP binding (4), forcing it to rely on its slow intrinsic GTP hydrolysis rate and thus favoring constant downstream signaling (5, 6).
Pharmacologic targeting of KRAS has been hindered by structural characteristics of the KRAS protein along with its high affinity for GTP (7). To overcome the difficulties associated with direct targeting of KRAS, indirect approaches have been devised to target downstream signaling pathways, post-translational modifications, and associated chaperone proteins (5). However, as discussed in greater detail below, these approaches have been laden with difficulty due to achieving pharmacologically effective doses of inhibitors or activation of compensatory signaling pathways.
In this review we focus on recent molecular insights that have enabled covalent targeting of the KRAS-G12C isoform, as well as the increasing recognition that understanding how oncogenic KRAS and its co-mutations shape the immune microenvironment will likely be critical to achieving durable therapeutic responses. While KRAS-G12C inhibitors have entered phase I trials as monotherapy, preclinical efficacy is enhanced in combination with additional pathway inhibitors or with ICB (8, 9). KRAS signaling and associated co-mutations also influence patterns of immune cell infiltration downstream of cytokines such as IL-6, and can promote T cell exclusion (10, 11). Thus, understanding the interplay between these different factors likely holds the key to precision KRAS-directed therapy that ultimately achieves durable response.
1.2. Oncogenic KRAS activation
The RAS gene family includes some of the most common oncogenes including KRAS, HRAS, and NRAS. KRAS is mutated in 20% of all cancer types, 90% of pancreatic cancers, 45% of colorectal cancers, and 25% of NSCLC (5, 12). While NRAS and HRAS mutations are less common than KRAS mutations, NRAS mutations have been found in 29% of melanomas and HRAS mutations in 5% and 6% of head and neck squamous and bladder cancers, respectively (12).
RAS proteins mediate extracellular signals from receptor tyrosine kinases (RTKs). The main downstream effectors of RAS include the mitogen-activated protein kinase (MAPK), the phosphatidylinositol 3-kinase (PI3K)/AKT/mTOR, and RAL signaling pathways. The RAS protein is a single-subunit small GTPase that switches between the GDP-bound state and the GTP-bound state. These two states are regulated by guanine nucleotide exchange factors (GEFs), including son-of-sevenless homologue 1 (SOS1), GRB2, SHP2, in addition to the aforementioned GAPs. GEFs catalyze the exchange of GDP for GTP, and GAPs promote the hydrolysis of GTP to GDP. The GTP-bound form of RAS activates RAF-MEK-ERK downstream signaling, promoting tumor cell survival and proliferation. The recognition that hotspot mutations in KRAS at G12, G13, and Q61 do not lock KRAS in the GTP-bound active state, but rather impair GAP mediated catalysis, forms the basis for the successful inhibition of KRAS-G12C by covalent inhibitors (see below). The frequency of hotspot mutations varies by tumor and isoform. G12 and G13 account for 83% and 14% of KRAS mutations, respectively, while Q61 accounts for 63% of NRAS mutations (5). Given its association with smoking, KRAS-G12C is the most common KRAS mutation in lung cancer, occurring in 40-50% of KRAS-mutant NSCLC (5, 13). On the other hand, KRAS-G12D is the most common mutation in pancreatic ductal adenocarcinoma and colorectal adenocarcinoma (5). While all of these mutations sustain GTP-bound activation, some mutations (excluding KRAS-G12C) also differentially affect intrinsic hydrolysis and exchange between GDP and GTP (6, 7).
1.3. Impact of pro-tumorigenic cytokines
KRAS-mutant NSCLC is characterized by evasion of antitumor immunity alongside inflammation that fuels tumor growth and oncogenic mutations, resulting in part from enhanced production of suppressive inflammatory cytokines (Figure 1) (14, 15). In addition to the major KRAS downstream pathways MAPK and PI3K/AKT/mTOR, RAL signaling and IL-1β can activate TBK1 to promote NF-κB and IL-6 mediated autocrine STAT3 pathway activation (16, 17). KRAS signaling and IL-1β pathway activation increases IL-6 secretion and further reinforce a positive feedback autocrine cytokine circuit through STAT3 mediated induction of the TBK1 homologue IKKε (16, 18). These effects are further magnified in the context of LKB1 inactivation as well as therapeutic MEK inhibition (10, 19). In KRAS-LKB1 genetically engineered mouse models, IL-6 neutralizing antibody or JAK/TBK1 inhibitor treatment inhibited tumor growth, though escape occurred due to rapid cellular transcriptional adaptation (10, 19).
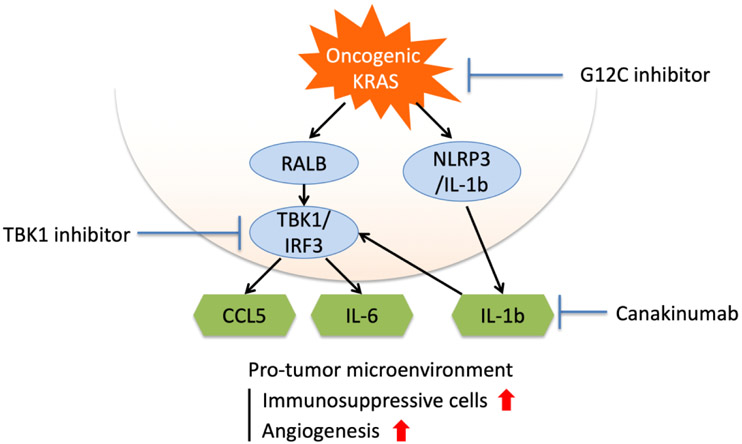
Figure 1.
