Abstract
Botulinum neurotoxins are considered as one of the most potent toxins and are produced by Clostridium botulinum. It is crucial to have a rapid and sensitive method to detect the bacterium Clostridium botulinum in food. In this study, a rapid detection assay of C. botulinum in food using loop-mediated isothermal amplification (LAMP) technology was developed. The optimal primers were identified among three sets of primers designed specifically based on the partial ntnh gene encoding nontoxic-nonhaemagglutinin (NTNH) for rapid detection of the target DNA in plasmids. The optimal temperature and reaction time of the LAMP assay were determined to be 64 °C and 60 min, respectively. The chemical kit could be assembled based on these optimized reaction conditions for quick, initial high-throughput screening of C. botulinum in food samples. The established LAMP assay showed high specificity and sensitivity in detecting the target DNA with a limit of 0.0001 pg/ul (i.e., ten times more sensitive than that of the PCR method) and an accuracy rate of 100%. This study demonstrated a potentially rapid, cost-effective, and easy-operating method to detect C. botulinum in food and clinical samples based on LAMP technology.
Keywords: Clostridium botulinum, botulinum neurotoxin, LAMP, ntnh, turbidity method, fluorescence method
1. Introduction
Botulinum poisoning, caused by a botulinum neurotoxin produced by Clostridium botulinum (a gram-positive anaerobic spore-forming bacterium), is a peripheral paralysis disease with a high mortality rate [1,2,3]. Botulism is generally categorized into four naturally occurring forms: foodborne botulism, infant botulism, wound botulism, and adult intestinal colonization [4]. Botulinum toxin (BoNT) is not produced by the metabolism of the bacterium itself. The non-toxic precursor toxin (a complex of neurotoxin and either a hemagglutinin or a non-hemagglutinating active protein) is first produced inside the bacterial cells and then released from the autolyzed bacteria into the intestinal tract, where the precursor toxin gains its toxic effects by interacting with trypsin or protein kinases produced by the bacteria [3]. The BoNTs cannot be degraded by gastric acid or digestive enzymes [5], while the toxins are absorbed through the intestine and then enter the blood, acting directly on the nucleus of the brain nerves by preventing the release of acetylcholine, hindering the transmission of nerve impulses and causing muscle paralysis [1].
BoNTs are generally categorized into seven types (A to G), possibly more [6], according to the serological characteristics of its toxin production [7], with more than 40 subtypes [8]. These BoNTs are classified as Category A agents by the Centers for Disease Control and Prevention (CDC) in the United States due to being the most potent of all biological toxins. They are proteins of ~150 kDa, generally existing as one of the structural components of progenitor toxic complexes, either as the M complex (~300 kDa) consisting of BoNT associated with a nontoxic-nonhemagglutinin (NTNH) protein (~150 kDa) or as the L and LL complexes (~500 and 900 kDa, respectively) in which the M complex associates with hemagglutinin proteins [9]. Human botulism is mainly caused by types A, B, and E. BoNTs show some specific geographical distribution in different countries and regions. For example, China has the highest frequency of type A, while type B is predominant in Europe and the eastern United States [10,11]. These different types of toxins express their toxic effects with different mechanisms. For example, Chellapandi and Prisilla studied the interaction between botulinum type A toxin and the intestinal tract to reveal the common toxin transcription and metabolic regulation mechanisms shared among the type A strains [12]. Specifically, the expression of BoNT in type A strains is controlled by the nutritional factors in the intestines. Studies also showed that the neurotoxin is particularly dangerous for children under one year of age, because the normal intestinal flora of infants is not strong enough to reject the botulinum completely, rendering the bacteria the opportunity to grow and reproduce, secrete toxins, and cause poisoning (i.e., infant botulism) [13,14]. The botulinum poisoning in animals is also commonly happening [15,16], including the outbreak of the type C toxin in birds in Incheon, South Korea [17], the identification of the botulinum spores in the honey produced in Republic of Serbia [18] and Poland [19], and the contamination of honey in the Republic of Kazakhstan [20].
Recent studies focused on the structure and the acting mechanisms of the BoNTs. For example, studies showed that the type E toxin is inactivated in low acid foods and phosphate-buffered saline [21]. Moreover, molecular simulation studies on the pathogenesis of the Clostridium botulinum strain ATCC3502 revealed the high degree similarity of its proteomes with humans [22], while a mathematical model was used to demonstrate that strain ATCC3502 was used to recognize the potential drug targets due to the lack of functional homologs of the neurotoxin in the host [23]. Studies also demonstrated the medical applications of these neurotoxins. For example, studies of the high-resolution crystal structure of binding domains A3 and A4 in the neurotoxin of C. botulinum revealed their subtle differences from other binding domains, providing opportunities of making novel specific therapeutic neurotoxins [24]. Furthermore, several types of biosensors were designed and commercialized based on the optical and electrochemical characteristics of detecting the BoNTs [25,26].
The most common form of botulism is foodborne, generally caused by consumption of contaminated food due to poor processing or temperature control of processed foods [27,28,29]. It is reported by the CDC that ~15% of the 145 botulism cases reported in 2011 were foodborne (CDC Report 2011). Due to the highly toxic nature and frequent occurrence of these neurotoxins, it is critical to screen and detect C. botulinum rapidly in order to prevent further outbreak of the pathogens [30,31,32,33,34,35,36]. Moreover, it is constantly demonstrated that it is imperative to have the kits available for quick, initial high-throughput screening of the presence of the pathogens and for further control of the potential pandemic of any infectious diseases, such as the well-known SARS outbreak in 2003 and the current COVID-19 outbreak. Therefore, major botulism outbreaks could be prevented by implementing prevention-based food safety controls, including the identification of C. botulinum in a timely manner. Traditionally, the enzyme-linked immunosorbent assay (ELISA) was used to detect C. botulinum neurotoxins in various samples [37,38]. To date, PCR has shown several advantages among the various methods used to detect the BoNT-producing bacteria [31,33,39,40,41,42,43]. For example, real-time fluorescent quantitative PCR technology was used to quickly identify the bacteria by detecting the gene bont coding for the neurotoxins with high sensitivity [30,39,44,45,46,47,48,49,50,51,52]. The PCR detection methods are generally favored due to their high speed, high sensitivity, high-throughput, and direct amplification and detection of DNA without the limitation by the bacterial survival status in the samples [33], while the disadvantages of PCR methods include the complicated procedures and requirements of professional operation, leading to their limited applications. Furthermore, the DNA extraction methods were optimized for the detection of C. botulinum in environmental samples [53].
The loop-mediated isothermal amplification (LAMP) technology is capable of amplifying DNA fragments under isothermal conditions [54]. Since its invention, the LAMP technology was significantly improved from many aspects and was established as the rapid, simple, and cost-effective alternative method to complement PCR techniques [55,56,57,58]. The LAMP reaction needs up to four or six primers (a pair of external primers, a pair of internal primers, and a pair of loop primers), designed specifically based on the conserved regions of the target sequence to identify specifically six or eight independent regions on the target sequence, and initiates a self-circulating chain displacement reaction with the Bst large fragment polymerase. A large amount of target DNA is synthesized in ~60 min under 60 to 65 °C with a by-product of white magnesium pyrophosphate precipitated [54,59]. Due to its high specificity, constant working temperature, high speed, and high sensitivity [60,61,62], the LAMP technology has been widely used (as reviewed in [63]) in detections of bacteria and viruses [64,65,66], drug resistance genes [57,58], parasites [67], and fetal gender identification [68]. Furthermore, the LAMP technology also showed great potential in rapidly detecting pathogenic bacteria in food. For example, the detection of artificial contamination of Bacillus cereus in cow milk was completed in ~20 min by using the real-time fluorescent LAMP [69].
The purpose of our study was to establish a novel laboratory method to detect C. botulinum (i.e., the bont gene clusters) rapidly in various types of food samples using the LAMP technology. Specifically, we designed and identified the optimal primers for the LAMP reactions to amplify a partial DNA fragment (557 bp) of the NTNH coding gene ntnh of the C. botulinum located immediately upstream of the gene bont, which encodes BoNT as the target sequence, in order to detect the bacteria (i.e., via the presence of the bont gene cluster), instead of using the bacterium itself (due to the lack of access to the most potent toxin-producing bacteria and the safety concern of culturing these bacteria in the laboratory). We further optimized the LAMP assay to determine the optimal temperature and reaction time of the LAMP reactions, determined the specificity and sensitivity of the primers designed for the LAMP assay, and verified the established LAMP method in various types of food samples. To our knowledge, this is the first report on the detection of C. botulinum based on the LAMP technology using gene ntnh as the DNA marker. We expected that the optimized working conditions of this LAMP assay could be implemented for further construction of kits for quick, initial high-throughput screening of food and clinical samples.
