Abstract
Macroautophagic clearance of cytosolic materials entails the initiation, growth, and closure of autophagosomes. Cargo triggers the assembly of a web of cargo receptors and core machinery. ATG9 vesicles seed the growing autophagosomal membrane, which is supplied by de novo phospholipid synthesis, phospholipid transport via ATG2 proteins, and lipid flipping by ATG9. Autophagosomes close via the ESCRT complexes. Here we review recent discoveries that illuminate the molecular mechanisms of autophagosome formation and discuss emerging questions in this rapidly developing field.
Introduction
Macroautophagy (henceforward, “autophagy”) is the means by which eukaryotic cells degrade and recycle large objects such as molecular aggregates, organelles, and intracellular pathogens 1. Autophagic dysfunction is central to many diseases, including viral and bacterial infections, cancer, and neurodegeneration diseases 2. Autophagy consists of the initiation and growth of a double membrane phagophore (a.k.a. “isolation membrane”) around the object to be degraded, followed by the sealing of the phagophore into the double membrane vesicle known as the autophagosome, and its fusion with the lysosome (Fig. 1). The parts list of proteins and protein complexes responsible for autophagy has been largely delineated 1, an accomplishment recognized by the Nobel Prize in Physiology or Medicine to Yoshinori Ohsumi. Yet how these complexes and proteins orchestrate membrane lipids through phagophore initiation, growth, and closure has been a mystery, of “origins unknown, biogenesis complex” 3. Within just the past two years, a great deal of light has now shone into the dark corners of this black box, further illuminating the nature of each of the main steps in phagophore formation (Fig. 1).
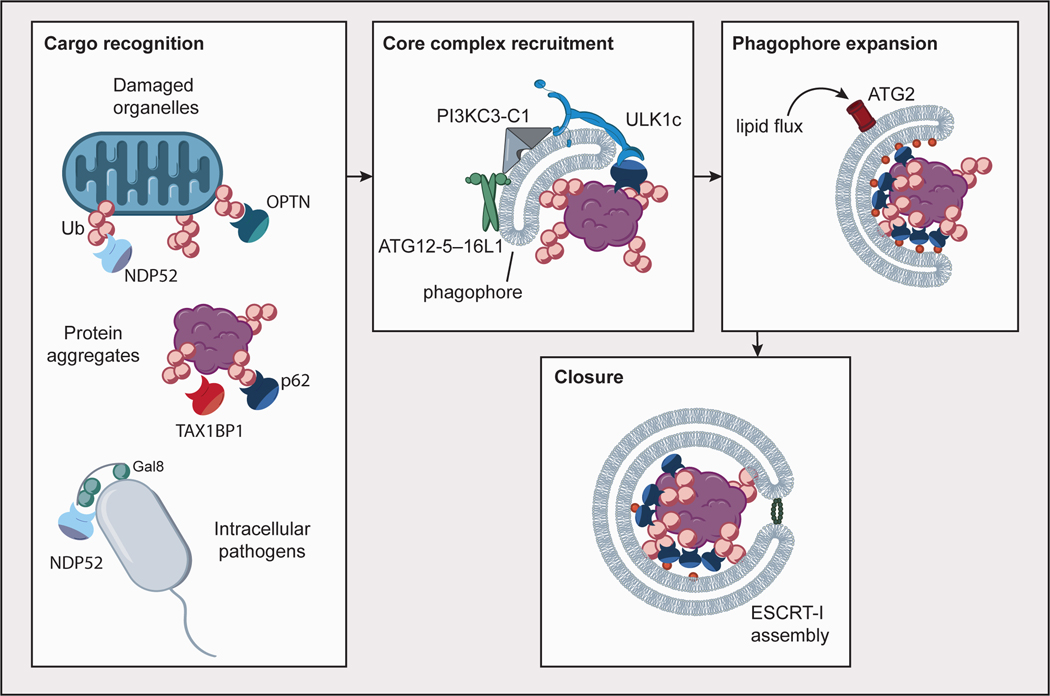
Fig. 1. Overview of autophagosome formation.
Autophagy cargoes such as damaged organelles, protein aggregates and intracellular pathogens are recognized by cargo receptors such as p62, NDP52, TAX1BP1, and OPTN, most commonly through modification with ubiquitin (Ub), as well as the presence of other signals such as Galectin-8. Core autophagy complexes (ULK1c, PI3KC3-C1, ATG12–ATG5–16L1) are recruited to the nascent phagophore through interactions with cargo receptors, the phagophore membrane, and one another in a positive feedback mechanism. Phagophore expansion is driven by lipid flux through ATG2. Autophagosome closure is mediated in part by assembly of ESCRT-I proteins in a ring at the aperture of the closing phagophore.
Bulk autophagy refers to the starvation-triggered non-specific engulfment and degradation of cytosolic material for the replenishment of biosynthetic precursors. Selective autophagy refers to the same process as targeted to particular substrates. While the fundamental machinery of autophagy was initially uncovered in yeast 4, this review will focus primarily on selective autophagy in mammalian cells. We will make reference to data from bulk autophagy in yeast in some cases, mainly where there are homologies between yeast and mammals that address knowledge gaps in the mammalian system. Other excellent reviews have covered some of the recent developments in autophagosome biogenesis 5, 6, yet progress continues at a rapid pace. This review covers the most recent developments from a mechanistic perspective, with an emphasis on discoveries emerging from reconstitution experiments.
