Abstract
Canavalia rosea (bay bean), distributing in coastal areas or islands in tropical and subtropical regions, is an extremophile halophyte with good adaptability to seawater and drought. Late embryogenesis abundant (LEA) proteins typically accumulate in response to various abiotic stresses, including dehydration, salinity, high temperature, and cold, or during the late stage of seed development. Abscisic acid-, stress-, and ripening-induced (ASR) genes are stress and developmentally regulated plant-specific genes. In this study, we reported the first comprehensive survey of the LEA and ASR gene superfamily in C. rosea. A total of 84 CrLEAs and three CrASRs were identified in C. rosea and classified into nine groups. All CrLEAs and CrASRs harbored the conserved motif for their family proteins. Our results revealed that the CrLEA genes were widely distributed in different chromosomes, and all of the CrLEA/CrASR genes showed wide expression features in different tissues in C. rosea plants. Additionally, we introduced 10 genes from different groups into yeast to assess the functions of the CrLEAs/CrASRs. These results contribute to our understanding of LEA/ASR genes from halophytes and provide robust candidate genes for functional investigations in plant species adapted to extreme environments.
Keywords: late embryogenesis abundant protein; abscisic acid-, stress-, and ripening-induced protein; salinity/alkaline; drought; Canavalia rosea (Sw.) DC
1. Introduction
Abiotic stresses, such as high temperature, high salinity/alkaline, extreme aridity, and cold or freezing, influence plant growth and often cause deficit in cellular water, thereby causing a series of changes, including biochemical alterations in gene expression, osmolytes, and the accumulation of specific proteins involved in the stress response. Late embryogenesis abundant proteins (LEAs) and abscisic acid-, stress-, and ripening-induced proteins (ASRs) are supposed to play crucial roles in the processes of drought resistance or other water-deficit stresses [1,2,3,4,5]. LEA genes have been characterized in plants ranging from algae to higher plants, as well as in invertebrates, fungi, and bacteria [6]. LEA proteins, which are intrinsically disordered proteins, often have the feature of high hydrophilicity and are identified as hydrophilins [7,8]. LEA proteins play protective roles during cell exposure to different abiotic stresses, and these roles are associated with antioxidant, metal ion binding, membrane and protein stabilization, hydration buffering, and DNA and RNA interaction properties [9,10]. Unlike LEA proteins, ASR proteins are plant specific and possess apparent features that are typically absent in Brassicaceae species [11]. Numerous reports have indicated that plant ASRs have important functions in response to water-deficit stress in many plant species, basically acting as transcription factors or chaperones [12,13]. Some previous reports also classified plant ASRs into the LEA superfamily [8,14].
LEAs are mainly composed of many hydrophilic amino acids with a repeated arrangement that forms a highly hydrophilic domain with high heat-stability characteristics [8]. The first plant LEA protein was initially isolated from cotton seeds during the late embryogenesis stage in 1983 [15]. Since then, numerous LEA members have been extensively characterized, and thousands of LEA sequences have been gathered in a dedicated database called LEAPdb [16]. In most plants, the LEA protein family consists of large and highly diverse polypeptides, and different reports have classified them into specific subfamilies based on various categorization features [17]. To date, most studies have widely accepted that based on the conserved motifs, amino acid composition, and phylogenetic relationships, plant LEA proteins can be classified into eight subgroups, including LEA_1, LEA_2, LEA_3, LEA_4, LEA_5, LEA_6, dehydrin (DHN), and seed maturation protein (SMP) [18,19].
LEA proteins are reported to be involved in response to various stress conditions and plant development. Many reports have indicated that the majority of plant LEA mRNAs are induced by various abiotic stresses, such as drought, high salinity, low temperature, oxidation, and osmotic stress [6,18,20]. Some reports have also indicated that in addition to transcriptional regulation, LEA proteins undergo translational or posttranslational modifications, reflecting the complexity of the regulation of these proteins at various levels [21,22]. Basically, LEA proteins are predicted to keep intrinsically disordered proteins in the fully hydrated state, which can then be folded in the water-deficit state and acquire α-helical structures. The ordered LEAs can act as molecular chaperones to bind enzymes, membranes, DNA/RNA, water or ions, and reactive oxygen species (ROS), therefore playing crucial roles in stabilizing proteins/membranes and maintaining cellular environmental homeostasis under abiotic stress [23]. Many studies have characterized several LEA gene families from a few sequenced genomes of plant species, particularly from plant species in specialized habitats, such as halophytes [24], xerophytes [25,26], heat- or freeze-tolerant plants [27,28], or cash crops with strong adversity resistance [24,25,26,27,28,29]. In general, the LEA families that have been identified and studied all contain dozens of members with similar structural and functional plasticity, with the typical evolutionary features and genomic diversification resulting from the adaptation of plants to water-limited environments [20].
The ASR proteins, by contrast, seem to be plant-specific with a small gene family [11,29]. ASRs also have a low molecular weight and exhibit heat-stable and highly hydrophilic characteristics in plants [30]. As observed for plant LEAs, many reports have found that ASRs are also functionally involved in abiotic stress responses, acting as molecular chaperons [31], osmotic adjustment proteins [32], metal-binding protein [33], and antioxidation or detoxification proteins [34,35]. In addition, some ASRs also possess unique functions as transcription factors [12,36]. Due to their highly similar biochemical properties and biological functions, some recent studies have also classified ASRs as LEA proteins in the LEA_7 group [11,37,38]. Numerous reports also indicate that plant ASR genes respond to ABA and abiotic stresses such as water deficit, salt, drought, cold, and osmotic stress [11,13].
Canavalia rosea (Fabaceae) is a wild legume occurring on coastal sand dunes and is mainly distributed in tropical and subtropical regions. In addition to the edible and medicinal characteristics of the mature bean [39], C. rosea also demonstrates excellent growth potential under saline-alkali and high-temperature arid coral reef environments, with great advantages in saline-alkali tolerance and drought resistance [40]. Therefore, as a typical symbiotic nitrogen-fixing legume, C. rosea constitutes a superior wild plant resource and plays important roles in the island greening, sand fixation, and revegetation on tropical and subtropical coral islands and coastal zones. There is no doubt that C. rosea has developed elaborate mechanisms to adapt to the high sanity/alkaline drought stress caused by saltwater intrusion or freshwater deficit in sand dunes or coral reefs, and thus the identification of stress-relevant genes in this species is quite necessary. In the present study, based on the genome sequence of C. rosea, we explored the number of LEA/ASR proteins and their families as well as their structural characterization, gene chromosomal location, and tissue- or habitat- specific expression patterns in C. rosea. The availability of a whole-genome sequence of C. rosea will facilitate genome-wide analysis for identifying the evolutionary relationships of C. rosea LEAs/ASRs with related leguminous species. Additionally, the genome provides a source for mining the genetic resources, including LEA/ASR genes, of C. rosea plants, which have evolved to survive under extreme stress.
2. Results
2.1. Identification of the C. rosea LEA Family and ASR Family
Based on the Hidden Markov model profile search and protein Blast research results, a total of 84 CrLEA and three CrASR members were identified and annotated from the C. rosea genome database. The set of C. sativa LEA proteins included three LEA_1 members, 60 LEA_2 members, six LEA_3 members, two LEA_4 members, two LEA_5 members, two LEA_6 members, five dehydrins, four SMP members, and three ASR members (Table 1 and Table S2). The predicted CrLEAs and CrASRs were named according to their subfamilies and the C. rosea gene nomenclature system (Table 1). The sequence information of all 84 CrLEA and three CrASR genes can be found in Table S2.
Table 1.
Description of CrLEAs identified from Canavalia rosea genome.
| Name | Gene Locus | Length (aa) | Mw (kD) | PI | Instability Index (II) |
Aliphatic Index (AI) |
GRAVY | Content of Disordered aa (%) |
Subcellular Localization * |
|---|---|---|---|---|---|---|---|---|---|
| CrLEA1-1 | 03T007960 | 174 | 17.92 | 9.13 | 11.32 | 35.00 | −1.060 | 83.91 | mito: 10, nucl: 4 |
| CrLEA1-2 | 04T011772 | 192 | 20.00 | 5.38 | 45.87 | 49.53 | −0.725 | 83.33 | nucl: 7, cyto: 3 |
| CrLEA1-3 | 05T014731 | 136 | 14.98 | 9.03 | 54.57 | 60.44 | −0.992 | 94.12 | nucl: 9, mito: 3 |
| CrLEA2-1 | 01T000091 | 294 | 32.86 | 10.00 | 57.75 | 79.18 | −0.266 | 25.51 | chlo: 3, cyto: 3, E.R.: 3 |
| CrLEA2-2 | 01T000357 | 210 | 22.61 | 9.47 | 39.79 | 88.24 | 0.250 | 10.48 | chlo: 5, mito: 5 |
| CrLEA2-3 | 01T000402 | 221 | 24.59 | 9.21 | 37.80 | 103.57 | 0.078 | 18.55 | chlo: 5, cyto: 5 |
| CrLEA2-4 | 01T001888 | 207 | 22.93 | 9.32 | 34.25 | 95.12 | 0.038 | 12.08 | plas: 8, cyto: 4 |
| CrLEA2-5 | 01T003219 | 230 | 25.26 | 9.21 | 34.53 | 111.39 | 0.283 | 16.96 | plas: 4.5, vacu: 4, nucl_plas: 3 |
| CrLEA2-6 | 02T004727 | 259 | 28.57 | 9.88 | 35.46 | 82.39 | −0.232 | 18.53 | chlo: 10, mito: 3 |
| CrLEA2-7 | 02T004857 | 262 | 29.07 | 9.80 | 50.89 | 84.35 | −0.331 | 22.14 | cyto: 7 |
| CrLEA2-8 | 02T005159 | 207 | 23.10 | 9.78 | 33.03 | 100.24 | −0.048 | 13.04 | chlo: 4, plas: 4 |
| CrLEA2-9 | 02T006010 | 192 | 20.74 | 9.28 | 13.81 | 106.98 | 0.272 | 6.25 | vacu: 4, E.R.: 3 |
| CrLEA2-10 | 02T006879 | 190 | 21.16 | 6.83 | 28.11 | 96.42 | −0.112 | 6.32 | nucl: 8, chlo: 3 |
| CrLEA2-11 | 02T006881 | 248 | 27.67 | 10.05 | 33.42 | 94.68 | −0.042 | 14.52 | cyto: 8 |
