Abstract
Background:
Beta amyloid (Aβ) peptide containing plaque aggregations in the brain are a hallmark of Alzheimer’s Disease (AD). However, Aβ is produced by cell types outside of the brain suggesting that the peptide may serve a broad physiologic purpose.
Objective:
Based upon our prior work documenting expression of amyloid β precursor protein (APP) in intestinal epithelium we hypothesized that salivary epithelium might also express APP and be a source of Aβ.
Methods:
To begin testing this idea, we compared human age-matched control and AD salivary glands to C57BL/6 wild type, AppNL–G–F, and APP/PS1 mice.
Results:
Both male and female AD, AppNL–G–F, and APP/PS1 glands demonstrated robust APP and Aβ immunoreactivity. Female AppNL–G–F mice had significantly higher levels of pilocarpine stimulated Aβ 1–42 compared to both wild type and APP/PS1 mice. No differences in male salivary Aβ levels were detected. No significant differences in total pilocarpine stimulated saliva volumes were observed in any group. Both male and female AppNL–G–F but not APP/PS1 mice demonstrated significant differences in oral microbiome phylum and genus abundance compared to wild type mice. Male, but not female, APP/PS1 and AppNL–G–F mice had significantly thinner molar enamel compared to their wild type counterparts.
Conclusion:
These data support the idea that oral microbiome changes exist during AD in addition to changes in salivary Aβ and oral health.
Keywords: microbiome, Alzheimer, amyloid, inflammation, saliva, biomarker
1. INTRODUCTION
Alzheimer’s disease (AD) is a neurodegenerative disease characterized by aggregations of amyloid-beta protein (Aβ) in the brain, as well as associated inflammatory changes including activation of macrophage and microglia. Aβ is a proteolytic cleavage product of the larger amyloid beta precursor protein, APP (1–5). Although APP is highly expressed in neurons, it is a ubiquitously expressed protein likely regulating a plethora of activities (6–13). For example, we and others have demonstrated expression of APP and secretion of Aβ from intestinal epithelial cells where it can be quantified from feces (8, 14, 15).
Diagnosis of AD is time-consuming and is often delayed until considerable neurodegeneration has occurred. Therefore, there is a clear need for sensitive, specific biomarkers of disease at the earliest possible stage. One possibility for diagnostic biomarker identification is saliva. For instance particular phospho-epitopes of tau protein are reportedly elevated in AD versus control saliva although there is some debate regarding changes in saliva levels of total tau in AD compared to controls (16–18). Similar salivary analyses of Aβ levels indicate an increase in Aβ 1–42 but not 1–40 in AD versus control patients (19–21). In addition, studies of salivary metabolite profiling by mass spectrometry have also demonstrated similar success in differentiating AD from control individuals (22–25).
In addition to the possibility of saliva serving as a source of biomarker profiling in AD there appears to be pathophysiology associated with the oral cavity during disease. For example, reduced salivary flow and altered pH has been reported in AD patients (26). A similar study comparing 28 AD and 35 control individuals demonstrated a selective decrease in submandibular versus parotid flow rate in AD patients (27). Dysphagia and aspiration pneumonia are serious medical conditions for late-stage AD patients. Increased risk for aspiration pneumonia include dysphagia, intubation, reduced gag reflex, and periodontal disease (28). There is some evidence that improved oral hygiene may decrease the risk of aspiration pneumonia in at-risk individuals such as AD patients (29). Finally, numerous studies demonstrate a positive correlation between periodontal disease, tooth loss, and overall poor oral health with risk and severity of AD (26, 30–43).
Based upon our prior identification of APP and Aβ expression in intestinal epithelium, we hypothesized that a similar biology occurred in salivary gland epithelium (8, 14, 15). To test this idea, saliva, salivary glands, and mandibles of male and female APP/PS1, AppNL–G–F, and C57BL/6 wild type mice, as well as human salivary gland tissue were collected and analyzed. Human AD submandibular salivary glands demonstrated ductal epithelial staining for both Aβ and APP correlating with similar findings in the AD mouse lines. Analysis of saliva obtained from the mice demonstrated a significant elevation in Aβ 1–42 levels only in female AppNL–G–F mice with no changes in saliva production in any group. Oral microbiome diversity analysis demonstrated significant differences between male and female AppNL–G–F compared to wild type mice. Finally, male but not female APP/PS1 and AppNL–G–F mice demonstrated reduced enamel compared to wild type controls.
2. MATERIALS AND METHODS
2.1. Antibodies and Reagents
Elite Vectastain ABC reagents, Vector VIP (SK-4600), biotinylated anti-rabbit (BA-1000), and mouse on mouse anti-mouse (MKB-2225) antibodies were purchased from Vector Laboratories Inc. (Burlingame, CA). APP (Y188) ab32136 rabbit monoclonal antibody that reacts with both mouse and human protein was purchased from Abcam Inc. (Cambridge, MA). Aβ anti-rabbit monoclonal antibody specific to human (8243) was obtained from Cell Signaling Technology Inc. (Danvers, MA).
2.2. Animals
Male and female mice were used (n=3–15). Mice used for microbiome, micro-CT analysis, and bacterial cultures were 5–10 months old. Immunohistochemistry and Aβ saliva ELISAs were performed from 10–15 month old mice. Wild type (WT) littermate control mice were C57BL/6. The triple mutation knock-in AppNL–G–F mice (KI:RBRC06344) were a gift from Drs. Takaomi C. Saido and Takashi Saito at the RIKEN Center for Brain Science, Japan (44). The AppNL–G–F mice have the Swedish “NL” mutation which promotes Aβ production, the Arctic “G” mutation promotes Aβ aggregation and reducing its degradation. The Iberian “F” mutation is responsible for increasing the Aβ42/Aβ40 ratio. The APP/PS1 mice express both the human Swedish APP and ΔE9 mutations in the PS1 gene resulting in over expression of human APP and secretion of human Aβ. The APP−/− mice have the APP gene knocked out of the entire organism by the insertion of a neomycin resistance cassette into the promoter region and Exon 1 of the APP gene. Animal use was approved by the University of North Dakota Institutional Animal Care and Use Committee (UND IACUC). C57BL/6 mice stock number 000664, APP/PS1 (B6.Cg-Tg(APPswe,PSEN1dE9)85Dbo) stock number 005864 and APP−/− (B6.129S7-APPtm1Dbo/J) stock number 004133, were all originally purchased from Jackson Laboratories (Bar Harbor, ME). Mice were provided food and water ad libitum and housed in the same room in a 12 h light/dark cycle. The investigation conforms to the National Research Council of the National Academies Guide for the Care and Use of Laboratory Animals (8th edition).
2.3. Saliva Collection for Microbiome
For colony-forming unit assays and ELISAs, male and female WT, AppNL–G–F, APP/PS1, and APP−/− mice were anesthetized via intraperitoneal injection with 75mg/kg ketamine (Henry Schein, Dublin OH) and 8mg/kg xylazine (Akorn Animal Health, Lake Forest, IL) and subcutaneously injected with 50μL of 0.5mg/mL of pilocarpine 151892 (MP Biomedicals, Santa Ana, CA) to induce salivation. Saliva drool was collected into Eppendorf tubes for 16 minutes from each mouse. Volume from each mouse was measured. For microbiome analysis, Helix buccal swabs SK-2S (Boca Scientific, Boca Raton, FL) were swabbed extensively along all of the surfaces inside the mouth for 1 minute. Swabs were placed in Helix Buccalfix stabilization buffer BFX-25 (Boca Scientific, Boca Raton, FL) and sent to RTL Genomics (Research and Testing Laboratory, Lubbock, TX) for 16S ribosomal RNA (rRNA) gene amplicons and analysis. Hypervariable regions V1 to V3 of 16S rRNA gene were amplified for sequencing in a reaction using HotStart Taq Master Mix Kit (Qiagen, Inc., Germantown, MD) with the universal primer set 27F/519R. The sequence data were analyzed using a standard microbial diversity analysis pipeline, which consisted of two major stages, denoising and chimera detection followed by microbial diversity analysis. The overall alpha diversity (which is determined by both richness and evenness, the distribution of abundance among distinct taxa) was studied. Shannon diversity analysis was used to characterize the abundance and evenness of the species present in a community and was also used as an index of evenness in this study. The number of different species or species richness in each experimental group was studied using Chao index. Principal coordinates analysis (PCoA) was performed to assess how different oral bacterial composition was across genotypes/sex (beta diversity). Measures of diversity were screened for group differences using two-way analysis of variance (ANOVA) multiple comparisons with uncorrected Fisher’s LSD.