The cascade of autophagy initiation in mammalian cells begins with the unc-51-like kinase 1 (ULK1) complex, consisting of its namesake, the ULK1 (or ULK2) protein kinase, the non-catalytic FIP200, ATG13, and ATG101 subunits (Box 1), whose counterpart in yeast is the Atg1 complex 7. The class III phosphatidylinositol 3-kinase complex I (PI3KC3-C1), consisting of VPS34, VPS15, BECN1, and ATG14, is activated early in autophagy initiation to generate phosphatidylinositol-3-phosphate (PI(3)P). PI(3)P enriched membranes recruit the downstream effector WIPIs (WD-repeat protein interacting with phosphoinositides), which in turn recruit and activate the ATG8/LC3 conjugation machinery 1, 8. The attachment of the ATG8 proteins of the LC3 and GABARAP subfamilies to the membrane lipid phosphatidylethanolamine (PE), termed LC3 lipidation, is a hallmark of autophagosome biogenesis. LC3 lipidation occurs via a ubiquitin-like conjugation cascade. The ubiquitin E1-like ATG7 and the E2-like ATG3 carry out the cognate reactions in the LC3 pathway. The ATG12–ATG5-ATG16L1 complex scaffolds transfer of LC3 from ATG3 to PE 9, 10. The role of the ATG12–ATG5-ATG16L1 is analogous to that of a RING domain ubiquitin E3 ligase, although there is no sequence homology between any of the subunits and ubiquitin E3 ligases. ATG2 transfers phospholipids from endoplasmic reticulum (ER) to the growing phagophore 11–13, whereas ATG9 translocates phospholipids from the cytoplasmic to the luminal leaflet, enabling phagophore expansion 14, 15. These above-mentioned proteins are sometimes referred to as the “core complexes” of autophagy.
Box 1– Autophagy core complexes and cargo receptors.
ULK1c:
Composed of ULK1 (Ser/Thr kinase), FIP200 (scaffold), ATG13, and ATG101. The kinase is activated by mTOR inhibition, and the complex is recruited to its sites of action by a subset of cargo receptors. The ULK2 kinase is partially redundant with ULK1. The orthologous complex in S. cerevisiae consists of the ULK1 ortholog Atg1, Atg13, and, unique to budding yeast, Atg17, Atg29, and Atg31.
PI3KC3-C1:
Class III PI 3-kinase complex I, comprised of VPS34 (lipid kinase), VPS15 (scaffold), ATG14 (scaffold, unique to complex I and to autophagy initiation), and BECN1 (regulatory subunit). ATG14 is unique to complex 1, and is replaced in the endosomal and late autophagy complex II by UVRAG. The catalytic subunit VPS34 phosphorylates phosphatidylinositol lipid headgroups to generate PI(3)P. The orthologous complex in S. cerevisiae consists of Vps34, Vps15, Atg14, and Vps30.
ATG12-ATG5-ATG16L1:
The ubiquitin-like ATG12 is covalently linked to ATG5 (by ATG10 and ATG7), which further associates non-covalently with ATG16L1. The coiled-coil protein ATG16L1 homodimerizes, forming a heterohexameric complex composed of two copies each of ATG12, ATG5, and ATG16L1. ATG16L2 is partially redundant with ATG16L1. The complex functions like an E3 ubiquitin ligase with respect to the conjugation of ATG8/LC3 family proteins to membrane phosphatidylethanolamine, although the subunits are not orthologs of ubiquitin E3s. The orthologous complex in S. cerevisiae consists of Atg12, Atg5, and Atg16.
ATG2:
ATG2A and ATG2B are high capacity, low specificity lipid transporters whose structure is centered on a long tunnel that carries phospholipids from their source, normally the ER, to the growing phagophore. The sole S. cerevisiae ortholog is Atg2.
ATG9:
ATG9A and ATG9B are the sole integral membrane proteins of the core autophagy machinery, and function as trimers. ATG9 vesicles are the seeds for autophagosome growth and the protein has phospholipid scramblase activity that equilibrates the lipid composition of the two leaflets of the bilayer in an ATP-independent manner. The sole S. cerevisiae ortholog is Atg9.
WIPIs:
Also known as PROPPINs, these β-propeller proteins bind to the lipids PI(3)P and PI(3,5)P2. In humans, the WIPI family comprises WIPI1, 2, 3, and 4. WIPIs target ATG16L1 and ATG2 to sites of autophagosome biogenesis. The S. cerevisiae orthologs are Atg18, Atg21, and Hsv2.
ATG8/LC3 family proteins:
Small ubiquitin-like proteins LC3A, LC3B, LC3C, GABARAP, GABARAPL1, GABARAPL2 which are covalently linked to phosphatidylethanolamine lipid headgroups on the phagophore membrane. Their ortholog is Atg8 in yeast. Most cargo receptors and most of the core autophagy proteins bind LC3 through LC3 interacting region (LIR) motifs.
Cargo receptors:
A group of proteins which selectively bind autophagy cargo (often, although not exclusively, through a ubiquitin-binding domain), which include, among many others, p62, NDP52, OPTN, NBR1, and TAX1BP1.
In this review, we will focus on a subset of selective autophagy pathways, including aggrephagy, mitophagy, and xenophagy, which are initiated by the ubiquitination of cargoes. These ubiquitinated substrates are recognized by a subset of cargo receptors, including p62 (sequestosome-1), NBR1, optineurin (OPTN), NDP52 and Tax1-binding protein 1 (TAX1BP1). All of these receptors contain a LC3-interaction region (LIR), a ubiquitin binding domain (UBD), and a dimerization/oligomerization domain 16–18. These cargo receptors connect cargo to the phagophore through their interaction with both clustered ubiquitin chains and membrane-conjugated LC3.
Initiation
“On demand” autophagosome biogenesis in selective autophagy
Until early 2019, the leading model in the field was that mammalian selective autophagy was initiated when autophagy receptors bridged cargo to pre-existing ATG8-positive membranes 19. The ATG8s themselves have been shown, however, to be non-essential for phagophore formation in at least one selective autophagy pathway, mitophagy 20. ATG8s do contribute to the rate of autophagosome formation and the control of autophagosome size in mitophagy 20, and in bulk autophagy, to the rate of degradation of the inner autophagosomal membrane in the lysosome 21. This suggested that autophagy receptors must be able to recruit and activate the core autophagy initiation machinery directly, bypassing the need for ATG8 proteins. Indeed, such a mechanism was identified almost two decades ago in the autophagy-like cytoplasm to vacuole targeting (Cvt) pathway of S. cerevisiae. In yeast, the aminopeptidase precursor prApe1 is sorted to the vacuole by the cargo receptor Atg19, which binds to Atg11 and in turn to the Atg1 core complex 22–24.
