Abstract
The development of therapeutic targets for COVID-19 relies on understanding the molecular mechanism of pathogenesis. Identifying genes or proteins involved in the infection mechanism is the key to shedding light on the complex molecular mechanisms. The combined effort of many laboratories distributed throughout the world has produced protein and genetic interactions. We integrated available results and obtained a host protein-protein interaction network composed of 1432 human proteins. Next, we performed network centrality analysis to identify critical proteins in the derived network. Finally, we performed a functional enrichment analysis of central proteins. We observed that the identified proteins are primarily associated with several crucial pathways, including cellular process, signaling transduction, neurodegenerative diseases. We focused on the proteins that are involved in human respiratory tract diseases. We highlighted many potential therapeutic targets, including RBX1, HSPA5, ITCH, RAB7A, RAB5A, RAB8A, PSMC5, CAPZB, CANX, IGF2R, and HSPA1A, which are central and also associated with multiple diseases.
Keywords: SARS-CoV-2, COVID-19, Protein-protein interaction, Centrality, Pathways, Disease
1. Introduction
The world is experiencing an unprecedented pandemic due to a massive outbreak of Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) infected viral disease, COVID-19. SARS-CoV-2 is a large enveloped coronavirus (family-Coronaviridae, subfamily-Coronavirinae) with non-segmented, single-stranded, and positive-sense RNA genomes (Wrapp et al., 2020), transmits rapidly through human to human contacts. Although SARS-CoV-2 is similar to other known coronaviruses, i.e., SARS-CoV or MERS-CoV (Perlman and Netland, 2009; De Groot et al., 2013; Weber et al., 2021), however it shows high rate of infection (Liu et al., 2020; Surveillances, 2020; Milano and Cannataro, 2020). Hence, there is a need to understand the disease pathogenesis of SARS-CoV-2 to develop effective therapies and vaccines.
The SARS-CoV-2 virus causes damage in multiple organs as the disease progresses from an asymptomatic phase to a life-threatening disease (Servick, 2020; Cho et al., 2013). Therefore, accurate molecular diagnosis of COVID-19 disease is essential by collecting the proper respiratory tract specimen (Whetton et al., 2020; Ortuso et al., 2021). In this context, the integrated analysis (Antonelli et al., 2019) of various data-sets, including clinical and imaging data, may explain, and hopefully predict, the longitudinal effects of SARS-CoV-2 infection (Tang et al., 2020; Kumar Das et al., 2021). In particular, many independent projects, throughout the world have studied in genomics and proteomics levels (Kumar Das et al., 2021), and then they integrated these outcomes with clinical case studies. These works have produced data about the effect of the infection at a molecular scale, evidencing genes and proteins' role, such as the interactions among viral and human proteins. Interactions between a host and its pathogen are primarily driven by interactions among the host proteins and pathogen proteins, referred to as host-pathogen protein-protein interaction (PPI) network. The SARS-CoV-2 virus-host interactome have been studied computationally, focusing various virulence factors that are influencing SARS-CoV-2 pathogenesis and interacting mechanism (Guzzi et al., 2020; Hoffmann et al., 2021; Messina et al., 2020; Li et al., 2021). Further, many recent works also used host-viral protein-protein interaction network as an input to elucidate potential drug targets or repurposed drug molecules (Beck et al., 2020, Das et al., 2021, Gordon et al., 2020, Zhou et al., 2020). Host-pathogen protein interactions provide important insights into the molecular mechanisms of pathogenecity (Memišević et al., 2015) and for understanding virulence factors influencing SARS-CoV-2 pathogenesis (Li et al., 2021; Thiel et al., 2003).
Protein-Protein Interactions (PPI) are usually modeled and analyzed with graph theory (Guzzi and Roy, 2020; Roy et al., 2019). In this formalism, the interactions are modeled as a graph whose nodes are proteins (or genes), and the edges are the interaction among them. Several studies have found that specific candidate proteins might play a crucial role (Li et al., 2013; Ferrari et al., 2018; Galicia et al., 2020; Lim et al., 2011). Protein-protein interaction networks are an essential ingredient for any systems-level understanding of cellular processes and modeling, and even drug discovery (Tucker et al., 2001; Thakur et al., 2015; Athanasios et al., 2017; Chautard et al., 2009; Nietzsche et al., 2016). The key genes/proteins involved in the different biological pathways can give valuable insight for in-depth characterization of disease progression (Lan et al., 2015; Safari-Alighiarloo et al., 2014; Wang et al., 2018; Jha et al., 2020). It is well accepted that all the viruses have evolved to target proteins that are central and have strong control over the human interactome (Jeong et al., 2001; Bösl et al., 2019; Albert et al., 2000; Navratil et al., 2011; Halehalli and Nagarajaram, 2015). Exploring the predicted interaction networks can suggest new directions for future experimental research and provide cross-species predictions for efficient interaction mapping (Xu and Li, 2006; Safari-Alighiarloo et al., 2014).
This study aimed to identify essential human host proteins based on topology analysis of the PPI of the host proteins targeted by SARS-CoV-2 proteins. We integrated available experimentally validated host-viral PPIs and obtained a set of host proteins that were participated in the integrated network. Network centrality analysis of those candidate host proteins were performed based on their interactions within host-host PPI. We performed enrichment analysis of the central host proteins to shed light on their significance in cellular, signaling, and disease pathways. The enriched host proteins, derived from above step were further analysized for their possible roles in any disease pathogenesis. Finally, a set of host proteins that having role in COVID-19 related diseases were reported as vulnerable proteins. The complete workflow of the current study is illustrated in Fig. 1.
Fig. 1.
The complete work-flow design of the current study.