2.4. Sample Collection
Mice were euthanized and submandibular salivary glands and mandibles were collected. Glands were fixed in 4% paraformaldehyde for 3 days followed by cryoprotection through two successive incubations in 30% sucrose then serially cryosectioned.
2.5. MicroCT Analysis
The right hemimandible from each mouse was scanned using a Scanco micro CT scanner 40 (Scanco Medical AG, Bassersdorf, Switzerland) at 55 kVp, 145 μA, integration time of 300 ms, and 12 μm isotropic voxel resolution. The first molar was identified as the first tooth seen after the incisor. Quantitative 3-D analysis of the first molar was performed after the region of interest was contoured and analyzed with the analysis program provided by Scanco Medical AG. The whole tooth including the enamel, pulp and dentin was contoured and analyzed. The enamel was identified as the bright white portion at the top of the tooth. All bright white portions of the tooth were contoured excluding the pulp and dentin. The pulp was identified as the lighter grey matter of the tooth. The pulp was contoured excluding the enamel and dentin. The dentin was identified as the dark portions within the pulp. The dentin was contoured excluding the pulp and enamel. The space between the tooth and the mandible was the last to be contoured and analyzed. The microCT scanner is calibrated regularly using known density phantoms as recommended by the manufacture and the recommended guidelines for microCT scanning were used.
2.6. Bacterial Cultures
Saliva collected from each mouse was diluted 100-fold and spread onto trypticase soy agar containing 5% sheep blood (BA plates, BD Biosciences, MD). Plates were incubated for 24 hours at 37°C. Total colony-forming units (CFUs) were counted and graphed.
2.7. Aβ enzyme-linked Immunosorbent Assay (ELISA)
Human Aβ 1–40 (EZHS40) and 1–42 (EZHS42) ELISA kits were purchased from MilliporeSigma (Burlington, MA). Saliva was centrifuged at 2,000 g for 10 min. Saliva ELISAs were performed according to manufacturer instructions.
2.8. Immunohistochemistry
Fixed submandibular glands were sectioned at 10μm thickness using a Leica CM1850 cryostat. Tissue sections were antigen retrieved using Vector citrate buffer H-3300 (Burlingame, CA) for Aβ immunostaining. Anti-Aβ antibody was from Cell Signaling Technology (8243) and the anti-APP antibody was from Abcam (ab32136). Both were used at 1:500. Antibody binding was visualized using the Vector VIP chromogen (Vector Laboratories, Burlingame, CA). Images were taken using an upright Leica DM1000 microscope and Leica DF320 digital camera system (Mannheim, Germany). Figures were made using Adobe Photoshop software (Adobe Systems, San Jose, CA).
2.9. Human Tissue
Paraffin-embedded submandibular gland tissue sections (10μm) from 5 male and 5 female AD and similar age controls were obtained from Banner Health (Sun City, AZ). The tissue was immunostained with anti-APP (Y188), anti-Aβ (Cell Signaling) or respective secondary only antibodies. Slides were antigen retrieved in 10mM Tris-1mM EDTA, pH 9, for 10min and rinsed with Tris-buffered saline (TBS). Antibody binding in the glands was visualized using the Vector VIP chromogen (Vector Laboratories, Burlingame, CA) reagent. Images were taken using an upright Leica DM1000 microscope and Leica DF320 digital camera system. Figures were made using Adobe Photoshop software.
2.10. Statistics
Data were analyzed using using GraphPad Prism version 8.0.0 for Windows (GraphPad Software, San Diego, California USA, www.GraphPad.com). Microbiome data was originally analyzed in R and further analysis used GraphPad Prism Software. Data are represented as mean values ±SEM. Statistical analyses were done using one-way ANOVA with Tukey’s post-hoc test. Differences less than p<0.05 were considered significant.
3. RESULTS
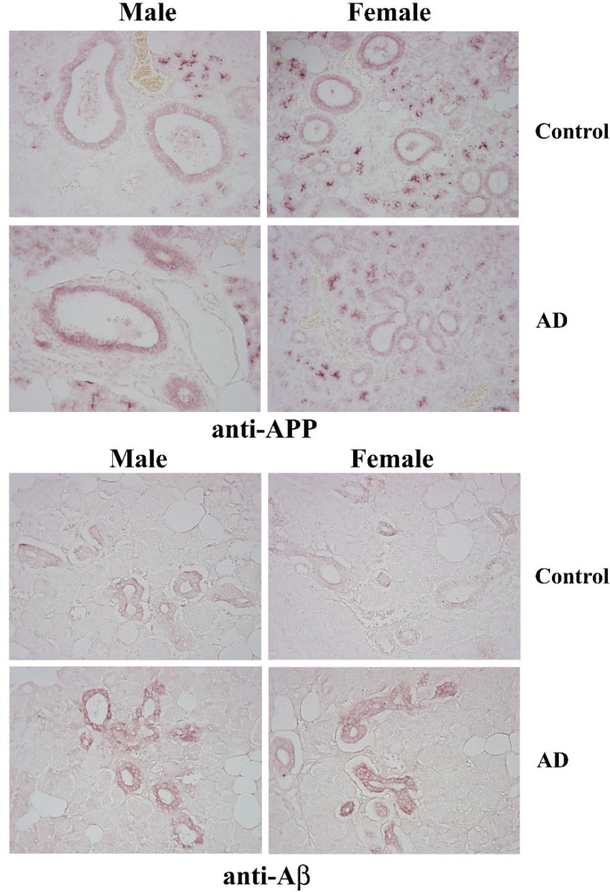
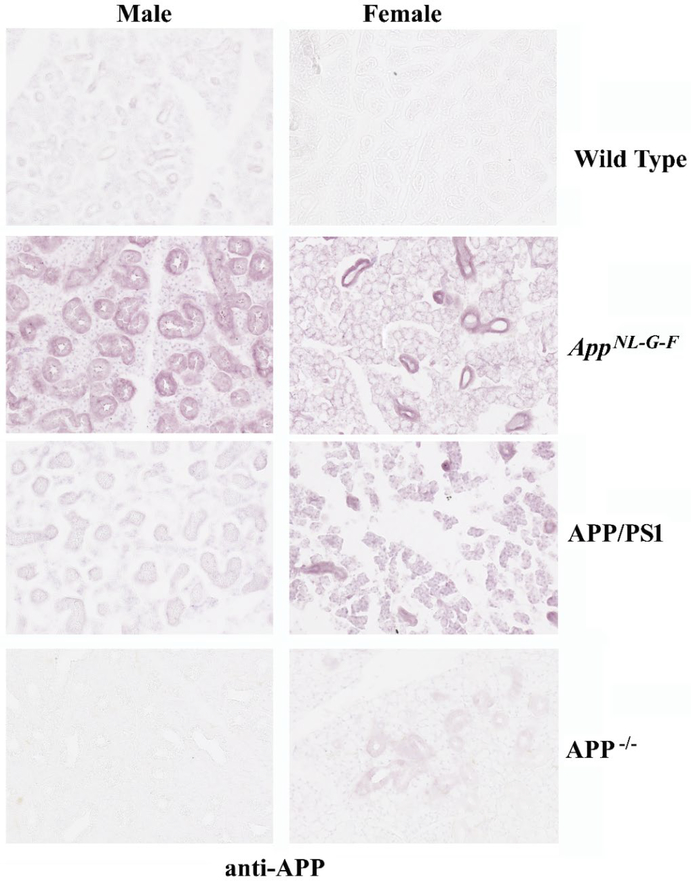
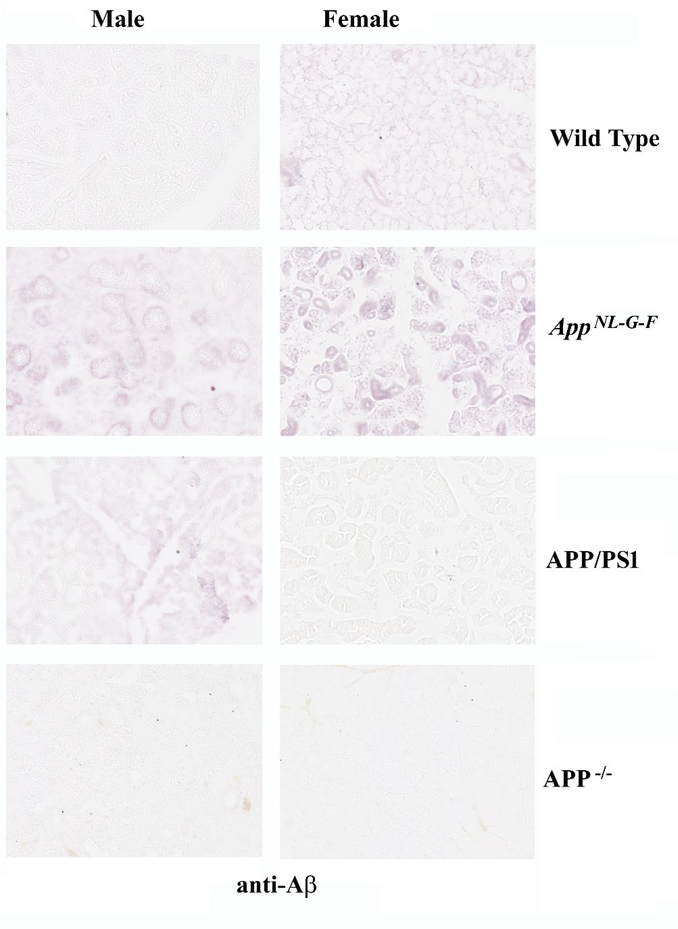
