Abstract
Simple Summary
Acute myeloid leukemia (AML) is a genetically heterogeneous disease. Clinical phenotypes of frequent mutations and their impact on patient outcome are well established. However, the role of rare mutations often remains elusive. We retrospectively analyzed 1529 newly diagnosed and intensively treated AML patients for mutations of BCOR and BCORL1. We report a distinct co-mutational pattern that suggests a role in disease progression rather than initiation, especially affecting mechanisms of DNA-methylation. Further, we found loss-of-function mutations of BCOR to be independent markers of poor outcomes in multivariable analysis. Therefore, loss-of-function mutations of BCOR need to be considered for AML management, as they may influence risk stratification and subsequent treatment allocation.
Abstract
Acute myeloid leukemia (AML) is characterized by recurrent genetic events. The BCL6 corepressor (BCOR) and its homolog, the BCL6 corepressor-like 1 (BCORL1), have been reported to be rare but recurrent mutations in AML. Previously, smaller studies have reported conflicting results regarding impacts on outcomes. Here, we retrospectively analyzed a large cohort of 1529 patients with newly diagnosed and intensively treated AML. BCOR and BCORL1 mutations were found in 71 (4.6%) and 53 patients (3.5%), respectively. Frequently co-mutated genes were DNTM3A, TET2 and RUNX1. Mutated BCORL1 and loss-of-function mutations of BCOR were significantly more common in the ELN2017 intermediate-risk group. Patients harboring loss-of-function mutations of BCOR had a significantly reduced median event-free survival (HR = 1.464 (95%-Confidence Interval (CI): 1.005–2.134), p = 0.047), relapse-free survival (HR = 1.904 (95%-CI: 1.163–3.117), p = 0.01), and trend for reduced overall survival (HR = 1.495 (95%-CI: 0.990–2.258), p = 0.056) in multivariable analysis. Our study establishes a novel role for loss-of-function mutations of BCOR regarding risk stratification in AML, which may influence treatment allocation.
Keywords: acute myeloid leukemia, BCOR, BCORL1, loss-of-function, risk stratification, survival
1. Introduction
Acute myeloid leukemia (AML) is a genetically heterogeneous disease [1]. In the last decade, next-generation sequencing (NGS) has been introduced in hematological practice, and with the use of myeloid gene panels, rare and recurrent mutations have been unveiled, with a majority of patients harboring more than one mutation even within defined AML entities [2]. The discovery of common recurrent mutations, such as NPM1 and FLT3-ITD, among others, has led to a better understanding of the molecular landscape of AML [1,3], with distinct consequences for prognostication and treatment allocation [4]. However, the role and potential impact of rare mutations on prognosis is not well understood, and warrants further studies to improve risk assessment, implying a more precise approach to AML treatment, as relapse and mortality rates are still unsatisfactory.
The BCL6 corepressor (BCOR) gene, and its homolog, the BCL6 corepressor-like 1 (BCORL1) gene, are located on chromosomes Xp11.4 and Xq26.1, respectively [5,6]. BCOR was originally described as a repressor of BCL6 [5], but also interacts with PCGF1, KDM2B, MLLT3, and IRF8, while BCORL1 interacts with PCGF1, KDM2B, CTBP1, and HDAC [7]. Both BCOR and BCORL1 are core proteins of the polycomb repressive complex PRC1.1. PRCs are essential for the maintenance of cell identity and cell differentiation, and their perturbance is an important factor in carcinogenesis [8]. PRC1.1 plays a role in epigenetic modification by adding an ubiquitin to histone H2A at lysine 119 [9]. This process (among others) leads to a silencing of Hox gene clusters [10], and mediates transcriptional repression [11]. BCOR is highly expressed in embryonic stem cells, where a role in maintaining the primed pluripotent state has been suggested [7,12]. As a mediator of stemness and differentiation, BCOR is involved in the development of B- and T-cells, as well as erythrocytes [13].
Since BCOR is vital in ectodermal and mesenchymal differentiation, germline loss-of-function mutations in BCOR lead to oculofaciocardiodental (OFCD) syndrome in heterozygous females, and prenatal death in hemizygous males [14]. Somatic mutations have been found in a variety of solid tumors, such as retinoblastoma, medulloblastoma, osteosarcoma, and hepatocellular carcinoma [15,16,17,18]. BCOR is an important factor in the regulation of myeloid cell proliferation and differentiation [12]. It has been described in hematologic entities such as aplastic anemia [19,20], chronic myelomonocytic leukemia [21], clonal hematopoiesis of indeterminate potential [22], myelodysplastic syndromes (MDS) [21,23,24,25], and AML [6,21,26,27,28]. In AML, mutations of BCOR (mBCOR) and BCORL1 (mBCORL1) both occur in 4–6% of patients, and an association with poor outcomes has been suggested [6,26,27,28]. However, given the rarity of these mutations, and the confinement of previous studies to smaller patient cohorts, further investigation of their impact on clinical phenotypes and outcomes in AML seems warranted. We here present the analysis on the impact of BCOR/BCORL1 mutational status on a large cohort of newly diagnosed and intensively treated AML patients.
2. Materials and Methods
2.1. Data Set
We retrospectively analyzed a multi-center cohort of 1529 AML patients. Eligibility criteria were newly diagnosed AML according to WHO definitions [29], age ≥ 18 years, and available biomaterial at diagnosis. All patients were treated with intensive regimens in the following clinical trials: AML96 [30], AML2003 [31], AML60+ [32], and SORAML [33] or were enrolled in the German Study Alliance Leukemia (SAL)’s AML registry (NCT03188874). Detailed information on treatment regimens is given in the respective references. All mentioned studies were carried out under the auspices of the SAL, and approved by the Institutional Review Board of the Dresden University of Technology (Dresden, Saxony, Germany). All participants gave their written, informed consent, in accordance with to the Declaration of Helsinki.
2.2. Definitions
AML was defined as de novo when neither previous malignancy nor previous treatment with chemo- and/or radiotherapy was reported. When myeloid neoplasms were documented prior to AML diagnosis, AML was defined as secondary (sAML). Prior exposure to chemo- and/or radiotherapy before the initial diagnosis defined therapy-associated AML (tMN). Early death was defined as death by any cause within 30 days of the initial diagnosis (ED30). Remission and survival criteria were defined according to ELN2017 recommendations [4].
2.3. Molecular Analysis
Next generation sequencing (NGS) using a TruSight Myeloid Sequencing Panel (Illumina, San Diego, CA, USA) was performed on pre-treatment bone marrow or peripheral blood, targeting 54 genes (Table S1) associated with myeloid neoplasms, including full coding exons of BCOR and BCORL1, as previously described in detail [34,35]. A DNeasy blood and tissue kit (Qiagen, Hilden, Germany) was used to extract DNA, and subsequent quantification was performed using a NanoDrop spectrophotometer. Pooled samples were sequenced paired-end (150 bp PE) using a NextSeq NGS instrument (Illumina). The SEQUENCE PILOT software package (JSI medical systems GmbH, Ettenheim, Germany) was used for sequence data alignment, variant calling, and filtering. A 5% variant allele frequency (VAF) cut-off was used. Genome-mapping algorithms were referenced to human genome build HG19. We compared VAFs of BCOR/BCORL1 mutations with VAFs of co-mutated drivers for dichotomization of dominant, subclonal, and secondary mutations. For putative subclonal mutations a minimum VAF difference of 10% was applied.
