Abstract
Coenzyme Q (CoQ), a redox-active lipid essential for oxidative phosphorylation, is synthesized by virtually all cells, but how eukaryotes make the universal CoQ head group precursor 4-hydroxybenzoate (4-HB) from tyrosine is unknown. The first and last steps of this pathway have been defined in Saccharomyces cerevisiae, but the intermediates and enzymes involved in converting 4-hydroxyphenylpyruvate (4-HPP) to 4-hydroxybenzaldehyde (4-HBz) have not been described. Here, we interrogate this pathway with genetic screens, targeted LC-MS, and chemical genetics. We identify three redundant aminotransferases (Bna3, Bat2, and Aat2) that support CoQ biosynthesis in the absence of the established pathway tyrosine aminotransferases, Aro8 and Aro9. We use isotope labeling to identify bona fide tyrosine catabolites, including 4-hydroxyphenylacetate (4-HPA) and 4-hydroxyphenyllactate (4-HPL). Additionally, we find multiple compounds that rescue this pathway when exogenously supplemented, most notably 4-hydroxyphenylacetaldehyde (4-HPAA) and 4-hydroxymandelate (4-HMA). Finally, we show that the Ehrlich pathway decarboxylase Aro10 is dispensable for 4-HB production. These results define new features of 4-HB synthesis in yeast, demonstrate the redundant nature of this pathway, and provide a foundation for further study.
Keywords: ubiquinone, coenzyme Q, biosynthesis, metabolism, mass spectrometry (MS), Saccharomyces cerevisiae, 4-hydroxybenzoate
Abbreviations: CoQ, coenzyme Q; 3,4-diHB, 3,4-dihydroxybenzoate; 3,4-diHMA, 3,4-dihydroxymandelate; 3,4-diHPA, 3,4-dihydroxyphenylacetate; 3,4-diHPAA, 3,4-dihydroxyphenylacetaldehyde; 4-HB, 4-hydroxybenzoate; 4-HBAL, 4-hydroxybenzyl alcohol; 4-HBz, 4-hydroxybenzaldehyde; 4-HMA, 4-hydroxymandelate; 4-HPA, 4-hydroxyphenylacetate; 4-HPAA, 4-hydroxyphenylacetaldehyde; 4-HPL, 4-hydroxyphenyllactate; 4-HPP, 4-hydroxyphenylpyruvate; EMS, ethyl methanesulfonate; LC-MS, liquid chromatography–mass spectrometry; pABA, para-aminobenzoic acid; p-coum, para-coumarate; PL, phenyllactate; PP, phenylpyruvate; PPHB, polyprenyl hydroxybenzoate
Coenzyme Q (ubiquinone or CoQ) is a ubiquitous redox-active lipid that functions as an essential component of the respiratory chain and acts as a potent lipophilic antioxidant (1, 2). CoQ is also an important redox cofactor for diverse processes, including pyrimidine synthesis and sulfide oxidation (3, 4). Consistent with these crucial cellular roles, defects in CoQ biosynthesis are associated with diverse human diseases, including cerebellar ataxia, nephrotic syndrome, and encephalomyopathy (5). A complete understanding of CoQ biosynthesis could aid clinical diagnoses for the many cases of CoQ deficiency still lacking an established genetic basis (6). Traditional methods including yeast complementation assays have identified many of the necessary genes for CoQ biosynthesis, but multiple key steps remain uncharacterized (7).
4-Hydroxybenzoate (4-HB) is the universal precursor for CoQ's fully substituted benzoquinone head group from bacteria to mammals (8, 9). In Saccharomyces cerevisiae, para-aminobenzoic acid (pABA) can also serve as a head group precursor (10, 11). pABA is synthesized from chorismate in two steps catalyzed by Abz1 and Abz2 (12, 13). The pathway for 4-HB production in eukaryotes has not been elucidated, but the first and last steps have been described recently. 4-Hydroxyphenylpyruvate (4-HPP) is a requisite early precursor made from tyrosine through aminotransferases Aro8 and Aro9 or de novo through the shikimate pathway (14). The last step in the pathway is the oxidation of 4-hydroxbenzaldehyde (4-HBz) to 4-HB, catalyzed by the broad-specificity aldehyde dehydrogenase Hfd1 (14, 15). However, the intermediates and enzymes involved in converting 4-HPP to 4-HBz are unidentified (Fig. 1A).
Figure 1.
Redundant aminotransferases convert tyrosine to 4-HPP to support 4-HB synthesis.A, schematic of 4-HB synthesis in budding yeast. B, respiratory growth rates of W303 wild-type (WT) and Δaro8Δaro9Δaro2 strains assayed in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol and supplemented with the indicated additives (mean ± SD, n = 3). C, CoQ6 levels from the indicated yeast strains grown in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol (mean ± SD, n = 3). D, light (12C) and heavy (13C9) 4-HPP levels from yeast grown in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol, supplemented with light (12C) or heavy (13C9) tyrosine (mean ± SD, n = 3). E, drop assay of Δaro8Δaro9 yeast transformed with the indicated plasmids and grown for 3 days on the indicated solid media. F, respiratory growth rates of the indicated yeast strains assayed in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol and supplemented with the indicated additives (mean ± SD, n = 3).
Established metabolic pathways provide hypotheses about the tyrosine to 4-HB pathway. The Ehrlich pathway is a general route to catabolize aromatic and branched-chain amino acids (16) (Fig. S1). After Aro8 and Aro9 convert tyrosine to 4-HPP, the aromatic α-keto acid decarboxylase Aro10 converts 4-HPP to 4-hydroxyphenylacetaldehyde (4-HPAA) (17). 4-HPAA is then reduced to tyrosol or oxidized to 4-hydroxyphenylacetate (4-HPA) depending on cellular needs (16), but it is unclear whether any of these intermediates contribute to 4-HB synthesis. Aspergillus niger readily converts 4-HPAA to 4-HB through a series of oxidations and decarboxylations resembling the mandelate pathway, although the enzymes involved are unidentified, and it is possible that S. cerevisiae converts 4-HPAA in a similar fashion (18, 19). There is also recent evidence for a nonoxidative route from 4-HPP to 4-HBz, as p-coumarate was shown to be incorporated into CoQ in both yeast and human cells (20). It has been proposed that p-coumarate could be hydrated and then cleaved by a carbon–carbon lyase to produce 4-HBz and acetate (14). Despite these promising hints, neither the oxidative nor nonoxidative pathway has been explored in detail. Here, we set out to identify intermediates and enzymes involved in converting tyrosine to 4-HB.
