Abstract
Background
Gestational diabetes mellitus (GDM), a common endocrine disorder with rising prevalence in pregnancy, has been reported to be associated with alteration of gut microbiota in recent years. However, the role of gut microbiome in GDM physiopathology remains unclear. This pilot study aims to characterize the alteration of gut microbiota in GDM on species-level resolution and evaluate the relationship with occurrence of GDM.
Methods
An analysis based on 16S rRNA microarray was performed on fecal samples obtained from 30 women with GDM and 28 healthy pregnant women.
Results
We found 54 and 141 differentially abundant taxa between GDM and control group at the genus and the species level respectively. Among GDM patients, Peptostreptococcus anaerobius was inversely correlated with fasting glucose while certain species (e.g., Aureimonas altamirensis, Kosakonia cowanii) were positively correlated with fasting glucose.
Conclusions
This study suggests that there are large amounts of differentially abundant taxa between GDM and control group at the genus and the species level. Some of these taxa were correlated with blood glucose level and might be used as biomarkers for diagnoses and therapeutic targets for probiotics or synbiotics.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-021-02207-0.
Keywords: Gestational diabetes mellitus, Gut microbiota, Bacterial species, 16S rRNA microarray
Introduction
Gestational diabetes mellitus (GDM) is defined as a glucose intolerance resulting in hyperglycemia of variable severity during pregnancy [1, 2]. It is associated with short-term obstetric and perinatal complications (e.g., preeclampsia, increased cesarean delivery rates, macrosomia, birth injury) and considered to increase the long-term health risks (e.g., cardiovascular, obesity and type 2 diabetes) for the mother and the offspring [3, 4]. Due to changes in hormones such as progesterone, estrogen and placental factors, insulin sensitivity naturally declines with advancing gestation during pregnancy. In this situation, a compensatory increase in insulin secretion usually maintains a normal glucose homeostasis. However, GDM occurs if pancreatic β-cells fail to meet the demand of insulin secretion [2].
GDM affects nearly 16.5% of pregnancies worldwide, and this number tends to increase as the obesity epidemic escalates [3]. The Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study involving 15 multinational centers reported a prevalence of GDM between 9.3 and 25.5% in the global population [5]. The GDM incidence in China is also alarming, with a recent systematic review implicated a pooled GDM prevalence of 14.8% in the Chinese population [6].
Intestinal bacteria have been suggested to play important roles in the host metabolism, especially in diabetes [7–9]. Thus, it is necessary to further explore the relationship between gut microbiota and GDM. Differential abundant taxa might be used as biomarkers for early diagnosis or targets for probiotic intervention. A number of studies have reported that the significant alterations of gut microbiota in women with GDM were associated with abnormal glucose metabolism [10–13]. Nonetheless, the link between gut microbiome and GDM is still ambiguous and needs to be clarified [14]. So far, only one study used the shotgun metagenomics sequencing method to analyze the microbial species in GDM [10], other published results were based on 16S rRNA sequencing technique that only covered taxa at genus level or above [15]. Given the limitation of most previous research on gut microbiota of GDM, we applied innovative microarray technique rather than sequencing platform for its prominent microbial species level resolution, which helped to better explain the gut dysbiosis [16, 17].
Method
Materials and methods
Study design and sample collection
A cohort study was conducted to analyze the role of intestinal microbiota in the development of GDM. The protocols of this study were approved by the Ethics Committee of The Third Affiliated Hospital of Guangzhou Medical University (GD2019–033). Informed, written consent was given by all volunteers in accordance with the protocol. In addition, all methods were carried out in accordance with the relevant guidelines and regulations. In our study, the pregnant women of 22–45 years of age were enrolled. Women who had the following criteria were excluded from our study: taking probiotics, use of antibiotics or other drugs within 1 month, complications of delivery pregnancy-induced hypertension, intestinal diseases, acute gastroenteritis, autoimmune, thyroid dysfunction, liver and kidney disease. Eventually, the study cohort consisted of 58 participants including 30 women with GDM and 28 health gravida, one of whom was twin pregnancy, the rest were singleton pregnancy.
