Abstract
Simple Summary
Machine learning (ML) is a form of artificial intelligence that could be used to enhance the efficiency of developing accurate prediction models for survival outcomes with cancer medicines, which is critical in informing disease prognosis and care planning. We used data from two recent clinical trials to develop and validate ML‐based clinical prediction models of the overall and progression‐free survival rates in patients with urothelial cancer initiating the immune checkpoint inhibitor (ICI) atezolizumab. We demonstrated that ML can efficiently develop an accurate prediction model of survival, enable an accurate prognostic risk classification, and provide realistic expectations of treatment outcomes in patients undergoing urothelial cancer-initiating ICIs therapy.
Abstract
Machine learning (ML) may enhance the efficiency of developing accurate prediction models for survival, which is critical in informing disease prognosis and care planning. This study aimed to develop an ML prediction model for survival outcomes in patients with urothelial cancer-initiating atezolizumab and to compare model performances when built using an expert-selected (curated) versus an all-in list (uncurated) of variables. Gradient-boosted machine (GBM), random forest, Cox-boosted, and penalised, generalised linear models (GLM) were evaluated for predicting overall survival (OS) and progression-free survival (PFS) outcomes. C-statistic (c) was utilised to evaluate model performance. The atezolizumab cohort in IMvigor210 was used for model training, and IMvigor211 was used for external model validation. The curated list consisted of 23 pretreatment factors, while the all-in list consisted of 75. Using the best-performing model, patients were stratified into risk tertiles. Kaplan–Meier analysis was used to estimate survival probabilities. On external validation, the curated list GBM model provided slightly higher OS discrimination (c = 0.71) than that of the random forest (c = 0.70), CoxBoost (c = 0.70), and GLM (c = 0.69) models. All models were equivalent in predicting PFS (c = 0.62). Expansion to the uncurated list was associated with worse OS discrimination (GBM c = 0.70; random forest c = 0.69; CoxBoost c = 0.69, and GLM c = 0.69). In the atezolizumab IMvigor211 cohort, the curated list GBM model discriminated 1-year OS probabilities for the low-, intermediate-, and high-risk groups at 66%, 40%, and 12%, respectively. The ML model discriminated urothelial-cancer patients with distinctly different survival risks, with the GBM applied to a curated list attaining the highest performance. Expansion to an all-in approach may harm model performance.
Keywords: machine learning, survival outcomes, immune checkpoint inhibitors, gradient boosting, random forest
1. Introduction
Urothelial cancer is an aggressive malignancy associated with about 200,000 global deaths annually and a 5-year survival rate of about 5% in the metastatic setting [1,2]. Immune-checkpoint inhibitors (ICIs) targeting the programmed death-1 (PD-1) pathway, such as atezolizumab and pembrolizumab, are an important emerging treatment option for metastatic urothelial cancer. Nonetheless, there is substantial heterogeneity in response to ICIs, and identifying individuals upfront that are most likely to respond to this treatment is a clinical priority [3].
Clinical prediction models of survival outcomes integrating clinicopathological predictors using data from large cohorts of patients may enable improved decision making and identify patients with different therapeutic prognoses [4]. While some prediction models for urothelial cancer exist in the literature [5,6,7], these models were developed using traditional statistical approaches (e.g., Cox regression), assessed a small number of predictors, and have insufficient evidence for clinical use, as predictors can differ significantly between cancer types or subtypes and treatments [8,9,10].
Machine learning (ML), a form of artificial intelligence, is an emerging alternative that may efficiently develop accurate clinical prediction models that can deal with high-dimensional data, and identify complex relationships between variables and outcomes that may be unidentifiable with traditional statistical approaches [11,12,13]. There were several recent breakthroughs demonstrating how using ML to rapidly interrogate complex data delivers a more efficient use of healthcare resources, including the detection of COVID-19 infection by ML interrogation of CT and X-ray images [14,15]. With respect to cancer, there are several ML-based algorithms that can process time-to-event survival outcome data, so identifying a suitable best-performing learning algorithm is critical to developing accurate prediction models of survival. To our knowledge, no study assessed ML-based algorithms’ ability to predict survival outcomes with immunotherapy in urothelial cancer. Furthermore, it is unclear whether developing ML prediction models using an all-in list (uncurated) of variables augments the discrimination performance of prediction models when compared to that of models developed with an expert preselected list (curated) (i.e., the traditional approach of selecting variables based on clinical knowledge and prior evidence). Therefore, the aims of this study were to develop and externally validate ML prediction models of survival outcomes for patients with urothelial-cancer-initiating atezolizumab and to compare the performance of ML models built with an expert-selected list (curated) versus an all-in list (uncurated) of variables.
