Abstract
Recently, the SARS-CoV-2 variants from the United Kingdom (UK), South Africa, and Brazil have received much attention for their increased infectivity, potentially high virulence, and possible threats to existing vaccines and antibody therapies. The question remains if there are other more infectious variants transmitted around the world. We carry out a large-scale study of 506,768 SARS-CoV-2 genome isolates from patients to identify many other rapidly growing mutations on the spike (S) protein receptor-binding domain (RBD). We reveal that essentially all 100 most observed mutations strengthen the binding between the RBD and the host angiotensin-converting enzyme 2 (ACE2), indicating the virus evolves toward more infectious variants. In particular, we discover new fast-growing RBD mutations N439K, S477N, S477R, and N501T that also enhance the RBD and ACE2 binding. We further unveil that mutation N501Y involved in United Kingdom (UK), South Africa, and Brazil variants may moderately weaken the binding between the RBD and many known antibodies, while mutations E484K and K417N found in South Africa and Brazilian variants, L452R and E484Q found in India variants, can potentially disrupt the binding between the RBD and many known antibodies. Among these RBD mutations, L452R is also now known as part of the California variant B.1.427. Finally, we hypothesize that RBD mutations that can simultaneously make SARS-CoV-2 more infectious and disrupt the existing antibodies, called vaccine escape mutations, will pose an imminent threat to the current crop of vaccines. A list of most likely vaccine escape mutations is given, including S494P, Q493L, K417N, F490S, F486L, R403K, E484K, L452R, K417T, F490L, E484Q, and A475S. Mutation T478K appears to make the Mexico variant B.1.1.222 the most infectious one. Our comprehensive genetic analysis and protein-protein binding study show that the genetic evolution of SARS-CoV-2 on the RBD, which may be regulated by host gene editing, viral proofreading, random genetic drift, and natural selection, gives rise to more infectious variants that will potentially compromise existing vaccines and antibody therapies.
Keywords: COVID-19, SARS-CoV-2, Mutation, Vaccine escape, Antibody, Binding affinity, Persistent homology, Deep learning
1. Introduction
Up to April 18, 2021, coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has taken 3,004,842 lives and infected 140,373,125 people according to the data from World Health Organization (WHO). The first complete SARS-CoV-2 genome sequence was deposited to the GenBank (Access number: NC_045512.2) on January 5, 2020. Thereafter, new SARS-Cov-2 genome sequences were accumulated rapidly at the GenBank and GISAID, which laid the foundations for analyzing the SARS-CoV-2 mutations, virulence, pathogenicity, antigenicity, and transmissibility. A complete SARS-CoV-2 genome is an unsegmented positive-sense single-stranded RNA virus, which encodes 29 structural and non-structural proteins (NSPs) by its 29,903 nucleotides. NSPs play vital roles in RNA replication, while structure proteins form the viral particle. There are four structural proteins on SARS-CoV-2, namely, spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins [[1], [2], [3], [4]]. Among them, the S protein with 1273 residues of SARS-CoV-2 has drawn much attention due to its critic role in viral infection and the development of vaccines and antibody drugs.
The SARS-CoV-2 enters the host cell by interacting between its S protein and the host angiotensin-converting enzyme 2 (ACE2), primed by host transmembrane protease, serine 2 (TMPRSS2) [5]. Such a process initiates the response from the host adaptive immune system, which generates antibodies to combat the invading virus. Therefore, the S protein of SARS-CoV-2 has become a target in the development of antibody therapies and vaccines. A major concern is the potential impacts of S protein mutations on viral infectivity, the existing vaccines, and antibody therapies.
The most well-known mechanism of mutations is the random genetic drift, which plays a role in the processes of transcription, translation, replication, etc. Compared with DNA viruses, RNA viruses are more prone to random mutations. Unlike other RNA viruses, such as influenza, SARS-CoV-2 has a genetic proofreading mechanism regulated by NSP14 and NSP12 (a.k.a RNA-dependent RNA polymerase) [6,7], which enables SARS-CoV-2 to have a higher fidelity in its replication. However, the host gene editing has been found to be the major source for existing SARS-CoV-2 mutations [8], counting for 65% of reported mutations. Therefore, the worldwide transmission of COVID-19 provides SARS-CoV-2 an abundant opportunity to experience fast mutations. Another important mechanism for SARS-CoV-2 evolution is natural selection, which makes the virus more infectious while less virulent, in general [9,10].
It has been established that the infectivity of different viral variants in host cells is proportional to the binding free energy (BFE) between the RBD of each variant and the ACE2 [5,[11], [12], [13], [14]]. Based on such a principle, it has been reported that mutations on the S protein have strengthened SARS-CoV-2 infectivity [15]. Whereas, virulent can be due to mutations on many SARS-CoV-2 proteins. The widely spread asymptomatic COVID-19 infection and transmission can be a result of mutation-induced virulent changes [16].
Recently, the United Kingdom (UK) variant B.1.1.7 (a.k.a 20I/501Y.V1) [17], the South Africa variant B.1.351 (a.k.a 20H/501Y.V2) [18], the Brazil(ian) variant P.1 (a.k.a 20J/501Y.V3) [19], and the India variant B.1.617 [20] have been circulating worldwide, including the United States (US) and Spain. These variants contain mutations on the S protein RBD and are widely speculated to make SARS-CoV-2 more infectious. Specifically, all three variants involve RBD mutation N501Y, whereas the South Africa and Brazil(ian) variants also contain RBD mutations E484K and K417N.
An important question is how these new variants will affect the vaccines and antibody drugs. Ideally, this question should be answered by experiments. However, SARS-CoV-2 has more than 28,000 unique single mutations, with nearly 7000 of them on the S protein, which are intractable for experimental means. In May 2020, an intensively validated topology-based neural network tree (TopNetTree) model [21] was employed to predict certain RBD mutations, including E484K, L452R, and K417N, would strengthen SARS-CoV-2 infectivity [15]. These predictions have been confirmed [[17], [18], [19]]. Additionally, all 451 new RBD mutations occurred since May 2020 were predicted as the most likely mutations in our work published online last May [15]. We also predicted a list of 625 unlikely RBD mutations [15] and currently, none of them has ever been observed. Recently, our TopNetTree model has been trained on SARS-CoV-2 datasets to accurately predict the S protein and ACE2 or antibody binding free energy changes induced by mutations [22]. A total of 31 disruptive mutations on S protein RBD has been reported as the potential mutations that would most likely disrupt the binding of S protein and essentially all the known SARS-CoV-2 antibodies had they ever occurred [22]. Therefore, tracking the growth rate of existing mutations on S protein RBD enables us to monitor the mutations that may impact the efficacy of the existing vaccines and antibody drugs. The study of fast-growing mutations also enables us to understand the SARS-CoV-2 evolutionary tendency and eventually predict future mutations.
The objective of this work is to track the fast-growing RBD mutations in pandemic-devastated countries and to analyze its evolutionary tendency around the world based on one of the most comprehensive data sets involving 506,768 SARS-CoV-2 genome sequences shown in the Mutation Tracker (https://users.math.msu.edu/users/weig/SARS-CoV-2_Mutation_Tracker.html). We found 6945 unique single mutations on the S protein and among them, 1024 occurred on the RBD. In terms of protein sequence, 100 of 651 non-degenerate mutations on the RBD were observed more than 28 times in the database and are regarded as significant mutations. We show that in addition to mutations N501Y, E484K, and K417N in the UK, South Africa, and Brazil(ian) variants, L452R, E484Q in the India variants, N439K, S477N, S477R, and N501T are also fast-growing mutations in 31 pandemic-devastated countries in the past few months. Using the TopNetTree model [21,22], we discover that essentially all 100 most observed mutations on the RBD are associated with the BFE strengthening of the binding of the RBD and ACE2 complex, resulting in more infectious SARS-CoV-2 variants. Considering mutation occurrence probability and ability to disrupt antibodies, we identify vaccine-escape and vaccine-weakening RBD mutations. The present finding suggests that S protein RBD mutations, in general, make the virus more infectious and are disruptive to the existing vaccines and antibody drugs.
2. Results
2.1. Gene-specific analysis on the S protein and the RBD
Driven by natural selection, random genetic drift, gene editing, host immune responses, etc. [9,10], viruses constantly evolve through mutations, which create genetic diversity and generates new variants. To have a good understanding of how the mutation will affect the infectivity, transmission, and virulence of SARS-CoV-2, it will be of great importance to study the mutations on SARS-CoV-2, particularly the S protein and its RBD, over a long time period. Therefore, in this work, we mainly focus on the mutations in S protein and S protein RBD. Here, a total of 28,507 unique single mutations has been decoded from 651,768 complete SARS-CoV-2 genome sequences.
Table 1 shows the distribution of 12 single-nucleotide polymorphism (SNP) types among 6945 unique mutations and 2,194,305 non-unique mutations on the S gene of SARS-CoV-2 worldwide. Symbols NU, NNU, RU, and RNU represent the number of unique mutations, the number of non-unique mutations, the ratio of 12 SNP types among unique mutation, and the ratio of 12 SNP types among non-unique mutations, respectively. It can be seen that A>G and C>T have a higher ratio in unique and non-unique cases, which may be related to the host immune response via APOBEC and ADAR gene editing as reported in [8]. Moreover, T>C has the highest mutation ratios among unique mutations. However, the ratio of T>C mutations among the non-unique mutations is not very high, indicating that T>C mutations do not commonly occur in the population.
Table 1.
