Abstract
The nuclear factor-erythroid 2 p45-related factor 2 (NRF2, also called Nfe2l2) and its cytoplasmic repressor, Kelch-like ECH-associated protein 1 (KEAP1), are major regulators of redox homeostasis controlling a multiple of genes for detoxification and cytoprotective enzymes. The NRF2/KEAP1 pathway is a fundamental signaling cascade responsible for the resistance of metabolic, oxidative stress, inflammation, and anticancer effects. Interestingly, a recent accumulation of evidence has indicated that NRF2 exhibits an aberrant activation in cancer. Evidence has shown that the NRF2/KEAP1 signaling pathway is associated with the proliferation of cancer cells and tumerigenesis through metabolic reprogramming. In this review, we provide an overview of the regulatory molecular mechanism of the NRF2/KEAP1 pathway against metabolic reprogramming in cancer, suggesting that the regulation of NRF2/KEAP1 axis might approach as a novel therapeutic strategy for cancers.
Keywords: NRF2, KEAP1, cancer metabolism, metabolic reprogramming
1. Introduction
Metabolic reprogramming is one of the characteristics of cancer [1,2]. Cancer metabolism plays an important role in tumorigenesis, which is strongly associated with pathways that regulate cell proliferation, stress response, genome stability, toxic responses, and bioenergetics [3]. These metabolic modifications not only increase key metabolic pathways such as glycolysis, pentose phosphate pathway (PPP), and glutaminolysis but also interact with multiple oncogenic signaling pathways such as phosphoinositide 3-kinase/protein kinase B- (PI3K/AKT), Ras-, p53-, Myc-, and reactive oxygen species (ROS)-related pathways [4,5,6,7,8,9]. One of the master regulators of cellular antioxidant response signaling, the nuclear factor erythroid 2-related factor 2 (NRF2)/Kelch-like-ECH-associated protein 1 (KEAP1) pathway affects several aspects of metabolic reprogramming. The NRF2/KEAP1 axis controls both the basal and stress-inducible expression and function of key metabolic components belonging to metabolic reactions such as glutathione biosynthesis and recycling, thioredoxin reductase, and thioredoxin signaling [10]. In addition, NRF2 controls key metabolic enzymes associated with the inhibition of adipogenesis, facilitation of flux through PPP, nicotinamide adenine dinucleotide phosphate (NADPH) regeneration, and increased purine biosynthesis [11,12,13,14,15,16,17,18,19,20]. It is noteworthy that the NRF2/KEAP1 signaling pathway has previously been shown to play a critical role in tumorigenesis and the correlation between redox and metabolism in cancer [12]. In this review, we summarize the components of the NRF2/KEAP1 pathway and the regulation of metabolic reprogramming by NRF2/KEAP1 axis. Furthermore, we discuss the potential strategies and therapeutic significance of NRF2/KEAP1 signaling pathway in cancer metabolism.
2. The Structure and Functions of NRF2/KEAP1
2.1. NRF2
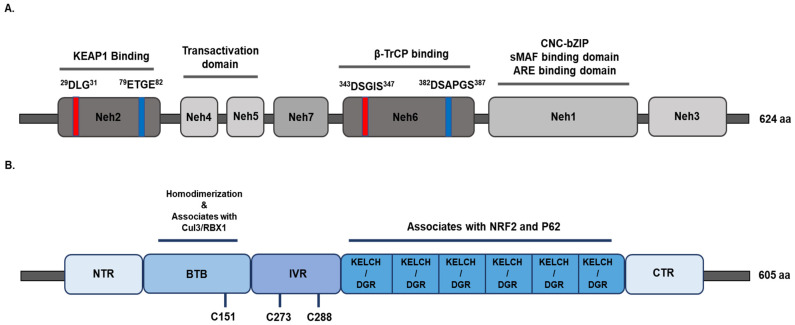
The NRF2 signaling pathway has a critical role in regulating cellular and tissue homeostasis and protecting cells against the management of oxidative and electrophilic stress [21,22,23]. NRF2 is encoded by the gene Nfe2l2 and belongs to a member of the Cap’n’collar (CNC) basic leucine zipper (bZIP) transcription factor family. It entails members having a conserved 43 amino acid homology region called the CNC, which contributes to the DNA-binding specificity of this family located at the N-terminal DNA-binding domain [24,25]. NRF2 is a modular protein and it consists of seven functional domains, known as the NRF2 ECH homology (Neh) domains Neh1–Neh7 [22,26,27,28]. As depicted in Figure 1A, the Neh1 domain contains a CNC-bZIP DNA-binding motif that allows NRF2 to dimerize with small Maf proteins and other transcription factors [25,27]. The Neh1 domain has also been reported to interact with UbcM2, an ubiquitin-conjugating enzyme, to regulate the stability of NRF2 [29]. The Neh2 domain is located in the N-terminus of NRF2, and it negatively controls the NRF2 through its DLG and ETGE motifs [30,31]. The two binding sites in Neh2, called DLG and ETGE motifs, help for NRF2 stability, and seven lysine residues are responsible for ubiquitin conjugation [30,32]. Importantly, the DLG and ETGE motifs bind with KEAP1, which is a substrate adaptor protein for the Cullin 3 (Cul3)-dependent E3 ubiquitin ligase complex that suppresses NRF2 by promoting its subsequent proteasomal degradation and ubiquitination [33,34,35]. The C-terminal Neh3 domain of NRF2 interacts with coactivators to facilitate the transactivation of NRF2 target genes. The Neh3 domain recruits chromo-ATPase/helicase DNA-binding protein family member CDH6, which functions as an NRF2 transcriptional coactivator [36]. The Neh4 and Neh5 domains are also important for the transactivation of NRF2 target genes and interact with cAMP response element-binding protein (CREB)-binding protein (CBP) and/or receptor-associated coactivator (RAC) [37,38]. The Neh6 domain negatively regulates NRF2 through DSGIS and DSAPGS motifs, β-transducin repeat-containing protein (β-TrCP). β-TrCP is a substrate adaptor for the S-phase kinase-associated protein 1 (SKP1)–Cul1–RING-box protein (Rbx1)/Roc1 ubiquitin ligase complex. DSGIS motif is phosphorylated by glycogen synthase kinase (GSK)-3β and increases the ability of β-TrCP to ubiquitinate NRF2 and promotes its rapid conversion [39,40,41]. The seventh Neh domain is known as Neh7 that mediates the repression of NRF2 interacting with the retinoic X receptor alpha (RXRα) and represses NRF2 target gene transcription [42].
Figure 1.
Domain structures of nuclear factor erythroid 2-related factor 2 (NRF2) and Kelch-like-ECH-associated protein 1 (KEAP1). (A) The relative positions of NRF2-ECH homology (Neh) domains, Neh1-Neh7, are indicated. The N-terminal Neh2 domain contains DLG and ETGE motifs, which interact with KEAP1. The Neh3, Neh4, and Neh5 domains are known as transactivation domains. The Neh6 domain contains the β-TrCP1 adaptor protein, which mediates proteasomal degradation. The c-terminal domain, Neh1, contains a CNC-bZIP, and it is responsible for heterodimerization with small MAF proteins (sMAFs). (B) KEAP1 consists of five domains that include the amino terminal region (NTR), a broad complex, tramtrack, bric-a-brac (BTB) domain, an intervening region (IVR), six Kelch domains, and the C-terminal region (CTR). The BTB domain is associated with CUL3-E3-ligase binding and the formation of keap1 homodimerization. The IVR contains several important cysteine residues that are responsible for modulating KEAP1-NRF2 activity. The Kelch/DGR domain is associated with NRF2 and P62, which is required for ETGE motifs. β-TrCP, β-transducin repeat-containing protein; CNC, cap‘n’collar; bZip, basic region leucine zipper; sMAFs, musculoaponeurotic fibrosarcoma protein; Cul3, Cullin 3.