2. Methods
2.1. Study Cohort
Individual participant data (IPD) from IMvigor210 (ClinicalTrials.gov Identifier: NCT02108652, data cut-off 4 July 2016) were used for model development (training set). IPD from the randomised atezolizumab arm of IMvigor211 (ClinicalTrials.gov Identifier: NCT02302807, data cut-off 13 March 2017) were used for external model validation (testing set). IMvigor210 was a single-arm Phase II trial in patients with locally advanced or metastatic urothelial cancer receiving atezolizumab 1200 mg IV every 3 weeks [2,16]. IMvigor211 was a Phase III trial in platinum-treated locally advanced or metastatic urothelial-cancer patients randomised to atezolizumab (1200 mg IV every 3 weeks) or chemotherapy (docetaxel (75 mg/m2 IV every 3 weeks), paclitaxel (175 mg/m2 IV every 3 weeks), or vinflunine (320 mg/m2 IV every 3 weeks) [17]. Data were accessed according to the Hoffmann–La Roche policy and made available through Vivli, Inc. (www.vivli.org).
IMvigor210 and IMvigor211 were conducted in accordance with the Guidelines for Good Clinical Practice and the Declaration of Helsinki [18]. Each participant signed and dated a written informed consent form before study enrolment [2,16,17]. Secondary analysis of deidentified IPD was classified as minimal-risk research and was confirmed as exempt from review by the Southern Adelaide Local Health Network, Office for Research and Ethics.
2.2. Predictors and Outcomes
The primary predicted outcome was overall survival (OS), with progression-free survival (PFS) as a secondary outcome. Primary study definitions of PFS were used, i.e., the independent review facility-assessed disease as per Response Evaluation Criteria in Solid Tumours (RECIST version 1.1) in IMvigor210, and the investigator-assessed disease as per RECIST (version 1.1) in IMvigor211.
Two lists of pretreatment variables were used to build the ML models: an expert preselected list (curated) and an all-in list (uncurated). The curated list included clinicopathological pretreatment factors (i.e., before the initiation of treatment) that were selected on the basis of biological plausibility, prior prognostic evidence, expert oncologist medical opinion, and being in line with prediction model guidelines for a minimum of 10 events per predictor variable [19]. The uncurated list included all pretreatment individual patient factors, subject to (1) being available in both training and validation data (≤10% missing), (2) available data definitions or reports for checking standardisation accuracy in a reasonable timeframe, and (3) within the top 15 most frequent comorbidities or concomitant medicines (i.e., an intention of the all-in approach was to speed up the model-building process rather than become stuck on needing an expert cancer researcher for data formatting and checking). Variables with missing values were imputed using approximate multiple imputation from the distribution of each variable conditional on all other variables as implemented in the transcan function from Hmisc package in R [20].
The curated list included 23 variables: (1) demographic factors (age, sex, race, body-mass index, and smoking status), (2) laboratory factors (levels of haemoglobin, albumin, white blood cells, C-reactive protein, neutrophil/lymphocyte ratio (NLR), platelet count, alkaline phosphatase, lactate dehydrogenase, and programmed death ligand 1 (PD-L1) expression on immune cells (PD-L1 ICs): PD-L1 IC0 (PD-L1 expression on <1%), PD-L1 IC1 (PD-L1 expression on ≥1% but <5%), and PD-L1IC2/3 (PD-L1 expression on ≥5) [2,16]), (3) disease or treatment factors (Eastern Cooperative Oncology Group Performance Status (ECOG-PS), disease stage (metastatic/locally advanced), the number of prior treatments, time since last chemotherapy, time since initial diagnosis, presence of liver metastasis, count of tumour sites, primary tumour site, and the presence of a urinary-tract infection.