The distribution of 12 SNP types among 6945 unique mutations and 2,194,305 non-unique mutations on the S gene of SARS-CoV-2 worldwide. NU is the number of unique mutations and NNU is the number of non-unique mutations. RU and RNU represent the ratios of 12 SNP types among unique and non-unique mutations. In this table, we bold the ratios that are greater than 10%.
| SNP type | Mutation type | NU | NNU | RU | RNU | SNP type | Mutation type | NU | NNU | RU | RNU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A>T | Transversion | 655 | 187,467 | 9.43% | 8.54% | C>T | Transition | 609 | 488,323 | 8.77% | 22.25% |
| A>C | Transversion | 567 | 12,914 | 8.16% | 0.59% | C>A | Transversion | 466 | 369,637 | 6.71% | 16.85% |
| A>G | Transition | 908 | 530,814 | 13.07% | 24.19% | C>G | Transversion | 269 | 3965 | 3.87% | 0.18% |
| T>A | Transversion | 589 | 6690 | 8.48% | 0.30% | G>T | Transversion | 523 | 111,949 | 7.53% | 5.10% |
| T>C | Transition | 976 | 60,918 | 14.05% | 2.78% | G>C | Transversion | 342 | 182,984 | 4.92% | 8.34% |
| T>G | Transversion | 498 | 179,748 | 7.17% | 8.19% | G>A | Transition | 543 | 58,896 | 7.82% | 2.68% |
Table 2 shows the distribution of 12 SNP types among 1024 unique mutations and 266,458 non-unique mutations on the spike RBD gene sequence of SARS-CoV-2 worldwide. To be noticed, compared to Table 1, the distribution of 12 SNP types acts differently on S protein RBD. The top 3 highest mutation ratios among non-unique mutations are A>T, G>A, and C>A, which indicating that these 3 types of mutations may have a higher impact on the transmission of SARS-CoV-2.
Table 2.
The distribution of 12 SNP types among 1024 unique mutations and 266,458 non-unique mutations on the spike RBD gene of SARS-CoV-2 worldwide. NU is the number of unique mutations and NNU is the number of non-unique mutations. RU and RNU represent the ratios of 12 SNP types among unique and non-unique mutations. In this table, we bold the ratios that are greater than 10%.
| SNP type | Mutation type | NU | NNU | RU | RNU | SNP type | Mutation type | NU | NNU | RU | RNU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A>T | Transversion | 84 | 170,165 | 8.20% | 63.86% | C>T | Transition | 90 | 11,562 | 8.79% | 4.34% |
| A>C | Transversion | 75 | 3685 | 7.32% | 1.38% | C>A | Transversion | 66 | 16,551 | 6.45% | 6.21% |
| A>G | Transition | 134 | 2310 | 13.09% | 0.87% | C>G | Transversion | 38 | 694 | 3.71% | 0.26% |
| T>A | Transversion | 89 | 890 | 8.69% | 0.33% | G>T | Transversion | 79 | 7419 | 7.71% | 2.78% |
| T>C | Transition | 161 | 7308 | 15.72% | 2.74% | G>C | Transversion | 47 | 907 | 4.59% | 0.34% |
| T>G | Transversion | 76 | 11,318 | 7.42% | 4.25% | G>A | Transition | 85 | 33,649 | 8.30% | 12.63% |
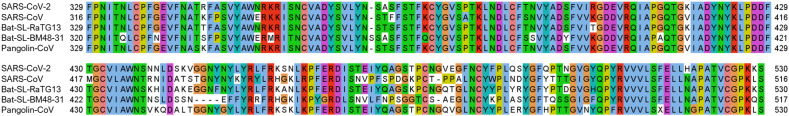
Fig. 1 is the 2D amino acid sequence alignment for the S protein RBD of SARS-CoV-2, Bat-SL-RaTG13, Pangolin-CoV, SARS-CoV, and Bat-SL-BM48-31. It can be seen that residues R346, N354, K417, N438, N440, S443, K444, V445, K458, N460, T478, S494, Q495, and Q498 located on the S protein RBD is not conservative, while the other residues are relatively conservative among different species.
Fig. 1.
2D sequence alignment for the S protein RBD of SARS-CoV-2, Bat-SL-RaTG13, Pangolin-CoV, SARS-CoV, and Bat-SL-BM48-31.
2.2. Impacts of SARS-CoV-2 spike RBD mutations on SARS-CoV-2 infectivity
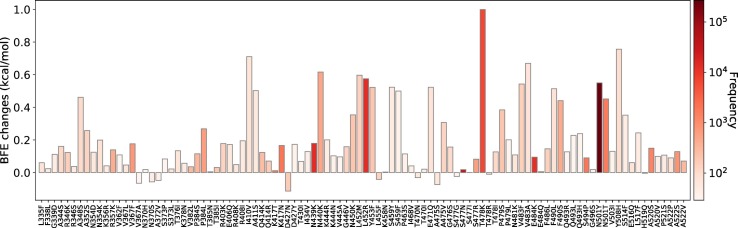
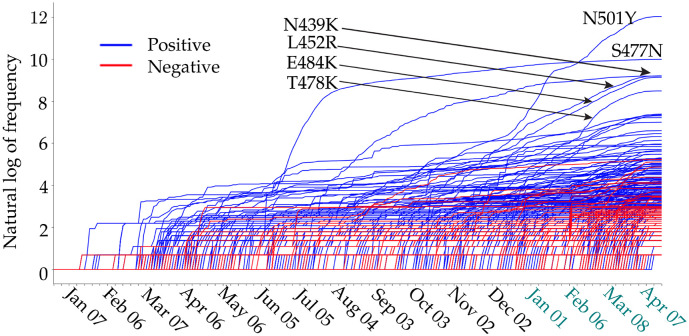
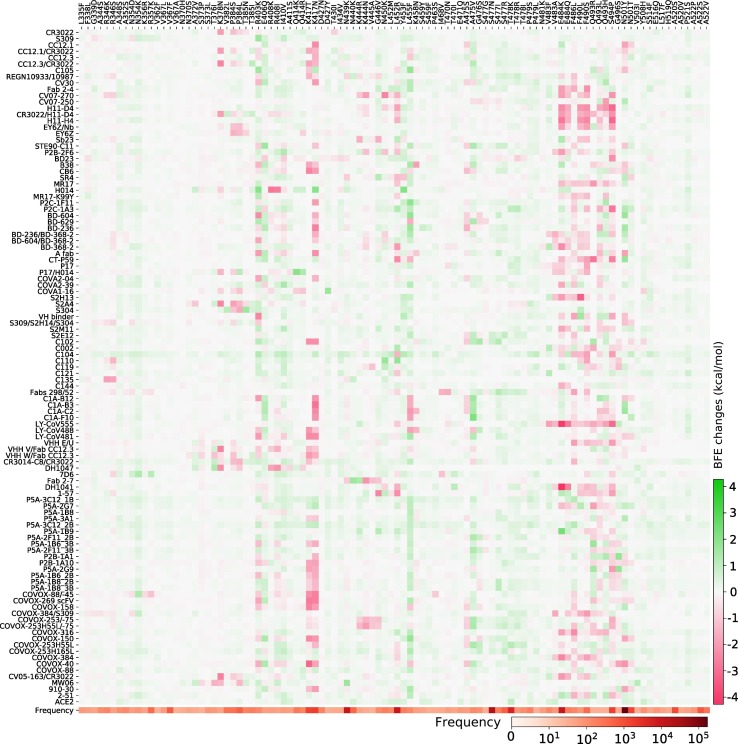
The RBD is located on the S1 domain of the S protein, which plays a vital role in binding with the human ACE2 to get entry into host cells. The mutations that are detected on the RBD may affect the binding process and lead to the BFE changes. In this section, we apply the TopNetTree model [22] to predict the mutation-induced BFE changes of RBD and ACE2. Fig. 2 illustrates the predicted BFE changes for S protein and human ACE2 induced by single-site mutations on the RBD. Here, we consider 100 most observed mutations. The bar plot of the other mutations on S RBD can be found in the Supporting Information. In this figure, a total of 100 most observed mutations are displayed. Among them, 9 mutations induce negligible negative BFE changes, while the other 91 mutations are binding-strengthening mutations. Mutation T478K has the largest BFE change which is nearly 1 kcal/mol. It may have made the Mexico variant B.1.1.222 the most infectious observed variant. To be noted, the residue T478 is not conservative among different species as illustrated in Fig. 1. The N501Y, S477N, L452R, N439K, and E484K mutations are the top mutations with significant frequencies. Among them, the N501Y and L452R mutations have relatively high BFE changes of 0.55 kcal/mol and 0.58 kcal/mol, respectively. Moreover, the frequency and predicted BFE changes are both at a high level for mutations N501T, Y508H. Fig. 3 illustrates the time evolution of 651 binding-strengthening (blue) and binding-weakening mutations (red) on the S protein RBD. Here, the y-axis reveals the natural log frequency of each mutation. Based on the our previous findings in [15], at this stage, 651 out of 1149 RBD mutations that we predicted as “most likely” mutations have been observed, and none of the 1912 “likely” and 625 “unlikely” mutations are tracked on the S protein RBD, suggesting the reliability of our model for predicting the BFE changes of S protein RBD and ACE2. Among 651 mutations that are detected on RBD, mutations N501Y, S477N, L452R, N439K, and E484K have the highest frequency up to April 18, 2021.
Fig. 2.
Illustration of SARS-CoV-2 mutation-induced BFE changes for the complexes of S protein and ACE2. Here, 100 most observed mutations on S RBD are illustrated.