2. Materials and Methods
2.1. Reagents and Instruments
Reagents (of analytical purity) Chelex-100 and betaine were purchased from Sigma, USA; manganese chloride, magnesium sulfate, potassium chloride, sodium hydroxide, EDTA, and ammonium sulfate from the Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China); Tris-HCl from the Shanghai Shengke Biotechnology Co., Ltd. (Shanghai, China); Triton X-100 from the Beijing Meilaibo (MyLab) Medical Technology Co., Ltd. (Beijing, China); dNTPs from Pharmacia, USA; NP-40 from Fluka, USA; 2× Tap MIX Kit from the Tiangen Biochemical Technology Co., Ltd. (Beijing, China); Agarose from Amresco, USA; LAMP DNA Amplification Kit, LAMP reaction tube, and calcein (the fluorescent detection reagent) from the Eiken Chemical Co., Ltd. (Tochigi, Japan); the real-time turbidity meter (LA-320C) from the Rongyan Chemical Co., Ltd. (Tokyo, Japan); thermostatic metal bath (HB-2) from the Wealtech Corporation (Sparks, NV, USA); Spectrophotometer (NanoQTM) from the CapitalBio Technology Co., Ltd. (Beijing, China); gel imaging system (Quantun -ST5) from the Gel imager VILBER (Beijing, China); and PCR thermal cycler (ETC811) from the Gene Amplifier Suzhou Dongsheng Xingye Scientific Instrument Co., Ltd. (Suzhou, China).
2.2. Selection of Target DNA Fragment
The NTNH coding gene ntnh of Clostridium botulinum was chosen as the target DNA marker for examination of the specificity tests [47,70,71]. Both the genes encoding BoNT and NTNH were previously used as the target DNA markers in detection of the bacterium C. botulinum, as indicated by the presence of the bont gene clusters [15,20,33,39,48,49,50,51,56,71,72]. Studies showed that the gene ntnh is a universally present component of bont gene clusters and is present in all bacterial strains producing BoNTs but absent from non-toxic strains [73,74]. Due to its highly toxic nature, the access to C. botulinum is extremely limited in China. A partial DNA fragment (557 bp) of gene ntnh identified by the bioinformatic analysis at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/ (accessed on 15 April 2021)) was artificially synthesized and purified by using the phosphoramidite method (Sangon Biotech, Shanghai, China). The nucleic acid sequence was subcloned to the linearized vector pMD18-T and then transformed into E. coli DH5α. The target DNA fragment was ligated into the plasmid pMD18-T (50 ng/μL) using the following system: T4 DNA Ligase 1 μL (Sangon Biotech, Shanghai, China), 10× ligation buffer 2 μL, DNA 2.14 μL (50 ng/μL), pMD18-T Vector 1 μL (50 ng/μL), H2O 13.86 μL, in a total volume of 20 μL, cultured under 16 °C for 4 h. The plasmids were then transformed into E. coli DH5α for storage. To confirm the successful transformation of the plasmids into E. coli DH5α, a pair of primers was designed to amplify the target sequence using PCR (forward primer 5′-GGCGAATCTTTGAGTATAGATG-3′ and reverse primer 5′-TTTCTACTATGTTTGCTCCTGG-3′) with the PrimeSTAR HS DNA Polymerase (Takara, Dalian, China). The PCR amplification procedure was as follows: The DNA template was pre-denatured for 5 min and denatured for 30 s at 94 °C, annealed for 50 s at 55 °C, extended for 40 s at 72 °C, and finally extended for 10 min at 72 °C after 35 cycles. The PCR products were examined using 1% agarose gel electrophoresis and sequenced using either the above forward or reverse primers (Sangon Biotech, Shanghai, China).
2.3. Design and Determination of the Optimal LAMP Primers
The LAMP primers based on the target DNA fragment of gene ntnh described above were designed using the LAMP primer design software Primer Explorer (http://primerexplorer.jp/e/, accessed on 12 July 2017). Three groups of LAMP reactions with three sets of primers (RD1, RD2, and RD3) were designed based on the six independent regions of the botulinum-specific target DNA sequence (Table 1). Two loop primers were designed to accelerate the amplification reactions (Shanghai Biotech Biotechnology Co., Ltd. Shanghai, China).
Table 1.
Clostridium botulinum Specific Primers Used in LAMP Reactions.
| Primer Sets | Primer Names and Types | DNA Sequences (5′ to 3′) |
|---|---|---|
| RD1 | F3 (outer forward primer) | AAGTATAAATTCCCCGGTAGA |
| B3 (outer backward primer) | GCTTGTAAGAACTTATCTTTTTCAC | |
| FIP (forward inner primer) | TGGGAGCAACCTTAAAAGCCTTAAAAAATGTTGTAGTAGTTAGAGCT | |
| BIP (backward inner primer) | GGTGGCGCCAGAGAGATATTAGAATCATATATTCCCCCATCAA | |
| LF (loop forward primer) | AAACCGTATCAGTTTTTCT | |
| LB (loop backward primer) | ATGGCGAATCTTTGAGTATA | |
| RD2 | F3 (outer forward primer) | GTTCTTACAAGCCATTATTACTTTG |
| B3 (outer backward primer) | AAGGAAATGGAATAGTACTTGAA | |
| FIP (forward inner primer) | ATCCATAAGGAAATGGAATAGCTGTAAAGAATTAATAGTACTAACGCTGG | |
| BIP (backward inner primer) | TATAGGTGGAGGGTATTATGCACCTTTATTAGATTTTGGTGCTGATCC | |
| RD3 | F3 (outer forward primer) | ATGATTCTAATTTTCTTTCACAAGA |
| B3 (outer backward primer) | GATTTTGGTGCTGATCCAA | |
| FIP (forward inner primer) | TCCCCAGCGTTAGTACTATTAATTCTGTGAAAAAGATAAGTTCTTACAAGC | |
| BIP (backward inner primer) | TGATTTCTACAGCTATTCCATTTCCTTCATATTAGGTGCATAATACCCTC |
2.4. LAMP Reactions
The 25-μL LAMP reaction mixtures contained 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100, 0.8 M Betaine, 8 mM MgSO4, 1.4 mM dNTP, 8U Bst DNA polymerase, 40 pmol primers FIP and BIP, 5 pmol primers F3 and B3, 20 pmol primers LB and LF, and 1 μL template DNA with varied concentrations based on different samples. The mixtures were set in the LAMP reaction tubes at a constant amplification temperature of 60–65 °C for 60 min. The optimal primers were selected based on the time of amplification of LAMP reaction and the concentration of the amplified DNA templates. Each reaction was performed in triplicates.
2.5. Detection of the Amplification Products of LAMP Reactions
The LAMP products were detected using two methods. First, the direct visual inspection was based on the change of the fluorescence of the LAMP reaction containing 1 µL of calcein [75,76]. A positive reaction was indicated by the color change from orange to green in the reaction, while a negative reaction was indicated by the failure of color change from orange to green. The advantage of the fluorescence detection was that the color change of the LAMP reactions was observed directly either by the naked eye under natural light or the UV light. Second, in the detection of turbidity method [22], the amplification products of the LAMP assay were examined using spectrophotometry. Specifically, the value of the optical density at 650 nm (OD650) of the magnesium pyrophosphate (Mg2P2O7) produced in the LAMP reactions was measured every 6 s using a Loopamp real-time turbidimeter (LA-320C, Eiken Chemical Co., Ltd., Tokyo, Japan) and was plotted against reaction time to generate the curve to determine the positive or negative reactions.
2.6. Determination of the Optimal Temperature for LAMP Reaction
The established LAMP reaction system using the optimal LAMP primers was used to identify the optimal temperature for the LAMP reactions. The temperature of the LAMP reactions was set to 60, 61, 62, 63, 64, 65, 66, and 67 °C, respectively, with the LAMP reaction time set to 60 min. The OD650 values of the reactions were measured every 6 s as described above.
2.7. Specificity of LAMP Reactions
The specificity tests were carried out based on the above established LAMP reaction system with the optimal primers and reaction temperature of 64 °C using a total of 30 species of bacteria, with 14 being pathogenic, 9 being non-toxic species in the genus of Clostridium, and 7 being taxonomically closely related to Clostridium (Table 2; CapitalBio Technology Co., Ltd., Beijing, China). The target DNA fragment ligated in the plasmids and the double-distilled water were used as the positive and reaction controls, respectively. These 30 strains of bacteria were provided by the CapitalBio Technology Co., Ltd. (Beijing, China). The culture and the extraction of genomic DNA of these bacteria were completed in the P3 laboratory of the CapitalBio Technology Co., Ltd. (Beijing, China) based on standard protocols. The genomic DNA of the bacteria was extracted using the Chelex method. Specifically, the equal volume of both the bacterial pellet in 200 µL phosphate-buffered saline (PBS) and the Chelex DNA extraction buffer (25 mM NaOH, 10 mM Tris-HCl, 1% Triton X-100, 1% NP-40, 0.1 mM EDTA, and 2% Chelex-100) were mixed and incubated at 100 °C for 10 min and cooled immediately on ice. The supernatant collected from the centrifugation at 14,000× g for 2 min was used as the template in further amplifications of the LAMP assay and PCR method. The genomic DNA was also prepared by 10-fold serial dilutions to give concentrations ranging from 1 ng/µL to 0.000001 pg/µL as needed.