The uncurated list contained 75 variables, composed of the curated-list variables and: (1) laboratory factors (derived NLR, eosinophil, neutrophils, alanine transaminase, aspartate aminotransferase, bilirubin, blood urea nitrogen, serum creatinine, estimated glomerular filtration rate, haemoglobin-to-platelet ratio, lymphocyte-to-monocyte ratio, platelet-to-lymphocyte ratio, and the total protein, calcium, potassium, sodium, magnesium, chloride, phosphate) and (2) disease or treatment factors (the presence of liver, brain, bone, lung or visceral metastasis, time since metastasis diagnosis, previous cystectomy, tumour stage at initial diagnosis, histologic characteristics, comorbidities (the presence of diabetes, hypertension, anaemia, renal failure, dyslipidaemia, fatigue, urinary-tract signs and symptoms, musculoskeletal pain, gastrointestinal and abdominal pain, generalised pain, constipation), and concomitant medicine use (opioids, antibiotics, proton-pump inhibitors, nonsteroidal anti-inflammatory drugs, statins, beta-blockers, calcium channel blockers, steroids, anticoagulants, anticonvulsant, bone-modulating agents, paracetamol, laxatives, vitamins, and minerals).
2.3. Machine-Learning Model Development
The patient cohort initiating atezolizumab in IMvigor210 was used for training the ML models. Four ML-based algorithms were evaluated: (1) gradient-boosted machine (GBM [21]), (2) random forest (RandomForestSRC [22]), (3) Cox-boosted model (CoxBoost [23]), and (4) penalised generalised linear models (GLM [24]). Hyperparameter tuning for ML algorithms was performed on the curated and uncurated lists by using grid search with 5-fold cross-validation with 10 repeats. Models were implemented in the ML package, mlr [25].
Model performance was externally validated using the independent IMvigor211 atezolizumab-treated patients. Model performance was assessed using the concordance statistic (c-statistic) [26] and calibration plots of observed versus predicted survival probabilities. The best-performing ML model was used to stratify patients into risk tertiles (low-, intermediate-, and high-risk prognostic groups). Survival probabilities of the risk groups were assessed using the Kaplan–Meier method. Relative variable importance in the best-performing ML model was evaluated using established methodologies [27,28]. All analyses were conducted in R version 3.6.2.
3. Results
3.1. Study Cohort
The training cohort consisted of 429 patients (239 OS events) who had initiated atezolizumab within IMvigor210. The validation cohort consisted of 467 patients (324 OS events) randomised to atezolizumab within IMvigor211. Patient characteristics are presented in Table S1. The median (95% CI) follow-up in IMvigor210 and IMvigor211 was 11 (11–12) and 17 (17–18) months, respectively.
3.2. Machine-Learning Model Development
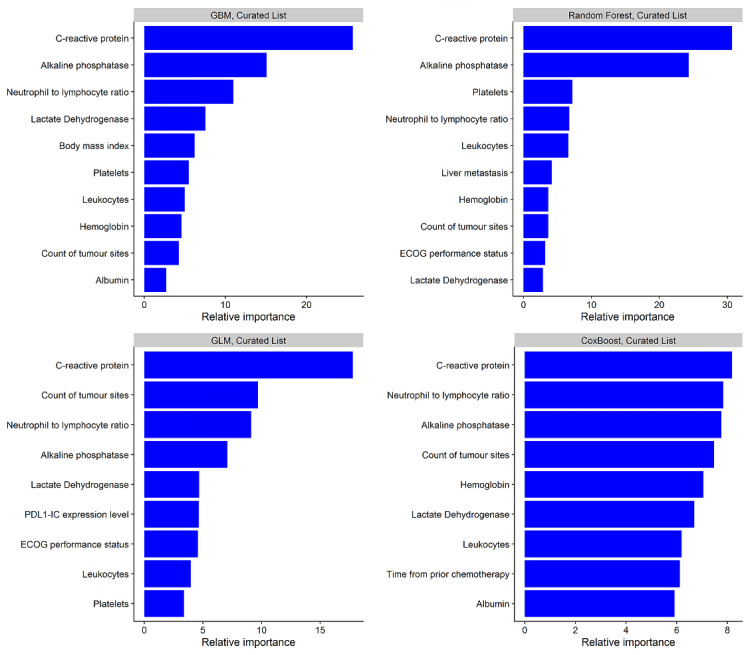
The search space and optimal hyperparameter values used for the developed ML models (both curated and uncurated versions) are presented in Table S2. The discrimination performance of the developed models on the training data is presented in Table S3 (both curated and uncurated versions). The top 10 most influential predictors of survival for each model (curated list versions) are presented in Figure 1, with C-reactive protein, alkaline phosphatase, neutrophil/lymphocyte ratio, lactate dehydrogenase, and the count of tumour sites among the most important variables in all constructed models. The top 10 most influential predictors from the developed models using the uncurated list are presented in Figure S1.