Fig. 3.
Illustration of the time evolution of 424 ACE2 binding-strengthening RBD mutations (blue) and 227 ACE2 binding-weakening RBD mutations (red) on the S protein RBD of SARS-CoV-2 from Jan 07, 2020 to April 18, 2021. The x-axis represents date and y-axis represents the natural log of frequency of each mutation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
It is important to those mutations that have been recorded with high frequency the beginning of 2021. Table 3 gives such information for top 40 mutations in 2021. It can be seen that mutations N501Y, L452R, T478K, N501T, N550K, F490S, V483F, L452M, and A348S have relatively high BFE changes of the binding of S protein and ACE2, suggesting that they may lead to more infectious variants.
Table 3.
List of top 40 high-frequency (HF) mutations and their corresponding BFE changes (unit: kcal/mol) of the binding of S protein and ACE2. Here, count shows the frequency occurred in 2021.
| Rank | HF mutation | Count | BFE change | Rank | HF mutation | Count | BFE change |
|---|---|---|---|---|---|---|---|
| Top 1 | N501Y | 168,801 | 0.5499 | Top 21 | N450K | 184 | 0.3535 |
| Top 2 | L452R | 9843 | 0.5752 | Top 22 | E484Q | 182 | 0.0057 |
| Top 3 | E484K | 9350 | 0.0946 | Top 23 | P330S | 182 | 0.0533 |
| Top 4 | S477N | 9276 | 0.018 | Top 24 | A522V | 179 | 0.0705 |
| Top 5 | N439K | 6056 | 0.1792 | Top 25 | D427N | 164 | −0.1133 |
| Top 6 | T478K | 4935 | 0.9994 | Top 26 | P479S | 153 | 0.3844 |
| Top 7 | K417N | 1634 | 0.1661 | Top 27 | V382L | 151 | 0.0355 |
| Top 8 | K417T | 1508 | 0.0116 | Top 28 | T385N | 151 | 0.0049 |
| Top 9 | S494P | 1483 | 0.0902 | Top 29 | Q414R | 143 | 0.0708 |
| Top 10 | N501T | 1295 | 0.4514 | Top 30 | R346K | 135 | 0.1234 |
| Top 11 | A520S | 819 | 0.1495 | Top 31 | T385I | 127 | 0.0314 |
| Top 12 | A522S | 621 | 0.1283 | Top 32 | R403K | 121 | 0.1778 |
| Top 13 | V367F | 536 | 0.1764 | Top 33 | L455F | 99 | −0.0415 |
| Top 14 | N440K | 432 | 0.6161 | Top 34 | V483F | 99 | 0.5428 |
| Top 15 | S477R | 394 | 0.082 | Top 35 | A475V | 96 | 0.3069 |
| Top 16 | P384L | 389 | 0.2681 | Top 36 | G446V | 86 | 0.1583 |
| Top 17 | R357K | 373 | 0.1393 | Top 37 | L452M | 83 | 0.5966 |
| Top 18 | F490S | 363 | 0.4406 | Top 38 | A348S | 82 | 0.4616 |
| Top 19 | P384S | 263 | 0.1151 | Top 39 | T478I | 81 | 0.1269 |
| Top 20 | Q414K | 224 | 0.1234 | Top 40 | A352S | 78 | 0.2576 |
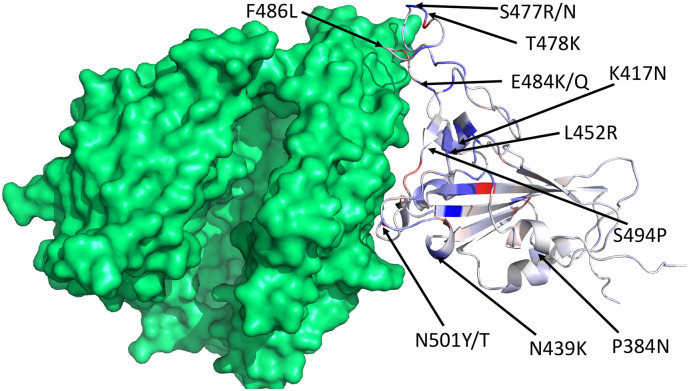
Fig. 4 shows the 3D structure of SARS-CoV-2 S protein RBD bound with ACE2. Here, we mark 13 mutations with either high frequency or high BFE changes. The blue and red colors represent the mutations that have positive and negative BFE changes, respectively. The darker the colour is, the larger the absolute value of the BFE change is. While mutations occur everywhere on the spike protein, the ones that are most important to COVID-19 infectivity and the efficacy of antibodies and vaccines are located at the interface between the spike protein and ACE2 or antibodies.
Fig. 4.
The 3D structure of SARS-CoV-2 S protein RBD bound with ACE2 (PDB ID: 6M0J). We choose blue and red colors to mark the binding-strengthening and binding-weakening mutations, respectively. Vaccine escape mutations described in Table 4 are labeled. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Impacts of SARS-CoV-2 spike RBD mutations on COVID-19 vaccines
It is be of paramount importance to track not only ACE2-binding-strengthening RBD mutations and FG mutations but also the antibody-binding-weakening RBD mutations. Our early work reported nearly 71% mutations on the S protein RBD will weaken the binding of S protein and antibodies, while 64.9% mutations on the RBD will strengthen the binding of S protein and ACE2, suggesting that these mutations may potentially enhance the infectivity of SARS-CoV-2 and make the existing antibodies less effective [22]. We call those mutations that weaken the binding of the S protein and most SARS-CoV-2 antibodies as antibody disrupting (AD) mutations [22]. Notably, most antibody disrupting mutations have negative BFE changes, suggesting that they will make the SARS-CoV-2 less infectious and thus, will not frequently occur due to natural selection. As a result, many of them may not be able to evade the existing vaccines in a population. Therefore, it is necessary to focus on the BFE changes of S protein and antibodies that are induced by 100 most observed mutations on S protein RBD.
In this work, we have collected a total of 106 antibodies. The detailed information of these 106 antibodies can be found in the Supporting Information. Fig. 5 shows the BFE changes for the S protein and 106 antibody complexes together with ACE2 following 100 most observed mutations on the S protein RBD. The red colour marks the mutation-induced negative BFE changes for the complexes of S protein and antibodies, which indicates that these mutations may weaken the binding and make the antibody less effective. Meanwhile, the green colour represents the positive BFE changes induced by mutations, which suggests that these mutations may strengthen the binding between S protein and antibodies. From Fig. 5, we can see that mutation E484K will disruptively weaken the binding of S protein with antibodies such as LY-CoV555 and DH1041, which are marked in dark red. Mutation S494P will disruptively weaken the binding of S protein with antibodies such as H11-D4, H11-H4, and LY-CoV555. Mutation K417N will disruptively weaken the binding of S protein with a large number of antibodies. Moreover, mutation N501Y will moderately weaken the binding of S protein with antibodies such as CC12.1/CR3022, COVOX-88/-45, COVOX-88, etc.
Fig. 5.
Illustration of SARS-CoV-2 S RBD 100 most observed mutations induced BFE changes for the complexes of S protein and 106 antibodies or ACE2. Here, red colour represents the negative changes that will weaken the binding, while the green colour shows the positive changes that will strengthen the binding. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Considering the impact of the possible calculation error, we set −0.3 kcal/mol as the threshold of the binding between S protein and antibodies induced by AD mutations. Specifically, we say a mutation is an AD mutation to the binding complex of S protein and antibody if its BFE change for the complex is less than 0.3 kcal/mol. We hypothesize that RBD mutations that can simultaneously strengthen the infectivity and disrupt the binding between the S protein and existing antibodies will pose imminent threats to the current crop of vaccines. We define a vaccine escape (VE) mutation as a high-frequency mutation that is an AD mutation for at least 24 (23%) different antibodies. We also define a vaccine-weakening (AW) mutation as a high-frequency mutation and AD mutation for 11 (10%) to 21 (20%) different antibodies.
Table 4 lists vaccine-escape (VE) and vaccine-weakening (VW) RBD mutations together with their corresponding BFE changes (unit: kcal/mol) of the binding between S protein and ACE2. The count represents the number of antibodies that will make a specific mutation to be an AD mutation. We can see that VE mutations F490S, L452R, VW mutations F490L, N501Y, V483A, and N501T have relatively high BFE changes of the binding of S protein and ACE2, suggesting that they are high-risk mutations. Moreover, L452R, N501Y, and N501T are also HF mutations, which should receive high attention.
Table 4.
List of vaccine escape (VE) and vaccine weakening (VW) Their corresponding BFE changes (unit: kcal/mol) of the binding of S protein and ACE2 are provided as well. Here, the count shows the number of antibodies that will make a specific mutation to be an AD mutation.
| VE mutation | BFE change | Count | VW mutation | BFE change | Count |
|---|---|---|---|---|---|
| S494P | 0.0902 | 50 | N501Y | 0.5499 | 21 |
| Q493L | 0.2279 | 43 | Q493R | 0.1271 | 21 |
| K417N | 0.1661 | 43 | R408I | 0.1949 | 19 |
| F490S | 0.4406 | 42 | Q493H | 0.2385 | 18 |
| F486L | 0.1456 | 41 | P384S | 0.1151 | 18 |
| R403K | 0.1778 | 34 | K378N | 0.0573 | 16 |
| E484K | 0.0946 | 31 | G496S | 0.0187 | 15 |
| L452R | 0.5752 | 28 | L455F | −0.0415 | 15 |
| K417T | 0.0116 | 28 | I410V | 0.7105 | 14 |
| F490L | 0.5139 | 25 | R346S | 0.0374 | 14 |
| E484Q | 0.0057 | 25 | V483A | 0.6695 | 13 |
| A475S | −0.0732 | 24 | K444N | 0.1024 | 12 |
| N501T | 0.4514 | 11 | |||
| P384L | 0.2681 | 11 |
2.4. Fast-growing mutations in COVID-19-devastated countries
In this section, we extract the 31 countries with the highest number of SNP profiles and analyze their mutations on S protein RBD, as illustrated in Table 5 . We can see that the BFE changes of S protein and ACE2 induced by mutations on the RBD are mostly positive, suggesting that the binding between ACE2 and S protein will be potentially strengthened in these 31 countries. This indicates that SARS-CoV-2 becomes more infectious, driven by most mutations on the receptor-binding domain.