2.4. Statistical Analysis
We compared categorical variables between groups using the chi-squared test, while continuous variables were compared using the Kruskal–Wallis test. The Kaplan–Meier method was used to estimate survival probabilities. The logrank test was used to compare survival time distributions between groups. Cox regression was used to estimate univariate and adjusted hazard ratios. For the binary endpoint of complete remission, logistic regression models were fitted to estimate univariate and adjusted odds ratios. A significance level of 0.05 was used to determine statistical significance. Calculations were performed in R 4.0.3.
3. Results
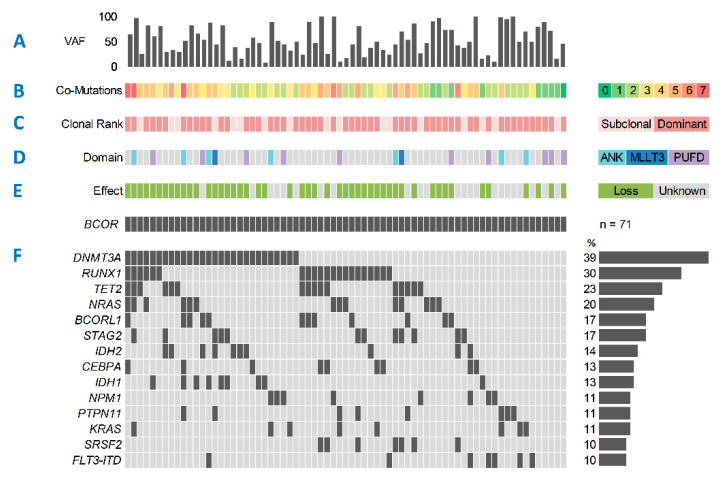
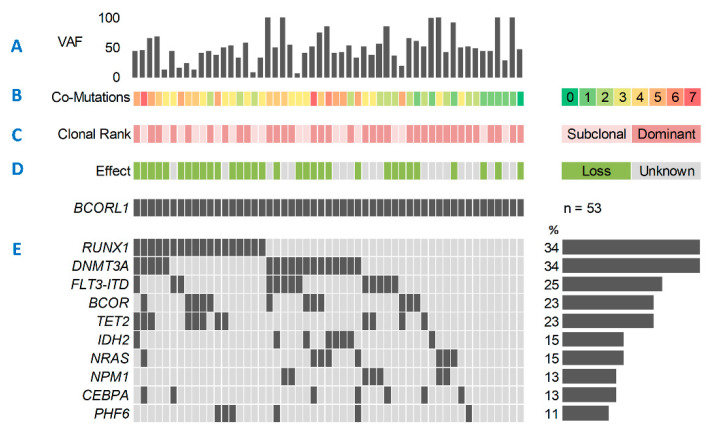
In the entire cohort (n = 1529), we found mBCOR in 71 (4.6%) and mBCORL1 in 53 (3.5%) patients. Twelve patients (0.8%) concomitantly harbored both mBCOR and mBCORL1. The median age for the entire cohort was 55 years (Interquartile range (IQR): 44–64). Median variant allele frequency (VAF) for mBCOR and mBCORL1 was 48% (range: 7–100%) and 47% (range: 5–100%), respectively (Figure 1A and Figure 2A). Most patients harboring mBCOR (87%) and mBCORL1 (83%) carried two or more additional mutations in other driver genes, while only one patient in each group had no other co-mutations detected by the panel (Figure 1B and Figure 2B). Both mBCOR and mBCORL1 were more frequently found in the dominant AML clone (76% and 68%, respectively, Figure 1C and Figure 2C). Mutations of BCOR were found in several exons and functional domains, including the MLLT3, ANK, and PUFD domains (Figure 1D). The majority (62%) of the mutations were single-nucleotide variants (SNVs). Most mutations had a loss-of-function (LOF) effect (frameshift or non-sense mutations, 68%, Figure 1E), likely resulting in a premature stop of transcription and inactivation of the BCOR protein. The remaining 32% of BCOR mutations exclusively comprised missense SNVs, with a presumed damaging effect for the majority of variants, as suggested by the PolyPhen-2 classifier [36].
Figure 1.
Mutational spectrum of BCOR-mutated AML. (A) BCOR variant allele frequency (VAF) percentages. (B) Number of co-mutations in addition to mutated BCOR. (C) Clonal rank of BCOR mutations determined by comparing BCOR VAF to co-mutated somatic driver variants. (D) Mutated BCOR domain, undetermined (grey). (E) Effect of mutated BCOR, loss-of-function (transcriptional stop, green), or unknown (no transcriptional stop, grey). (F) Most common co-mutations. Mutations with a frequency of <10% not shown: ASXL1, GATA2, WT1, SF3B1, U2AF1, TP53, ZRSR2, IKZF1, KIT, ETV6, EZH2, FLT3-TKD, JAK2, PHF6, CBL, CBLB, CSF3R, NOTCH1, PDGFRA, SMC3.
Figure 2.
Mutational spectrum of BCORL1-mutated AML. (A) BCORL1 variant allele frequency (VAF) percentages. (B) Number of co-mutations in addition to mutated BCORL1. (C) Clonal rank of BCORL1 mutations determined by comparing BCORL1 VAF to co-mutated somatic driver variants. (D) Effect of mutated BCORL1, loss-of-function (transcriptional stop, green), or unknown (no transcriptional stop, grey). (E) Most common co-mutations. Mutations with a frequency of <10% not shown: FLT3-TKD, IDH1, SRSF2, KIT, TP53, PTPN11, SF3B1, U2AF1, WT1, ZRSR2, STAG2, ETV6, EZH2, GATA2, KDM6A, ASXL1, CBL, KRAS, MYD88, SETBP1, JAK2, SMC3.
However, due to the unknown effect of protein function, these mutations were classified as UFs (unknown functions) for sub-analysis. The co-mutational landscape of mBCOR AML (Figure 1F) was characterized by high rates of mutation in DNMT3A (39%), RUNX1 (30%), TET2 (23%), NRAS (20%), BCORL1 (17%), and STAG2 (17%). Low mutation frequencies were detected for other genes frequently mutated in AML, such as FLT3 (ITD: 10%, TKD: 3%), NPM1 (11%), and TP53 (4%). Similar to mBCOR, in mBCORL1 AML the majority of mutations had a loss-of-function effect (60%, Figure 2D) and were mostly SNVs (62%). The most common co-mutations (Figure 2E) were DNMT3A (34%), RUNX1 (34%), FLT3-ITD (25%), BCOR (23%), and TET2 (23%).
Mutations of BCORL1 were more prevalent in females than in males (mBCORL1 female 62.3%, male: 37.7%; wild-type BCORL1 female: 47.2%, male: 52.8%; p = 0.044), while for mBCOR no such association was observed. For both mBCOR and mBCORL1 no statistically significant associations were detected for age at diagnosis, the presence of a complex karyotype, hemoglobin levels, or platelet counts. With respect to disease status, the rate of sAML was significantly higher amongst patients harboring mBCOR than amongst wtBCOR patients (21.7% vs. 11%, p = 0.023), while no specific association with mutation type (LOF or UF) or mBCORL1 was found regarding AML type. Outcomes did not differ for patients with sAML and mBCOR or mBCORL1 compared to patients with de novo AML. In the ELN2017 intermediate-risk group we found a higher proportion of LOF of mBCOR compared to UF mBCOR and wtBCOR (LOF mBCOR: 86.4% vs. UF mBCOR: 28.6% vs. wtBCOR: 38.8%, p < 0.001), as well as mBCORL1 compared to wtBCORL1 (70% vs. 39.1%, p < 0.001). Median white blood cell (WBC) counts in LOF mBCOR AML were significantly lower compared to UF mBCOR and wtBCOR (p < 0.001), while mBCORL1 showed no significant association with WBC. Table 1 and Table 2 summarize patient characteristics for mBCOR (LOF and UF) and mBCORL1, respectively.