2.2. KEAP1
KEAP1, a substrate adaptor for a Cul3-containing E3 ubiquitin ligase, interacts with NRF2 and controls the stability of NRF2. KEAP1 possesses five domains (Figure 1B), including N-terminal region (NTR), the Broad complex Tramtrack and Bric-à-Brac (BTB) domain, the intervening region (IVR), the Kelch domain/double glycine repeat (DGR), and the C-terminal region (CTR) [43]. The BTB domain binds with Cul3 and mediates KEAP1 homodimerization that is critical for ubiquitination and the proteasomal degradation of NRF2 [44,45]. Additionally, the BTB domain consists of cysteine residue Cys151 that is associated with oxidative stress level [46]. The IVR domain contains reactive cysteine residues such as Cys257, Cys273, Cys288, and Cys297 that have been proposed to promote KEAP1-dependent NRF2 ubiquitination [47]. The Kelch/DGR domain includes six Kelch repeats that interact with the binding of KEAP1 to the ETGE or DLG motifs located within the Neh2 domain of NRF2 [48,49,50]. Three functional domains of KEAP1 play critical roles in mediating NRF2 repression and ubiquitination.
2.3. Regulation of the Stability of NRF2/KEAP1 Complex
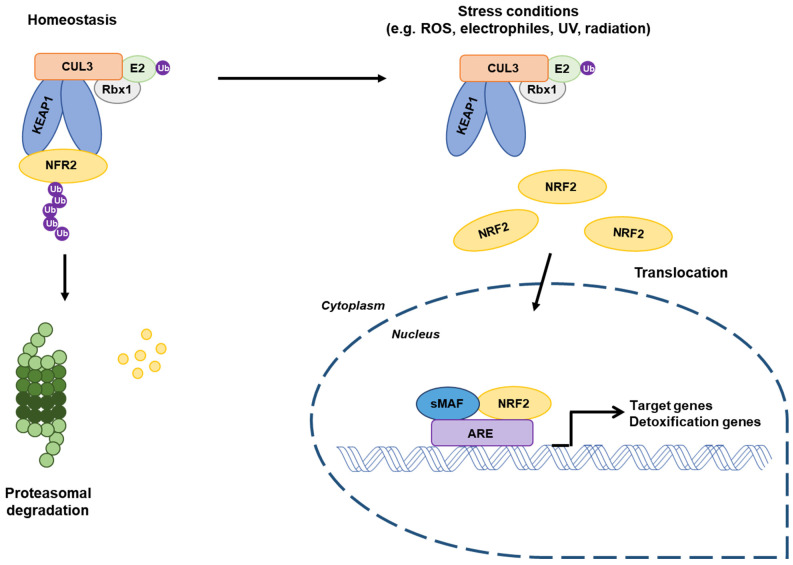
KEAP1 is one of the major regulators of intracellular levels of NRF2. Under basal conditions, NRF2 is repressed by proteasomal degradation mediated by KEAP1. NRF2 is primarily localized in a cytosol and binds with KEAP1 as a dimer via the KEAP1 Kelch domain and ETGE/DLG motifs of NRF2 (Figure 1B), and it promotes NRF2 ubiquitination, leading to a subsequent proteolysis. Therefore, KEAP1 tightly regulates the expression of NRF2 to a low level in order to avoid the unnecessary expression of its target genes [30,51,52]. However, KEAP1 is exposed by ROS, electrophiles, or another stressor at major cysteine residues, leading to conformational change in the KEAP1/Cul3/RBX/NRF2 complex and loss of NRF2 ubiquitination [47,49]. As a consequence, NRF2 dissociates from KEAP1 and translocates into the nucleus, where it heterodimerizes with small Maf proteins (sMAFs) and subsequently binds to the antioxidant responsive elements (AREs) located in the promoter of NRF2 target genes (Figure 2) [28,51,52,53]. An additional signaling pathway for the KEAP1-independent regulation of non-canonical NRF2 stability has been revealed. The β-TrCP–SKP1–RBX1–CUL1 E3 ubiquitin ligase complex regulates NRF2 for proteasomal degradation upon GSK-3β-dependent phosphorylation within the Neh6 domain of NRF2 [40].
Figure 2.
Regulation of NRF2 molecular mechanisms. Under basal conditions, NRF2 binds to its suppressor KEAP1 in the cytosol and interacts with the Cul3-RBX1 E3 ubiquitin ligase that constantly leads to NRF2 ubiquitination and proteasomal degradation. Under stressed conditions, conformational changes in KEAP1 and dissociation of NRF2 occur. Stabilized NRF2 translocates into the nucleus and forms a dimer with sMAFs proteins. The complex binds to antioxidant responsive elements sequences, promoting the transcription of target genes. RBX1, RING-box protein. Adapted from Jaramillo and Zhang [54].
2.4. Functions of NRF2 as a Transcription Factor
NRF2 coordinately regulates coding for detoxification or metabolic enzymes, antioxidants, NADPH regeneration enzymes, and multi drug-metabolizing enzymes. NRF2 controls the redox status of glutathione (GSH) homeostasis by directly regulating two subunits of the glutamate–cysteine ligase catalytic (GCLC) and modifier (GCLM) subunits involved in glutathione biosynthesis [55,56]. Moreover, several ROS-detoxifying enzymes such as glutathione peroxidase 2 (GPX2), glutathione S-transferases (GSTA1,2,3,5, GSTM 1–3, and GSTP1) [57,58], but also a GSH-based antioxidant system, thioredoxin 1 [55], thioredoxin reductase 1 [59,60,61], and thioredoxin-inhibitor are transcriptionally regulated by NRF2 [62,63,64]. Furthermore, NRF2 plays an important role in xenobiotics and drug detoxification by controlling the expression of drug-metabolizing enzymes while requiring NADPH as a cofactor. The expression of NADPH-generating enzymes such as glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconate dehydrogenase (PGD), isocitrate dehydrogenase 1 (IDH1), and malic enzyme 1 (ME1) are also regulated by NRF2 [18,20,65]. Together, NRF2 activity ensures the expression of enzymes against xenobiotics and oxidative stress as well as catalyzing reductive reactions. Evidence has shown that NRF2 regulates more than 200 genes involved in cellular processes, cytoprotection, metabolism, and gene transcription, as listed in Table 1.
Table 1.
A list of genes regulated by NRF2.
