Abstract
Circadian clocks temporally coordinate physiology and align it with geophysical time, enabling diverse lifeforms to anticipate daily environmental cycles. In complex organisms, clock function originates from the molecular oscillator within each cell and builds upwards anatomically into an organism-wide system. Recent advances have transformed our understanding of how clocks are connected to achieve coherence across tissues. Circadian misalignment, often imposed in modern society, disrupts coordination among clocks and has been linked to diseases from metabolic syndrome to cancer. Thus, uncovering the physiological circuits whereby biological clocks achieve coherence will inform on both challenges and opportunities in human health.
Summary sentence:
In complex organisms, circadian rhythms are the manifestation of communicating cell and tissue clocks.
Introduction
Life has evolved as the Earth rotates around its axis, establishing light-dark as a perpetual cycle. As a result, biological mechanisms that monitor geophysical time are conserved across species. In fact, all photosensitive life forms harbor a timed molecular oscillator, or clock, which emerged in cyanobacteria ~2.1 billion years ago. Around that time, a day on Earth was ~6 hr shorter, thus these prokaryotes likely exhibited rhythms with an 18 hr period (1). Over time, as the Moon slowly drifted away from Earth, days lengthened to 24 hr and species including mammals emerged with the ~24 hr circadian rhythms that are recognized today.
Simple observation brought forth the idea that circadian rhythms are intrinsically generated and therefore genetically encoded. The daily opening and closing of Mimosa pudica leaves persists when the plant is sealed in a photo-impenetrable box. Moreover, circadian wheel running activity of rodents continues when they are placed in constant darkness. Similarly, humans in isolation units, devoid of environmental influences, maintain rhythmicity (2). This intrinsic ability reveals that organisms not only react to timed environmental cues, but also anticipate them. The prevailing opinions for how clock-generated anticipation imparts a selective fitness include energetic efficiency, temporal separation of anabolic and catabolic processes and the coordination of damaging and reparative cellular events. In humans, circadian alignment is associated with health, and misalignment with the propensity for disease.
In prokaryotes, time keeping is a task for a single cell directly exposed to the environment, and synchronization of intracellular processes to geophysical time, referred to as entrainment, is directly achieved. For complex multicellular organisms, wherein tissues reside internally, timing must be coordinated across the organism. Interestingly, the peripheral organs of some vertebrates entrain directly to light, as is the case in zebrafish (3). In mammals, however, most cells lack photosensitivity and need specialized circuits to communicate circadian information.
Experiments in rodents pinpointed the suprachiasmatic nucleus (SCN) as the anatomical structure of the central circadian clock (4). The subsequent discovery that mammalian core clock genes are expressed in most cells opened the possibility that all cells and tissues harbor their own clocks. The development of genetic tools for live imaging of clock proteins and tissue-specific clock disruption enabled the findings that peripheral tissues have clock functionality (5–7). Here, we review recent progress in our understanding of how cell and tissue clocks communicate in mammals, with reference to others species where appropriate.
Clock oscillators: from cells to organism
The core molecular oscillator is a transcription-translation feedback loop (Figure 1). The activators circadian locomotor output cycles protein kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1) heterodimerize to recognize E-box motifs and regulate the expression of thousands of genes, named clock-controlled genes (CCGs) or output genes (8). Among these, the repressors period (Per) and cryptochrome (Cry) are translated and assemble into repressor complexes that inhibit CLOCK:BMAL1 function. The activity and proteasomal degradation of repressor complexes is tightly controlled by posttranslational modifications, and its clearance alleviates CLOCK:BMAL1 inhibition for the start of another cycle (9). Interlocking of this primary loop with additional activator-repressor pairs, for example nuclear receptor subfamily 1 group D (REV-ERB) and RAR-related orphan receptor (ROR), allows for differentially phased cycles of transcription according to the distribution of dedicated motifs, including E-box, D-box and ROR elements, across the genome (10). These interconnected loops, which exhibit inherent ~24 hr periodicity, rely on nucleosome dynamics, long-range promoter-enhancer interactions and a variety of co-factors and chromatin remodelers (10, 11).
Figure 1. Extracellular signals and the molecular clock.
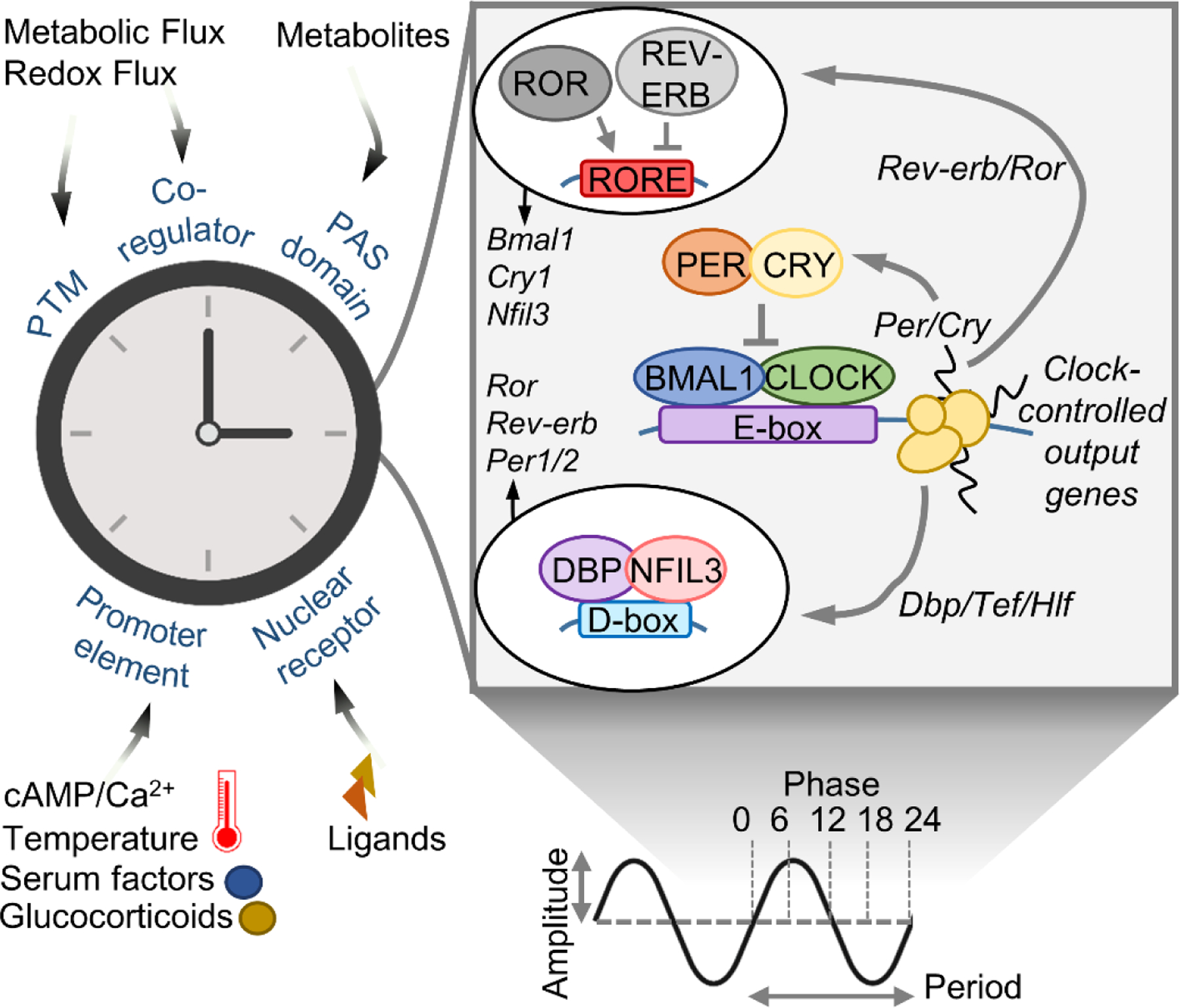