The mammalian counterpart of the Atg1 complex is the ULK1 complex, and the counterpart of the Atg11 scaffolding subunit in yeast is FIP200 in mammals (Box 1). The C-terminal domain of Atg11 binds to the Atg19 receptor in yeast 25, which suggested that the corresponding C-terminal “Claw” 26 domain of FIP200 might bind to autophagy receptors in mammals. The receptor p62 was found to contain a FIP200 interacting region (FIR) that has weak homology to the Atg11 binding region of Atg19, and overlaps with the p62 LIR 26. The affinity of the p62 FIR for the FIP200 Claw increases when the FIR is phosphorylated 26. The p62 phospho-FIR:FIP200 interaction is sufficient to recruit the ULK1 complex to sites of condensed cargo, and so initiate autophagosome biogenesis 26 (Fig. 2a).
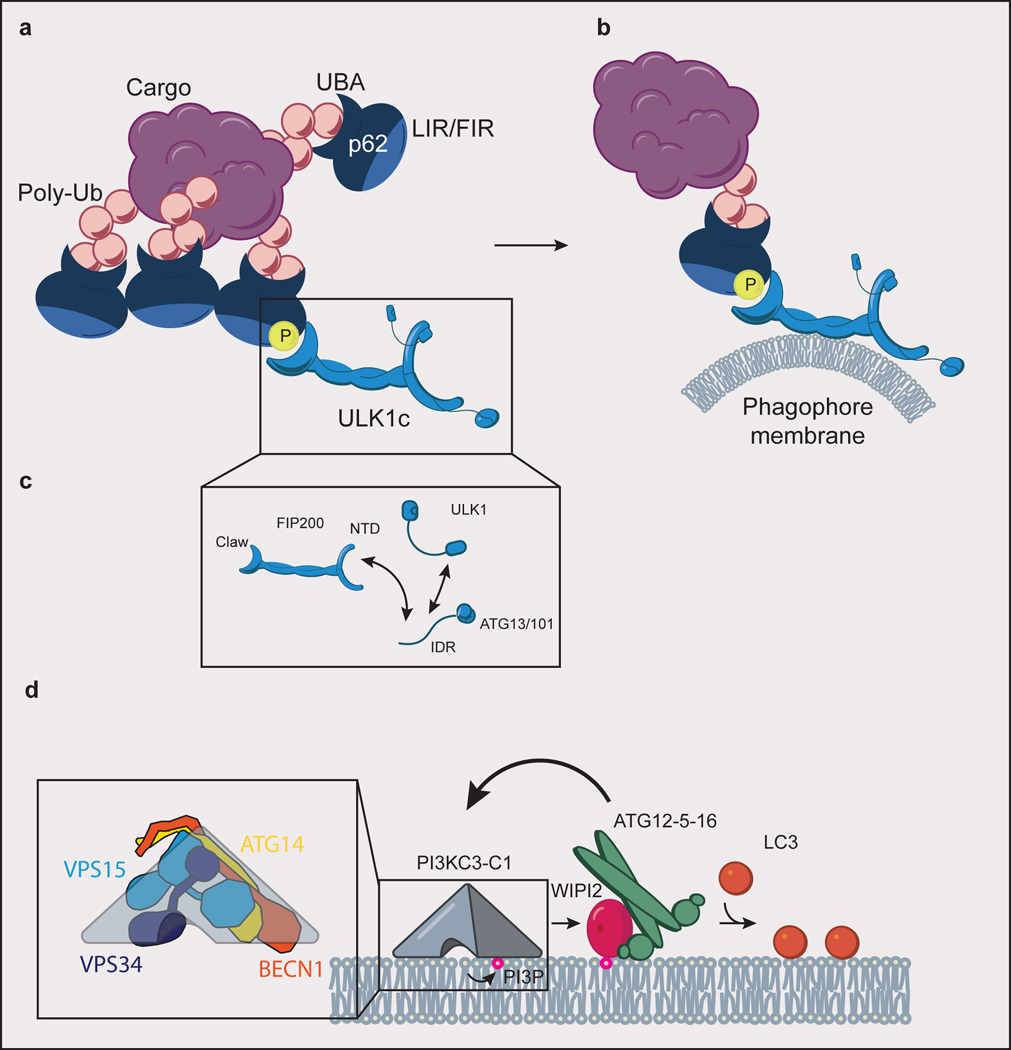
Fig. 2. Autophagy initiation.
(a) ULK1c is recruited to p62 condensate cargo through an interaction of the FIP200 claw domain with phospho-p62. p62 binds to ubiquitinated (Ub) cargo via its UBA domain, to LC3 via its LIR motif, and to FIP200 of the ULK1c via its FIR motif. (b) Interaction of ULK1c with p62 condensates stimulates “on demand” phagophore membrane generation. (c) The subunits of ULK1c (FIP200, ULK1, ATG13, ATG13/101) are held together in large part through a network of interacting IDRs. (d) PI3KC3-C1 (inset: comprised of ATG14, BECN1, VPS15, and VPS34) phosphorylates phosphatidylinositol lipid headgroups to form PI(3)P, which recruits WIPI proteins to the phagophore membrane. WIPI2 subsequently recruits ATG12–5–16 to the phagophore in a positive feedback loop with PI3KC3-C1. ATG12–5–16 covalently links LC3 to phosphatidylethanolamine lipid headgroups (orange circles) in the phagophore membrane.