The 2-h 75 g oral glucose tolerance test (OGTT) was examined in women in their third trimester (24–28 gestation weeks) according to the International Association of the Diabetes and Pregnancy Study Groups (IADPSG) criteria for GDM [18, 19]. The participants were required to eat no less than 250 g carbohydrates 3 days before the OGTT test and at least 8 h-overnight fast before collecting the venous blood. Anthropometric measurements, basic information and the family history were obtained at the same day. Pregnant women were diagnosed with GDM if one or more of the following OGTT result were met: fasting plasma glucose (FPG), ≥5.1 mmol/liter; 1 h glucose, ≥10.0 mmol/liter; 2 h glucose, ≥8.5 mmol/liter [20]. Overweight was defined as body mass index (BMI) ≥ 25.0 kg/m2 and obese was defined as BMI ≥ 28.0 kg/m2 [21]. After instructed by pre-recorded video, the participants were required to collect at least 500 mg stool samples into sterilized sample tubes with preservative solution (Halgen, China) within 24–48 h before OGTT or dietary adjustment after diagnosis of GDM. Bacterial DNA was extracted using Halgen Stool Isolation kit (Halgen, China) according to the manufacturer’s protocol. Then DNA amplification and array hybridization were performed following the methods detailed in the previous study [16]. Venous blood was collected in the fasting state for metabolic biomarkers. Plasma glucose was analyzed by Cobas 8000 modular analyzer (Roche Diagnostics Ltd., Rotkreuz, Switzerland). Body weight and high was verified by the pregnant women’s self-report. Pregnancy trimester were determined by first semester ultrasound and last menstrual period.
Bacterial DNA extraction and microarray hybridization
Bacterial DNA was extracted from the stool samples by using the Stool DNA Extraction Kit (Halgen, China) according to the product instruction manual. DNA amplification and labeling were carried out following the previously published methods [16] before array hybridization experiments. According to our previous study [16], DNA products were hybridized with probes of the microarray (Halgen, China) designed for the entire variable regions of bacterial 16S rRNA. The relative abundance of each bacterial species was measured by the mean of the Cy5/Cy3 ratios of the corresponding species-specific probes.
Data analysis
The clinical features were analyzed using Pearson chi-square test or Fisher’s exact test as appropriate for categorical variables and independent Student’s t-test or Rank sum test was used as appropriate for continuous variables. Correlations were calculated using Spearman’s rank correlations. The microarray data was produced by the specialize program transforming the hybridization signal to relative abundance value following the methods described in our previous publication [16]. The α-diversity was calculated using and QIIME software [22] with default parameters and further compared by Wilcoxon rank-sum test. PCoA and NMDS analyses were performed by QIIME modules and visualized by R packages (version 3.5.2). Linear Discriminant Analysis (LDA) Effect Size (LEfSe) [23] analysis was performed to identify taxonomic biomarkers that characterize the differences between pregnant women with and without GDM. The p-value for each species were calculated by Wilcoxon test.
Result
Characteristics of subject recruitment
A total of 58 stool samples were collected from 30 women with GDM and 28 normoglycemic female control subjects. As shown in Table 1, there are no significant difference between two groups in demographic characteristics including height, weight, BMI before pregnancy, gestational weight gain, and the gestational week at examination. As expected, markers of glucose tolerance such as fasting plasma glucose (FPG), 75 g OGTT 1-h postprandial glucose (1 h-PG) and 2-h postprandial glucose (2 h-PG) were higher in the GDM group than in the control group (Student’s t-test P value: 0.011, 0.001, 0.001, respectively).
Table 1.