Table 2.
Thirty Strains of Bacteria Selected for Specificity Test of LAMP Reactions.
| Number of Bacteria | Species of Bacteria |
|---|---|
| 1 | Shigella flexneri ATCC12022 * |
| 2 | Escherichia coli ATCC25922 * |
| 3 | Yersinia enterocolitica ATCC23715 * |
| 4 | Enterococcus faecalis ATCC29212 * |
| 5 | Pseudomonas aeruginosa 10211 * |
| 6 | Staphylococcus aureus 1897/2722 * |
| 7 | Streptococcus dysgalactiae ATCC12388 * |
| 8 | Bacillus cereus ATCC11778 * |
| 9 | Streptococcus pneumoniae 31002 * |
| 10 | Enterococcus faecium ATCC29212 * |
| 11 | Vibrio Parahemolyticus ATCC17802 * |
| 12 | Staphylococcus epidermidis 1.2429 * |
| 13 | Salmonella enterica ATCC14028 * |
| 14 | Streptococcus uberis ATCC700407 * |
| 15 | Clostridium tetani ATCC19406 |
| 16 | Clostridium perfringens NCTC8798 |
| 17 | Clostridium difficile ATCC9689 |
| 18 | Clostridium beijerinckii NCIMB8052 |
| 19 | Clostridium acetobutylicum ATCC824 |
| 20 | Clostridium sporogenes ATCC19404 |
| 21 | Clostridium sordellii ATCC9714 |
| 22 | Clostridium butyricum ATCC25755 |
| 23 | Clostridium tertium CICC10820 |
| 24 | Bntorobater sakazakii ATCC29544 |
| 25 | Clostridium bifermentans CICC22952 |
| 26 | Clostridium thermosaccharolyticum ATCC7956 |
| 27 | Bacillus stearothermophilus ATCC7953 |
| 28 | Bacillus thuringiensis LSZ9408 |
| 29 | Bacillus subtilis ATCC6633 |
| 30 | Bacillus megaterium DSM90 |
Symbol “*” after the strain names indicates a pathogenic species. All these strains were obtained from the CapitalBio Technology Co., Ltd. (Beijing, China).
2.8. Sensitivity of LAMP Assay and PCR Method
The sensitivity of the above established LAMP assay was examined using target DNA fragment with 10-fold serial dilutions after quantification. Double-distilled water was used as a reaction control. The amplified products of LAMP reaction were detected using both the fluorescence and the turbidity methods, as described above. The mixtures of the 25-µL volume PCR contained 12.5 µL Taq mix reagents, 1 pmol primers F3 and B3, and 1 µL DNA template with 10-fold serial dilutions. The PCR amplification cycles included: pre-denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, and a final extension at 72 °C for 7 min. Agarose (1%) gel electrophoresis under 120 V for 35 min was used to examine the PCR-amplified products, which were then stained with ethidium bromide and imaged to identify the sensitivity of PCR method in the detection of Clostridium botulinum.
2.9. Validation of the Optimized LAMP Assay and Potential Kit Assembly
The established LAMP assay was evaluated using a total of 24 samples, including eight samples of Korean kimchi (i.e., preserved cabbage) and two samples of preserved meat (pork), bacon, sour meat (pork), salami (pork), ham (pork), fish product (canned sardine), soybean paste, and dairy product (powdered cow milk), respectively, purchased from the local grocery stores with the target DNA fragment as the positive control. Genomic DNA was extracted from these samples using various types of DNA extraction kits (e.g., Qiagen, Takara, Sigma-Aldrich, and Fisher Scientific) according to manufacturer’s instructions (Table 3). The amplification products of LAMP reactions were detected using the turbidity method, as described above. The overall performance of this LAMP assay was assessed using the national food safety standards in China (GB/T 4789.12-2016). The samples showing the positive LAMP reactions were further confirmed by 16S rRNA sequencing, and the 16S rRNA DNA sequences were used to carry out Basic Local Alignment Search Tool (BLAST) analysis at NCBI (https://www.ncbi.nlm.nih.gov/ accessed on 15 April 2021). Specifically, the concentration and purity of the genomic DNA were determined using the spectrophotometer (NanoDrop 2000) and 1% agarose gel electrophoresis. The mixtures of 50-μL volume of PCR contained PrimeSTAR HS DNA Polymerase 25 μL, 16S rRNA universal primer-27F (5′-AGAGTTTCCTTGGCTCAG-3′) 1 μL and primer-1492R (5′-ACGGHTACCTTGTTTGGACTT-3′) 1 μL, genomic DNA 1 μL, and double-distilled H2O 22 μL. The PCR amplification procedure was as follows: 94 °C pre-denaturation for 5 min, followed by 35 cycles of 94 °C denaturation for 30 s, 55 °C annealing for 50 s, 72 °C extension for 40 s, and a final extension of 72 °C for 10 min. The PCR product (5 μL) was examined using 1% agarose gel electrophoresis. The purified PCR products were sequenced using either primer-27F or primer-1492R by the CapitalBio Technology Co., Ltd. (Beijing, China). The construction of the chemical kits for the rapid detection of Clostridium botulinum in the food or clinical samples was recommended based on the optimized conditions of the LAMP reaction established in our study.
Table 3.
A Total of 184 Food Samples Tested Using the LAMP Assay Established in This Study.
| Sample | Genomic DNA Concentration (ng/μL) |
|---|---|
| Korean kimchi 1–16 | 1.8–45.2 |
| Preserved meat (pork) 1–8 | 143.5–154.6 |
| Bacon 1–8 | 89.5–124.2 |
| Sour meat (pork) 1–8 | 123.5–180.1 |
| Salami (pork) 1–8 | 125.2–165.6 |
| Ham (pork) 1–8 | 68.5–95.6 |
| Fish product (canned sardine) 1–8 | 105.5–127.3 |
| Soybean paste 1–8 | 176.2–275.2 |
| Dairy product (powdered cow milk) 1–8 | 125.8–151.1 |
| Pork rib 1–8 | 106.2–143.5 |
| Fish product (canned anchovy) 1–8 | 51.8–70.1 |
| Fish product (canned tuna) 1–8 | 19.6–26.9 |
| Fish product (canned salmon) 1–8 | 82.5–95.4 |
| Fish product (canned yellow croaker) 1–8 | 29.8–40.1 |
| Stinky tofu 1–8 | 215.1–235.7 |
| Dried tofu 1–8 | 135.9–158.7 |
| Vegetarian meat 1–8 | 60.8–73.5 |
| Vegetarian cow stomach 1–8 | 40.2–51.1 |
| Vegetarian ham 1–8 | 74.6–92.7 |
| Quail egg (canned) 1–8 | 28.9–41.7 |
| Chicken foot (canned) 1–8 | 50.2–67.1 |
| Honey 1–8 | 95.2–105.2 |
3. Results
3.1. The Optimal Primers for LAMP Reactions
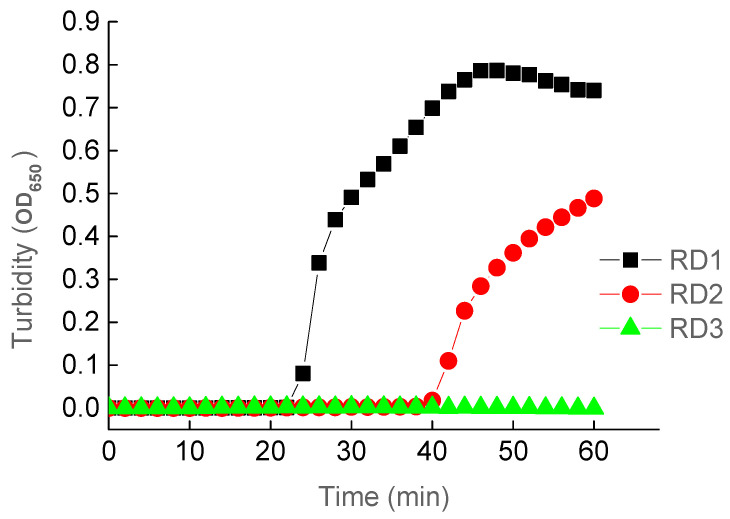
The results of the LAMP assays using primer sets RD1 and RD2 showed positive reactions, while RD3 failed to amplify the target DNA fragment (Figure 1). Primer set RD1 was identified as the most optimal because this set of primers was faster (~20 min) than RD2 to amplify the target DNA. The higher efficiency of the LAMP reactions using RD1 was indicated by the higher OD650 values (~45 min) than that of RD2 measuring the magnesium pyrophosphate (Mg2P2O7) generated in the amplifications of the target DNA fragment. Therefore, the optimal primer set RD1 was used in the subsequent LAMP reactions. These results demonstrated that the use of loop primers in RD1 significantly shortened the amplification time by ~50%. Previous studies showed similar results—that the application of loop primers decreased the LAMP reaction time [77]. These studies further demonstrated the importance of using the loop primers in the LAMP reactions.