| CrLEA2-12 | 02T007016 | 233 | 26.24 | 9.41 | 48.70 | 97.38 | −0.033 | 21.46 | cyto: 8 |
| CrLEA2-13 | 02T007186 | 250 | 27.17 | 9.34 | 36.97 | 90.88 | −0.004 | 16.4 | chlo: 8, mito: 4 |
| CrLEA2-14 | 03T008517 | 227 | 25.78 | 9.40 | 36.41 | 89.30 | −0.131 | 7.49 | golg: 4.5, golg_plas: 4.5, plas: 3.5, E.R.: 3 |
| CrLEA2-15 | 03T008518 | 228 | 26.55 | 9.48 | 37.63 | 81.18 | −0.291 | 10.09 | chlo: 10, pero: 3 |
| CrLEA2-16 | 03T008519 | 209 | 23.77 | 9.71 | 36.98 | 97.32 | 0.018 | 7.18 | cyto: 11 |
| CrLEA2-17 | 03T009222 | 197 | 21.48 | 9.76 | 31.71 | 110.71 | 0.356 | 7.61 | chlo: 6, E.R.: 4 |
| CrLEA2-18 | 03T009270 | 235 | 26.49 | 8.98 | 33.86 | 103.19 | 0.329 | 6.81 | cyto: 10 |
| CrLEA2-19 | 03T010271 | 222 | 24.12 | 9.64 | 24.57 | 121.08 | 0.232 | 15.77 | nucl: 6 |
| CrLEA2-20 | 03T010272 | 195 | 21.68 | 6.95 | 23.09 | 100.41 | 0.074 | 6.67 | E.R.: 4, mito: 3, extr: 3 |
| CrLEA2-21 | 03T010452 | 176 | 19.07 | 5.86 | 26.80 | 113.47 | 0.181 | 6.25 | plas: 6, vacu: 5 |
| CrLEA2-22 | 04T011627 | 200 | 23.20 | 9.40 | 35.70 | 104.65 | 0.115 | 0.5 | extr: 3, vacu: 3, E.R.: 3 |
| CrLEA2-23 | 04T011636 | 228 | 26.46 | 9.57 | 34.97 | 81.67 | −0.233 | 11.84 | chlo: 8 |
| CrLEA2-24 | 04T011638 | 210 | 23.82 | 9.63 | 47.73 | 101.52 | 0.099 | 6.19 | cyto: 7 |
| CrLEA2-25 | 04T012994 | 249 | 27.84 | 9.90 | 38.97 | 101.37 | −0.087 | 20.48 | cyto: 7, nucl: 3, E.R.: 3 |
| CrLEA2-26 | 04T013772 | 225 | 25.61 | 9.42 | 41.04 | 76.62 | −0.399 | 11.11 | chlo: 14 |
| CrLEA2-27 | 05T014592 | 322 | 35.41 | 10.11 | 58.09 | 71.71 | −0.333 | 31.68 | chlo: 3, vacu: 3 |
| CrLEA2-28 | 05T016584 | 272 | 29.42 | 9.60 | 42.56 | 84.19 | −0.214 | 32.35 | nucl: 5, chlo: 4, plas: 3 |
| CrLEA2-29 | 05T016770 | 202 | 23.15 | 9.75 | 44.59 | 107.13 | 0.049 | 11.39 | chlo: 5, mito: 3 |
| CrLEA2-30 | 06T017171 | 233 | 26.69 | 10.13 | 39.27 | 102.45 | −0.009 | 19.31 | cyto: 5, plas: 4 |
| CrLEA2-31 | 06T017436 | 320 | 35.70 | 4.80 | 25.33 | 91.97 | −0.437 | 16.25 | cyto: 9 |
| CrLEA2-32 | 06T017480 | 273 | 30.26 | 9.43 | 50.16 | 89.56 | −0.200 | 19.41 | cyto: 4, chlo: 3, plas: 3 |
| CrLEA2-33 | 06T018775 | 257 | 28.95 | 10.17 | 49.92 | 99.69 | −0.162 | 23.35 | nucl: 6, cyto: 5 |
| CrLEA2-34 | 07T020998 | 211 | 24.12 | 9.10 | 30.10 | 95.59 | 0.045 | 6.64 | extr: 5, chlo: 3, E.R.: 3 |
| CrLEA2-35 | 07T020999 | 499 | 57.65 | 9.51 | 31.38 | 89.80 | −0.086 | 2.2 | plas: 7.5, golg_plas: 6, golg: 3.5 |
| CrLEA2-36 | 07T021162 | 247 | 27.83 | 9.15 | 54.28 | 97.41 | −0.057 | 13.77 | plas: 4, golg: 3 |
| CrLEA2-37 | 07T021185 | 308 | 34.10 | 9.58 | 62.03 | 62.27 | −0.514 | 25.32 | chlo: 3, plas: 3, E.R.: 3 |
| CrLEA2-38 | 08T022453 | 223 | 24.65 | 9.50 | 27.00 | 97.89 | 0.155 | 15.25 | chlo: 5, cyto: 3 |
| CrLEA2-39 | 08T022523 | 219 | 24.64 | 9.74 | 53.37 | 106.80 | 0.185 | 5.94 | E.R.: 5 |
| CrLEA2-40 | 08T022549 | 252 | 27.66 | 6.83 | 38.09 | 110.32 | 0.311 | 5.56 | cyto: 7 |
| CrLEA2-41 | 08T023188 | 317 | 35.77 | 9.67 | 57.68 | 82.93 | −0.391 | 36.59 | chlo: 4, nucl: 3, mito: 3 |
| CrLEA2-42 | 09T024657 | 244 | 27.90 | 7.50 | 40.73 | 100.20 | 0.075 | 18.03 | chlo: 6 |
| CrLEA2-43 | 09T024732 | 184 | 19.82 | 9.68 | 21.51 | 121.25 | 0.497 | 4.89 | vacu: 5, extr: 3 |
| CrLEA2-44 | 09T024733 | 186 | 20.56 | 8.99 | 14.50 | 107.37 | 0.354 | 3.76 | extr: 4, vacu: 4 |
| CrLEA2-45 | 09T024798 | 220 | 24.57 | 8.97 | 38.64 | 104.95 | 0.119 | 6.82 | chlo: 4 |
| CrLEA2-46 | 09T025174 | 255 | 27.99 | 10.30 | 36.23 | 87.22 | −0.138 | 18.04 | chlo: 8, mito: 5 |
| CrLEA2-47 | 09T025255 | 308 | 34.13 | 9.93 | 58.73 | 72.37 | −0.347 | 29.55 | chlo: 4, E.R.: 3 |
| CrLEA2-48 | 09T025271 | 228 | 25.83 | 9.63 | 28.59 | 105.39 | 0.153 | 10.96 | vacu: 3 |
| CrLEA2-49 | 10T025889 | 190 | 21.52 | 9.52 | 31.74 | 88.79 | −0.412 | 9.47 | vacu: 5, extr: 3, E.R.: 3 |
| CrLEA2-50 | 10T025950 | 185 | 20.54 | 8.80 | 27.57 | 114.27 | 0.268 | 4.86 | extr: 3, vacu: 3, E.R.: 3 |
| CrLEA2-51 | 10T026128 | 183 | 20.17 | 9.67 | 34.72 | 89.40 | 0.371 | 5.46 | chlo: 7, vacu: 3 |
| CrLEA2-52 | 10T026178 | 192 | 21.44 | 8.41 | 50.06 | 97.40 | 0.068 | 18.23 | cyto: 7.5, cyto_nucl: 6, nucl: 3.5 |
| CrLEA2-53 | 10T026179 | 204 | 22.68 | 10.14 | 39.68 | 94.61 | −0.012 | 10.29 | chlo: 6, E.R.: 4 |
| CrLEA2-54 | 10T026917 | 221 | 24.71 | 9.14 | 41.43 | 97.47 | −0.089 | 18.55 | cyto: 7, cysk: 4 |
| CrLEA2-55 | 11T027970 | 382 | 42.52 | 4.97 | 23.26 | 96.83 | −0.283 | 17.02 | cyto: 8, nucl: 3 |
| CrLEA2-56 | 11T028314 | 258 | 29.48 | 10.11 | 52.91 | 92.91 | −0.151 | 20.93 | chlo: 11 |
| CrLEA2-57 | 11T028577 | 244 | 27.78 | 10.10 | 47.89 | 104.18 | −0.164 | 18.85 | cyto: 5, nucl: 3 |
| CrLEA2-58 | 11T029314 | 264 | 29.23 | 9.27 | 46.36 | 88.98 | −0.062 | 18.56 | cyto: 8, chlo: 4 |
| CrLEA2-59 | 11T029316 | 264 | 29.19 | 9.11 | 47.73 | 90.45 | −0.031 | 18.18 | cyto: 8, chlo: 3 |
| CrLEA2-60 | 11T029318 | 264 | 29.43 | 9.23 | 48.32 | 83.45 | −0.146 | 18.18 | cyto: 7.5, cyto_nucl: 4.5, chlo: 4 |
| CrLEA3-1 | 01T003524 | 106 | 11.73 | 10.09 | 56.38 | 89.25 | −0.325 | 46.23 | chlo: 12 |
| CrLEA3-2 | 02T007058 | 84 | 9.69 | 9.34 | 40.26 | 68.57 | −0.664 | 39.29 | chlo: 5, mito: 4, nucl: 3 |
| CrLEA3-3 | 03T008662 | 97 | 10.57 | 9.91 | 43.60 | 76.39 | −0.300 | 47.42 | cyto: 7, chlo: 6 |
| CrLEA3-4 | 04T012522 | 97 | 10.20 | 9.70 | 27.95 | 77.63 | −0.252 | 46.39 | chlo: 13 |
| CrLEA3-5 | 04T013832 | 101 | 11.26 | 9.25 | 44.98 | 66.63 | −0.655 | 48.51 | mito: 8, chlo: 3 |
| CrLEA3-6 | 05T015908 | 105 | 11.32 | 9.88 | 60.57 | 69.62 | −0.456 | 54.29 | cyto: 8, mito: 5 |
| CrLEA4-1 | 03T008401 | 310 | 34.21 | 7.05 | 28.99 | 49.00 | −1.093 | 65.16 | chlo: 6, nucl: 6 |
| CrLEA4-2 | 04T011481 | 427 | 46.51 | 6.04 | 36.13 | 43.89 | −1.296 | 77.52 | nucl: 12 |
| CrLEA5-1 | 02T006409 | 95 | 10.26 | 5.87 | 48.75 | 41.16 | −1.355 | 87.37 | nucl: 6, cyto: 6 |
| CrLEA5-2 | 03T010121 | 158 | 17.54 | 9.44 | 49.86 | 48.73 | −1.237 | 72.78 | chlo: 9, nucl: 5 |
| CrLEA6-1 | 02T003983 | 82 | 9.20 | 9.20 | 47.60 | 54.76 | −1.194 | 73.17 | nucl: 6, mito: 5 |
| CrLEA6-2 | 05T014654 | 457 | 50.38 | 9.10 | 56.56 | 52.32 | −0.962 | 71.12 | nucl: 13 |
| CrDHN1 | 01T000101 | 80 | 8.58 | 6.49 | 24.98 | 51.12 | −1.068 | 48.75 | nucl: 5 |
| CrDHN2 | 02T003923 | 215 | 24.36 | 5.25 | 57.49 | 51.63 | −1.520 | 79.07 | nucl: 12 |
| CrDHN3 | 02T003929 | 190 | 19.95 | 8.81 | 30.05 | 39.11 | −1.043 | 82.63 | nucl: 13 |
| CrDHN4 | 06T019348 | 220 | 25.01 | 5.45 | 43.12 | 47.41 | −1.636 | 80.91 | nucl: 11 |
| CrDHN5 | 07T021123 | 194 | 19.47 | 6.28 | −5.30 | 16.65 | −1.215 | 72.16 | nucl: 9 |
| CrSMP1 | 04T011095 | 203 | 20.80 | 4.97 | 28.89 | 82.81 | −0.185 | 17.24 | cyto: 7, chlo: 6 |
| CrSMP2 | 04T012849 | 278 | 28.83 | 5.20 | 34.95 | 78.35 | −0.357 | 25.9 | cyto: 6, chlo: 4 |
| CrSMP3 | 04T013711 | 258 | 26.15 | 4.77 | 36.17 | 74.65 | −0.286 | 17.05 | cyto: 5, chlo: 4 |
| CrSMP4 | 09T023499 | 284 | 29.44 | 4.90 | 28.98 | 79.82 | −0.137 | 25 | chlo: 8 |
| CrASR1 | 04T013503 | 235 | 25.92 | 5.79 | 40.95 | 28.00 | −1.449 | 90.64 | nucl: 6, cyto: 3 |
| CrASR2 | 07T020512 | 130 | 14.98 | 6.34 | 42.06 | 47.46 | −1.321 | 80.77 | mito: 5, nucl: 4, cyto: 3 |
| CrASR3 | 07T020519 | 114 | 13.09 | 6.41 | 36.67 | 51.58 | −1.341 | 79.82 | mito: 5, nucl: 4, cyto: 3 |
The physicochemical parameters, including molecular weight (kDa) and pI, of each CrLEA proteins were calculated using the compute pI/Mw tool of ExPASy (http://www.expasy.org/tools/, accessed on 1 March 2021). GRAVY (grand average of hydropathy) values were calculated using the PROTPARAM tool (http://web.expasy.org/protparam/, accessed on 1 March 2021). The contents of disordered amino acids (%) in each CrLEA/ASR were calculated according to the online program PrDOS (Protein DisOrder prediction System, http://prdos.hgc.jp/cgi-bin/top.cgi, accessed on 1 March 2021), with the false positive rate (FP) = 5%, and also the correspondingly disorder profile plots for each protein were shown in Figure S2. Subcellular location prediction was conducted using the WoLF_PSORT (https://www.genscript.com/wolf-psort.html, accessed on 1 March 2021). nucl: Nucleus; plas: Plasma membrane; cyto: Cytoplasm; mito: Mitochondria; ER: Endoplasmic reticulum; pero: Peroxisomal; chlo: Chloroplast; golg: Golgi; vacu: Tonoplast membrane; cysk: Cytoskeleton; extr: Extracellular region; secr: Secretory. * The scores predicted by WoLF PSORT below 3 were ignored. The scores represent the probabilities of the subcellular localization.
The physiochemical properties were assessed using a series of bioinformatics programs. In this large and varied family, the protein lengths of all CrLEAs and CrASRs ranged from 82 to 499 aa, with a molecular weight (MW) ranging from 8.58 (CrDHN1) to 57.65 (CrLEA2-35) kDa (Table 1). The theoretical isoelectric points (PI) of the CrLEAs and CrASRs ranged from 4.77 (CrSMP3) to 10.30 (CrLEA2-46), and most of them (67 members, 75%) were considered to be basic (pI > 7). The instability index ranged between 62.03 (CrLEA2-37) and −5.30 (CrDHN5), and averaged around 39, and a small number of members (38 members, 43%) had a high instability index (II) (>40), which indicated that most of these members might be stable. Most CrLEAs and CrASRs presented calculated grand average of hydropathy (GRAVY) values of less than 0, implying that these proteins are quite hydrophilic. Conversely, the aliphatic index (AI) assessment showed that most members had lower values, indicating that only a small number of these proteins appeared to be lipophilic or hydrophobic. We calculated the contents of disordered amino acids in all CrLEAs and CrASRs according to the prediction of PrDOS, and the results indicated that most of CrLEA_2s and all five CrSMPs had the lower values (<30%) than other CrLEAs or CrASRs (>40% in most members) (Table 1), which is coincided with LEA_2 subfamily has been considered atypical because of containing more hydrophobic amino acids and possessing more defined secondary structure in solution than the other LEA subfamilies [20]. We also summarized the 3D structures of all CrLEAs and CrASRs (Figure S1), and the results showed that all of CrLEA_2s comprised several β-sheets and presented defined secondary structure, while the other CrLEAs and CrASRs, except that two CrLEA_4s contained two α-helices, showed disordered structures, which was in complete accordance with the features of intrinsically disordered proteins (IDPs) [23]. The detailed disorder profile plots of all CrLEAs and CrASRs predicted by PrDOS program were also summarized in Supplementary Materials Figure S2.