Enhancing pro-inflammatory signaling in KRAS-driven lung cancer. Immunosuppressive signaling pathways downstream of mutant KRAS and opportunities for pharmacologic inhibition to suppress cytokine signaling.
Constitutive generation of IL-1β by activation of inflammasomes in the lung can promote chronic inflammation and tumorigenesis. Indeed, depletion of GATA2, a regulator of IL-1β, has been reported to inhibit tumor growth in a mouse model of KRAS-mutant NSCLC (20). The inhibition of the NLRP3 pathway, a mediator of the inflammasome and IL-1β release, has also been reported to inhibit cell proliferation and migration in KRAS-mutant lung cancer cell lines (18). Canakinumab is a humanized anti-IL-1β monoclonal antibody which antagonizes its activity. In a serendipitous observation, canakinumab treatment strongly reduced the incidence of lung cancer in patients treated for atherosclerosis on the CANTOS trial (21). KRAS mutations were not specifically studied in the CANTOS trial, however, the frequency of a smoking history in a population with atherosclerosis was high and it is presumed that these patients have a high frequency of KRAS mutations. These protective effects were dose-dependent, strongly supporting that IL-1β is involved in lung cancer carcinogenesis in humans and highlighting the therapeutic potential of its blockade.
KRAS mutations can also enhance pro-tumorigenic immune interactions via metabolic reprogramming, as elegantly shown in models of the pancreatic cancer stroma (22). Blocking the upregulation of cytokine receptors on tumor cells could suppress interactions with invading Th2 cells in the microenvironment, offering additional therapeutic targets to enhance antitumor immunity in KRAS-driven cancers.
2. RAS Targeted Therapy
2.1. Drugging the undruggable
While therapies targeting KRAS have been tested since the early 2000s, there has yet to be an approved agent. However, the identification of a covalent binding pocket in the KRAS-G12C isoform (7) led to a paradigm shift and emergence of the first direct KRAS inhibitors with promising clinical data and predicted integration into clinical practice. Review of previous strategies for indirect KRAS inhibition, along with recent molecular breakthroughs enabling direct targeting, will inform development of additional KRAS inhibitory strategies and combination with other targeted and immune treatments.
2.2. Farnesyltransferase inhibitors
While directly targeting RAS initially proved difficult, inhibiting post-translational modification of RAS showed early promise. RAS proteins undergo post-translational modification by prenylation, allowing them to translocate to the cell membrane for activation. The number of isoprene units covalently bound to a free thiol of cysteine determines the type of prenylation: While HRAS, NRAS, and KRAS all undergo farnesylation (three isoprenes), NRAS and KRAS can also undergo geranylgeranylation (four isoprenes) (12). Therapies that inhibit these post-translational modifications lock RAS isoforms in an inactive confirmation.
Although farnesyltransferase inhibitors advanced to phase III trials, they failed to meet primary endpoints (23). Preclinical work sheds light on potential mechanisms of resistance, which may have been obscured by over-reliance on HRAS mutant models in the development of farnesyltransferase inhibitors. Farnesyltransferase is critical for RAS anchoring to the membrane in HRAS mutant tumors, but geranylgeranyl transferase type I governs the process in the absence of farnesyltransferase in KRAS and NRAS mutant tumors, identifying a potential mechanism of resistance to farnesyltransferase inhibitors (23, 24). These findings raise the possibility of future combination therapies to inhibit these post-translational modifications in KRAS and NRAS mutant tumors.
2.3. Targeting RAS-activators
The critical cycling from the inactive GDP-bound state to the active GTP-bound confirmation nominated the catalyst of this cycle, the GEF SOS1, as a therapeutic target. In addition to its role as a GEF, SOS1 binds to GTP-coupled KRAS to promote a positive feedback loop via engagement of activating downstream signals. The selective SOS1 inhibitor BI-3406 demonstrated efficacy in xenograft models of KRAS-mutant cancers and showed synergy with MEK inhibitors to prevent acquired resistance (25). A phase I clinical trial (NCT04111458) has opened to test the analog BI 1701963 as monotherapy, and in combination with trametinib in advanced KRAS-mutant solid tumors.
SHP2 is a phosphatase that mediates activation of KRAS by RTKs. While the precise function of SHP2 has not yet been determined, it appears to bind GRB2 and SOS to mediate downstream activation of RAS (26). Thus, SHP2 inhibition favors the GDP bound state of KRAS by preventing effective GTP loading. SHP2 inhibitors are also being tested in combination with MEK inhibitions to prevent acquired resistance (27). The combination of MEK inhibitors and SHP099 (SHP2 inhibitor) showed efficacy in xenograft and genetically engineered models of KRAS-mutant cancers such as pancreatic, lung, and ovarian cancers, as well as KRAS-expressed triple-negative breast cancer (27-29). Inhibition of KRAS codon 12 mutant cell proliferation has been observed with the selective SHP2 allosteric inhibitor RMC-4550 (26). Inhibition of codon 13 and 61 mutants, however, was not observed. These results suggest that each KRAS mutation conveys a different degree of intrinsic dependence on individual GEFs.