| General Biochemical Function | Gene Symbol | Name | Refs |
|---|---|---|---|
| Biotransformation and detoxification (Phase I, II, III) | ABCB6 | ATP-binding cassette, subfamily B (MDR/Tap) member 6 | [59] |
| ABCC1 | ATP-binding cassette, subfamily C (CFTR/MRP) | [59] | |
| ADH7 | Alcohol dehydrogenase class 4 mu/ sigma chain | [59] | |
| CBR1 | Carbonyl reductase 1 | [60] | |
| CYP1B1 | Cytochrome P450 | [59] | |
| EPHX1 | Epoxide hydrolase 1, microsomal | [59,60] | |
| UGT1A1 | UDP Glucuronosyltransferase 1 | [60] | |
| Anti-oxidant | GCLC | Glutamate–cysteine ligase, catalytic subunit | [66] |
| GCLM | Glutamate–cysteine ligase, modifier subunit | [66] | |
| GPX1 | Glutathione peroxidase 1 | [60] | |
| GSR1 | Glutathione reductase 1 | [60] | |
| PRDX1 | Peroxiredoxin 1 | [59] | |
| SRXN1 | Sulfiredoxin 1 | [59] | |
| TXN1 | Thioredoxin | [59] | |
| Carbohydrate metabolism and NADPH generation | G6PD | Glucose-6-phosphate dehydrogenase | [61,62,67] |
| HDK1 | Hexokinase domain containing 1 | [60] | |
| IDH1 | NADP-dependent isocitrate dehydrogenase | [59] | |
| ME1 | Malic enzyme 1 | [66] | |
| PGD | 6-phosphogluconate dehydrogenase | [66] | |
| TALDO1 | Transaldolase | [60] | |
| TKT | Transketolase isoform 1 | [60] | |
| Lipid metabolism | ACOT7 | Acetyl-CoA thioesterase 7 | [19] |
| ACOX1 | Acetyl-CoA oxidase 1 | [19] | |
| SCD2 | Stearoyl-CoA desaturase-2 | [19] | |
| Heme and iron metabolism | BLVRA | Biliverdin reductase A | [60] |
| BLVRB | Biliverdin reductase B | [60] | |
| FTH1 | Ferritin, heavy polypeptide | [60] | |
| FTL1 | Ferritin, light polypeptide | [60] | |
| HMOX1 | Heme oxygenase 1 | [60] | |
| Proteasomal degradation | ATF4 | Activating transcription factor-4 | [68] |
| PSMA1 | Proteasome subunit alpha type-1 | [68] | |
| PSMB5 | Proteasome subunit beta type-5 | [68] | |
| SQSTM1 | Sequestosome 1 (p62) | [65] | |
| Autophagy | ATG5 | Autophagy protein 5 | [59] |
| ATG7 | Autophagy protein 7 | [59] | |
| LC3B | Microtubule-associated protein 1A/1B-light chain 3B | [67] | |
| Apoptosis | BCL2 | B-cell lymphoma 2 | [69] |
3. NRF2 /KEAP1 Axis-Mediated Metabolic Reprogramming in Cancer
One of the classical pathways against metabolic reprogramming is the Warburg effect or aerobic glycolysis [70]. In normal cells, glycolysis is a physiological response to hypoxia. However, unlike normal cells, cancer cells constitutively increase glucose uptake and catabolize glucose into lactate regardless of oxygen availability [5,71]. Accelerating glycolytic flux allows producing ATP as well as fulfilling the metabolic demands of proliferating cells [70,71]. These metabolic properties of the cancer cells increase the synthesis of DNA and lipid [72]. Despite the aberrant activation of the cancers, it appears to involve a general induction against several pathways that support key functions such as redox balance, anabolic, and catabolic [73]. The NRF2/KEAP1 signaling pathway has been reported as a transcription factor that activates the antioxidant genes. However, more evidence reveals that the NRF2/KEAP1 signaling pathway is correlated with metabolic reprogramming in various cancer cells through several mechanisms [11,74]. It is becoming clear that the NRF2/KEAP1 pathway plays an important role in exerting the metabolic reprogramming of cancer cells through a transcriptional program inducing the proliferation of cancer cells and malignant progression. In addition, NRF2 supports intermediate metabolism through glutaminolysis [75], thereby generating an imbalance in metabolic processes such as the biosynthesis of amino acids [20] and nucleotides [17]. This section provides information on the role of the NRF2/KEAP1 axis in the regulation of the cancer metabolism against the cancer redox homeostasis and metabolic mechanisms.
3.1. The Role of NRF2/KEAP1 in Cellular Metabolism
3.1.1. PI3K/AKT Signaling Pathway
NRF2 plays a critical role in the proliferation of cancer cells via metabolic reprogramming. When growth factors are stimulated in normal cells, PI3K signaling activation and its downstream AKT and mammalian rapamycin target (mTOR) promote programming including increased activity glycolytic flux as well as fatty acid synthesis [76]. However, in the oncogenic pathway, the PI3K/AKT pathway serves as a major proliferative signal by interacting with the NRF2 signaling. The PI3K signaling pathway has been reported to control the regulation of NRF2 signaling independently of KEAP1 [77,78,79]. When insulin-like growth factor (IGF) receptor is activated, PI3K catalyzes the phosphorylation of the lipid phosphatidylinositol 4,5-bisphophate (PIP2) to produce phosphatidylinositol 3,4,5-triphosphate (PIP3). The generation of PIP3 is important for the AKT activity, which mediates downstream signaling events including the inhibition of the GSK-3β [80,81]. GSK-3β is a key mediator that is inhibited by AKT-mediated phosphorylation [82], and NRF2 is phosphorylated by GSK-3β, enabling its recognition by β-TrCP that in turn marks NRF2 for ubiquitination regardless of the mediated by engaging the KEAP1/CUL3 complex [39,41,83]. When the GSK-3β is inactivated by phosphorylation, resulting in NRF2 accumulation through inhibition of KEAP1-independent degradation and increased abundance in NRF2, it promotes the activation of metabolic genes as well as anabolic metabolism, especially in the presence of active PI3K–AKT signaling [17]. Evidence shows that the PI3K pathway contributes to the activation of NRF2 in several contexts. The PI3K is antagonized by the tumor suppressor phosphatase and tensin homolog (PTEN) [84]. When the concentration of PTEN is low, AKT is activated, while GSK-3β is inhibited. Inhibition of GSK-3β has been reported to reduce NRF2 phosphorylation; subsequently, NRF2 escapes KEAP1-independent and β-TrCP-CUL1-dependent degradation in the nucleus. Thus, deletion of PTEN has been observed to increase the nuclear accumulation of NRF2 and the expression of NRF2 target gene [17]. Similarly, deletion of KEAP1 and PTEN leads to significantly increased NRF2 level in liver cells [85] and contributes to the tumorigenic potent on PTEN-deleted prostate cancer cells [86]. Additionally, inhibition of the PI3K/AKT pathway markedly reduces endogenous NRF2 protein and enzymes in KEAP1-mutant lung cancer cells and KEAP1-deficient mouse embryonic fibroblasts (MEFs) [40]. Taken together, activation of the PI3K/AKT signaling pathway increases the nuclear translocation of NRF2, independent of KEAP1, and allows NRF2 to promote metabolic reprogramming and increase cell proliferation.
3.1.2. p62
Previous studies have demonstrated that NRF2 and p62/Sequestosome 1 protein (p62/SQSTM) can regulate the activity of NRF2 [83,87,88]. p62/SQSTM1 is a scaffold protein that regulates selective cytoplasmic aggregators of ubiquitinated proteins and organelles for degradation through the autophagy pathway. p62 contains an STGE motif that is similar to the NRF2 ETGE motif; it directly interacts with the Kelch domain of KEAP1 and the accumulation of p62 triggers serine phosphorylation of an STGE motif [83,88]. This phosphorylation enhances p62/KEAP1 interaction, aggregation, and autophagic degradation, which results in the stabilization of NRF2. However, this adaptive response can become pathological under circumstances of impaired autophagy, thereby triggering the accumulation of cytoplasmic protein inclusions in a p62- and NRF2-dependent manner [89,90,91]. The phosphorylation of p62 at S349 activates NRF2 and directly targets not only glucose metabolism to the glucuronate pathway but also glutamine metabolism to glutathione synthesis [92]. This is relevant, given that the amplification of the p62 gene and aberrant accumulation of phosphorylated p62 protein have been implicated in the acceleration of cancer development. In hepatocellular carcinoma cells, these changes confer the resistance against anti-cancer drugs and trigger cell proliferation. Furthermore, phosphorylated p62 has been reported to accumulate in tumor regions positive for hepatitis C virus [93].
3.1.3. AMP-Activated Protein Kinase
AMP-activated protein kinase (AMPK) is a master regulator of metabolism and energy homeostasis in normal cells. In times of energy deprivation, the levels of ATP are decreased, and NRF2 deficiency may lead to AMPK activation. AMPK activation has been known to lead to the upregulation of glucose, fatty acid uptake, activation of autophagy, and an increase in ATP levels [89]. Several studies report that the AMPK promotes the activation of NRF2 [94], and a recent study revealed that the AMPK phosphorylates on serine 588 of NRF2 [95]. The activation of AMPK regulates the phosphorylation and inhibition of GSK-3β [90], which promotes the degradation of NRF2 through β-TRCP/CUL1 pathway. Therefore, AMPK regulates the nuclear localization and stabilization of NRF2.