The subcellular localization prediction revealed that almost all of the CrLEA and CrASR proteins were present in all subcellular compartments, including the nucleus, cytoplasm, chloroplast, and mitochondria, and even extracellular regions or secretory pathways. Exceptionally, the dehydrin subfamily was more likely to be located in the cell nucleus, and the SMP subfamily was more likely to exist in the cytoplasm. Regarding the CrASR members, they were predicted locating both in the nucleus and in the cytoplasm (Table 1), which is consistent with their possible functions being transcription factors or chaperones, as previous reports in other species [11,12,14].
2.2. Phylogenetic Analysis of CrLEA Proteins and CrASR Proteins
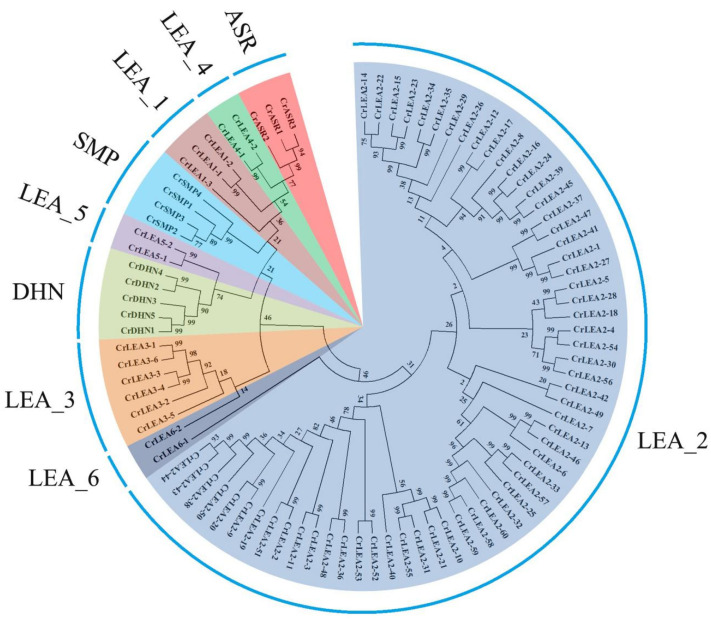
To systematically classify the C. rosea LEA and ASR genes and uncover the evolutionary relationships among this diverse gene family, an unrooted phylogenetic tree of 84 CrLEA and three CrASR members was constructed with MEGA6 using neighbor-joining analysis based on the sequences of the CrLEA and CrASR proteins (Figure 1). The members of this superfamily, including a total of 87 members, clustered together according to their Pfam domains and were classified into two main branches: The LEA_2 subfamily was the largest, with only one distinctive member, CrLEA6-1; the other LEA_1, LEA_3, LEA_4, LEA_5, SMP, DHN, ASR members, and CrLEA6-2 formed another branch. Interestingly, according to the evolutionary relationships, the CrASR subfamily showed a closer relationship with the LEA_4 and LEA_1 subfamilies. There were 29 sister gene pairs in the evolutionary tree, with a bootstrap support value >90% (Figure 1). Among them, nine sub-branches, including CrLEA2-16/CrLEA2-24/CrLEA2-39/CrLEA2-45, CrLEA2-37/CrLEA2-47/CrLEA2-41/CrLEA2-1/CrLEA2-27, CrLEA2-13/CrLEA2-46/CrLEA2-6, CrLEA2-33/CrLEA2-57/CrLEA2-25/CrLEA2-32/CrLEA2-60/CrLEA2-58/CrLEA2-59, CrLEA2-10/CrLEA2-21/CrLEA2-31/CrLEA2-55, CrLEA2-50/CrLEA2-38/CrLEA2-43/CrLEA2-44, CrLEA3-4/CrLEA3-3/CrLEA3-6/CrLEA3-1, CrDHN1/CrDHN5/CrDHN3/CrDHN2/CrDHN4, and CrASR2/CrASR1/CrASR3, all showed bootstrap support values over 90% on each branch (Figure 1).
Figure 1.
Phylogenetic analysis of the LEA and ASR proteins in Canavalia rosea. LEA and ASR subfamilies were distinguished by different colors. The unrooted tree was generated by ClustalW in MEGA6 with the sequences of the 87 C. rosea LEA and ASR proteins.
As plant-specific ASRs have been considered as a small gene family independent from the plant LEA family in some previous reports [11], here we also constructed a single CrASR phylogenetic tree using MEGA6 (Figure S3). The sequence information of all plant ASR proteins is presented in Table S2. Our result indicated that the ASR members did not show strict evolutionary relationships within the same species, or even within the same family. The sequences varied vastly in the same species ASRs, which indicated that this family was not particularly evolutionarily conserved (Figure S3).
2.3. Gene Structures and Conserved Motifs of CrLEAs and CrASRs in C. rosea
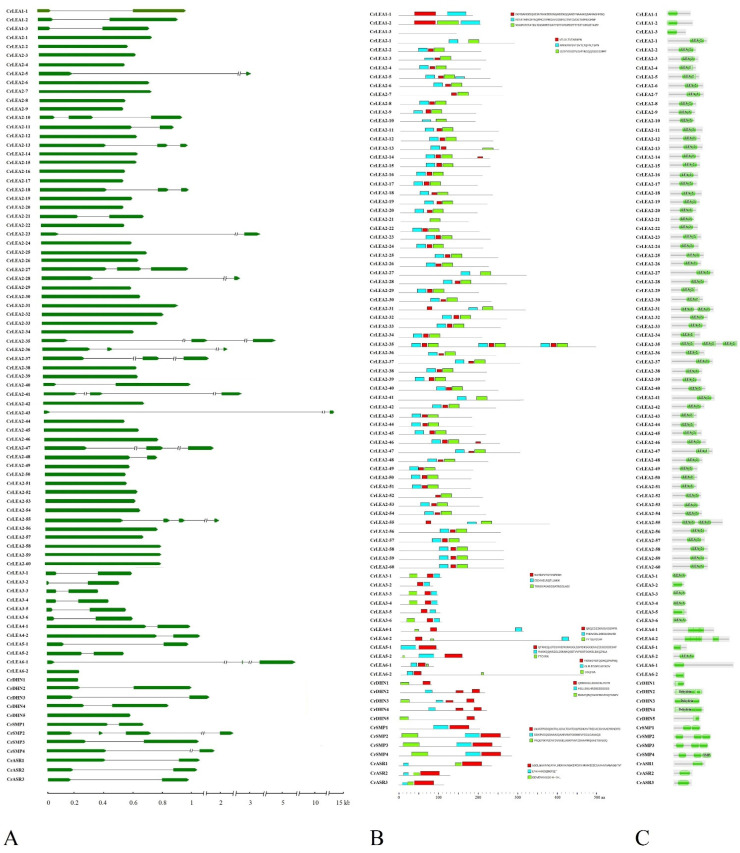
The structure of genes should be of important reference significance for estimating their evolutionary relationships and functional expression patterns, and the conserved motif analyses of proteins are greatly valuable for determining their biochemical functions. To investigate the structural characteristics of CrLEAs and CrASRs, the exon–intron structures (Figure 2A) and conserved motifs (Figure 2B) of 87 member genes or proteins were analyzed. Gene structure analysis showed that the majority of the CrLEAs and CrASRs contained 0 or one intron, and only 12 CrLEA genes (CrLEA2-10, CrLEA2-13, CrLEA2-18, CrLEA2-27, CrLEA2-35, CrLEA2-36, CrLEA2-37, CrLEA2-41, CrLEA2-47, CrLEA2-55, CrLEA6-1, and CrSMP2) possessed two or three introns (Figure 2A). The CrLEA_2 subfamily had the most intronless members, which might indicate that these members probably evolved recently through retrotransposon processes under a certain degree of evolutionary pressure imposed by natural selection. Meanwhile, in each subfamily, some members possessed similar exon–intron structures and intron and exon lengths, which also indicated that these genes possessed closer evolutionary relationships than the other members.
Figure 2.
Exon–intron structures of CrLEA/CrASR genes and conserved motifs in CrLEA/CrASR proteins. (A) The lengths and positions of introns and exons were shown with black lines and thick green arrows for total of CrLEA/CrASR genes ordered by their names. (B) Conserved motifs of C. rosea CrLEA/CrASR proteins in the same subfamily, and these motifs were identified using Multiple EM for Motif Elicitation (MEME) and boxes with different colors represent different motifs. (C) Predications of CrLEA/CrASR proteins by the Pfam database (http://pfam.xfam.org/, accessed on 1 March 2021). The conserved pfam domains were marked with green boxes.
The conserved motifs of the CrLEA and CrASR proteins were analyzed and compared (Figure 2B). Due to the smaller molecular lengths of this protein superfamily, we separately analyzed three independent conserved motifs in each CrLEA and CrASR subfamily. These conserved motifs are listed in Table S3. The results suggested that members of each subfamily possessed similar specific conserved motifs, implying functional specificities of different subfamily proteins. We also compared the conserved motifs with the Pfam domain searched by the Pfam database, and the preliminary results suggested that the conserved motif prediction was consistent with the Pfam domain localization in most CrLEA or CrASR members (Figure 2C).
2.4. Chromosomal Locations and Evolutionary Characterization of CrLEAs and CrASRs
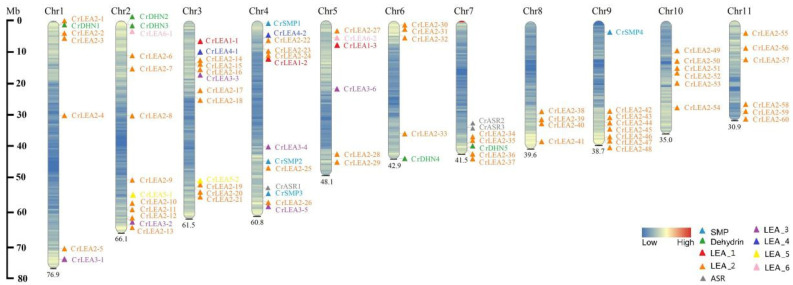
To further investigate the evolutionary relationships and genomic position of the CrLEA/CrASR superfamily, we mapped their chromosomal locations according to their gene locus in Table 1. Most of the CrLEAs/CrASRs were extensively and evenly distributed on the 11 chromosomes. High-density LEA_2 gene clusters were identified in certain chromosomal regions, including chromosomes 2, 3, 7, 8, 9, 10, and 11 (Figure 3), which indicated that these cluster-chromosomal locations of the CrLEA_2 genes may be the result of gene duplication.
Figure 3.
Chromosomal distribution of CrLEA/CrASR genes in C. rosea. Different colors showed different subfamilies, and the scale of the chromosome is in millions of bases (Mb).
Gene family expansion occurs via three mechanisms: Tandem duplication, segmental duplication, and whole-genome duplication (WGD). Tandem duplication and segmental duplication are essential for the evolution of gene families in order to adapt to varying environmental conditions. The results showed that 19 pairs of segmentally duplicated genes and seven pairs of tandem duplicated genes were identified in 84 C. rosea LEA genes (Table 2), and one tandem duplication gene cluster was found on the end of the 11th chromosome (CrLEA2-58/CrLEA2-59/CrLEA2-60) (Figure 3).
Table 2.
Ka/Ks analysis and duplicated date calculation for CrLEA genes.
| Duplicated Pair | Duplicate Type | Ka | Ks | Ka/Ks | Positive Selection |
|---|---|---|---|---|---|
| CrLEA1-1/CrLEA1-2 | Segmental | 0.391 | 1.260 | 0.310 | No |
| CrLEA2-2/CrLEA2-51 | Segmental | 0.125 | 0.992 | 0.126 | No |
| CrLEA2-4/CrLEA2-54 | Segmental | 0.235 | 0.911 | 0.258 | No |
| CrLEA2-5/CrLEA2-28 | Segmental | 0.267 | 1.221 | 0.219 | No |
| CrLEA2-9/CrLEA2-20 | Segmental | 0.343 | 0.858 | 0.400 | No |
| CrLEA2-10/CrLEA2-21 | Segmental | 0.268 | 0.981 | 0.273 | No |
| CrLEA2-12/CrLEA2-17 | Segmental | 0.215 | 0.704 | 0.305 | No |
| CrLEA2-13/CrLEA2-46 | Segmental | 0.232 | 0.967 | 0.240 | No |
| CrLEA2-15/CrLEA2-23 | Segmental | 0.253 | 0.857 | 0.295 | No |
| CrLEA2-16/CrLEA2-24 | Segmental | 0.105 | 1.042 | 0.101 | No |
| CrLEA2-30/CrLEA2-56 | Segmental | 0.187 | 0.761 | 0.246 | No |
| CrLEA2-31/CrLEA2-55 | Segmental | 0.102 | 0.449 | 0.227 | No |
| CrLEA2-33/CrLEA2-57 | Segmental | 0.157 | 0.564 | 0.278 | No |
| CrLEA2-38/CrLEA2-44 | Segmental | 0.320 | 0.591 | 0.541 | No |
| CrLEA2-39/CrLEA2-45 | Segmental | 0.174 | 0.757 | 0.230 | No |
| CrLEA3-1/CrLEA3-6 | Segmental | 0.278 | 0.692 | 0.402 | No |
| CrLEA3-3/CrLEA3-4 | Segmental | 0.138 | 0.669 | 0.206 | No |
| CrLEA4-1/CrLEA4-2 | Segmental | 0.419 | 1.700 | 0.246 | No |
| CrLEA5-1/CrLEA5-2 | Segmental | 0.086 | 0.702 | 0.123 | No |
| CrLEA2-14/CrLEA2-15 | Tandem | 0.488 | 1.489 | 0.328 | No |
| CrLEA2-19/CrLEA2-20 | Tandem | 0.906 | n/c a | n/c | No |
| CrLEA2-34/CrLEA2-35 | Tandem | 0.141 | 0.384 | 0.367 | No |
| CrLEA2-43/CrLEA2-44 | Tandem | 0.270 | 0.360 | 0.750 | No |
| CrLEA2-52/CrLEA2-53 | Tandem | 0.560 | 0.937 | 0.598 | No |
| CrLEA2-58/CrLEA2-59 | Tandem | 0.003 | 0.020 | 0.150 | No |
| CrLEA2-59/CrLEA2-60 | Tandem | 0.056 | 0.126 | 0.444 | No |
a: Ks cannot be calculated due to saturation of substitutions.