FIR motif interactions are not the only mechanism for ULK1 core complex recruitment by cargo receptors. FIP200 binds to another autophagy receptor, NDP52, as revealed by yeast two-hybrid screens 27, 28. The SKICH domain of NDP52 interacts with the coiled-coil domain of FIP200 28, in contrast to the FIR-Claw interaction described above for p62. The FIP200-NDP52 interaction is capable of driving both xenophagy 28 and mitophagy 29. NDP52 engagement induces a conformational change in FIP200 that promotes membrane binding of the ULK1 complex 30. In xenophagy, TBK1 and the TBK1 adaptor SINTBAD/NAP1 are also required 28. In mitophagy, TBK1 facilitates FIP200 recruitment by NDP52, but chemically directed recruitment of FIP200 can bypass the need for cargo-NDP52 engagement, demonstrating that ULK1 complex recruitment is sufficient to trigger autophagosome biogenesis 29. These findings, coupled with the results described above for p62, show that at least two receptors trigger autophagy initiation by recruiting the ULK1 complex to its site of action. This paradigm is not restricted to the ULK1 complex. A third autophagy receptor, OPTN, was proposed to interact directly with ATG9A to initiate mitophagy 31, potentially bypassing FIP200. The targeting of ATG9A and the ULK1 complex by different cargo receptors could explain the long-standing observation that these components could be targeted independently of each other and ATG8s to sites of mitophagy initiation 32. It will be interesting to see if other core components besides the ULK1 complex and ATG9, such as PI3KC3-C1, the ATG16L1 complex, or WIPIs, can be directly recruited by autophagy receptors. The emerging paradigm is thus that autophagic membranes are made “on demand” when cargo, via the recruitment of core autophagy components by cargo receptors, activates the upstream core complexes (Fig. 2b).
Scaffolding the phagophore in bulk autophagy
In selective autophagy, the cargo itself templates the shape of the phagophore 33. In bulk autophagy, the mechanism of scaffolding is less clear. First principles of membrane elasticity dictate that in free solution an expanding membrane sheet will fold into the shape of a cup 34, and that crescent-shaped autophagy proteins lower the free energy barrier to promote this process 35. Whether membrane elasticity alone can drive phagophore shaping in the cytosol without a template is uncertain. p62 was initially identified as the receptor for aggrephagy 36, however, recent studies show that p62 can form phase-separated condensates with polyubiquitin 37, 38. 15 nm-wide filaments form from purified p62 in vitro 39, and in the presence of model ubiquitinated cargo, condense into 0.5–2.0 μm clusters 37. These clusters have many of the properties of phase-separated condensates, including high sphericity and a high propensity to fuse 37. Condensed p62 is capable of recruiting the ULK1 complex 26, explaining how uptake of phase-separated condensates could occur. In cells these condensates are similar in size to other selective autophagy substrates 40. Another receptor, NBR1, can directly promote the formation of p62 condensates 41. Thus, at least in cases where there is clear evidence for cargo condensation, bulk autophagy may be actually be “condensate-phagy” or “fluid-phagy” 42. A membrane elasticity model for the phagophore was recently shown to be compatible with engulfment of fluid p62 condensates, consistent with this idea 43. This would be just one of the many flavors of selective autophagy, with phagophore growth scaffolded by the size and shape of the condensate.
A multivalent web of low affinity interactions directs initiation
An early and influential model for autophagy initiation based on yeast genetics suggested that the core complexes were recruited in strict order in starvation-induced autophagy, beginning with the FIP200 counterpart Atg17, and culminating with the Atg8 lipidation machinery 44. Live cell imaging, however, found that ULK1 and the ATG5 component of ATG8 lipidation machinery arrive simultaneously in mammalian starvation-induced autophagy 45. Evidence is growing that a network of multiple high and low affinity interactions stabilizes the network of autophagy initiation complexes at the nascent phagophore. This contributes to the redundancy and robustness of the process.
The ULK1 complex (Atg1 complex in yeast) best illustrates how a multiplicity of interactions can drive autophagy initiation. ULK1, FIP200, and ATG13 subunits of the ULK1 complex all contain larger intrinsically disordered regions (IDRs) 46. Indeed, the majority of ATG13 consists of a C-terminal IDR. The key contacts between ATG13-FIP200 and ATG13-ULK1 are driven by the ATG13 IDR, which forms the main unifying element of the overall complex 47. The organization of the complex is therefore structurally loose 48, 49 (Fig. 2c). While the individual subunits are required for canonical autophagy, their assembly is not always strictly required. In starvation induced autophagy, removal of the ATG13 IDR binding sites for FIP200 and ULK1 has moderate to no phenotype 50, 51. This implies that the ULK1 complex subunits have enough redundant interactions with other components of the autophagy initiation system for them carry out their functions without the strict need to actually assemble into the canonical ULK1 complex.
The Atg1 complex of S. cerevisiae is also organized principally by the IDR of its Atg13 subunit, which interacts at multiple sites 52, 53 within the dimeric banana-shaped Atg17 subcomplex 54. Atg13 can bridge multiple Atg17 molecules 53. The capacity of Atg13 to bridge multiple Atg17 dimers drives phase separation of Atg1 42, 53, at least in vitro. It takes only ~15–50 copies each of Atg13 and Atg17 to initiate a phagophore 55, 56. The interpretation of mesoscopic phase separation for biomolecular process involving only a few tens of molecules is an interesting question that is not fully resolved 57. Many of the physical properties of condensates that are important in biology, such as their ability to sequester their contents and fuse with one another, do not apply on the scale of a handful of molecules 57. Other aspects, notably assembly through a network of multiple low affinity interactions, are relevant across all scales.
A circuit involving PI3KC3-C1, WIPI2, and the ATG16L1 complex provides another example of cooperativity in autophagy initiation. Upon its recruitment to sites of autophagy initiation, PI3KC3-C1 phosphorylates phosphatidylinositol (PI) to PI(3)P. Membrane bound PI(3)P then recruits WIPI proteins to the membrane (Fig. 2d). At least one of the WIPIs, WIPI2, is key to recruiting the ATG16L1 complex to membranes, and thereby triggering ATG8 lipidation 58. Reconstitution of the PI3KC3-C1, WIPI2, and ATG16L1 circuit revealed additional weaker interactions between WIPI2 and PI3KC3-C1 that promote positive feedback to drive the net ATG8s lipidation reaction forward 59. The presence of ubiquitinated cargo, cargo receptors, and the ULK1 complex further enhances the rate of ATG8 lipidation 60.