Figure 1.
LAMP Reactions Using Three Sets of Primers (RD1, RD2, and RD3).
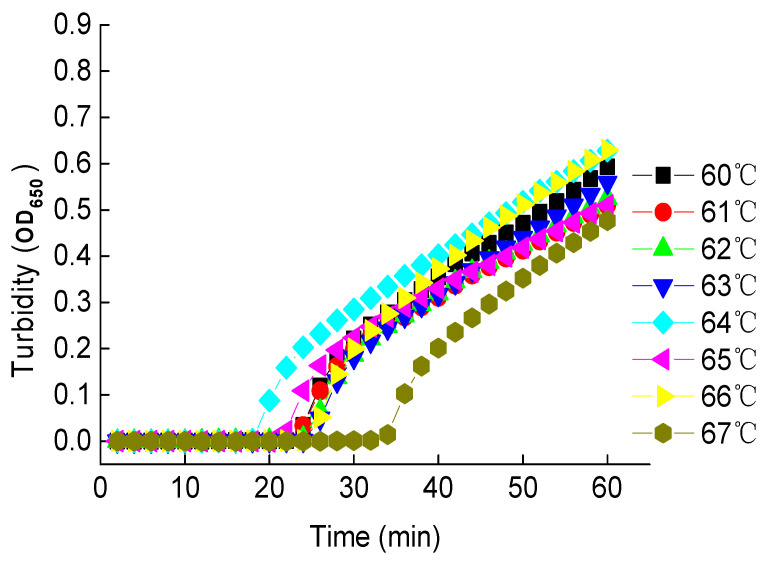
3.2. The Optimal Temperature for LAMP Reactions
Using the optimal primer set RD1, LAMP reactions under various temperatures were carried out to determine the optimal temperature for LAMP assays. Results showed that the fastest amplification (~18 min) was obtained under 64 °C with a reaction time of 60 min, while the slowest amplification (~35 min) was observed under 67 °C (Figure 2). The amplification times fell between 20 and 24 min under other temperatures investigated. The highest OD650 values were observed in 60 min under all of these temperatures. Therefore, the optimal temperature and reaction time were set to 64 °C and 60 min, respectively, for further LAMP reactions. This temperature fell into the general range of optimal temperatures from 60 to 70 °C, while this reaction time was generally used in LAMP reactions. It was noted that plots showing the OD650 values vs. reaction time did not reach the plateau in 60 min. It was expected the amplification reactions would continue given the extended reaction time and the sufficient amount of ingredients loaded in the LAMP reaction tube. However, in 60 min, the concentration of magnesium pyrophosphate precipitated was sufficient enough to indicate the positive amplification of the LAMP reaction; therefore, it was not necessary to extend the reaction time beyond 60 min.
Figure 2.
The LAMP Reactions Under Different Temperatures.
3.3. Specificity of the LAMP Assay
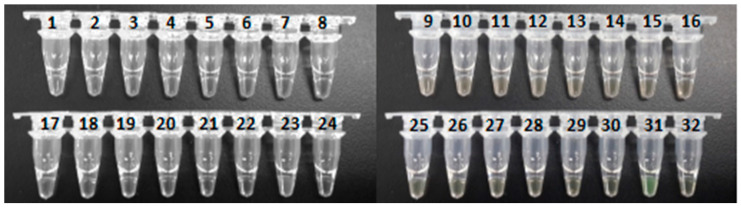
The specificity of the LAMP assay was examined by using the plasmid containing the target DNA fragment as the positive control and the double-distilled water as the reaction control, with the LAMP reaction time set to 60 min and reaction temperature of 64 °C. As expected, results showed that the 30 non-botulinum strains of bacteria and the reaction control did not show the amplification of the target DNA fragment, which was amplified in the positive control in 20 min using primer set RD1 (Figure 3). The results of the specificity experiments were also examined using the fluorescence method (Figure 4). Again, it was clearly observed that only the positive control changed color from orange to green. These results of both turbidity method and fluorescence visual inspection demonstrated the high specificity of the established LAMP assay under the optimized reaction conditions.
Figure 3.
Specificity of the LAMP Assay Examined Using the Turbidity Method. Symbols of Black Squares Indicate the Positive Control Containing the Target DNA Fragment. No Amplification Is Observed in the 30 Strains of Bacteria (Table 2) and the Reaction Control
Figure 4.
Specificity of the LAMP Assay Examined Using the Visible Fluorescence Method. The Numbers of the Samples Are the Same as Those Listed in Table 2. Samples 31 and 32 Are Positive and Reaction Controls, Respectively.
Previous studies using PCR technology extensively utilized the ntnh gene as the DNA marker to detect the bont gene clusters with high specificity and sensitivity [15,19,20,48,50,51,70]. Studies using LAMP technology to detect the BoNT-producing Clostridium botulinum types A and B showed high specificity with the bont genes [56]. In comparison to the study by Sakuma et al. [56], we used the partial DNA fragment of gene ntnh as the DNA marker to detect the bont gene clusters because the ntnh gene is located immediately upstream of the gene bont. These results indicated that both the BoNT and NTNH genes are appropriate DNA markers to be used in either the PCR technology or the LAMP reaction to detect Clostridium botulinum. It was noted that because the target DNA fragment was artificially synthesized based on the conservative region of the ntnh gene, the specificity of these results was further confirmed by the 16S rRNA sequencing in the verification below.
3.4. Sensitivity of the LAMP Assay and PCR
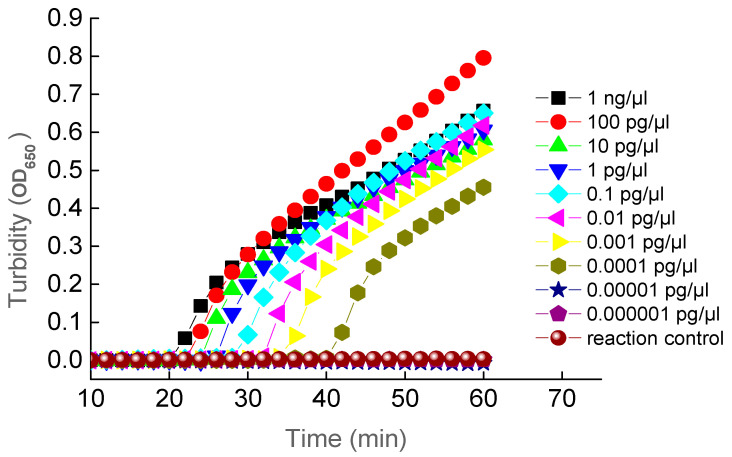
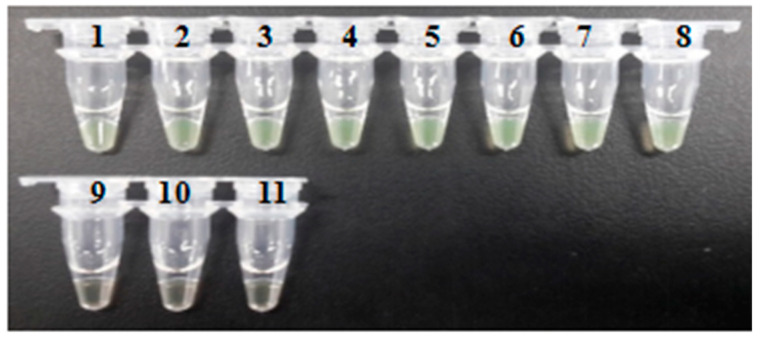
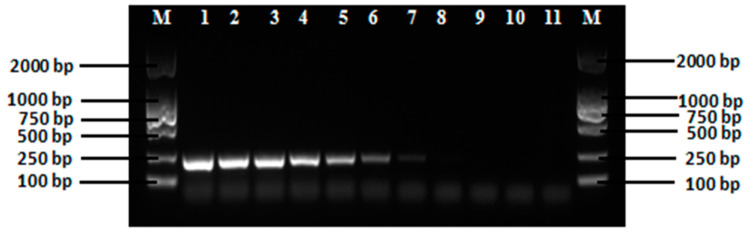
The sensitivity of the established LAMP reactions was examined by diluting the target DNA to a series of concentrations ranging from 1 ng/µL to 0.000001 pg/µL. These results were examined using both the turbidity method (Figure 5) and the visual fluorescence method (Figure 6) showing that the lowest concentration of the target DNA being amplified by LAMP reactions was 0.0001 pg/µL. The sensitivity of PCR in detecting the target DNA fragment was carried out using primers F3 and B3 with an expected length of amplified DNA fragment of 222 bp. Results showed that the lowest concentration of the target DNA being amplified was 0.001 pg/µL (Figure 7). These data demonstrated that the PCR method showed sensitivity ten times lower than that of the LAMP assay. These results demonstrated that the LAMP assay showed greater sensitivity than that of the PCR method. Previous studies reported similar results with comparable sensitivity as our research. The LAMP technology has generally shown increased sensitivity in comparison to the PCR method [54,56,57,76,78,79].