3.1.4. Crosstalk between Mitochondrial Metabolism
Mitochondria are major organelles responsible for ATP synthesis and cellular processes. In addition, it controls tricarboxylic acid (TCA) cycle, calcium, and ROS homeostasis, fatty acids, and amino acids metabolism. It is noteworthy that the mitochondrial dysfunction is caused by metabolic rewiring [91]. The NRF2-dependent interactions in metabolism and homeostasis affect mitochondrial function. Recently, it has been shown that NRF2 is activated by aberrant accumulation of the TCA cycle. In the absence of NRF2, glucose oxidation and the flowing of substrate into the TCA cycle are diminished [91]. In contrast, the constitutive activation of NRF2 has been observed to induce the glucose oxidation and flowing of substrate into the TCA cycle [96]. ATP levels were also observed to be decreased in NRF2-deficient MEFs, and on the contrary, the constitutive activation of NRF2 increased the levels of ATP [97]. Similarly, the silencing of NRF2 caused the decrease of ATP production and oxygen consumption in human colon cancer cells [98]. In particular, inactivation of fumarate hydratase (FH) triggers the accumulation of the TCA cycle intermediates, fumarate and succinate, which lead to an interruption in KEAP1/NRF2 binding [99,100]. Fumarate has been reported to interact with cysteine residues within the KEAP1 protein upon the activation of NRF2 [99,100]. Furthermore, a loss of FH locus has been shown to lead to an aggressive form of renal cancer in hereditary leiomyomatosis and renal cell carcinoma patients [100,101,102,103]. Collectively, these findings suggest that the NRF2 interacts dependently with the axis of cancer metabolism and mitochondrial function. In addition, KEAP1 was found to be associated with mitochondrial interfaces. Interestingly, it has been revealed that KEAP1 exists in close proximity to the mitochondria and interacts with the mitochondrial outer membrane histidine phosphatase, PGAM5 [104,105,106]. The depletion of PGAM5 or NRF2 causes an inhibition of mitochondrial retrograde trafficking, due to activation of the KEAP1–cullin-3 E3 ubiquitin complex and rescued degradation of Miro2, which is a mitochondrial GTPase that links mitochondria to microtubules [104,105]. Furthermore, the PGAM5–KEAP1 complex induces oxeiptosis, a caspase-independent cell death program, under high ROS generated [107]. Whereas, under the unstressed conditions, KEAP1 is important role for the maintenance of mitochondrial homeostasis with p62 and Rbx1 through mitochondrial ubiquitination in liver disease [108].
3.2. Modulation of Metabolic Processes by NRF2/KEAP1 Signaling
3.2.1. Pentose Phosphate Pathway
Glucose is an important source of cellular energy. It enters the cells through glucose transporters, and its metabolites serve as substrates for biosynthetic processes. Moreover, glucose provides other metabolic intermediates for biosynthetic pathways, such as PPP. The pentose phosphate pathway (also called the phosphogluconate pathway and the hexose monophosphate shunt) consists of the oxidative branch and the non-oxidative branch [109]. All the branches need the ribose-5-phosphate and NADPH for the changing demands of the cells. In the oxidative phase of PPP, the G6PD and PGD catalyze the reaction. Entry into the oxidative arm is catalyzed by G6PD, and G6PD has been known to be controlled by NRF2 [17]. It determines the flux of glucose through PPP and biosynthetic reactions. In another branch, the non-oxidative branch of PPP, NRF2 positively regulates the expression of transaldolase 1 (TALDO1) and transketolase (TKT) [17,19]. NRF2 plays a crucial role in promoting the proliferation of cancer cells and metabolism process in lung cancer cells, including the direct transcriptional regulation of PPP-related enzymes such as G6PD, PGD, TKT, and TALDO1, which are responsible for NADPH regeneration [17]. The G6PD, PGD, TKT, and TALDO1 support glucose flux and generate purines, which are building blocks of DNA and RNA, which help to accelerate proliferation in cancer cells. The ribose-5-phosphate is a major product of the PPP, and it is essential for the biosynthesis of nucleotides. The ribose-5-phosphate contributes the sugar group to nucleotides and ultimately forms the sugar backbone in DNA. The ribose-5-phosphate is metabolized to phosphoribosyl–pyrophosphate, which is a common precursor for both purine and pyrimidine nucleotides. Mitsuishi et al. has reported that NRF2 enhances the expression of purine base synthesis by the indirect regulation of phosphoribosyl–pyrophosphate amidotransferase, which catalyzes the rate-limiting step in the de novo purine biosynthesis, and methylenetetrahydrofolate dehydrogenase 2, which is a nuclear-encoded mitochondrial bifunctional enzyme with methylenetetrahydrofolate dehydrogenase and methenyltetrahydrofolate cyclohydrolase activities, thus initiating the creation of the purine ring [17]. Indeed, the biosynthesis of purine has been shown to be affected by the activity of NRF2, and it has been observed to flow continuously through the PPP. Moreover, carbon flux is increased in KEAP1-knockout MEFs and decreased in their NRF2-knockout counterparts.
3.2.2. Amino Acid Metabolism
Accumulating evidence shows that NRF2 regulates the intracellular pool of amino acids by coordinating several molecular pathways such as biosynthesis, absorption, proliferation, cancer metabolic reprogramming, and redox balance. NRF2 influences not only metabolism but also intracellular concentrations of the cysteine/glutamate transporter system. Several studies have focused on the interaction between NRF2 and xCT, which is a transmembrane antiporter coded by the SLC7A11 gene, which is increased in several cancers and has been known to mediate the extrusion of glutamate and support the redox homeostasis [110,111,112]. NRF2 was found to increase not only the expression of the SLC7A11 but also the activity of xCT in breast cancer cells [110]. In contrast, silenced NRF2 suppressed both xCT and glutamate export in breast cancer cells [112]. Moreover, the correlation of NRF2 and SLC7A11 has been revealed from the result in almost 950 cancer cell lines [113,114]. Indeed, glucose starvation induced the overexpression of SLC7A11 and subsequent upregulation of glucose dependence for cell survival through the NRF2- and activating transcription factor 4 (ATF4)-dependent transcription in renal cancer cells [110]. ATF4 is a transcription factor that plays an important role in amino acid deprivation, metabolic stress, and ER stress. ATF4 targeted by NRF2 has been known to induce the expression of xCT promoter [115]. In a separate study, NRF2 controlled the transcription of key enzymes through ATF4 activation, which is involved in serine/glycine biosynthesis in non-small-cell lung carcinoma cells [116]. Indeed, this study shows that the activation of NRF2 and ATF4 is associated with the poorer prognosis in the lung cancer patients [116]. Moreover, it is revealed that the KRAS-dependent regulation of the ATF4 mechanism via PI3K/AKT signaling pathway required the activation of NRF2, suggesting that the activation of these axis mechanisms is related to the expression of amino acid transporters such as SLC1A5, SLC38A2, SLC7A5, SLC7A1, and SLC7A11 genes [117]. In agreement with this, NRF2 can transcriptionally promote the expression of ATF4 and the coding for amino acid transporters (AATs) involved in the import of proline, tryptophan, alanine, glycine, and glutamine in the colon cancer cells [118]. Furthermore, it is worthy that the inhibition of AATs may trigger the apoptosis in autophagy-deficient colorectal cancer cells but not wild-type colorectal cancer cells upon glutamine withdrawal [118].
In additionally, KEAP1 is involved in cysteine/glutamate metabolism. When metabolic and redox response pathways are activated, KEAP1 requires significant energy as well as metabolic substrates such as carbon and sulfur, leading to depletion of the TCA cycle intermediates [111,119,120,121,122]. Moreover, the loss of the functional mutant-KEAP1 increased the dependence on glutamine in human KRAS-driven lung adenoma cell lines. KEAP1-mutant cells reduced the intracellular glutamate pool by increasing glutamate consumption for GSH synthesis and exporting glutamate via anti-porter xCT in exchange for cysteine [75,118]. These studies indicate that the NRF2/KEAP1 signaling pathway could regulate amino acid metabolism in malignant tumors.