As expected from other studies of RTK vertical pathway inhibition, blocking KRAS effectors can synergize with direct KRAS targeting. In KRAS-G12C NSCLC and PDAC cell lines, adding a SHP2 inhibitor to a KRAS covalent inhibitor increased CD8-positive T cells in the TME while decreasing myeloid suppressor cells and enhancing the effect of PD-1 inhibitors (9, 29). These results suggest that SHP2 inhibition can also influence the tumor immune microenvironment.
2.4. Targeting RAS effectors: MAPK, PI3K/AKT/mTOR and TBK1/JAK
Efforts to inhibit KRAS activity have focused on the primary downstream signaling pathways for RAS family proteins: MAPK, PI3K/AKT/mTOR and TBK1/JAK. Initial approaches to target the MAPK signaling pathway were carried out using the BRAF V600E inhibitors vemurafenib and dabrafenib, developed to treat melanoma (30). However, BRAF-V600E inhibitors proved unsuccessful in KRAS-mutant tumors due to the preponderance of BRAF heterodimers (BRAF and CRAF). Additionally, in KRAS-mutant models, BRAF-V600E inhibitors resulted in paradoxical activation of ERK (31, 32). MEK inhibitors were able to overcome this positive feedback loop, suggesting the possibility of single-agent activity to target KRAS effector function. While a number of MEK inhibitors have entered clinical trials for KRAS-mutant tumors, none have demonstrated significant efficacy (33, 34). However, MEK inhibitors may prove more effective in combination with other agents. In combination with docetaxel, the selective allosteric MEK1 and MEK2 inhibitor selumetinib showed promise in a phase II trial of second-line treatment in KRAS-mutant NSCLC. However, the combination failed to show efficacy as compared to docetaxel monotherapy in a phase III trial (SELECT-1; NCT01933932) (35). Concurrent inhibition of multiple signaling pathways downstream of KRAS was also considered a promising therapeutic strategy. In a preclinical model, the combination of MEK and PI3K inhibitors was found to inhibit tumor growth in KRAS-mutant lung cancer (36). Concurrent inhibition of MAPK and TBK1 also showed promising results in preclinical model (19). However, combination treatment MEK inhibitors and PI3K inhibitors, as well as the combination of MEK inhibitors and TBK1 inhibitors, failed in clinical trials because of dose-limiting toxicities from MEK inhibitor effects on normal cells (37, 38) and inadequate dosing to inhibit TBK1, for example (39).
2.5. KRAS-G12C covalent inhibitors
The high affinity of KRAS for GTP prevents the steric hindrance approaches that have proven successful with TKIs. Agents that bind to multiple sites of RAS (40) and inhibit the dimerization of KRAS (41), were tried and failed. A breakthrough came by considering KRAS mutations separately, with identification of a previously unknown binding pocket in KRAS-G12C (7). Compounds that covalently bind this pocket trap the enzyme its inactive, GDP-bound state. Viewed more broadly, in contrast with kinase inhibitors that bind the active protein conformation, these drugs can inhibit KRAS activity by binding the nonfunctional state (42). As expected, binding of these agents to GDP-bound KRAS-G12C inhibits activation of downstream signaling such as RAF. Of note, these treatments bind only to GDP-bound KRAS-G12C and not to wild-type KRAS. In addition, although 75% of KRAS-G12C is GTP-bound, this covalent approach is effective due to the fact that KRAS maintains intrinsic GTPase activity (5, 43).
Since the initial observation by Ostrem et al., a number of KRAS-G12C inhibitors have undergone preclinical and clinical development. Targeting this mutation with the covalent binding agent ARS-1620 showed initial promise in animal models (43, 44). AMG 510 was the first agent with reported success in clinical trials for KRAS-mutant cancers. In preclinical studies, AMG 510 inhibited cell proliferation in KRAS-G12C-carrying cell lines and slowed xenograft tumor growth (45). This study additionally showed synergistic effects with chemotherapy (carboplatin), ICB (PD-1 inhibitor), and vertical pathway inhibition with MEK inhibitors. Phase I trials also showed promising results, in particular with NSCLC patients (46). Among the fifty-nine NSCLC patients receiving AMG 510, 32.2% had a confirmed objective response and 88.1% achieved disease control. Of the forty-two CRC patients, 7.1% had a confirmed objective response while 73.8% achieved disease control (46). The FDA granted a fast-track designation to AMG 510 for patients with previously treated metastatic NSCLC harboring KRAS-G12C mutations, with several clinical trials ongoing to assess long-term safety and efficacy, alone or in combination with chemotherapy, targeted or immune therapies, compared with chemotherapy alone (NCT03600883, NCT04303780, NCT04625647, NCT04185883).