Clinical characteristic and OGTT results of GDM and control groups
| Characteristic | Women with GDM (n = 30) | Normoglycemic women (n = 28) | P value |
|---|---|---|---|
| Age (year) | 32.80 (5.22) | 30.29 (4.50) | 0.055 |
| Ht (cm) | 159.43 (5.47) | 157.43 (3.89) | 0.112 |
| Wt (kg) pregnancy | 57.78 (10.06) | 53.62 (9.43) | 0.098 |
| BMI (kg/m2) pregnancy | 22.68 (3.45) | 21.61 (3.53) | 0.173 |
| Overweight or Obese (BMI ≥ 28 kg/m2) pregnancy | 3 (10.00%) | 1 (3.60%) | 0.650 |
| Gestational weight gain (kg) at OGTT | 42.44 (7.24) | 38.98 (6.05) | 0.061 |
| Fasting glucose (mmol/liter) | 4.56 (0.48) | 4.28 (0.33) | 0.011* |
| 1 h-PG (mmol/liter) | 10.32 (0.81) | 7.32 (1.27) | 0.001* |
| 2 h-PG (mmol/liter) | 8.74 (1.32) | 6.45 (0.65) | 0.001* |
| Gestational wk. (kg) at examination | 25.4 (1.01) | 25.22 (1.58) | 0.599 |
The P value calculated by Chi-Squared test or t test. “*” mean p < 0.05. Overweight or Obese rate (BMI ≥ 28 kg/m2) pregnancy was shown as n (%), other results were shown as mean (SD)
Overall differences between GDM and control group
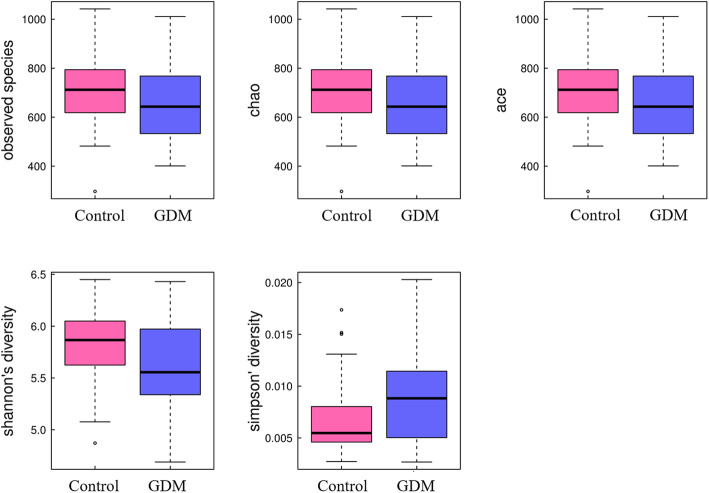
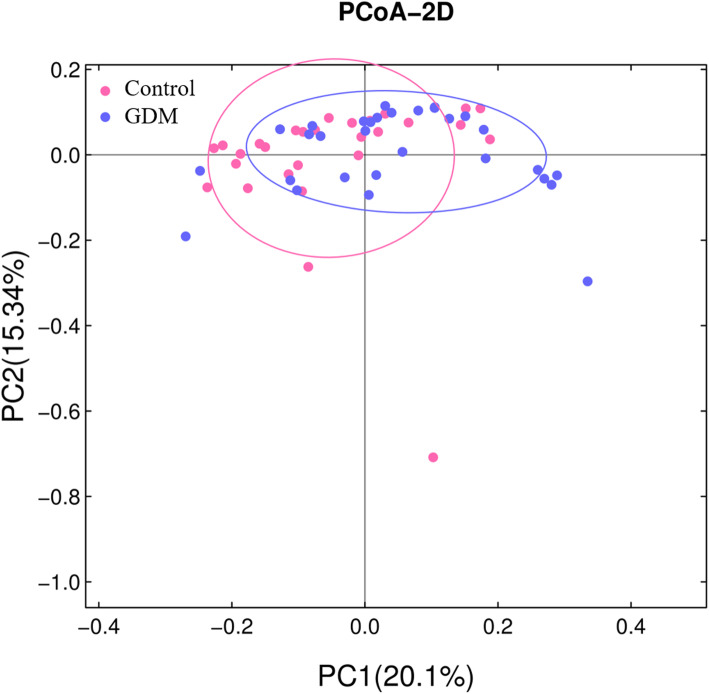
A total of 1234 unique OTUs were identified in all samples, including 1117 OTUs shared by both groups. We found that the α-diversity of GDM group were slightly but not significantly lower than that of control group (Fig. 1). On the other hand, Bray-Curtis distance-based analysis of β-diversity showed a partial separation of GDM and control groups, indicating that the microbiota structure differed between GDM and normoglycemic women (Fig. 2, ADONIS statistic: R2 = 0.03384, P value =0.017).
Fig. 1.
The α-diversity of gut microbiome in GDM and control groups. The α-diversity of GDM group were lower albeit not statistically significance, Wilcoxon rank-sum test P value for Observed species, Chao, Ace, Shannon and Simpson are 0.144, 0.144, 0.144, 0.075, 0.067, respectively
Fig. 2.