Growth
Feeding lipids to the autophagosomal membrane
Autophagosomes are remarkable for their ability to grow essentially de novo in a matter of minutes 61. The nature of the lipid supply for this massive growth, estimated to require the transfer of 4,000 phospholipid molecules per second 62, has been one of the most enduring mysteries in the field 3. The answer was provided by research into the Atg2 protein, which was identified in 2001 63, 64 but whose function has been elusive until recently. Human ATG2A is rod-shaped and, in its complex with WIPI4, capable of tethering membranes 65, 66. ATG2 has sequence homology to the VPS13 proteins, a family of lipid transporters 67. These observations lead three teams to the seminal discovery that the yeast Atg2 and human ATG2A protein is the lipid transporter linking growing phagophores to membrane sources such as the ER 11–13 (Fig. 3). The lipid transport rates measured in vitro are sufficient for a plausible ~102 molecules of ATG2 per phagophore to meet the demands of membrane growth 68. The cryo-EM structure of a thermophilic yeast Vps13 revealed a long and wide hydrophobic tunnel running the length of this rod-like protein 69, explaining how members of the Vps13/Atg2 protein family could serve as a “firehose for phospholipids” 62.
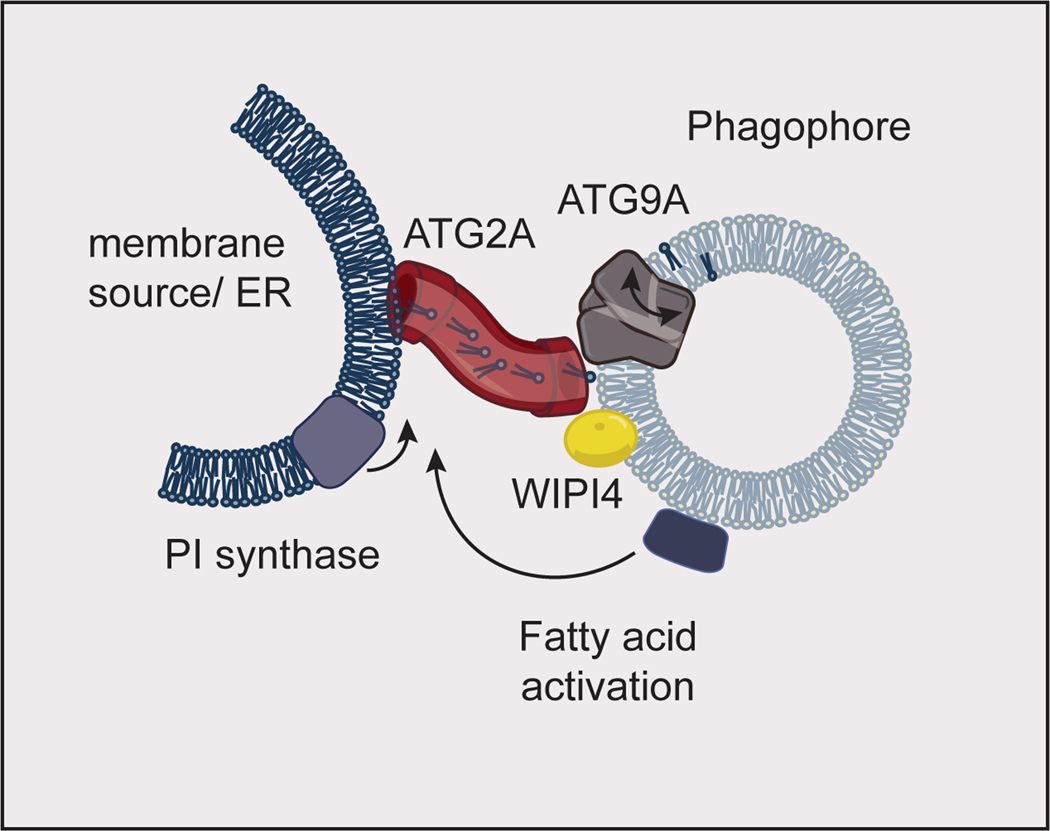
Fig. 3. Autophagosome growth.
Fatty acid activation at the phagophore feeds phospholipid synthesis by phosphatidylinositol (PI) synthase at the membrane source for the autophagosome. This phospholipid synthesis drives lipid flux through ATG2A (which binds WIPI4 on the phagophore) into the growing phagophore membrane. ATG9A aids in phagophore growth by scrambling lipids between the inner and outer leaflets of the phagophore. The model shown here is inspired by data obtained in yeast. The proteins implicated in autophagosome growth are conserved between yeast and humans, and human proteins are shown here. ER: Endoplasmic Reticulum.
The Atg2 “firehose” model for autophagosome expansion immediately raised several questions. One major question was how to resolve the potential imbalance created by the rapid delivery of phospholipids to the outer leaflet of the phagophore. Atg9 (ATG9A/B in mammals) is the sole integral membrane protein of the core autophagy machinery 70, and its function or functions have a major question in its own right. The cryo-EM structure of ATG9A 71 revealed a trimeric membrane embedded assembly riddled with cavities suggestive of a potential role in lipid transport. Two subsequent cryo-EM structural papers revealed in greater detail the presence of hydrophilic channels on the surface of yeast Atg9 and human ATG9A 14, 15. The channels run the length of the membrane spanning surface of Atg9, a feature characteristic of the scramblases that equilibrate phospholipids between the two leaflets of the membrane bilayer. These studies and another went on to confirm that Atg9 and ATG9A have phospholipid scramblase activity in vitro. Phospholipids in wild-type but not atg9Δ yeast are distributed symmetrically, consistent with Atg9 scramblase activity against multiple phospholipid species 72. Taken together, the findings for Atg2 and Atg9 explain how phospholipids are directed into the growing phagophore (Fig. 3).