Based upon our prior efforts documenting APP expression in gastrointestinal epithelium, we assessed whether APP was similarly expressed in salivary gland ductal epithelium (14). Comparison of age-matched male and female control and AD submandibular glands demonstrated robust APP immunoreactivity in the ductal epithelium, as expected (Fig. 1). There was not clear difference in staining intensity across any of the groups. However, anti-Aβ immunostaining demonstrated robust ductal epithelial staining in both the male and female AD glands compared to controls (Fig. 1). In order to validate the relevance of our transgenic mouse models of AD, we next immunostained submandibular glands from C57BL/6 wild type mice with anti-APP and Aβ antibodies to compare to the immunoreactivity in two different transgenic mouse models of AD, AppNL–G–F and APP/PS1, as well as APP−/− mice. Similar to the human tissue, APP immunoreactivity localized primarily to ductal epithelium in both sexes and all lines with the most robust pattern of staining observed in the female AppNL–G–F mice (Fig. 2). However, the female APP−/− tissue demonstrated faint immunoreactivity suggesting some cross-reactivity exists for the APP immunostaining (Fig. 2). Immunostaining for AΒ in the mouse salivary glands demonstrated immunoreactivity similar to human AD glands with localization surrounding ducts in both male and female AppNL–G–F mice (Fig. 3). APP/PS1 male mice demonstrated similar Aβ immunoreactivity with no recognizable staining in female APP/PS1 mice or either sex of wild type or APP−/− mouse glands (Fig 3).
Fig. 1. APP and Aβ immunoreactivities were present in human submandibular salivary glands.
Submandibular gland sections from age and gender matched AD and non-demented controls sections were immunostained using anti-APP and Aβ antibodies. Antibody binding was visualized using Vector VIP as the chromogen. Representative images of 10μm sections at 20X are shown (n=5).
Fig. 2. Murine submandibular salivary glands displayed APP immunoreactivity.
Submandibular glands were collected from C57BL6 wild type (WT), AppNL–G–F, APP/PS1, and APP−/− male and female mice, fixed in 4% paraformaldehyde, and serially sectioned (10μm. Sections were immunostained using anti-APP antibodies. Antibody binding was visualized using Vector VIP as the chromogen. Representative images at 20X are shown from n=2–4 C57BL6 wild type (WT), AppNL–G–F, APP/PS1 mice with a male and female APP−/− gland used as an antibody specificity control.
Fig. 3. Aβ immunoreactivity was observed in murine salivary glands.
Submandibular glands were collected from C57BL/6 wild type (WT), AppNL–G–F, APP/PS1, and APP−/− male and female mice, fixed in 4% paraformaldehyde, and serially sectioned (10μm). Sections were immunostained using anti-Aβ antibody and antibody binding was visualized using Vector VIP as the chromogen. Representative images at 20X are shown from n=2–4 C57BL/6 wild type (WT), AppNL–G–F, APP/PS1 mice with a male and female APP−/− gland used as an antibody specificity control.
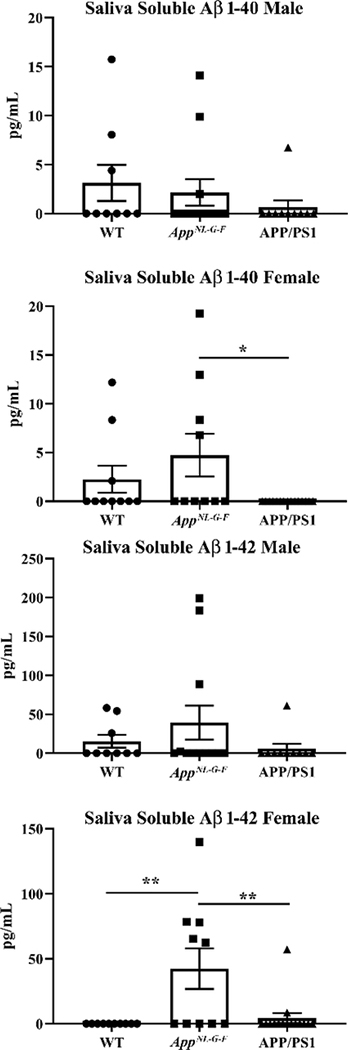
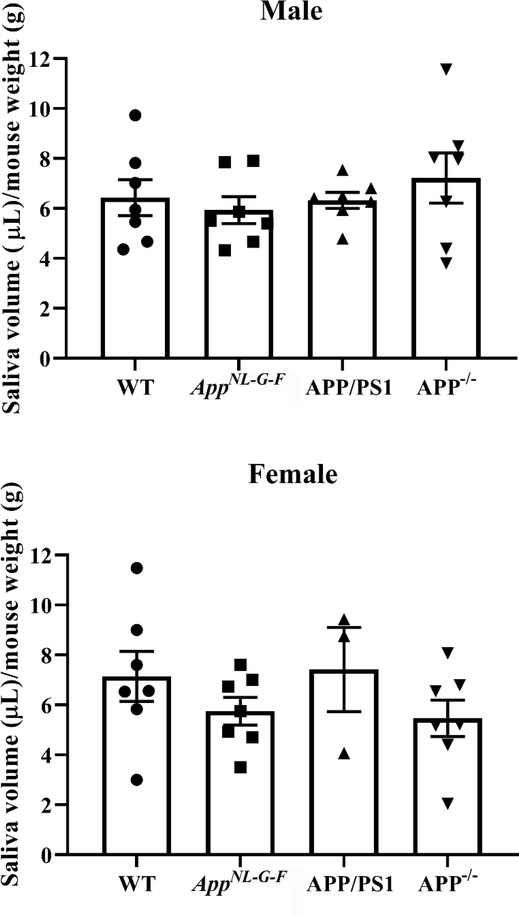
Our prior work demonstrated the ability of intestinal epithelial cells to secrete Aβ (15). To ascertain whether a similar biology was relevant in the salivary glands we collected pilocarpine-induced saliva from wild type, AppNL–G–F, and APP/PS1 mice for quantifying Aβ levels. Although a human specific Aβ ELISA reagent was used, we did observe significant variability in detectable levels of Aβ including within wild type samples suggesting some species cross-reactivity or non-specificity existed (Fig. 4). Neither female nor male levels of Aβ 1–40 significantly differed between wild type and AppNL–G–F mice although levels were higher in female AppNL–G–F compared to APP/PS1 mice (Fig. 4). Male mice demonstrated no significant differences Aβ 1–42 levels across strains (Fig. 4). However, female AppNL–G–F mice demonstrated significantly elevated Aβ 1–42 levels compared to both wild type and APP/PS1 mice. This increase is consistent with prior characterization of this line with a bias towards preferential Aβ 1–42 production (44). In an effort to correlate the appearance of Aβ immunoreactivity and presence of Aβ in saliva with the function of the salivary glands, we next quantified the total volume of saliva produced from both sexes and all mouse lines after pilocarpine-stimulated salivation. APP−/− mice were included to assess whether APP expression regulated saliva production. There were no significant differences in saliva production in females or males compare to their respective wild type mice (Fig. 5).