Table 1.
Patient characteristics for mBCOR.
| Parameter | wtBCOR | mBCOR-LOF | mBCOR–UF | p-Value |
|---|---|---|---|---|
| N. of patients | 1458 | 48 | 23 | |
| Age, median (IQR) | 55 (44–64) | 53 (45–65) | 61 (51–69) | 0.165 |
| Sex, n (%) | 0.150 | |||
| Female | 689 (47.3) | 26 (54.2) | 15 (65.2) | |
| Male | 769 (52.7) | 22 (45.8) | 8 (34.8) | |
| AML type, n (%) | 0.065 | |||
| De novo | 1235 (85.6) | 37 (78.7) | 15 (68.2) | |
| sAML | 158 (11) | 9 (19.1) | 6 (27.3) | |
| tMN | 49 (3.4) | 1 (2.1) | 1 (4.5) | |
| ELN2017, n (%) | <0.001 | |||
| Favorable | 561 (41.7) | 1 (2.3) | 8 (38.1) | |
| Intermediate | 522 (38.8) | 38 (86.4) | 6 (28.6) | |
| Adverse | 261 (19.4) | 5 (11.4) | 7 (33.3) | |
| Complex karyotype, n (%) | 0.111 | |||
| No | 1178 (87.8) | 44 (93.6) | 19 (82.6) | |
| Yes | 164 (12.2) | 3 (6.4) | 4 (17.4) | |
| FLT3-ITD, n (%) | 0.022 | |||
| wt | 1114 (76.9) | 44 (93.6) | 19 (82.6) | |
| m | 334 (23.1) | 3 (6.4) | 4 (17.4) | |
| NPM1, n (%) | <0.001 | |||
| wt | 961 (67) | 45 (97.8) | 16 (69.6) | |
| m | 474 (33) | 1 (2.2) | 7 (30.4) | |
| WBC, median (IQR) in GPt/L | 20.85 (4.96–56.4) | 4 (1.82–16.88) | 16.5 (6.46–34.65) | <0.001 |
| Hb, median (IQR) in mmol/L | 5.9 (5.0–7.0) | 6.15 (5.25–7.78) | 5.59 (5.06–6.01) | 0.211 |
| Platelets, median (IQR) in GPt/L | 50 (27.0–92.0) | 54 (25.5–106) | 73 (41.5–118.5) | 0.176 |
| LDH, median (IQR) in mmol/L | 457 (290.25–795.0) | 306 (223.65–458.3) | 407 (254.5–897) | 0.001 |
| BM blasts (%), median (IQR) | 63 (44–79) | 58.75 (48.88–72.12) | 61 (45–72.73) | 0.874 |
| PB blasts (%), median (IQR) | 42 (13–75) | 19 (6–59) | 39 (19.75–70) | 0.076 |
wt: wild-type; m-: mutated; LOF: loss-of-function effect (transcriptional stop); UF: unknown effect (no transcriptional stop); sAML: secondary acute myeloid leukemia; tMN: therapy-related acute myeloid leukemia; ELN2017: European Leukemia Net 2017; WBC: white blood cells; BM: bone marrow; PB: peripheral blood; n/N: number; IQR: interquartile range.
Table 2.
Patient characteristics for mBCORL1.
| Parameter | wtBCORL1 | mBCORL1 | p-Value |
|---|---|---|---|
| N. of patients | 1476 | 53 | |
| Age, median (IQR) | 55 (44–65) | 52 (43–62) | 0.177 |
| Sex, n (%) | 0.044 | ||
| Female | 697 (47.2) | 33 (62.3) | |
| Male | 779 (52.8) | 20 (37.7) | |
| AML type, n (%) | 0.621 | ||
| De novo | 1244 (85.3) | 43 (82.7) | |
| sAML | 167 (11.4) | 6 (11.5) | |
| tMN | 48 (3.3) | 3 (5.8) | |
| ELN2017, n (%) | <0.001 | ||
| Favorable | 564 (41.5) | 6 (12) | |
| Intermediate | 531 (39.1) | 35 (70) | |
| Adverse | 264 (19.4) | 9 (18) | |
| Complex karyotype, n (%) | 0.975 | ||
| No | 1192 (88) | 39 (86.7) | |
| yes | 163 (12) | 6 (13.3) | |
| FLT3-ITD, n (%) | 0.782 | ||
| wt | 1138 (77.6) | 39 (75) | |
| m | 328 (22.4) | 13 (25) | |
| NPM1, n (%) | 0.006 | ||
| wt | 977 (67.3) | 45 (86.5) | |
| m | 475 (32.7) | 7 (13.5) | |
| WBC, median (IQR) in GPt/L | 20.09 (4.8–55.27) | 14.35 (3.55–54.95) | 0.340 |
| Hb, median (IQR) in mmol/L | 5.9 (5.0–7.0) | 6.2 (5.3–7.0) | 0.341 |
| Platelets, median (IQR) in GPt/L | 51 (27.0–93.3) | 50.5 (30.0–90.5) | 0.727 |
| LDH, median (IQR) in mmol/L | 452 (288.0–794.8) | 333 (254.0–621.0) | 0.053 |
| BM blasts (%), median (IQR) | 63 (44–79) | 65.25 (48.88–82.75) | 0.299 |
| PB blasts (%), median (IQR) | 40.5 (12–74) | 48.5 (15–78) | 0.419 |
wt: wild-type; m-: mutated; sAML: secondary acute myeloid leukemia; tMN: therapy-related acute myeloid leukemia; ELN2017: European Leukemia Net 2017; WBC: white blood cells; BM: bone marrow; PB: peripheral blood; n/N: number; IQR: interquartile range.