Figure 1.
Relative importance of top 10 variables for predicting survival using machine-learning models built with the curated variable list. GLM: generalised linear model with regularisation. ECOG: Eastern Cooperative Oncology Group. PDL-1-IC: Programmed death-ligand 1 gene expression level on immune cells.
On external validation, the GBM algorithm provided slightly higher OS discrimination (c = 0.71) than that of random forest (c = 0.70), CoxBoost (c = 0.70), and GLM (c = 0.69) models (curated list versions) (Table 1). All models were equivalent in predicting PFS (c = 0.62). Expansion to the uncurated list was generally associated with slightly worse discrimination for the GBM (OS c = 0.70, PFS c = 0.62), random forest (OS c= 0.69, PFS c = 0.61), CoxBoost (OS c= 0.69, PFS c = 0.61), and GLM (OS c = 0.69, PFS c = 0.61) models (Table 1).
Table 1.
Discrimination performance on validation set for overall survival (OS) and progression-free survival (PFS).
| OS | PFS | |||
|---|---|---|---|---|
| Curated list | Uncurated list | Curated list | Uncurated list | |
| Learner | C-statistics | C-statistics | C-statistics | C-statistics |
| Gradient-boosted machine | 0.71 | 0.70 | 0.62 | 0.62 |
| Random forest | 0.70 | 0.69 | 0.62 | 0.61 |
| Cox-boosted | 0.70 | 0.69 | 0.62 | 0.61 |
| generalised linear model | 0.69 | 0.69 | 0.62 | 0.61 |
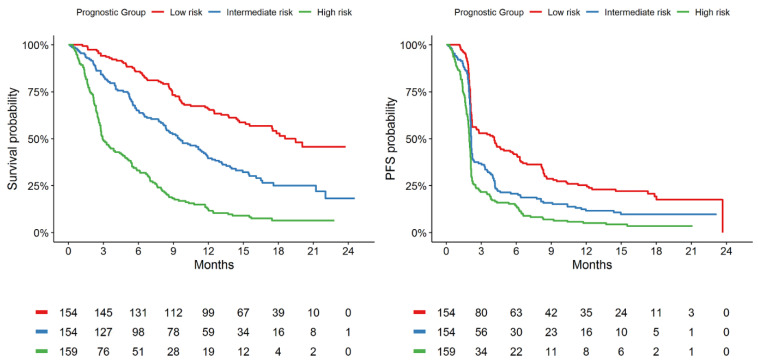
On the basis of these observations, the GBM model built with the curated list was selected for further evaluation as it provided the highest and most consistent discrimination performance among the evaluated ML algorithms. On validation, the GBM model (curated-list version) was observed to be well-calibrated for OS and PFS prediction (Figure S2). The GBM model (curated-list version) was observed to discriminate 1-year OS probabilities of 66%, 40%, and 12% for the the low-, intermediate-, and high-risk groups, respectively, defined from the randomised atezolizumab patients of IMvigor211, while 1-year PFS probabilities of 25%, 12%, and 5% were observed for the defined risk groups (Table 2). Kaplan–Meir plots of OS and PFS probabilities are presented in Figure 2.
Table 2.