Atg9 vesicles are the seeds of autophagosome biogenesis
The functions of ATG9 vesicles have been one of the leading questions in autophagosome biogenesis. As characterized in yeast, trans-Golgi network (TGN)-derived Atg9 vesicles are required for autophagy initiation 73, 74. The size and number of these vesicles fail to account for the amount of lipid needed to make a phagophore by several orders of magnitude, however. This led to the hypothesis that Atg9 vesicles are not the membrane source, but rather the seeds of phagophore growth. A recent reconstitution of yeast autophagy initiation including the cargo receptor Atg19, the Atg11 scaffold (a FIP200 counterpart in yeast), PI3KC3-C1, the Atg8 lipidation system, Atg2, and Atg9, confirmed that Atg9 vesicles can function as seeds for phagophore growth 75. In this system, Atg19 and Atg11 linked a model cargo to the rest of the machinery. Recruitment of the lipid transporter Atg2 was sufficient to catalyze lipid transfer from donor vesicles. Newly transferred phosphatidylethanolamine (PE) was capable of serving as an acceptor for Atg8 lipidation by the conjugation machinery. A key point is that lipid transfer can occur upstream of Atg8 lipidation. This is consistent with the findings in mammalian cells that ATG8 conjugation machinery proteins are not essential for phagophore initiation 20, 21, 76, and that ATG8s lipidation can be bypassed by cargo receptor TAX1BP1-driven FIP200 clustering 77. Phagophore seeding by ATG9 vesicles thus seems likely to be operative in mammals as well.
The autophagosome itself supplies fatty acids for its growth
ATG2 and ATG9 are not ATPases nor are they known to be coupled to other energy inputs, therefore their lipid transfer and scramblase activities must be driven thermodynamically by a concentration gradient. This gradient appears most likely to be established by de novo phospholipid synthesis. In yeast, the acyl CoA-synthetase Faa1 is localized to the phagophore, and its presence is required for phagophore growth 78. Isotopic labeling was used to track fatty acids activated by phagophore-localized Faa1 and show that these fatty acids are incorporated into phospholipids which can then be returned to the phagophore by Atg2 78. Consistent with this idea, mammalian autophagy initiation occurs at ER sites enriched in PI synthase-enriched domains 79, and choline phospholipid synthesis was shown to be required for phagophore growth in cancer cells 80. Fatty acid activation in the phagophore thus appears to be a key step in powering phagophore growth (Fig. 3).
Closure
As the phagophore grows and engulfs its substrate, eventually the double membrane converges until only a single unclosed membrane neck is left. The open neck allows communication between the cytosol and the interior. Until the neck is closed, is not possible to acidify the interior and so degrade its contents. Membrane necks whose interior is connected to the cytosol, such as the neck of the closing phagophore, are resolved by the ESCRT machinery 81, 82. Neurodegeneration-linked ESCRT-related phenotypes in autophagy were noted more than a decade ago 83–86. Only recently has it been possible to create assays that pinpoint the closure step in autophagy 87–89 (Fig. 4a). Human ESCRT-I is required for autophagosome closure, in particular the VPS37A subunit 90. It is not yet known how the ESCRTs are recruited to the closing phagophore, although it presumably somehow involves VPS37A. Once recruited to its site of action, ESCRT-I forms a ring-shaped assembly of ~12 copies of the complex that helps recruit downstream ESCRTs and coordinates closure 91 (Fig. 4b). On large cargoes such as bacteria and mitochondria, autophagy may initiate at more than one site, and the ESCRT might also be involved in piecing several membrane fragments into a single phagophore (Fig. 4c), as demonstrated in nuclear envelope reformation 82. Key questions remain about how closure is correctly synchronized with the termination of growth, how ESCRTs are targeted to the phagophore neck, and how lysosomal fusion is forestalled until closure finishes.
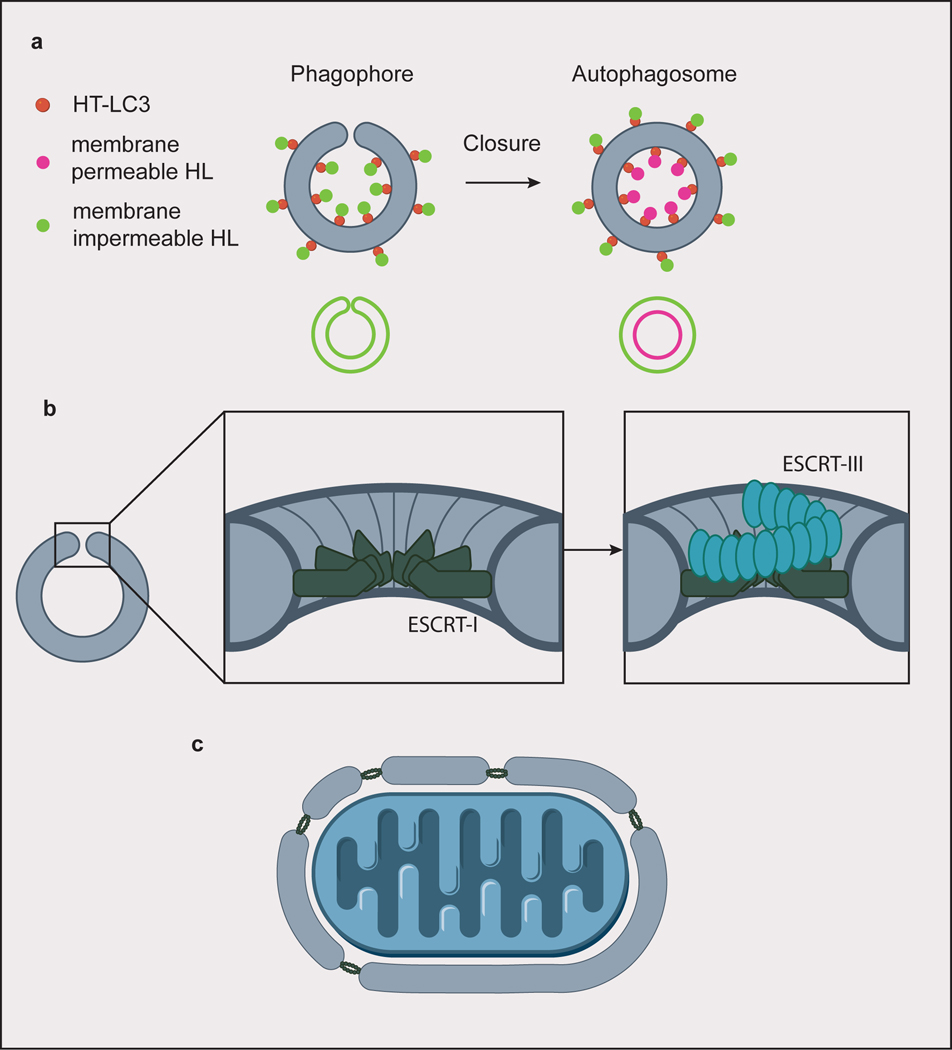
Fig. 4. Autophagosome closure.