Results
Redundant tyrosine aminotransferase activity supports 4-HB synthesis
To study the fate of 4-HPP in 4-HB synthesis, we constructed a W303 Δaro8Δaro9Δaro2 strain as a negative control, hypothesizing that it would be unable to produce 4-HPP (Fig. 1A). Respiratory growth was only moderately impaired in –pABA glycerol media (Fig. 1B), and we observed only a moderate CoQ deficiency (Fig. 1C), consistent with BY4741 Δaro8Δaro9Δaro2 (14). We asked how Δaro8Δaro9Δaro2 yeast can make CoQ, given that both routes to 4-HPP synthesis (tyrosine aminotransferase and shikimate pathway) are presumably blocked. We reasoned that there is either redundant tyrosine aminotransferase activity present in this strain or a 4-HPP-independent route for 4-HB synthesis. To differentiate between these two models, we measured 4-HPP production in Δaro8Δaro9Δaro2 yeast. There was a small amount of 4-HPP made in this strain (Fig. 1D), which is likely sufficient to support 4-HB synthesis. Yeast possesses 12 α-amino acid aminotransferases including Aro8 and Aro9, and we hypothesized that the other ten of these enzymes are the most likely candidates for redundant tyrosine aminotransferase activity. Aminotransferases are typically able to catalyze transamination in both directions (albeit with different efficiencies), as demonstrated by the ability of both Aro8 and Aro9 to interconvert tyrosine and 4-HPP. Therefore, to test this hypothesis, we individually overexpressed all 12 α-amino acid aminotransferases in an Δaro8Δaro9 strain, which is auxotrophic for tyrosine because it cannot make enough de novo tyrosine from 4-HPP to support protein synthesis (21). Only Aro8, Aro9, Bna3, Bat2, and Aat2 were sufficient to confer tyrosine prototrophy, indicating that these enzymes can interconvert tyrosine and 4-HPP in vivo (Fig. 1E). Deleting Bna3, Bat2, and Aat2 from the Δaro8Δaro9Δaro2 strain blocked respiratory growth, which was rescuable with 4-HB supplementation (Fig. 1F). The Δaro8Δaro9Δaro2Δbna3Δbat2 and Δaro8Δaro9Δaro2Δaat2 strains are respiratory-competent, suggesting that BNA3, BAT2, and AAT2 must all be deleted to block CoQ synthesis. We also observed that 4-HB supplementation did not fully rescue Δaat2 strains, indicating that Aat2 is required for normal respiratory growth independent of 4-HB production. Altogether, these results establish that five aminotransferases—Aro8, Aro9, Bat2, Bna3, and Aat2—redundantly convert tyrosine to 4-HPP in vivo and emphasize the minute amount of 4-HPP production required for CoQ synthesis.
HFD1 is likely the only gene singly required for 4-HB production
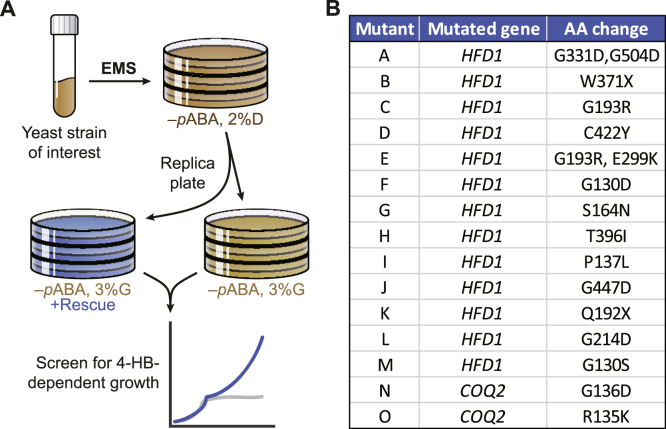
Thus far, HFD1 is the only known gene required for converting 4-HPP to 4-HB (Fig. 1A). In an attempt to discover other genes, we performed forward-genetic screens by mutagenizing yeast with ethyl methanesulfonate (EMS) and isolating colonies requiring exogenous 4-HB for respiratory growth in media lacking pABA (Fig. 2A). In total, we performed eight of these screens with various sensitized background strains that collectively yielded 15 mutant strains deficient in 4-HB synthesis (Fig. 2B; Table 1). Thirteen of these mutants harbor missense mutations in HFD1, and two harbor missense mutations in COQ2, which encodes a 4-HB polyprenyltransferase that functions downstream of 4-HB synthesis (22). Notably, both mutated Coq2 residues reside in its putative 4-HB binding site (23), and therefore these mutant strains likely require 4-HB supplementation to overcome decreased 4-HB affinity and catalytic efficiency. Our inability to detect mutations in additional genes while consistently recovering HFD1 suggests that other enzymes involved in 4-HB synthesis are likely either essential for growth under our screening conditions or perform a redundant function in the pathway. This redundancy could arise from multiple enzymes performing each step (like the aminotransferases above), the existence of multiple distinct pathways, or both.
Figure 2.
Forward-genetic screens consistently identify HFD1 as the only gene required for 4-HB production.A, general schematic of forward-genetic yeast screens for genes involved in 4-HB synthesis. B, summary of mutants and identified mutations recovered from eight forward-genetic screens.
Table 1.