Effect size and 1-year survival probability for gradient-boosted machine (GBM) stratified prognostic groups on validation cohort within IMvigor211.
| Prognostic Group | OS | PFS | ||
|---|---|---|---|---|
| HR (95% CI) (p < 0.001) |
1-year survival probability (95% CI) | HR (95% CI) (p < 0.001) |
1-year PFS probability (95% CI) | |
| Low risk | 1.00 | 66% (59–74) | 1.00 | 25% (19–33) |
| Intermediate risk | 2.09 (1.55–2.81) | 40% (33–48) | 1.54 (1.21–1.98) | 12% (7–18) |
| High risk | 5.09 (3.81–6.81) | 12% (8–19) | 2.38 (1.87–3.04) | 5% (3–10) |
Figure 2.
Kaplan–Meier estimates indicating differences in OS and PFS according to stratified machine-learning prognostic groups on validation cohort within IMvigor211 using curated-list GBM model.
4. Discussion
In this study, we used data from two large, high-quality clinical trials and evaluated several ML algorithms in the development of prediction models of survival for patients with urothelial-cancer-initiating atezolizumab. The GBM algorithm consistently provided the highest OS validation performance using both the expert-selected (curated) and the all-in (uncurated) variable lists. However, expansion to the all-in list was associated with slightly worse discrimination performance compared to that in the expert-selected subset. The GBM model (curated list) was able to discriminate patients into three prognostic risk groups with distinct survival outcomes. To our knowledge, this is the first study to use ML‑based approaches to develop and validate a survival-prediction model in patients with urothelial-cancer-initiating immunotherapy, and the first study to compare the performance of ML models built with an expert-selected variable list versus an all-in approach.
ML was applied to cancer diagnosis and risk assessment, but minimally explored for predicting personalised survival outcomes with emerging ICIs [12,29,30,31]. Recently, Hopkins et al. assessed 24 predictor variables to develop an OS prediction model for patients with nonsmall-cell lung cancer (n = 797) treated with atezolizumab, and model-validation performance using the random-forest approach (c = 0.77) was found to be superior to the GLM (0.76) and ctree (c = 0.69) models [4]. Comparatively, our study evaluated a wider range of ML algorithms and externally validated them using a large independent cohort of patients.
In addition to comparing ML algorithms in a new cancer-treatment modality, this study demonstrates that ML is proficient at identifying important predictors of treatment outcomes with ICIs in urothelial cancer. In this analysis, ML identified C-reactive protein, alkaline phosphatase, neutrophil/lymphocyte ratio, lactate dehydrogenase, and the count of tumour sites among the most important variables in all constructed models, in agreement with previous research assessing atezolizumab therapeutic outcomes in nonsmall-cell lung cancer [4,32,33]. Further, the developed model may be able to facilitate accurate risk stratification based on individual patient characteristics. For example, on external validation in the atezolizumab arm of IMvigor211, the GBM model had prediction performance consistent with a strongly performing model (c = 0.71) [8,34], and it was able to discriminate patients into low-, intermediate-, and high-risk groups with estimated 1-year OS probabilities of 66%, 40%, and 12%, respectively. This demonstrates the potential of ML prediction models to inform treatment decisions and provide more realistic expectations for treatment outcomes with patients initiating ICIs.
Expansion to the all-in (uncurated) variable-list approach resulted in slightly worse prediction performance. The slight deterioration in performance may have been due to the presence of noninformative variables that ultimately cause model overfitting or uncertainty [35]. While the all-in (dump-and-play) approach has the potential to enable biostatisticians to begin model building without expert input, the time required for artificial intelligence to tune and fit the model was substantially longer than the time required to tune the model using the curated list with fewer variables. Ultimately, it was our experience that reducing the variable list with expert help both improved model performance and saved time from a computational perspective.
A strength of this analysis was the completeness and quality of the large contemporary immunotherapy dataset that was used to train and then externally validate model discrimination and calibration performance. In addition, we studied two outcomes, OS and PFS, and we were able to confirm the insights about ML performance for each outcome. Regarding the all-in list, it is possible that some variables were not collected in the IMvigor210 and IMvigor211 trials, and the nature of clinical-trial inclusion criteria can limit the generalisability of data distributions when compared to routine care. As the model developed and validated in this study used data from the IMvigor210 and IMvigor211 trials, the training and validation cohorts were restricted to patients with urothelial-cancer-initiating atezolizumab monotherapy. Confirming the performance of ML prediction models for other ICIs, ICI combination therapy, anticancer-medicine classes, lines of therapy, and cancer types is an important future direction.