3.2.3. Lipid Metabolism
As for lipid metabolism, NRF2 positively regulates catabolic metabolism that is involved in the degradation of phospholipids and triglycerides, and enzymes involved in fatty acids oxidation. NRF2 has been reported to control the efficiency of fatty acids oxidation by regulating the expression of the carnitine palmitoyltransferase isoforms (CPT1 and CPT2) within mitochondria and two peroxisomal enzymes, acyl-CoA oxidase 1 and 2 (ACOX1 and ACOX2), which are related in lipids beta-oxidation [16]. In KEAP1-wild-type and/or -knockout MEFs, the acceleration of fatty acid synthesis not only increased ATP production but also stimulated respiration [16]. Moreover, the absence of NRF2 has been reported to significantly decrease the efficiency of fatty acids oxidation [16]. Conversely, NRF2 has been shown to suppress the anabolic processes associated with lipid biosynthesis, fatty acid desaturation, and fatty acid transport [12,123]. These findings suggest that NRF2 may negatively regulate lipid biosynthesis and reduce the consumption of NADPH in cancer cells. In the murine models, hepatic mRNA levels of ATP-citrate lyase, acetyl-CoA carboxylase 1, fatty acid synthase, stearoyl CoA desaturase 1, and fatty acid elongase were downregulated by the activation of NRF2 and contrary suppressed in NRF2-knockout mice [16,19]. NRF2 transcriptionally regulates fatty acids oxidation-related genes and activates the degradation of damaged lipids, thereby reducing the form of NADPH in cancer cells. Taken together, these studies demonstrate that the NRF2/KEAP1 signaling pathway can regulate lipid metabolism such as lipid biosynthesis and fatty acid oxidation, respectively.
3.2.4. Iron Metabolism
NRF2 is well known to play a key role in iron homeostasis. NRF2 controls the intracellular levels of the HO-1 enzyme and the storage of iron through the regulation of ferritin. Notably, NRF2 regulates the synthesis of heme, which is metabolized from heme to iron and biliverdin. NRF2 controls the expression of biliverdin reductase that metabolizes biliverdin to bilirubin and ferrochelatase, which is excreted as waste [19,124]. On the other side, heme biosynthesis needs the amino acid glycine through the NRF2–ATF4 serine biosynthesis pathway [20]. Accelerated iron synthesis during cancer development can induce carcinogenesis, cancer progression, and metastasis formation. The intracellular levels of the iron exporter ferroportin have been reported to be markedly downregulated in breast cancer cells, which is associated with accelerated cancer progression [125]. Furthermore, NRF2 has been reported to transcriptionally regulate proteins involved in iron and heme metabolism such as ferritin, ferrochlatase, heme-responsive gene 1, ferroportin, etc. [126]. This evidence suggests that NRF2 might play an important role in the cytoprotection and metabolic regulation through iron metabolism.
4. Conclusions
The NRF2/KEAP1 axis plays a major role in the cellular regulation of redox homeostasis, mitochondrial physiology, autophagy, proteostasis, immune system, and metabolism. The NRF2/KEAP1 complex is mediated by activating stimulation, interaction with other transcription factors, activators or repressors, and crosstalk with other signaling pathways. At the center of a complex regulatory network, the NRF2/KEAP1 pathway is emerging as a critical regulator of metabolism in cancer cells as its interactions with the metabolism-related pathway including the PI3K/AKT/mTOR pathway, p62 pathway, AMPK, and TCA cycle have been revealed. In addition, the NRF2/KEAP1 axis contributes to the several metabolic processes in cancers and the production of metabolites that promote cell proliferation and survival. In particular, the constitutive overexpression of NRF2 accelerates the proliferation of cancer cells, which is the result of the reprogramming of intracellular anabolic and catabolic metabolism. Understanding more integrated NRF2/KEAP1-mediated cancer metabolism may facilitate the discovery of new anti-cancer treatment strategies through cancer metabolic reprogramming.
Abbreviations
| AATs | Amino acid transporters |
| ACOX | Acyl-CoA oxidase |
| AKT | Protein kinase B |
| AMPK | AMP-activated protein kinase |
| ARE | Antioxidant responsive element |
| ATF4 | Activating transcription factor 4 |
| BTB | Broad complex, tramtrack, bric-a-brac |
| bZIP | Basic leucine zipper |
| CBP | CREB-binding protein |
| CNC | Cap‘n’collar |
| CPT | Carnitine palmitoyltransferase isoforms |
| CREB | cAMP response element binding protein |
| CTR | C-terminal region |
| Cul3 | Cullin 3 |
| DGR | Double glycine repeat |
| FH | Fumarate hydratase |
| G6PD | Glucose-6- phosphate dehydrogenase |
| GCL | Glutamate–cysteine ligase |
| GPX | Glutathione peroxidase |
| GSH | Glutathione |
| GSK | Glycogen synthase kinase |
| GST | Glutathione S-transferases |
| IGF | Insulin-like growth factor |
| IVR | intervening region |
| KEAP1 | Kelch-like-ECH-associated protein 1 |
| MEF | Mouse embryonic fibroblast |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NRF2 | Nuclear factor erythroid 2-related factor 2 |
| NTR | Amino terminal region |
| p62/SQSTM | p62/Sequestosome 1 protein |
| PGD | Phosphogluconate dehydrogenase |
| PI3K | Phosphoinositide 3-kinase |
| PIP2 | Lipid phosphatidylinositol 4,5-bisphophate |
| PIP3 | Phosphatidylinositol 3,4,5-triphosphate |
| PPP | Pentose phosphate pathway |
| PTEN | Phosphatase and tensin homolog |
| RAC | Receptor-associated coactivator |
| RBX1 | RING-box protein |
| ROS | Reactive oxygen species |
| RXRα | Retinoic X receptor alpha |
| SKP1 | S-phase kinase-associated protein 1 |
| sMAF | Small musculoaponeurotic fibrosarcoma protein |
| TALDO1 | Transaldolase 1 |
| TCA | Tricarboxylic acid |
| TKT | Transketolase |
| β-TrCP | β-transducin repeat-containing protein |
Author Contributions
Conceptualization, M.-Y.S., K.-S.C. and E.-H.K.; writing—original draft preparation, M.-Y.S.; writing—review and editing, M.-Y.S., D.-Y.L., K.-S.C. and E.-H.K.; visualization, M.-Y.S.; supervision, E.-H.K.; project administration, E.-H.K.; funding acquisition, E.-H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Basic Science Research Program grant (No. 2020R1F1A 1076477 to E.-H.K. and 2020R1l1A3066367 to K.-S.C.) from National Research Foundation (NRF), Republic of Korea.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hanahan D., Weinberg R.A. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Pavlova N.N., Thompson C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu W.L., Papagiannakopoulos T. The Pleiotropic Role of the KEAP1/NRF2 Pathway in Cancer. Annu. Rev. Cancer Biol. 2020;4:413–435. doi: 10.1146/annurev-cancerbio-030518-055627. [DOI] [Google Scholar]
- 4.Bensinger S.J., Christofk H.R. New aspects of the Warburg effect in cancer cell biology. Semin. Cell Dev. Biol. 2012;23:352–361. doi: 10.1016/j.semcdb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 6.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 7.Ward P.S., Thompson C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirschey M.D., DeBerardinis R.J., Diehl A.M.E., Drew J.E., Frezza C., Green M.F., Jones L.W., Ko Y.H., Le A., Lea M.A., et al. Dysregulated metabolism contributes to oncogenesis. Semin. Cancer Biol. 2015;35:S129–S150. doi: 10.1016/j.semcancer.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson C.B. Metabolic enzymes as oncogenes or tumor suppressors. N. Engl. J. Med. 2009;360:813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes J.D., McMahon M. NRF2 and KEAP1 mutations: Permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Menegon S., Columbano A., Giordano S. The Dual Roles of NRF2 in Cancer. Trends Mol. Med. 2016;22:578–593. doi: 10.1016/j.molmed.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Chartoumpekis D.V., Wakabayashi N., Kensler T.W. Keap1/Nrf2 pathway in the frontiers of cancer and non-cancer cell metabolism. Biochem. Soc. Trans. 2015;43:639–644. doi: 10.1042/BST20150049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boroughs L.K., DeBerardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panieri E., Santoro M. ROS homeostasis and metabolism: A dangerous liason in cancer cells. Cell Death Dis. 2016;7:e2253. doi: 10.1038/cddis.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludtmann M.H., Angelova P.R., Zhang Y., Abramov A.Y., Dinkova-Kostova A.T. Nrf2 affects the efficiency of mitochondrial fatty acid oxidation. Biochem. J. 2014;457:415–424. doi: 10.1042/BJ20130863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki H., Sato H., Kuriyama-Matsumura K., Sato K., Maebara K., Wang H., Tamba M., Itoh K., Yamamoto M., Bannai S. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 2002;277:44765–44771. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]