Details of forward-genetic screens
| Mutant | Background strain | Control plate additive | Rescue plate additive | Control growth rate (mOD/min) | Control growth s.d. | Rescue growth rate (mOD/min) | Rescue growth s.d. | Mutated gene | AA change |
|---|---|---|---|---|---|---|---|---|---|
| A | BY4742 Δaro2Δaro10 | Water | 4-HB (10 μM) | −0.002 | 0.002 | 0.038 | 0.005 | HFD1 | G331D,G504D |
| B | BY4742 Δaro2Δaro10 | Water | 4-HB (10 μM) | 0.003 | 0.003 | 0.099 | 0.012 | HFD1 | W371X |
| C | BY4742 Δaro2Δaro10Δpdc5 | Water | 4-HB (10 μM) | 0.011 | 0.007 | 0.107 | 0.013 | HFD1 | G193R |
| D | BY4742 Δaro10Δpdc5 | Water | 4-HB (10 μM) | 0.005 | 0.005 | 0.308 | 0.024 | HFD1 | C422Y |
| E | BY4742 Δaro10Δpdc5Δpdc6Δabz1 | Water | 4-HB (10 μM) | 0.008 | 0.006 | 0.121 | 0.053 | HFD1 | G193R, E299K |
| F | BY4742 Δaro10Δpdc5Δpdc6Δabz1 | Water | 4-HB (10 μM) | 0 | 0.003 | 0.048 | 0.025 | HFD1 | G130D |
| G | W303 Δcoq6 + Coq6 G386A/N388D | 3,4-diHMA (1 mM) | 3,4-diHB (100 μM) | 0.042 | 0.017 | 0.449 | 0.052 | HFD1 | S164N |
| H | W303 Δcoq6 + Coq6 G386A/N388D | 3,4-diHMA (100 μM) | 3,4-diHB (30 μM) | 0.003 | 0.011 | 0.148 | 0.02 | HFD1 | T396I |
| I | W303 Δcoq6 + Coq6 G386A/N388D | 3,4-diHMA (100 μM) | 3,4-diHB (30 μM) | −0.004 | 0.002 | 0.45 | 0.038 | HFD1 | P137L |
| J | W303 Δaro8Δaro9Δaro2Δgly1 | Water | 4-HB (10 μM) | 0.017 | 0.151 | HFD1 | G447D | ||
| K | W303 Δaro8Δaro9Δaro2Δbat2Δbna3Δyer152c | Water | 4-HB (10 μM) | 0.007 | 0.001 | 0.12 | 0.009 | HFD1 | Q192X |
| L | W303 Δaro8Δaro9Δaro2Δbat2Δbna3Δyer152c | Water | 4-HB (10 μM) | 0 | 0.001 | 0.249 | 0.004 | HFD1 | G214D |
| M | W303 Δaro8Δaro9Δaro2Δbat2Δbna3Δyer152c | Water | 4-HB (10 μM) | 0.017 | 0.006 | 0.137 | 0.02 | HFD1 | G130S |
| N | W303 Δaro8Δaro9Δaro2Δbat2 | Water | 4-HB (10 μM) | 0.002 | 0.002 | 0.019 | 0.008 | COQ2 | G136D |
| O | W303 Δaro8Δaro9Δaro2Δgly1 | Water | 4-HB (10 μM) | −0.004 | 0.004 | 0.014 | 0.001 | COQ2 | R135K |
Targeted LC-MS identifies tyrosine catabolites
Because our extensive genetic screens did not identify new pathway enzymes, we next set out to map the intermediates between 4-HPP and 4-HBz by identifying tyrosine catabolites and 4-HB precursors. To determine whether potential pathway intermediates are bona fide tyrosine catabolites, we supplemented yeast with heavy (13C-labeled) tyrosine, extracted metabolites, and performed targeted liquid chromatography–mass spectrometry (LC-MS). In wild-type (WT) yeast, we observed multiple heavy-labeled compounds after supplementing with heavy tyrosine, some of which (e.g., 4-HPP, 4-HBz, 4-HB) have already been linked to 4-HB synthesis (Fig. 3A). Among the other catabolites we identified, the most abundant were tyrosol, 4-hydroxyphenylacetate (4-HPA), and 4-hydroxyphenyllactate (4-HPL). Phenylalanine catabolites phenylpyruvate (PP) and phenyllactate (PL) were not labeled, consistent with the documented inability of yeast to interconvert tyrosine and phenylalanine (24) (Fig. 3A). We constructed a model of the tyrosine to 4-HB pathway using our absolute quantification data from WT yeast, highlighting the low steady-state levels of 4-HBz and 4-HB compared with earlier intermediates (Fig. 3B).
Figure 3.
Targeted LC-MS identifies tyrosine catabolites.A, LC-MS absolute quantification of targeted light (12C) and heavy (13C) compounds from yeast grown in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol, supplemented with light (12C) or heavy (13C9) tyrosine (mean ± SD, n = 3). B and C, schematic of tyrosine catabolites in WT (B) or Δhfd1 (C) yeast. Circle areas are proportional to extract concentrations for each compound. D, respiratory growth rates of the indicated yeast strains assayed in-pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol and supplemented with the indicated additives (mean ± SD, n = 3). E and F, ratios of 4-HPP/4-HPL (E) or PP/PL (F) from the indicated yeast strains grown in -pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol.
Respiratory deficiency decreases 4-HPP/4-HPL ratio
Next, we repeated this targeted LC-MS approach with Δhfd1 yeast to see if any upstream intermediates accumulate in this strain. 4-HB was heavily decreased, as expected, but we noted an unexpected decrease in 4-HPP, 4-HPA, and 4-HBz relative to WT yeast (Fig. 3C). A possible explanation for these changes is the concomitant increase in 4-HPL abundance in Δhfd1. This altered 4-HPP/4-HPL ratio may be due specifically to loss of Hfd1 or may be the result of general respiratory deficiency. In the latter case, reducing 4-HPP to 4-HPL could act as an electron sink to regenerate depleted oxidized redox cofactors such as NAD+ in a manner analogous to the reduction of pyruvate to lactate (25). To distinguish between these possibilities, we performed targeted LC-MS on Δhfd1 yeast, whose respiratory deficiency could be rescued with pABA, and Δcoq2 yeast, a respiratory-deficient control (Fig. 3D). The 4-HPP/4-HPL ratio was sharply decreased in Δcoq2 yeast, matching Δhfd1, and rescuing Δhfd1 respiratory growth with pABA restored the 4-HPP/4-HPL ratio (Fig. 3E). Similar results were observed with the analogous phenylalanine catabolites PP and PL, demonstrating that these changes are not specific to tyrosine metabolism (Fig. 3F). These data suggest that the perturbed 4-HPP/4-HPL ratio in Δhfd1 stems from an indirect effect of respiratory deficiency rather than the lack of Hfd1, explaining the counterintuitive observation that upstream intermediates decrease in abundance in Δhfd1 yeast. More broadly, this result suggests that 4-HPP and PP have underappreciated roles in buffering the cellular redox state during perturbed cellular respiration.
Proposed intermediates rescue yeast in vivo
Having identified intermediates derived from tyrosine, we next asked which of these compounds can be converted to 4-HB in vivo. Potential 4-HB precursors were added to Δaro8Δaro9Δaro2 yeast to determine whether they could rescue this strain's respiratory growth defect. Of the tested compounds, 4-HPAA and 4-hydroxymandelate (4-HMA) rescued most strongly (Fig. 4A). Notably, 4-HPA and 4-HPL, compounds that were abundantly labeled in the targeted LC-MS assay, did not rescue respiratory growth. We verified that 4-HPAA and 4-HMA do not rescue Δhfd1 yeast, demonstrating that their conversion to 4-HB depends on Hfd1 activity and that there is no appreciable 4-HB contamination in these compound stocks (Fig. 4B).