5. Conclusions
Using two large, contemporary clinical trials, we demonstrated that the GBM algorithm, applied to an expert-selected variable list, attained the highest validation performance for OS prediction. This model was further demonstrated capable of discriminating urothelial-cancer patients initiating atezolizumab with distinctly different survival outcomes. Using an all-in list of variables as opposed to an expertly selected list may harm discrimination performance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13092001/s1, Figure S1: Relative importance of the top 10 variables for predicting survival using the uncurated variable list. GBM: gradient boosted machine, GLM: generalized linear model with regularization, Figure S2: Calibration plots (Kaplan-Meier observed versus model predicted probabilities) of overall survival (OS) and progression free survival (PFS) on validation data using the GBM model constructed using curated variables list, Table S1: Pre-treatment patient characteristics by atezolizumab study cohort, Table S2: Hyperparameter tuning of machine learning algorithms on overall survival data using the primary and extended lists, Table S3: Discrimination performance on IMvigor210 training cohort for overall survival and progression free survival.
Author Contributions
Conception and design: A.Y.A., A.M.H., and M.J.S.; development of methodology: A.Y.A., A.M.H., G.K., A.R., and M.J.S.; acquisition of data: A.Y.A., A.M.H., A.R., and M.J.S.; analysis and interpretation of data (e.g., statistical analysis, biostatistics, and computational analysis): A.Y.A., A.M.H., G.K., R.A.M., A.R., and M.J.S.; writing, review, and/or revision of the manuscript: A.Y.A., A.M.H., G.K., R.A.M., A.R., and M.J.S.; administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): A.Y.A., A.M.H., R.A.M., A.R., and M.J.S.; study supervision: A.Y.A., A.M.H., R.A.M., A.R., and M.J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Flinders Foundation Health Seed Grant RPF20/877. R.A.M: A.R. and M.J.S. are supported by the Beat Cancer Research Fellowships from Cancer Council South Australia. A.M.H is supported by a Postdoctoral Fellowship from the National Breast Cancer Foundation, Australia (PF-17-007). The data access and salary of A.Y.A. were supported by funding from an Australian Tour de Cure Early Career Research Grant (RSP-155-18/19).
Institutional Review Board Statement
Secondary analysis of anonymised clinical-trial data was confirmed as negligible-risk research by the Southern Adelaide Local Health Network, Office for Research and Ethics, and was exempt from review.
Informed Consent Statement
Not applicable to this independent, secondary analysis study.
Data Availability Statement
Data were accessed according to Roche’s policy and process for clinical study data sharing and is available for request at https://vivli.org/.
Conflicts of Interest
R.A.M., A.R., and M.J.S. report investigator-initiated project grants from Pfizer outside the submitted work. A.Y.A., J.B., G.K., and A.M.H. have no conflicts of interest to disclose. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.International Agency for Research on Cancer WHO. Cancer Incidence and Mortality Worldwide. [(accessed on 16 July 2020)]; Available online: https://gco.iarc.fr.