MRTX849 is another covalent KRAS-G12C inhibitor currently undergoing phase I and II trials. Preclinical studies demonstrated that MRTX849 could suppress proliferation in cell lines and caused tumor regression in 65% of KRAS-G12C patient-derived xenograft models (47). MRTX849 treatment led to partial responses in 3/6 NSCLC patients and 1/4 CRC patients, with stable disease in the remaining patients with NSCLC and CRC (47-49). CRISPR screening was used to identify potential combination therapies, with RTKs and components of the mTOR pathway as hits (9). CRISPR/Cas9 screening also identified potential mechanisms of acquired resistance to MRTX849, including alterations in cell cycle genes and loss of KEAP1 or NRAS (9). MRTX849 is being studied in combination with ICB, TKIs, and the SHP2 inhibitor TNO155 in ongoing clinical trials compared with chemotherapy (NCT04613596, NCT04685135, NCT04330664). A similar covalent inhibitor JNJ-74699157 has completed recruitment, with results expected soon (NCT04006301)(5).
3. Immunotherapy in RAS-Mutant Cancers
3.1. Immune checkpoint blockade
ICB has transformed care for patients with non-small cell lung cancers, and colorectal cancers with microsatellite instability (50-52). Checkpoint inhibitors, which bind and inhibit the actions of PD-1, PD-L1, and CTLA-4 inhibitory molecules on tumor and immune cells, unleash cytotoxic T-cells to generate antitumor immunity.
3.2. ICB in RAS-mutant NSCLC
Amongst cancers harboring KRAS mutations, NSCLC is the most commonly and successfully treated with ICB. PD-1, PD-L1, and CTLA-4 antibodies are now approved for NSCLC treatment as single agents, in combination, and as a partner to chemotherapy. KEYNOTE-42 showed the efficacy of pembrolizumab monotherapy as first-line treatment in NSCLC with high PD-L1 compared to platinum-doublet chemotherapy, leading to the use of PD-L1 as a predictive biomarker (53). Subsequent trials demonstrated the efficacy of ICB in combination with chemotherapy in patients both with and without PD-L1 expression (54, 55). Nivolumab plus ipilimumab is also FDA-approved for patients with NSCLC whose tumors express PD-L1(≥1%) (56). In addition to PD-L1 expression, tumor mutation burden and tumor infiltrating lymphocytes (TILs) have been reported as predictive factors for ICB (56, 57), but these factors alone are insufficient as predictive biomarkers in current clinical practice.
While KRAS-mutant NSCLC responds better to ICB than NSCLC harboring EGFR mutations or ALK fusions, which respond poorly (58-60), the wide variation in response suggests other influences (50, 52). NSCLCs with KRAS mutations are more frequently PD-L1 positive and are associated with induced MEK-mediated downstream signaling (61). Co-mutations also influence NSCLC response to ICB (52). KRAS-mutant NSCLC can be divided into categories based on co-occurring mutations: TP53 (KP) and STK11/LKB-1 (KL). LKB-1 mutations carry a poor prognosis in NSCLC (62), and KP and KL tumors demonstrate distinct immune response gene signatures (11). KP tumors are more likely to exhibit an interferon-driven inflammatory response, which correlates with improved response to ICB (11). While KP tumors had a 35.7% response rate, KL tumors only exhibited a 7.4% response rate. Another study suggested that progression-free survival and overall survival were also significantly shorter in KL tumors, which demonstrated few PD-L1 positive tumor cells and CD8-positive T-cells (52). Although STK11/LKB1 loss is associated with low PD-L1, response to ICB in PD-L1 positive KL remained inferior to KP. As discussed above, KL tumors produce abundant IL-6, resulting in an immunosuppressive environment (10). TMB is higher in KP as compared to KRAS wild-type tumors (with or without TP53 mutations), which may also influence ICB response (63). These results indicate that other mechanisms in addition to PD-L1 may contribute to the poor response to ICB observed in KL tumors.
4. Combined KRAS G12C Inhibition and Innate Immune Targeting
4.1. Inhibiting the immune suppressive TME
While KRAS-G12C inhibitors have shown promising results, especially in NSCLC, targeted agents often fail to cure patients due to acquired resistance. While ICB combinations are underway, the fact that KRAS mutations create an immune TME favorable for tumor growth suggests that other combination therapies with agents capable of reshaping TME may be needed. Early trial data demonstrate that KRAS-G12C inhibitors lack severe side effects compared with MAPK or PI3K inhibition because of their mutation-specific mechanism. Combination of TBK inhibition or IL-1β inhibition may thus be an additional option with KRAS-G12C inhibitors (Figure 1). The low incidence of adverse effects with KRAS-G12C inhibitors may enable sufficient TBK inhibition to have a therapeutic effect. Inhibitors of IL-1β have the potential to break positive feed-forward loops in the TME, as discussed above. IL-1β inhibitors may prove especially effective in early lung cancer because they are likely to affect tumor initiation based on the results of the CANTOS trial. These combinations offer hope of enhancing the direction inhibition of KRAS by reversing the growth promoting TME that likely promotes KRAS-G12C inhibitor escape
4.2. cGAS-STING pathway activation
Cytokines such as IL-6 and IL-1β, which induce chronic inflammation, play an important role in tumorigenesis, while the accumulation of immune cells such as CD8-positive T cells is relevant to the treatment of advanced lung cancer. KL tumors have fewer Tumor Infiltrating Lymphocytes (TILs) compared to KP tumors (64). KRAS-G12C inhibitors have been reported to be effective in combination with ICB and MEK inhibitors (45), and are also expected to be effective in combination with therapies that increase TILs in KL tumors.