Figure 4.
Novel 4-HB precursors rescue respiratory growth of CoQ-deficient yeast.A–C, respiratory growth rates of the indicated yeast strains assayed in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol and supplemented with the indicated additives (mean ± SD, n = 3). D, updated schematic of 4-HB synthesis.
To further validate these findings, we performed rescue experiments in a Coq6 “bypass” strain. Coq6 is an essential enzyme in the CoQ biosynthesis pathway that hydroxylates the CoQ precursor polyprenyl hydroxybenzoate (PPHB) at the C3 position. However, a strain containing an activity-dead Coq6 mutant (G386A, N388D) can be bypassed with a “pre-hydroxylated” 3,4-dihydroxybenzoate (3,4-diHB) (26). We tested whether 3,4-dihydroxy analogs of various hypothesized 4-HB precursors could also rescue the Coq6-dead strain. 3,4-dihydroxyphenylacetaldehyde (3,4-diHPAA) and 3,4-dihydroxymandelate (3,4-diHMA) rescued respiratory growth while 3,4-dihydroxyphenylacetate (3,4-diHPA) did not, consistent with the results with the 4-hydroxy compounds in Δaro8Δaro9Δaro2 (Fig. 4C). Once again, Hfd1 was required for conversion to 4-HB.
Paradoxically, the most abundant tyrosine catabolites observed with targeted LC-MS (4-HPA and 4-HPL) did not rescue our 4-HB-deficient strains, while the compounds that rescued best (4-HPAA and 4-HMA) were not observed and verified as tyrosine catabolites (Fig. 4D). A possible explanation is that compounds that we observe at steady state might be rate-limiting or unproductive intermediates, while compounds we do not observe might be converted too quickly to accumulate. Of note, we were unable to measure 4-HPAA in the LC-MS assay, so it is unclear if a pool of 4-HPAA is present in vivo.
Aro10 is dispensable for 4-HB synthesis
Multiple compounds including 4-HPAA, 4-HPA, and 4-HMA are likely positioned downstream of the Ehrlich pathway aromatic α-keto acid decarboxylase Aro10 (Fig. 4D; Fig. S1). To further examine whether these compounds support 4-HB synthesis, we deleted Aro10 and the potentially redundant decarboxylases Pdc5 and Pdc6 (27). The Δaro10Δpdc5Δpdc6 strain had elevated levels of 4-HPP, the putative substrate of Aro10 in tyrosine catabolism (Fig. 5A). 4-HPL levels were similarly increased (Fig. 5B), supporting the hypothesis that 4-HPL exists in an equilibrium with 4-HPP as an electron sink. 4-HPA and tyrosol, intermediates likely downstream of the Aro10 product 4-HPAA, were sharply decreased but not eliminated, providing further evidence that Aro10 decarboxylates 4-HPP to 4-HPAA in WT yeast (Fig. 5, C and D). However, the effect of these knockouts on 4-HB synthesis was minimal, as 4-HBz and 4-HB levels were unchanged or even increased (Fig. 5, E and F). This demonstrates that Aro10 is dispensable for 4-HB synthesis and suggests that either the smaller pool of 4-HPAA in Δaro10Δpdc5Δpdc6 is sufficient to make the same amount of 4-HB or that 4-HPAA is simply not converted to 4-HB.
Figure 5.
Ehrlich pathway 4-HPP decarboxylase Aro10 is not required for 4-HB synthesis.A–F, relative levels of light (12C) and heavy (13C) compounds from yeast grown in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol, supplemented with light (12C) or heavy (13C9) tyrosine (mean ± SD, n = 3). G, respiratory growth rates of the indicated yeast strains assayed in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol and supplemented with the indicated additives (mean ± SD, n = 3). H, CoQ6 levels from the indicated yeast strains grown in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol (mean ± SD, n = 3). I, respiratory growth rates of the indicated yeast strains assayed in –pABA media containing 0.1% (w/v) glucose and 3% (w/v) glycerol and supplemented with the indicated additives (mean ± SD, n = 3). J, CoQ6 levels from the indicated yeast strains grown in –pABA, –Ura media containing 0.1% (w/v) glucose and 3% (w/v) glycerol (mean ± SD, n = 3).
If 4-HPAA is converted to 4-HB, we reasoned that Δaro8Δaro9Δaro2 yeast, which has decreased levels of 4-HPP and downstream compounds, might be sensitive to Aro10 deletion. However, deleting Aro10 and Pdc5 from Δaro8Δaro9Δaro2 did not affect respiratory growth (Fig. 5G) or CoQ levels (Fig. 5H). We also hypothesized that overexpressing Aro10 in Δaro8Δaro9Δaro2 yeast might help or harm 4-HB synthesis if 4-HPP conversion to 4-HPAA is productive or unproductive, respectively. However, we observe no change in respiratory growth (Fig. 5I) or CoQ levels (Fig. 5J) upon Aro10 overexpression. Overall, these results call into question the recent claim that Aro10 is involved in 4-HBz synthesis in S. cerevisiae (28, 29).
Discussion
Studies of genetic robustness in yeast suggest that, even though only 10–20% of genes are essential, most genes are involved in functions that can only be interrogated with multiple genetic or environmental perturbations (30, 31). Our results demonstrate that the yeast tyrosine-to-4-HB pathway contains significant redundancy and indicate that multiple gene knockouts are required to ablate steps prior to Hfd1's conversion of 4-HBz to 4-HB. For example, the first reaction in the pathway, transamination of tyrosine to 4-HPP, is catalyzed primarily by Aro8 and Aro9, but we observe that deleting these two enzymes is not sufficient to block this step. We determine that three additional aminotransferases—Bat2, Bna3, and Aat2—are sufficient to perform this reaction in vivo and that deletion all five of these enzymes is required to block 4-HPP production from tyrosine. Thus, yeast α-amino acid aminotransferases have a broader substrate specificity than previously thought, as five of 12 of these enzymes accept tyrosine as a substrate in vivo. The last step in the 4-HB synthesis pathway is catalyzed by Hfd1, another broad-specificity enzyme whose substrates range from small aromatic aldehydes (4-HBz in CoQ synthesis) to long-chain fatty aldehydes (hexadecanal in sphingolipid catabolism) (14, 15). These examples suggest that other pathway enzymes might also have broad substrate specificities, and we expect that the relatively low amount of 4-HB required for respiratory growth means that even “side activities” could suffice.