- 2.Balar A.V., Galsky M.D., Rosenberg J., Powles T., Petrylak D.P., Bellmunt J., Loriot Y., Necchi A., Hoffman-Censits J., Perez-Gracia J.L., et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: A single-arm, multicentre, phase 2 trial. Lancet. 2017;389:67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park Y.-J., Kuen D.-S., Chung Y. Future prospects of immune checkpoint blockade in cancer: From response prediction to overcoming resistance. Exp. Mol. Med. 2018;50:1–13. doi: 10.1038/s12276-018-0130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopkins A.M., Kichenadasse G., Garrett-Mayer E., Karapetis C.S., Rowland A., Sorich M.J. Development and Validation of a Prognostic Model for Patients with Advanced Lung Cancer Treated with the Immune Checkpoint Inhibitor Atezolizumab. Clin. Cancer Res. 2020;26:3280–3286. doi: 10.1158/1078-0432.CCR-19-2968. [DOI] [PubMed] [Google Scholar]
- 5.Bellmunt J., Choueiri T.K., Fougeray R., Schutz F.A., Salhi Y., Winquist E., Culine S., Von Der Maase H., Vaughn D.J., Rosenberg J.E. Prognostic Factors in Patients With Advanced Transitional Cell Carcinoma of the Urothelial Tract Experiencing Treatment Failure With Platinum-Containing Regimens. J. Clin. Oncol. 2010;28:1850–1855. doi: 10.1200/JCO.2009.25.4599. [DOI] [PubMed] [Google Scholar]
- 6.Sonpavde G., Pond G.R., Rosenberg J.E., Bajorin D.F., Choueiri T.K., Necchi A., Di Lorenzo G., Bellmunt J. Improved 5-Factor Prognostic Classification of Patients Receiving Salvage Systemic Therapy for Advanced Urothelial Carcinoma. J. Urol. 2016;195:277–282. doi: 10.1016/j.juro.2015.07.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonpavde G., Manitz J., Gao C., Tayama D., Kaiser C., Hennessy D., Makari D., Gupta A., Abdullah S.E., Niegisch G., et al. Five-Factor Prognostic Model for Survival of Post-Platinum Patients with Metastatic Urothelial Carcinoma Receiving PD-L1 Inhibitors. J. Urol. 2020;204:1173–1179. doi: 10.1097/JU.0000000000001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Révész D., Engelhardt E.G., Tamminga J.J., Schramel F.M.N.H., Onwuteaka-Philipsen B., Van De Garde E.M.W., Steyerberg E.W., Jansma E.P., De Vet H.C.W., Coupé V.M.H. Decision support systems for incurable non-small cell lung cancer: A systematic review. BMC Med. Inform. Decis. Mak. 2017;17:1–12. doi: 10.1186/s12911-017-0542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins G.S., Le Manach Y. Small data sets to develop and validate prognostic models are problematic. Eur. J. Cancer. 2016;54:167–168. doi: 10.1016/j.ejca.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 10.Mahar A.L., Compton C., McShane L.M., Halabi S., Asamura H., Rami-Porta R., Groome P.A. Refining Prognosis in Lung Cancer. J. Thorac. Oncol. 2015;10:1576–1589. doi: 10.1097/JTO.0000000000000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz J.A., Wishart D.S. Applications of Machine Learning in Cancer Prediction and Prognosis. Cancer Inform. 2006;2:59–77. doi: 10.1177/117693510600200030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kourou K., Exarchos T.P., Exarchos K.P., Karamouzis M.V., Fotiadis D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhys H.I. Machine Learning with R, the Tidyverse, and MLR. Manning Publications; New York, NY, USA: 2020. [Google Scholar]
- 14.Alshazly H., Linse C., Barth E., Martinetz T. Explainable COVID-19 Detection Using Chest CT Scans and Deep Learning. Sensors. 2021;21:455. doi: 10.3390/s21020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg J.E., Hoffman-Censits J., Powles T., van der Heijden M.S., Balar A.V., Necchi A., Dawson N., O’Donnell P.H., Balmanoukian A., Loriot Y., et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powles T., Durán I., Van Der Heijden M.S., Loriot Y., Vogelzang N.J., De Giorgi U., Oudard S., Retz M.M., Castellano D., Bamias A., et al. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): A multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2018;391:748–757. doi: 10.1016/S0140-6736(17)33297-X. [DOI] [PubMed] [Google Scholar]
- 18.World Medical Association Declaration of Helsinki Recommendations guiding physicians in biomedical research involving human subjects. Cardiovasc. Res. 1997;35:2–3. [PubMed] [Google Scholar]
- 19.Steyerberg E.W., Harell F.E., Jr. Regression Modeling Strategies: With Applications, to Linear Models, Logistic and Ordinal Regression, and Survival Analysis, 2nd ed. Heidelberg: Springer. Biom. 2016;72:1006–1007. doi: 10.1111/biom.12569. [DOI] [Google Scholar]
- 20.Harrell F.E., Jr., with Contributions from Charles Dupont and Many Others Hmisc: Harrell Miscellaneous. R Package Version 4.5. [(accessed on 19 March 2021)];2021 Available online: https://cran.r-project.org/web/packages/Hmisc/index.html.