We reported that the expression of Stimulator of Interferon Genes (STING), which is important for innate immunity, is suppressed in KL tumors (64). Antitumor immunity proceeds in a stepwise fashion starting with innate immune recognition of cancer cells and subsequent activation of cytotoxic T-lymphocytes. The activation of STING results from cytoplasmic dsDNA recognition by the enzyme cGAS, leading to production of the cyclic dinucleotide second messenger 2’3’-cGAMP. During activation, TBK1 and IRF3 undergo cascade phosphorylation (65). Initiation of the STING-TBK1-IRF3 pathway promotes the secretion of type I interferons and cytokines including CXCL10, eliciting T-cell recruitment. Silencing of STING in KL tumors thus prevents IRF3 engagement and is responsible for enhancing pro-tumorigenic IL-6 production downstream of TBK1. Thus, restoring STING expression in KL cells rewires cytokine production towards an anti-tumorigenic interferon response (64).
Activating the STING-TBK1-IRF3 signaling in cancer cells by enhancing cytoplasmic DNA accumulation is being explored from a therapeutic perspective. Indeed, the accumulation of cytoplasmic DNA from radiotherapy and DNA-damaging agents activates the cGAS-STING pathway, leading to antitumor immunity (66, 67). Other strategies for activating the STING pathway include the use of PARP inhibitors against BRCA-mutant tumors to promote genomic instability and the accumulation of cytoplasmic DNA (68). Taxanes, such as paclitaxel, also activate the STING pathway (69). While promising results from preclinical studies show that direct injection of cyclic dinucleotide STING agonists can enhance immunogenicity and restrict tumor growth in mice, results from clinical trials are thus far disappointing, possibly due to delivery and pharmacodynamic issues (65).
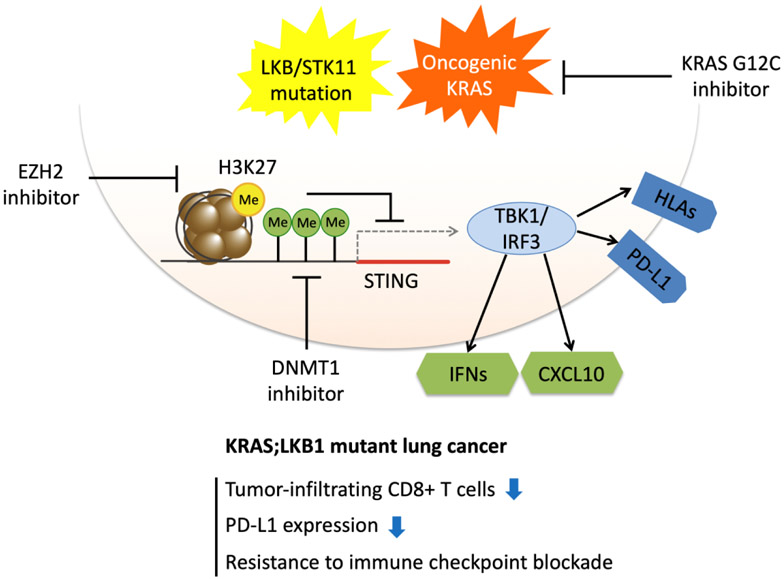
Most prior research has focused on activating STING in immune cells, and clinical trials of STING agonists have been conducted in combination with ICB. Yet effective therapeutic combinations may require restoration of tumor cell STING expression, as we have shown for KL tumors. Indeed, we demonstrated that STING expression is silenced in KL NSCLC by epigenetic mechanisms involving EZH2 and DNMT1 (64). Since KRAS-G12C inhibitors are effective in patients with KL tumors (47), they may act in part by increasing interferon-associated cytokine release to promote T cell infiltration (8), Thus priming STING re-expression by treatment with EZH2 and/or DNMT1 inhibitors may also enhance response to KRAS-G12C inhibitors and limit resistance to treatment by restoring immunogenicity, even in advanced stages of disease (Figure 2).
Figure 2.
Activating STING in KRAS/LKB1-mutant NSCLC to synergize with KRAS-G12C inhibition. STING re-expression by enhancer of zeste homolog 2 (EZH2) and/or DNA methyltransferase 1 (DNMT1) inhibitors in KRAS/LKB1-mutant NSCLC. KRAS-G12C inhibition prevents downstream signaling and may release immunogenic antigens while STING re-expression enhances immunogenicity through TBK1-IRF3 signaling. STING = stimulator of interferon genes; IFN = interferon; HLA = human leukocyte antigen; Me signifies histone methylation to suppress gene transcription.
4. Conclusions
For tumors with KRAS mutations, future treatments will focus on specific isoforms to build on the recent success of KRAS-G12C inhibitors. ICB monotherapy has proven relatively ineffective for certain tumors harboring KRAS mutations, and additional combinations may be necessary to reverse the immunosuppressive KRAS TME.