Table 5.
The statistical analysis of mutations on S protein RBD of 31 countries with large sequencing data. Nseq is the number of sequences in each country. NU-RBD is the number of unique mutations on RBD and NNU-RBD is the number of non-unique mutations on RBD. Npositive and Nnegative represent the number of unique single mutations that will respectively result in positive and negative BFE changes of S protein and ACE2 induced by mutations on S protein RBD.
| Country (Country code) | Nseq | NU | NNU | Npositive | Nnegative |
|---|---|---|---|---|---|
| United Kingdom (UK) | 174,372 | 297 | 98,015 | 234 | 63 |
| United States (USA) | 127,809 | 352 | 44,660 | 252 | 100 |
| Denmark (DK) | 29,689 | 94 | 9628 | 81 | 13 |
| Germany (DE) | 18,778 | 324 | 16,033 | 207 | 117 |
| Canada (CA) | 13,050 | 64 | 1180 | 55 | 9 |
| Netherlands (NL) | 12,293 | 86 | 7824 | 74 | 12 |
| Sweden (SE) | 12,183 | 54 | 8346 | 51 | 3 |
| Switzerland (CH) | 10,257 | 70 | 5623 | 62 | 8 |
| Australia (AU) | 9822 | 41 | 7654 | 34 | 7 |
| France (FR) | 8945 | 76 | 6925 | 64 | 12 |
| Belgium (BE) | 7057 | 68 | 4806 | 63 | 5 |
| Italy (IT) | 6568 | 62 | 4056 | 58 | 4 |
| Spain (ES) | 6435 | 75 | 2340 | 61 | 14 |
| Ireland (IE) | 4193 | 41 | 3498 | 38 | 3 |
| Brazil (BR) | 3914 | 39 | 2899 | 32 | 7 |
| Iceland (IS) | 3868 | 13 | 158 | 13 | 0 |
| India (IN) | 3728 | 53 | 342 | 48 | 5 |
| Luxembourg (LU) | 3719 | 36 | 2224 | 33 | 3 |
| Norway (NO) | 3271 | 27 | 1374 | 26 | 1 |
| Poland (PL) | 3102 | 40 | 2505 | 34 | 6 |
| Mexico (MX) | 2908 | 48 | 1715 | 46 | 2 |
| Portugal (PT) | 2625 | 34 | 1370 | 31 | 3 |
| Latvia (LV) | 2391 | 21 | 761 | 20 | 1 |
| Lithuania (LT) | 2001 | 22 | 1052 | 21 | 1 |
| Slovenia (SI) | 1831 | 27 | 1543 | 20 | 7 |
| Finland (FI) | 1734 | 24 | 784 | 21 | 3 |
| Turkey (TR) | 1729 | 33 | 1126 | 32 | 1 |
| Czech Republic (CZ) | 1685 | 24 | 1339 | 22 | 2 |
| United Arab Emirates (AE) | 1581 | 21 | 80 | 21 | 0 |
| Austria (AT) | 1580 | 25 | 815 | 22 | 3 |
| Singapore (SG) | 1423 | 22 | 319 | 21 | 1 |
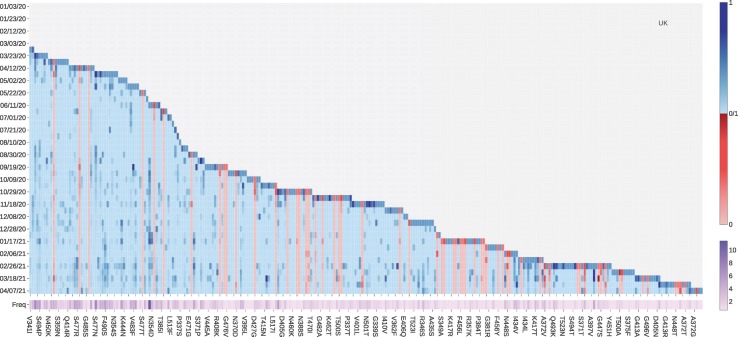
Tracking the binding-strengthening mutations will play a vital role in the development of anti-virus drugs, antibody drugs, and vaccines. Therefore, we calculate the growth rate of mutations on the RBD on a 10-day average, aiming to monitor the binding-strengthening mutations that have rapid growth over time. Fig. 6 illustrates the log growth rate and log frequency of mutations on the S protein RBD in the United Kingdom on a 10-day average. The blue and red colors respectively represent the positive and negative BFE changes induced by a specific mutation, and the purple colour represents the log frequency of a specific mutation. The darker the colour is, the higher the log growth rate/log frequency will be. For a better view, please check the HTML file in our Supporting Information. From Fig. 6, we can see that the N501Y mutation with a positive BFE change have a relatively high growth rate since early September 2020, which consist with the news that a new strain B.1.1.7 (also known as 20I/501Y.V1) in the United Kingdom has the potential to increase the pandemic trajectory [23]. Moreover, mutations V367F, E484K, N354D, and S373L with positive BFE changes also have a relatively higher mutation rate since early 2021, indicating that these four mutations may strengthen the binding of ACE2 and the S protein RBD, and potentially increase the infectivity of SARS-CoV-2. From Fig. 5, vaccine escape mutation E484K has dramatically disruptive effects on antibodies such as H11-H4, LY-CoV555, and DH1041.
Fig. 6.
The log growth rate and log frequency of mutations on S protein RBD in the United Kingdom. The blue and red colors respectively represent the binding-strengthening and binding-weakening mutations on RBD. The darker blue/red means the binding-strengthening/binding-weakening mutations with a higher growth rate in a specific 10-day period. The darker purple represents the mutation with a higher log frequency. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
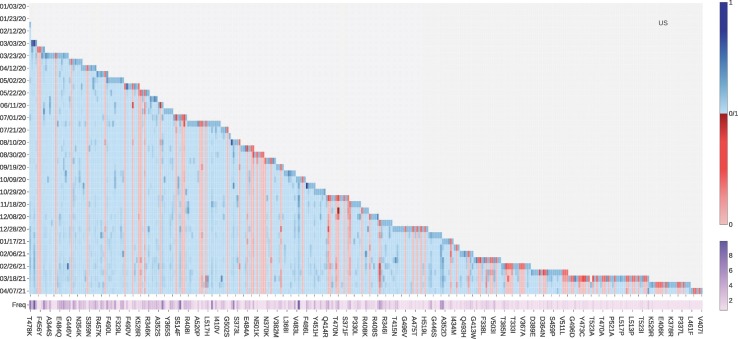
Fig. 7 illustrates the log growth rate and log frequency of mutations on S protein RBD in the United States on a 10-day average. Similar to the United Kingdom, the VW mutation N501Y and VE mutation E484K recently have a high log growth rate. Additionally, ACE2 binding-strengthening mutations T385I, N439K, S477R, and L452R also have a high log growth rate since late 2020. To be noted, L452R is a VE mutation and HF mutation that had been reported as the key mutation that linked to COVID-19 outbreaks in California on January 17, 2021 [24].
Fig. 7.