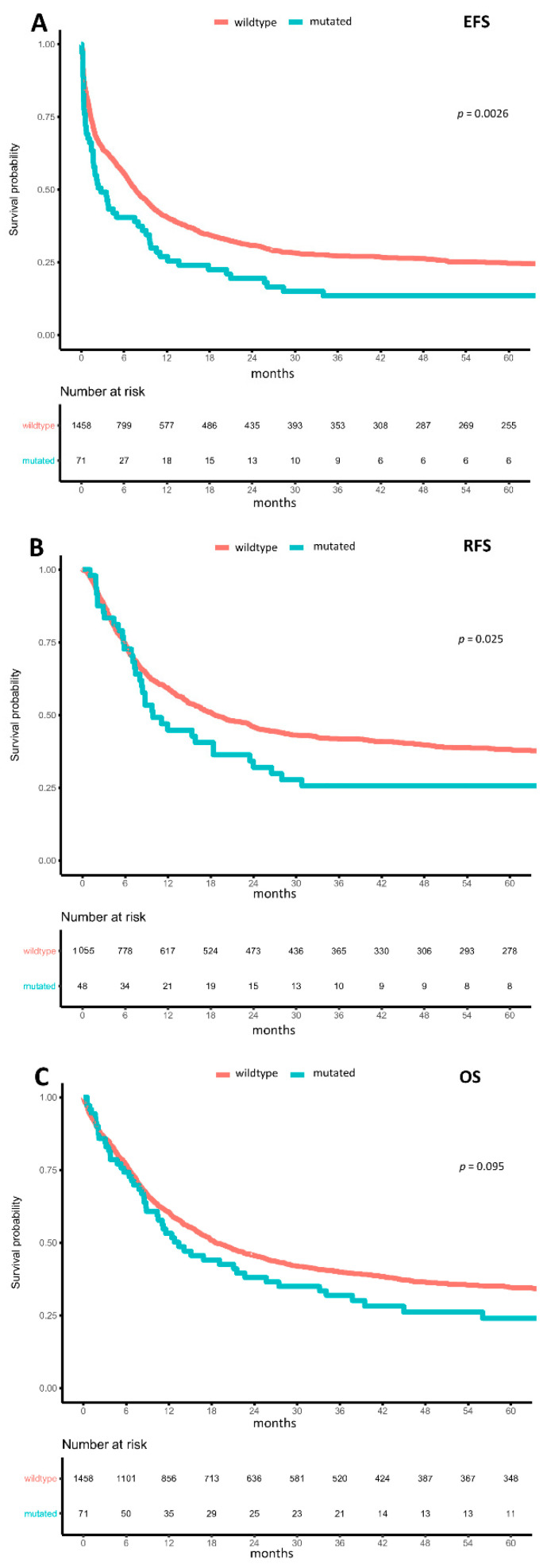
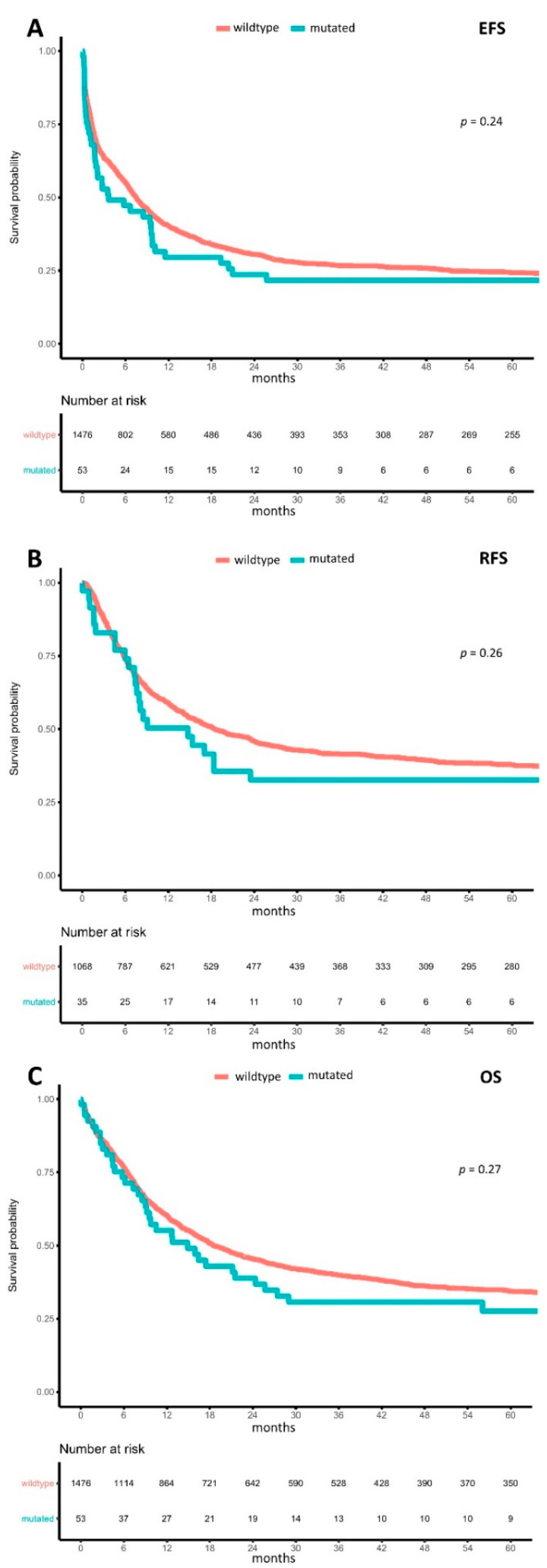
With respect to clinical outcomes, mBCOR and mBCORL1 were not associated with the rate of complete remission (CR) (mBCOR: OR = 0.781 (95%-CI: 0.469–1.301), p = 0.342 and mBCORL1: OR = 0.795 (95%-CI: 0.442–1.432), p = 0.445) or with ED30 (mBCOR: OR = 0.679 (95%-CI: 0.209–2.199), p = 0.518 and mBCORL1: OR = 1.288 (95%-CI: 0.454–3.649), p = 0.634). In contrast, mBCOR was associated with lower median measures of event-free survival (EFS) (2.8 months (95%-CI: 1.7–8.5) vs. 7.6 months (95%-CI: 6.8–8.5), HR = 1.485 (95%-CI: 1.147–1.922), p = 0.003; Figure 3A), relapse-free survival (RFS) (9.9 months (95%-CI: 8.1–24.0) vs. 18.7 months (95%-CI: 16.3–23.3), HR = 1.452 (95%-CI: 1.047–2.014), p = 0.026; Figure 3B), and overall survival (OS) (13.4 months (95%-CI: 10.4–27.4) vs. 18.5 months (95%-CI: 16.8–21.4), HR = 1.452 (95%-CI: 0.959–1.677), p = 0.095; Figure 3C) in a univariate analysis. In a multivariable model adjusted for age, AML type (de novo, sAML, tMN), and ELN2017 risk, mBCOR in general was not independently associated with EFS (HR = 1.243 (95%-CI: 0.89–1.736), p = 0.202), RFS (HR = 1.407 (95%-CI: 0.93–2.129), p = 0.106), and OS (HR = 1.216 (95%-CI: 0.846–1.748), p = 0.292).
Figure 3.
Impact of BCOR mutations on outcomes. Kaplan–Meier plots for (A) event-free survival, (B) relapse-free survival, and (C) overall survival in AML, with wild-type (orange) and mutated (turquoise) BCOR compared using the logrank test.