We used two strategies to identify metabolic intermediates that could be participating in 4-HB synthesis. First, we observed bona fide tyrosine catabolites with isotopic labeling, including 4-HPL, 4-HPA, and tyrosol. Second, we identified compounds that boost respiratory growth when added exogenously, including 4-HPAA and 4-HMA. Surprisingly, we found that the heavy tyrosine catabolites that accumulate in vivo do not match the compounds that rescue yeast when supplemented. Further work will be required to fully resolve this paradox, but it is possible that the compounds that accumulate at steady state are simply “waste” or “cul-de-sac” products. This notion is consistent with the putative roles of tyrosol and 4-HPA as Ehrlich pathway end products (16) and our hypothesis that 4-HPL exists in a separate equilibrium with 4-HPP. Regardless, assigning intermediates to this pathway with certainty will require additional genetic or biochemical evidence.
S. cerevisiae underwent an ancient whole-genome duplication (WGD) event that resulted in >1000 paralogous genes, contributing to genetic robustness (32, 33). Therefore, it may be beneficial to study 4-HB synthesis in model organisms with less genomic redundancy. To this end, work is underway in our laboratory to study this pathway in yeast species that have not undergone WGD. Pathways from phenylalanine to 4-HB have been identified in plants, but these do not account for the entirety of 4-HB synthesis in A. thaliana (34, 35). It will also be important to study 4-HB synthesis in mammalian cells, given the importance of CoQ to human health. 4-HB is present in human blood at ∼2 μM (36), and it is unclear whether different cell types produce their own 4-HB or obtain it from the bloodstream. We have shown that the human aldehyde dehydrogenase ALDH3A1 rescues CoQ synthesis in Δhfd1 yeast (15), but it is unknown whether human cells convert 4-HBz to 4-HB.
In summary, we show that the S. cerevisiae 4-HB synthesis pathway is robust to single genetic alterations. We demonstrate that deleting three additional enzymes is required to block the transamination of tyrosine to 4-HPP in the absence of established enzymes Aro8 and Aro9. We expect that the subsequent steps are also redundant, and our isotopic labeling and chemical genetics approaches nominate several new metabolic intermediates that may aid in identifying the responsible enzymes. From this work in yeast, we suggest that unresolved cases of human CoQ deficiency could result from mutations in cytosolic aminotransferases or oxidoreductases, with or without assigned biochemical functions.
Experimental procedures
Yeast strains and cultures
S. cerevisiae haploid W303 (MATa his3 leu2 met15 trp1 ura3) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) yeast were used. Yeast deletion strains were generated using standard homologous recombination or CRISPR-mediated methods. For homologous recombination, open reading frames were replaced with the KanMX6, HygMX6, or NatMX6 cassette as previously described (37). CRISPR-mediated deletions were performed as described (38, 39). 20-mer guide sequences were designed with the ATUM CRISPR gRNA design tool (https://www.atum.bio/eCommerce/cas9/input) and cloned into pRCC-K, and 500 ng of the guide-inserted pRCC-K was used per yeast transformation. Donor DNA was 300 pmol of an 80-nt Ultramer consisting of 40 bp upstream and 40 bp downstream of the ORF (for scarless deletions) or ∼6 μg of PCR-amplified Longtine cassette with flanking homology 40 bp upstream and 40 bp downstream of the ORF (for cassette-replacement deletions). Deletions were confirmed by a PCR assay or Sanger sequencing.
Rich media contained yeast extract (1% w/v, DOT Scientific), peptone (2% w/v, DOT Scientific), and the indicated carbon source. Synthetic complete (and dropout) media contained drop-out mix (US Biological), yeast nitrogen base (with ammonium sulfate and without amino acids) (US Biological), and the indicated carbon source. –pABA (and dropout) media contained Complete Supplement Mixture (Formedium), Yeast Nitrogen Base without amino acids and without pABA (Formedium), and the indicated carbon source. Rich media were sterilized by autoclave (20 min exposure), and synthetic media were sterilized by filtration (0.22 μm pore size).
Respiratory growth assays
Individual colonies of S. cerevisiae were used to inoculate pABA-limited media (2% w/v glucose, 100 nM pABA) starter cultures, which were incubated for 24 h (30 °C, 230 rpm). Yeast were swapped into –pABA media with glucose (0.1%, w/v), glycerol (3%, w/v), and MES (50 mM, pH 5.6) at an initial density of 5 × 106 cells/ml with indicated additives. The cultures were incubated (30 °C, 1140 rpm) in an Epoch2 plate reader (BioTek) in a sterile 96-well polystyrene round-bottom microwell plate (Thermo) with a Breathe-Easy cover seal (Diversified Biotech). Optical density readings (A600) were obtained every 10 min, and growth rates were calculated with Gen5 v3.02.2 software (BioTek), excluding time points before the diauxic shift and during stationary phase growth.
Drop assay
Individual colonies of yeast transformed with p415 GPD constructs were used to inoculate starter cultures (–Leu, –pABA, 2% w/v glucose, 1 μM pABA), which were incubated overnight (30 °C, 230 rpm). Cells were spun down (21,000g, 2 min) and resuspended in water. Serial dilutions of yeast (105, 104, 103, 102, or 10 cells) were dropped onto 1.5% w/v agar plates (–Leu, 2% w/v glucose or –Tyr, –Leu, –pABA, 2% w/v glucose, 1 μM pABA) and incubated (30 °C, 3 days).
Forward-genetic screens
Individual colonies of yeast were used to inoculate YEPD starter cultures, which were incubated overnight. 1.0 × 108 cells were pelleted, washed once with sterile water, and resuspended in 2.5 ml of 100 mM sodium phosphate buffer, pH 7.0. Ethyl methanesulfonate (EMS) (80 μl) was added, and cells were incubated (90 min, 30 °C, 230 rpm). Cells were washed thrice with sodium thiosulfate (5% w/v) to inactivate EMS. Cells were resuspended in water, and 1.0 × 104 cells were plated on pABA-limited (2% w/v glucose, 100 nM pABA) plates. After 3 days, cells were replica-plated onto –pABA (3% glycerol, w/v) plates with 0 μM or 10 μM 4-HB (Sigma), or other rescue compounds if indicated in Table 1. Colonies that grew on plates with rescue compound were picked into YEPD overnight cultures and struck on YEPD plates, and respiratory growth phenotypes were confirmed with plate reader growth assays (see above). Mutations were identified with Sanger sequencing.