Bray Curtis Principal Coordinate (PCoA) analysis for gut microbiome of GDM and control groups. The two components of Bray Curtis PCoA plot explained 20.1 and 15.34% of the variants. ADONIS statistic: R2 = 0.03384, P = 0.017
Bacterial species with differential abundance
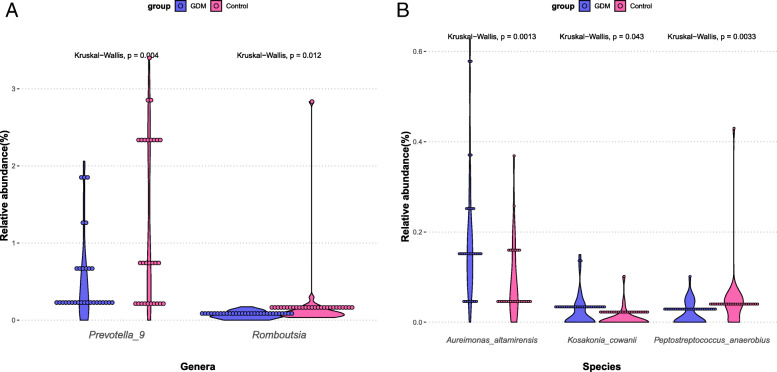
At the genus level, we found 54 genera with differential abundance between GDM and control group (Additional file 1), including 42 genera depleted in GDM (e.g., Prevotella and Romboutsia) (Fig. 3a). At the species level, there were 37 species significantly enriched in GDM patients such as, Corynebacterium spp. (Corynebacterium appendicis, Corynebacterium coyleae, Corynebacterium durum, Corynebacterium frankenforstense, Corynebacterium freneyi, Corynebacterium glaucum, Corynebacterium kroppenstedtii, Corynebacterium xerosis), Lactobacillus spp. (Lactobacillus ceti, Lactobacillus sanfranciscensis, Lactobacillus vaccinostercus) and Blautia hydrogenotrophica. Meanwhile, we also found 104 species enriched in normoglycemic pregnant women such as Bacteroides spp. (Bacteroides acidifaciens, Bacteroides intestinalis, Bacteroides nordii, Bacteroides plebeius, Bacteroides salyersiae), Bacillus spp. (Bacillus anthracis, Bacillus clausii, Bacillus idriensis, Bacillus massilioanorexius), Bifidobacterium spp. (Bifidobacterium bifidum, Bifidobacterium gallicum, Bifidobacterium longum), Clostridium spp. (Clostridium pasteurianum, Clostridium saccharogumia), Eubacterium spp. (Eubacterium hallii, Eubacterium multiforme), Prevotella spp. (Prevotella falsenii, Prevotella maculosa, Prevotella nigrescens, Prevotea oris, Prevotella paludivivens, Prevotella stercorea) (Fig. 3b). More details can be found in Additional file 2.
Fig. 3.
The relative abundance of part of differentially abundant taxa in GDM and control groups. a Part of the differentially abundant genera between GDM and control groups. b Part of the differentially abundant species between GDM and control groups
Species correlated with glucose tolerance in GDM
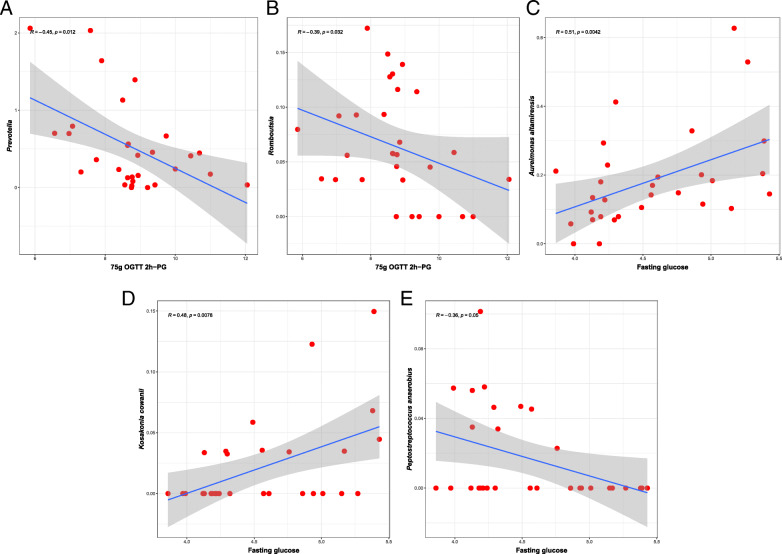
To further understand the relationship between gut microbiome and GDM, we evaluated the correlations between the differentially abundant taxa and clinical traits. At genus level, Prevotella and Romboutsia were negatively correlated with 2 h-PG in75 g OGTT in GDM group. At species level, we found some species positively correlated with fasting glucose in GDM patients, such as Aureimonas altamirensis, Kosakonia cowanii. On the other hand, Peptostreptococcus anaerobius was negatively correlated with fasting glucose in GDM group (Fig. 4, Additional file 3). These correlations were consistent with the taxonomy abundance difference between two groups and implied the potential critical role in GDM pathophysiology.