(a) A screen for factors involved in autophagosome closure identified ESCRT-I protein VPS37A by using membrane permeable/impermeable Halo ligand (HL) dyes to selectively label Halo tagged (HT) LC3. Hits were identified from cells whose phagophores only stained with the impermeable dye, indicating failure to close the autophagosomes. Closed autophagosomes stain with both the membrane permeable and impermeable dyes. (b) ESCRT-I forms a dodecameric ring assembly at the rim of the closing autophagosome. In canonical ESCRT-mediated membrane scission, subsequent helical assembly of ESCRT-III constricts the membrane rim. (c) For larger cargoes, ESCRTs could be involved in piecing together membranes to form a single, large phagophore.
Conclusions and outlook
The findings outlined above highlight what we consider the most important developments in autophagy mechanisms over the past two years. Seven years ago, a comprehensive review concluded that “There remain, however, many unanswered questions about the precise protein and lipid composition of the autophagic membranes, the mechanisms behind their shape, how they detach from the ER and how the autophagosome is finally closed.” Over the just past two years, the field has made major inroads into answering these questions. It is truly time for the textbooks to be rewritten. Of course, these developments raise many new questions. Now that the individual steps of initiation, growth, and closure are becoming much better understood, questions arise as to their coordination with each other and with other cell processes. What are the cues to transition from initiation to growth, and growth to closure? As our understanding becomes more quantitative, we can begin to ask what the rate limiting reactions at each step are. As we understand these parameters, we can begin to rationally modify these pathways, leveraging the basic insights described above to develop potential pro-autophagic therapies.
Acknowledgements
This work was supported by HFSP (RGP0026/2017 to J.H.H.) and NIH R01 GM111730 (J.H.H.).
Footnotes
Conflicts of interest
J.H.H. is a co-founder of Casma Therapeutics.
References
- 1.Mizushima N, Yoshimori T & Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27, 107–132 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Levine B & Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 176, 11–42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamb CA, Yoshimori T & Tooze SA The autophagosome: origins unknown, biogenesis complex. Nature Reviews Molecular Cell Biology 14, 759 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Wen X & Klionsky DJ An overview of macroautophagy in yeast. Journal of Molecular Biology 428, 1681–1699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakatogawa H. Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol 21, 439–458 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Melia TJ, Lystad AH & Simonsen A. Autophagosome biogenesis: From membrane growth to closure. J Cell Biol 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zachari M & Ganley IG The mammalian ULK1 complex and autophagy initiation. Essays in biochemistry 61, 585–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurley JH & Young LN Mechanisms of Autophagy Initiation. Annual Review of Biochemistry 86, 225–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanada T et al. The Atg12-Atg5 Conjugate Has a Novel E3-like Activity for Protein Lipidation in Autophagy. Journal of Biological Chemistry 282, 37298–37302 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Ichimura Y et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Maeda S, Otomo C & Otomo T. The autophagic membrane tether ATG2A transfers lipids between membranes. Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osawa T et al. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol 26, 281–288 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Valverde DP et al. ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 218, 1787–1798 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda S et al. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matoba K et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol (2020). [DOI] [PubMed] [Google Scholar]
- 16.Birgisdottir AB, Lamark T & Johansen T. The LIR motif - crucial for selective autophagy. J Cell Sci 126, 3237–3247 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Kirkin V, McEwan DG, Novak I & Dikic I. A Role for Ubiquitin in Selective Autophagy. Mol. Cell 34, 259–269 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Kirkin V & Rogov VV A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol Cell 76, 268–285 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Johansen T & Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen TN et al. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol 215, 857–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuboyama K et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036–1041 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Kim J et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. Journal of Cell Biology 153, 381–396 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shintani T, Huang WP, Stromhaug PE & Klionsky DJ Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev.Cell 3, 825–837 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamber RA, Shoemaker CJ & Denic V. Receptor-Bound Targets of Selective Autophagy Use a Scaffold Protein to Activate the Atg1 Kinase. Mol. Cell 59, 372–381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yorimitsu T & Klionsky DJ Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Molecular Biology of the Cell 16, 1593–1605 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turco E et al. FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol Cell 74, 330–346 e311 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J et al. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol 7, 536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravenhill BJ et al. The Cargo Receptor NDP52 Initiates Selective Autophagy by Recruiting the ULK Complex to Cytosol-Invading Bacteria. Mol Cell 74, 320–329 e326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vargas JNS et al. Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol Cell 74, 347–362 e346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi X, Chang C, Yokom AL, Jensen LE & Hurley JH The autophagy adaptor NDP52 and the FIP200 coiled-coil allosterically activate ULK1 complex membrane recruitment. Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamano K et al. Critical role of mitochondrial ubiquitination and the OPTN-ATG9A axis in mitophagy. J Cell Biol 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itakura E, Kishi-Itakura C, Koyama-Honda I & Mizushima N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. Journal of Cell Science 125, 1488–1499 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Zaffagnini G & Martens S. Mechanisms of Selective Autophagy. Journal of Molecular Biology 428, 1714–1724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knorr RL, Dimova R & Lipowsky R. Curvature of Double-Membrane Organelles Generated by Changes in Membrane Size and Composition. PLoS One 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahrami AH, Lin MG, Ren X, Hurley JH & Hummer G. Scaffolding the cup-shaped double membrane in autophagy. PLoS Comput Biol 13, e1005817 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjorkoy G et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. Journal of Cell Biology In press (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaffagnini G et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J 37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun D, Wu R, Zheng J, Li P & Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res 28, 405–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciuffa R et al. The Selective Autophagy Receptor p62 Forms a Flexible Filamentous Helical Scaffold. Cell Reports 11, 748–758 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Kageyama S et al. p62/SQSTM1-droplet serves as a platform for autophagosome formation and anti-oxidative stress response. Nat Commun 12, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sánchez-Martín P et al. NBR1-mediated p62-liquid droplets enhance the Keap1-Nrf2 system. EMBO Rep 21, e48902 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujioka Y et al. Phase separation organizes the site of autophagosome formation. Nature 578, 301–305 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Agudo-Canalejo J et al. Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature, doi: 10.1038/s41586-41020-42992-41583 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Suzuki K, Kubota Y, Sekito T & Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12, 209–218 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Koyama-Honda I, Itakura E, Fujiwara TK & Mizushima N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 9, epub ahead of print (2013). [DOI] [PubMed] [Google Scholar]