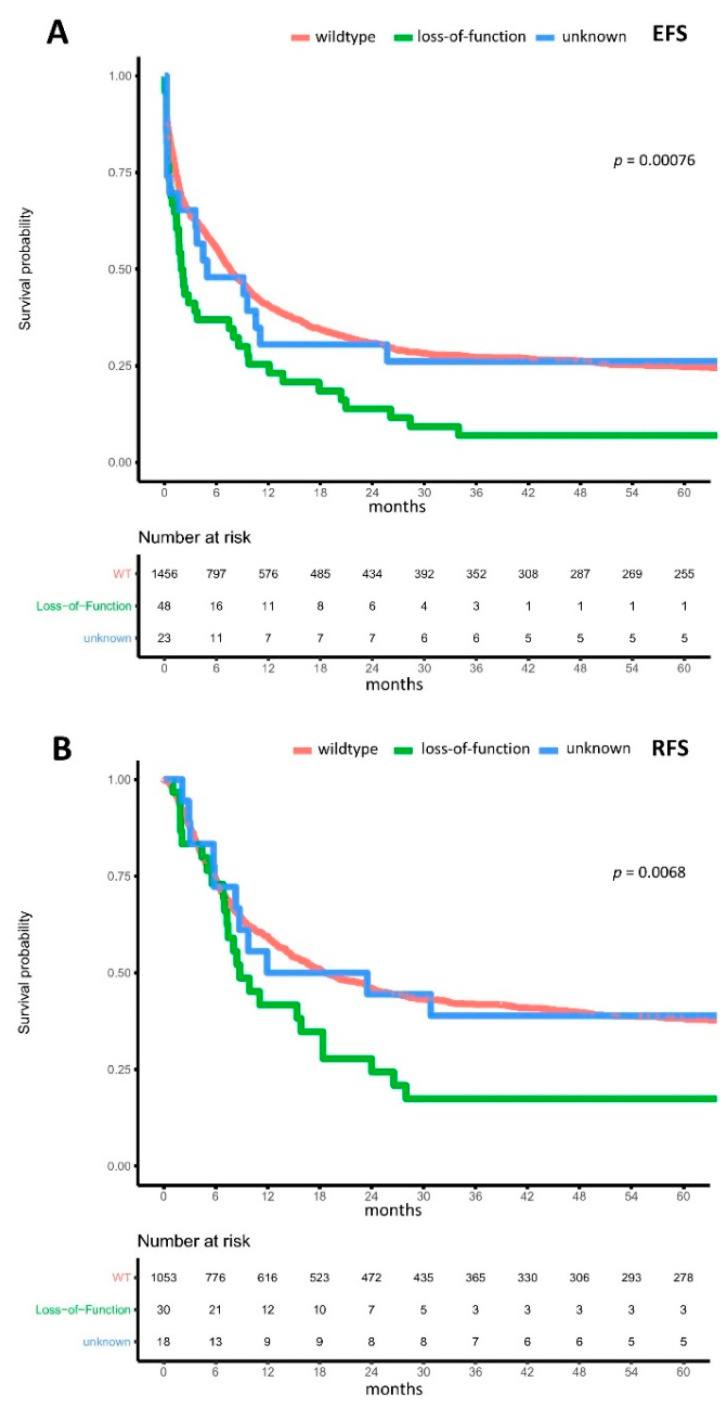
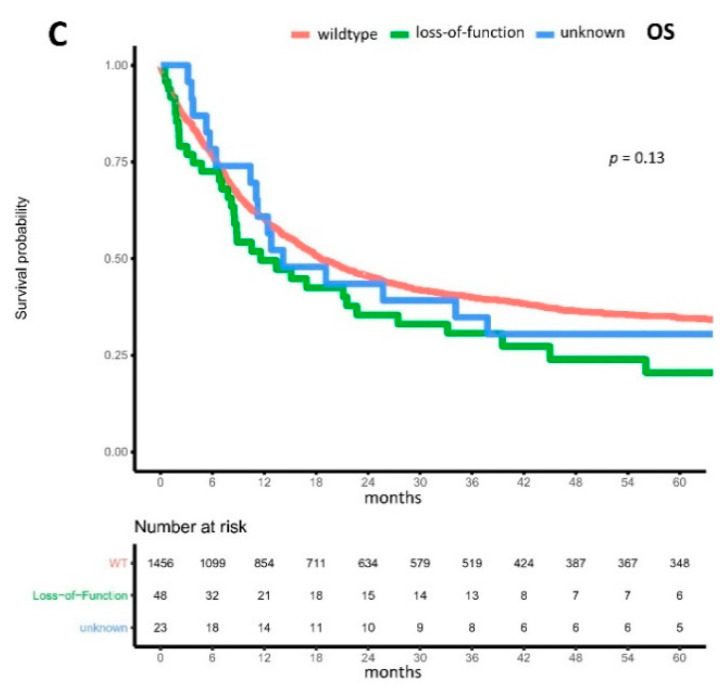
For patients harboring LOF mBCOR, compared to UF mBCOR and wtBCOR, we found in both univariate and multivariable analysis significantly reduced median EFS (LOF mBCOR: 1.9 months (95%-CI: 1.4–8.0) vs. UF variant of mBCOR: 4.9 months (95%-CI: 1.676–n.a.) vs. wild-type BCOR: 7.5 months (95%-CI: 6.8–8.5), multivariable HR of LOF compared to wtBCOR = 1.464 (95%-CI: 1.005–2.134), p = 0.047, Figure 4A) and RFS (LOF mBCOR: 8.8 months (95%-CI: 7.3–24.0) vs. UF variant of mBCOR: 17.7 months (95%-CI: 8.317–n.a.) vs. wild-type BCOR: 18.7 months (95%-CI: 16.3–23.3), multivariable HR of LOF compared to wtBCOR = 1.904 (95%-CI: 1.163–3.117), p = 0.01, Figure 4B). Regarding OS, we found significantly reduced median OS for patients with LOF mBCOR variants in univariate analysis (LOF mBCOR: 11.6 months (95%-CI: 8.5–33.2) vs. UF variant of mBCOR: 14.2 months (95%-CI: 11.045–n.a.) vs. wild-type BCOR: 18.4 months (95%-CI: 16.7–21.4), HR of LOF compared to wtBCOR = 1.409 (95%-CI: 1.010–1.965), p = 0.044), and a strong trend for reduced median OS in a multivariable analysis (HR = 1.495 (95%-CI: 0.990–2.258), p = 0.056, Figure 4C). Additionally, adjusting for co-mutations in DNMT3A, TET2, and RUNX1 did not significantly affect HR. However, in order to precisely account for multiple interactions, an even larger cohort is needed, due to the rarity of LOF BCOR.
Figure 4.
Impact of BCOR loss-of-function mutations on outcomes. Kaplan–Meier plots for (A) event-free survival, (B) relapse-free survival, and (C) overall survival in AML, with mutations of BCOR resulting in a transcriptional stop and loss-of-function (green), BCOR mutations without transcriptional stop resulting in unknown function (blue), and wild-type BCOR (orange) compared using the logrank test.
In AML with mBCORL1 in general compared to wtBCORL1, median EFS (3.6 months (95%-CI: 1.9–9.8) vs. 7.5 months (95%-CI: 6.7–8.3), HR = 1.204 (95%-CI: 0.885–1.639), p = 0.236; Figure 5A), median RFS (14.8 months (95%-CI 7.6–66.6) vs. 18.5 months (95%-CI: 15.9–23.3), HR = 1.263 (95%-CI: 0.84–1.897), p = 0.263; Figure 5B) and median OS (14.9 months (95%-CI: 9.2–27.4) vs. 18.5 months (95%-CI: 16.7–21.4), HR = 1.105 (95%-CI: 0.714–1.71), p = 0.654; Figure 5C) were both lower, although none of these differences were statistically significant. Univariate analysis of LOF mBCORL1 showed significantly reduced EFS (LOF mBCORL1: 3.5 months (95%-CI: 1.1–10.1) vs. UF mBCORL1: 6.2 months (95%-CI: 1.9–not reached) vs. wtBCORL1: 7.5 months (95%-CI: 6.7–8.3), HR = 1.521 (95%-CI: 1.045–2.213), p = 0.028); however, in a multivariable analysis adjusting for age, AML type, and ELN2017 risk, this was not statistically significant.
Figure 5.