Simplified scheme of the molecular clock/oscillator. Right – the core transcriptional activators circadian locomotor output cycles protein kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1), together with the repressors period (PER) and cryptochrome (CRY) and interlocked auxiliary loops (regulating D-boxes and ROR elements), control a near 24-hr cycle of each other’s expression as well as clock controlled output genes (CCGs). Left – extracellular signals that engage the clock through various means to set period, phase, amplitude and output. PTM – posttranslational modification; cAMP – cyclic adenosine monophosphate; PAS – Per-Arnt-Sim; ROR –RAR-related orphan receptor; REV-ERB – nuclear receptor subfamily 1 group D; DBP – albumin D-box binding protein; Tef – thyrotroph embryonic factor; Hlf – hepatic leukemia factor; NFIL3 – nuclear factor, interleukin 3 regulated.
Sensing and responding to the extracellular environment is a built-in feature of the clock that shapes its period, phase, amplitude and output (Figure 1). CLOCK, BMAL1 and PER proteins contain per-ARNT-sim (PAS) domains, structures that respond to metabolites and xenobiotics (12, 13). The nuclear receptors REV-ERB and ROR are activated by hormonal ligands and metabolites such as heme and sterols (14, 15). Clock and clock-controlled gene promoters contain DNA elements that integrate intracellular signals resulting from variations in calcium, cyclic adenosine monophosphate (cAMP), growth factors, temperature and hormones (16–20). Additional layers of regulation arise from various posttranslational modifications on clock proteins, as well as interaction with nutrient- and redox-responsive co-regulators (21–25).
The self-sustaining nature of the clock is conceptually remarkable, allowing individual cells to function as cell-autonomous oscillators. Single-cell imaging of Per2-luciferase in low density cultures shows that dispersed neurons and fibroblasts have a wide range of periods and randomly distributed phases, evidence that each oscillator is independent from the next (26, 27). Increasing cell density, however, increases the number of rhythmic cells and induces weak local coupling of period and phase between neighboring cells (28, 29). Thus, as tonic inputs from the population reach a critical level, a collective strengthening of rhythmicity leads to enhanced synchrony.
The combined regulation of clock inputs and outputs forms the basis for coordination and reinforcement between individual oscillators, i.e. coupling. Whereas synchronization of cells aligns phases and resets periods, coupling engenders a population rhythm that is more precise, more robust and more resilient than its constitutive cells (30). Both SCN and peripheral tissue explants remain synchronized longer than their dispersed cells, illustrating coupling of individual oscillators (6, 27, 31). Owing to neuronal circuitry, SCN explants oscillate in a manner that appears to perpetuate indefinitely. By contrast, cells of peripheral tissues desynchronize over time, presumably from a lack of reinforcing synaptic mechanisms, rendering them inherently less coupled (6). Desynchronizing explants show tissue-specificity in oscillator uncoupling; cells of bone and tendon mostly lose phase coherence over time, yet in heart tissue this is accompanied by loss of amplitude of individual cells (32, 33).
Coupling oscillators in the brain
The brain receives photic information through the retino-hypothalamic tract, axonal projections of melanopsin-expressing, intrinsically photosensitive retinal ganglion cells (ipRGCs) that relay non-image-forming light signals from the eye to the SCN. Each day, internal time is aligned with external time through the photic resetting of the SCN clock. Acute depolarization by light induces synaptic mechanisms that lead to the activation of a transcriptional program involving cAMP-responsive element-binding protein (CREB)-mediated induction of immediate early genes, clock genes such as Per1 and Per2 (34) and chromatin remodeling (35).
Coupling of SCN neurons has been extensively studied (36). Emerging evidence indicates that the clock circuitry connects astrocytes and neurons (Figure 2). Astrocytes harbor their own distinct molecular clock and pattern of intracellular calcium trafficking, which oscillate in anti-phase to neurons, peaking during the inactive phase (37). Astrocytes constrain the period of the SCN because their endogenous period is several hours shorter than neurons (37, 38). Remarkably, restoring Cry1 expression specifically in astrocytes of Cry1-null mice reinstates molecular and behavioral rhythms, revealing that astrocytes are sufficient time-keeping (39). The neurotransmitter glutamate couples the clock across the two cell types (37, 39). Blocking Connexin 43 hemichannels – and consequently astrocytic release of glutamate – results in desynchronized SCN slices (39). A scenario has emerged whereby glutamate acts rhythmically on pre-synaptic neurons through N-methyl-D-aspartate (NMDA) receptors to depolarize γ-aminobutyric acid-specific (GABAergic) post-synaptic neurons and engage the clock through CREB-mediated activation of Per2 (37). Extra-synaptic GABAA receptors and astrocytic GABA transporter 3 (GAT3) also contribute to this interplay. A 2h GABA pulse can entrain cultured neurons through the GABAA receptor, and GABAergic antagonists rescue behavior of mice lacking astrocytic Bmal1 (40). How coupling signals are relayed to the clock in astrocytes is unclear, but in vitro experiments show the neurotransmitters glutamate, serotonin and dopamine induce Per1 gene expression in astrocytes through extracellular regulated kinase (ERK) signaling (41).