Fig. 4. Soluble Aβ was detectable in AppNL–G–F saliva.
Male and female mice were anesthetized and injected with pilocarpine (0.5mg/mL) to induce salivation. Saliva was collected for 16 minutes from each mouse. Aβ1–40 and Aβ1–42 ELISAs were performed to quantify levels of soluble Aβ in saliva from WT, AppNL–G–F, and APP/PS1 mice. Data are graphed as mean values +/− SEM, (n=9–15). Statistical differences were calculated by one-way ANOVA with Tukey post-hoc analysis, *p<0.05, **p<0.01.
Fig. 5. WT, AppNL–G–F, APP/PS1, and APP−/− mice had comparable saliva volume.
Male and female mice were anesthetized and injected with pilocarpine (0.5mg/mL) to induce salivation. Saliva was collected for 16 minutes. Saliva volumes were normalized to the respective mouse weight in grams. Statistical differences were calculated by one-way ANOVA with Tukey post-hoc analysis. Data are graphed as mean values +/− SEM (n=3–7).
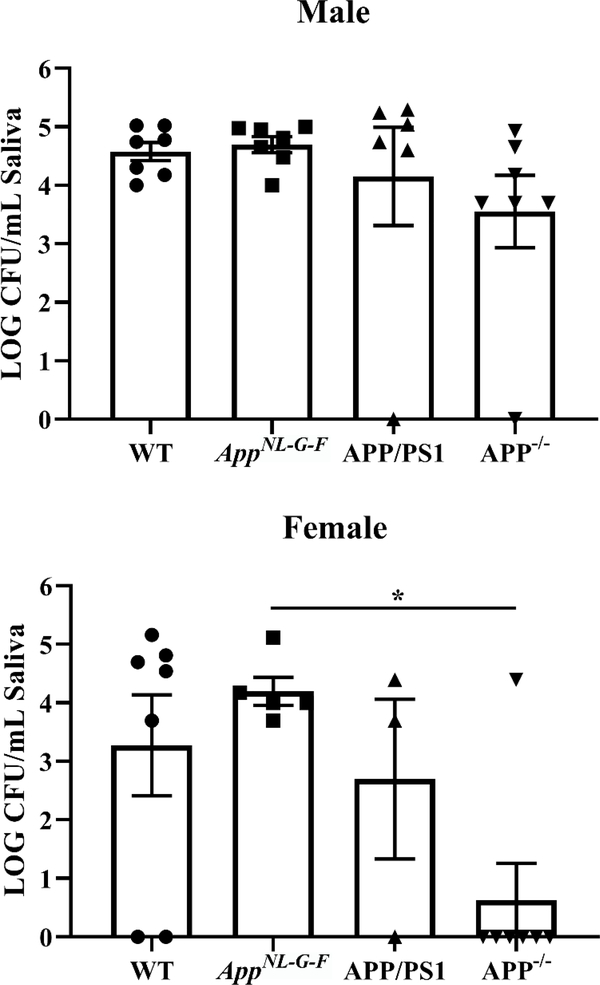
Based upon prior work demonstrating that anti-microbial properties of Aβ (45–47), we assessed whether there may be differences in the oral microbiome in the mice in correlation with the presence of Aβ. Once again, we elected to include APP−/− mice as a negative control for effects due to APP expression and processing. Saliva was plated onto blood agar plates as a preliminary assessment of aerobic growth differences across sex and mouse line. Only female APP−/− mice differed from AppNL–G–F mice with significantly fewer colony-forming units (Fig. 6).
Fig. 6. APP−/− mice had altered oral bacterial growth compared to AppNL–G–F mice.
Pilocarpine-stimulated saliva collected from male and female WT, AppNL–G–F, APP/PS1, and APP−/− mice was plated onto blood agar plates. Numbers of colonies were counted at 24 hours and normalized per volume of saliva from each condition. Data are graphed as mean values +/− SEM. Statistical differences were calculated by one-way ANOVA with Tukey post-hoc analysis, *p<0.05 ( n=3–7).