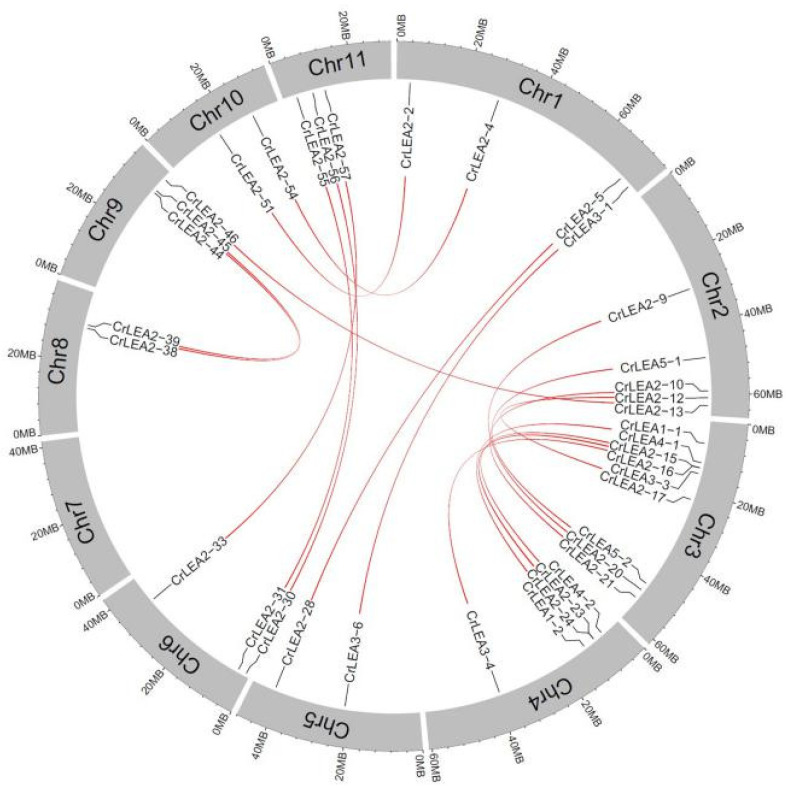
Synonymous (Ks) and nonsynonymous (Ka) values were calculated to explore the selective pressures on duplicated CrLEAs based on all of the nucleotide sequences of CrLEAs. In general, when the ratio is greater than 1, the replicated gene is under positive selection, whereas the replicated gene is neutrally selected when the ratio is equal to 1, and the replicated gene is under purifying selection when the ratio is less than 1. The results revealed that most of the CrLEAs possessed Ka/Ks ratios greater than 0.1. The Ka/Ks for a paralogous gene pair of CrLEAs was 0.101–0.750 with a mean value of ~0.307. These results indicated that they appeared to have undergone extensive purifying selection during evolution and might preferentially conserve function and structure under selective pressure (Table 2). The distribution of segmentally duplicated CrLEA genes in the C. rosea chromosomes is simply illustrated in Figure 4.
Figure 4.
Segmental duplication of CrLEA/CrASR genes in C. rosea genome.
2.5. Cis-Regulatory Element Analyses of CrLEAs and CrASRs
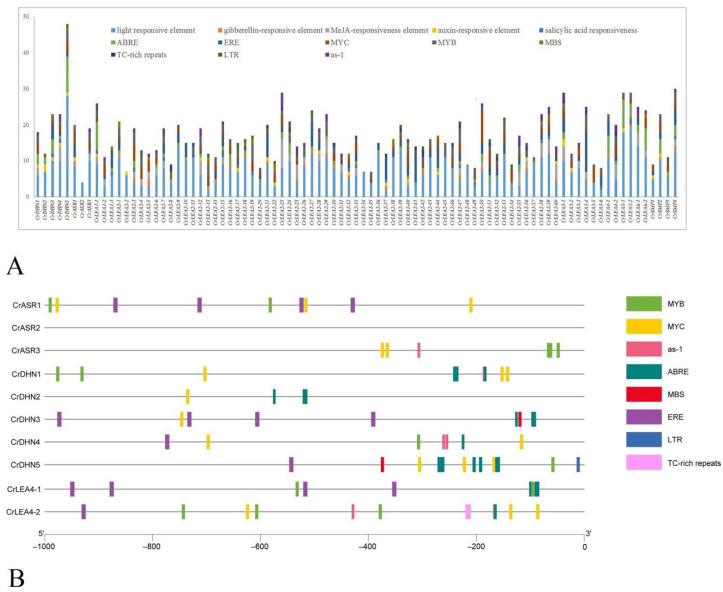
The CrLEA members showed a duplication-prone pattern (Table 2, Figure 4), and in some subfamily members, the CrLEA/CrASR members often possessed similar gene structures and conserved amino acid motifs (Figure 2), which suggested that these members might have functional redundancy or superposition. This is also a process of adaptive evolution in which plant species promote their adaptability and improve their survival under extreme environments or special habitats by altering their DNA. Compared with the numbers or structures of genes, the promoter regions of functional genes showed more variability, which also created a more elaborate and more efficient regulatory mechanism for exercising the biological functions of the genes. To further understand the potential functions and regulatory mechanisms of CrLEAs and CrASRs, especially for exploring the possible roles of this gene family for the adaptation of C. rosea to high salinity and drought, we analyzed the promoter regions of the upstream 1000 bp of 87 putative CrLEAs and CrASRs, which are believed to play important roles in regulating the spatial and temporal expression of genes. In general, a total of 13 cis-regulatory elements were summarized in this study, including light response elements, gibberellin-responsive elements, MeJA-responsive elements, auxin response elements, salicylic acid responsiveness elements, ABRE, ERE, MYC, MYB and MBS, TC-rich repeats, LTR, and as-1. The results are shown in Figure 5A, and the interpretation and localization of these cis-regulatory elements are provided in Table S4.
Figure 5.
Prediction of cis-regulatory elements in the 1000-bp upstream regulatory regions of CrLEA/CrASR genes. (A) Summaries of the twelve cis-regulatory elements in the 87 CrLEA/CrASR promoter regions. (B) Distribution of the eight cis-regulatory elements (MYB, MYC, as-1, ABRE, MBS, ERE, LTR, and TC-rich repeat) in the 10 CrLEA/CrASR genes promoter regions. The scale bar represents 200 bp.
In addition, ASRs and dehydrins have been reported to be crucial in the tolerance to different abiotic stresses, including drought, high salinity/alkaline, or extreme temperatures [11,41]. The LEA_4 subfamily has been suggested to have a close association with the evolution of desiccation tolerance in plants [20]. In this study, we paid special attention to the promoter regions of CrASRs (three members), CrDHNs (five members), and CrLEA_4s (two members). We summarized the abiotic stress-related cis-regulatory elements (including MYB, MYC, as-1, ABRE, MBS, ERE, LTR, and TC-rich repeats) within these 10 CrASR and CrLEA promoter regions (Figure 5B). The categories and numbers of these elements suggested that the mechanisms regulating CrASR and CrLEA expression are involved in stress responses. As these genes had a high number of cis-regulatory elements related to ABA and drought stress responses, they were also selected for further functional analysis to obtain their detailed functions and regulatory mechanisms.
2.6. Expression Profiles of CrLEAs and CrASRs in Different Tissues and Plants Residing in Different Habitats
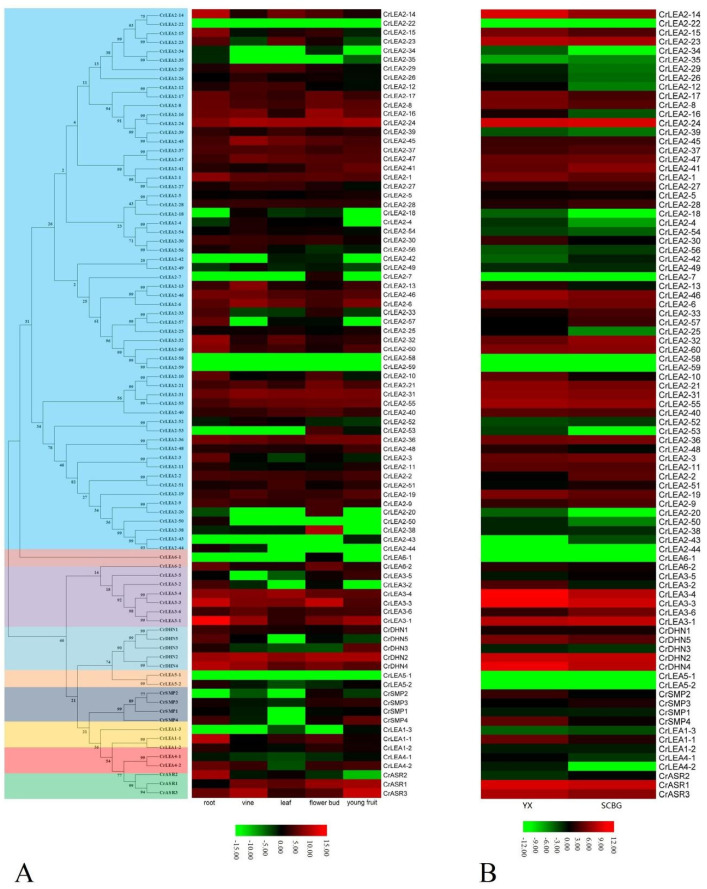
To obtain the expression patterns of the CrLEA and CrASR family members in different tissues, we selected five tissue types, including the roots, vines, young leaves, flowering buds, and young fruits gathered from SCBG, for detailed RNA-Seq analysis. As shown in Figure 6A, there were only four gene members (CrLEA2-22, CrLEA2-58, CrLEA2-59, and CrLEA5-1) whose transcripts could not be detected in all five tissues, which indicated that these genes were probably not expressed or only showed very low transcriptional levels. Furthermore, in combination with the CrLEA gene duplication, we did not find obvious expressive similarity between the duplicated gene pairs (Figure 6A, Table 2), which indicated that these genes underwent genetic evolution mainly related to the regulation of gene expression patterns before their basic functions exhibited significant differentiation. Generally, the RNA-Seq analysis using different tissues showed that CrLEAs and CrASRs have tissue expression specificity, and some showed relatively higher expression levels in all tested tissues, while some were even undetectable (Figure 6A).
Figure 6.
Heatmaps showing (A) the expression levels of the 87 CrLEA/CrASR genes in the root, vine, leaf, flower bud, and young fruit of C. rosea plant and (B) expression differences of the 87 CrLEA/CrASR genes in mature C. rosea leaves planting in South China Botanical Garden (SCBG) and in Yongxing Island (YX). On the far left is the phylogenetic tree showing the different subfamilies by different colored boxes.
We further analyzed the transcriptional differences of the CrLEAs and CrASRs between the mature leaves collected from two habitats: SCBG and YX Island. As we can see from Figure 6B, except for the eight undetectable CrLEAs, including CrLEA2-7, CrLEA2-22, CrLEA2-44, CrLEA2-58, CrLEA2-59, CrLEA5-1, CrLEA5-2, and CrLEA6-1, a great amount of the other members (52 CrLEAs and CrASRs) showed higher expression in the YX sample than in the SCBG sample, except that 27 members were opposites, including CrLEA1-3, CrLEA2-2, CrLEA2-3, CrLEA2-9, CrLEA2-24, CrLEA2-27, CrLEA2-28, CrLEA2-32, CrLEA2-33, CrLEA2-35, CrLEA2-36, CrLEA2-37, CrLEA2-41, CrLEA2-42, CrLEA2-43, CrLEA2-45, CrLEA2-47, CrLEA2-51, CrLEA2-52, CrLEA2-56, CrLEA2-57, CrLEA2-60, CrLEA3-1, CrLEA3-5, CrLEA3-6, CrSMP3, and CrASR2. Given the extreme heat, drought, and salt/alkaline environment in YX Island, this suggested that their biological functions are closely related to the adaptation to coral reef habitats.