Figure 2. Coupling of clocks in neurons and astrocytes.
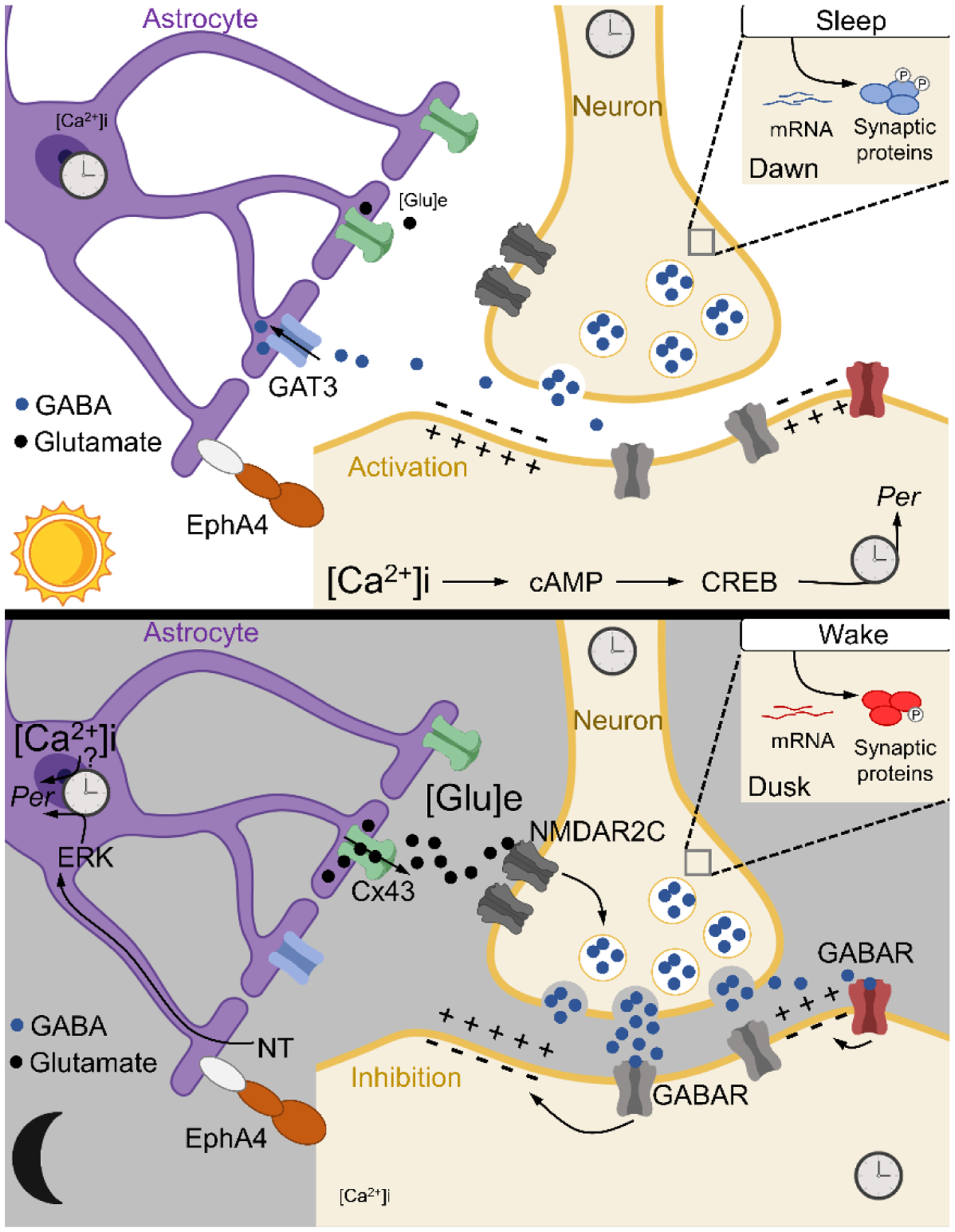
Astrocytes harbor autonomous clocks that operate in anti-phase to neurons. During daytime, astrocytic glutamate release is suppressed, which inhibits presynaptic terminals to alleviate GABA, stimulating electrical activity and Ca2+-induced CREB activation leading to Per2 gene expression. At nighttime, astrocytes release glutamate, which acts on presynaptic terminals to enhance GABA, in effect inhibiting electrical activity and Per2. Regulation of Per2 in this manner links rhythmic astrocytic signals to the clock in neurons. Other components implicated in this process are GAT3, an astrocytic GABA transporter, and EphA4, an adhesion molecule that physically connects astrocytes and neurons and the connexin 43 (Cx43) hemichannel. Upper right inlet – the sleep-wake cycle drives oscillation of synaptic proteins and their phosphorylation, another layer of circadian regulation. GABA – γ-aminobutyric acid; cAMP – cyclic adenosine monophosphate; CREB – cAMP response element-binding protein; Per2 – period 2; GAT3 – GABA transporter type 3; EphA4 – EPH receptor A4; Glu – glutamate; ERK – extracellular signal-regulated kinase; NT – neurotransmitter; NMDAR2C – N-methyl-D-aspartate receptor 2 c; GABAR – GABA receptor.
Coupling of peripheral oscillators
Not yet fully explored, coupling mechanisms in non-neural tissues have been described in plants and animals (42, 43). In mice, the cochlear clock is anatomically divided by regions. Per2 expression initiates from the hair cell and spiral ganglion neurons of the apex and travels toward the base (42), effectively creating clock ‘regions’ with sequential phases, positioned along the length of the cochlear axis. These discrete regions are maintained when action potentials or gap junctions are inhibited, but lost by blockade of voltage-gated potassium channels or extracellular calcium, pointing to non-neuronal coupling mechanisms (42). This tonotopical arrangement of cochlear clock may mediate time-dependent sensitivity to different auditory frequencies.