5. Expert opinion
Current treatments for KRAS-mutant tumors depend on the specific isoform and co-mutations. The early success of KRAS-G12C inhibitors, both in suppressing tumor growth and activating antitumor immunity in historically suppressed TMEs, gives hope for therapeutic breakthroughs to help large percentages of patients with NSCLC. The precise role of these new agents in the NSCLC armamentarium remains unclear. In order to gain first-line approval and maximize response, they will likely need to be combined with other targeted and immune therapies. Indeed, as discussed above, trials are underway combining covalent KRAS G12C inhibitors with inhibitors of PD-1, PD-L1, SHP2, MEK, EGFR, CDK, mTOR, and HER2. This panoply of targets, while promising and based on rigorous mechanistic studies, does risk restricting statistical power to identify the most promising combinations and may add years to the development timetable.
Previous trials for SHP2 and MEK inhibitors showed modest activity, suggesting possible benefit in combination with direct KRAS targeting. The known toxicity profile and experience in NSCLC nominates these agents for rapid clinical development in combination with covalent KRAS G12C inhibitors. Toxicity may prove to be a major obstacle, as MAPK signaling is critical for normal cellular function especially in the skin and GI tract, and downstream inhibitors are notoriously unpopular with patients. Furthermore, while dual vertical pathway inhibition has precedent in other oncogene driven lung cancers (EGFR, ALK, BRAF mutations), acquired resistance remains a fundamental barrier. In contrast, combinations to enhance antitumor immunity could lead to durable long-term responses and even cures by eliminating “persister” cells.
In our view, therapeutic combinations to enhance the observed inflammatory activation from KRAS G12C inhibition represents one of the most promising approaches. Combining KRAS G12C inhibitors with PD-1/PD-L1 checkpoint inhibitors is the most straightforward initial approach and may ease the integration into first line therapy since many patients with KRAS-mutant NSCLC would receive these agents as standard of care. Concerns remain regarding overlapping side effects, including the possibility of pneumonitis, though emerging clinical data for AMG 510 and MRTX849 will allow for a better assessment of this risk. Combinations targeting immune signaling components such as IL-1β or the cGAS-STING pathway, which regulate the suppressed immune response in KL tumors, show potential to enhance response rates from KRAS-G12C inhibitors and prevent acquired resistance. These approaches are earlier in development and would likely apply to select patient subsets. Treatments that generate cytosolic DNA, such as chemotherapy or PARP inhibitors, represent an alternate approach to activate innate immune signaling and may integrate nicely with existing practice for patients scheduled to receive (or already benefiting from) cytotoxic therapy. Epigenetic treatments to restore STING expression, such as EZH2 or DNMT inhibitors, should also be tested in future trials, though their activity and efficacy in NSCLC is currently unproven.
A better understanding of the direct immunosuppressive effects of KRAS, as well as the importance of molecular context in specific tumor types, will allow for combination therapies to reverse KRAS immunosuppression and activate antitumor immunity. Ongoing translational efforts epitomized by the development of AMG 510 and MRTX849 will allow for real-time re-evaluation of therapeutic combinations as they progress through trials. To that end, reliable functional assays that reflect patient clinical responses (i.e. organoids and other ex vivo culture systems), can maximize the knowledge gained from each patient enrolled on trial. As we enter a new era in lung cancer treatment, we hope that the previously parallel development of targeted and immune therapeutics can merge to overcome the weaknesses of each and extend survival for patients with these challenging cancers.
Article Highlights.
KRAS downstream pathways enhance production of suppressive inflammatory cytokines such as IL-6 and IL-1β and create an immune TME favorable for tumor growth.
KRAS mutation isoform and co-mutations are critical to designing targeted therapies, as evidenced by the recent clinical success of KRAS-G12C inhibitors.
KRAS-G12C inhibitors showed promising results in clinical trials for NSCLC and showed synergistic effects with other treatments such as chemotherapy, ICB, and MEK inhibitors in preclinical models.
KRAS-G12C inhibitors and targeting inflammatory cytokine immune signaling such as IL-1β have the potential to enhance response by altering the TME.
STING is suppressed in KL NSCLC, and restoring STING could induce T cell infiltration. Combining KRAS-G12C inhibitors and activators of the cGAS-STING pathway may have synergistic effects.
Acknowledgments
Funding
This work was supported by NIH R01CA190294 (DAB), the Parker Institute for Cancer Immunotherapy (DAB), Schaubert Family Funds (DAB), and Gross-Loh Research Fellowship (EHK), Uehara Memorial Foundation Research Fellowship (TT), and Lilly Oncology Fellowship Program (TT).