2. Materials and method
2.1. SARS-CoV-2 interacting (human) host proteins
We used recently reported host proteins that were physically verified using Affinity purification mass spectrometry (AP-MS) for their interactions with SARS-CoV-2 proteins (Gordon et al., 2020; Li et al., 2021; Stukalov et al., 2020; Cannataro and Harrison, 2021). The used host-viral protein interactions are also available in BioGRID1 (Stark et al., 2006). A total of 2489 host-viral interactions (consisting of 1432 unique host proteins interacting with 30 SARS-CoV-2 viral proteins) were obtained. In Fig. 2(a), we provided the number of interacting host proteins for each viral protein. It is to be noted that the majority of the host proteins are targeted by any specific viral protein (Fig. 2(b)).
Fig. 2.
The quantitative information of host-viral interactions. (a) The abundance (percentage) of collected interacting human host protein for different SARS-CoV-2 viral proteins; (b) A host-viral interaction network pattern, where few host proteins interactions can be seen associated with multiple viral proteins although most of interactions are virus protein specific unique (more details can be seen from Supplementary-A).
2.2. Reconstruction of host PPI network
Starting with the host proteins that are interacting with the virus proteins, we reconstructed the host PPI sub-network by querying the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, Version 10.02 ) (Szklarczyk et al., 2010).
The host PPI sub-network is then visualized using Cytoscape,3 a general platform for complex network analysis and visualization (Shannon et al., 2003). We also used Cytoscape to obtain giant components of the host PPI network and calculated various network centrality scores.
2.3. Centrality analysis of host PPI sub-network
In network analysis, indicators of centrality identify the most critical nodes in the network (Bonacich, 1987). The centrality measure are used to characterize each node and edge in the PPI network. The degree measure is the most intuitive for topology analysis of the PPI network. Several other crucial factors that can influence network links are betweenness centrality, closeness centrality, clustering coefficient, topological coefficient, and neighborhood connectivity.
-
(i)
Degree centrality(Dc): The degree centrality (simply degree) of a node n in a network is the number of direct neighbours of n. The densely connected nodes in a PPI network are considered as hub nodes (Han et al., 2004).
-
(ii)
Betweenness centrality(Bc): Betweenness centrality quantifies the number of times a node acts as a bridge along the shortest path between two other nodes (Yoon et al., 2006). The betweenness centrality of a node n is defined as:
| (1) |
where σ st is the total number of shortest paths from node s to node t and σ st(n) is the number of those paths that pass through n.
-
(iii)
Closeness centrality(Cc): Closeness centrality is a way of detecting nodes that are able to spread information very efficiently through the network (Newman, 2005). It can be calculated as:
| (2) |
where L(m, n) is the length of shortest path between node n and m, and m denotes any other nodes that are reachable to node n.
-
(iv)
Average shortest-path length(Sp): The shortest-path length between two nodes (say n and m) is defined as the number of minimum steps required reach node n from m!(Mao and Zhang, 2013). The average shortest path length of node n is the average value of all pair of nodes shortest path from the node n.
-
(v)
Clustering coefficient(Ccoef): Clustering coefficient is a measure of the degree to which nodes in a graph tend to cluster together (Barabasi and Oltvai, 2004). In undirected networks, the clustering coefficient of a node n is defined as:
| (3) |
where k n is the number of neighbors of n and e n is the number of connected pairs between all neighbors of n.
-
(vi)
Topological coefficient(Tc): Topological coefficient is a relative measure for the extent to which a node shares neighbors with other nodes (Goldberg and Roth, 2003). The topological coefficient T c of a node n with k n neighbors is computed as follows:
| (4) |
where J(n, m) is defined for all nodes m that share at least one neighbor with n, and the value J(n, m) is the number of neighbors shared between the nodes n and m, plus one if there is a direct link between n and m.
-
(vii)
Neighborhood connectivity(Nc): Neighborhood connectivity of a node n is defined as the average connectivity of all neighbors of n (Maslov and Sneppen, 2002). The neighborhood connectivity distribution gives the average of the neighborhood connectivities of all nodes n with k neighbors for k = 0, 1, ⋯n-1.
We used NetworkAnalyzer (Shannon et al., 2003) to calculate the above centrality scores. NetworkAnalyzer calculates C c (Closeness centrality) as the reciprocal of the average shortest path length. So, high C c means highly central, and thus low S p.
2.4. Gene ontology and pathway enrichment analysis
We performed enrichment analysis to determine the set of significant genes/proteins in different functional and biological pathways. We used KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa and Goto, 2000) for elucidating pathway enrichment of a host protein and Gene Ontology (GO) for the assessment of protein functions (Ashburner et al., 2000). In this study, we performed GO and KEGG analysis using Enrichr software,4 a web-based suite of gene list enrichment analysis tools (Kuleshov et al., 2016).
2.5. Gene-disease association network
Complex diseases are caused by a group of genes known as disease genes. More often, a gene can participate in various disease conditions (Goh et al., 2007; W. T. C. C. Consortium, 2007). It helps unravel the disease pathogenesis, which helps in disease diagnosis, treatment, and disease prevention. We obtained gene-disease association network from DisGeNET (v7.0) database,5 which contains 1,134,942 gene-disease associations (GDAs), between 21,671 genes and 30,170 disease (Piñero et al., 2020). From this database, we considered curated gene-disease associations only.
3. Results and discussion
Here, we report the outcomes of intermediate steps to reach our objective of isolating key host proteins followed by their significance analysis.