The log growth rate and log frequency of mutations on S protein RBD in the United States. The blue and red colors respectively represent the binding-strengthening and binding-weakening mutations on RBD. The darker blue/red means the binding-strengthening/binding-weakening mutations with a higher growth rate in a specific 10-day period. The darker purple represents the mutation with a higher log frequency. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
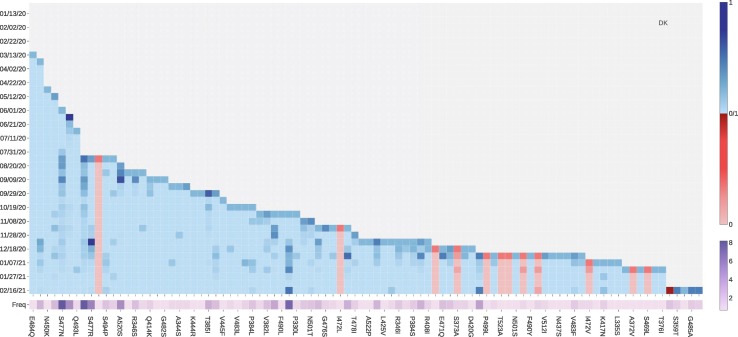
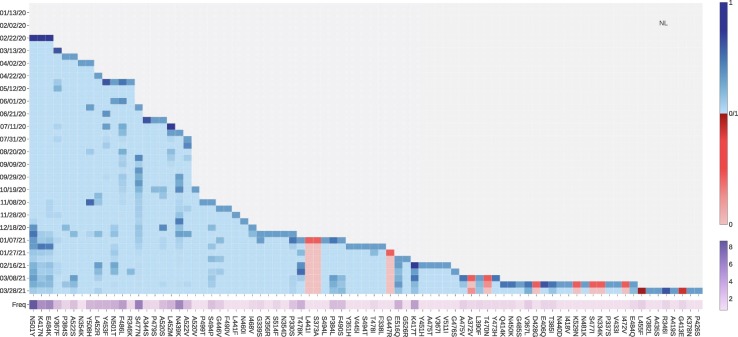
Fig. 8 tracks the fast-growing mutations in Denmark. ACE2 binding-strengthening mutation L452R has a fast-growing tendency since December 8, 2020. From Table 4, mutation L452R may disrupt the binding of 28 existing antibodies with S protein. Binding-strengthening mutation S477N has a high growth rate from late July to early December. Mutation S477R that induce the positive BFE changes has a very rapid growth between November 28, 2020, to December 08, 2020, while the number of S447R mutations has recently not increased rapidly. To be noted, neither S477R nor S477N has much negative effect on the existing antibodies. The number of ACE2 binding-strengthening mutation N439K has kept a high growth rate since early August. However, the increasing rate of the N439K mutation slows down recently. As first reported in the United Kingdom, the N501Y mutation also has a fast-growing tendency since early December 2020, making the SARS-CoV-2 more infectious. A similar pattern can also be observed in Netherlands, Switzerland, Norway, and Sweden. Moreover, as shown in Fig. 9 , four ACE2 binding-strengthening mutations have a rapid growth since late December 2020: N501Y, K417N, E484K, and P479S. Among them, K417N and E484K are both VE mutations with relatively high BFE changes, suggesting that researchers should keep tracking these mutations in the following months in Denmark. Furthermore, the B.1.351 lineage (also known as 20H/501Y.V2) was first identified in Nelson Mandela Bay, South Africa, which can be traced back to the beginning of October 2020, carries K417N, E484K, and N501Y on S protein RBD.
Fig. 8.
The log growth rate and log frequency of mutations on S protein RBD in the Denmark. The blue and red colors respectively represent the binding-strengthening and binding-weakening mutations on RBD. The darker blue/red means the binding-strengthening/binding-weakening mutations with a higher growth rate in a specific 10-day period. The darker purple represents the mutation with a higher log frequency. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
The log growth rate and log frequency of mutations on S protein RBD in the Netherlands. The blue and red colors respectively represent the binding-strengthening and binding-weakening mutations on RBD. The darker blue/red means the binding-strengthening/binding-weakening mutations with a higher growth rate in a specific 10-day period. The darker purple represents the mutation with a higher log frequency. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
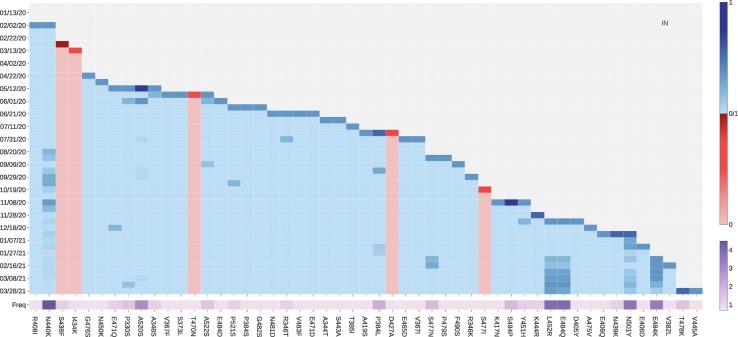
ACE2 binding-strengthening mutations in India include N440K, L452R, E484Q, N501Y, and E484K (see Fig. 10 ). It is worth to mention that except for N440K, all the ACE2 binding-strengthen mutations in India are either VE or VW mutations and have rapidly grown since February 06, 2021. Moreover, India variant B.1.617 has a ‘double mutation’ L452R and E484Q that are more infectious and vaccine evading, indicating that India's dire COVID-19 situation.
Fig. 10.
The log growth rate and log frequency of mutations on S protein RBD in India. The blue and red colors respectively represent the binding-strengthening and binding-weakening mutations on RBD. The darker blue/red means the binding-strengthening/binding-weakening mutations with a higher growth rate in a specific 10-day period. The darker purple represents the mutation with a higher log frequency. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
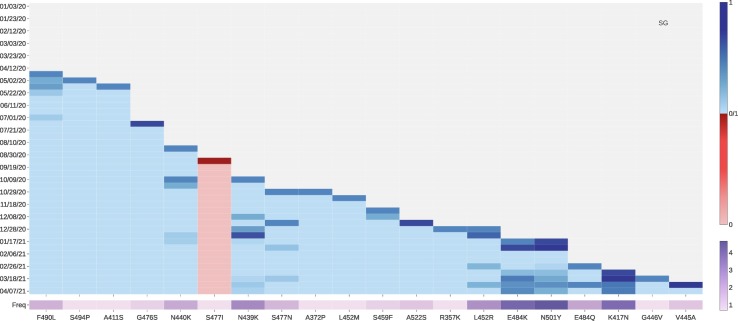
Singapore also has ACE2 binding-strengthening mutations K417N, E484K, N501Y, S477N, and L452R, as those found in other countries. Moreover, one ACE2 binding-strengthening mutation N440K with a high frequency has a relatively high growth rate since 2021 (See Fig. 11 ). Notably, the growth rate of mutation E484Q increases at the middle March of 2021. Considering the recent emergence of ‘double mutation’ L452R and E484Q in India, Singapore needs to pay more attention to tracking new variant B.1.617.
Fig. 11.
The log growth rate and log frequency of mutations on S protein RBD in Singapore. The blue and red colors respectively represent the binding-strengthening and binding-weakening mutations on RBD. The darker blue/red means the binding-strengthening/binding-weakening mutations with a higher growth rate in a specific 10-day period. The darker purple represents the mutation with a higher log frequency. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
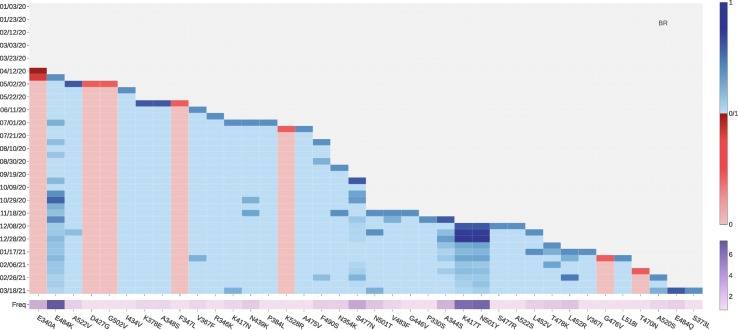
The National Institute of Infectious Diseases (NIID) in Japan first reported that four travelers from Brazil sampled a branch of the B.1.1.28 lineage called P.1 variant (also known as 20J/501Y.V3) [25]. This variant contains three mutations in the S protein RBD: VE mutation K417T, VE mutation E484K, and VW mutation N501Y. All of them are all ACE2 binding-strengthening mutations with a fast growth rate since late December 2020, as illustrated in Fig. 12 .
Fig. 12.
The log growth rate and log frequency of mutations on S protein RBD in Brazil. The blue and red colors respectively represent the binding-strengthening and binding-weakening mutations on RBD. The darker blue/red means the binding-strengthening/binding-weakening mutations with a higher growth rate in a specific 10-day period. The darker purple represents the mutation with a higher log frequency. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
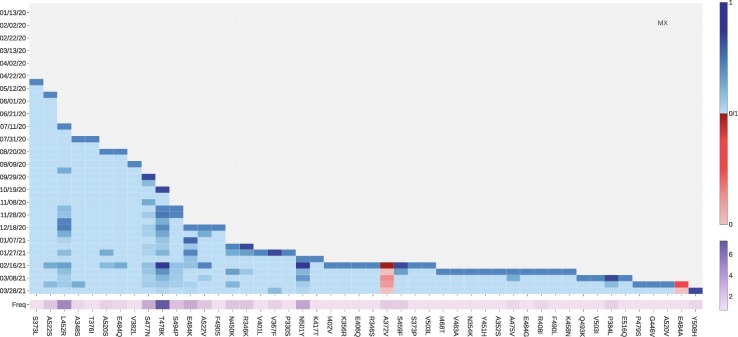
From analyzing the SNP profiles in Mexico, we notice that 6 ACE2 binding-strengthening mutations, L452R, S477N, T478K, S494P, E484K, and A552V, have a rapid growth since late October 2020. Among them, T478K is part of the Mexico variant B.1.1.222 and has the highest growth rate since late October 2020. Fig. 2 shows that T478K leads to the highest increase in ACE2-S protein RBD BFE change, indicating that fast-growing mutation T478K may potentially make the SARS-CoV-2 more transmissible and infectious. However, T478K does not pose a problem to antibodies.
2.5. Discussion
The BFE changes following 551 non-degenerate mutations on the S protein RBD are presented in Figs. S1-S5 of the Supporting information. These plots highlight the magnitude disparity in BFE changes induced by binding-strengthening mutations and binding-weakening mutations. Such a large disparity indicates that SARS-CoV-2 is evolutionarily quite advance with respect to human infection. Figs. S6-S27 of the Supporting information provide the log growth rate and log frequency of mutations on S protein RBD in the Germany, Canada, Sweden, Switzerland, Australia, France, Belgium, Italy, Spain, Ireland, Iceland, Luxembourg, Norway, Poland, Portugal, Latvia, Lithuania, Slovenia, Finland, Turkey, Czechia, United Arab Emirates, and Austria. Table 6 shows the most significant mutations on S protein RBD of 31 countries with large sequencing data. This information, together with those given in Fig. 6, Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11, Fig. 12, Fig. 13 , shows that, in addition to well-known mutations E484K, K417N, and N501Y, mutations N439K, L452R, S477N, S477R, and N501T are also ACE2 binding-strengthening mutations that have a high growth rate recently with high frequency. Tracking the growth rate tendency on a 10-day average for a long time enables us to detect the mutations that may strengthen the binding between S protein and ACE2, which will guide the development of vaccines and antibody therapies.