Fig. 4.
The caption was rephrased as follow: Fig. 4. Correlation between part of differential abundant taxa with glucose tolerance indicators. a Prevotella and b Romboutsia genus was negatively correlated with 75 g OGTT 2 h-PG in women with GDM. c Aureimonas altamirensis and d Kosakonia cowanii was positively correlated with fasting glucose level in women with GDM. e Peptostreptococcus anaerobius was negatively correlated with fasting glucose level in women with GDM
Discussion
The correlation between alterations of gut microbiome and GDM has been repeatedly reported [10–13, 24]. However, there is little knowledge about the composition of gut microbiota in GDM at the species level. By comparing 30 GDM patients and 28 normoglycemic pregnancy, we found the alterations in gut bacterial species and further explored their association with glucose intolerance. Our results provided more specified insights into the pathology of GDM.
By analyzing the differential bacterial species between two groups, we observed the beneficial acetate and lactate-producing bacteria (e.g., Bifidobacterium spp.) and butyrate-producing bacteria (e.g., Eubacterium spp.) depleted in GDM patients. These results were in line with previous in-vitro studies suggesting that short-chain fatty acids (SCFAs) may be able to improve insulin sensitivity and prevent inflammation induced by sterile or bacterial inflammation [25, 26]. Moreover, we found that Blautia hydrogenotrophica was more abundant in women with GDM. Blautia was reported to be the dominant genus with enriched abundance in glucose-intolerant individuals [27] and associate with high BMI [28]. Our finding was consistent with previous study reporting the elevated abundance of Blautia in GDM patients [4].
Previous studies showed that elevated Prevotella genus may contribute to impair gut permeability in women with GDM by increasing mucin oligosaccharide degradation [28–30]. In contrast, our results suggested the depletion of Prevotella genus and some Prevotella spp. in GDM. The Prevotella genus was found to be highly prevalent in non-Westerners who consume a plant-rich diet [31, 32]. Moreover, our results showed the negative association between the Prevotella genus and glucose intolerance indicator such as 2 h-PG in 75 g OGTT. It has been shown that Prevotella genus can improve glucose metabolism stimulated by the intake of prebiotics [33]. Thus, we speculated that the enriched Prevotella spp. including Prevotella falsenii, Prevotella maculosa, Prevotella nigrescens, Prevotella oris, Prevotella paludivivens and Prevotella stercorea may contribute to the higher fiber-rich diet consumption and indicates their beneficial roles in glucose metabolism. However, a minority of Prevotella spp., such as Prevotella aurantiaca was enriched in GDM, this may be explained by the high species and function diversity of Prevotella genus [34]. In general, these results shed more light on the ambiguous role of Prevotella genus within the intestinal microbiota and their effects on the host. Likewise, we also found that several Bacteroides spp. were enriched in GDM women but the majority of Bacteroides genus are enriched in control group. Similar situation also existed in other genera in our study. Thus, the association of gut microbiome at the genus level may cause oversimplified vision that does not take into account sub-genus diversity.
Additionally, our results showed that the reduced Romboutsia genus was associated with higher 2 h-PG in 75 g OGTT in GDM patients. Mangifesta et al. reported that Romboutsia genus was more abundant in healthy mucosa samples compared with that from polyps-associated or colorectal cancer tissue, which indicated that the depletion of this genus was associated with disease condition [35]. We assumed that decreased Romboutsia genus may be involved in the occurrence of GDM via changing the gastrointestinal mucosa permeability.
Furthermore, some species were positively correlated with fasting glucose in GDM patients, such as Aureimonas altamirensis, Kosakonia cowanii. These species were reported to be potential pathogens of infection, as a risk factor for causing GDM via chronic low-grade inflammation [36–39]. Whereas Peptostreptococcus anaerobius was negatively correlated with fasting glucose in GDM group. Previous findings provided supportive evidence that the abundance of P. anaerobius was significantly increased in type 2 diabetes after weight loss intervention compared with lean and obese controls [40]. Although P. anaerobius was considered to be widely distributed in human gut microbiota contributing to systemic infection [40], our results implied that P. anaerobius might play a crucial role in balancing glucose metabolism. These findings need further investigation to determine the role of above discussed taxonomy in GDM.