3.1. Deriving PPI network for candidate host proteins
Our list of host proteins consists of 1432 distinct proteins that are targeted by SARS-CoV-2 during COVID-19. We rebuilt the PPI network centered around our candidate proteins using STRING DB. There are 7076 edges in the derived PPI network. We derived PPI by keeping only the interactions whose confidence scores were at least 0.7 (high confidence). The derived PPI network was then analyzed using Cytoscape. We identified the big connected component (also called gain/main component) of the PPI network. After discarding all disconnected components in the PPI network, we considered the giant component of the PPI network with 1111 nodes (approximately 78% of total candidate proteins) and 7043 edges (Fig. 3 ).
Fig. 3.
The gain (connected) component of PPI network obtained from whole PPI network. The network is consisting of 1111 unique nodes (proteins) and 7043 interactions edges (more details can be seen from Supplementary-A).
3.2. Network topology analysis of gain component
We performed the topological analysis of the gain component using NetworkAnalyzer (Shannon et al., 2003). The degree distribution of all the candidate proteins in the gain component showed that the majority of the proteins in the gain component exhibit a higher degree of connectivity (Fig. 4 ). Few proteins with degree (shown within parentheses) more than 50 are CDK1(73), PPP2R1A(65), NOP56(60), POLR2B(60), RAB1A(59), RBX1(58), SKIV2L2(57), NAPA(57), RPS14(56), STX5(54), TGOLN2(54), TCEB1(53), DCTN2(53), TCEB2(52), HSPA9(51), and GNB2L1(50).
Fig. 4.

The degree distribution of all 1111 nodes (proteins) in the gain component of PPI network. The X-axis indicates degree distribution, whereas Y-axis shows relative frequency distributions.
The histogram analysis of all the centrality measures (discussed in Section 2.3) showing non-random distribution (Supplementary-B). Out of seven centrality mesures we considered only those measures which were aligning in their decissions. To select the candidate centrality measures, we computed Pearson's correlation among all centrality scores (Table 1 ). The correlation between degree centrality (D c) and closeness centrality (C c) observed to be the highest (r = 0.759) in comparison to other measures. Although, we observed correlation between D C and neighbourhood centrality (N c) is the third-highest (r = 0.557), but N c and B c showed less correlative (r = 0.1101). Overall, we observed that the correlation scores among the three centrality measures (D C, B c, C c) are quite similar. Therefore, we selected them in subsequent analysis. We identified 373 proteins in these criteria, which are considered highly central proteins (above the median score for all three selected parameters). When we considered all measures, we found only six common proteins (GEMIN4, DDX20, GOLGA3, FKBP15, PMPCA, and AK4) above the median score in each category of centrality measurement, which was the reason for selecting three centrality measures for our downstream analysis.
Table 1.
Correlation analysis among all centrality parameters computed for 1111 proteins. The correlation score (>0.5) among the three centrality measures (Dc,Bc,Cc) are similar (highlighted in bold).
| Dc | Bc | Ccoef | Tc | Nc | |
|---|---|---|---|---|---|
| Bc | 0.603 | 1 | |||
| Ccoef | 0.209 | −0.168 | 1 | ||
| Tc | −0.32 | −0.283 | 0.451 | 1 | |
| Nc | 0.557 | 0.1101 | 0.346 | −0.139 | 1 |
| Cc | 0.759 | 0.5146 | 0.213 | −0.329 | 0.684 |
3.3. Pathway and gene ontology enrichment analysis of highly central proteins
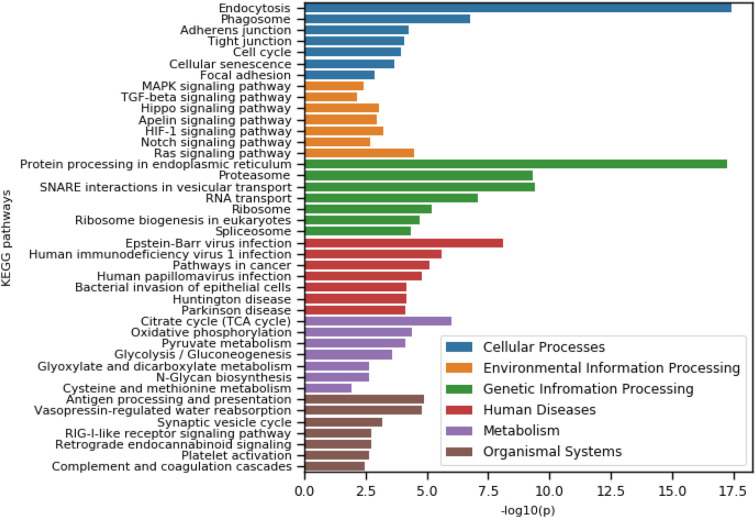
We performed KEGG pathway analysis of selected 373 highly central proteins. We obtained a total of 84 enriched KEGG pathways within the significant level (adj − p < .05). The enriched pathways are involved in Cellular Processes (9), Environmental Information Processing (9), Genetic Information Processing (13), Human Disease (31), Organismal Systems (15), and Metabolism (7). The top seven pathways in each category were shown in Fig. 5 .
Fig. 5.
The top 7 enriched pathways from each category of KEGG pathways. In each category, pathways are shown ordered by − log 10(p) value.