Plasmid cloning
Expression plasmids were cloned with standard restriction enzyme cloning methods. ORF-specific primers were used to amplify genes from W303 yeast genomic DNA, amplicons were ligated into digested p415 GPD plasmid (40), and ligation products were transformed into E. coli DH5α chemically competent cells (NEB). Plasmids were isolated from transformants, and Sanger sequencing was used to identify those containing the correct insertion.
Targeted HPLC-ECD for yeast CoQ6
CoQ measurements were performed generally as described (39). Individual colonies of S. cerevisiae were used to inoculate pABA-limited media (2% w/v glucose, 100 nM pABA) starter cultures, which were incubated for 24 h (30 °C, 230 rpm). Yeast was swapped into –pABA media with glucose (0.1%, w/v) and glycerol (3%, w/v) and incubated for 24 h (30 °C, 230 rpm). 1 × 108 cells were collected by centrifugation (4000g, 5 min), resuspended in 1 ml water, transferred to 1.5 ml screw-cap tubes, and centrifuged (21,000g, 1 min). In total, 50 μl of KCl (150 mM) and 100 μl of glass beads (0.5 mm; RPI) were added; 600 μl of ice-cold methanol (with 1 μM CoQ8 internal standard) was added, and samples were vortexed (10 min, 4 °C). In total, 400 μl of petroleum ether was added to extract lipids, and samples were vortexed (3 min, 4 °C) and centrifuged (1000g, 3 min) to separate phases. The petroleum ether (upper) layer was collected, and the extraction was repeated with another round of petroleum ether (400 μl), vortexing (3 min, 4 °C), and centrifugation (1000g, 3 min). The petroleum ether layers were pooled and dried under argon. Lipids were resuspended in 2-propanol (25 μl) and transferred to amber glass vials (Sigma; QSertVial, 12 × 32 mm, 0.3 ml). Sodium borohydride (1 μl of 10 mM in 2-propanol) was added to reduce quinones, and samples were vortexed briefly and incubated (5–10 min). Methanol (24 μl) was added to remove excess sodium borohydride, and samples were vortexed briefly and incubated (5–10 min). Samples were briefly flushed with nitrogen gas.
Samples were analyzed by reverse-phase high-pressure liquid chromatography with electrochemical detection (HPLC-ECD) using a C18 column (Thermo Scientific, Betasil C18, 100 × 2.1 mm, particle size 3 μm) at a flow rate of 0.3 ml/min with a mobile phase of 75% methanol, 20% 2-propanol, and 5% ammonium acetate (1 M, pH 4.4). After separation on the column, quinones were quantified on ECD detector (Thermo Scientific ECD3000-RS) equipped with 6020RS omni Coulometric Guarding Cell "E1" and 6011RS ultra Analytical Cell "E2" and "E3." To prevent premature quinone oxidation, the E1 guarding electrode was set to −200 mV. Measurements were made using the analytical E2 electrode operating at 600 mV after complete oxidation of the quinone sample, and E3 electrode (600 mV) was used to ensure that total signal was recorded on the E2 cell. For each experiment, a CoQ6 standard in 2-propanol was also prepared with sodium borohydride and methanol treatment, and different volumes were injected to make a standard curve. Quinones were quantified by integrating respective peaks using the Chromeleon 7.2.10 software (Thermo) and normalized to CoQ8 internal standard.
Targeted LC-MS
Individual colonies of S. cerevisiae were used to inoculate pABA-limited media (2% w/v glucose, 100 nM pABA) starter cultures, which were incubated for 24 h (30 °C, 230 rpm). 1.2 × 107 cells were transferred to 60 ml of light (–pABA, 0.1% w/v glucose, 3% w/v glycerol, 10 nM pABA) or heavy (–pABA, –Tyr, 0.1% w/v glucose, 3% w/v glycerol, 10 nM pABA, 276 μM 13C9,15N-Tyr) growth media and incubated for 24 h (30 °C, 230 rpm). 5 × 108 cells were isolated by rapid vacuum filtration onto a nylon filter membrane (0.45 μm pore size, Millipore) using a Glass Microanalysis Filter Holder (Millipore), briefly washed with phosphate buffered saline (3 ml), and immediately submerged into extraction solvent (ACN/MeOH/H2O 2:2:1 v/v/v, 1.5 ml, −20 °C) containing internal standards (100 nM 3-hydroxybenzoate and 100 nM 3-hydroxyphenylhydracrylate) in plastic screw-cap tubes. Tubes with the extraction solvent, nylon filter, and yeast were stored at −80 °C prior to analysis.
Samples were vortexed for 30 s twice, and the nylon membrane filters were removed with tweezers prior to centrifuging (13,000g, 5 min, 4 °C). Supernatants (1 ml) were transferred to new 2 ml Eppendorf tubes with the addition of an internal standard (m-coumaric acid) to 20 μM. Samples were dried in a vacuum concentrator. Once dried, samples were reconstituted in 100 μl H2O. 2 ml Eppendorf tubes containing resuspended samples were vortexed 15 s prior to transfer to autosampler glass inserts. Standards were prepared from 25 or 50 mM stock solutions of each compound and diluted to bracket the concentration range 800 to 0.391 μM (for tyrosine), 500 to 0.244 μM (for phenylalanine), 100 to 0.049 μM (for 4-hydroxymandelic acid, 3-phenyllactic acid, and tyrosol), and 8 to 0.004 μM (for remaining compounds) with 12 1:1 dilution steps.