Impact of BCORL1 mutations on outcome. Kaplan–Meier plots for (A) event-free survival, (B) relapse-free survival, and (C) overall survival in AML, with wild-type (orange) and mutated (turquoise) BCORL1 compared using the logrank test.
We did not find statistically significant differences in mBCOR and mBCORL1 regarding outcomes in different ELN2017 risk groups. Since few patients with mBCOR and mBCORL1 were in the ELN2017 favorable (n = 9 and n = 6, respectively) or ELN2017 adverse-risk groups (n = 12 and n = 6, respectively), we focused on the ELN2017 intermediate-risk group (n = 44 and n = 35, respectively). In ELN2017 intermediate-risk AML with mBCOR, median EFS and OS did not differ compared to wtBCOR, while there was a trend of lower median RFS in a univariate model (11.9 months (95%-CI: 7.0–24.0) vs. 14.8 months (95%-CI: 12.5–20.3), HR = 1.446 (95%-CI: 0.977–2.139), p = 0.065), as well as in a multivariable model adjusted for age and AML type (HR = 1.637 (95%-CI: 0.995–2.693), p = 0.052). In ELN2017 intermediate-risk AML with mBCORL1, median EFS and RFS did not differ, but there was a trend of lower median OS in a univariate model (14.9 months (95%-CI: 7.3–24.4) vs. 17.0 months (95%-CI: 14.0–20.6), HR = 1.401 (95%-CI: 0.954–2.058), p = 0.086), while in a multivariable model adjusted for age and AML type this difference was not statistically significant. There was no significant association between LOF of mBCOR or mBCORL1 and ELN2017 risk groups regarding outcomes.
4. Discussion
We analyzed a large cohort of newly diagnosed and intensively treated AML patients according to their mutational status of BCOR and BCORL1. The respective proportions of mBCOR and mBCORL1 in the cohort were comparable to those reported in recent studies [6,21,26,27]. We found both mBCOR with LOF and mBCORL1 to be more prevalent in patients in the ELN2017 intermediate-risk group [4]. The proportion of sAML amongst patients harboring mBCOR was significantly higher than in their wild-type counterparts, confirming previous reports [37]. Mutations of BCORL1 were significantly more prevalent in females than in males, as has been previously suggested [27], and patients harboring mBCOR with LOF had significantly lower WBC, and a trend for lower peripheral blood blast counts. Mutations of BCOR frequently co-occurred with mutations of DNMT3A, RUNX1, TET2, NRAS, and BCORL1. Since both BCOR and DNMT3A function as epigenetic modifiers [7,38], a synergistic role in leukemogenesis has been reported recently [39,40]. In murine models, the co-occurrence of mBCOR and mutations of TET2, another epigenetic modifier [41] frequently associated with mBCOR in our cohort, has been reported to induce MDS [25,42]. This may further reinforce the notion that BCOR’s interaction with other DNA methylators plays a crucial role in leukemogenesis. RUNX1 and BCOR both play essential roles in the proliferation and differentiation of myeloid cells [10,43]. Therefore, the interplay of mBCOR and other essential regulators of normal myeloid development appears to be a factor that may promote leukemogenesis. Recent studies suggest, however, that perturbations of BCOR function alone do not suffice to induce malignant transformation [10,21,44], and thus co-mutations appear to be needed in order to drive leukemogenesis or, alternatively, sequential, secondary acquisition of LOF in BCOR following an oncogenic event, e.g., MDS driver mutation triggers disease progression towards AML, as suggested by the higher frequency of sAML among mBCOR patients. Accordingly, in our cohort of patients with mBCOR AML, the majority had at least two co-mutations, while only one patient harbored no other co-mutations targeted by our panel (Figure 1B). Similarly to mBCOR, co-mutations of mBCORL1 were RUNX1, DNMT3A, and TET2, as well as BCOR and FLT3-ITD, which were only rarely co-mutations of mBCOR. Again, the majority of patients harboring mBCORL1 had at least two other co-mutations, and only one patient had no other co-mutations revealed by our panel (Figure 2B). As for mBCOR, this suggests a potential interplay of impaired BCORL1 function with other dysfunctional mechanisms of DNA methylation, cell differentiation, and signal transduction in leukemogenesis.
Regarding outcomes, we found no statistically significant differences between patients with mBCOR and mBCORL1 regarding CR rate and ED30, compared to wild-type patients. Interestingly, while we observed lower median EFS, RFS, and OS for both mBCOR and mBCORL1 in general, only LOF mutations of mBCOR were associated with significantly reduced EFS and RFS and a trend of reduced OS in multivariable testing adjusted for age, AML type, and ELN2017 risk. In MDS, Abuhadra et al. [23] recently reported significantly reduced OS for patients with frameshift mutations of BCOR, while general mutation status did not affect OS. Previous studies of mBCOR in AML have reported poorer outcomes to often be associated with distinct co-mutations; however, a significant association of LOF mutations of BCOR as independent markers of poor outcomes has not yet been reported in AML. Terada et al. [27] reported reduced OS in AML patients who were younger than 65 years, wild-type FLT3, and had intermediate-risk cytogenetics. Grossmann et al. [26] reported a higher prevalence of mBCOR and a trend of reduced OS in normal-karyotype AML with no mutations in NPM1, FLT3-ITD, CEBPA, or MLL-PTD. However, in a validation cohort no association between mBCOR and OS was observed. Nevertheless, in both cohorts reduced EFS was detected [26]. Recently, Eisfeld et al. [45] reported reduced survival for patients in the ELN2017 favorable-risk group who harbored mutations in BCOR or SETBP1. Our findings underline the complexity of the mutational landscape of AML, where even mutational variants of rare mutations have to be considered in order to determine their clinical and prognostic effects. Future work needs to focus on the implementation of LOF mutations of BCOR in risk stratification tools for AML management.
5. Conclusions
In conclusion, both mBCOR and mBCORL1 are rare but recurrent mutations in AML. While previous studies suggested poor outcomes for mBCOR in AML, especially in the context of co-occurring mutations, we found loss-of-function mutations of mBCOR to be independent markers of poor outcomes in AML, while mBCORL1 was not significantly associated with outcomes in multivariable testing.
Acknowledgments
This work was carried out under the auspices of the German SAL. We thank all involved patients, nurses, laboratory technicians, and physicians for their contributions. Open Access Funding by the Publication Fund of the TU Dresden.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13092095/s1, Table S1: Summary of the 54 genes targeted by the TruSight Myeloid Sequencing Panel.