Our current study mainly focused on the proteins that are involved in cellular process, signaling transduction, and human disease (viral and neurodegenerative) pathways. The most affected thirty-one (31) pathways were considered in the context of COVID-19 disease (Seif et al., 2020; Ganesan et al., 2019; Luo et al., 2020; Grimes and Grimes, 2020). There are nine enriched pathways in cellular process (Endocytosis, Phagosome, Adherens junction, Tight junction, Cell cycle, Cellular senescence, Focal adhesion, Regulation of actin cytoskeleton, Lysosome), nine enriched pathways in Environmental Information Processing-signaling transduction (Ras signaling pathway, HIF-1 signaling pathway, Hippo signaling pathway, Apelin signaling pathway, MAPK signaling pathway, TGF-beta signaling pathway, AMPK signaling pathway, NF-kappa B signaling pathway), nine enriched pathways from human viral disease sub-category (Human immunodeficiency virus-1 infection, Human papillomavirus infection, Human cytomegalovirus infection, Hepatitis B, Human T-cell leukemia virus-1 infection, Influenza A, Hepatitis C, Measles) and four enriched pathways from neurodegenerative disease with sub-category (Huntington disease, Parkinson's disease, Alzheimer's disease, Prion diseases). A total of 141 distinct proteins (out of 373) were obtained from these pathways, which were then ranked based on their presence in selected enriched pathways. We found that 79 proteins were associated with our candidate pathways. All these proteins were then further studied for disease-gene association in the next.
We also performed gene set enrichment analysis (Gene ontology). It is observed that the selected genes are primarily involved in Biological processes (Supplementary-C). The top ten terms in each category of gene ontology (Biological processes (BF), Molecular Function (MF), and Cellular Component (CC)) were shown in Fig. 6 that includes neutrophil-mediated immunity (GO:0002446), neutrophil activation involved in immune response (GO:0002283) and viral process (GO:0016032) from BP category; dolichyl-diphosphooligosaccharide-protein glycotransferase activity (GO:0004579), GDP binding (GO:0019003), cadherin binding (GO:0045296) and ATPase activity (GO:0016887) from MF category; and focal adhesion (GO:0005925) from CC category.
Fig. 6.
The top 10 enriched terms in each category of gene ontology (BP-Biological process, MF-Molecular function, CC-Cellular component). In each category, terms are shown ordered by log10(combined score) value.
3.4. Analysis of disease-gene associations
The shortlisted 141 proteins involved in four significant pathways (cellular process, signaling transduction, viral and neurodegenerative)were further screened by looking into their association with COVID-19 related diseases. We particularly focused on three highly influential diseases during COVID-19, namely cardiovascular, respiratory tract (Wu et al., 2020; Clerkin et al., 2020; Konturek et al., 2020; Cannataro et al., 2005) and immune system disease (Melenotte et al., 2020; Chowdhury et al., 2020). To obtain disease-gene association, we used DisGeNET database (Piñero et al., 2020) and selected CURETED source only. We found a total of 64 proteins (out of 141) playing roles in various COVID-19 related diseases such as Asthma, Pneumonitis, Pneumonia, Influenza, Lung diseases, Cardiomyopathies, Coronary, Arteriosclerosis, Coronary Artery Disease, Heart failure, HIV Infections, and others.(Supplementary-D). We compared proteins involved in all three disease categories and individual diseases in each category (Fig. 7 ). A total of 119, 37, and 48 unique diseases (total count-204) and 44, 17, and 24 distinct proteins are associated with the Cardiovascular, Respiratory, and Immune system disease category. Interestingly, we found few proteins that are associated with all three disease categories (AREG, CAV1, IFIH1, PARP1, PLAU, TGFB1, ATM, B2M, DDX58, ENO1, HSPA5, PRKDC, STAT6, TGFBR1, and TGFBR2). The top few proteins which are associated with ten or more diseases are PLAU(59), TGFB1(29), CAV1(17), PARP1(17), TGFBR2(13), ATP2A2(11), AREG(10), FASN(10), IFIH1(10), and ITGB1(10). Finally, we listed a total of 64 unique genes/proteins and their various quantitative scores (degree, disease count (out of 204), disease type count (out of 3), and pathway count (out of 31)) in Table 2 .
Fig. 7.
Comparison of three groups of disease categories (Cardiovascular, Respiratory, Immune system) using venn-diagram. (a) based on number of proteins count in each category; (b) based on number of disease associated proteins (curated from database) among the observed proteins in each category.
Table 2.
The shortlisted 64 host proteins with their degree in host-host PPI , disease count (out of 204), disease type count (out of 3), and pathway count (out of 31). Some of the proteins that are known to be targeted by other viruses were also highlighted.