LC-MS/MS analyses were performed using a randomized sample order. An Ultimate HPG-3400RS pump and WPS-3000RS autosampler (Thermo Fisher) were mated to an ACQUITY UPLC HSS T3 column (2.1 × 150 mm, 1.8 μm particle diameter, Waters Corporation) with an ACQUITY UPLC HSS T3 VanGuard Pre-Column (2.1 × 5 mm, 1.8 μm particle diameter, Waters Corporation). The autosampler was maintained at 4 °C. Mobile phase A was 0.1% aqueous acetic acid and mobile phase B was ACN. The column was heated to 35 °C. Mobile phase B was increased from 0% to 25% over 12.5 min at a flow rate of 0.4 ml/min and then increased to 80% over 1 min along with an increase in flow rate to 0.5 ml/min. Mobile phase B was held at 80% for 2 min before being decreased to 0% over 1 min. Mobile phase B was held at 0% for 6 min for column re-equilibration before next injection. The LC system was coupled to a TSQ Quantiva Triple Quadrupole mass spectrometer (Thermo Scientific) by a heated ESI source. The ion transfer tube and vaporizer temperatures were kept at 350 °C. Sheath Gas was set to 28 units, Auxiliary Gas to 14 units, and Sweep Gas to 0 unit. Negative and positive spray voltages were time-dependent and set to 3600V and 3800V respectively. For targeted analysis, the MS was operated in single reaction monitoring (SRM) mode acquiring scheduled, targeted scans to quantify selected compound transitions. At least one transition was used for each compound. All transitions, collision energies, and RF Lens voltages were previously optimized by infusion of each compound. The retention time window for each compound or groups of compounds was optimized from prior experiments. MS acquisition parameters were 0.7 FWHM resolution for Q1 and 1.2 FWHM for Q3, 0.6 s cycle time, 1.5 mTorr CID gas, and 20 s Chrom Filter.
LC-MS/MS data were analyzed as described (41). Raw files were processed using Xcalibur Quan Browser (v4.0.50, Thermo Scientific) with results exported and further processed using Microsoft Excel 2010. Quantification was achieved through use of an external calibration curve constructed from the analysis of aqueous standard mixtures. Signal was not linear across the entire measured range for some compounds, and so calibrants were selected to bracket all samples, when possible, with a minimum of five calibrant levels. For highly abundant compounds, more dilute samples were used for quantification to stay below the upper limit of the calibration range.
Statistical analysis
All experiments were performed in at least biological triplicate, unless stated otherwise. In all cases, "mean" refers to arithmetic mean, and "SD" refers to sample standard deviation. Statistical analyses were performed using Microsoft Excel. In all cases, n represents biological replicates of an experiment.
Data availability
All data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
J. J. C. is a consultant for Thermo Fisher Scientific.
Acknowledgments
We would like to thank Pagliarini lab members for helpful discussions, Matt Stefely for figure assistance, and Jodi Nunnari for providing the Coq6-dead yeast strain.
Author contributions
K. P. R. and D. J. P. conceptualization; K. P. R., J. J. C., and D. J. P. funding acquisition; K. P. R., A. J., S. E. J., T. R. R., and J. D. R investigation; K. P. R., T. R. R., and J. D. R. methodology; D. J. P. supervision; K. P. R. and D. J. P. writing—original draft; K. P. R., A. J., S. E. J., T. R. R., J. D. R., J. J. C., and D. J. P. writing—review and editing.
Funding and additional information
This work was supported by National Institutes of Health (NIH) grant R35GM131795 and funds from the BJC Investigator Program (to D. J. P.), NIH T32AG000213 and a National Science Foundation Graduate Research Fellowship DGE-1747503 (to K. P. R.), NIH P41GM108538 (to J. J. C. and D. J. P.), and U.S. Department of Energy grant DE-SC0018409. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Ronald Wek
Supporting information
References
- 1.Lester R.L., Crane F.L. The natural occurrence of coenzyme Q and related compounds. J. Biol. Chem. 1959;234:2169–2175. [PubMed] [Google Scholar]
- 2.Turunen M., Olsson J., Dallner G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Munier-Lehmann H., Vidalain P.O., Tangy F., Janin Y.L. On dihydroorotate dehydrogenases and their inhibitors and uses. J. Med. Chem. 2013;56:3148–3167. doi: 10.1021/jm301848w. [DOI] [PubMed] [Google Scholar]
- 4.Ziosi M., Di Meo I., Kleiner G., Gao X.H., Barca E., Sanchez-Quintero M.J., Tadesse S., Jiang H., Qiao C., Rodenburg R.J., Scalais E., Schuelke M., Willard B., Hatzoglou M., Tiranti V. Coenzyme Q deficiency causes impairment of the sulfide oxidation pathway. EMBO Mol. Med. 2017;9:96–111. doi: 10.15252/emmm.201606356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acosta M.J., Vazquez Fonseca L., Desbats M.A., Cerqua C., Zordan R., Trevisson E., Salviati L. Coenzyme Q biosynthesis in health and disease. Biochim. Biophys. Acta. 2016;1857:1079–1085. doi: 10.1016/j.bbabio.2016.03.036. [DOI] [PubMed] [Google Scholar]
- 6.Salviati L., Trevisson E., Doimo M., Navas P. Primary coenzyme Q10 deficiency. In: Pagon R.A., Adam M.P., Ardinger H.H., editors. GeneReviews [Internet] University of Washington, Seattle; Seattle, WA: 2017. [Google Scholar]
- 7.Stefely J.A., Pagliarini D.J. Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem. Sci. 2017;42:824–843. doi: 10.1016/j.tibs.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olson R.E., Bentley R., Aiyar A.S., Dialameh G.H., Gold P.H., Ramsey V.G., Springer C.M. Benzoate derivatives as intermediates in the biosynthesis of coenzyme Q in the rat. J. Biol. Chem. 1963;238:3146–3148. [PubMed] [Google Scholar]
- 9.Cox G.B., Gibson F. Biosynthesis of vitamin K and ubiquinone. Relation to the shikimic acid pathway in Escherichia coli. Biochim. Biophys. Acta. 1964;93:204–206. doi: 10.1016/0304-4165(64)90285-5. [DOI] [PubMed] [Google Scholar]
- 10.Pierrel F., Hamelin O., Douki T., Kieffer-Jaquinod S., Muhlenhoff U., Ozeir M., Lill R., Fontecave M. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 2010;17:449–459. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 11.Marbois B., Xie L.X., Choi S., Hirano K., Hyman K., Clarke C.F. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2010;285:27827–27838. doi: 10.1074/jbc.M110.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edman J.C., Goldstein A.L., Erbe J.G. Para-aminobenzoate synthase gene of Saccharomyces cerevisiae encodes a bifunctional enzyme. Yeast. 1993;9:669–675. doi: 10.1002/yea.320090613. [DOI] [PubMed] [Google Scholar]