- 46.Mei Y et al. Intrinsically disordered regions in autophagy proteins. Proteins 82, 565–578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung CH et al. ULK-Atg13-FIP200 Complexes Mediate mTOR Signaling to the Autophagy Machinery. Molecular Biology of the Cell 20, 1992–2003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin MG & Hurley JH Structure and function of the ULK1 complex in autophagy. Curr. Opin. Cell Biol. 39, 61–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi X et al. ULK complex organization in autophagy by a C-shaped FIP200 N-terminal domain dimer. J Cell Biol 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallot-Hieke N et al. Systematic analysis of ATG13 domain requirements for autophagy induction. Autophagy 14, 743–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hieke N et al. Expression of a ULK1/2 binding-deficient ATG13 variant can partially restore autophagic activity in ATG13-deficient cells. Autophagy 11, 1471–1483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujioka Y et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 21, 513–521 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto H et al. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev. Cell 38, 86–99 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Ragusa MJ, Stanley RE & Hurley JH Architecture of the Atg17 Complex as a Scaffold for Autophagosome Biogenesis. Cell 151, 1501–1512 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geng J, Baba M, Nair U & Klionsky DJ Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol 182, 129–140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin MG, Schoneberg J, Davies CW, Ren X & Hurley JH The dynamic Atg13-free conformation of the Atg1 EAT domain is required for phagophore expansion. Mol Biol Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyon AS, Peeples WB & Rosen MK A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dooley HC et al. WIPI2 Links LC3 Conjugation with PI3P, Autophagosome Formation, and Pathogen Clearance by Recruiting Atg12–5-16L1. Mol. Cell 55, 238–252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fracchiolla D, Chang C, Hurley JH & Martens S. A PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J Cell Biol 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang C et al. Reconstitution of cargo-induced LC3 lipidation in mammalian selective autophagy. biorxiv 10.1101/2021.01.08.425958 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Axe EL et al. Autophagosome formation from compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. Journal of Cell Biology 182, 685–701 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prinz WA & Hurley JH A firehose for phospholipids. J Cell Biol 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shintani T, Suzuki K, Kamada Y, Noda T & Ohsumi Y. Apg2p functions in autophagosome formation on the perivacuolar structure. Journal of Biological Chemistry 276, 30452–30460 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Wang CW et al. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. Journal of Biological Chemistry 276, 30442–30451 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng JX et al. Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy 13, 1870–1883 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chowdhury S et al. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc Natl Acad Sci U S A 115, E9792–E9801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar N et al. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol 217, 3625–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.von Bulow S & Hummer G. Kinetics of Atg2-mediated lipid transfer from the ER can account for phagophore expansion. bioRxiv, 2020.2005.2012.090977 (2020). [Google Scholar]
- 69.Li P, Lees JA, Lusk CP & Reinisch KM Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J Cell Biol 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noda T et al. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. Journal of Cell Biology 148, 465–479 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guardia CM et al. Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery. Cell Rep 31, 107837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orii M, Tsuji T, Ogasawara Y & Fujimoto T. Transmembrane phospholipid translocation mediated by Atg9 is involved in autophagosome formation. J Cell Biol 220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mari M et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. Journal of Cell Biology 190, 1005–1022 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamamoto H et al. Atg9 Vesicles are an Important Membrane Source During Early Steps of Autophagosome Formation. Journal of Cell Biology 198, 219–233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sawa-Makarska J et al. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science 369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kishi-Itakura C, Koyama-Honda I, Itakura E & Mizushima N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J Cell Sci 127, 4089–4102 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Ohnstad AE et al. Receptor-mediated clustering of FIP200 bypasses the role of LC3 lipidation in autophagy. Embo j, e104948 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schutter M, Giavalisco P, Brodesser S & Graef M. Local Fatty Acid Channeling into Phospholipid Synthesis Drives Phagophore Expansion during Autophagy. Cell 180, 135–149 e114 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Nishimura T et al. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. Embo j 36, 1719–1735 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andrejeva G et al. De novo phosphatidylcholine synthesis is required for autophagosome membrane formation and maintenance during autophagy. Autophagy 16, 1044–1060 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hurley JH The ESCRT complexes. Critical Reviews in Biochemistry and Molecular Biology 45, 463–487 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vietri M, Radulovic M & Stenmark H. The many functions of ESCRTs. Nat Rev Mol Cell Biol 21, 25–42 (2020). [DOI] [PubMed] [Google Scholar]
- 83.Skibinski G et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat.Genet. 37, 806–808 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Parkinson N et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology 67, 1074–1077 (2006). [DOI] [PubMed] [Google Scholar]
- 85.Lee JA, Beigneux A, Ahmad ST, Young SG & Gao FB ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17, 1561–1567 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Filimonenko M et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. Journal of Cell Biology 179, 485–500 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takahashi Y et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat Commun 9, 2855 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhen Y et al. ESCRT-mediated phagophore sealing during mitophagy. Autophagy, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhou F et al. Rab5-dependent autophagosome closure by ESCRT. J Cell Biol 218, 1908–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takahashi Y et al. VPS37A directs ESCRT recruitment for phagophore closure. J Cell Biol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Flower TG et al. A helical assembly of human ESCRT-I scaffolds reverse-topology membrane scission. Nat Struct Mol Biol 27, 570–580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]