2.7. Expression Profiles of CrLEAs and CrASRs in Response to Different Stressors and the ABA Treatment
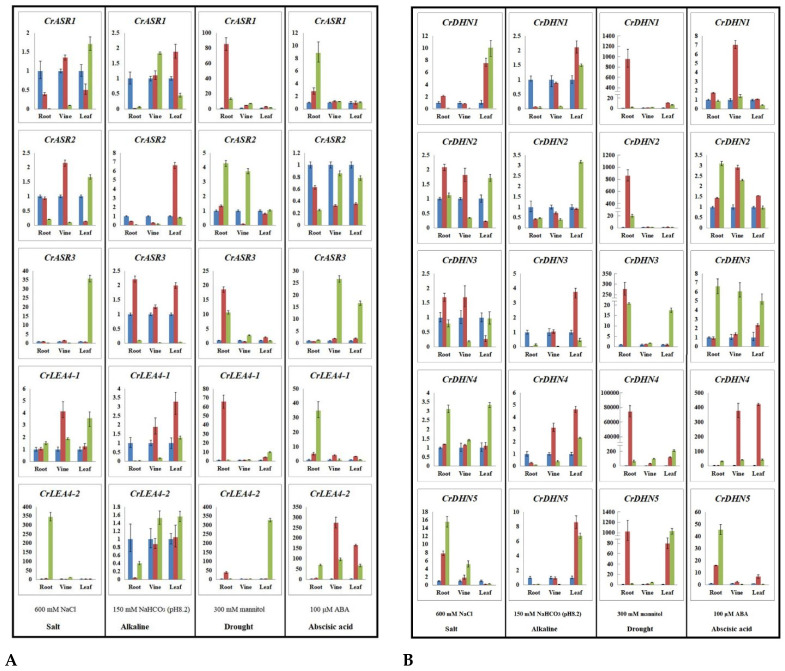
Based on reports on the different LEA numbers involved in abiotic stresses [8,11,20,41,42,43,44,45], with reference to the RNA-Seq results and the promoter region analysis, we further identified 10 genes (including three CrASRs, five CrDHNs, and two CrLEA_4s) for expression analysis in different C. rosea tissues. Our aim was to examine the spatio-temporal patterns of these genes under various abiotic stress conditions and ABA treatment, from which we simulated the stressful conditions of the tropical coral reef in the laboratory, as far as possible. It has been proved that plant ASRs are closely related with water deficit stress and drought tolerance [11,41], and the dehydrins play fundamental roles in plant response and adaptation to salinity and dehydration stresses [8,42]. LEA_4 genes, also known as group 3 [8], have been proved being strongly associated with drought tolerance in basal and angiosperm resurrection plants via ABA signaling pathway [20]. The RNA-Seq analysis showed that nine of these 10 genes displayed higher expression levels in the YX leaf sample than in the SCBG leaf sample (except for CrASR2). The qRT-PCR analysis showed that all 10 genes were induced by different stresses or ABA in different organs to various degrees, while their expression levels were specific and variable (Figure 7). The mannitol stress (simulated drought) and ABA treatment caused significantly higher expression induction than the high salinity and alkaline treatments. Generally, alkaline stress showed a slighter effect than the other three treatments. CrASR1 showed obviously elevated expression in the C. rosea roots under mannitol or ABA challenges, while CrASR3 showed marked expression induction in the C. rosea leaves or vines in response to high salt or ABA treatments. CrASR3 expression was also induced by mannitol in the roots (Figure 7A). Distinctively, CrLEA4-1 and CrLEA4-2 were significantly induced in all C. rosea tissues with the mannitol stress and ABA treatment, and the transcripts of CrLEA4-2 showed a significant increase in the roots of C. rosea under high salt treatment (Figure 7A). Regarding the five dehydrin genes in C. rosea, all of the CrDHNs, without exception, had strong responses to mannitol and showed extremely significant expression induction in the root tissues of the C. rosea plants, while their expression patterns in response to the other three treatments (including high salinity, alkaline, and ABA) showed different degrees of induction or suppression (Figure 7B). Based on the above analysis, the results of the qRT-PCR are basically consistent with the results of the RNA-Seq analysis pertaining to the different habitats, and we propose that these genes may play important roles in the response of C. rosea to abiotic stress and adaptation to the extreme environment in the coral reef.
Figure 7.
Quantitative RT-PCR detection of the expression levels of the 10 CrLEA/CrASR genes responding to different stresses (600 mM NaCl, 150 mM NaHCO3, 300 mM mannitol, or 100 mM ABA) in C. rosea seedling plants. (A) 3 CrASRs and 2 CrLEA_4s. (B) 5 CrDHNs. Relative expression values were calculated using the 2−ΔCt method with housekeeping gene CrEF-α as reference gene.
2.8. Abiotic Stress Tolerance of Yeast Heterologously Expressing CrLEAs and CrASRs
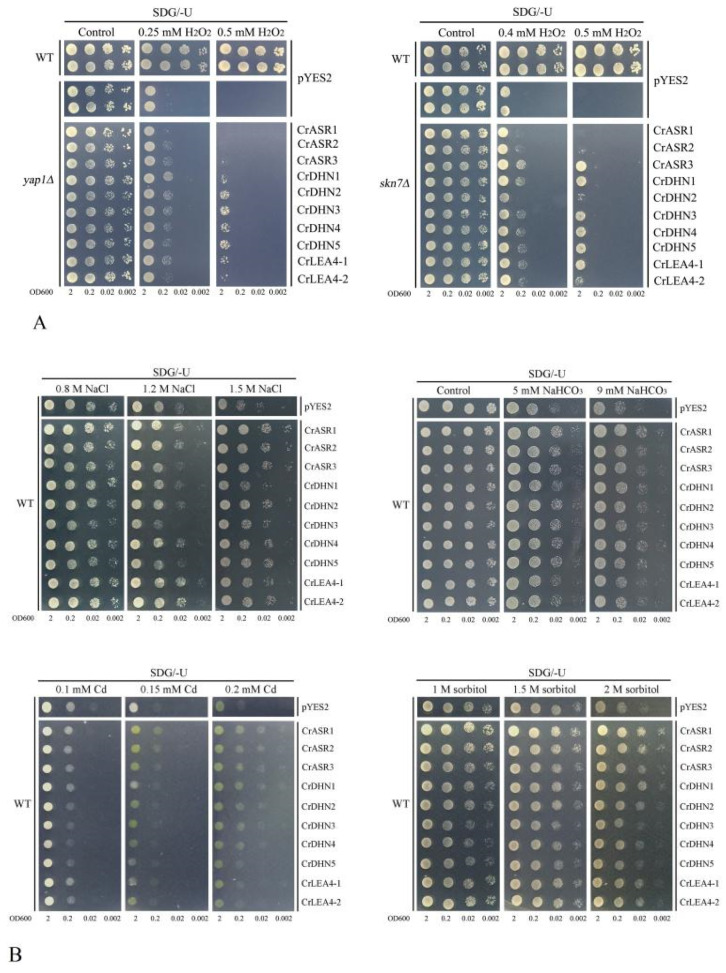
Combined with our transcriptional analyses and promoter regions’ prediction, we could conclude preliminarily that these 10 CrLEAs and CrASRs (three CrASRs, five CrDHNs, and two CrLEA_4s) are closely related with the adaptation to abiotic stresses in C. rosea, especially to high salinity, alkaline, or water deficit, etc. To identify their potential roles in vivo, we performed a series of heterogeneous expression assays of the above 10 genes in a yeast system for functional stress tolerance investigation. For the antioxidation tolerance test, 10 CrLEAs and CrASRs were introduced into two H2O2-sensitive mutant strains yap1Δ and skn7Δ, with the corresponding wild-type (WT) yeast BY4741 and two mutant strains transformed with the empty vector pYES2 as controls. As we can see from Figure 8A, with the exception of CrASR1, the rest of the nine C. rosea genes all showed varying degrees of increased tolerance to H2O2. This indicated that these nine genes all possessed some antioxidation activities (Figure 8A). Due to the highly hydrophilic features and putative antioxidative abilities of CrLEAs and CrASRs, we also tested several other abiotic stress tolerances in the yeast WT strain (Figure 8B, upper). In Figure 8B with respect to salt stress, the majority of the above 10 genes showed elevated salinity tolerance in yeast than empty vector pYES2, while the effects varied. The expression of all 10 genes elevated the alkaline stress tolerance of the yeast. We assessed the cadmium (Cd) tolerance of WT yeast expressing CrLEAs and CrASRs, mainly based on their antioxidative abilities (Figure 8A) as well as the speculations of some reports that ASR or dehydrin proteins possess the ability to bind metals due to their possession of His-rich motifs [33,46,47]. Our results indicated that at least three CrASRs and four CrDHNs (including CrDHN1, CrDHN2, CrDHN3, and CrDHN4) all presented a certain degree of enhanced Cd tolerance in yeast (Figure 8B, lower). Furthermore, most of the 10 CrLEAs and CrASRs could increase the high osmotic stress tolerance of yeast when expressed in the WT strain (Figure 8B, lower). Although these yeast stress tolerance results are preliminary, and a few members even showed no significant effects against some stress challenges, there is still need for further functional identification in plants using transgenic systems.
Figure 8.
The tolerance confirmations of the 3 CrASRs, 5 CrDHNs, and 2 CrLEA_4s heteroexpression in yeast. (A) H2O2 oxidative stress tolerance confirmations in yeast. (B) Salt, alkaline, cadmium, and high osmotic tolerance confirmations in yeast. The stress factors with different concentrations were showed in figures.
2.9. Analysis of the Transcriptional Activation Activity of CrASRs
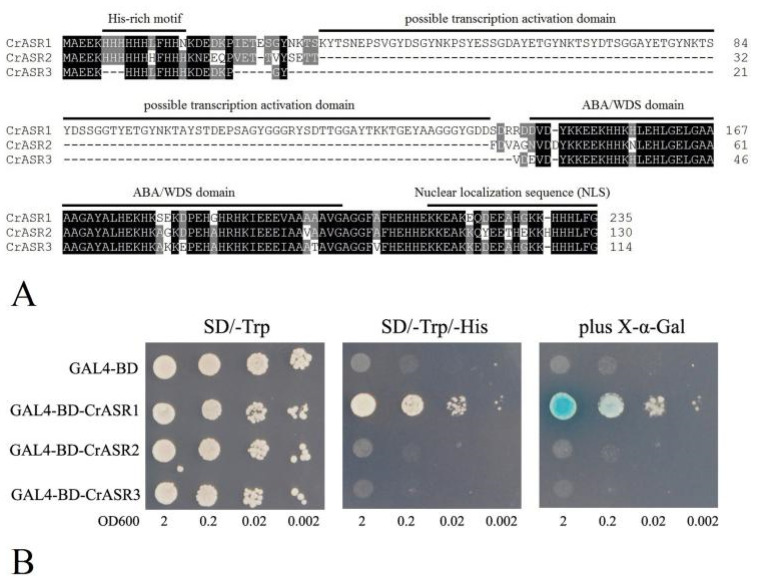
Plant-specific ASRs are typically small gene families comprising several members, and some ASRs have been confirmed to act as transcription factors, with their N-terminals possessing transcription activation ability [48], although in some ASR members, this domain is obviously absent. We first aligned the sequences of three CrASRs, which indicated that only CrASR1 had the possible N-terminal transcription activation domain and could act as a transcription factor (Figure 9A). To further assess our speculation, the complete coding regions of all three CrASR cDNAs were ligated in-frame with the GAL4 DNA binding domain of the pGBKT7 vector and transformed in yeast in this study, with the empty vector GAL4-BD (pGBKT7-BD) as the negative control. The yeast growth on the SD medium lacking tryptophan (SD/-Trp) was normal and even, while the growth on the SD medium lacking tryptophan and histidine (SD/-Trp/-His) was inhibited. Only the yeast cells containing GAL4-BD-CrASR1 exhibited normal growth, and the LacZ staining assay of β-galactosidase activity was positive (Figure 9B), which indicated that CrASR1 showed transcription activation activity and might be a transcription factor.
Figure 9.
CrASRs sequence alignment and GAL4 DNA binding domain-CrASRs fusion analyses for transactivation activity in yeast. (A) Sequence alignment of CrASR1, CrASR2, and CrASR3. The N-terminal His-rich motif, possible transcription activation domain, ABA/WDS domain, and C-terminal nuclear localization sequence were marked with black bold lines. (B) The GAL4 DNA binding domain was fused with 3 CrASRs and transformed into the yeast strain AH109 containing the His3 and LacZ reporter genes. Analysis of β-galactosidase activity of the relative yeast strains on plates. The yeast culture (OD600 to 2) was serially diluted to OD600 values of 0.2, 0.02 and 0.002, and then the 2-μL yeast liquid was spotted onto SD plates and cultured for 2 days at 30 °C.