| Gene/Protein | Degree in PPI network | Pathway count | Disease category | Disease count | Known target virus |
|---|---|---|---|---|---|
| ADAM17 | 10 | 2 | Immune | 1 | |
| ALDOA | 26 | 1 | Cardiovascular | 1 | |
| AP3B1 | 17 | 1 | Respiratory | 2 | |
| AREG | 20 | 2 | Respiratory, Cardiovascular, Immune | 10 | |
| ATM | 29 | 6 | Cardiovascular, Immune | 8 | |
| ATP2A2 | 10 | 1 | Cardiovascular | 11 | |
| ATP6 | 19 | 5 | Cardiovascular | 1 | Human SARS coronavirus, Bovine papillomavirus type 1, Human papillomavirus type 16 |
| ATR | 17 | 5 | Respiratory | 2 | Human adenovirus 5 |
| B2M | 25 | 4 | Cardiovascular, Immune | 4 | Hepatitis C virus genotype 1b (isolate Con1) |
| CANX | 31 | 2 | Cardiovascular | 1 | |
| CAPZB | 34 | 1 | Cardiovascular | 1 | |
| CAV1 | 22 | 2 | Respiratory, Cardiovascular, Immune | 17 | Poliovirus type 1 (strain Sabin) |
| CD44 | 19 | 1 | Immune | 1 | |
| COX2 | 9 | 3 | Cardiovascular | 1 | |
| CRKL | 11 | 5 | Cardiovascular | 4 | |
| DDX58 | 10 | 6 | Respiratory, Cardiovascular | 2 | |
| ENO1 | 13 | 1 | Cardiovascular, Immune | 2 | |
| EPHA2 | 9 | 2 | Cardiovascular | 5 | |
| FASN | 9 | 1 | Cardiovascular | 10 | |
| GAPDH | 23 | 2 | Cardiovascular | 1 | Hepatitis C virus genotype 1b (isolate Con1), Epstein-Barr virus (strain GD1) |
| GLA | 16 | 1 | Cardiovascular | 5 | |
| GNAQ | 14 | 5 | Cardiovascular | 1 | |
| GUSB | 16 | 1 | Immune | 1 | |
| HDAC2 | 12 | 5 | Respiratory | 2 | Human herpesvirus 1 (strain 17), Human papillomavirus type 16, Human papillomavirus type 31 |
| HLA-A | 15 | 8 | Immune | 5 | Epstein-Barr virus (strain GD1), Human papillomavirus type 16 |
| HLA-C | 14 | 8 | Immune | 7 | |
| HMGCR | 11 | 1 | Immune | 4 | |
| HSPA1A | 30 | 5 | Cardiovascular | 1 | Epstein-Barr virus (strain GD1) |
| HSPA5 | 46 | 1 | Respiratory, Cardiovascular | 3 | Epstein-Barr virus (strain GD1) |
| IFIH1 | 10 | 3 | Respiratory, Cardiovascular, Immune | 10 | Sendai virus (strain Fushimi) |
| IGF2R | 31 | 2 | Respiratory | 1 | |
| ITCH | 46 | 1 | Immune | 1 | Epstein-Barr virus (strain B95-8) |
| ITGA6 | 10 | 3 | Immune | 1 | |
| ITGB1 | 29 | 5 | Cardiovascular | 10 | Hepatitis C virus genotype 1b (isolate Con1) |
| JAK2 | 22 | 2 | Cardiovascular | 6 | |
| LDHA | 14 | 1 | Cardiovascular | 4 | |
| LDLR | 20 | 2 | Cardiovascular | 4 | |
| MET | 18 | 4 | Respiratory | 1 | |
| NDUFS2 | 20 | 3 | Cardiovascular | 3 | |
| NF1 | 16 | 2 | Cardiovascular | 3 | |
| NOTCH1 | 23 | 3 | Cardiovascular | 4 | Hepatitis C virus genotype 1b (isolate Con1) |
| NOTCH2 | 11 | 2 | Cardiovascular | 1 | |
| NOTCH3 | 15 | 3 | Cardiovascular | 2 | |
| PARP1 | 14 | 1 | Respiratory, Cardiovascular, Immune | 17 | Human herpesvirus 1 (strain 17) |
| PCNA | 25 | 3 | Immune | 1 | Human herpesvirus 1 (strain 17) |
| PDIA3 | 14 | 3 | Cardiovascular | 1 | |
| PLAU | 24 | 1 | Respiratory, Cardiovascular, Immune | 59 | |
| PPP1CB | 14 | 4 | Cardiovascular | 2 | |
| PRKDC | 15 | 1 | Respiratory, Immune | 3 | Human herpesvirus 1 (strain 17) |
| PSMC5 | 34 | 1 | Immune | 7 | Human adenovirus 5, Human adenovirus 12, Simian virus 40 |
| PSMD6 | 24 | 1 | Immune | 2 | |
| PTPN11 | 22 | 1 | Cardiovascular | 6 | |
| RAB5A | 40 | 3 | Cardiovascular | 1 | |
| RAB7A | 41 | 2 | Cardiovascular | 1 | |
| RAB8A | 40 | 3 | Immune | 1 | |
| RBX1 | 58 | 4 | Immune | 5 | |
| SERPINE1 | 16 | 4 | Cardiovascular | 8 | |
| SLC9A1 | 12 | 2 | Cardiovascular | 7 | |
| SORT1 | 14 | 1 | Cardiovascular | 3 | |
| STAT6 | 11 | 1 | Respiratory, Immune | 4 | |
| TGFB1 | 29 | 7 | Respiratory, Cardiovascular, Immune | 29 | Hepatitis C virus genotype 1b (isolate Con1) |
| TGFBR1 | 17 | 9 | Respiratory, Cardiovascular | 6 | |
| TGFBR2 | 16 | 8 | Respiratory, Cardiovascular | 13 | |
| XPO1 | 25 | 2 | Cardiovascular | 2 |
3.5. Viral proteins targeting key host proteins
We next looked into the source viral-host PPI network (refer Fig. 2) to identify the viral proteins that were targeting shortlisted 64 disease-associated proteins. We found 25 SARS-CoV-2 proteins were interacting 64 key host proteins (Fig. 8 ).Among 25 SARS-CoV-2 viral proteins, eight are accessory proteins (Orf3a, Orf7b, Orf6, Orf7a, Orf7b, Orf8, Orf9b, Orf10), four structural proteins (E,M,N,S) and thirteen non-structural poly-proteins (nsp1, nsp10, nsp12, nsp13, nsp14, nsp2, nsp3, nsp4, nsp5, nsp6, nsp7, nsp8, nsp9). It was observed that several host proteins were interacting with a single viral protein. Very few host proteins were interacting with more than one viral protein. The viral protein, Orf7b interacts with a maximum number of target host proteins, followed by Orf3a and M protein. Further, five host proteins were found to interact with both Orf3a and Orf7b.
Fig. 8.
The interaction network represents the most influential host protein and viral protein. The network is consisting of sixty-four (64) host proteins interacting with twenty-five (25) SARS-CoV-2 viral proteins. The yellow colour node represents the viral proteins in the network, whereas the green one represents the host proteins.