Figure 5.
Sensitivity of LAMP Assay under Various Concentrations of the Target DNA Examined Using Turbidity Method.
Figure 6.
Sensitivity of the LAMP Assay Examined Using Visible Fluorescence Method. The Concentrations of the Target DNA in Samples 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 are 1 ng/µL, 100 pg/µL, 10 pg/µL, 1 pg/µL, 0.1 pg/µL, 0.01 pg/µL, 0.001 pg/µL, 0.0001 pg/µL, 0.00001 pg/µL, and 0.000001 pg/µL, Respectively. Sample 11 is the Reaction Control.
Figure 7.
Sensitivity of PCR using Primers F3 and B3 with the Target DNA Fragment of 222 bp. The Concentrations of the Target DNA in Lanes 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 are 1 ng/µL, 100 pg/µL, 10 pg/µL, 1 pg/µL, 0.1 pg/µL, 0.01 pg/µL, 0.001 pg/µL, 0.0001 pg/µL, 0.00001 pg/µL, and 0.000001 pg/µL, Respectively. Sample 11 is the Reaction Control. Lane M Contains DNA Markers.
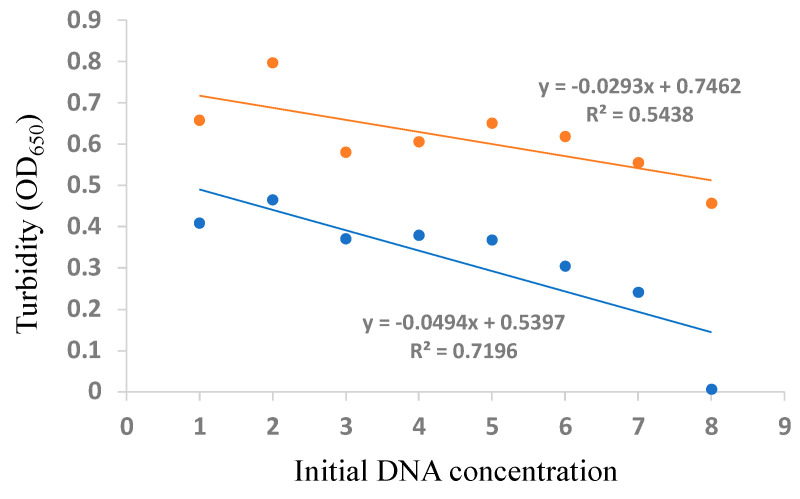
The calibration curves of C. botulinum detection based on the initial DNA concentration vs. the turbidity in the LAMP reaction at 40 and 60 min were constructed (Figure 8).
Figure 8.
Calibration Curves of Clostridium botulinum Detection Based on the Initial DNA Concentration vs. Turbidity in the LAMP Reaction at 60 min (orange dots and line) and 40 min (blue dots and line). The Initial DNA Concentrations of 1,000; 100; 10; 1; 0.1; 0.01; 0.001; and 0.0001 pg/µL are Normalized Based on “log(DNA concentration) + 5” Corresponding to Labels of 1–9, Respectively, on the x-axis.
These studies demonstrated clearly that the LAMP assay contained more advantages over the PCR method (Table 4). First, the LAMP assay is operated in an isothermal condition, while the PCR is carried out in a temperature-cycling environment, which is always time-consuming and requires high precision of the PCR instruments. Moreover, the LAMP method is evaluated using the turbidity method in a positive reaction by real-time examination of the reaction mixture with direct visual fluorescence detection without performing the agarose gel electrophoresis. In comparison to the previous studies taking 24 h for the real-time PCR assay to screen enrichment cultures of bacterial strains using ntnh gene as the target DNA markers [48], our LAMP assay was completed with a positive detection in less than 90 min, including the extraction of DNA samples (below). Second, the specificity and sensitivity are greatly increased by the LAMP method. This is because the LAMP assay uses the specifically designed primers to recognize the target sequences of four or six of the six to eight independent regions in the target DNA sequence, while PCR primers recognize only two independent regions of the target sequence. Third, studies also showed that the purification of DNA samples is generally not required in the LAMP reactions because it is less susceptible to different components in the samples than the PCR method [80]. Although it was noted that the LAMP method may show a high rate of false positive reactions due to its high amplification efficiency of target DNA fragment [81], this type of potential contamination could be avoided by spatially separating the reagent preparation and the LAMP reactions. We note the limitation of our study. Specifically, the current study was conducted using a relatively small sample size. Therefore, for better comparing the sensitivity and specificity of our proposed method to the PCR approach, we suggest additional experiments in the future with larger sample sizes and a wider array of food products for further comparing the external validity of these testing methods.
Table 4.
Comparison of the Efficiency of LAMP and PCR Methods in Detecting Clostridium botulinum in Food Samples.
| Condition | LAMP | PCR |
|---|---|---|
| Temperature of reaction | Isothermal | Cycling |
| Time of completion | Short (~90 min) | Long (~24 h) |
| Detection method | Direct examination of turbidity in a positive reaction with visual fluorescence | Requirement of high precision of detection on the agarose gel electrophoresis |
| Specificity and sensitivity | Increased by specifically designed primers to recognize four or six independent regions in the target DNA sequence | PCR primers recognize only two independent regions of the target sequence |
| Purification of DNA sample | Not required due to its less susceptibility | Required |
3.5. Verification of the Established LAMP Assay
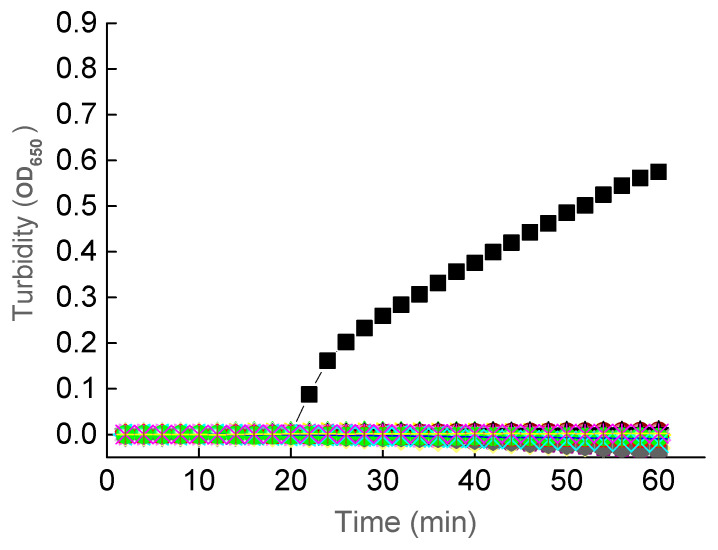
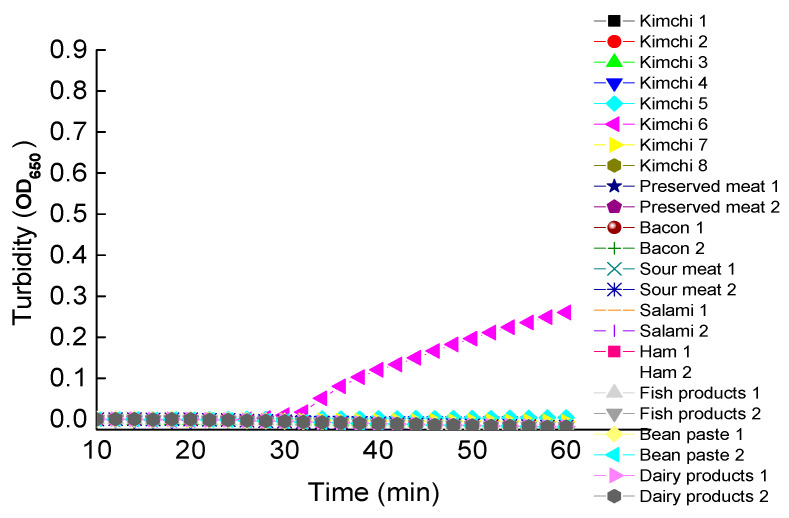
Using the established LAMP assay in this study, a group of 184 food samples purchased from the local grocery stores was tested for the detection of Clostridium botulinum (Figure 9; many samples with the absence of C. botulinum not shown). Results showed that the amplification (~25 min) was detected in one of the eight samples of Korean kimchi (i.e., sample 6), indicating the possible contamination or presence of C. botulinum. The possible presence of C. botulinum in this sample of Korean kimchi and the absence in other tested samples are probably due to the different procedures of processing these different types of food. Specifically, Korean kimchi is generally made with substantial exposure to the open air without complete sterilization. Overall, the entire procedure of the LAMP assay was completed with a positive detection in less than 90 min, including the extraction of DNA samples. This sample was further confirmed by 16S rRNA sequencing and the BLAST at NCBI. Results of the nucleotide BLAST search in the NCBI database showed that the 16S rRNA sequence (of 1445 bp in length) derived from the positive Korean kimchi sample 6 was closely related to those of various strains of C. botulinum (e.g., NCTC 13319, CFSAN064329, DFPST0029, B515, B609, B305, B742, and F1425) among many of the top hits of the BLAST search with homology over 99%. These results demonstrated the 100% compliance rate based on the food safety standard in China (GB/T 4789.12-2016). It was noted that this LAMP assay could be further verified by using clinical samples. However, due to our limited access to these samples, these verifications could not be completed in this study. It was previously demonstrated constantly that it is crucial to have easy-operating, cost-effective, and reliable kits available for initial high-throughput screening of the pathogens and for further control of the potential pandemic of any infectious diseases, such as the SARS outbreak in 2003 and the current COVID-19 outbreak. Although there are some commercial kits available for the detection of Clostridium botulinum (e.g., https://www.coleparmer.com/; https://www.bc-diagnostics.com/; and https://www.kitpcr.com/ accessed on 15 December 2020) [82], limitations of these commercial kits include either limited number of types of toxins being detected with low sensitivity or the involvement of PCR-based operations. Therefore, our development of the novel LAMP assay for quick detection of Clostridium botulinum is practically important.