Footnotes
Declaration of interest
D.A.Barbie is a consultant for Qiagen/N-of-One and Tango Therapeutics, has received research support from Bristol Myers Squibb, Novartis, Lilly, Gilead Sciences, and is a founder and shareholder in Xsphera Biosciences. EH Knelson has a Sponsored Research Agreement with Takeda Pharmaceuticals. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers
- 1.Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research N. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32(2):185–203 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu S, Jang H, Nussinov R, Zhang J. The Structural Basis of Oncogenic Mutations G12, G13 and Q61 in Small GTPase K-Ras4B. Sci Rep. 2016;6:21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore AR, Rosenberg SC, McCormick F, Malek S. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov. 2020;19(8):533–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol Cancer Res. 2015;13(9):1325–35. [DOI] [PubMed] [Google Scholar]
- 7.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–51.**Identification of an allosteric binding pocket in GDP-bound KRAS G12C, leading to development of the first successful KRAS targeted therapies
- 8.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019.**Early demonstration of clinical activity for KRAS G12C covalent inhibitors and identification of immune effects
- 9.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRASG12C Inhibitor, MRTX849, Provides Insight Toward Therapeutic Susceptibility of KRAS Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, et al. STK11/LKB1 Deficiency Promotes Neutrophil Recruitment and Proinflammatory Cytokine Production to Suppress T-cell Activity in the Lung Tumor Microenvironment. Cancer Res. 2016;76(5):999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5(8):860–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nature Reviews Clinical Oncology. 2018;15(11):709–20. [DOI] [PubMed] [Google Scholar]
- 13.Yu HA, Sima CS, Shen R, Kass S, Gainor J, Shaw A, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol. 2015;10(3):431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ancrile B, Lim KH, Counter CM. Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes Dev. 2007;21(14):1714–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462(7269):104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Z, Aref AR, Cohoon TJ, Barbie TU, Imamura Y, Yang S, et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov. 2014;4(4):452–65.*Demonstration that KRAS signaling promotes a positive feedback autocrine cytokine circuit through TBK1
- 17.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbie TU, Alexe G, Aref AR, Li S, Zhu Z, Zhang X, et al. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J Clin Invest. 2014;124(12):5411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitajima S, Asahina H, Chen T, Guo S, Quiceno LG, Cavanaugh JD, et al. Overcoming Resistance to Dual Innate Immune and MEK Inhibition Downstream of KRAS. Cancer Cell. 2018;34(3):439–52 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149(3):642–55. [DOI] [PubMed] [Google Scholar]
- 21.Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ, et al. Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. The Lancet. 2017;390(10105):1833–42.*First clinical trial demonstrating benefit of IL-1β inhibition in tumorigenesis
- 22.Dey P, Li J, Zhang J, Chaurasiya S, Strom A, Wang H, et al. Oncogenic KRAS-Driven Metabolic Reprogramming in Pancreatic Cancer Cells Utilizes Cytokines from the Tumor Microenvironment. Cancer Discov. 2020;10(4):608–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11(11):775–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basso AD, Kirschmeier P, Bishop WR. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J Lipid Res. 2006;47(1):15–31. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann MH, Gmachl M, Ramharter J, Savarese F, Gerlach D, Marszalek JR, et al. BI-3406, a Potent and Selective SOS1–KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition. Cancer Discovery. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol. 2018;20(9):1064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fedele C, Ran H, Diskin B, Wei W, Jen J, Geer MJ, et al. SHP2 Inhibition Prevents Adaptive Resistance to MEK Inhibitors in Multiple Cancer Models. Cancer Discovery. 2018;8(10):1237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong GS, Zhou J, Liu JB, Wu Z, Xu X, Li T, et al. Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat Med. 2018;24(7):968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fedele C, Li S, Teng KW, Foster CJR, Peng D, Ran H, et al. SHP2 inhibition diminishes KRASG12C cycling and promotes tumor microenvironment remodeling. J Exp Med. 2021;218(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371(20):1877–88. [DOI] [PubMed] [Google Scholar]
- 31.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–5. [DOI] [PubMed] [Google Scholar]
- 32.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blumenschein GR Jr., Smit EF, Planchard D, Kim DW, Cadranel J, De Pas T, et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)dagger. Ann Oncol. 2015;26(5):894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmer L, Barlesi F, Martinez-Garcia M, Dieras V, Schellens JH, Spano JP, et al. Phase I expansion and pharmacodynamic study of the oral MEK inhibitor RO4987655 (CH4987655) in selected patients with advanced cancer with RAS-RAF mutations. Clin Cancer Res. 2014;20(16):4251–61. [DOI] [PubMed] [Google Scholar]
- 35.Janne PA, van den Heuvel MM, Barlesi F, Cobo M, Mazieres J, Crino L, et al. Selumetinib Plus Docetaxel Compared With Docetaxel Alone and Progression-Free Survival in Patients With KRAS-Mutant Advanced Non-Small Cell Lung Cancer: The SELECT-1 Randomized Clinical Trial. JAMA. 2017;317(18):1844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bedard PL, Tabernero J, Janku F, Wainberg ZA, Paz-Ares L, Vansteenkiste J, et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin Cancer Res. 2015;21(4):730–8. [DOI] [PubMed] [Google Scholar]