Fibroblast and neuronal rhythms depend on membrane potential and intracellular calcium, signals that can be shared across cells through gap junctions (44), which are debated as universal clock couplers. One such mechanism exists in Drosophila melanogaster – cycling Innexin (INX) gap junctions drive a rhythm of blood brain barrier permeability by regulating the flow of Mg2+ ions between glial cell layers. As a result, nighttime (compared to daytime) administration of the anti-epileptic drug phenytoin promotes accumulation in the brain and is more efficacious (45). This gating of access to the brain, if conserved in mammals, is highly relevant to the timing of drug delivery for neurological disorders. In other tissue-contexts, physical contacts between cells are often dispensable for rhythms, and released factors appear to be more dominant signals for coupling (46–48).
Organ to system level clock function
Why does the behavior of individual peripheral oscillators change in tissues? Experiments in vitro that reconstruct tissue-like environments implicate the cell cycle, extracellular matrix (ECM) and diversity of local cell types (49, 50). For instance, inhibiting the cell cycle increases coherence of fibroblast oscillations in vitro. Hepatocytes cultured on ECM components have more robust clock gene rhythms that, over time, lose amplitude rather than their phase relationships (synchrony) with other cells. Pancreatic islets contain several cell types that release locally acting entrainment factors (31, 48, 51). In the liver, single nucleus sequencing experiments demonstrate that cellular rhythms can arise from communication between different cell types. In adult mice, hepatocyte-specific knockout of REV-ERBα and REV-ERBβ, and in effect clock function, reprograms rhythmic features of neighboring endothelial and Kupffer cells, including enhancers, transcripts and metabolites, despite an intact clock in those cells (52). Bioinformatic analysis identifies intercellular ligand-receptor pairs that might enable the passage of circadian signals from one cell to the next, perhaps to integrate cell-type-specific functional niches.
Coupling of cellular oscillators constitutes a tissue clock, likewise coupled tissue oscillators constitute an organismal clock. Local clocks in peripheral tissues are coupled to central clocks in the brain. Lesions that ablate the SCN, or genetic deletion of the SCN clock, attenuates amplitudes and alters phases of the liver, heart, kidney, white adipose tissue, pancreas, adrenal gland and epidermis (53–58). Casein kinase 1 delta and epsilon (CK1δ, ε) mutations, introduced specifically into GABA neurons of mice, induce an abnormally long circadian period in the brain to which peripheral tissues entrain despite harboring an endogenous 24 hr period (59). Coupling to the SCN has also been demonstrated by grafting fibroblasts in the mouse. Remarkably, cells with ablation of Per1 and thereby a defective clock, readily gain normal rhythmicity when grafted in a wild type mouse (60).
Peripheral clocks may connect to each other independently of the central SCN pacemaker. Intriguingly, under constant light conditions, brain clocks and locomotor activity are arrhythmic, yet peripheral clock oscillations can persist – a finding suggestive of coupling between peripheral tissues (61). Supporting this idea, Liver-specific deletion of Bmal1 subtly increases the amplitude of clock gene expression in muscle (7) and streptozoticin-induced diabetic mice, which lack pancreatic β cell clocks and rhythmic production of insulin, have livers that are slightly phase advanced and slightly less robust (49). These results suggest coupling to a certain extent, but may also be indicative of systemic metabolic communication. High-throughput metabolomics has revealed a staggering number of metabolites oscillating in serum, underscoring the multitude of options for the spreading of molecular information (62). Loss of circadian coherence among serum metabolites may contribute to certain pathological conditions. For example, in a genetic mouse model of lung adenocarcinoma, distal tumors impose a profound rewiring of hepatic transcriptional and metabolic programs (63).
Synchrony and Homeostasis
In 1849, physiologist Claude Bernard defined homeostasis as a state of steady chemical and physical equilibrium, essential for maintenance of living organisms as well as their capacity of adaptation to a changing environment (64). Accordingly, homeostasis is based on the coordinated balance among tissues and organs, to insure systemic control of physiology. In this context, the discovery that clocks are present in all vertebrate tissues (5, 6, 48, 65), effectively generating a circadian network, provided evidence that circadian rhythms represent the quintessential example of homeostatic control (Figure 3 and Table 1) (66, 67). Consistent with homeostatic failure as a basis of many diseases (22, 63), clock malfunctioning is linked to cardiovascular disorders, metabolic perturbations, neurological disorders and increased risk of cancer (54, 55, 68).
Figure 3. Connections of the mammalian clock system.