Table 6.
Most significant mutations on S protein RBD of 31 countries with large sequencing data.
| Country | Most significant mutations |
|---|---|
| United Kingdom | N439K, S477N, S494P, and N501Y, |
| United States | A520S, N501Y, S494P, E484K, S477N, N501T, and L452R |
| Denmark | S477N, Y453F, S477R, N439K, and N501Y |
| Germany | N439K, S477N, and N501Y |
| Canada | R357K, E484K, and L452R |
| Netherlands | N501Y, K417N, E484K, F486L, S477N, N439K, and K417T |
| Sweden | E484K, S477N, N439K, N501Y, and K417N |
| Switzerland | N439K, S477N, N501Y, Q414K, N450K, L452R, and T478K |
| Australia | S477N, N501Y, L452R, L455F, N439K, and N501T |
| France | S477N, N439K, L452R, A522S, E484K, N501Y, and K417T |
| Belgium | N501Y, S477N, E484K, N450K, K417N, and K417T |
| Italy | N439K, S477N, L452R, E484K, N501Y, K417T, N440K, and Q414K |
| Spain | S477N, N501Y, S494P, and E484K |
| Ireland | N439K, N501Y, and E484K |
| Brazil | E484K, K417T, and N501Y |
| Iceland | S477N, N439K, and E406Q |
| India | N440K, A520S, P384L, S477N, S494P, L452R, E484Q, N501Y, and E484K |
| Luxembourg | S477N, N439K, and N501Y |
| Norway | N439K, S477N, A520S, and N501Y |
| Poland | N439K, S477N, A522S, N501Y, F494P |
| Mexico | L452R and T478K |
| Portugal | S477N, L452R, and N501Y |
| Latvia | E484K, N501Y, N439K, V367F, A522V, S494P, and K417N |
| Lithuania | V362F, N439K, N501Y, S477N, S490L, L452R, S477I, and E471Q |
| Slovenia | N439K, S477R, S477N, N501Y, K356R, and E484K |
| Finland | P384L, S477N, N439K, A352S, and N501Y |
| Turkey | S477N, N501Y, K417N, N501T, and E484K |
| Czech Republic | S459Y, N439K, S477N, N501Y, E484K, and K417N |
| United Arab Emirates | N501Y, N440K, S477N, N439K, E484K, and K417N |
| Austria | S477N, N439K and N501Y |
| Singapore | F490L, N440K, N439K, S477N, L452R, E484K, N501Y, and K417N |
Fig. 13.
The log growth rate and log frequency of mutations on S protein RBD in Mexico. The blue and red colors respectively represent the binding-strengthening and binding-weakening mutations on RBD. The darker blue/red means the binding-strengthening/binding-weakening mutations with a higher growth rate in a specific 10-day period. The darker purple represents the mutation with a higher log frequency. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Based on our study of mutation impacts on 106 antibodies [22], we found that the E484K mutation may cause a dramatically disruptive effect on antibodies such as H11-D4, P2B—2F6, Fab 2-4, H11-H4, COVA2-39, BD368-2, VH binder, S2M11, S2H13, CV07-270, P2C-1A3, P17, etc, which is consistent with the finding that E484K may affect neutralization by some polyclonal and monoclonal antibodies [26,27]. Mutation N501Y could weaken antibodies B38, A fab, CC12.1, VH binder, S309 S2H12 S304, C1A—B12, 910 30, STE90-C11, COVOX-150, COVOX-40, COVOX-88, and COVOX-269. Mutation N501T could weaken antibodies B38, CC12.1, S309 S2H12 S304, etc. Both E484 and N501 are coil residues on the RBD. Similarly, mutation K417N, which is a helix-residue of the RBD, could weaken antibodies B38, CB6, CV30, CC12.1, COVA2-04, BD-604, BD-236, A fab, P2C—1F11, C1A—B12, C1A—B3, C1A—F10, C1A—C2, etc. [22]. It is interesting to understand whether newly identified fast-growing mutations N439K, L452R, S477R, and E484K are also disruptive to vaccines and antibodies. By checking the results reported early [22], we note that mutation L452R may make antibodies such as H11-D4, P2B—2F6, SR4, MR17, MR17-K99Y, H11-H4, BD-368-2, CV07-270, Fabs 298 52, CT-P59, etc., ineffective. However, mutation N439K is not as disruptive as E484K, K417N, N501Y, and N501T. It may weaken the binding of antibody SR4 and others. S477N can slightly weaken antibodies BD23 and CV07-250. Mutation S477R may even enhance the binding of most antibodies to the RBD. Finally, mutation E484Q may weaken the binding of many antibodies (such as LY-CoV555, DH1047, H11-H4, H11-D4, and CV07-270) in complex with S protein.
3. Methods
3.1. Data collection and pre-processing
The first complete SARS-CoV-2 genome sequence was released on the GenBank (Access number: NC_045512.2) on January 5, 2020, by Zhang's group at Fudan University [28]. Since then, the rapid increment of the complete genome sequences is kept depositing to the GISAID database [29]. In this work, a total of 506,768 complete SARS-CoV-2 genome sequences with high coverage and exact submission date are downloaded from the GISAID database [29] (https://www.gisaid.org/) as of April 18, 2021. We take the NC_045512.2 as the reference genome, and the multiple sequence alignment (MSA) will be applied by Clustal Omega [30] with default parameters, which results in 506,768 SNP profiles. There are 106 antibodies or antibody combinations discussed with their corresponding PDB ID provided in the Supporting information.
3.2. The growth rate of mutations
Assume we have N SNP profiles, which have a total of M n non-unique mutations and M u unique mutations (M u ≤ M n). Let ΔN i be the number of the increment of a particular mutation during the ith 10-day period, and N i be the total number of a particular mutation.
Let the number of a particular mutation in the jth day of the ith 10-day period to be N i j, where 1 ≤ i ≤ 10. Let the ΔN i = N i 10 − N i 1 be the number of the increment of a particular mutation during the ith 10-day period. Then the growth rate of a particular mutation in the ith 10-day period will be defined as
| (1) |
Moreover, the natural logarithm growth rate of a particular mutation in the ith 10-day period will be defined as
| (2) |
3.3. TopNetTree model for protein-protein interaction (PPI) binding free energy changes upon mutation
Mutation-induced protein-protein binding free energy (BFE) changes are an important approach for understanding the impact of mutations on protein-protein interactions (PPIs) and viral infectivity [31]. A variety of advanced methods has been developed [31,32]. The topology-based network tree (TopNetTree) model [15,21] is applied to predict mutation-induced BFE changes of PPIs in this work. TopNetTree model was implemented by integrating the topological representation and network tree (NetTree) to predict the BFE changes (ΔΔG) of PPIs following mutations [21]. The structural complexity of protein-protein complexes is simplified by algebraic topology [[33], [34], [35]] and is represented as the vital biological information in terms of topological invariants. NetTree integrates the advantages of convolutional neural networks (CNN) and gradient-boosting trees (GBT), such that CNN is treated as an intermediate model that converts vectorized element- and site-specific persistent homology features into a higher-level abstract feature, and GBT uses the upstream features and other biochemistry features for prediction. The performance test of tenfold cross-validation on the dataset (SKEMPI 2.0 [36]) was carried out using gradient boosted regression tree (GBRTs). The errors with the SKEMPI 2.0 dataset are 0.85 in terms of Pearson correlation coefficient (R p) and 1.11 kcal/mol in terms of the root mean square error (RMSE) [21].
3.3.1. Training sets for TopNetTree model
The TopNetTree model is trained by several important training sets. The most important dataset which provides the information for BFE changes upon mutations in the SKEMPI 2.0 dataset [36]. The SKEMPI 2.0 is an updated version of the SKEMPI database, which contains new mutations and data from other three databases: AB-Bind [37], PROXiMATE [38], and dbMPIKT [39]. There are 7085 elements including single- and multi-point mutations in SKEMPI 2.0. 4169 variants in 319 different protein complexes are filtered as single-point mutations are used for TopNetTree model training. Moreover, SARS-CoV-2 related datasets are also included to improve the prediction accuracy after a label transformation. They are all deep mutation enrichment ratio data, mutational scanning data of ACE2 binding to the receptor-binding domain (RBD) of the S protein (including 2223 training samples) [40], mutational scanning data of RBD binding to ACE2 (including 3783 and 1539 training samples, respectively) [41,42], and mutational scanning data of RBD binding to CTC-445.2 and of CTC-445.2 binding to the RBD (including 1539 and 2831 training samples, respectively) [42]. The validation results for this SARS-CoV-2 TopNetTree model on SARS-CoV-2 related test set can be found in the literature [22].
3.3.2. Topology-based feature generation of PPIs
Persistent homology, a branch of algebraic topology, is a powerful method for simplifying the structural complexity of macromolecules [[33], [34], [35]]. To construct topological data analysis on protein-protein interactions, we first preset the constructions for a PPI complex into various subsets.
-
1.
: atoms of the mutation sites.