3. Discussion
Due to the significant roles of LEA and ASR proteins in water deficit stress responses and ROS scavenging abilities, they are believed to act in multiple developmental processes and in response to various stresses, as indicated in a number of previous reports [1,2,3,9]. As C. rosea is a halophyte with the typical features of high salt/alkaline and drought tolerance, it is necessary to systematically investigate the potential role of LEA/ASRs, especially given the lack of studies on LEA/ASR genes in legumes (Fabaceae). As C. rosea generally occurs in tropical and subtropical coastal regions, water shortages are one of the main impacts of its environmental surroundings. It is believed that C. rosea has developed a series of sophisticated mechanisms at multiple levels to cope with stress, including morphological, physiological, and genetic changes and adaptations, ultimately regulating the expression of stress-responsive genes through complex networks. The downstream responsive genes include some water stress-related genes, such as LEAs or ASRs, aquaporins, or ROS-producing and scavenging genes.
LEA protein genes, which were first identified on account of their marked transcript accumulation in embryos for coping with rapid dehydration during seed maturation [15], were later found to be induced in vegetative plant tissues under environmental stress conditions. ASR genes, which encode hydrophilic proteins or transcription factors, were first identified from a tomato cDNA library and participated in response to water-stressed conditions both in stressed leaves and in ripe fruits [41]. LEA proteins constitute a large multigene family that is closely related to the response to abiotic stresses in multiple plant species and that protects cells against water deficit caused by drought and other stresses. While LEAs are not just confined to Plantae, ASRs are exclusive to the plant kingdom (but are absent in Brassicaceae). Both LEAs and ASRs are known to participate in multiple developmental processes and in response to various stresses, mainly water shortage challenges. In the present study, we conducted whole-genome scanning in C. rosea, and a total of 84 CrLEAs and three CrASRs were identified (Table 1). The number of LEA or ASR genes in C. rosea was compared with other plant species [11,20], and the number of different subfamilies of LEA/ASR genes from other legume species was summarized (Table S5). We found that the numbers of LEA/ASR genes in all of these typical diploid legume species were similar, while the soybean (Glycine max) genome contained obviously more (143), probably due to a whole-genome duplication event in the distant past. Among them, the LEA_2 subfamily (PF03168) possessed the largest member number and the greatest variability in all plant species (Figure 1, Table S5), which might imply a diversified functionality of this atypical LEA subfamily [20].
All of these CrLEA/CrASR proteins possess common characteristics, including small molecular weights, an abundance of hydrophilic amino acids, and coding genes that are intronless or contain few introns (Table 1, Figure 2). Most LEA proteins are termed hydrophilins mainly due to their unifying and outstanding feature of high hydrophilicity and a high content of Gly and small amino acids such as Ala and Ser [8]. Previous research indicated that some LEA proteins present a high degree of unordered structure in solution and are considered IDPs [23]. The high hydrophilicity and intrinsically disordered features of LEAs facilitate their protective functions by promoting associations with membrane surfaces or protein partners for protection, and by sequestering H2O, ROS, ions, or other small molecules for alleviating damage or toxicity [23]. Except for some LEA_2 members, most of CrLEA/CrASR proteins showed negative GRAVY scores (<0) (Table 1), indicating that these proteins have strong hydrophilicity and could offer pivotal protective roles under rapid and severe dehydration in plants. Subcellular localization prediction revealed that the CrLEA/CrASR proteins were present in all subcellular compartments, which is consistent with the functional prediction that LEA/ASR proteins in principal groups are ubiquitous within cells and are required in all cellular compartments responding to abiotic stress [49]. We also predicted the 3D structures and calculated the disordered amino acid contents of all CrLEA/CrASR proteins, and the results indicated that except CrLEA_2 and CrSMP subfamilies, the other proteins showed obviously disordered structures (Figures S1 and S2). Furthermore, previous studies have indicated that plant stress-responsive genes without or with few introns could reduce the time required from transcription to translation, therefore providing good adaptation abilities for plants responding to changes in environments and habitats [46]. Combined with the CrLEA/CrASR gene structure and protein motif, we can infer that CrLEA/CrASR proteins are evolutionarily conserved, and their functions have group specificity.
Gene duplication plays a crucial role in the expansion of gene families and is a major way in which genomes can be reshaped, therefore promoting organismal adaptive evolution to the environment [47]. Based on the evolutionary characterization of CrLEAs and CrASRs, we found that most segmental and tandem duplications occurred in the CrLEA_2 subfamily, and similar characteristics have also been reported for LEA genes in other plant species [20], which suggests that the LEA_2 subfamily contains the most diverse LEA members in plants. LEA_2 is considered as an atypical LEA protein because it possesses more hydrophobic amino acids and a more defined secondary structure compared with other LEA subfamily members [20]. Additionally, also we found that this subfamily contained the only tandem duplicate cluster in C. rosea (Table 2), indicating that tandem duplications have contributed significantly to the expansion and diversification of the large LEA_2 family in C. rosea. The other segmental duplications occurring in CrLEA_1, CrLEA_3, CrLEA_4, and CrLEA_5 indicated that these LEA genes might participate in the adaptive evolution of C. rosea to water shortage stresses by increasing their gene numbers.
As LEAs have been confirmed to endow plants with a variety of abiotic and biological stress tolerances, LEA families often have multiple copies of LEA genes in various plant species for that very reason. Correspondingly, even within the same LEA subfamily, our transcriptome data showed that the expression of different members was specific (Figure 6), which is probably derived from the influence of the promoter regions. Our statistical analysis results of the promotors of CrLEAs/CrASRs showed that most of the promoter regions contained cis-regulatory elements, such as ABRE, MYC, MYB, MBS, TC-rich repeats, and several hormone-related responsive elements, suggesting that these genes could be regulated or affected by different stresses (Figure 5A). We also analyzed 10 promoter regions (including three CrASRs, five CrDHNs, and two CrLEA_4s) in detail. We found that the promoter regions showed more variability than the gene coding region, which corresponds with their gene-specific expression pattern (Figure 5B and Figure 6). The contribution of single CrLEA or CrASR genes to the stress tolerance and environmental adaptation of C. rosea needs to be further explored.
An investigation of the natural habitat of C. rosea indicates that this species possesses significant growth potential and is an adaptable pioneer species that can be used for island greening, sand fixation, and the ecological restoration of coral islands and coastal zones in tropical or subtropical regions [40]. We firstly examined the expression of the CrLEA and CrASR genes in the different tissues and developmental periods using RNA-Seq. The expression profiles revealed spatial variations in the expression of CrLEAs or CrASRs in different organs (Figure 6A). Our further habitat-specific RNA-Seq data indicated that most of the CrLEA/CrASR genes had greater expression levels in coastal C. rosea (YX) than in inland C. rosea (SCBG), while some were quite the opposite (Figure 6B). Our results suggested that the differential expression of CrLEA/CrASR genes might be an adaptive mechanism for dealing with intracellular and extracellular water-deficit signals in C. rosea plants, which is associated with different water loss strategies in different habitats and, in the longer term, may also function in the biological adaptability to changes in environmental factors and different habitats.
We also used qRT-PCR to investigate the expression patterns of 10 candidate genes in C. rosea plants under salt, alkaline, high osmotic treatment, and ABA stresses. Detailed expression profiles of these 10 CrLEA/CrASR genes revealed that most were significantly up-regulated after the high osmotic stress (NaCl or mannitol) or ABA treatment, while the alkaline stress showed the least impact on the expression changes of candidate genes (Figure 7). This result is consistent with several previous reports of LEA genes in other plant species, in which their expression was also greatly affected by abiotic stress challenges and hormone treatments [19,48,50]. The relevant roles of ASRs, DHNs, or the LEA_4s subfamily genes during plant stress tolerance have been reported in earlier studies using various techniques [11,20,42]. Here both the RNA-Seq and the qRT-PCR results indicated that these genes might be vital factors influencing the adaptability of C. rosea plants to salt/alkaline and drought stresses in tropical or subtropical coastal regions.
The accumulation of LEA proteins in cells is crucial for the response of organisms to different abiotic stresses. Numerous reports have demonstrated that overexpressing LEA genes in different organisms resulted in improved salinity and dehydration tolerance. For example, Ipomoea pes-caprae is a perennial herbaceous vine plant distributed mainly on sandy beaches or in sunny positions on the roadside in tropical and sub-tropical regions. It exhibits excellent salt tolerance and drought resistance, and the induction of IpLEAs in yeast obviously improved the salt and oxidative stress tolerance of yeast clones [51]. Prior studies revealed that some LEA genes from Dendrobium officinale were able to enhance the cellular tolerance to high temperature and salt stresses of E. coli cells [49]. For ASR genes, our previous research also demonstrated that an I. pes-caprae ASR gene, IpASR, could improve salinity and drought tolerance in transgenic E. coli and Arabidopsis [45]. In summary, LEA and ASR genes are pertinent research subjects in plant stress physiology and are mainly involved in drought or salinity tolerance in plants. Here, we identified and characterized 10 CrLEA/CrASR genes in C. rosea for the first time and performed functional verification in yeast to investigate bioactivities for abiotic stress resistance. Based on our results, nine of the candidate genes increased the yeast cell resistance to H2O2, showing individual differences (Figure 8A). The majority of the 10 tested genes elevated the high salinity, alkaline, and high osmotic stress tolerance of yeast compared with the empty vector control, while some even showed heavy metal Cd tolerance, but not significantly so (Figure 8B). Our results demonstrated that these CrLEA/CrASR genes may play protective roles in cells under water-deficit conditions caused by high salt/alkaline and high osmotic stresses, and they also could improve cell survival via their antioxidant capacities or metal chelating abilities.
Although CrASR1 did not present antioxidant properties in yeast, it is a highly hydrophilic protein with a relatively low GRAVY value score (Table 1). Unlike CrASR2 or CrASR3, CrASR1 has a remarkable property in that it contains a Gly-rich domain in its N-terminus (Figure 9A). According to our previous reports [35,45], it is uncertain whether this type of ASR protein could activate transcription and act as a transcription factor, while our result on the transcriptional activation assay in yeast indicated that CrASR1 did exhibit transcription factor activity, yet CrASR2 and CrASR3 had exactly the opposite effect (Figure 9B). This indicated that CrASR1 could act as a transcriptional regulator together with chaperone-like proteins or hydrophilins, like other LEA/ASR proteins [11,34]. Previous studies have proved that tomato ASR1 is a drought stress-responsive transcription factor, and its consensus ASR1-binding motif was enriched in some specific tomato genomic loci, possibly containing several aquaporin genes [12,52]. A recent study on CaASR1 in another Solanaceae species, Capsicum annuum, also provided evidence that CaASR1 could interact with the transcription factor CabZIP63 and is a positive regulator of the defense response of pepper to the pathogen R. solanacearum, probably by acting as a transcription activator [53]. Our results strongly suggest that CrASR1 showed quite different features from the other two CrASRs or CrLEAs (Figure 8 and Figure 9), and this protein might fulfill an important function in salt/drought tolerance and regulate the environmental adaptation of C. rosea to tropical coastal regions, although more research is needed for further clarification.
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
C. rosea plants growing in Yongxing Island (YX, 16°83′93′′ N, 112°34′00′′ E) and South China Botanical Garden (SCBG, 23°18′76′′ N, 113°37′02′′ E) were used in this study. To analyze the tissue-specific transcriptional patterns of the CrLEA and CrASR genes, the roots, stems, leaves, flowers, and fruits were sampled from C. rosea plants grown in SCBG. To investigate the involvement of the CrLEA and CrASR genes in the adaptation to a high-temperature island climate, coastal saline environment, and seasonal drought environment, adult plant leaves were also gathered from C. rosea growing on both YX Island and SCBG.
The responses of the CrLEA and CrASR genes to different stresses and hormone treatments were investigated. Seedlings of C. rosea germinated from seeds in a soil/vermiculite mixture for 30 d were subjected to treatments. In brief, for high salinity stress, the C. rosea seedlings were removed from the pots and carefully washed with distilled water to remove soil from the roots, following which they were transferred into 600 mM NaCl solution; for salt-alkaline stress, the cleaned C. rosea seedlings were soaked in a 150 mM NaHCO3 (pH 8.2) solution; for the drought treatment, the seedlings were soaked in a 300 mM mannitol solution; and for abscisic acid (ABA) treatment, a freshly prepared working solution of 100 μM exogenous ABA was sprayed onto the leaves of the C. rosea seedlings. The second and/or third mature leaves from the shoot apexes were collected at 0, 2, and 24 h during the stress treatments, and the starting point (0 h) was used as the control. All samples were immediately frozen in liquid nitrogen and stored at −80 °C for subsequent gene expression analysis. Three independent biological replicates were conducted.