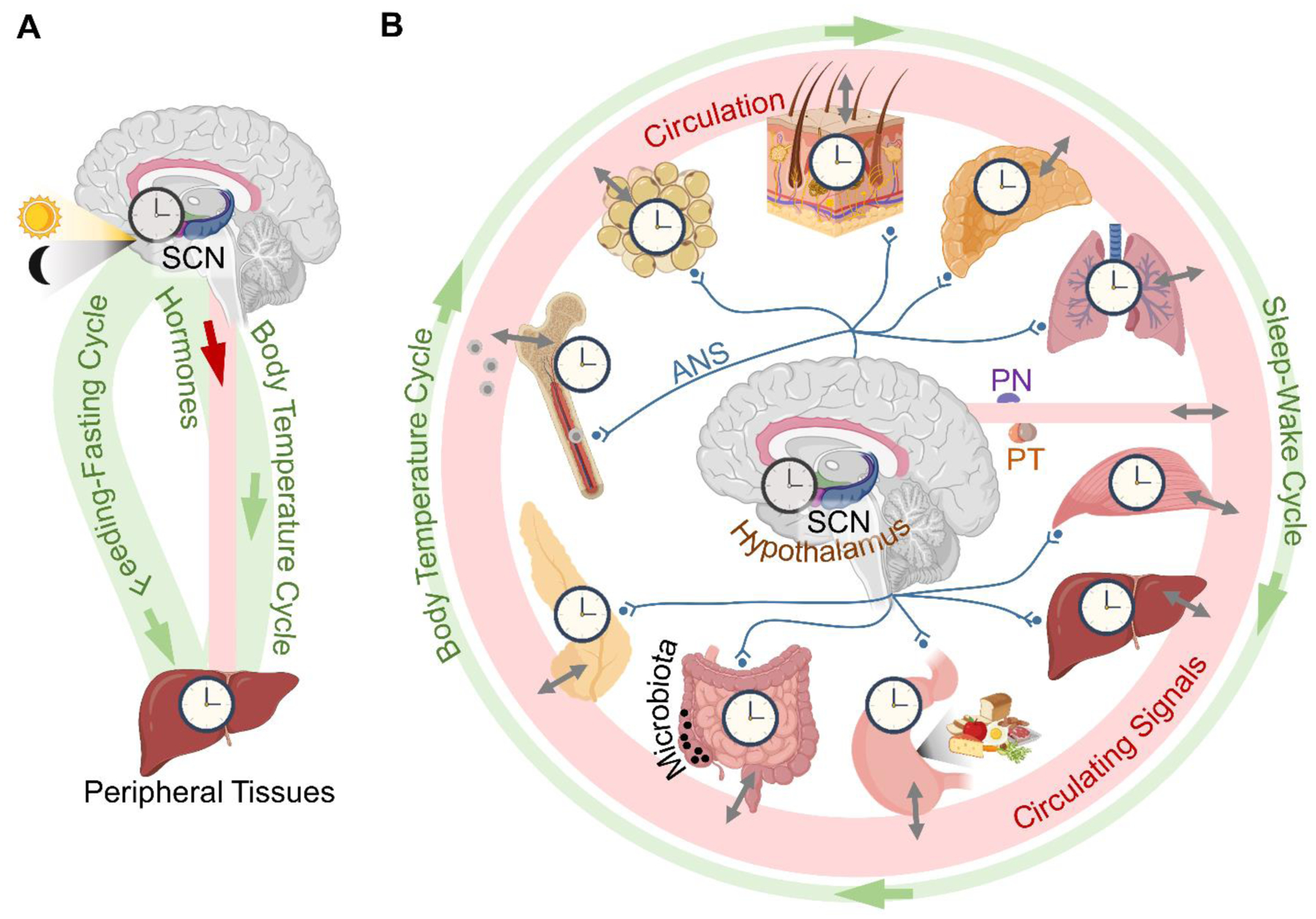
(A) A conceptual model based on knowledge from the turn of the century. (B) A current model of the system. In addition to the suprachiasmatic nucleus (SCN), other distinct hypothalamic clocks control daily energy homeostasis. The autonomic nervous system (ANS) differentially innervates peripheral tissues and modulates their clocks by adjusting sympathetic tone. All clocks are connected via the circulatory system by an ever-growing list of synchronizing factors, many of which are under local circadian control in peripheral tissues. See also Table 1 for circadian factors, their signaling pathways and clock targets within the system. PN – pineal gland; PT – pituitary gland.
Table 1. Communicating signals within the mammalian clock network.
Question mark indicates an undefined mechanism or target.
| Tissue | Signal | Mediator | Clock Target | Refs |
|---|---|---|---|---|
| Brain | Hypothalamic Neurotransmitters and Neuropeptides | Synaptic receptors, cAMP, PKA, CREB, ERK, PRDX, mTOR, Fos, Jun, DUSP4, NMDAR2C, GABAAR | Per, Cry | 36, 40 |
| Autonomic Nervous System | adrenergic, cholinergic receptors, MAPK, PKA | Per1 | 72 | |
| Sleep-Wake, Feed-Fast Cycles | Metabolic hormones, Nutrient flux, Coupled energy sensors (AMPK, SIRTs, PGClα), CO2, O2 | CLOCK:BMAL1, PER2, REV-ERBα | 22 | |
| Temperature Cycle | HSF1 | PER2, Per2 | 74, 75 | |
| Pituitary Gland | Vasopressin | AVPR1–3, CREB | Per1–2 | 66 |
| Pineal Gland | Melatonin | PKC | Per1–2 | 66 |
| Thyroid | Thyroxine; Triiodothyronine | Thyroid hormone receptor | ? | 66 |
| Adrenal Gland | Corticosterone | Glucocorticoid Receptor | CRY1–2, CLOCK, Clock gene GREs | 66 |
| Aldosterone | Mineralocorticoid receptor | Clock gene GREs | 66 | |
| Lung | O2 | HIF1α | CRY1, PER2, BMAL1 | 25 |
| CO2 | ? | ? | 76, 77 | |
| Adipose | Leptin | Leptin receptor | ? | 105, 106 |
| Adiponectin | AdipoR1–2 | ? | 82 | |
| Liver | FGF21 | β-Klotho | ? | 87 |
| Angptl8 | PirB, NF-κB, p38, Akt | Per1 | 83 | |
| IGF-1 | GSK-3β | Bmal1 | 99 | |
| RBP4 | ? | ? | 86 | |
| Skeletal Muscle | IL-6, IL-8, MCP-1 | ? | ? | 65 |
| Pancreas | Insulin | PTEN, mTORC1, Akt, miR24, 29, 30 | BMAL1 phosphorylation, PER, Per | 89, 48 |
| Glucagon | adenylate cyclase | ? | 48, 94 | |
| Somatostatin | SSTR2–3 | ? | 94 | |
| GLP-1 | GLP1R | ? | 94 | |
| GI Tract | Oxyntomodulin | Glucagon Receptor, CREB | Per | 85 |
| Ghrelin | GHSR | ? | 84 |
cAMP - cyclic adenosine monophosphate; PKA - protein kinase A; CREB - cAMP response element-binding protein; ERK - extracellular signal-regulated kinase; PRDX - peroxiredoxin; mTOR - mammalian target of rapamycin; DUSP4 - dual specificity protein phosphatase 4; N-methyl-D-aspartate receptor 2C; gamma-aminobutyric acid A receptor; Per - period; Cry - cryptochrome; MAPK - mitogen-activated protein kinase; AMPK - 5’ adenosine monophosphate-activated kinase; SIRT - sirtuin; PGC1α - peroxisome proliferator-activated receptor gamma coactivator 1-alpha; CLOCK - circadian locomotor output cycles kaput; BMAL1 - brain and muscle ARNT-Like 1; REV-ERBα - nuclear receptor subfamily 1, group D, member 1; HSF1 - heat shock factor 1; MRTF - myocardin-related transcription factor; PER2 - period 2; AVPR - arginine vasopressin receptor; PKC - protein kinase C; GRE - glucocorticoid response element; HIF1α - hypoxia-inducible factor 1-alpha; CRY1 - cryptochrome 1; AdipoR - adiponectin receptor; FGF21 - fibroblast growth factor 21; Angptl8 - angiopoietin-related protein 8; PirB - murine paired immunoglobulin receptor B; NF-κB - nuclear factor kappa-light-chain-enhancer of activated B cells; Akt - protein kinase B; IGF-1 - insulin-like growth factor 1; GSK-3β - glycogen synthase kinase 3 beta; RBP4 - retinol binding protein 4; IL-6 - interleukin 6; IL-8 - interleukin 8; CCL2 - chemokine (C-C motif) ligand 2; PTEN - phosphatase and tensin homolog; mTORC1 - mTOR complex 1; SSTR - somatostatin receptor; GLP-1 - glucagon-like peptide-1; GLP1R - GLP1 receptor; GHSR - growth hormone secretagogue receptor; SCFA - shot chain fatty acid; HDAC3 - histone deacetylase 3; STAT3 - signal transduction and activator of transcription 3; ILC3 - type 3 innate lymphoid cells; NFIL3 - nuclear factor interleukin 3 regulated.