Figure 9.
The LAMP Reactions of Eight Samples of Korean Kimchi and Two Samples of Preserved Meat (Pork), Bacon, Sour Meat (Pork), Salami (Pork), Ham (Pork), Fish Product (Canned Sardine), Soybean Paste, and Dairy Product (Powdered Cow Milk), Respectively. The Genomic DNA Concentrations of These Samples are Given in Table 3.
4. Conclusions
Development of a fast and reliable protocol to detect pathogens is always crucial for early diagnosis and control of a potential outbreak. We established a novel LAMP assay to detect the BoNT-producing bacterium Clostridium botulinum quickly. Furthermore, this method is easy to operate and cost-effective, showing high specificity and sensitivity in comparison to the PCR method. In our establishment of this novel detecting method, instead of using the bacterium itself (due to lack of access to the most potent toxin-producing bacteria), we identified a partial DNA fragment (557 bp) of the NTNH coding gene ntnh of the Clostridium botulinum located immediately upstream of the gene coding for BoNT as the target sequence for examination of the specificity and sensitivity of the LAMP assay. The optimal primers, temperature, and reaction time were determined to be RD1, 64 °C, and 60 min, respectively. The validity of the method was confirmed by using this LAMP assay to detect the presence of Clostridium botulinum in one of the 16 Korean kimchi samples tested. These optimized conditions for the LAMP assay could be adopted to make chemical kits for quick initial high-throughput screening of Clostridium botulinum in food and clinical settings.
Author Contributions
Conceptualization, Y.C., H.L., and L.Y.; methodology, L.W., R.S., and J.E.S.S.; software, Y.C. and H.L.; validation, Y.C., H.L., and L.Y.; formal analysis, Y.C., H.L., and L.Y.; investigation, L.W., R.S., and J.E.S.S.; resources, Y.C. and H.L.; data curation, Y.C., H.L., and L.Y.; writing—original draft preparation, Y.C., H.L., L.Y., L.W., R.S., J.E.S.S., and F.S.; writing—review and editing, Y.C., H.L., and F.S.; supervision, Y.C.; project administration, Y.C., H.L., and F.S.; funding acquisition, Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key project of the Department of Science and Technology of Jilin Province, China (20150204064NY). The APC was funded by H.L.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shukla H.D., Sharma S.K. Clostridium botulinum: A bug with beauty and Weapon. Crit. Rev. Microbiol. 2005;31:11–18. doi: 10.1080/10408410590912952. [DOI] [PubMed] [Google Scholar]
- 2.Dutta S.R., Passi D., Singh M., Singh P., Sharma S., Sharma A. Botulinum toxin the poison that heals: A brief review. Natl. J. Maxillofac Surg. 2016;7:10–16. doi: 10.4103/0975-5950.196133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulain B., Popoff M.R. Why are botulinum neurotoxin-producing bacteria so diverse and botulinum neurotoxins so toxic? Toxins. 2019;11:34. doi: 10.3390/toxins11010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sobel J. Botulism. Clin. Infect. Dis. 2005;41:1167–1173. doi: 10.1086/444507. [DOI] [PubMed] [Google Scholar]
- 5.Gu S., Rumpel S., Zhou J., Strotmeier J., Bigalke H., Perry K., Shoemaker C.B., Rummel A., Jin R. Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science. 2012;335:977–981. doi: 10.1126/science.1214270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang S., Masuyer G., Zhang J., Shen Y., Lundin D., Henriksson L., Miyashita S., Martrnez-Carranza M., Dong M., Stenmark P. Identification and characterization of a novel botulinum neurotoxin. Nat. Commun. 2017;8:14130. doi: 10.1038/ncomms14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatheway C.L. In: Closridium botulinum: Ecology and Control in Foods. Hauschild A.H.W., Dodds K.L., editors. Marcel Dekker, Inc.; New York, NY, USA: 1993. pp. 3–20. [Google Scholar]
- 8.Peck M.W., Smith T.J., Anniballi F., Austin J.W., Bano L., Bradshaw M., Cuervo P., Cheng L.W., Derman Y., Dorner B.G., et al. Historical perspectives and guidelines for botulinum neurotoxin subtype nomenclature. Toxins. 2017;9:38. doi: 10.3390/toxins9010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kukreja R.V., Singh B.R. Comparative role of neurotoxin-associated proteins in the structural stability and endopeptidase activity of botulinum neurotoxin complex types A and E. Biochemistry. 2007;46:14316–14324. doi: 10.1021/bi701564f. [DOI] [PubMed] [Google Scholar]
- 10.Popoff M.R. Ecology of neurotoxigenic strains of clostridia. Curr. Top. Microbiol. Immunol. 1995;195:1–29. doi: 10.1007/978-3-642-85173-5_1. [DOI] [PubMed] [Google Scholar]
- 11.Hatheway C.L. Toxigenic clostridia. Clin. Microbiol. Rev. 1990;3:66–98. doi: 10.1128/CMR.3.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chellapandi P., Prisilla A. Clostridium botulinum type A-virulome-gut interactions: A systems biology insight. Hum. Microbiome J. 2018;7–8:15–22. doi: 10.1016/j.humic.2018.01.003. [DOI] [Google Scholar]
- 13.Cagan E., Peker E., Dogan M., Caksen H. Infant botulism. Eur. J. Med. 2010;42:92–94. doi: 10.5152/eajm.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xin W., Huang Y., Ji B., Li P., Wu Y., Liu J., Wang X., Yang H., Kang L., Gao S., et al. Identification and characterization of Clorstridium botulinum strains associated with an infant botulism case in China. Anaerobe. 2019;55:1–7. doi: 10.1016/j.anaerobe.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 15.Grenda T., Kwiatek K. Application of molecular-biology methods to the diagnosis of botulism in mallard ducks. Bull. Vet. Inst. Pulawy. 2009;53:365–368. [Google Scholar]
- 16.Grenda T., Kukier E., Goldsztejn M., Kozieł N., Kwiatek K. Botulism: Current problem in veterinary medicine. Med. Weter. 2016;72:152–156. [Google Scholar]
- 17.Son K., Kim Y.K., Woo C., Wang S.J., Kim Y., Oem J.K., Jheong W., Jeong J. Minimizing an outbreak of avian botulism (Clostridium botulinum type C) in Incheon, South Korea. J. Vet. Med. Sci. 2018;80:553–556. doi: 10.1292/jvms.17-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matović K., Mišić D., Karabasil N., Nedić N., Dmitrić M., Jevtić G. Clostridium botulinum spores in European honey bees from Serbia. J. Apic. Res. 2019;58:420–426. doi: 10.1080/00218839.2018.1560654. [DOI] [Google Scholar]
- 19.Grenda T., Grabczak M., Sieradzki Z., Kwiatek K., Pohorecka K., Skubida M., Bober A. Clostridium botulinum spores in Polish honey samples. J. Vet. Sci. 2018;19:635–642. doi: 10.4142/jvs.2018.19.5.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mustafina R., Maikanov B., Wiśniewski J., Tracz M., Anusz K., Grenda T., Kukier E., Goldsztejn M., Kwiatek K. Contamination of honey produced in the Republic of Kazakhstan with Clostridium botulinum. Bull. Vet. Inst. Pulawy. 2015;59:241–246. doi: 10.1515/bvip-2015-0036. [DOI] [Google Scholar]
- 21.Maier M.B., Schweiger T., Lenz C.A., Vogel R.F. Inactivation of non-proteolytic Clostridium botulinum type E in low-acid foods and phosphate buffer by heat and pressure. PLoS ONE. 2018;13:e0200102. doi: 10.1371/journal.pone.0200102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhardwaj T., Haque S., Somvanshi P. In silico identification of molecular mimics involved in the pathogenesis of Clostridium botulinum ATCC 3502 strain. Microb. Pathog. 2018;121:238–244. doi: 10.1016/j.micpath.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Bhardwaj T., Somvanshi P. A computational approach using mathematical modeling to assess the peptidoglycan biosynthesis of Clostridium botulinum ATCC 3502 for potential drug targets. Gene Rep. 2018;12:179–186. doi: 10.1016/j.genrep.2018.07.002. [DOI] [Google Scholar]