Applying the innovative microarray technique rather than the sequencing platform, we found large amounts of differentially abundant taxa between GDM and control group at the species level resolution, which broadens our understanding of gut dysbiosis in GDM women. However, some limitations of this study need to be addressed. First, due to the small sample size and single-centered nature, external validation is recommended for future study. Second, we cannot entirely exclude the influence of potential confounders such as diet and gravidity history on gut microbiota. Well-controlled clinical studies addressing potential confounders will be needed to validate our findings.
Conclusion
This study suggests that there are large amounts of differentially abundant taxa between GDM and control group at the genus and the species level. Some of these taxa were correlated with blood glucose level and might be used as biomarkers for diagnoses and therapeutic targets for probiotics or synbiotics.
Supplementary Information
Additional file 1. The differential taxonomy at genus level between the two groups, Wilcoxon test P value < 0.05.
Additional file 2. The differential taxonomy at species level between the two groups, Wilcoxon test P value < 0.05.
Additional file 3. Differential abundant Genera and Species correlated with glucose tolerance indicators in GDM patients, Spearman’s rank correlations < 0.05.
Acknowledgements
We thank the hospital and laboratory staff, volunteers for participating in this study.
Abbreviations
- GDM
Gestational diabetes mellitus
- OGTT
Oral glucose tolerance test
- SCFAs
Short-chain fatty acids
- FPG
Fasting plasma glucose
- 1 h-PG
1-h postprandial glucose
- 2 h-PG
2-h postprandial glucose
Authors’ contributions
Q.L., K.W., Y.L. and F.C. designed the study, performed general coordination. F.C., Y.G. and K.W. analyzed and interpreted the microbiota data. F.C., Y.L., W.H. and W.W. collected the samples and performed the examination. F.C., Y. G and K.W. were the major contributors in writing and revising the manuscript. All authors reviewed and approved the final manuscript.
Funding
This study is supported by the Research Fund for Lin He’s Academician Workstation of New Medicine and Clinical Translation; Basic and Applied Basic Research Foundation of Guangdong Province (No.2020A1515011347); Innovation and Entrepreneurship (Employment) Education Project of Guangzhou Universities (No.2020PT105).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The protocol of the study was approved by the Ethics Committee of The Third Affiliated Hospital of Guangzhou Medical University (GD2019–033). The participation was voluntary for each person involved in this study. Before launching the study, each volunteer signed an informed consent which contained the following information: (1) aims and tasks of the study; (2) protocol of the study; (3) criteria for inclusion in the study; (4) duties of a volunteer participating in the study; (5) benefits for taking part in the study; (6) potential risks, inconvenience, and adverse effects. The volunteers could refuse participation in the study at any stage without any consequences.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fang Chen and Yu Gan contributed equally to this work.
Contributor Information
Kejian Wang, Email: kejian-wang@foxmail.com.
Qing Li, Email: 81292522@163.com.
References
- 1.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 2.Baz B, Riveline JP, Gautier JF. Endocrinology of pregnancy: gestational diabetes mellitus: definition, aetiological and clinical aspects. Eur J Endocrinol. 2016;174(2):R43–R51. doi: 10.1530/EJE-15-0378. [DOI] [PubMed] [Google Scholar]
- 3.Plows JF, Stanley JL, Baker PN, Reynolds CM, Vickers MH. The pathophysiology of gestational diabetes mellitus. Int J Mol Sci. 2018;19(11):3342. [DOI] [PMC free article] [PubMed]
- 4.Ye G, Zhang L, Wang M, Chen Y, Gu S, Wang K, Leng J, Gu Y, Xie X. The gut microbiota in women suffering from gestational diabetes mellitus with the failure of glycemic control by lifestyle modification. J Diabetes Res. 2019;2019:6081248. doi: 10.1155/2019/6081248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sacks DA, Hadden DR, Maresh M, Deerochanawong C, Dyer AR, Metzger BE, Lowe LP, Coustan DR, Hod M, Oats JJ, et al. Frequency of gestational diabetes mellitus at collaborating centers based on IADPSG consensus panel-recommended criteria: the hyperglycemia and adverse pregnancy outcome (HAPO) study. Diabetes Care. 2012;35(3):526–528. doi: 10.2337/dc11-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao C, Sun X, Lu L, Liu F, Yuan J. Prevalence of gestational diabetes mellitus in mainland China: a systematic review and meta-analysis. J Diabetes Investig. 2019;10(1):154–162. doi: 10.1111/jdi.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harsch IA, Konturek PC. The role of gut microbiota in obesity and type 2 and type 1 diabetes mellitus: new insights into “old” diseases. Med Sci. 2018;6(2):32. [DOI] [PMC free article] [PubMed]