Specialized circuitry senses and relays circadian information in the brain and body. The SCN clock is able to keep time in the absence of external timing cues, as supported by seminal SCN-lesion experiments in rodents. Recently, formal demonstration of this ability has been made possible by genetic disruption of clock function, which leaves the anatomical structure of the SCN intact. Without Bmal1 in the SCN, peripheral clocks remain synchronized under light-dark conditions, and only upon release into constant darkness do they gradually desynchronize (53–55).
Interactions among hypothalamic neurons pass circadian signals from the SCN to distal regions of the brain and on to peripheral organs. Per2 expression follows dense projections from the SCN core region to the shell region, with modulation by a minority of local neuropeptide-expressing neurons (34). The shell, and core to a lesser extent, innervate and receive innervation from other hypothalamic nuclei with roles in systemic metabolic homeostasis and their own functionally distinct clocks, including the ventromedial hypothalamus (VMH), dorsomedial hypothalamus (DMH) and arcuate nucleus (ARC) (69–71). For example, VMH Bmal1 controls daily thermogenesis in brown adipose tissue via adrenergic, sympathetic neuronal connections (71). Rhythmic secreted factors and neuronal activity from the hypothalamus further act on the pineal and pituitary neuroendocrine centers to coordinate 24 hr systemic homeostasis (72).
Systemic rhythms blanket clocks
Autonomic nervous system efferents, circulating endocrine factors and the sleep-wake and feeding-fasting cycles are entrainment cues for peripheral tissues. Experimental joining of the bloodstreams of SCN-lesioned and intact mice rescues clock gene oscillations in the liver and kidney of the arrhythmic host. There is no rescue for skeletal muscle, heart or spleen, implicating blood-borne signals for the entrainment of some tissues and neural pathways for others (73). The sleep-wake cycle sets whole-organism-level systemic rhythms. Mammalian body temperature fluctuates just 2–3°C over the circadian cycle, yet this small oscillation is sufficient to synchronize clock gene expression and phase shift clocks in a heat shock factor (HSF)-dependent manner (74). In the mouse, these responses are mediated transcriptionally, via heat shock responsive elements in the Per2 gene promoter, as well as post-transcriptionally, as PER2 function can be driven by systemic cues in the absence of other clock components (75). Locomotor activity cycles drive daily tissue oxygenation; varying O2 levels within the physiological range synchronizes muscle cells through hypoxia-inducible factor 1 alpha (HIF1α), an oxygen sensor coupled to the clock (25, 76, 77). Periods of feeding and fasting generate a rhythm of CO2, another blood gas which modulates clock phase (76). Feeding and fasting also drive a general rhythm of nutrient flux that has pronounced effects on the clock through coupled energy and redox sensors (21, 78). Engaged simultaneously, systemic cues recruit and align peripheral clocks.
The influence of behavioral cycles is not restricted to the periphery but dictates brain rhythms as well. In forebrain synapses, while the temporal accumulation of mRNAs is unaffected, constantly high sleep pressure over the circadian cycle disrupts rhythmic protein accumulation and phosphorylation (79, 80). Effectively then, neuronal resident clocks control the priming of synaptic terminals with mRNA, while the sleep-wake cycle drives translation of proteins and their functional status.
Peripherally-derived timing cues: layers of synchrony
Peripheral clocks govern additional layers of control which parallel ‘top-down’ regulation. The number of bona fide entrainment factors that emanate from peripheral tissues has grown substantially (Table 1). Ex vivo experiments demonstrate the cell- and tissue-autonomous, rhythmic secretion of insulin and glucagon from pancreatic β and α cells, various myokines from human skeletal myotubes, and glucocorticoids from the adrenals (48, 65, 81). Other organs that secrete circadian factors include the gut, stomach, liver and adipose tissue (82–85) (Figure 3).
In vivo, many factors have been implicated in the entrainment to feeding time. Both familiar and additional signal transduction pathways that converge on the clock have been found (86, 87). The hepatokine angiopoietin-related protein 8 (ANGPTL8) and its receptor paired immunoglobulin-like receptor (PIRB) trigger a phosphorylation cascade involving mitogen-activated protein kinase (MAPK), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-ΚB) and protein kinase B (AKT) that induces Per1 gene expression (83). Downstream signaling of glucagon and gut-secreted oxyntomodulin is reminiscent of the SCN, wherein cAMP signaling activates CREB at clock gene promoters (85). The growth and metabolism integrator mammalian target of rapamycin (mTOR) has emerged as a ubiquitous means to signal to the clock. mTOR complexes contribute to photic entrainment and synchronization of the SCN. Outside the brain, mTOR activation shortens the period and enhances amplitude of hepatocytes and adipocytes, and alters clock protein levels in fibroblasts (88). In the liver, mTOR complex 1 is involved in translating pre-existing Per2 mRNA (89).