Author Contributions
Conceptualization, J.-N.E., S.S. (Sebastian Stasik), C.T. and J.M.M.; Data curation, M.K.; Formal analysis, J.-N.E., S.S. (Sebastian Stasik), M.K., C.R., A.K., S.S. (Sebastian Scholl), A.H., M.C., T.H.B., R.N. (Ralph Naumann), B.S., V.K., H.E., M.S., A.B., A.N., K.S.-E., C.S., S.W.K., R.H., M.H., N.F., R.N. (Richard Noppeney), U.K., C.D.B., M.K., Z.R., U.P., W.E.B., J.M., H.S., C.M.-T., G.E., F.S., F.K., J.S., M.B., C.T. and J.M.M.; Investigation, J.-N.E., S.S. (Sebastian Stasik), M.K., C.R., A.K., S.S. (Sebastian Scholl), A.H., M.C., T.H.B., R.N.(Ralph Naumann), B.S., V.K., H.E., M.S., A.B., A.N., K.S.E., C.S., S.W.K., R.H., M.H., N.F., R.N. (Richard Noppeney), U.K., C.D.B., M.K., Z.R., U.P., W.E.B., J.M., H.S., C.M.-T., G.E., F.S., F.K., J.S., M.B., C.T. and J.M.M.; Methodology, J.-N.E., S.S. (Sebastian Stasik), M.K., C.T. and J.M.M.; Project administration, M.B., C.T. and J.M.M.; Resources, J.-N.E., S.S. (Sebastian Stasik), M.K., C.R., A.K., S.S. (Sebastian Scholl), A.H., M.C., T.H.B., R.N. (Ralph Naumann), B.S., V.K., H.E., M.S., A.B., A.N., K.S.E., C.S., S.W.K., R.H., M.H., N.F., R.N. (Richard Noppeney), U.K., C.D.B., M.K., Z.R., U.P., W.E.B., J.M.., H.S., C.M.-T., G.E., F.S., F.K., J.S., M.B., C.T. and J.M.M.; Supervision, C.T. and J.M.M.; Validation, J.-N.E., S.S. (Sebastian Stasik), C.T. and J.M.M.; Visualization, J.-N.E. and S.S. (Sebastian Stasik); Writing—original draft, J.-N.E.; Writing—review & editing, S.S. (Sebastian Stasik), M.K., C.R., A.K., S.S. (Sebastian Scholl), A.H., M.C., T.H.B., R.N. (Ralph Naumann), B.S., V.K., H.E., M.S., A.B., A.N., K.S.E., C.S., S.W.K., R.H., M.H., N.F., R.N. (Richard Noppeney), U.K., C.D.B., M.K., Z.R., U.P., W.E.B., J.M., H.S., C.M.-T., G.E., F.S., F.K., J.S., M.B., C.T. and J.M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the guidelines of the Declaration of Helsinki. This study was carried out under the auspices of the German Study Alliance Leukemia (SAL). All mentioned studies were approved by the Institutional Review Board of the Dresden University of Technology (approval number: EK 98032010).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy issues.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Papaemmanuil E., Gerstung M., Bullinger L., Gaidzik V.I., Paschka P., Roberts N.D., Potter N.E., Heuser M., Thol F., Bolli N., et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shumilov E., Flach J., Kohlmann A., Banz Y., Bonadies N., Fiedler M., Pabst T., Bacher U. Current status and trends in the diagnostics of AML and MDS. Blood Rev. 2018;32:508–519. doi: 10.1016/j.blre.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Bullinger L., Döhner K., Döhner H. Genomics of Acute Myeloid Leukemia Diagnosis and Pathways. J. Clin. Oncol. 2017;35:934–946. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- 4.Döhner H., Estey E., Grimwade D., Amadori S., Appelbaum F.R., Büchner T., Dombret H., Ebert B.L., Fenaux P., Larson R.A., et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huynh K.D., Fischle W., Verdin E., Bardwell V.J. BCoR, a novel corepressor involved in BCL-6 repression. Genome Res. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 6.Li M., Collins R., Jiao Y., Ouillette P., Bixby D., Erba H., Vogelstein B., Kinzler K.W., Papadopoulos N., Malek S.N. Somatic mutations in the transcriptional corepressor gene BCORL1 in adult acute myelogenous leukemia. Blood. 2011;118:5914–5917. doi: 10.1182/blood-2011-05-356204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Astolfi A., Fiore M., Melchionda F., Indio V., Bertuccio S.N., Pession A. BCOR involvement in cancer. Epigenomics. 2019;11:835–855. doi: 10.2217/epi-2018-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sparmann A., Van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 9.Simon J.A., Kingston R.E. Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Q., Gearhart M.D., Gery S., Shojaee S., Yang H., Sun H., Lin D.-C., Bai J.-W., Mead M., Zhao Z., et al. BCOR regulates myeloid cell proliferation and differentiation. Leukemia. 2016;30:1155–1165. doi: 10.1038/leu.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gearhart M.D., Corcoran C.M., Wamstad J.A., Bardwell V.J. Polycomb Group and SCF Ubiquitin Ligases Are Found in a Novel BCOR Complex That Is Recruited to BCL6 Targets. Mol. Cell. Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z., Gearhart M.D., Lee Y.-W., Kumar I., Ramazanov B., Zhang Y., Hernandez C., Lu A.Y., Neuenkirchen N., Deng J., et al. A Non-canonical BCOR-PRC1.1 Complex Represses Differentiation Programs in Human ESCs. Cell Stem Cell. 2018;22:235–251.e9. doi: 10.1016/j.stem.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wamstad J.A., Corcoran C.M., Keating A.M., Bardwell V.J. Role of the Transcriptional Corepressor Bcor in Embryonic Stem Cell Differentiation and Early Embryonic Development. PLoS ONE. 2008;3:e2814. doi: 10.1371/journal.pone.0002814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng D., Thakker N., Corcoran C.M., Donnai D., Perveen R., Schneider A., Hadley D.W., Tifft C.J., Zhang L., Wilkie A.O.M., et al. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat. Genet. 2004;36:411–416. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., Benavente C.A., McEvoy J., Flores-Otero J., Ding L., Chen X., Ulyanov A., Wu G., Wilson M.W., Wang J., et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nat. Cell Biol. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pugh T.J., Weeraratne S.D., Archer T.C., Krummel D.A.P., Auclair D., Bochicchio J., Carneiro M.O., Carter S.L., Cibulskis K., Erlich R.L., et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nat. Cell Biol. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierron G., Tirode F., Lucchesi C., Reynaud S., Ballet S., Cohen-Gogo S., Perrin V., Coindre J.-M., Delattre O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat. Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- 18.Totoki Y., Tatsuno K., Yamamoto S., Arai Y., Hosoda F., Ishikawa S., Tsutsumi S., Sonoda K., Totsuka H., Shirakihara T., et al. High-resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 2011;43:464–469. doi: 10.1038/ng.804. [DOI] [PubMed] [Google Scholar]
- 19.Marsh J.C.W., Mufti G. Clinical significance of acquired somatic mutations in aplastic anaemia. Int. J. Hematol. 2016;104:159–167. doi: 10.1007/s12185-016-1972-8. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa S. Clonal hematopoiesis in acquired aplastic anemia. Blood. 2016;128:337–347. doi: 10.1182/blood-2016-01-636381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damm F., Chesnais V., Nagata Y., Yoshida K., Scourzic L., Okuno Y., Itzykson R., Sanada M., Shiraishi Y., Gelsi-Boyer V., et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013;122:3169–3177. doi: 10.1182/blood-2012-11-469619. [DOI] [PubMed] [Google Scholar]