We looked further for any other viruses that are targeting our 64 host proteins. We mined VirusMINT (Chatr-Aryamontri et al., 2009), a virus-host association database, to find the other related viral diseases. We found that the majority of the highlighted host proteins were also targeted by Hepatitis C virus genotype 1b, Poliovirus Type 1, Human herpesvirus 1, Human papillomavirus type 16 & 31, Simian virus 40, Sendai virus, Human adenovirus 5 & 12, Epstein-Barr virus, Human SARS coronavirus. Bovine papillomavirus type 1, and Epstein-Barr virus (Table 2). Further, a few genes (AREG, PLAU, MET, NF1, JAK2, SERPINE1) were observed to be improtant drug targets and associated with various signaling pathways as reported in recent research (Das et al., 2021). These proteins might be highly essential and need to put utmost importance on developing host-directed antiviral therapies for COVID-19.
4. Conclusion
In this study, we analyzed the human (host) protein-protein interaction network targeted by SARS-CoV-2 . Using topology and pathway enrichment analysis of important host PPI network , we reported 64 potential key SARS-CoV-2 interacting host proteins associated with disease pathways, like respiratory, cardiovascular, and immune system diseases. Among them, we highlighted a set of highly central host proteins, such as RBX1, HSPA5, ITCH, RAB7A, RAB5A, RAB8A, PSMC5, CAPZB, CANX, IGF2R, and HSPA1A, which might influence the whole PPI network. Some of the key host proteins are known to target different viruses and may be highly important for therapeutic solution for controlling COVID-19 severeness. We strongly believe that the highlighted key proteins are promising drug targets, which might play a crucial role in inhibiting COVID-19 progression.
Author contributions
Jayanta Kumar Das and Swarup Roy conceived and designed the study. Swarup Roy and Pietro H. Guzzi supervised the study. Jayanta Kumar Das performed the computational analysis and wrote the original draft, which is finally edited by Swarup Roy and Pietro H. Guzzi. All authors approved the final version of the draft.
Declaration of Competing Interest
We would like to declare that we do not have any conflicts of interest in connection with the submitted work.
Acknowledgements
This work has been partially supported by DST-ICPS Data Science program [DST/ICPS/Cluster/Data Science/General] from Govt. of India and VQA PON from Italian Ministry of Economic Development (MISE). The authors thank to Dr. Lawrence M. Nogee and Dr. Mohan Kumar Krishnan for their constructive suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104921.
Appendix A. Supplementary data
Supplementary-A
Supplementary-B
Supplementary-C
Supplementary-D
References
- Albert R., Jeong H., Barabási A.-L. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- Antonelli L., Guarracino M.R., Maddalena L., Sangiovanni M. Integrating imaging and omics data: a review. Biomed. Signal Process. Control. 2019;52:264–280. [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasios A., Charalampos V., Vasileios T., et al. Protein-protein interaction (ppi) network: recent advances in drug discovery. Curr. Drug Metab. 2017;18:5–10. doi: 10.2174/138920021801170119204832. [DOI] [PubMed] [Google Scholar]
- Barabasi A.-L., Oltvai Z.N. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (sars-cov-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacich P. Power and centrality: a family of measures. Am. J. Sociol. 1987;92:1170–1182. [Google Scholar]
- Bösl K., Ianevski A., Than T.T., Andersen P.I., Kuivanen S., Teppor M., Zusinaite E., Dumpis U., Vitkauskiene A., Cox R.J., et al. Common nodes of virus–host interaction revealed through an integrated network analysis. Front. Immunol. 2019;10:2186. doi: 10.3389/fimmu.2019.02186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannataro M, Guzzi PH, Mazza T, Tradigo G, Veltri P. Preprocessing of mass spectrometry proteomics data on the grid. 18th IEEE Symposium on Computer-Based Medical Systems (CBMS’05) 2005:554–559. doi: 10.1109/CBMS.2005.87. [DOI] [Google Scholar]
- Cannataro M., Harrison A. 2021. Bioinformatics Helping to Mitigate the Impact of Covid-19–Editorial. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A., Ceol A., Peluso D., Nardozza A., Panni S., Sacco F., Tinti M., Smolyar A., Castagnoli L., Vidal M., et al. Virusmint: a viral protein interaction database. Nucleic Acids Res. 2009;37:D669–D673. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chautard E., Thierry-Mieg N., Ricard-Blum S. Interaction networks: from protein functions to drug discovery. a review. Pathol. Biol. 2009;57:324–333. doi: 10.1016/j.patbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Cho Y.-R., Mina M., Lu Y., Kwon N., Guzzi P.H. M-finder: uncovering functionally associated proteins from interactome data integrated with go annotations. Proteome Sci. 2013;11:1–12. doi: 10.1186/1477-5956-11-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M.A., Hossain N., Kashem M.A., Shahid M.A., Alam A. Immune response in covid-19: a review. J. Infect. Public Health. 2020;13(11):1619–1629. doi: 10.1016/j.jiph.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkin K.J., Fried J.A., Raikhelkar J., Sayer G., Griffin J.M., Masoumi A., Jain S.S., Burkhoff D., Kumaraiah D., Rabbani L., et al. Covid-19 and cardiovascular disease. Circulation. 2020;141:1648–1655. doi: 10.1161/CIRCULATIONAHA.120.046941. [DOI] [PubMed] [Google Scholar]