Organism-wide homeostasis requires the proper timing and output of a network of clocks, a concept not limited to metabolism. One example is the homing of leukocytes. At the onset of light, the SCN activates sympathetic nerves of the bone marrow, releasing into the blood hematopoietic stem cells that give rise to circulating leukocytes (90). The clocks in leukocytes, synchronized by systemic redox flux, time the cell surface presentation of migratory factors whilst the clock in the endothelium enables the leukocyte adhesion cascade by timed-presentation of the cell surface homing factors vascular cell adhesion protein 1 (VCAM-1) and intercellular adhesion molecule 1 (ICAM-1) (91, 92). As a result, immigration of leukocytes into target tissues proceeds at the onset of dark until blood count falls and bone marrow mechanisms are reset. The extent to which innate and adaptive immunity are systemically regulated by the clock system is gaining appreciation (93).
How synchrony within peripheral tissues is established remains poorly understood, yet work in the pancreatic islet demonstrates how an array of local paracrine factors can push and pull on the phases of resident cells. Dispersed islet cells from rodents and humans display robust rhythms comparable to intact islets, indicating that physical contacts are dispensable for synchrony. When α and β cells are cultured separately, 4 hrs separates the peaks of insulin and glucagon secretion, however this gap shortens to 2 hrs in a mixed population (48). This relationship is established by cell-type-specific expression of receptors and signaling pathway activity such that glucagon induces high amplitude Per2 oscillations in β cells but not in α cells, whereas insulin can synchronize both α and β cells. δ cells factor into this phase relationship through release of somatostatin (94). In vivo ~8 hrs separate the peaks of glucagon and insulin, timing that is ultimately set by additional neuronal, feeding and hormonal timing cues.
An interesting aspect of clock network synchrony is its apparent redundancy, with myriad cues engaged simultaneously. Removing a single rhythmic cue often has modest, if not negligible, effects on synchrony of the system. Subjecting mice to arrhythmic feeding regimes has no effect on the phase or amplitude of the liver clock, a highly feeding-sensitive organ (95). This is likely an inherent feature aimed at reinforcing stability. A variety of cues may also exist to coordinate specific clock output across tissues. For instance, adrenalectomy, which eliminates the glucocorticoid rhythm, has a negligible effect on the molecular clock, yet renders certain glucocorticoid target genes arrhythmic in the liver and adipose tissue (16, 96, 97).
Closing the circuit: peripheral feedback to the brain
The SCN is unperturbed by manipulations that have profound effects on peripheral clocks, such as day-time feeding (98). However, the central clock is housed in the hypothalamus, the neuroendocrine center that senses and integrates systemic energy metabolism. The early discovery that muscle-specific overexpression of BMAL1 in an otherwise clock-less mouse can partially rescue circadian locomotor activity raised the possibility of feedback from peripheral to central clocks (99, 100). The liver clock has been shown to regulate locomotion in mice, specifically a bout of activity immediately prior to food availability, referred to as food anticipatory activity (FAA). FAA continues in SCN-lesioned mice, but is abolished by liver-specific deletion of Per2, demonstrating that this behavior requires a peripherally-derived clock signal (101). Restoring the deficit of circulating β-hydroxybutyrate left behind by loss of Per2 partially rescues corticosterone levels and FAA, however liver-specific overexpression of Per2 in Per2−/− mice fails to, thus intact clock function in other tissues, likely non-SCN brain regions, is required for this response.
The brain relies on peripheral tissues to support metabolite levels, an aspect of systemic metabolism that can be observed in the cortex by liver-specific clock disruption and in the hypothalamus by adipose clock disruption (102, 103). Mice with adipose-specific ablation of Bmal1 exhibit obesity, concomitant with reduced energy expenditure, increased food intake during the rest phase, and lower concentrations of polyunsaturated fatty acids (PUFAs). Feeding these mutant mice a diet rich in PUFAs restores energy homeostasis, including body weight, food intake and PUFA content in the hypothalamus, thus linking the adipocyte clock to hypothalamic clocks through circulating fatty acids (103). Adipose-secreted proteins, referred to as adipokines, also constitute brain-body feedback circuits; a large existing literature describes the multi-level circadian regulation of leptin and adiponectin (103–106).
Circadian information flows from the SCN to other hypothalamic nuclei, yet this flow is not unidirectional, as the SCN also receives synaptic inputs from the ARC and DMH (69, 107). Physically disrupting the neuronal connectivity between the SCN and ARC results in ARC desynchronization and loss of locomotor activity rhythms, circulating corticosterone levels and body temperature. Moreover, without connections to the ARC, glucose administration fails to induce c-Fos expression in the SCN, suggesting that hypothalamic nuclei can relay metabolic feedback to the SCN (70). Considering that inverted feeding disrupts clock gene rhythms in extra-SCN nuclei, it is tempting to speculate that this interplay finely tunes central clock function based on peripheral clock status (98).
Inputs that determine clock output
Genome-wide profiling experiments in rodents and primates show a substantial portion (up to ~15%) of the transcriptome oscillates (108, 109). Remarkably, considering there is only a small overlap of oscillating genes in different tissues, it is thought that most of the mammalian genome has the potential to undergo circadian regulation (108, 109). How the same core clock transcription factors (TFs) impart tissue-specificity is an active area of research. Data points to both intrinsic and extrinsic signals that aid in dictating clock output. An important note is that output autonomy varies dramatically across tissues. For example, the olfactory bulb is able to drive daily rhythms in olfactory responsivity even with the SCN ablated (110), whereas peripheral clocks are semi-autonomous and rely on the function of the clock in many tissues (53, 54).
Connectivity of central and peripheral clocks
Work in flies and mice is beginning to reveal the connectivity of clock systems for peripheral and metabolic tissues (Figure 4) (111–113). Disrupting specifically the local clock in liver, muscle or adipose tissue ablates the majority of mRNA and protein oscillations, yet a subset of mRNAs continues to oscillate. Thus, certain rhythms result from distal clock function, or yet unidentified external cues (75, 103, 114). Importantly, in the absence of all other clocks in vivo, genetic reconstitution of hepatic and epidermal clocks is able to drive ~10–20% of the transcriptional and metabolic output, demonstrating their autonomy as well as their inherent limitation (53, 54). Interestingly, epidermal autonomous transcripts overlap with those that escape aging-associated reprogramming (115), suggesting that the mechanisms which enable full circadian output across tissues can be lost even if the local clock machinery appears unperturbed.