4.2. Identification of LEA/ASR Genes in the C. rosea Genome and Phylogenetic Analysis of CrLEA/CrASR Superfamily Proteins
The putative CrLEA and CrASR gene sequences were collected from the genome database of C. rosea. All of the C. rosea proteins were first identified using DIAMOND [54] and InterProscan (https://www.ebi.ac.uk/interpro/search/sequence/, accessed on 1 March 2021) to assess the conserved domains and motifs (e < 1 × 10−5), following which they were annotated using the Pfam database (http://pfam.xfam.org/, accessed on 1 March 2021). The Pfam ID PF03760 (LEA_1), PF03168 (LEA_2), PF03242 (LEA_3), PF02987 (LEA_4), PF00477 (LEA_5), PF10714 (LEA_6), PF00257 (dehydrin, DHN), PF04927 (SMP), and PF02496 (ASR) were used to search the CrLEAs and CrASRs, and the putative sequences of CrLEA and CrASR proteins were identified and submitted to SMART (http://smart.embl-heidelberg.de/, accessed on 1 March 2021) and the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 1 March 2021) to confirm the presence of the LEA or ASR domains. Finally, the selected CrLEAs and CrASRs were named based on their sequence homology with other known plant LEAs or ASRs and the C. rosea genome annotations. An unrooted neighbor-joining phylogenetic tree was created based on multiple protein sequence alignments of all identified CrLEAs and CrASRs from C. rosea using Clustal X 2.0 and MEGA 6 with 1000 bootstrap replicates. The obtained LEA and ASR nucleotide and protein sequences from C. rosea are listed in Table S1. The gene structure analysis for CrLEAs and CrASRs was displayed with GSDS (http://gsds.cbi.pku.edu.cn/, accessed on 1 March 2021).
4.3. Analysis of Protein-Conserved Motifs and Biochemical Features of CrLEAs/ASRs
To investigate the characteristics of the CrLEA and CrASR proteins, the molecular weight (MW), theoretical isoelectric point (pI), and grand average of hydropathy (GRAVY) were predicted using the ProtParam tool (http://web.expasy.org/protparam/, accessed on 1 March 2021). Furthermore, the subcellular localization predictions for these CsLEA proteins were carried out using the WoLF_PSORT tool (http://www.genscript.com/wolf-psort.html, accessed on 1 March 2021). The contents of disordered amino acids (%) in each CrLEA/ASR were calculated according to the online program PrDOS (Protein DisOrder prediction System, http://prdos.hgc.jp/cgi-bin/top.cgi, accessed on 22 April 2021). The deduced amino acid sequences of the CrLEAs and CrASRs were analyzed by the Multiple Expectation Maximization for Motif elicitation (MEME) tool (http://meme-suite.org/index.html, accessed on 1 March 2021) to identify conserved domains and motifs of each subgroup of these proteins. The selection of the maximum number of motifs was set to 3, with a minimum width of 11 and a maximum width of 50 amino acids, and an e-value < 1 × 10−8. The Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index, accessed on 1 March 2021) was used for homology modelling of the three-dimensional (3D) structure of all CrLEA and CrASR proteins.
4.4. Gene Duplication and Collinearity Analysis of CrLEAs
The CrLEA and CrASR genes were mapped onto C. rosea chromosomes according to the positional information of these genes in the C. rosea genome database and were displayed using MapInspect software (http://mapinspect.apponic.com/, accessed on 1 March 2021). Gene segmental duplications were assessed using MCScanX software (http://chibba.pgml.uga.edu/mcscan2/, accessed on 1 March 2021), and tandem duplications were identified manually. The number of synonymous substitutions per synonymous site (Ka), the number of non-synonymous substitutions per non-synonymous site (Ks), and the p-value from Fisher’s exact test of neutrality were calculated using the Nei-Gojobori model with 1,000 bootstrap replicates [55]. A Ka/Ks ratio < 1 indicates purifying selection, a Ka/Ks ratio = 1 indicates neutral selection, and a Ka/Ks ratio > 1 indicates positive selection. The gene segmental duplications of the CrLEAs were visualized using the online shinyCircos software (https://venyao.xyz/shinyCircos/, accessed on 1 March 2021).
4.5. Promoter Sequence Profiling of CrLEAs/ASRs
The promoter regions (1000 bp upstream from the translation start site) of all CrLEA and CrASR genes were retrieved from the genome database of C. rosea for further analysis of the cis-regulatory elements and motifs by querying them through the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 1 March 2021). The stress- and hormone-related cis-regulatory elements included light response elements, gibberellin-responsive elements, methyl jasmonate (MeJA)-responsive elements, auxin response elements, salicylic acid responsiveness elements, ABA response elements (ABRE), ethylene response elements (ERE), MYC transcription factor binding elements (MYC), MYB transcription factor binding elements (MYB and MBS), TC-rich repeats, LTR, and as-1. These elements are believed to be involved in plant responses to dehydration, low temperature, salt stress, and other abiotic stresses (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 1 March 2021).
4.6. RNA-Seq of Different C. rosea Tissues
The C. rosea RNA-Seq datasets were constructed using Illumina HiSeq X sequencing technology. In brief, seven different tissues from C. rosea plants (root, vine, young leaf, flower bud, and young silique samples collected from C. rosea plants growing in SCBG; mature leaf samples from C. rosea growing in SCBG and on YX Island) were examined using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed on 1 March 2021) based on the primary 40 Gb clean reads and were mapped to the C. rosea reference genome using Tophat v2.0.10 (http://tophat.cbcb.umd.edu/, accessed on 1 March 2021). The heatmaps showing the CrLEA and CrASR gene expression profiles were generated using TBtools [56], principally adopting the fragments per kilobase of transcript per million mapped reads (FPKM) values, and the expression levels (log2 of FPKM values) of these genes were visualized.
4.7. Expression Analysis by Quantitative Reverse Transcription (qRT) PCR of CrLEAs/ASRs under Different Stress Treatments
The transcript abundance of several CrLEA and CrASR genes was further investigated using qRT-PCR. Total RNA was extracted from the C. rosea seedling roots, vines, and leaves under the stress/ABA treatment and unstressed treatment using an RNA plant extraction kit (Magen, Biotech, Guangzhou, China) according to the manufacturer’s instructions, and approximately 1 μg of purified total RNA was reverse transcribed into cDNA in a 20 μL reaction volume using AMV reverse transcriptase (TransGen Biotech, Beijing, China) according to the supplier’s instructions. In brief, total RNA was extracted from C. rosea seedling tissues under the stress/ABA treatments and reverse transcribed into cDNA. Quantitative RT-PCR was conducted using the LightCycler480 system (Roche, Basel, Switzerland) and TransStart Tip Green qPCR SuperMix (TransGen). All of the gene expression data obtained via qRT-PCR were normalized to the expression of CrEF-α (Table S1). The primers used for qRT-PCR (CrEF-αRTF/CrEF-αRTR for the reference gene and other CrLEA- or CrASR-specific primer pairs) are listed in Supplementary Materials Table S1.
4.8. Cloning of CrLEA/ASR cDNAs and Heterologous Expression in Yeast
To clone the CrLEA/ASR cDNAs, total RNA isolation and cDNA synthesis was performed as described above. The full-length CrLEA/ASR cDNA was PCR-amplified from C. rosea leaf cDNA using the gene specific primers listed in Supplementary Materials Table S1. The construction of the yeast expression vector pYES2-CrLEAs/ASRs were constructed with in-fusion technique (BD In-Fusion PCR cloning Kit, Takara Bio USA, Mountain View, CA, USA) according to the manufacturer’s instruction. In short, the PCR fragments were inserted into the BamHI and EcoRI sites of pYES2 and after sequencing confirmation, these recombinant expression vectors were transformed into different yeast strains to verify the stress tolerance functions. The wild-type (WT) yeast (Saccharomyces cerevisiae) strain BY4741 (Y00000, MATa; ura3Δ0; leu2Δ0; his3Δ1; met15Δ0) and the two H2O2-sensitive mutant strains yap1Δ (Y00569, BY4741; MATa; ura3Δ0; leu2Δ0; his3Δ1; met15Δ0; YML007w::kanMX4) and skn7Δ (Y02900, BY4741; MATa; ura3Δ0; leu2Δ0; his3Δ1; met15Δ0; YHR206w::kanMX4) were obtained from Euroscarf (Frankfurt, Germany). The various yeast strains were transformed using the LiOAc/PEG technique, and uracil complementation was used for selection. A synthetic defined (SD) yeast medium without uracil (SD-Ura) containing 2% (w/v) galactose (Gal) was used as the yeast induction medium (SDG-Ura) during the salt/alkaline tolerance and oxidative stress experiments on solid medium for the localization observations. The concentrations of the different stress factors are indicated in the figure legends.
4.9. Transcriptional Activity Analysis of CrASRs in Yeast
The open reading frames (ORFs) of three CrASRs were cloned into the vector pGBKT7 (Clontech, Mountain View, CA, USA) for transcription activation analysis with the yeast two-hybrid assay according to the manufacturer’s introduction (Clontech, CA, USA). In brief, the full CDSs of CrASRs were amplified from C. rosea leaves cDNA using the corresponding primers as listed in Supplementary Materials Table S1. Then all PCR products were inserted into GAL4-DBD vector pGBKT7 at EcoRI site and sequenced. These constructs along with the negative control pGBKT7 (containing the binding domain, BD) were transformed into yeast strain AH109 using the LiOAc/PEG method. The yeast clones were cultured in liquid SD-Trp medium to an OD600 value of 2, after which they were diluted using a gradient dilution (1:10, 1:100, and 1:1000). Two-microliter yeast cultures were spotted onto the corresponding synthetically defined (SD/-Trp and SD/-Trp/-Leu) medium plates for 2 days at 30 °C. Yeast transformation and determination of blue/white colonies were conducted according to the instructions of the manufacturer (Clontech), and X-α-Gal was used as a substrate for the reporter gene MEL1. The primers used in construction of pGBKT7 vectors for CrASRs transactivation activity assay in yeast are listed in Supplementary Materials Table S1.
4.10. Statistical Analysis
All the experiments in this study were repeated three times independently, with the results shown as the mean ± SD (n ≥ 3). Pairwise differences between means were analyzed using Student’s t-tests in Excel 2010 (Microsoft Corporation, Albuquerque, NM, USA).
5. Conclusions
In summary, this study is the first to systematically summarize members of the LEA and ASR gene family in a leguminous halophyte, C. rosea. In total, 84 CrLEA genes and three CrASR genes were identified in the C. rosea genome and were classified into nine subgroups. Chromosomal mapping and synteny analysis revealed that these members are distributed in all 11 chromosomes of C. rosea, with several gene tandem and segmental duplication patterns for LEA gene expansion in the C. rosea genome. The CrLEA/CrASR members within the same subfamily are highly conserved in both gene structures and protein motifs. The CrLEA/CrASR superfamily was involved in the adaptation of C. rosea to different habitats, and 10 of the CrLEA/CrASR members showed obvious differences in gene expression in response to salt/alkaline stress, high osmotic stress, or ABA treatment. This systematic study provides new information on the LEA/ASR superfamily in C. rosea and further expands our understanding of the association of CrLEA/CrASR genes with natural ecological adaptability and abiotic stress responses in C. rosea. Our findings should also inform the genetic improvement of other legume plants or crops and provide candidate stress-resistance genes for future research.
Acknowledgments
We sincerely thank several unknown workers who provided us Canavalia rosea plants in South China Botanical Garden (Guangzhou city, China) and C. rosea seeds gathered from Hainan province, China.
Abbreviations
| LEA | late embryogenesis abundant |
| ASR | abscisic acid-, stress-, and ripening-induced |
| LEAPdb | database for the late embryogenesis abundant proteins |
| DHN | dehydrin |
| SMP | seed maturation protein |
| ROS | reactive oxygen species |
| IDP | intrinsically disordered protein |
| WGD | whole-genome duplication |
| WT | wild type |
| YX | Yongxing Island |
| SCBG | South China Botanical Garden |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/article/10.3390/ijms22094554/s1.