- Das Jayanta Kumar, Roy Swarup, Chakraborty Subhadip, et al. A Scheme for Inferring Viral-Host Associations based on Codon Usage Patterns Identifies the Most Affected Signaling Pathways during COVID-19. Journal of Biomedical Informatics. 2021;118:103801. doi: 10.1016/j.jbi.2021.103801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., et al. Commentary: Middle east respiratory syndrome coronavirus (mers-cov): announcement of the coronavirus study group. J. Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Kia D.A., Tomkins J.E., Hardy J., Wood N.W., Lovering R.C., Lewis P.A., Manzoni C. Stratification of candidate genes for parkinson’s disease using weighted protein-protein interaction network analysis. BMC Genomics. 2018;19:1–8. doi: 10.1186/s12864-018-4804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galicia J.C., Guzzi P.H., Giorgi F.M., Khan A.A. Predicting the response of the dental pulp to sars-cov2 infection: a transcriptome-wide effect cross-analysis. Genes Immun. 2020;21:360–363. doi: 10.1038/s41435-020-00112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan H., Balasubramanian V., Iyer M., Venugopal A., Subramaniam M.D., Cho S.-G., Vellingiri B. mtor signalling pathway-a root cause for idiopathic autism? BMB Rep. 2019;52:424. doi: 10.5483/BMBRep.2019.52.7.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K.-I., Cusick M.E., Valle D., Childs B., Vidal M., Barabási A.-L. The human disease network. Proc. Natl. Acad. Sci. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg D.S., Roth F.P. Assessing experimentally derived interactions in a small world. Proc. Natl. Acad. Sci. 2003;100:4372–4376. doi: 10.1073/pnas.0735871100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A sars-cov-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes J.M., Grimes K.V. p38 mapk inhibition: a promising therapeutic approach for covid-19. J. Mol. Cell. Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi P.H., Roy S. Academic Press; 2020. Biological Network Analysis: Trends, Approaches, Graph Theory, and Algorithms. [Google Scholar]
- Guzzi P.H., Mercatelli D., Ceraolo C., Giorgi F.M. Master regulator analysis of the sars-cov-2/human interactome. J. Clin. Med. 2020;9:982. doi: 10.3390/jcm9040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halehalli R.R., Nagarajaram H.A. Molecular principles of human virus protein–protein interactions. Bioinformatics. 2015;31:1025–1033. doi: 10.1093/bioinformatics/btu763. [DOI] [PubMed] [Google Scholar]
- Han J.-D.J., Bertin N., Hao T., Goldberg D.S., Berriz G.F., Zhang L.V., Dupuy D., Walhout A.J., Cusick M.E., Roth F.P., et al. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- Hoffmann H.-H., Sánchez-Rivera F.J., Schneider W.M., Luna J.M., Soto-Feliciano Y.M., Ashbrook A.W., Le Pen J., Leal A.A., Ricardo-Lax I., Michailidis E., et al. Functional interrogation of a sars-cov-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29(2):267–280. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Mason S.P., Barabási A.-L., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jha M., Roy S., Kalita J.K. Prioritizing disease biomarkers using functional module based network analysis: a multilayer consensus driven scheme. Comput. Biol. Med. 2020;126:104023. doi: 10.1016/j.compbiomed.2020.104023. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. Kegg: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konturek P.C., Harsch I., Neurath M., Zopf Y. Covid-19-more than respiratory disease: a gastroenterologist’s perspective. J. Physiol. Pharmacol. 2020;71:765–767. doi: 10.26402/jpp.2020.2.02. [DOI] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Das J., Tradigo G., Veltri P., Guzzi P.H., Roy S. Data science in unveiling covid-19 pathogenesis and diagnosis: evolutionary origin to drug repurposing. Brief. Bioinform. 2021;22:855–872. doi: 10.1093/bib/bbaa420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Wang J., Li M., Peng W., Wu F. Computational approaches for prioritizing candidate disease genes based on ppi networks. Tsinghua Sci. Technol. 2015;20:500–512. [Google Scholar]
- Li W., Chen L., He W., Li W., Qu X., Liang B., Gao Q., Feng C., Jia X., Lv Y., et al. Prioritizing disease candidate proteins in cardiomyopathy-specific protein-protein interaction networks based on “guilt by association” analysis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing sars-cov-2 pathogenesis. Medicine. 2021;2(1):99–112. doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing sars-cov-2 pathogenesis. Medicine. 2021;2:99–112. doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D., Kim N.-K., Park H.-S., Lee S.-H., Cho Y.-M., Oh S.J., Kim T.-H., Kim H. Identification of candidate genes related to bovine marbling using protein-protein interaction networks. Int. J. Biol. Sci. 2011;7:992. doi: 10.7150/ijbs.7.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of covid-19 is higher compared to sars coronavirus. J. Travel Med. 2020;27(2) doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Li Y.-X., Jiang L.-J., Chen Q., Wang T., Ye D.-W. Targeting jak-stat signaling to control cytokine release syndrome in covid-19. Trends Pharmacol. Sci. 2020;41(8):531–543. doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G., Zhang N. Analysis of average shortest-path length of scale-free network. J. Appl. Math. 2013;2013 [Google Scholar]