- 21.Greenwell B., Boehmke B., Cunningham J., Developers G. GBM: Generalized Boosted Regression Models. R Package Version2.1.5. [(accessed on 19 March 2021)];2019 Available online: https://cran.r-project.org/web/packages/gbm/index.html.
- 22.Ishwaran H., Kogalur U.B., Kogalur M.U.B. randomForestSRC: Fast Unified Random Forests for Survival, Regression, and Classification (RF-SRC) Version 2.11 2021. [(accessed on 19 March 2021)]; Available online: https://cran.r-project.org/web/packages/randomForestSRC/index.html.
- 23.Binder H. CoxBoost: Cox Models by Likelihood Based Boosting for a Single Survival Endpoint or Competing Risks. R Package Version 1.4. [(accessed on 20 March 2021)]; Available online: http://cran.nexr.com/web/packages/CoxBoost/index.html.
- 24.Friedman J., Hastie T., Tibshirani R., Narasimhan R., Tay K., Simon N., Qian J. Glmnet: Lasso and Elastic-Net Regularized Generalized Linear Models. R Package Version 1.4. [(accessed on 20 March 2021)]; Available online: https://cran.r-project.org/web/packages/glmnet/index.html.
- 25.Bischl B., Lang M., Kotthoff L., Schiffner J., Richter J., Studerus E., Casalicchio G., Jones Z.M. mlr: Machine Learning in R. J. Mach. Learn. Res. 2016;17:5938–5942. [Google Scholar]
- 26.Harrell F.E., Lee K.L., Mark D.B. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Friedman J.H., Meulman J.J. Multiple additive regression trees with application in epidemiology. Stat. Med. 2003;22:1365–1381. doi: 10.1002/sim.1501. [DOI] [PubMed] [Google Scholar]
- 28.Ishwaran H. Variable importance in binary regression trees and forests. Electron. J. Stat. 2007;1:519–537. doi: 10.1214/07-EJS039. [DOI] [Google Scholar]
- 29.Yue W., Wang Z., Chen H., Payne A., Liu X. Machine Learning with Applications in Breast Cancer Diagnosis and Prognosis. Designs. 2018;2:13. doi: 10.3390/designs2020013. [DOI] [Google Scholar]
- 30.Wang P., Li Y., Reddy C.K. Machine Learning for Survival Analysis. ACM Comput. Surv. 2019;51:1–36. doi: 10.1145/3214306. [DOI] [Google Scholar]
- 31.Ashley M.H., Adel S., Sasha Z., Chris S.K., Andrew R., Michael J.S., Hopkins A.M., Shahnam A., Zhang S., Karapetis C.S., et al. Prognostic model of survival outcomes in non-small cell lung cancer patients initiated on afatinib: Pooled analysis of clinical trial data. Cancer Biol. Med. 2019;16:341–349. doi: 10.20892/j.issn.2095-3941.2018.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopkins A., Kichenadasse G., Abuhelwa A., McKinnon R., Rowland A., Sorich M. Value of the Lung Immune Prognostic Index in Patients with Non-Small Cell Lung Cancer Initiating First-Line Atezolizumab Combination Therapy: Subgroup Analysis of the IMPOWER150 Trial. Cancers. 2021;13:1176. doi: 10.3390/cancers13051176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorich M.J., Rowland A., Karapetis C.S., Hopkins A.M. Evaluation of the Lung Immune Prognostic Index for Prediction of Survival and Response in Patients Treated With Atezolizumab for NSCLC: Pooled Analysis of Clinical Trials. J. Thorac. Oncol. 2019;14:1440–1446. doi: 10.1016/j.jtho.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Modi N.D., Sorich M.J., Rowland A., Logan J.M., McKinnon R.A., Kichenadasse G., Wiese M.D., Hopkins A.M. A literature review of treatment-specific clinical prediction models in patients with breast cancer. Crit. Rev. Oncol. 2020;148:102908. doi: 10.1016/j.critrevonc.2020.102908. [DOI] [PubMed] [Google Scholar]
- 35.Kuhn M., Johnson K. Applied Predictive Modeling. 1st ed. Springer; New York, NY, USA: 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were accessed according to Roche’s policy and process for clinical study data sharing and is available for request at https://vivli.org/.