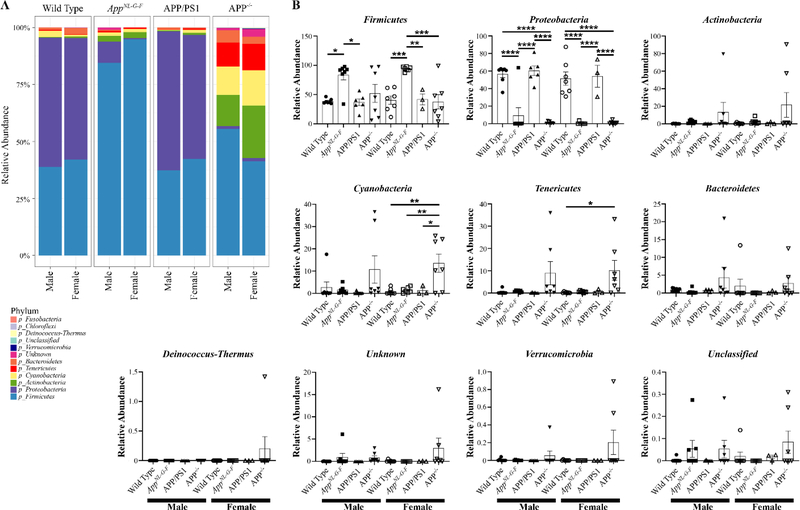
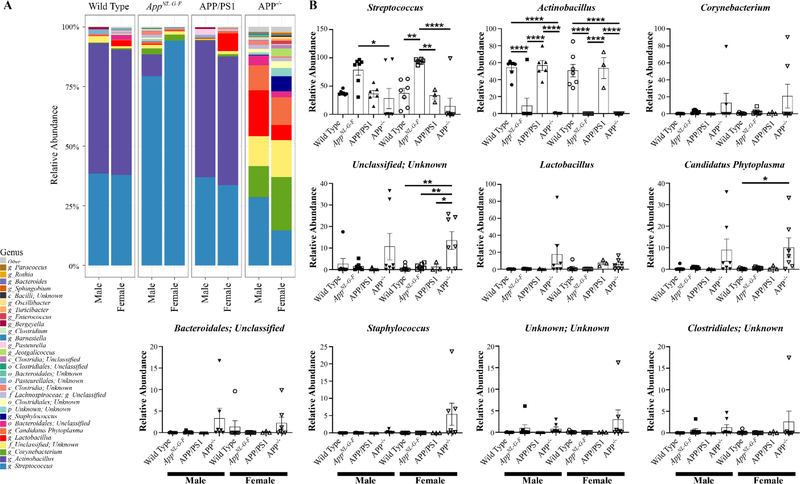
To improve the chances of observing differences in the oral microbiomes of the mice, we next performed 16S rRNA sequencing from oral swabs. The alpha diversity, variance within the sample groups, was determined from both Shannon and Chao1 indices (Supplemental Fig. 1). There was not a dramatic difference in Shannon diversity among either sex compared to wild type controls (Supplemental Fig. 1). The richness of species estimated by the Chao1 also demonstrated no significant differences of any genotype compared to respective wild type controls (Supplemental Fig. 1). Interestingly, female APP−/− mice had significantly reduced species richness compare to their respective male APP−/− mice (Supplemental Fig. 1). Male and female mice in each genotype demonstrated remarkably similar microbial composition (Supplemental Fig. 2). Although wild type and APP/PS1 mice had very similar microbial composition, APP−/− and AppNL–G–F mice demonstrated dramatically different compositions from all other genotypes (Supplemental Fig. 2). Following up on this, a comparison of the most abundant phyla across groups revealed once again similarity between wild type and APP/PS1 mice of both sexes (Fig. 7). However, AppNL–G–F mice differed from wild type controls considerably, showing increased Firmicutes and decreased Proteobacteria in both sexes compared to their respective wild type counterparts (Fig. 7). Male and female APP−/− mice also demonstrated uniqueness with significantly elevated Tenericutes compared to wild type mice (Fig. 7). Genus abundance comparison similarly demonstrated significantly elevated Streptococcus and decreased Actinobacillus in AppNL–G–F compared to wild type mice in both sexes (Fig. 8). APP−/− mice also demonstrated consistent differences from wild type mice with a significant increase in Candidatus Phytoplasma in both sexes (Fig. 8).
Fig. 7. Phyla of the oral microbiome differed between mouse lines.
Oral swabs were taken from male and female WT, AppNL–G–F, APP/PS1, and APP−/− mice for 16S rRNA sequencing. A heat map of the relative abundance of the 12 most common phyla is shown. Statistical differences were calculated using one-way ANOVA with Tukey post-hoc test. Data are graphed as mean values +/− SEM, n=3–7. *p < 0.05, **p < 0.01, ***p < 0.001, ***p < 0.0001.
Fig. 8. Several genera differences exist in the oral microbiome between the mouse lines.
Oral swabs were taken from male and female WT, AppNL–G–F, APP/PS1, and APP−/− mice for 16S rRNA sequencing. A heat map of the relative abundance of the 30 most abundant genera. Statistical differences were calculated using one-way ANOVA with Tukey post-hoc test, *p < 0.05, **p < 0.01, ***p < 0.001, ***p < 0.0001. Data are graphed as mean values +/− SEM, n=3–7.
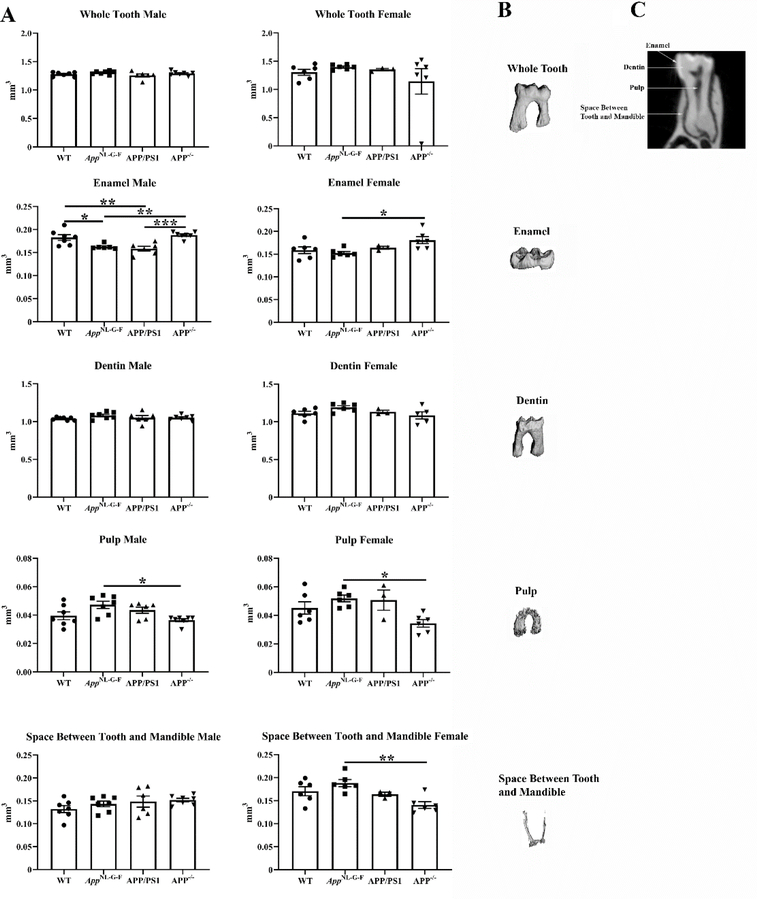
Since there were such significant changes in microbial diversity and abundance across the mouse lines, we assessed oral health by quantifying molar x-rays from each condition. Interestingly, both male AppNL–G–F and APP/PS1 mice demonstrated significantly reduced tooth enamel compared to wild type controls with no difference among females (Fig. 9).
Fig. 9. Male AppNL–G–F and APP/PS1 mice had decreased tooth enamel.
The right hemimandibles from male and female WT, AppNL–G–F, APP/PS1, and APP−/− mice were scanned. The first molar from each mouse including the enamel, dentin, pulp, and the space between the tooth and mandible was contoured and analyzed. (A) Data are graphed as mean values +/− SEM, n=3–7. Statistical differences were calculated by one-way ANOVA with Tukey post-hoc analysis, *p < 0.05, **p < 0.01, ***p < 0.001. Representative images of a (B) molar and (C) discrete subregions for quantitation are shown.
4. DISCUSSION
We demonstrated that human and mouse salivary gland ductal epithelium express APP similar to our prior work examining intestinal epithelium (14). Of the two AD mouse lines, only the female AppNL–G–F mice demonstrated a corresponding elevation in saliva Aβ 1–42 levels. However, in contrast to the APP/PS1 mouse line, both male and female AppNL–G–F mice demonstrated a significant increase in oral microbiome diversity compared to wild type mice. Interestingly, male but not female APP/PS1 and AppNL–G–F mice both demonstrated enamel thinning compared to wild type controls. These data confirm that particular AD mouse models can be used to model human saliva Aβ secretion albeit in a sex-selective fashion. In addition, our findings indicate that the oral microbiome differs between AD and control mice in a transgenic line dependent fashion. Collectively these findings correlate with numerous human findings indicating that saliva may be a source of biomarker discovery in AD (19–25) and extend recent reports of fecal microbiome differences between AD and controls to suggest similar differences may occur in the oral microbiome (48–56).