- 38.Tolcher AW, Khan K, Ong M, Banerji U, Papadimitrakopoulou V, Gandara DR, et al. Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clin Cancer Res. 2015;21(4):739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbie DA, Spira A, Kelly K, Humeniuk R, Kawashima J, Kong S, et al. Phase 1B Study of Momelotinib Combined With Trametinib in Metastatic, Kirsten Rat Sarcoma Viral Oncogene Homolog-Mutated Non-Small-Cell Lung Cancer After Platinum-Based Chemotherapy Treatment Failure. Clin Lung Cancer. 2018;19(6):e853–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welsch ME, Kaplan A, Chambers JM, Stokes ME, Bos PH, Zask A, et al. Multivalent Small-Molecule Pan-RAS Inhibitors. Cell. 2017;168(5):878–89.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, et al. Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol. 2017;13(1):62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nussinov R, Jang H, Gursoy A, Keskin O, Gaponenko V. Inhibition of Nonfunctional Ras. Cell Chem Biol. 2021;28(2):121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell. 2018;172(3):578–89 e17. [DOI] [PubMed] [Google Scholar]
- 44.Molina-Arcas M, Moore C, Rana S, van Maldegem F, Mugarza E, Romero-Clavijo P, et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci Transl Med. 2019;11(510). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575(7781):217–23. [DOI] [PubMed] [Google Scholar]
- 46.Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med. 2020;383(13):1207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020;10(1):54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jänne PA, Rybkin II, Spira AI, Riely GJ, Papadopoulos KP, Sabari JK, et al. KRYSTAL-1: Activity and Safety of Adagrasib (MRTX849) in Advanced/ Metastatic Non–Small-Cell Lung Cancer (NSCLC) Harboring KRAS G12C Mutation. European Journal of Cancer. 2020;138:S1–S2. [Google Scholar]
- 49.Johnson ML, Ou SHI, Barve M, Rybkin II, Papadopoulos KP, Leal TA, et al. KRYSTAL-1: Activity and Safety of Adagrasib (MRTX849) in Patients with Colorectal Cancer (CRC) and Other Solid Tumors Harboring a KRAS G12C Mutation. European Journal of Cancer. 2020;138:S2. [Google Scholar]
- 50.Mazieres J, Drilon A, Lusque A, Mhanna L, Cortot AB, Mezquita L, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol. 2019;30(8):1321–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. New England Journal of Medicine. 2015;372(26):2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discovery. 2018;8(7):822–35.*Demonstration that LKB1 mutations in NSCLC predict poor outcome with immune checkpoint blockade.
- 53.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823–33. [DOI] [PubMed] [Google Scholar]
- 54.Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378(22):2078–92. [DOI] [PubMed] [Google Scholar]
- 55.Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N Engl J Med. 2018;378(24):2288–301. [DOI] [PubMed] [Google Scholar]
- 56.Hellmann MD, Paz-Ares L, Bernabe Caro R, Zurawski B, Kim SW, Carcereny Costa E, et al. Nivolumab plus Ipilimumab in Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2019;381(21):2020–31. [DOI] [PubMed] [Google Scholar]
- 57.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. The Lancet Oncology. 2016;17(12):e542–e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi K, Nakachi I, Naoki K, Satomi R, Nakamura M, Inoue T, et al. Real-world Efficacy and Safety of Nivolumab for Advanced Non-Small-cell Lung Cancer: A Retrospective Multicenter Analysis. Clin Lung Cancer. 2018;19(3):e349–e58. [DOI] [PubMed] [Google Scholar]
- 59.Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z, et al. EGFR Mutations and ALK Rearrangements Are Associated with Low Response Rates to PD-1 Pathway Blockade in Non-Small Cell Lung Cancer: A Retrospective Analysis. Clin Cancer Res. 2016;22(18):4585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herbst RS, Baas P, Kim D-W, Felip E, Pérez-Gracia JL, Han J-Y, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. The Lancet. 2016;387(10027):1540–50. [DOI] [PubMed] [Google Scholar]
- 61.Coelho MA, de Carne Trecesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, et al. Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity. 2017;47(6):1083–99 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papillon-Cavanagh S, Doshi P, Dobrin R, Szustakowski J, Walsh AM. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open. 2020;5(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao G, Liao W, Ma Q, Zhang B, Chen Y, Wang Y. KRAS G12D mutation predicts lower TMB and drives immune suppression in lung adenocarcinoma. Lung Cancer. 2020;149:41–5. [DOI] [PubMed] [Google Scholar]
- 64.Kitajima S, Ivanova E, Guo S, Yoshida R, Campisi M, Sundararaman SK, et al. Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer. Cancer Discov. 2019;9(1):34–45.**LKB1 loss suppresses STING expression by epigenetic silencing in KRAS mutant NSCLC.
- 65.Kwon J, Bakhoum SF. The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 2020;10(1):26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng L, Liang H, Fu S, Weichselbaum RR, Fu YX. From DNA Damage to Nucleic Acid Sensing: A Strategy to Enhance Radiation Therapy. Clin Cancer Res. 2016;22(1):20–5. [DOI] [PubMed] [Google Scholar]
- 67.Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov. 2019;9(5):646–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding L, Kim HJ, Wang Q, Kearns M, Jiang T, Ohlson CE, et al. PARP Inhibition Elicits STING-Dependent Antitumor Immunity in Brca1-Deficient Ovarian Cancer. Cell Rep. 2018;25(11):2972–80 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lohard S, Bourgeois N, Maillet L, Gautier F, Fetiveau A, Lasla H, et al. STING-dependent paracriny shapes apoptotic priming of breast tumors in response to anti-mitotic treatment. Nat Commun. 2020;11(1):259. [DOI] [PMC free article] [PubMed] [Google Scholar]