- 24.Davies J.R., Rees J., Liu S.M., Acharya K.R. High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains. J. Struct. Biol. 2018;202:113–117. doi: 10.1016/j.jsb.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Eivazzadeh-Keihan R., Pashazadeh-Panahi P., Baradaran B., de la Guardia M., Hejazi M., Sohrabi H., Mokhtarzadeh A., Maleki A. Recent progress in optical and electrochemical biosensors for sensing of Clostridium botulinum neurotoxin. Trends Anal. Chem. 2018;103:184–197. doi: 10.1016/j.trac.2018.03.019. [DOI] [Google Scholar]
- 26.Walper S.A., Aragoneś G.L., Sapsford K.E., Brown C.W., III, Rowland C.E., Breger J.C., Medintz I.L. Detecting biothreat agents: From current diagnostics to developing sensor technologies. ACS Sens. 2018;3:1894–2024. doi: 10.1021/acssensors.8b00420. [DOI] [PubMed] [Google Scholar]
- 27.Wangroongsarb P., Kohda T., Jittaprasartsin C., Suthivarakom K., Kamthalang T., Umeda K., Sawanpanyalert P., Kozaki S., Ikuta K. Molecular characterization of Clostridium botulinum isolates from foodborne outbreaks in Thailand, 2010. PLoS ONE. 2014;9:e77792. doi: 10.1371/journal.pone.0077792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter A.T., Peck M.W. Genomes, neurotoxins and biology of Clostridium botulinum Group I and Group II. Res. Microbiol. 2015;166:303–317. doi: 10.1016/j.resmic.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scalfaro C., Auricchio B., De Medici D., Anniballi F. Foodborne botulism: An evolving public health challenge. Infect. Dis. 2019;51:97–101. doi: 10.1080/23744235.2018.1524584. [DOI] [PubMed] [Google Scholar]
- 30.Lindstrom M., Korkeala H. Laboratory diagnostics of botulism. Clin. Microbiol. Rev. 2006;19:298–314. doi: 10.1128/CMR.19.2.298-314.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grenda T., Kukier E., Kwiatek K. Methods and difficulties in detection of Clostridium botulinum and its toxins. Pol. J. Vet. Sci. 2014;17:195–205. doi: 10.2478/pjvs-2014-0029. [DOI] [PubMed] [Google Scholar]
- 32.Maslanka S.E., Lúquez C., Dykes J.K., Tepp W.H., Pier C.L., Pellett S., Raphael B.H., Kalb S.R., Barr J.R., Rao A., et al. A novel botulinum neurotoxin, previously reported as serotype H, has a hybrid-like structure with regions of similarity to the structures of serotypes A and F and is neutralized with serotype A antitoxin. J. Infect. Dis. 2016;213:379–385. doi: 10.1093/infdis/jiv327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thirunavukkarasu N., Johnson E., Pillai S., Hodge D., Stanker L., Wentz T., Singh B., Venkateswaran K., McNutt P., Adler M., et al. Botulinum neurotoxin detection methods for public health response and surveillance. Front. Bioeng. Biotechnol. 2018;6:80. doi: 10.3389/fbioe.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao A.K., Lin N.H., Griese S.E., Chatham-Stephens K., Badell M.L., Sobel J. Clinical Criteria to Trigger Suspicion for Botulism: An evidence-based tool to facilitate timely recognition of suspected cases during sporadic events and outbreaks. Clin. Infect. Dis. 2018;2018(Suppl. 1):66. doi: 10.1093/cid/cix814. [DOI] [PubMed] [Google Scholar]
- 35.Hobbs R.J., Thomas C.A., Haliwell J., Gwenin C.D. Rapid detection of botulinum neurotoxins—A review. Toxins. 2019;11:418. doi: 10.3390/toxins11070418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nepal M.R., Jeong T.C. Alternative methods for testing botulinum toxin: Current status and future perspectives. Biomol. Ther. 2020;28:302–310. doi: 10.4062/biomolther.2019.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma S.K., Ferreira J.L., Eblen B.S., Whiting R.C. Detection of type A, B, E, and F Clostridium botulinum neurotoxins in foods by using an amplified enzyme-linked immunosorbent assay with digoxigenin-labeled antibodies. Appl. Environ. Microbiol. 2006;72:1231–1238. doi: 10.1128/AEM.72.2.1231-1238.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maslanka S.E., Lúquez C., Raphael B.H., Dykes J.K., Joseph L.A. Utility of botulinum toxin ELISA A, B, E, F kits for clinical laboratory investigations of human botulism. Botulinum J. 2011;2:72–92. doi: 10.1504/TBJ.2011.041817. [DOI] [Google Scholar]
- 39.De Medici D., Anniballi F., Wyatt G.M., Lindstrom M., Messelhaußer U., Aldus C.F., Delibato E., Korkeala H., Peck M.W., Fenicia L. Multiplex PCR for detection of botulinum neurotoxin-producing Clostridia in clinical, food, and environmental samples. Appl. Environ. Microbiol. 2009;75:6457–6461. doi: 10.1128/AEM.00805-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh A.K., Stanker L.H., Sharma S.K. Botulinum neurotoxin: Where are we with detection technologies? Crit. Rev. Microbiol. 2013;39:43–56. doi: 10.3109/1040841X.2012.691457. [DOI] [PubMed] [Google Scholar]
- 41.Koh C.Y., Schaff U.Y., Piccini M.E., Stanker L.H., Cheng L.W., Ravichandran E., Singh B.R., Sommer G., Singh A.K. Centrifugal microfluidic platform for ultrasensitive detection of botulinum toxin. Anal. Chem. 2015;87:922–928. doi: 10.1021/ac504054u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babrak L., Lin A., Stanker L.H., McGarvey J., Hnasko R. Rapid microfluidic assay for the detection of botulinum neurotoxin in animal sera. Toxins. 2016;8:13. doi: 10.3390/toxins8010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng W.L., Land M.K., Tam C., Brandon D.L., Stanker H.L. Technologies for Detecting Botulinum Neurotoxins in Biological and Environmental Matrices. InTech; London, UK: 2016. pp. 125–144. [Google Scholar]
- 44.Huang Y., Shi Y., Ye C., Xu X. Establishment of real-time PCR assays for rapid detection of Clostridium botulinum type A and B. Dis. Surveill. 2019;34:844–848. [Google Scholar]
- 45.Wu L., Li Y., Yan A., Jiang Z. Detection of Clostridium botulinum type A and B strains in food poisoning sample by real-time fluorescent quantitative PCR. Chin. J. Health Lab. Technol. 2018;28:163–165. [Google Scholar]
- 46.Lindstrom M., Keto R., Markkula A., Nevas M., Hielm S., Korkeala H. Multiplex PCR assay for detection and identification of Clostridium botulinum types A, B, E, and F in food and fecal material. Appl. Environ. Microbiol. 2001;67:5694–5699. doi: 10.1128/AEM.67.12.5694-5699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fach P., Micheau P., Mazuet C., Perelle S., Popoff M. Development of real-time PCR tests for detecting botulinum neurotoxins A, B, E, F producing Clostridium botulinum, Clostridium baratii and Clostridium butyricum. J. Appl. Microbiol. 2009;107:465–473. doi: 10.1111/j.1365-2672.2009.04215.x. [DOI] [PubMed] [Google Scholar]
- 48.Raphael B.H., Anreadis J.D. Real-time PCR detection of the nontoxic nonhemagglutinin gene as a rapid screening method for bacterial isolates harboring the botulinum neurotoxin (A-G) gene complex. J. Microbiol. Methods. 2007;71:343–346. doi: 10.1016/j.mimet.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 49.Williamson C.H.D., Vazquez A.J., Hill K., Smith T.J., Nottingham R., Stone N.E., Sobek C.J., Cocking J.H., Fernández R.A., Caballero P.A., et al. Differentiating botulinum neurotoxin-producing Clostridia with a simple, multiplex PCR assay. Appl. Environ. Microbiol. 2017;83:e00806-17. doi: 10.1128/AEM.00806-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hill B.J., Skerry J.C., Smith T.J., Arnon S.S., Douek D.C. Universal and specific quantitative detection of botulinum neurotoxin genes. BMC Microbiol. 2010;10:267. doi: 10.1186/1471-2180-10-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maikanov B., Mustafina R., Auteleyeva L., Wisniewski J., Anusz K., Grenda T., Kwiatek K., Goldsztejn M., Grabczak M. Clostridium botulinum and Clostridium perfringens occurrence in Kazakh honey samples. Toxins. 2019;11:472. doi: 10.3390/toxins11080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peck M.W., Plowman J., Aldus C.F., Wyatt G.M., Izurieta W.P., Stringer S.C., Barker G.C. Development and application of a new method for specific and sensitive enumeration of spores of nonproteolytic Clostridium botulinum types B, E, and F in foods and food materials. Appl. Environ. Microbiol. 2010;76:6607–6614. doi: 10.1128/AEM.01007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Maréchal C., Fourour S., Ballan V., Rouxel S., Souillard R., Chemaly M. Detection of Clostridium botulinum group III in environmental samples from farms by real-time PCR using four commercial DNA extraction kits. BMC Res. Notes. 2018;11:441. doi: 10.1186/s13104-018-3549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W., Huang S., Liu N., Dong D., Yang Z., Tang Y., Ma W., He X., Ao D., Xu Y., et al. Establishment of an accurate and fast detection method using molecular beacons in loop-mediated isothermal amplification assay. Sci. Rep. 2017;7:40125. doi: 10.1038/srep40125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakuma T., Kurosaki Y., Fujinami Y., Takizawa T., Yasuda J. Rapid and simple detection of Clostridium botulinum types A and B by loop-mediated isothermal amplification. J. Appl. Microbiol. 2009;106:1252–1259. doi: 10.1111/j.1365-2672.2008.04084.x. [DOI] [PubMed] [Google Scholar]