The presence of APP and Aβ immunoreactivity in the salivary glands of both mice and humans suggests that the salivary Aβ is a result of local production and secretion of Aβ by the ductal epithelium. Indeed, we have demonstrated a similar process in our prior work examining colonic epithelium (15). Secretion in this fashion would be analogous to similar well-characterized anti-microbial protein in saliva (57–59). Secretion of Aβ as one of numerous anti-microbial peptides in saliva (60) would be consistent with its reported ability to modulate bacterial growth (45–47). It is unclear whether Aβ secretion is regulated in this mechanism and will require future work. On the other hand, it is also possible that Aβ in saliva is a reflection of blood levels of the peptide and salivary secretion is a ultrafiltrate of blood Aβ. Additional studies to define a relationship between blood and saliva Aβ levels in comparison to localized salivary gland production are needed to understand the role of the peptide in saliva. Regardless of the method of Aβ production, at least for female AppNL–G–F mice, the mouse findings replicate observations from human AD patients demonstrating elevated Aβ 1–42 in saliva (19–21).
It is unclear why female AppNL–G–F but not APP/PS1 mice demonstrated increased salivary Aβ 1–42 levels or why no differences existed in male Aβ 1–42 saliva levels across the lines. Our prior work comparing brain Aβ levels between demonstrated elevated Aβ 1–42 levels in females versus males at 6 months of age in AppNL–G–F but not APP/PS1 mice (61). This suggests that female mice from this line have overall elevated Aβ 1–42 levels throughout the body and saliva peptide levels correlate with changes in the brain. This is generally consistent with original reports of preferentially increased brain Aβ 1–42 production in AppNL–G–F mice (44). This may support the ultrafiltrate hypothesis explaining the presence of saliva Aβ although mechanisms of local salivary gland Aβ production cannot be excluded. Although it is presently unknown, it is possible that the lack of elevated Aβ levels in saliva of APP/PS1 mice is due to differences in the transgenic lines. Use of the physiologic APP mouse promoter for driving production in the knock-in AppNL–G–F mice may be responsible for regulating localized salivary gland production rather than saliva Aβ simply being an excreted source of blood peptide.
The most remarkable difference observed was the significant oral microbiome diversity changes observed in both male and female AppNL–G–F mice compared to wild type controls. Surprisingly, the APP/PS1 mice did not differ dramatically from controls. Since these changes did not correlate with the absence or presence of saliva Aβ, the relationship instead appears to be related to the particular transgenic line. Of particular interest was the significant increase in Streptococcus and decrease in Actinobacillus genera in AppNL–G–F mice compared to controls. Although the Streptococcus genera are associated with a healthy oral microbiome, particular species including Streptococcus mutans are important in caries formation due, in part, to its acidogenicity and acid tolerance (62). A particular Actinobacillus, Aggregatibacter (formerly Actinobacillus) actinomycetemcomitans, is a common periodontitis associated bacteria (63). Further work will be needed to identify the particular species changes in these two genera in the AppNL–G–F compared to wild type mice. Interestingly, Aggregatibacter actinomycetemcomitans induces toxicity of Streptococcus mutans in vitro correlating precisely with the inverse relationship we observed between these genera in AppNL–G–F mice (64). Since the bacterial changes did not correlate with changes in saliva Aβ across sex it is unclear why bacterial changes are characteristic of disease in the AppNL–G–F mice. It is intriguing to speculate that microbial changes may not be a reflection of altered Aβ production but instead related to the function of APP in the epithelium. There is a well-characterized relationship between the phenotype of intestinal epithelium and the growth of gut bacteria for instance, that may also apply to the oral cavity (65).
There was an intriguing thinning of molar enamel thickness in male but not female APP/PS1 and AppNL–G–F mice compared to controls. Numerous human studies demonstrate a positive risk relationship between periodontitis and AD (26, 30, 34–36, 59) and the presence of some indication of decreased tooth health is generally consistent with oral cavity dysfunction in the AD lines. Moreover, this change in male but not female teeth was consistent for both APP/PS1 and AppNL–G–F mice providing a positive relationship between genetic predisposition for AD and tooth integrity that was sex-dependent. It is possible that older mice from either line would eventually have demonstrated a significant relationship between periodontitis and brain changes such as plaque load and gliosis and additional study is needed. Indeed, others have shown that oral infection of periodontitis-associated bacteria is capable of stimulating brain changes related to AD (66). Future work will need to compare oral microbiome, microCT, and saliva changes at distinct aging time points to contrast the temporal relationship of changes during disease and age. This may identify any age-associated changes in saliva Aβ production as well as any mechanistic linkage to changes in the oral microbiome and oral health.
CONCLUSION
We have observed a variety of sex and strain-dependent changes comparing two different AD mouse lines to wild type control mice when examining the oral cavity and saliva production. Our findings are overall consistent with the idea that Aβ is secreted in saliva, perhaps in a regulated, sex-selective fashion, as a biomarker of AD. In addition, oral microbial dysbiosis, at least in AppNL–G–F mice, Appears to be a characteristic of disease similar to changes that have been reported from feces.
HUMAN AND ANIMAL RIGHTS
Animal use was approved by the University of North Dakota Institutional Animal Care and Use Committee protocol number: 1806–10c. Work was performed in compliance with the US National Research Council’s Guide for the Care and Use of Laboratory Animals.
Supplementary Material
ACKNOWLEDGMENTS
We are most grateful to Ms. Kim Michelsen, USDA ARS, Grand Forks Human Nutrition Research Center, for her technical assistance performing the MicroCT scanning and analysis. Histological services were provided by Ms. Beth Ann DeMontigny and Ms. Donna Laturnus of the UND Histology Core Facility supported by NIH/NIGMS awards P20GM113123, U54GM128729, and UND SMHS funds. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
We are grateful to the Banner Sun Health Research Institute Brain and Body Donation Program of Sun City, Arizona for the provision of human brain and salivary glands. The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026 National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute of Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05–901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research.
We are grateful to Takashi Saito and Takaomi C. Saido from the Riken Brain Institute for providing the triple mutation knock-in AppNL–G–F mice.
FUNDING
This work was supported by NIH 1R01AG042819, NIH R01AG048993, NIH RF1AG069378, and NIH P20GM103442. Histological services were provided by the UND Histology Core Facility supported by the NIH/NIGMS awards P20GM113123, U54GM128729, and UND SMHS funds. The work of JJC was supported by the U.S. Department of Agriculture, Agricultural Research Service project plan #3062–51000-053–00D. USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely to provide specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AVAILABILITY OF DATA AND MATERIALS
All relevant data is contained within the manuscript.