- Maslov S., Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- Melenotte C., Silvin A., Goubet A.-G., Lahmar I., Dubuisson A., Zumla A., Raoult D., Merad M., Gachot B., Hénon C., et al. Immune responses during covid-19 infection. OncoImmunology. 2020;9:1807836. doi: 10.1080/2162402X.2020.1807836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memišević V., Zavaljevski N., Rajagopala S.V., Kwon K., Pieper R., DeShazer D., Reifman J., Wallqvist A. Mining host-pathogen protein interactions to characterize burkholderia mallei infectivity mechanisms. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina F., Giombini E., Agrati C., Vairo F., Bartoli T.A., Al Moghazi S., Piacentini M., Locatelli F., Kobinger G., Maeurer M., et al. Covid-19: viral–host interactome analyzed by network based-approach model to study pathogenesis of sars-cov-2 infection. J. Transl. Med. 2020;18:1–10. doi: 10.1186/s12967-020-02405-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milano M., Cannataro M. Statistical and network-based analysis of italian covid-19 data: communities detection and temporal evolution. Int. J. Environ. Res. Public Health. 2020;17:4182. doi: 10.3390/ijerph17124182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navratil V., de Chassey B., Combe C.R., Lotteau V. When the human viral infectome and diseasome networks collide: towards a systems biology platform for the aetiology of human diseases. BMC Syst. Biol. 2011;5:13. doi: 10.1186/1752-0509-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.E. A measure of betweenness centrality based on random walks. Soc. Networks. 2005;27:39–54. [Google Scholar]
- Nietzsche M., Landgraf R., Tohge T., Börnke F. A protein–protein interaction network linking the energy-sensor kinase snrk1 to multiple signaling pathways in arabidopsis thaliana. Curr. Plant Biol. 2016;5:36–44. [Google Scholar]
- Ortuso F., Mercatelli D., Guzzi P.H., Giorgi F.M. Structural genetics of circulating variants affecting the sars-cov-2 spike/human ace2 complex. J. Biomol. Struct. Dyn. 2021:1–11. doi: 10.1080/07391102.2021.1886175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-sars: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J., Ramrez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I. The disgenet knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Manners H.N., Elmsallati A., Kalita J.K. In: Encyclopedia of Bioinformatics and Computational Biology. Ranganathan S., Gribskov M., Nakai K., Schönbach C., editors. Vol. 1. Elsevier; 2019. Alignment of protein-protein interaction networks; pp. 997–1015. [DOI] [Google Scholar]
- Safari-Alighiarloo N., Taghizadeh M., Rezaei-Tavirani M., Goliaei B., Peyvandi A.A. Vol. 7. 2014. Protein-protein Interaction Networks (ppi) and Complex Diseases, Gastroenterology and Hepatology From Bed to Bench; p. 17. [PMC free article] [PubMed] [Google Scholar]
- Seif F., Aazami H., Khoshmirsafa M., Kamali M., Mohsenzadegan M., Pornour M., Mansouri D. Jak inhibition as a new treatment strategy for patients with covid-19. Int. Arch. Allergy Immunol. 2020;181:467–475. doi: 10.1159/000508247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servick K. For survivors of severe covid-19, beating the virus is just the beginning. Science. 2020;8 doi: 10.1126/science.abc1486. [DOI] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Breitkreutz B.-J., Reguly T., Boucher L., Breitkreutz A., Tyers M. Biogrid: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukalov A., Girault V., Grass V., Bergant V., Karayel O., Urban C., Haas D.A., Huang Y., Oubraham L., Wang A., et al. Multi-level proteomics reveals host-perturbation strategies of sars-cov-2 and sars-cov. Biorxiv. 2020 doi: 10.1101/2020.06.17.156455. [DOI] [PubMed] [Google Scholar]
- Surveillances V. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (covid-19)—China, 2020. China CDC Week. 2020;2:113–122. [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The string database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2010;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.-W., Schmitz J.E., Persing D.H., Stratton C.W. Laboratory diagnosis of covid-19: current issues and challenges. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur S., Dhiman M., Tell G., Mantha A.K. A review on protein–protein interaction network of ape1/ref-1 and its associated biological functions. Cell Biochem. Funct. 2015;33:101–112. doi: 10.1002/cbf.3100. [DOI] [PubMed] [Google Scholar]
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weißbrich B., Snijder E.J., Rabenau H., Doerr H.W., et al. Mechanisms and enzymes involved in sars coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- Tucker C.L., Gera J.F., Uetz P. Towards an understanding of complex protein networks. Trends Cell Biol. 2001;11:102–106. doi: 10.1016/s0962-8924(00)01902-4. [DOI] [PubMed] [Google Scholar]
- W. T. C. C. Consortium, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang Y., Huang Q., Li C. Integrated bioinformatics analysis reveals key candidate genes and pathways in breast cancer. Mol. Med. Rep. 2018;17:8091–8100. doi: 10.3892/mmr.2018.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G.M., Hong C., Palmer N.P., Avillach P., Murphy S.N., Gutiérrez-Sacristán A., Xia Z., Serret-Larmande A., Neuraz A., Omenn G.S., et al. International comparisons of harmonized laboratory value trajectories to predict severe covid-19: leveraging the 4ce collaborative across 342 hospitals and 6 countries: a retrospective cohort study. medRxiv. 2021 doi: 10.1101/2020.12.16.20247684. [DOI] [Google Scholar]
- Whetton A.D., Preston G.W., Abubeker S., Geifman N. Proteomics and informatics for understanding phases and identifying biomarkers in covid-19 disease. J. Proteome Res. 2020;19:4219–4232. doi: 10.1021/acs.jproteome.0c00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-em structure of the 2019-ncov spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in china. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li Y. Discovering disease-genes by topological features in human protein–protein interaction network. Bioinformatics. 2006;22:2800–2805. doi: 10.1093/bioinformatics/btl467. [DOI] [PubMed] [Google Scholar]
- Yoon J., Blumer A., Lee K. An algorithm for modularity analysis of directed and weighted biological networks based on edge-betweenness centrality. Bioinformatics. 2006;22:3106–3108. doi: 10.1093/bioinformatics/btl533. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-ncov/sars-cov-2. Cell Discov. 2020;6:1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary-A
Supplementary-B
Supplementary-C
Supplementary-D