Author Contributions
S.J. and M.Z. conceived and designed the experiments; R.L. and M.Z. conducted most of the experiment, collected the data; T.Z. helped with data processing and bioinformatics analysis; Z.W. and Q.M. helped in sequence analyses; M.Z. and R.L. drafted the manuscript; S.J. and M.Z. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the NSFC-Guangdong Province Union Funds (U1701246), the National Natural Sciences Foundation of China (31570257), the Guangdong Science and Technology Program (2019B121201005), and the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0408). The funders had no roles in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. However, most of the data is shown in Supplementary files.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cao J., Li X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum) Planta. 2014;241:757–772. doi: 10.1007/s00425-014-2215-y. [DOI] [PubMed] [Google Scholar]
- 2.Pedrosa A.M., Martins C.D.P.S., Gonçalves L.P., Costa M.G.C. Late Embryogenesis Abundant (LEA) Constitutes a Large and Diverse Family of Proteins Involved in Development and Abiotic Stress Responses in Sweet Orange (Citrus sinensis L. Osb.) PLoS ONE. 2015;10:e0145785. doi: 10.1371/journal.pone.0145785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W., Gao T., Chen J., Yang J., Huang H., Yu Y. The late embryogenesis abundant gene family in tea plant (Camellia sinensis): Genome-wide characterization and expression analysis in response to cold and dehydration stress. Plant Physiol. Biochem. 2019;135:277–286. doi: 10.1016/j.plaphy.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Liu H., Xing M., Yang W., Mu X., Wang X., Lu F., Wang Y., Zhang L. Genome-wide identification of and functional in-sights into the late embryogenesis abundant (LEA) gene family in bread wheat (Triticum aestivum) Sci. Rep. 2019;9:13375. doi: 10.1038/s41598-019-49759-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hand S.C., Menze M.A., Toner M., Boswell L., Moore D. LEA Proteins During Water Stress: Not Just for Plants Anymore. Annu. Rev. Physiol. 2011;73:115–134. doi: 10.1146/annurev-physiol-012110-142203. [DOI] [PubMed] [Google Scholar]
- 6.Reyes J.L., Campos F., Wei H., Arora R., Yang Y., Karlson D.T., Covarrubias A.A. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 2008;31:1781–1790. doi: 10.1111/j.1365-3040.2008.01879.x. [DOI] [PubMed] [Google Scholar]
- 7.Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A.A. The Enigmatic LEA Proteins and Other Hydrophilins. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wise M.J., Tunnacliffe A. POPP the question: What do LEA proteins do? Trends Plant Sci. 2004;9:13–17. doi: 10.1016/j.tplants.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Tunnacliffe A., Wise M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94:791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- 10.González R.M., Iusem N.D. Twenty years of research on Asr (ABA-stress-ripening) genes and proteins. Planta. 2014;239:941–949. doi: 10.1007/s00425-014-2039-9. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez P.G., Carrari F. ASR1 transcription factor and its role in metabolism. Plant Signal. Behav. 2015;10:e992751. doi: 10.4161/15592324.2014.992751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Dong Y., Li C., Pan Y., Yu J. SiASR4, the Target Gene of SiARDP from Setaria italica, Improves Abiotic Stress Adaption in Plants. Front. Plant Sci. 2017;7:2053. doi: 10.3389/fpls.2016.02053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrari F., Fernie A.R., Iusem N.D. Heard it through the grapevine? ABA and sugar cross-talk: The ASR story. Trends Plant Sci. 2004;9:57–59. doi: 10.1016/j.tplants.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Galau G.A., Chlan C.A., Dure L. Developmental biochemistry of cottonseed embryogenesis and germination. Plant Mol. Biol. 1983;2:189–198. doi: 10.1007/BF01578378. [DOI] [PubMed] [Google Scholar]
- 15.Hunault G., Jaspard E. LEAPdb: A database for the late embryogenesis abundant proteins. BMC Genom. 2010;11:221. doi: 10.1186/1471-2164-11-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Candat A., Paszkiewicz G., Neveu M., Gautier R., Logan D.C., Avelange-Macherel M.-H., Macherel D. The Ubiquitous Distribution of Late Embryogenesis Abundant Proteins across Cell Compartments in Arabidopsis Offers Tailored Protection against Abiotic Stress. Plant Cell. 2014;26:3148–3166. doi: 10.1105/tpc.114.127316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hundertmark M., Hincha D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thali-ana. BMC Genom. 2008;9:118. doi: 10.1186/1471-2164-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu M., Tong Q., Wang Y., Wang Z., Xu G., Elias G.K., Li S., Liang Z. Transcriptomic Analysis of the Grapevine LEA Gene Family in Response to Osmotic and Cold Stress Reveals a Key Role for VamDHN3. Plant Cell Physiol. 2020;61:775–786. doi: 10.1093/pcp/pcaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Artur M.A.S., Zhao T., Ligterink W., Schranz E., Hilhorst H.W.M. Dissecting the genomic diversification of late embryo-genesis abundant (LEA) protein gene families in plants. Genome Biol. Evol. 2019;11:459–471. doi: 10.1093/gbe/evy248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatelain E., Hundertmark M., Leprince O., Le Gall S., Satour P., Deligny-Penninck S., Rogniaux H., Buitink J. Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 2012;35:1440–1455. doi: 10.1111/j.1365-3040.2012.02501.x. [DOI] [PubMed] [Google Scholar]
- 21.Verdier J., Lalanne D., Pelletier S., Torres-Jerez I., Righetti K., Bandyopadhyay K., Leprince O., Chatelain E., Vu B.L., Gouzy J., et al. A regulatory network-based approach dissects late maturation processes re-lated to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 2013;163:757–774. doi: 10.1104/pp.113.222380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun X., Rikkerink E.H., Jones W.T., Uversky V.N. Multifarious Roles of Intrinsic Disorder in Proteins Illustrate Its Broad Impact on Plant Biology. Plant Cell. 2013;25:38–55. doi: 10.1105/tpc.112.106062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y.P., Giorgi F.M., Lohse M., Kvederaviciute K., Klages S., Usadel B., Meskiene I., Reinhardt R., Hincha D.K. Tran-scriptome sequencing and microarray design for functional genomics in the extremophile Arabidopsis relative Thellungiella salsuginea (Eutrema salsugineum) BMC Genom. 2013;14:793. doi: 10.1186/1471-2164-14-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muvunyi B.P., Yan Q., Wu F., Min X., Yan Z.Z., Kanzana G., Wang Y., Zhang J. Mining Late Embryogenesis Abundant (LEA) Family Genes in Cleistogenes songorica, a Xerophyte Perennial Desert Plant. Int. J. Mol. Sci. 2018;19:3430. doi: 10.3390/ijms19113430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Artur M.A.S., Rienstra J., Dennis T.J., Farrant J.M., Ligterink W., Hilhorst H. Structural plasticity of intrinsically disor-dered LEA proteins from Xerophyta schlechteri provides protection in vitro and in vivo. Front. Plant. Sci. 2019;10:1272. doi: 10.3389/fpls.2019.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagaraju M., Kumar S.A., Reddy P.S., Kumar A., Rao D.M., Kishor P.B.K. Genome-scale identification, classification, and tissue specific expression analysis of late embryogenesis abundant (LEA) genes under abiotic stress conditions in Sorghum bicolor L. PLoS ONE. 2019;14:e0209980. doi: 10.1371/journal.pone.0209980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J., Lan T. Functional characterization of the late embryogenesis abundant (LEA) protein gene family from Pinus tabu-liformis (Pinaceae) in Escherichia coli. Sci. Rep. 2016;6:14467. doi: 10.1038/srep19467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golan I., Dominguez P.G., Konrad Z., Shkolnik-Inbar D., Carrari F., Bar-Zvi D. Tomato ABSCISIC ACID STRESS RIP-ENING (ASR) gene family revisited. PLoS ONE. 2014;9:e107117. doi: 10.1371/journal.pone.0107117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamdi K., Salladini E., O’Brien D.P., Brier S., Chenal A., Yacoubi I., Longhi S. Structural disorder and induced folding within two cereal, ABA stress and ripening (ASR) proteins. Sci. Rep. 2017;7:1–21. doi: 10.1038/s41598-017-15299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konrad Z., Bar-Zvi D. Synergism between the chaperone-like activity of the stress regulated ASR1 protein and the osmolyte glycine-betaine. Planta. 2008;227:1213–1219. doi: 10.1007/s00425-008-0693-5. [DOI] [PubMed] [Google Scholar]
- 31.Dai J.R., Liu B., Feng D.R., Liu H.Y., He Y.M., Qi K.B., Wang H.B., Wang J.F. MpAsr encodes an intrinsically unstruc-tured protein and enhances osmotic tolerance in transgenic Arabidopsis. Plant Cell Rep. 2011;30:1219–1230. doi: 10.1007/s00299-011-1030-1. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J., Zhu Q., Yu H., Li L., Zhang G., Chen X., Jiang M., Tan M. Comprehensive Analysis of the Cadmium Tolerance of Abscisic Acid-, Stress- and Ripening-Induced Proteins (ASRs) in Maize. Int. J. Mol. Sci. 2019;20:133. doi: 10.3390/ijms20010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Li Y., Yin Z., Jiang J., Zhang M., Guo X., Ye Z., Zhao Y., Xiong H., Zhang Z., et al. OsASR5enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2signalling in rice. Plant Biotechnol. J. 2016;15:183–196. doi: 10.1111/pbi.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye Y., Lin R., Su H., Chen H., Luo M., Yang L., Zhang M. The functional identification of glycine-rich TtASR from Tetra-gonia tetragonoides (Pall.) Kuntze involving in plant abiotic stress tolerance. Plant Physiol. Biochem. 2019;143:212–223. doi: 10.1016/j.plaphy.2019.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Hu W., Huang C., Deng X., Zhou S., Chen L., Li Y., Wang C., Ma Z., Yuan Q., Wang Y., et al. TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 2013;36:1449–1464. doi: 10.1111/pce.12074. [DOI] [PubMed] [Google Scholar]
- 36.Battaglia M., Covarrubias A.A. Late Embryogenesis Abundant (LEA) proteins in legumes. Front. Plant Sci. 2013;4:190. doi: 10.3389/fpls.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wetzler D.E., Fuchs Wightman F., Bucci H.A., Rinaldi J., Caramelo J.J., Iusem N.D., Ricardi M.M. Conformational plas-ticity of the intrinsically disordered protein ASR1 modulates its function as a drought stress-responsive gene. PLoS ONE. 2018;13:e0202808. doi: 10.1371/journal.pone.0202808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niveditha V.R., Sridhar K.R. Antioxidant activity of raw, cooked and Rhizopus oligosporus fermented beans of Canavalia of coastal sand dunes of Southwest India. J. Food Sci. Technol. 2012;51:3253–3260. doi: 10.1007/s13197-012-0830-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J., Liu N., Ren H., Jian S. Physiology and biochemical characteristics of Canavalia maritime under stress. J. Trop. Subtrop. Bot. 2019;27:157–163. [Google Scholar]
- 40.Iusem N.D., Bartholomew D.M., Hitz W.D., Scolnik P.A. Tomato (Lycopersicon esculentum) Transcript Induced by Water Deficit and Ripening. Plant Physiol. 1993;102:1353–1354. doi: 10.1104/pp.102.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanin M., Brini F., Ebel C., Toda Y., Takeda S., Masmoudi K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal Behav. 2011;6:1503–1509. doi: 10.4161/psb.6.10.17088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mu P., Feng D., Su J., Zhang Y., Dai J., Jin H., Liu B., He Y., Qi K., Wang H., et al. Cu2+ triggers reversible aggrega-tion of a disordered His-rich dehydrin MpDhn12 from Musa paradisiaca. J. Biochem. 2011;150:491–499. doi: 10.1093/jb/mvr082. [DOI] [PubMed] [Google Scholar]
- 43.Li R.H., Liu G.B., Wang H., Zheng Y.Z. Effects of Fe3+ and Zn2+ on the structural and thermodynamic properties of a soy-bean ASR protein. Biosci. Biotechnol. Biochem. 2013;77:475–481. doi: 10.1271/bbb.120666. [DOI] [PubMed] [Google Scholar]
- 44.Zheng J.-X., Zhang H., Su H.-X., Xia K.-F., Jian S.-G., Zhang M. Ipomoea pes-caprae IpASR Improves Salinity and Drought Tolerance in Transgenic Escherichia coli and Arabidopsis. Int. J. Mol. Sci. 2018;19:2252. doi: 10.3390/ijms19082252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shu Y., Liu Y., Zhang J., Song L., Guo C. Genome-wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant. Sci. 2016;6:1247. doi: 10.3389/fpls.2015.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das Laha S., Dutta S., Schäffner A.R., Das M. Gene duplication and stress genomics in Brassicas: Current understanding and future prospects. J. Plant Physiol. 2020;255:153293. doi: 10.1016/j.jplph.2020.153293. [DOI] [PubMed] [Google Scholar]
- 47.Wu C., Hu W., Yan Y., Tie W., Ding Z., Guo J., He G. The late embryogenesis abundant protein family in cassava (Mani-hot esculenta Crantz): Genome-wide characterization and expression during abiotic stress. Molecules. 2018;23:1196. doi: 10.3390/molecules23051196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling H., Zeng X., Guo S. Functional insights into the late embryogenesis abundant (LEA) protein family from Dendrobium officinale (Orchidaceae) using an Escherichia coli system. Sci. Rep. 2016;6:39693. doi: 10.1038/srep39693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y., Li C., Zhang B., Yi J., Yang Y., Kong C., Lei C., Gong M. The role of the late embryogenesis-abundant (LEA) protein family in development and the abiotic stress response: A comprehensive expression analysis of potato (Solanum tu-berosum) Genes. 2019;10:148. doi: 10.3390/genes10020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang M., Zhang H., Zheng J.-X., Mo H., Xia K.-F., Jian S.-G. Functional Identification of Salt-Stress-Related Genes Using the FOX Hunting System from Ipomoea pes-caprae. Int. J. Mol. Sci. 2018;19:3446. doi: 10.3390/ijms19113446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ricardi M.M., González R.M., Zhong S., Domínguez P.G., Duffy T., Turjanski P.G., Salgado Salter J.D., Alleva K., Carra-ri F., Giovannoni J.J., et al. Genome-wide data (ChIP-seq) enabled identification of cell wall-related and aquaporin genes as targets of tomato ASR1, a drought stress-responsive transcription factor. BMC Plant Biol. 2014;14:29. doi: 10.1186/1471-2229-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J., Shen L., Yang S., Guan D., He S. CaASR1 promotes salicylic acid- but represses jasmonic acid-dependent sig-naling to enhance the resistance of Capsicum annuum to bacterial wilt by modulating CabZIP63. J. Exp. Bot. 2020;71:6538–6554. doi: 10.1093/jxb/eraa350. [DOI] [PubMed] [Google Scholar]
- 53.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 54.Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitu-tions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 55.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An integrative toolkit developed for in-teractive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 56.Gietz D., Jean A.S., Woods R.A., Schiestl R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. However, most of the data is shown in Supplementary files.