REFERENCES
- 1.Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D, et al. The Swedish mutation causes early-onset Alzheimer’s disease by beta-secretase cleavage within the secretory pathway. Nature medicine. 1995; 1(12): 1291–6. [DOI] [PubMed] [Google Scholar]
- 2.Evin G, Cappai R, Li QX, Culvenor JG, Small DH, Beyreuther K, et al. Candidate gamma-secretases in the generation of the carboxyl terminus of the Alzheimer’s disease beta A4 amyloid: possible involvement of cathepsin D. Biochemistry. 1995;34(43):14185–92. [DOI] [PubMed] [Google Scholar]
- 3.Citron M, Teplow DB, Selkoe DJ. Generation of amyloid beta protein from its precursor is sequence specific. Neuron. 1995;14(3):661–70. [DOI] [PubMed] [Google Scholar]
- 4.Higaki J, Quon D, Zhong Z, Cordell B. Inhibition of beta-amyloid formation identifies proteolytic precursors and subcellular site of catabolism. Neuron. 1995; 14(3):651–9. [DOI] [PubMed] [Google Scholar]
- 5.Haass C, Hung AY, Schlossmacher MG, Teplow DB, Selkoe DJ. beta-Amyloid peptide and a 3-kDa fragment are derived by distinct cellular mechanisms. The Journal of biological chemistry. 1993;268(5):3021–4. [PubMed] [Google Scholar]
- 6.Puig KL, Combs CK. Expression and function of APP and its metabolites outside the central nervous system. Experimental gerontology. 2013;48(7):608–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akaaboune M, Allinquant B, Farza H, Roy K, Magoul R, Fiszman M, et al. Developmental regulation of amyloid precursor protein at the neuromuscular junction in mouse skeletal muscle. Molecular and cellular neurosciences. 2000;15(4):355–67. [DOI] [PubMed] [Google Scholar]
- 8.Galloway S, Jian L, Johnsen R, Chew S, Mamo JC. beta-amyloid or its precursor protein is found in epithelial cells of the small intestine and is stimulated by high-fat feeding. The Journal of nutritional biochemistry. 2007;18(4):279–84. [DOI] [PubMed] [Google Scholar]
- 9.Herzog V, Kirfel G, Siemes C, Schmitz A. Biological roles of APP in the epidermis. European journal of cell biology. 2004;83(11–12):613–24. [DOI] [PubMed] [Google Scholar]
- 10.Lee YH, Tharp WG, Maple RL, Nair S, Permana PA, Pratley RE. Amyloid precursor protein expression is upregulated in adipocytes in obesity. Obesity. 2008;16(7):1493–500. [DOI] [PubMed] [Google Scholar]
- 11.Sandbrink R, Masters CL, Beyreuther K. Beta A4-amyloid protein precursor mRNA isoforms without exon 15 are ubiquitously expressed in rat tissues including brain, but not in neurons. The Journal of biological chemistry. 1994;269(2):1510–7. [PubMed] [Google Scholar]
- 12.Selkoe DJ, Podlisny MB, Joachim CL, Vickers EA, Lee G, Fritz LC, et al. Beta-amyloid precursor protein of Alzheimer disease occurs as 110- to 135-kilodalton membrane-associated proteins in neural and nonneural tissues. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(19):7341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada T, Sasaki H, Dohura K, Goto I, Sakaki Y. Structure and expression of the alternatively-spliced forms of mRNA for the mouse homolog of Alzheimer’s disease amyloid beta protein precursor. Biochemical and biophysical research communications. 1989;158(3):906–12. [DOI] [PubMed] [Google Scholar]
- 14.Puig KL, Lutz BM, Urquhart SA, Rebel AA, Zhou X, Manocha GD, et al. Overexpression of mutant amyloid-beta protein precursor and presenilin 1 modulates enteric nervous system. Journal of Alzheimer’s disease : JAD. 2015;44(4):1263–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puig KL, Manocha GD, Combs CK. Amyloid precursor protein mediated changes in intestinal epithelial phenotype in vitro. PloS one. 2015;10(3):e0119534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashton NJ, Ide M, Scholl M, Blennow K, Lovestone S, Hye A, et al. No association of salivary total tau concentration with Alzheimer’s disease. Neurobiology of aging. 2018;70:125–7. [DOI] [PubMed] [Google Scholar]
- 17.Pekeles H, Qureshi HY, Paudel HK, Schipper HM, Gornistky M, Chertkow H. Development and validation of a salivary tau biomarker in Alzheimer’s disease. Alzheimer’s & dementia (Amsterdam, Netherlands). 2019;11:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi M, Sui YT, Peskind ER, Li G, Hwang H, Devic I, et al. Salivary tau species are potential biomarkers of Alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2011;27(2):299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bermejo-Pareja F, Antequera D, Vargas T, Molina JA, Carro E. Saliva levels of Abeta1–42 as potential biomarker of Alzheimer’s disease: a pilot study. BMC neurology. 2010;10:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee M, Guo JP, Kennedy K, McGeer EG, McGeer PL. A Method for Diagnosing Alzheimer’s Disease Based on Salivary Amyloid-beta Protein 42 Levels. Journal of Alzheimer’s disease : JAD. 2017;55(3): 1175–82. [DOI] [PubMed] [Google Scholar]
- 21.Sabbagh MN, Shi J, Lee M, Arnold L, Al-Hasan Y, Heim J, et al. Salivary beta amyloid protein levels are detectable and differentiate patients with Alzheimer’s disease dementia from normal controls: preliminary findings. BMC neurology. 2018;18(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Figueira J, Jonsson P, Nordin Adolfsson A, Adolfsson R, Nyberg L, Ohman A. NMR analysis of the human saliva metabolome distinguishes dementia patients from matched controls. Molecular bioSystems. 2016;12(8):2562–71. [DOI] [PubMed] [Google Scholar]
- 23.Huan T, Tran T, Zheng J, Sapkota S, MacDonald SW, Camicioli R, et al. Metabolomics Analyses of Saliva Detect Novel Biomarkers of Alzheimer’s Disease. Journal of Alzheimer’s disease : JAD. 2018;65(4):1401–16. [DOI] [PubMed] [Google Scholar]
- 24.Ralbovsky NM, Halamkova L, Wall K, Anderson-Hanley C, Lednev IK. Screening for Alzheimer’s Disease Using Saliva: A New APProach Based on Machine Learning and Raman Hyperspectroscopy. Journal of Alzheimer’s disease : JAD. 2019;71(4): 1351–9. [DOI] [PubMed] [Google Scholar]
- 25.Yilmaz A, Geddes T, Han B, Bahado-Singh RO, Wilson GD, Imam K, et al. Diagnostic Biomarkers of Alzheimer’s Disease as Identified in Saliva using 1H NMR-Based Metabolomics. Journal of Alzheimer’s disease : JAD. 2017;58(2):355–9. [DOI] [PubMed] [Google Scholar]
- 26.Aragon F, Zea-Sevilla MA, Montero J, Sancho P, Corral R, Tejedor C, et al. Oral health in Alzheimer’s disease: a multicenter case-control study. Clinical oral investigations. 2018;22(9):3061–70. [DOI] [PubMed] [Google Scholar]
- 27.Ship JA, DeCarli C, Friedland RP, Baum BJ. Diminished submandibular salivary flow in dementia of the Alzheimer type. Journal of gerontology. 1990;45(2):M61–6. [DOI] [PubMed] [Google Scholar]
- 28.Kalia M Dysphagia and aspiration pneumonia in patients with Alzheimer’s disease. Metabolism: clinical and experimental. 2003;52(10 Suppl 2):36–8. [DOI] [PubMed] [Google Scholar]
- 29.Scannapieco FA, Cantos A. Oral inflammation and infection, and chronic medical diseases: implications for the elderly. Periodontology 2000. 2016;72(1):153–75. [DOI] [PubMed] [Google Scholar]
- 30.Chen CK, Wu YT, Chang YC. Association between chronic periodontitis and the risk of Alzheimer’s disease: a retrospective, population-based, matched-cohort study. Alzheimer’s research & therapy. 2017;9(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foley NC, Affoo RH, Siqueira WL, Martin RE. A Systematic Review Examining the Oral Health Status of Persons with Dementia. JDR clinical and translational research. 2017;2(4):330–42. [DOI] [PubMed] [Google Scholar]
- 32.Hatipoglu MG, Kabay SC, Guven G. The clinical evaluation of the oral status in Alzheimer-type dementia patients. Gerodontology. 2011;28(4):302–6. [DOI] [PubMed] [Google Scholar]
- 33.Holmer J, Eriksdotter M, Schultzberg M, Pussinen PJ, Buhlin K. Association between periodontitis and risk of Alzheimer’s disease, mild cognitive impairment and subjective cognitive decline: A case-control study. Journal of clinical periodontology. 2018;45(11):1287–98. [DOI] [PubMed] [Google Scholar]