- 13.Botet J., Mateos L., Revuelta J.L., Santos M.A. A chemogenomic screening of sulfanilamide-hypersensitive Saccharomyces cerevisiae mutants uncovers ABZ2, the gene encoding a fungal aminodeoxychorismate lyase. Eukaryot. Cell. 2007;6:2102–2111. doi: 10.1128/EC.00266-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payet L.A., Leroux M., Willison J.C., Kihara A., Pelosi L., Pierrel F. Mechanistic details of early steps in coenzyme Q biosynthesis pathway in yeast. Cell Chem. Biol. 2016;23:1241–1250. doi: 10.1016/j.chembiol.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Stefely J.A., Kwiecien N.W., Freiberger E.C., Richards A.L., Jochem A., Rush M.J.P., Ulbrich A., Robinson K.P., Hutchins P.D., Veling M.T., Guo X., Kemmerer Z.A., Connors K.J., Trujillo E.A., Sokol J. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat. Biotechnol. 2016;34:1191–1197. doi: 10.1038/nbt.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazelwood L.A., Daran J.M., van Maris A.J., Pronk J.T., Dickinson J.R. The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008;74:2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kneen M.M., Stan R., Yep A., Tyler R.P., Saehuan C., McLeish M.J. Characterization of a thiamin diphosphate-dependent phenylpyruvate decarboxylase from Saccharomyces cerevisiae. FEBS J. 2011;278:1842–1853. doi: 10.1111/j.1742-4658.2011.08103.x. [DOI] [PubMed] [Google Scholar]
- 18.Kishore G., Sugumaran M., Vaidyanathan C.S. Metabolism of Dl-(+/-)-phenylalanine by Aspergillus-Niger. J. Bacteriol. 1976;128:182–191. doi: 10.1128/jb.128.1.182-191.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramakrishna Rao D.N., Vaidyanathan C.S. Metabolism of mandelic acid by Neurospora crassa. Can. J. Microbiol. 1977;23:1496–1499. doi: 10.1139/m77-222. [DOI] [PubMed] [Google Scholar]
- 20.Xie L.X., Williams K.J., He C.H., Weng E., Khong S., Rose T.E., Kwon O., Bensinger S.J., Marbois B.N., Clarke C.F. Resveratrol and para-coumarate serve as ring precursors for coenzyme Q biosynthesis. J. Lipid Res. 2015;56:909–919. doi: 10.1194/jlr.M057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urrestarazu A., Vissers S., Iraqui I., Grenson M. Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol. Gen. Genet. 1998;257:230–237. doi: 10.1007/s004380050643. [DOI] [PubMed] [Google Scholar]
- 22.Ashby M.N., Kutsunai S.Y., Ackerman S., Tzagoloff A., Edwards P.A. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:polyprenyltransferase. J. Biol. Chem. 1992;267:4128–4136. [PubMed] [Google Scholar]
- 23.Cheng W., Li W. Structural insights into ubiquinone biosynthesis in membranes. Science. 2014;343:878–881. doi: 10.1126/science.1246774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braus G.H. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: A model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 1991;55:349–370. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 26.Ozeir M., Muhlenhoff U., Webert H., Lill R., Fontecave M., Pierrel F. Coenzyme Q biosynthesis: Coq6 is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem. Biol. 2011;18:1134–1142. doi: 10.1016/j.chembiol.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Romagnoli G., Luttik M.A., Kotter P., Pronk J.T., Daran J.M. Substrate specificity of thiamine pyrophosphate-dependent 2-oxo-acid decarboxylases in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012;78:7538–7548. doi: 10.1128/AEM.01675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valera M.J., Boido E., Ramos J.C., Manta E., Radi R., Dellacassa E., Carrau F. The mandelate pathway, an alternative to the PAL pathway for the synthesis of benzenoids in yeast. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.00701-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valera M.J., Zeida A., Boido E., Beltran G., Torija M.J., Mas A., Radi R., Dellacassa E., Carrau F. Genetic and transcriptomic evidences suggest ARO10 genes are involved in benzenoid biosynthesis by yeast. Yeast. 2020;37:427–435. doi: 10.1002/yea.3508. [DOI] [PubMed] [Google Scholar]
- 30.Deutscher D., Meilijson I., Kupiec M., Ruppin E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat. Genet. 2006;38:993–998. doi: 10.1038/ng1856. [DOI] [PubMed] [Google Scholar]
- 31.Harrison R., Papp B., Pal C., Oliver S.G., Delneri D. Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2307–2312. doi: 10.1073/pnas.0607153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon J.L., Byrne K.P., Wolfe K.H. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu Z., Steinmetz L.M., Gu X., Scharfe C., Davis R.W., Li W.H. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 34.Block A., Widhalm J.R., Fatihi A., Cahoon R.E., Wamboldt Y., Elowsky C., Mackenzie S.A., Cahoon E.B., Chapple C., Dudareva N., Basset G.J. The origin and biosynthesis of the benzenoid moiety of ubiquinone (coenzyme Q) in arabidopsis. Plant Cell. 2014;26:1938–1948. doi: 10.1105/tpc.114.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soubeyrand E., Johnson T.S., Latimer S., Block A., Kim J., Colquhoun T.A., Butelli E., Martin C., Wilson M.A., Basset G.J. The peroxidative cleavage of Kaempferol contributes to the biosynthesis of the benzenoid moiety of ubiquinone in plants. Plant Cell. 2018;30:2910–2921. doi: 10.1105/tpc.18.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loke W.M., Jenner A.M., Proudfoot J.M., McKinley A.J., Hodgson J.M., Halliwell B., Croft K.D. A metabolite profiling approach to identify biomarkers of flavonoid intake in humans. J. Nutr. 2009;139:2309–2314. doi: 10.3945/jn.109.113613. [DOI] [PubMed] [Google Scholar]
- 37.Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Generoso W.C., Gottardi M., Oreb M., Boles E. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J. Microbiol. Methods. 2016;127:203–205. doi: 10.1016/j.mimet.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 39.Kemmerer Z.A., Robinson K.P., Schmitz J.M., Paulson B.R., Jochem A., Hutchins P.D., Coon J.J., Pagliarini D.J. UbiB proteins regulate cellular CoQ distribution. bioRxiv. 2020 doi: 10.1101/2020.12.09.418202. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Serate J., Xie D., Gajbhiye S., Kulzer P., Sanford G., Russell J.D., McGee M., Foster C., Coon J.J., Landick R., Sato T.K. Production of hydrolysates from unmilled AFEX-pretreated switchgrass and comparative fermentation with Zymomonas mobilis. Bioresour. Technol. Rep. 2020;11 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.