- 57.Liu W., Zou D., Li Y., Wang X., He X., Wei X., Shao C., Li X., Shang W., Yu K., et al. Sensitive and rapid detection of the New Delhi metallo-beta-lactamase gene by loop-mediated isothermal amplification. J. Clin. Microbiol. 2012;50:1580–1585. doi: 10.1128/JCM.06647-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X., Liu W., Wang J., Zou D., Wang X., Yang Z., Yin Z., Gui Q., Shang W., Li H., et al. Rapid detection of Trichinella spiralis larvae in muscles by loop-mediated isothermal amplification. Int. J. Parasitol. 2012;42:1119–1126. doi: 10.1016/j.ijpara.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 59.Huang S., Xie T., Yan J. Principle and application of loop-mediated isothermal amplification. Chin. J. Biol. 2011;24:1511–1513. [Google Scholar]
- 60.Wang X.R., Wu l.F., Wang Y., Ma Y.Y., Chen F.H., Qu H.L. Rapid detection of Staphlococcus aureus by loop-mediated isothermal amplification. Appl. Biochem. Biotechnol. 2015;175:882–891. doi: 10.1007/s12010-014-1328-x. [DOI] [PubMed] [Google Scholar]
- 61.Tirloni E., Bernardi C., Drago S., Stampone G., Pomillio F., Cattaneo P., Stella S. Evaluation of a loop-mediated isothermal amplification method for the detection of Listeria monocytogenes in dairy food. Ital. J. Food Saf. 2017;6:6890. doi: 10.4081/ijfs.2017.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yaqing H., Wenping Z., Zhiyi Y., Xionghu W., Shouyi Y., Hong Y., Yingchun D., Guifang H. Detection of human Enterovirus 71 reverse transcription loop-mediated isothermal amplification (RT-LAMP) Lett. Appl. Microbiol. 2012;54:233–239. doi: 10.1111/j.1472-765X.2011.03198.x. [DOI] [PubMed] [Google Scholar]
- 63.Becherer L., Borst N., Bakheit M., Frischmann S., Zengerle R., von Stetten F. Loop-mediated isothermal amplification (LAMP)—Review and classification of methods for sequence-specific detection. Anal. Methods. 2020;12:717–746. doi: 10.1039/C9AY02246E. [DOI] [Google Scholar]
- 64.Yamazaki W., Kumeda Y., Misawa N., Nakaguchi Y., Nishibuchi M. Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of the tdh and trh genes of Vibrio parahaemolyticus and related Vibrio species. Appl. Environ. Microbiol. 2010;76:820–828. doi: 10.1128/AEM.02284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fukuda S., Sasaki Y., Seno M. Rapid and sensitive detection of norovirus genomes in oysters by a two-step isothermal amplification assay system combining nucleic acid sequence-based amplification and reverse transcription-loop-mediated isothermal amplification assays. Appl. Environ. Microbiol. 2008;74:3912–3914. doi: 10.1128/AEM.00127-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Q., Domesie K.J., Ge B. Loop-mediated isothermal amplification for Salmonella detection in food and feed: Current applications and future directions. Foodborne Pathog. Dis. 2018;15:309–331. doi: 10.1089/fpd.2018.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kubota R., Jenkins D.M. Real-time duplex applications of loop-mediated amplification (LAMP) by assimilating probes. Int. J. Mol. Sci. 2015;16:4786–4799. doi: 10.3390/ijms16034786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Almasi M.A., Almasi G. Loop-mediated isothermal amplification (LAMP) for embryo sex determination in pregnant women at eight weeks of pregnancy. J. Reprod. Infertil. 2017;18:197–204. [PMC free article] [PubMed] [Google Scholar]
- 69.Jia Y., Fu B., Wang Y., Ma X., Zhang X., Yuan N., Zhang W. Detection of Bacillus cereus in milk by real-time fluorescence loop-mediated isothermal amplification method. Food Sci. 2016;37:184–189. [Google Scholar]
- 70.Hutson R.A., Zhou Y., Collins M.D., Johnson E.A., Hatheway C.L., Sugiyama H. Genetic characterization of Clostridium botulinum type A containing silent type B neurotoxin gene sequences. J. Biol. Chem. 1996;271:10786–10792. doi: 10.1074/jbc.271.18.10786. [DOI] [PubMed] [Google Scholar]
- 71.Woudstra C., Maréchal C.L., Souillard R., Bayon-Auboyer M.H., Anniballi F., Auricchio B., De Medici D., Bano L., Koene M., Sansonetti M.H., et al. Molecular gene profiling of Clostridium botulinum Group III and its detection in naturally contaminated samples originatig from various European countries. Appl. Environ. Microbiol. 2015;81:1495–2505. doi: 10.1128/AEM.03915-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grenda T., Kukier E., Goldsztejn M., Kwiatek K., Kozieł N. Optimisation of protocol for Clostridium botulinum detection in mink feed. Bull. Vet. Inst. Pulawy. 2015;59:377–382. doi: 10.1515/bvip-2015-0056. [DOI] [Google Scholar]
- 73.East A.K., Collins M.D. Conserved structure of genes encoding components of botulinum neurotoxin complex M and the sequence of the gene coding for the nontoxic component in nonproteolytic Clostridium botulinum type F. Curr. Microbiol. 1994;29:69–77. doi: 10.1007/BF01575751. [DOI] [PubMed] [Google Scholar]
- 74.Hill K.K., Smith T.J., Helma C.H., Ticknor L.O., Foley B.T., Svensson R.T., Brown J.L., Johnson E.A., Smith L.A., Okinaka R.T., et al. Genetic diversity among botulinum neurotoxin-producing clostridial strains. J. Bacteriol. 2007;189:818–832. doi: 10.1128/JB.01180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui Q., Liu W., Yang Z., Lu S., Yuan J., Li Y. Quick detection of L2 type cephalosporinase beta lactamase gene in Stenotrophomonas maltophilia by loop-mediated isothermal amplification. Mil. Med. Sci. 2013;37:943–946. [Google Scholar]
- 76.Tian X., Feng J., Wang Y. Direct loop-mediated isothermal amplification assay for on-site dtection of Staphylococcus aureus. FEMS Microbiol. Lett. 2018;365:fny092. doi: 10.1093/femsle/fny092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagamine K., Hase T., Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes. 2002;16:223. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 78.Wu X., Lv B., Jiang W., Bai L., Wu G., Wang J., Wang R., Pan A., Tang X. Rapid detection technique of duck-derived components in beef and sheep products by LAMP. J. Food Sci. Biotechnol. 2019;38:126–133. [Google Scholar]
- 79.Fan R.H., Huang M., Zhong H., Ye X., Luo Y. Establishment of RT-LAMP assay for detection of Freesia mosaic virus. Exp. Method Technol. 2019;45:153–156. [Google Scholar]
- 80.Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Bioph. Meth. 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 81.Mori Y., Nagamine K., Tomita N., Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 82.Sharma S.K., Eblen B.S., Bull R.L., Burr D.H., Whiting R.C. Evaluation of lateral-flow Clostridium botulinum neurotoxin detection kits for food analysis. Appl. Environ. Microbiol. 2005;71:3935–3941. doi: 10.1128/AEM.71.7.3935-3941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.