- 34.Ide M, Harris M, Stevens A, Sussams R, Hopkins V, Culliford D, et al. Periodontitis and Cognitive Decline in Alzheimer’s Disease. PloS one. 2016;11(3):e0151081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leira Y, Dominguez C, Seoane J, Seoane-Romero J, Pias-Peleteiro JM, Takkouche B, et al. Is Periodontal Disease Associated with Alzheimer’s Disease? A Systematic Review with Meta-Analysis. Neuroepidemiology. 2017;48(1–2):21–31. [DOI] [PubMed] [Google Scholar]
- 36.Martande SS, Pradeep AR, Singh SP, Kumari M, Suke DK, Raju AP, et al. Periodontal health condition in patients with Alzheimer’s disease. American journal of Alzheimer’s disease and other dementias. 2014;29(6):498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minn YK, Suk SH, Park H, Cheong JS, Yang H, Lee S, et al. Tooth loss is associated with brain white matter change and silent infarction among adults without dementia and stroke. Journal of Korean medical science. 2013;28(6):929–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okamoto N, Morikawa M, Tomioka K, Yanagi M, Amano N, Kurumatani N. Association between tooth loss and the development of mild memory impairment in the elderly: the Fujiwara-kyo Study. Journal of Alzheimer’s disease : JAD. 2015;44(3):777–86. [DOI] [PubMed] [Google Scholar]
- 39.Oue H, Miyamoto Y, Koretake K, Okada S, Doi K, Jung CG, et al. Tooth loss might not alter molecular pathogenesis in an aged transgenic Alzheimer’s disease model mouse. Gerodontology. 2016;33(3):308–14. [DOI] [PubMed] [Google Scholar]
- 40.Oue H, Miyamoto Y, Okada S, Koretake K, Jung CG, Michikawa M, et al. Tooth loss induces memory impairment and neuronal cell loss in APP transgenic mice. Behavioural brain research. 2013;252:318–25. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi K, Ohara T, Furuta M, Takeshita T, Shibata Y, Hata J, et al. Tooth Loss and Risk of Dementia in the Community: the Hisayama Study. Journal of the American Geriatrics Society. 2017;65(5):e95–e100. [DOI] [PubMed] [Google Scholar]
- 42.Tiisanoja A, Syrjala AM, Tertsonen M, Komulainen K, Pesonen P, Knuuttila M, et al. Oral diseases and inflammatory burden and Alzheimer’s disease among subjects aged 75 years or older. Special care in dentistry : official publication of the American Association of Hospital Dentists, the Academy of Dentistry for the Handicapped, and the American Society for Geriatric Dentistry. 2019;39(2):158–65. [DOI] [PubMed] [Google Scholar]
- 43.Kamer AR, Pirraglia E, Tsui W, Rusinek H, Vallabhajosula S, Mosconi L, et al. Periodontal disease associates with higher brain amyloid load in normal elderly. Neurobiology of aging. 2015;36(2):627–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saito T, Matsuba Y, Mihira N, Takano J, Nilsson P, Itohara S, et al. Single APP knock-in mouse models of Alzheimer’s disease. Nature neuroscience. 2014;17(5):661–3. [DOI] [PubMed] [Google Scholar]
- 45.Kumar DK, Choi SH, Washicosky KJ, Eimer WA, Tucker S, Ghofrani J, et al. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Science translational medicine. 2016;8(340):340ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moir RD, Lathe R, Tanzi RE. The antimicrobial protection hypothesis of Alzheimer’s disease. Alzheimer’s & dementia : the journal of the Alzheimer’s Association. 2018;14(12):1602–14. [DOI] [PubMed] [Google Scholar]
- 47.Soscia SJ, Kirby JE, Washicosky KJ, Tucker SM, Ingelsson M, Hyman B, et al. The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PloS one. 2010;5(3):e9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akbari E, Asemi Z, Daneshvar Kakhaki R, Bahmani F, Kouchaki E, Tamtaji OR, et al. Effect of Probiotic Supplementation on Cognitive Function and Metabolic Status in Alzheimer’s Disease: A Randomized, Double-Blind and Controlled Trial. Front Aging Neurosci. 2016;8:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nimgampalle M, Kuna Y. Anti-Alzheimer Properties of Probiotic, Lactobacillus plantarum MTCC 1325 in Alzheimer’s Disease induced Albino Rats. Journal of clinical and diagnostic research : JCDR. 2017;11(8):KC01–KC5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dinan TG, Stilling RM, Stanton C, Cryan JF. Collective unconscious: how gut microbes shape human behavior. J Psychiatr Res. 2015;63:1–9. [DOI] [PubMed] [Google Scholar]
- 51.Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, Johnson SC, et al. Gut microbiome alterations in Alzheimer’s disease. Sci Rep. 2017;7(1): 13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balin BJ, Little CS, Hammond CJ, APPelt DM, Whittum-Hudson JA, Gerard HC, et al. Chlamydophila pneumoniae and the etiology of late-onset Alzheimer’s disease. J Alzheimers Dis. 2008;13(4):371–80. [DOI] [PubMed] [Google Scholar]
- 53.Harach T, Marungruang N, Duthilleul N, Cheatham V, Mc Coy KD, Frisoni G, et al. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci Rep. 2017;7:41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pistollato F, Sumalla Cano S, Elio I, Masias Vergara M, Giampieri F, Battino M. Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr Rev. 2016;74(10):624–34. [DOI] [PubMed] [Google Scholar]
- 55.Austin SA, Combs CK. Amyloid precursor protein mediates monocyte adhesion in AD tissue and apoE(−)/(−) mice. Neurobiol Aging. 2010;31(11): 1854–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu SC, Cao ZS, Chang KM, Juang JL. Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nat Commun. 2017;8(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimizu K, Hanaoka Y, Akama T, Kono I. Ageing and free-living daily physical activity effects on salivary beta-defensin 2 secretion. Journal of sports sciences. 2017;35(7):617–23. [DOI] [PubMed] [Google Scholar]
- 58.Gillum TL, Kuennen MR, Castillo MN, Williams NL, Jordan-Patterson AT. Exercise, but not acute sleep loss, increases salivary antimicrobial protein secretion. Journal of strength and conditioning research. 2015;29(5):1359–66. [DOI] [PubMed] [Google Scholar]
- 59.Malcolm J, Sherriff A, Lappin DF, Ramage G, Conway DI, Macpherson LM, et al. Salivary antimicrobial proteins associate with age-related changes in streptococcal composition in dental plaque. Molecular oral microbiology. 2014;29(6):284–93. [DOI] [PubMed] [Google Scholar]
- 60.Jourdain ML, Velard F, Pierrard L, Sergheraert J, Gangloff SC, Braux J. Cationic antimicrobial peptides and periodontal physiopathology: A systematic review. Journal of periodontal research. 2019;54(6):589–600. [DOI] [PubMed] [Google Scholar]
- 61.Manocha GD, Floden AM, Miller NM, Smith AJ, Nagamoto-Combs K, Saito T, et al. Temporal progression of Alzheimer’s disease in brains and intestines of transgenic mice. Neurobiology of aging. 2019;81:166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanz M, Beighton D, Curtis MA, Cury JA, Dige I, Dommisch H, et al. Role of microbial biofilms in the maintenance of oral health and in the development of dental caries and periodontal diseases. Consensus report of group 1 of the Joint EFP/ORCA workshop on the boundaries between caries and periodontal disease. Journal of clinical periodontology. 2017;44 Suppl 18:S5–S11. [DOI] [PubMed] [Google Scholar]
- 63.Sakurai K, Wang D, Suzuki J, Umeda M, Nagasawa T, Izumi Y, et al. High incidence of actinobacillus actinomycetemcomitans infection in acute coronary syndrome. International heart journal. 2007;48(6):663–75. [DOI] [PubMed] [Google Scholar]
- 64.Fine DH, Furgang D, Goldman D. Saliva from subjects harboring Actinobacillus actinomycetemcomitans kills Streptococcus mutans in vitro. Journal of periodontology. 2007;78(3):518–26. [DOI] [PubMed] [Google Scholar]
- 65.Litvak Y, Byndloss MX, Baumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362(6418). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ilievski V, Zuchowska PK, Green SJ, Toth PT, Ragozzino ME, Le K, et al. Chronic oral application of a periodontal pathogen results in brain inflammation, neurodegeneration and amyloid beta production in wild type mice. PloS one. 2018;13(10):e0204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.