-
2.
: atoms in the neighborhood of the mutation site within a cut-off distance r.
-
3.
: antibody atoms within r of the binding site.
-
4.
: antigen atoms within r of the binding site.
-
5.
: atoms in the system that has atoms of element type E. The distance matrix is specially designed such that it excludes the interactions between the atoms form the same set. For interactions between atoms ai and a j in set and/or set ℬ, the modified distance is defined as
| (3) |
where D e(a i, a j) is the Euclidian distance between a i and a j.
In algebraic topology, different molecular atoms can be constructed as points presented by v 0, v 1, v 2, …, v k as k + 1 affinely independent points in simplicial complex. A simplicial complex is a finite collection of sets of points K = {σ i}, and σ i are called linear combinations of these points in ℝn (n ≥ k). To construct a simplicial complex, the Vietoris-Rips (VR) complex and alpha complex, which are widely used for point clouds, are applied in this model [34]. The boundary operator for a k-simplex would transfer a k-simplex to a k − 1-simplex. Consequently, the algebraic construction to connect a sequence of complexes by boundary maps is called a chain complex
and the kth homology group is the quotient group defined by
| (4) |
Then the Betti numbers are defined by the ranks of kth homology group H k which counts k-dimensional invariants, especially, β 0 = rank (H 0) reflects the number of connected components, β 1 = rank (H 1) reflects the number of loops, and β 2 = rank (H 2) reveals the number of voids or cavities. Together, the set of Betti numbers {β 0, β 1, β 2, ⋯} indicates the intrinsic topological property of a system.
Persistent homology is devised to track the multiscale topological information over different scales along a filtration [34] and is significantly important for constructing feature vectors for the machine learning method. Features generated by binned barcode vectorization can reflect the strength of atom bonds, van der Waals interactions, and can be easily incorporated into a CNN, which captures and discriminates local patterns. Another method of vectorization is to get the statistics of bar lengths, birth values, and death values, such as sum, maximum, minimum, mean, and standard derivation. This method is applied to vectorize Betti-1 (H 1) and Betti-2 (H 2) barcodes obtained from alpha complex filtration based on the fact that higher-dimensional barcodes are sparser than H 0 barcodes.
3.3.3. Machine learning models
It is very challenging to predict binding affinity changes following mutation for PPIs due to the complex dataset and 3D structures. A hybrid machine learning algorithm that integrates a CNN and GBT is designed to overcome difficulties, such that partial topologically simplified descriptions are converted into concise features by the CNN module and a GBT module is trained on the whole feature set for a robust predictor with effective control of overfitting [21]. The gradient boosting tree (GBT) method produces a prediction model as an ensemble method which is a class of machine learning algorithms. It builds a popular module for regression and classification problems from weak learners. By the assumption that the individual learners are likely to make different mistakes, the method using a summation of the weak learners to eliminate the overall error. Furthermore, a decision tree is added to the ensemble depending on the current prediction error on the training dataset. Therefore, this method (a topology-based GBT or TopGBT) is relatively robust against hyperparameter tuning and overfitting, especially for a moderate number of features. The GBT is shown for its robustness against overfitting, good performance for moderately small data sizes, and model interpretability. The current work uses the package provided by scikit-learn (v 0.23.0) [43]. A supervised CNN model with the PPI ΔΔG as labels is trained for extracting high-level features from H 0 barcodes. Once the model is set up, the flatten layer neural outputs of CNN are feed into a GBT model to rank their importance. Based on the importance, an ordered subset of CNN-trained features is combined with features constructed from high-dimensional topological barcodes, H 1 and H 2 into the final GBT model.
4. Conclusion
Understanding the evolution trend of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and estimating its threats to the existing vaccines and antibody drugs are of paramount importance to the current battle against coronavirus disease 2019 (COVID-19). To this end, we carry out a unique analysis of mutations on the spike (S) protein receptor-binding domain (RBD). Our study is based on comprehensive 506,768 SARS-CoV-2 genome isolates recorded on the Mutation Tracker (https://users.math.msu.edu/users/weig/SARS-CoV-2_Mutation_Tracker.html). There are 6945 unique single mutations and 2,194,305 non-unique mutations on the S protein gene. Therefore, an average genome sample has 2.6 mutations on the S protein but new samples have increasingly more mutations. In terms of the protein sequence, 651 non-degenerate mutations occurred on the RBD. However, most of these RBD mutations have a relatively low frequency, leaving 100 most observed mutations that have been detected more than 28 times in the database. We track fast-growing (FG) RBD mutations in 31 pandemic-devastated countries, including the UK, the US, Singapore, Spain, India, Brazil, etc. To avoid random low-frequency mutations, we pursue this task by analyzing the 10-day growth rate of 100 most observed RBD mutations. We show that four fast-growing mutations N439K, S477N, S477R, and N501T in addition to all known infectious variants containing N501Y, L452R, E484Q, E484K, and K417N, deserve the world's attention.
Additionally, we reveal that essentially all the 100 most observed mutations on the RBD strengthen the RBD binding with the host angiotensin-converting enzyme 2 (ACE2), based on a cutting-edge topology-based neural network tree (TopNetTree) model trained on SARS-CoV-2 experimental datasets [21,22]. More specifically, we found that mutations N501Y, E484K, and K417N in the United Kingdom (UK), South Africa, or Brazil variants, L452R and E484Q in the India, as well as mutations N439K, S477N, S477R, and N501T are all associated with the enhancement of the BFE of the S protein and ACE2, confirming the earlier speculation. This result suggests that SARS-CoV-2 has evolved into more infectious strains due to the wide-spread transmission.
Finally, the early finding shows that more 70% mutations would weaken the efficacy of known antibodies [22]. We report that rapidly growing mutations S494P, Q493L, K417N, F486L, F490S, R403K, E484K, K417T, L452R, E484Q, A475S, and F490L are more likely to disrupt existing vaccines and many antibody drugs. While mutations Q493R, R408I, Q493H, P384S, and N501T can also be disruptive, but mutations N439K, V367F, and S477R are not as disruptive as other rapidly growing ones. Note that L452R in the California variant B.1.427 is as infectious as N501Y and as disruptive as E484K. We have predicted vaccine escape mutations that are not only fast-growing but also can disrupt many existing vaccines. We have also identified vaccine weakening mutations as fast-growing RBD mutations that will weaken the binding between the S protein and many existing antibodies. A list of vaccine escape and vaccine weakening RBD mutations is predicted. We unveil that regulated by host gene editing, viral proofreading, random genetic drift, and natural selection, the mutations on the S protein RBD tend to disrupt the existing antibodies and vaccines and increase the transmission and infectivity of SARS-CoV-2.
Data and model availability
The SARS-CoV-2 SNP data in the world is available at Mutation Tracker. The information of 106 antibodies with their corresponding PDB IDs can be found in the Section S2 of the Supporting information. The SARS-CoV-2 S protein RBD SNP data in 31 countries can be downloaded from the Supplementary data. The TopNetTree model is available at TopNetTree. The related training datasets are described in Section 3.3.3.