- 19.Wu K.C., Cui J.Y., Klaassen C.D. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol. Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeNicola G.M., Chen P.H., Mullarky E., Sudderth J.A., Hu Z., Wu D., Tang H., Xie Y., Asara J.M., Huffman K.E., et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 2015;47:1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uruno A., Motohashi H. The Keap1-Nrf2 system as an in vivo sensor for electrophiles. Nitric Oxide. 2011;25:153–160. doi: 10.1016/j.niox.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Baird L., Dinkova-Kostova A. The cytoprotective role of the Keap1–Nrf2 pathway. Arch. Toxicol. 2011;85:241–272. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- 23.Vomund S., Schafer A., Parnham M.J., Brune B., von Knethen A. Nrf2, the Master Regulator of Anti-Oxidative Responses. Int. J. Mol. Sci. 2017;18:2772. doi: 10.3390/ijms18122772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loboda A., Damulewicz M., Pyza E., Jozkowicz A., Dulak J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: An evolutionarily conserved mechanism. Cell. Mol. Life Sci. 2016;73:3221–3247. doi: 10.1007/s00018-016-2223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moi P., Chan K., Asunis I., Cao A., Kan Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes J.D., McMahon M., Chowdhry S., Dinkova-Kostova A.T. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 27.Namani A., Li Y., Wang X.J., Tang X. Modulation of NRF2 signaling pathway by nuclear receptors: Implications for cancer. Biochim. Biophys. Acta. 2014;1843:1875–1885. doi: 10.1016/j.bbamcr.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Telkoparan-Akillilar P., Suzen S., Saso L. Pharmacological Applications of Nrf2 Inhibitors as Potential Antineoplastic Drugs. Int. J. Mol. Sci. 2019;20:2025. doi: 10.3390/ijms20082025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen L., Plafker K.S., Starnes A., Cook M., Klevit R.E., Plafker S.M. The ubiquitin-conjugating enzyme, UbcM2, is restricted to monoubiquitylation by a two-fold mechanism that involves backside residues of E2 and Lys48 of ubiquitin. Biochemistry. 2014;53:4004–4014. doi: 10.1021/bi500072v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong K.I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukutomi T., Takagi K., Mizushima T., Ohuchi N., Yamamoto M. Kinetic, Thermodynamic, and Structural Characterizations of the Association between Nrf2-DLGex Degron and Keap1. Mol. Cell. Biol. 2014;34:832. doi: 10.1128/MCB.01191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Gordon G.B. A strategy for cancer prevention: Stimulation of the Nrf2-ARE signaling pathway. Mol. Cancer Ther. 2004;3:885. [PubMed] [Google Scholar]
- 33.Cullinan S.B., Gordan J.D., Jin J., Harper J.W., Diehl J.A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furukawa M., Xiong Y. BTB Protein Keap1 Targets Antioxidant Transcription Factor Nrf2 for Ubiquitination by the Cullin 3-Roc1 Ligase. Mol. Cell. Biol. 2005;25:162. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D., Lo S.-C., Cross J., Templeton D., Hannink M. Keap1 Is a Redox-Regulated Substrate Adaptor Protein for a Cul3-Dependent Ubiquitin Ligase Complex. Mol. Cell. Biol. 2005;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nioi P., Nguyen T., Sherratt P.J., Pickett C.B. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol. Cell. Biol. 2005;25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh Y., Itoh K., Yoshida E., Miyagishi M., Fukamizu A., Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim J.H., Yu S., Chen J.D., Kong A.N. The nuclear cofactor RAC3/AIB1/SRC-3 enhances Nrf2 signaling by interacting with transactivation domains. Oncogene. 2013;32:514–527. doi: 10.1038/onc.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rada P., Rojo A.I., Evrard-Todeschi N., Innamorato N.G., Cotte A., Jaworski T., Tobon-Velasco J.C., Devijver H., Garcia-Mayoral M.F., Van Leuven F., et al. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol. Cell. Biol. 2012;32:3486–3499. doi: 10.1128/MCB.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H., Liu K., Geng M., Gao P., Wu X., Hai Y., Li Y., Li Y., Luo L., Hayes J.D., et al. RXRα Inhibits the NRF2-ARE Signaling Pathway through a Direct Interaction with the Neh7 Domain of NRF2. Cancer Res. 2013;73:3097. doi: 10.1158/0008-5472.CAN-12-3386. [DOI] [PubMed] [Google Scholar]
- 43.Lee J.S., Surh Y.J. Nrf2 as a novel molecular target for chemoprevention. Cancer Lett. 2005;224:171–184. doi: 10.1016/j.canlet.2004.09.042. [DOI] [PubMed] [Google Scholar]
- 44.Zipper L.M., Mulcahy R.T. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 45.Lo S.C., Li X., Henzl M.T., Beamer L.J., Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cleasby A., Yon J., Day P.J., Richardson C., Tickle I.J., Williams P.A., Callahan J.F., Carr R., Concha N., Kerns J.K., et al. Structure of the BTB domain of Keap1 and its interaction with the triterpenoid antagonist CDDO. PLoS ONE. 2014;9:e98896. doi: 10.1371/journal.pone.0098896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto T., Suzuki T., Kobayashi A., Wakabayashi J., Maher J., Motohashi H., Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canning P., Sorrell F.J., Bullock A.N. Structural basis of Keap1 interactions with Nrf2. Free Radic. Biol. Med. 2015;88:101–107. doi: 10.1016/j.freeradbiomed.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004;279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 51.Rushmore T.H., Morton M.R., Pickett C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–11639. doi: 10.1016/S0021-9258(18)99004-6. [DOI] [PubMed] [Google Scholar]
- 52.Katsuoka F., Motohashi H., Ishii T., Aburatani H., Engel J.D., Yamamoto M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol. Cell. Biol. 2005;25:8044–8051. doi: 10.1128/MCB.25.18.8044-8051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katsuoka F., Yamazaki H., Yamamoto M. Small Maf deficiency recapitulates the liver phenotypes of Nrf1- and Nrf2-deficient mice. Genes Cells. 2016;21:1309–1319. doi: 10.1111/gtc.12445. [DOI] [PubMed] [Google Scholar]
- 54.Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hawkes H.J., Karlenius T.C., Tonissen K.F. Regulation of the human thioredoxin gene promoter and its key substrates: A study of functional and putative regulatory elements. Biochim. Biophys. Acta. 2014;1840:303–314. doi: 10.1016/j.bbagen.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 56.Higgins L.G., Kelleher M.O., Eggleston I.M., Itoh K., Yamamoto M., Hayes J.D. Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol. Appl. Pharmacol. 2009;237:267–280. doi: 10.1016/j.taap.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Hayes J.D., Ashford M.L. Nrf2 orchestrates fuel partitioning for cell proliferation. Cell Metab. 2012;16:139–141. doi: 10.1016/j.cmet.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 58.Reichard J.F., Motz G.T., Puga A. Heme oxygenase-1 induction by NRF2 requires inactivation of the transcriptional repressor BACH1. Nucleic Acids Res. 2007;35:7074–7086. doi: 10.1093/nar/gkm638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C., Shyr C., Wakabayashi N., Kensler T.W., Wasserman W.W., et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agyeman A.S., Chaerkady R., Shaw P.G., Davidson N.E., Visvanathan K., Pandey A., Kensler T.W. Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Res. Treat. 