- 22.Steensma D.P. Clinical consequences of clonal hematopoiesis of indeterminate potential. Hematology. 2018;2018:264–269. doi: 10.1182/asheducation-2018.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abuhadra N., Mukherjee S., Al-Issa K., Adema V., Hirsch C.M., Advani A., Przychodzen B., Makhoul A., Awada H., Maciejewski J.P., et al. BCOR and BCORL1 mutations in myelodysplastic syndromes (MDS): Clonal architecture and impact on outcomes. Leuk. Lymphoma. 2019;60:1587–1590. doi: 10.1080/10428194.2018.1543885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montalban-Bravo G., Takahashi K., Patel K., Wang F., Xingzhi S., Nogueras G.M., Huang X., Pierola A.A., Jabbour E., Colla S., et al. Impact of the number of mutations in survival and response outcomes to hypomethylating agents in patients with myelodysplastic syndromes or myelodysplastic/myeloproliferative neoplasms. Oncotarget. 2018;9:9714–9727. doi: 10.18632/oncotarget.23882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tara S., Isshiki Y., Nakajima-Takagi Y., Oshima M., Aoyama K., Tanaka T., Shinoda D., Koide S., Saraya A., Miyagi S., et al. Bcor insufficiency promotes initiation and progression of myelodysplastic syndrome. Blood. 2018;132:2470–2483. doi: 10.1182/blood-2018-01-827964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossmann V., Tiacci E., Holmes A.B., Kohlmann A., Martelli M.P., Kern W., Spanhol-Rosseto A., Klein H.-U., Dugas M., Schindela S., et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood. 2011;118:6153–6163. doi: 10.1182/blood-2011-07-365320. [DOI] [PubMed] [Google Scholar]
- 27.Terada K., Yamaguchi H., Ueki T., Usuki K., Kobayashi Y., Tajika K., Gomi S., Kurosawa S., Saito R., Furuta Y., et al. Usefulness ofBCORgene mutation as a prognostic factor in acute myeloid leukemia with intermediate cytogenetic prognosis. Genes Chromosom. Cancer. 2018;57:401–408. doi: 10.1002/gcc.22542. [DOI] [PubMed] [Google Scholar]
- 28.De Rooij J.D., Heuvel-Eibrink M.M.V.D., Hermkens M.C., Verboon L.J., Arentsen-Peters S.T.C.J.M., Fornerod M., Baruchel A., Stary J., Reinhardt D., De Haas V., et al. BCOR and BCORL1 mutations in pediatric acute myeloid leukemia. Hematology. 2015;100:e194–e195. doi: 10.3324/haematol.2014.117796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 30.Röllig C., Thiede C., Gramatzki M., Aulitzky W., Bodenstein H., Bornhäuser M., Platzbecker U., Stuhlmann R., Schuler U., Soucek S., et al. A novel prognostic model in elderly patients with acute myeloid leukemia: Results of 909 patients entered into the prospective AML96 trial. Blood. 2010;116:971–978. doi: 10.1182/blood-2010-01-267302. [DOI] [PubMed] [Google Scholar]
- 31.Schaich M., Parmentier S., Kramer M., Illmer T., Stölzel F., Röllig C., Thiede C., Hänel M., Schäfer-Eckart K., Aulitzky W., et al. High-Dose Cytarabine Consolidation with or Without Additional Amsacrine and Mitoxantrone in Acute Myeloid Leukemia: Results of the Prospective Randomized AML2003 Trial. J. Clin. Oncol. 2013;31:2094–2102. doi: 10.1200/JCO.2012.46.4743. [DOI] [PubMed] [Google Scholar]
- 32.Röllig C., Kramer M., Gabrecht M., Hänel M., Herbst R., Kaiser U., Schmitz N., Kullmer J., Fetscher S., Link H., et al. Intermediate-dose cytarabine plus mitoxantrone versus standard-dose cytarabine plus daunorubicin for acute myeloid leukemia in elderly patients. Ann. Oncol. 2018;29:973–978. doi: 10.1093/annonc/mdy030. [DOI] [PubMed] [Google Scholar]
- 33.Röllig C., Serve H., Hüttmann A., Noppeney R., Müller-Tidow C., Krug U., Baldus C.D., Brandts C.H., Kunzmann V., Einsele H., et al. Addition of sorafenib versus placebo to standard therapy in patients aged 60 years or younger with newly diagnosed acute myeloid leukaemia (SORAML): A multicentre, phase 2, randomised controlled trial. Lancet Oncol. 2015;16:1691–1699. doi: 10.1016/S1470-2045(15)00362-9. [DOI] [PubMed] [Google Scholar]
- 34.Gebhard C., Glatz D., Schwarzfischer L., Wimmer J., Stasik S., Nuetzel M., Heudobler D., Andreesen R., Ehninger G., Thiede C., et al. Profiling of aberrant DNA methylation in acute myeloid leukemia reveals subclasses of CG-rich regions with epigenetic or genetic association. Leukemia. 2019;33:26–36. doi: 10.1038/s41375-018-0165-2. [DOI] [PubMed] [Google Scholar]
- 35.Stasik S., Schuster C., Ortlepp C., Platzbecker U., Bornhauser M., Schetelig J., Ehninger G., Folprecht G., Thiede C. An optimized targeted Next-Generation Sequencing approach for sensitive detection of single nucleotide variants. Biomol. Detect. Quantif. 2018;15:6–12. doi: 10.1016/j.bdq.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;76:7–20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nazha A., Zarzour A., Al-Issa K., Radivoyevitch T., Carraway H.E., Hirsch C.M., Przychodzen B., Patel B.J., Clemente M., Sanikommu S.R., et al. The complexity of interpreting genomic data in patients with acute myeloid leukemia. Blood Cancer J. 2016;6:e510. doi: 10.1038/bcj.2016.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ley T.J., Ding L., Walter M.J., McLellan M.D., Lamprecht T.L., Larson D.E., Kandoth C., Payton J.E., Baty J., Welch J.J., et al. DNMT3AMutations in Acute Myeloid Leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sportoletti P., Sorcini D., Guzman A.G., Reyes J.M., Stella A., Marra A., Sartori S., Brunetti L., Rossi R., Del Papa B., et al. Bcor deficiency perturbs erythro-megakaryopoiesis and cooperates with Dnmt3a loss in acute erythroid leukemia onset in mice. Leukemia. 2020:1–15. doi: 10.1038/s41375-020-01075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M., Yang C., Zhang L., Schaar D.G. Molecular Mutations and Their Cooccurrences in Cytogenetically Normal Acute Myeloid Leukemia. Stem Cells Int. 2017;2017:6962379. doi: 10.1155/2017/6962379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito S., D’Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nat. Cell Biol. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malcovati L., Papaemmanuil E., Ambaglio I., Elena C., Gallì A., Della Porta M.G., Travaglino E., Pietra D., Pascutto C., Ubezio M., et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood. 2014;124:1513–1521. doi: 10.1182/blood-2014-03-560227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ichikawa M., Yoshimi A., Nakagawa M., Nishimoto N., Watanabe-Okochi N., Kurokawa M. A role for RUNX1 in hematopoiesis and myeloid leukemia. Int. J. Hematol. 2013;97:726–734. doi: 10.1007/s12185-013-1347-3. [DOI] [PubMed] [Google Scholar]
- 44.Kelly M.J., So J., Rogers A.J., Gregory G., Li J., Zethoven M., Gearhart M.D., Bardwell V.J., Johnstone R.W., Vervoort S.J., et al. Bcor loss perturbs myeloid differentiation and promotes leukaemogenesis. Nat. Commun. 2019;10:1–14. doi: 10.1038/s41467-019-09250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eisfeld A.-K., Kohlschmidt J., Mims A., Nicolet D., Walker C.J., Blachly J.S., Carroll A.J., Papaioannou D., Kolitz J.E., Powell B.E., et al. Additional gene mutations may refine the 2017 European LeukemiaNet classification in adult patients with de novo acute myeloid leukemia aged <60 Years. Leukemia. 2020;34:3215–3227. doi: 10.1038/s41375-020-0872-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy issues.