Acknowledgment
This work was supported in part by NIH grant GM126189, NSF grants DMS-2052983, DMS-1761320, and IIS-1900473, NASA grant 80NSSC21M0023 , Michigan Economic Development Corporation, George Mason University award PD45722, Bristol-Myers Squibb 65109, and Pfizer. The authors thank The IBM TJ Watson Research Center, The COVID-19 High Performance Computing Consortium, NVIDIA, and MSU HPCC for computational assistance. RW thanks Dr. Changchuan Yin for useful discussion.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygeno.2021.05.006.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Michel Christian Jean, Mayer Claudine, Poch Olivier, Thompson Julie Dawn. 2020. Characterization of accessory genes in coronavirus genomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helmy Yosra A., Fawzy Mohamed, Elaswad Ahmed, Sobieh Ahmed, Kenney Scott P., Shehata Awad A. The covid-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J. Clin. Med. 2020;9(4):1225. doi: 10.3390/jcm9041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naqvi Ahmad Abu Turab, Fatima Kisa, Mohammad Taj, Fatima Urooj, Singh Indrakant K., Singh Archana, Atif Shaikh Muhammad, Hariprasad Gururao, Hasan Gulam Mustafa, Hassan Md Imtaiyaz. Insights into sars-cov-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta (BBA) Mol. Bas. Dis. 2020:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jingfang Mu, Fang Yaohui, Yang Qi, Shu Ting, Wang An, Huang Muhan, Liang Jin, Deng Fei, Yang Qiu, Zhou Xi. Sars-cov-2 n protein antagonizes type i interferon signaling by suppressing phosphorylation and nuclear translocation of stat1 and stat2. Cell Discov. 2020;6(1):1–4. doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann Markus, Kleine-Weber Hannah, Schroeder Simon, Krüger Nadine, Herrler Tanja, Erichsen Sandra, Schiergens Tobias S., Herrler Georg, Wu Nai-Huei, Nitsche Andreas, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sevajol Marion, Subissi Lorenzo, Decroly Etienne, Canard Bruno, Imbert Isabelle. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferron François, Subissi Lorenzo, De Morais Ana Theresa Silveira, Le Nhung Thi Tuyet, Sevajol Marion, Gluais Laure, Decroly Etienne, Vonrhein Clemens, Bricogne Gérard, Canard Bruno, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. 2018;115(2) doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Rui, Hozumi Yuta, Zheng Yong-Hui, Yin Changchuan, Wei Guo-Wei. Host immune response driving SARS-CoV-2 evolution. Viruses. 2020;12(10):1095. doi: 10.3390/v12101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanjuán Rafael, Domingo-Calap Pilar. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016;73(23):4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grubaugh Nathan D., Hanage William P., Rasmussen Angela L. Making sense of mutation: what D614G means for the COVID-19 pandemic remains unclear. Cell. 2020;182(4):794–795. doi: 10.1016/j.cell.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Wendong, Shi Zhengli, Yu Meng, Ren Wuze, Smith Craig, Epstein Jonathan H., Wang Hanzhong, Crameri Gary, Hu Zhihong, Zhang Huajun, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 12.Xiu-Xia Qu, Hao Pei, Song Xi-Jun, Jiang Si-Ming, Liu Yan-Xia, Wang Pei-Gang, Rao Xi, Song Huai-Dong, Wang Sheng-Yue, Zuo Yu, et al. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280(33):29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song Huai-Dong, Chang-Chun Tu, Zhang Guo-Wei, Wang Sheng-Yue, Zheng Kui, Lei Lian-Cheng, Chen Qiu-Xia, Gao Yu-Wei, Zhou Hui-Qiong, Xiang Hua, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. 2005;102(7):2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walls Alexandra C., Park Young-Jun, Tortorici M. Alejandra, Wall Abigail, McGuire Andrew T., Veesler David. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Jiahui, Wang Rui, Wang Menglun, Wei Guo-Wei. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Rui, Chen Jiahui, Hozumi Yuta, Yin Changchuan, Wei Guo-Wei. Decoding asymptomatic covid-19 infection and transmission. J. Phys. Chem. Lett. 2020;11(23):10007–10015. doi: 10.1021/acs.jpclett.0c02765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Julian W., Tambyah Paul A., Hui David S.C. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2020;82:E27–E28. doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulenga Mwenda, Ngonda Saasa, Nyambe Sinyange, George Busby, Peter J Chipimo, Jason Hendry, Otridah Kapona, Samuel Yingst, Jonas Z Hines, Peter Minchella, et al. Detection of b. 1.351 sars-cov-2 variant strain—zambia, december 2020. 2021. [DOI] [PMC free article] [PubMed]
- 19.Faria Nuno R., Claro Ingra Morales, Candido Darlan, Franco L.A. Moyses, Andrade Pamela S., Coletti Thais M., Silva Camila A.M., Sales Flavia C., Manuli Erika R., Aguiar Renato S., et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological. 2021 https://www.icpcovid.com/sites/default/files/2021-01/Ep%20102-1%20Genomic%20characterisation%20of%20an%20emergent%20SARS-CoV-2%20lineage%20in%20Manaus%20Genomic%20Epidemiology%20-%20Virological.pdf [Google Scholar]
- 20.Cherian Sarah, Potdar Varsha, Jadhav Santosh, Yadav Pragya, Gupta Nivedita, Das Mousmi, Das Soumitra, Agarwal Anurag, Singh Sujeet, Abraham Priya, et al. Convergent evolution of SARS-CoV-2 spike mutations, L452rR, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv. 2021 doi: 10.1101/2021.04.22.440932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Menglun, Cang Zixuan, Wei Guo-Wei. A topology-based network tree for the prediction of protein–protein binding affinity changes following mutation. Nat. Machine Intellig. 2020;2(2):116–123. doi: 10.1038/s42256-020-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Jiahui, Gao Kaifu, Wang Rui, Wei Guowei. Prediction and mitigation of mutation threats to COVID-19 vaccines and antibody therapies. Chem. Sci. 2021 doi: 10.1039/D1SC01203G. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galloway Summer E., Paul Prabasaj, MacCannell Duncan R., Johansson Michael A., Brooks John T., MacNeil Adam, Slayton Rachel B., Tong Suxiang, Silk Benjamin J., Armstrong Gregory L., et al. Emergence of sars-cov-2 b. 1.1. 7 lineage—united states, december 29, 2020–january 12, 2021. Morbid. Mort. Weekly Rep. 2021;70(3):95. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Wenjuan, Davis Brian, Chen Stephanie S., Martinez Jorge Sincuir, Plummer Jasmine T., Vail Eric. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA. 2021;325(13):1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felipe Naveca, Cristiano da Costa, Valdinete Nascimento, Victor Souza, André Corado, Fernanda Nascimento, Ágatha Costa, Débora Duarte, George Silva, Matilde Meja, et al. Sars-cov-2 reinfection by the new variant of concern (voc) p. 1 in amazonas, brazil. virological. org. Preprint available at: https://virological.org/t/sars-cov-2-reinfection-by-thenew-variant-of-concern-voc-p-1-in-amazonas-brazil/596. Available at: https://virological.org/t/sars-cov-2-reinfection-by-the-new-variant-of-concern-voc-p-1-in-amazonas-brazil/596, 2021.
- 26.Weisblum Yiska, Schmidt Fabian, Zhang Fengwen, DaSilva Justin, Poston Daniel, Lorenzi Julio C.C., Muecksch Frauke, Rutkowska Magdalena, Hoffmann Hans-Heinrich, Michailidis Eleftherios, et al. Escape from neutralizing antibodies by sars-cov-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paola Cristina Resende, João Felipe Bezerra, Romero Henrique Teixeira de Vasconcelos, Ighor Arantes, Luciana Appolinario, Ana Carolina Mendonça, Anna Carolina Paixao, Ana Carolina Duarte Rodrigues, Thauane Silva, Alice Sampaio Rocha, et al. Spike e484k mutation in the first sars-cov-2 reinfection case confirmed in Brazil, 2020. January, 10:2021, 2021.
- 28.Wu Fan, Zhao Su, Yu Bin, Chen Yan-Mei, Wang Wen, Song Zhi-Gang, Hu Yi, Tao Zhao-Wu, Tian Jun-Hua, Pei Yuan-Yuan, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shu Yuelong, McCauley John. GISAID: global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sievers Fabian, Higgins Desmond G. Multiple Sequence Alignment Methods. Springer; 2014. Clustal omega, accurate alignment of very large numbers of sequences; pp. 105–116. [DOI] [PubMed] [Google Scholar]
- 31.Li Gen, Pahari Swagata, Murthy Adithya Krishna, Liang Siqi, Fragoza Robert, Yu Haiyuan, Alexov Emil. SAAMBE-SEQ: a sequence-based method for predicting mutation effect on protein–protein binding affinity. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodrigues Carlos H.M., Myung Yoochan, Pires Douglas E.V., Ascher David B. mcsm-ppi2: predicting the effects of mutations on protein–protein interactions. Nucleic Acids Res. 2019;47(W1):W338–W344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlsson Gunnar. Topology and data. Bull. Am. Math. Soc. 2009;46(2):255–308. [Google Scholar]
- 34.Edelsbrunner Herbert, Letscher David, Zomorodian Afra. Proceedings 41st Annual Symposium on Foundations of Computer Science. IEEE; 2000. Topological persistence and simplification; pp. 454–463. [Google Scholar]
- 35.Xia Kelin, Wei Guo-Wei. Persistent homology analysis of protein structure, flexibility, and folding. Int. J. Numer. Meth. Biomed. Eng. 2014;30(8):814–844. doi: 10.1002/cnm.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jankauskaitė Justina, Jiménez-Garca Brian, Dapkūnas Justas, Fernández-Recio Juan, Moal Iain H. Skempi 2.0: an updated benchmark of changes in protein–protein binding energy, kinetics and thermodynamics upon mutation. Bioinformatics. 2019;35(3):462–469. doi: 10.1093/bioinformatics/bty635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirin Sarah, Apgar James R., Bennett Eric M., Keating Amy E. AB-Bind: antibody binding mutational database for computational affinity predictions. Protein Sci. 2016;25(2):393–409. doi: 10.1002/pro.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jemimah Sherlyn, Yugandhar K., Gromiha M. Michael. Proximate: a database of mutant protein–protein complex thermodynamics and kinetics. Bioinformatics. 2017;33(17):2787–2788. doi: 10.1093/bioinformatics/btx312. [DOI] [PubMed] [Google Scholar]
- 39.Liu Quanya, Chen Peng, Wang Bing, Zhang Jun, Li Jinyan. dbmpikt: a database of kinetic and thermodynamic mutant protein interactions. BMC Bioinform. 2018;19(1):1–7. doi: 10.1186/s12859-018-2493-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erik Procko. The sequence of human ace2 is suboptimal for binding the s spike protein of sars coronavirus 2. BioRxiv, 2020.
- 41.Starr Tyler N., Greaney Allison J., Hilton Sarah K., Ellis Daniel, Crawford Katharine H.D., Dingens Adam S., Navarro Mary Jane, Bowen John E., Tortorici M. Alejandra, Walls Alexandra C., et al. Deep mutational scanning of sars-cov-2 receptor binding domain reveals constraints on folding and ace2 binding. Cell. 2020;182(5):1295–1310. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linsky Thomas W., Vergara Renan, Codina Nuria, Nelson Jorgen W., Walker Matthew J., Su Wen, Barnes Christopher O., Hsiang Tien-Ying, Esser-Nobis Katharina, Yu Kevin, et al. De novo design of potent and resilient hace2 decoys to neutralize sars-cov-2. Science. 2020;370(6521):1208–1214. doi: 10.1126/science.abe0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedregosa Fabian, Varoquaux Gaël, Gramfort Alexandre, Michel Vincent, Thirion Bertrand, Grisel Olivier, Blondel Mathieu, Prettenhofer Peter, Weiss Ron, Dubourg Vincent, et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2