2012;132:175–187. doi: 10.1007/s10549-011-1536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F., Xue P., Pi J., Kleeberger S.R., Bell D.A. Identification of novel NRF2-regulated genes by ChIP-Seq: Influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yates M.S., Tran Q.T., Dolan P.M., Osburn W.O., Shin S., McCulloch C.C., Silkworth J.B., Taguchi K., Yamamoto M., Williams C.R., et al. Genetic versus chemoprotective activation of Nrf2 signaling: Overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis. 2009;30:1024–1031. doi: 10.1093/carcin/bgp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maher J.M., Dieter M.Z., Aleksunes L.M., Slitt A.L., Guo G., Tanaka Y., Scheffer G.L., Chan J.Y., Manautou J.E., Chen Y., et al. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46:1597–1610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- 64.Digaleh H., Kiaei M., Khodagholi F. Nrf2 and Nrf1 signaling and ER stress crosstalk: Implication for proteasomal degradation and autophagy. Cell. Mol. Life Sci. 2013;70:4681–4694. doi: 10.1007/s00018-013-1409-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gureev A.P., Shaforostova E.A., Popov V.N. Regulation of Mitochondrial Biogenesis as a Way for Active Longevity: Interaction Between the Nrf2 and PGC-1alpha Signaling Pathways. Front. Genet. 2019;10:435. doi: 10.3389/fgene.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacLeod A.K., McMahon M., Plummer S.M., Higgins L.G., Penning T.M., Igarashi K., Hayes J.D. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: Demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 2009;30:1571–1580. doi: 10.1093/carcin/bgp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang Z., Hu B., Zang F., Wang J., Zhang X., Chen H. Nrf2 drives oxidative stress-induced autophagy in nucleus pulposus cells via a Keap1/Nrf2/p62 feedback loop to protect intervertebral disc from degeneration. Cell Death Dis. 2019;10:510. doi: 10.1038/s41419-019-1701-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuda O., Brezinova M., Silhavy J., Landa V., Zidek V., Dodia C., Kreuchwig F., Vrbacky M., Balas L., Durand T., et al. Nrf2-Mediated Antioxidant Defense and Peroxiredoxin 6 Are Linked to Biosynthesis of Palmitic Acid Ester of 9-Hydroxystearic Acid. Diabetes. 2018;67:1190–1199. doi: 10.2337/db17-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niture S.K., Jaiswal A.K. Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J. Biol. Chem. 2012;287:9873–9886. doi: 10.1074/jbc.M111.312694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 71.Weinhouse S., Warburg O., Burk D., Schade A.L. On Respiratory Impairment in Cancer Cells. Science. 1956;124:267–272. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- 72.Baenke F., Peck B., Miess H., Schulze A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Models Mech. 2013;6:1353–1363. doi: 10.1242/dmm.011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cantor J.R., Sabatini D.M. Cancer cell metabolism: One hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taguchi K., Yamamoto M. The KEAP1-NRF2 System in Cancer. Front. Oncol. 2017;7:85. doi: 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Romero R., Sayin V.I., Davidson S.M., Bauer M.R., Singh S.X., LeBoeuf S.E., Karakousi T.R., Ellis D.C., Bhutkar A., Sanchez-Rivera F.J., et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017;23:1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dibble C.C., Manning B.D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vivanco I., Sawyers C.L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 78.Martin D., Rojo A.I., Salinas M., Diaz R., Gallardo G., Alam J., De Galarreta C.M., Cuadrado A. Regulation of heme oxygenase-1 expression through the phosphatidylinositol 3-kinase/Akt pathway and the Nrf2 transcription factor in response to the antioxidant phytochemical carnosol. J. Biol. Chem. 2004;279:8919–8929. doi: 10.1074/jbc.M309660200. [DOI] [PubMed] [Google Scholar]
- 79.Li M.H., Cha Y.N., Surh Y.J. Peroxynitrite induces HO-1 expression via PI3K/Akt-dependent activation of NF-E2-related factor 2 in PC12 cells. Free Radic. Biol. Med. 2006;41:1079–1091. doi: 10.1016/j.freeradbiomed.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 80.Klippel A., Kavanaugh W.M., Pot D., Williams L.T. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol. Cell. Biol. 1997;17:338–344. doi: 10.1128/MCB.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Franke T.F., Kaplan D.R., Cantley L.C., Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 82.Salazar M., Rojo A.I., Velasco D., de Sagarra R.M., Cuadrado A. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J. Biol. Chem. 2006;281:14841–14851. doi: 10.1074/jbc.M513737200. [DOI] [PubMed] [Google Scholar]
- 83.Copple I.M., Lister A., Obeng A.D., Kitteringham N.R., Jenkins R.E., Layfield R., Foster B.J., Goldring C.E., Park B.K. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J. Biol. Chem. 2010;285:16782–16788. doi: 10.1074/jbc.M109.096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maehama T., Dixon J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 85.Taguchi K., Hirano I., Itoh T., Tanaka M., Miyajima A., Suzuki A., Motohashi H., Yamamoto M. Nrf2 enhances cholangiocyte expansion in Pten-deficient livers. Mol. Cell. Biol. 2014;34:900–913. doi: 10.1128/MCB.01384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rojo A.I., Rada P., Mendiola M., Ortega-Molina A., Wojdyla K., Rogowska-Wrzesinska A., Hardisson D., Serrano M., Cuadrado A. The PTEN/NRF2 axis promotes human carcinogenesis. Antioxid. Redox Signal. 2014;21:2498–2514. doi: 10.1089/ars.2014.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 88.Jain A., Lamark T., Sjottem E., Larsen K.B., Awuh J.A., Overvatn A., McMahon M., Hayes J.D., Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mihaylova M.M., Shaw R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Horike N., Sakoda H., Kushiyama A., Ono H., Fujishiro M., Kamata H., Nishiyama K., Uchijima Y., Kurihara Y., Kurihara H., et al. AMP-activated protein kinase activation increases phosphorylation of glycogen synthase kinase 3beta and thereby reduces cAMP-responsive element transcriptional activity and phosphoenolpyruvate carboxykinase C gene expression in the liver. J. Biol. Chem. 2008;283:33902–33910. doi: 10.1074/jbc.M802537200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Negrette-Guzman M., Huerta-Yepez S., Vega M.I., Leon-Contreras J.C., Hernandez-Pando R., Medina-Campos O.N., Rodriguez E., Tapia E., Pedraza-Chaverri J. Sulforaphane induces differential modulation of mitochondrial biogenesis and dynamics in normal cells and tumor cells. Food Chem. Toxicol. 2017;100:90–102. doi: 10.1016/j.fct.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 92.Saito T., Ichimura Y., Taguchi K., Suzuki T., Mizushima T., Takagi K., Hirose Y., Nagahashi M., Iso T., Fukutomi T., et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat. Commun. 2016;7:12030. doi: 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu B., Fang M., He Z., Cui D., Jia S., Lin Q.X.X., Xu X., Zhou T., Liu W. Hepatitis B virus stimulates G6PD expression through HBx-mediated Nrf2 activation. Cell Death Dis. 2015;6:e1980. doi: 10.1038/cddis.2015.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zimmermann K., Baldinger J., Mayerhofer B., Atanasov A.G., Dirsch V.M., Heiss E.H. Activated AMPK boosts the Nrf2/HO-1 signaling axis--A role for the unfolded protein response. Free Radic. Biol. Med. 2015;88:417–426. doi: 10.1016/j.freeradbiomed.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Joo M.S., Kim W.D., Lee K.Y., Kim J.H., Koo J.H., Kim S.G. AMPK Facilitates Nuclear Accumulation of Nrf2 by Phosphorylating at Serine 550. Mol. Cell. Biol. 2016;36:1931–1942. doi: 10.1128/MCB.00118-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Singh A., Happel C., Manna S.K., Acquaah-Mensah G., Carrerero J., Kumar S., Nasipuri P., Krausz K.W., Wakabayashi N., Dewi R., et al. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J. Clin. Investig. 2013;123:2921–2934. doi: 10.1172/JCI66353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holmstrom K.M., Baird L., Zhang Y., Hargreaves I., Chalasani A., Land J.M., Stanyer L., Yamamoto M., Dinkova-Kostova A.T., Abramov A.Y. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol. Open. 2013;2:761–770. doi: 10.1242/bio.20134853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim T.H., Hur E.G., Kang S.J., Kim J.A., Thapa D., Lee Y.M., Ku S.K., Jung Y., Kwak M.K. NRF2 blockade suppresses colon tumor angiogenesis by inhibiting hypoxia-induced activation of HIF-1alpha. Cancer Res. 2011;71:2260–2275. doi: 10.1158/0008-5472.CAN-10-3007. [DOI] [PubMed] [Google Scholar]