- 8.Kahn SE, Cooper ME, Del Prato S. Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present, and future. Lancet. 2014;383(9922):1068–1083. doi: 10.1016/S0140-6736(13)62154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shan K, Qu H, Zhou K, Wang L, Zhu C, Chen H, Gu Z, Cui J, Fu G, Li J, et al. Distinct gut microbiota induced by different fat-to-sugar-ratio high-energy diets share similar pro-obesity genetic and metabolite profiles in prediabetic mice. mSystems. 2019;4(5):e00219–19. [DOI] [PMC free article] [PubMed]
- 10.Kuang YS, Lu JH, Li SH, Li JH, Yuan MY, He JR, Chen NN, Xiao WQ, Shen SY, Qiu L, et al. Connections between the human gut microbiome and gestational diabetes mellitus. GigaScience. 2017;6(8):1–12. doi: 10.1093/gigascience/gix058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mokkala K, Houttu N, Vahlberg T, Munukka E, Ronnemaa T, Laitinen K. Gut microbiota aberrations precede diagnosis of gestational diabetes mellitus. Acta Diabetol. 2017;54(12):1147–1149. doi: 10.1007/s00592-017-1056-0. [DOI] [PubMed] [Google Scholar]
- 12.Crusell MKW, Hansen TH, Nielsen T, Allin KH, Ruhlemann MC, Damm P, Vestergaard H, Rorbye C, Jorgensen NR, Christiansen OB, et al. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome. 2018;6(1):89. doi: 10.1186/s40168-018-0472-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma S, You Y, Huang L, Long S, Zhang J, Guo C, Zhang N, Wu X, Xiao Y, Tan H. Alterations in gut microbiota of gestational Diabetes patients during the first trimester of pregnancy. Front Cell Infect Microbiol. 2020;10:58. doi: 10.3389/fcimb.2020.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasain Z, Mokhtar NM, Kamaruddin NA, Mohamed Ismail NA, Razalli NH, Gnanou JV, Raja Ali RA. Gut microbiota and gestational Diabetes mellitus: a review of host-gut microbiota interactions and their therapeutic potential. Front Cell Infect Microbiol. 2020;10:188. doi: 10.3389/fcimb.2020.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10(1):5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Zou Y, Ruan M, Chang L, Chen X, Wang S, Yang W, Zhang L, Guo Y, Chen Y, Zhang Y, He H, Gan Y, Wang K, Zhu X. Pediatric acute lymphoblastic leukemia patients exhibit distinctive alterations in the gut microbiota. Front Cell Infect Microbiol. 2020;10:558799. doi: 10.3389/fcimb.2020.558799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu Q, Li J, Shi Z, Chen Y, Lin L, Li J, Wang H, Yan J, Zhou Q, Li X, Li L, Zhou J, He Z. HuMiChip2 for strain level identification and functional profiling of human microbiomes. Appl Microbiol Biotechnol. 2017;101(1):423–435. doi: 10.1007/s00253-016-7910-0. [DOI] [PubMed] [Google Scholar]
- 18.American Diabetes A Diagnosis and classification of diabetes mellitus. Diabetes Care. 2012;35(Suppl 1):S64–S71. doi: 10.2337/dc12-s064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Association of D. Pregnancy Study Groups Consensus P. Metzger BE, Gabbe SG, Persson B, Buchanan TA, Catalano PA, Damm P, Dyer AR, Leiva A, et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33(3):676–682. doi: 10.2337/dc09-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.American Diabetes A. Standards of medical care in diabetes--2014. Diabetes Care. 2014;37(Suppl 1):S14–S80. doi: 10.2337/dc14-S014. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Lu FC, Department of Disease Control Ministry of Health PRC The guidelines for prevention and control of overweight and obesity in Chinese adults. Biomed Environ Sci. 2004;17(Suppl):1–36. [PubMed] [Google Scholar]