- 99.Kinch L., Grishin N.V., Brugarolas J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2. Cancer Cell. 2011;20:418–420. doi: 10.1016/j.ccr.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ooi A., Wong J.C., Petillo D., Roossien D., Perrier-Trudova V., Whitten D., Min B.W., Tan M.H., Zhang Z., Yang X.J., et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20:511–523. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 101.Ooi A., Furge K. Fumarate hydratase inactivation in renal tumors: HIF1α, NRF2, and “cryptic targets” of transcription factors. Chin. J. Cancer. 2012;31:413–420. doi: 10.5732/cjc.012.10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alam N.A., Rowan A.J., Wortham N.C., Pollard P.J., Mitchell M., Tyrer J.P., Barclay E., Calonje E., Manek S., Adams S.J., et al. Genetic and functional analyses of FH mutations in multiple cutaneous and uterine leiomyomatosis, hereditary leiomyomatosis and renal cancer, and fumarate hydratase deficiency. Hum. Mol. Genet. 2003;12:1241–1252. doi: 10.1093/hmg/ddg148. [DOI] [PubMed] [Google Scholar]
- 103.Adam J., Hatipoglu E., O’Flaherty L., Ternette N., Sahgal N., Lockstone H., Baban D., Nye E., Stamp G.W., Wolhuter K., et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: Roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O’Mealey G.B., Plafker K.S., Berry W.L., Janknecht R., Chan J.Y., Plafker S.M. A PGAM5-KEAP1-Nrf2 complex is required for stress-induced mitochondrial retrograde trafficking. J. Cell Sci. 2017;130:3467–3480. doi: 10.1242/jcs.203216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lo S.-C., Hannink M. PGAM5, a Bcl-XL-interacting Protein, Is a Novel Substrate for the Redox-regulated Keap1-dependent Ubiquitin Ligase Complex. J. Biol. Chem. 2006;281:37893–37903. doi: 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- 106.Lee D.F., Kuo H.P., Liu M., Chou C.K., Xia W., Du Y., Shen J., Chen C.T., Huo L., Hsu M.C., et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol. Cell. 2009;36:131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Holze C., Michaudel C., Mackowiak C., Haas D.A., Benda C., Hubel P., Pennemann F.L., Schnepf D., Wettmarshausen J., Braun M., et al. Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat. Immunol. 2018;19:130–140. doi: 10.1038/s41590-017-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamada T., Murata D., Adachi Y., Itoh K., Kameoka S., Igarashi A., Kato T., Araki Y., Huganir R.L., Dawson T.M., et al. Mitochondrial Stasis Reveals p62-Mediated Ubiquitination in Parkin-Independent Mitophagy and Mitigates Nonalcoholic Fatty Liver Disease. Cell Metab. 2018;28:588–604.e5. doi: 10.1016/j.cmet.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang P., Du W., Wu M. Regulation of the pentose phosphate pathway in cancer. Protein Cell. 2014;5:592–602. doi: 10.1007/s13238-014-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koppula P., Zhang Y., Shi J., Li W., Gan B. The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J. Biol. Chem. 2017;292:14240–14249. doi: 10.1074/jbc.M117.798405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sayin V.I., LeBoeuf S.E., Singh S.X., Davidson S.M., Biancur D., Guzelhan B.S., Alvarez S.W., Wu W.L., Karakousi T.R., Zavitsanou A.M., et al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. eLife. 2017;6:e28083. doi: 10.7554/eLife.28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J., Pavlova N.N., Thompson C.B. Cancer cell metabolism: The essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36:1302–1315. doi: 10.15252/embj.201696151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khamari R., Trinh A., Gabert P.E., Corazao-Rozas P., Riveros-Cruz S., Balayssac S., Malet-Martino M., Dekiouk S., Joncquel Chevalier Curt M., Maboudou P., et al. Glucose metabolism and NRF2 coordinate the antioxidant response in melanoma resistant to MAPK inhibitors. Cell Death Dis. 2018;9:325. doi: 10.1038/s41419-018-0340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.LeBoeuf S.E., Wu W.L., Karakousi T.R., Karadal B., Jackson S.R., Davidson S.M., Wong K.K., Koralov S.B., Sayin V.I., Papagiannakopoulos T. Activation of Oxidative Stress Response in Cancer Generates a Druggable Dependency on Exogenous Non-essential Amino Acids. Cell Metab. 2020;31:339–350.e4. doi: 10.1016/j.cmet.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sato H., Nomura S., Maebara K., Sato K., Tamba M., Bannai S. Transcriptional control of cystine/glutamate transporter gene by amino acid deprivation. Biochem. Biophys. Res. Commun. 2004;325:109–116. doi: 10.1016/j.bbrc.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 116.Ye P., Mimura J., Okada T., Sato H., Liu T., Maruyama A., Ohyama C., Itoh K. Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition. Mol. Cell. Biol. 2014;34:3421–3434. doi: 10.1128/MCB.00221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gwinn D.M., Lee A.G., Briones-Martin-Del-Campo M., Conn C.S., Simpson D.R., Scott A.I., Le A., Cowan T.M., Ruggero D., Sweet-Cordero E.A. Oncogenic KRAS Regulates Amino Acid Homeostasis and Asparagine Biosynthesis via ATF4 and Alters Sensitivity to L-Asparaginase. Cancer Cell. 2018;33:91–107.e6. doi: 10.1016/j.ccell.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shin C.-S., Mishra P., Watrous J.D., Carelli V., D’Aurelio M., Jain M., Chan D.C. The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat. Commun. 2017;8:15074. doi: 10.1038/ncomms15074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kang Y.P., Torrente L., Falzone A., Elkins C.M., Liu M., Asara J.M., Dibble C.C., DeNicola G.M. Cysteine dioxygenase 1 is a metabolic liability for non-small cell lung cancer. eLife. 2019;8:e45572. doi: 10.7554/eLife.45572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mills E.L., Ryan D.G., Prag H.A., Dikovskaya D., Menon D., Zaslona Z., Jedrychowski M.P., Costa A.S.H., Higgins M., Hams E., et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bollong M.J., Lee G., Coukos J.S., Yun H., Zambaldo C., Chang J.W., Chin E.N., Ahmad I., Chatterjee A.K., Lairson L.L., et al. A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling. Nature. 2018;562:600–604. doi: 10.1038/s41586-018-0622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen P.H., Smith T.J., Wu J., Siesser P.F., Bisnett B.J., Khan F., Hogue M., Soderblom E., Tang F., Marks J.R., et al. Glycosylation of KEAP1 links nutrient sensing to redox stress signaling. EMBO J. 2017;36:2233–2250. doi: 10.15252/embj.201696113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lim J.K.M., Delaidelli A., Minaker S.W., Zhang H.-F., Colovic M., Yang H., Negri G.L., von Karstedt S., Lockwood W.W., Schaffer P., et al. Cystine/glutamate antiporter xCT (SLC7A11) facilitates oncogenic RAS transformation by preserving intracellular redox balance. Proc. Natl. Acad. Sci. USA. 2019;116:9433–9442. doi: 10.1073/pnas.1821323116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xue D., Zhou C., Shi Y., Lu H., Xu R., He X. Nuclear transcription factor Nrf2 suppresses prostate cancer cells growth and migration through upregulating ferroportin. Oncotarget. 2016;7:78804–78812. doi: 10.18632/oncotarget.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kong Y., Hu L., Lu K., Wang Y., Xie Y., Gao L., Yang G., Xie B., He W., Chen G., et al. Ferroportin downregulation promotes cell proliferation by modulating the Nrf2-miR-17-5p axis in multiple myeloma. Cell Death Dis. 2019;10:624. doi: 10.1038/s41419-019-1854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kerins M.J., Ooi A. The Roles of NRF2 in Modulating Cellular Iron Homeostasis. Antioxid. Redox Signal. 2018;29:1756–1773. doi: 10.1089/ars.2017.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]