- 22.Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics. 2011;Chapter 10:Unit 10 17. doi: 10.1002/0471250953.bi1007s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrocino I, Ponzo V, Gambino R, Zarovska A, Leone F, Monzeglio C, Goitre I, Rosato R, Romano A, Grassi G, Broglio F, Cassader M, Cocolin L, Bo S. Changes in the gut microbiota composition during pregnancy in patients with gestational diabetes mellitus (GDM) Sci Rep. 2018;8(1):12216. doi: 10.1038/s41598-018-30735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy R, Nguyen-Ngo C, Lappas M. Short-chain fatty acids as novel therapeutics for gestational diabetes. J Mol Endocrinol. 2020;65(2):21–34. doi: 10.1530/JME-20-0094. [DOI] [PubMed] [Google Scholar]
- 26.Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing Colon Bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egshatyan L, Kashtanova D, Popenko A, Tkacheva O, Tyakht A, Alexeev D, Karamnova N, Kostryukova E, Babenko V, Vakhitova M, Boytsov S. Gut microbiota and diet in patients with different glucose tolerance. Endocr Connect. 2016;5(1):1–9. doi: 10.1530/EC-15-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ottosson F, Brunkwall L, Ericson U, Nilsson PM, Almgren P, Fernandez C, Melander O, Orho-Melander M. Connection between BMI-related plasma metabolite profile and gut microbiota. J Clin Endocrinol Metab. 2018;103(4):1491–1501. doi: 10.1210/jc.2017-02114. [DOI] [PubMed] [Google Scholar]
- 29.Fugmann M, Breier M, Rottenkolber M, Banning F, Ferrari U, Sacco V, Grallert H, Parhofer KG, Seissler J, Clavel T, Lechner A. The stool microbiota of insulin resistant women with recent gestational diabetes, a high risk group for type 2 diabetes. Sci Rep. 2015;5(1):13212. doi: 10.1038/srep13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Pan LL, Lv S, Yang Q, Zhang H, Chen W, Lv Z, Sun J. Alterations of gut microbiota and blood Lipidome in gestational Diabetes mellitus with hyperlipidemia. Front Physiol. 2019;10:1015. doi: 10.3389/fphys.2019.01015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcon O, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1(3):e1500183. [DOI] [PMC free article] [PubMed]
- 32.Martinez I, Stegen JC, Maldonado-Gomez MX, Eren AM, Siba PM, Greenhill AR, Walter J. The gut microbiota of rural Papua new guineans: composition, diversity patterns, and ecological processes. Cell Rep. 2015;11(4):527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 33.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Bjorck I, Backhed F. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22(6):971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Ley RE. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol. 2016;13(2):69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 35.Mangifesta M, Mancabelli L, Milani C, Gaiani F, de Angelis N, de Angelis GL, van Sinderen D, Ventura M, Turroni F. Mucosal microbiota of intestinal polyps reveals putative biomarkers of colorectal cancer. Sci Rep. 2018;8(1):13974. doi: 10.1038/s41598-018-32413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendes RE, Denys GA, Fritsche TR, Jones RN. Case report of Aurantimonas altamirensis bloodstream infection. J Clin Microbiol. 2009;47(2):514–515. doi: 10.1128/JCM.02171-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eshaghi A, Shahinas D, Patel SN, Kus JV. First draft genome sequence of Aureimonas altamirensis, isolated from patient blood culture. FEMS Microbiol Lett. 2015;362(6):fnv016. [DOI] [PubMed]
- 38.Kim N, Hwang JH, Cho YG, Kim DS, Lee HS, Lee J. First case of Aureimonas altamirensis bacteremia in Korea. Ann Lab Med. 2019;39(6):587–589. doi: 10.3343/alm.2019.39.6.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mardaneh J, Soltan-Dallal MM. Isolation and identification of E. cowanii from powdered infant formula in NICU and determination of antimicrobial susceptibility of isolates. Iran J Pediatr. 2014;24(3):261–266. [PMC free article] [PubMed] [Google Scholar]
- 40.Remely M, Hippe B, Zanner J, Aumueller E, Brath H, Haslberger AG. Gut microbiota of obese, type 2 diabetic individuals is enriched in Faecalibacterium prausnitzii, Akkermansia muciniphila and Peptostreptococcus anaerobius after weight loss. Endocr Metab Immune Disord Drug Targets. 2016;16(2):99–106. doi: 10.2174/1871530316666160831093813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The differential taxonomy at genus level between the two groups, Wilcoxon test P value < 0.05.
Additional file 2. The differential taxonomy at species level between the two groups, Wilcoxon test P value < 0.05.
Additional file 3. Differential abundant Genera and Species correlated with glucose tolerance indicators in GDM patients, Spearman’s rank correlations < 0.05.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.