Figure 4. Inputs determining peripheral clock output.
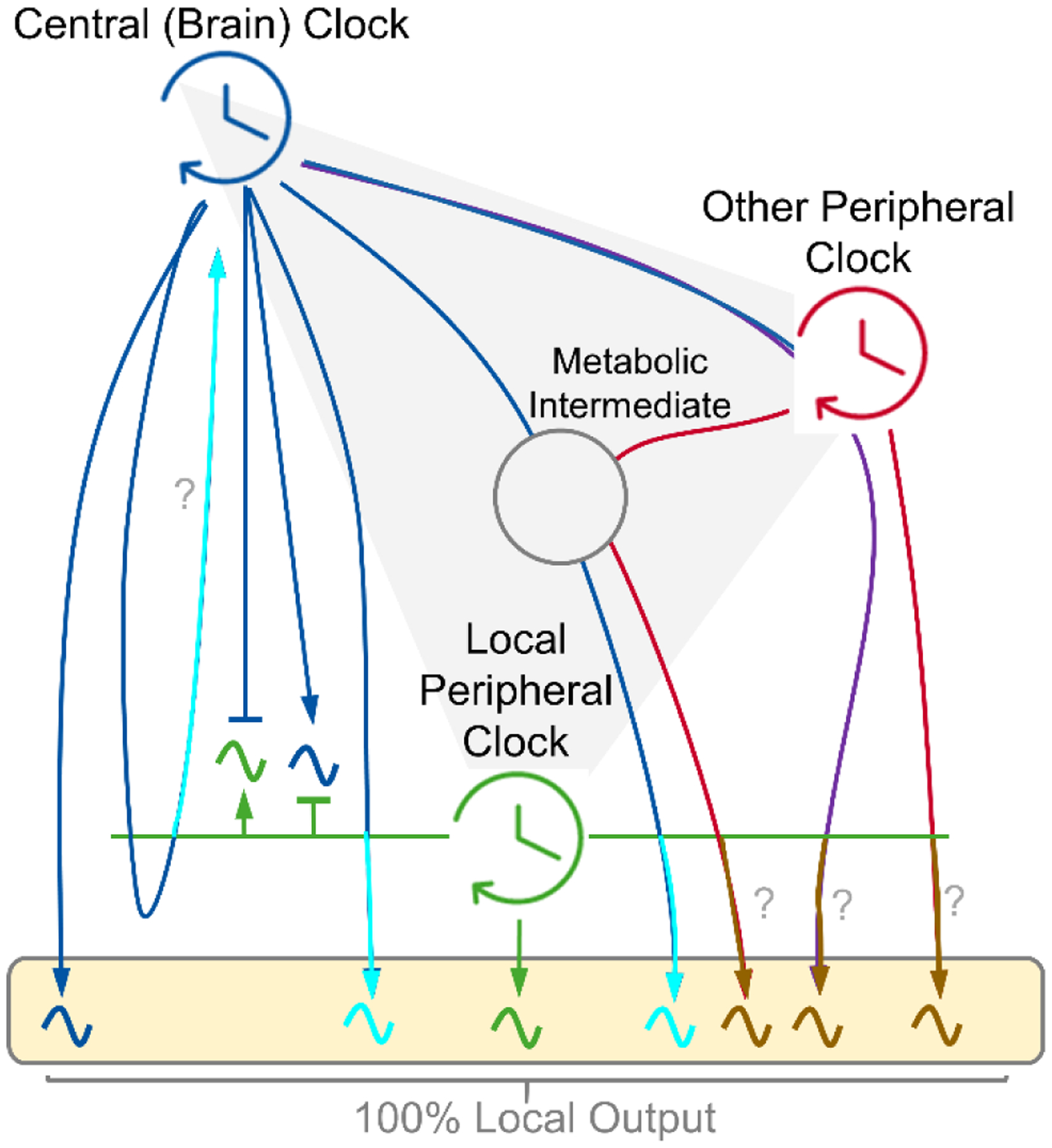
Conceptual model showing the drivers of circadian output of a peripheral metabolic tissue. Arrows indicate a driving circadian signal. The box depicts 100% of the local oscillatory output. Local clocks exhibit autonomy in driving a small fraction of output (green) (54) (53). Central (brain) clock-controlled cycles, feeding-fasting and sleep-wake, can drive rhythms independently or presumably in cooperation with the local clock (blue and turquoise). Peripheral clocks may influence rhythmicity of each other in a central clock dependent or independent manner (red, purple and brown). Many circadian signals likely synergize across tissues and intermediates (e.g. gut microbiota) to shape the full rhythmic output, with mechanisms which remain elusive (gray). It is unclear if the local clock is required for certain connections (?).
What inputs, then, dictate rhythmic outputs (Figure 5)? Imposing an arrhythmic feeding paradigm on mice disrupts ~70% of the rhythmic transcriptome in the liver, as well as rhythms of key metabolic genes in skeletal muscle and brown adipose tissue, hence feeding-fasting provides a driving input to metabolic tissues (95, 116–120). To what extent feeding signals directly regulate target tissues remains unclear. Metabolic intermediaries, such as the gut microbiota, are likely contributors. Rhythmic abundance of gut bacteria is driven by feeding and fasting, and microbiota-depletion reprograms the rhythmic transcriptome of both the intestinal epithelium and liver, demonstrating its wide-ranging influence (121). Loss of inputs including the feeding rhythm, microbiota or non-hepatic Bmal1 results in hundreds of de novo oscillating genes in the liver, therefore inputs seemingly inhibit portions of the local output (53, 54, 95, 121). An important aside is that loss of the aforementioned inputs does not alter core clock function in any overt manner, despite the requirement of the local clock for most rhythmic output. Hence, clock machinery might effectively integrate circadian signals within the system to regulate output. This concept awaits formal demonstration.
Figure 5. Multi-level control of clock output and communication.
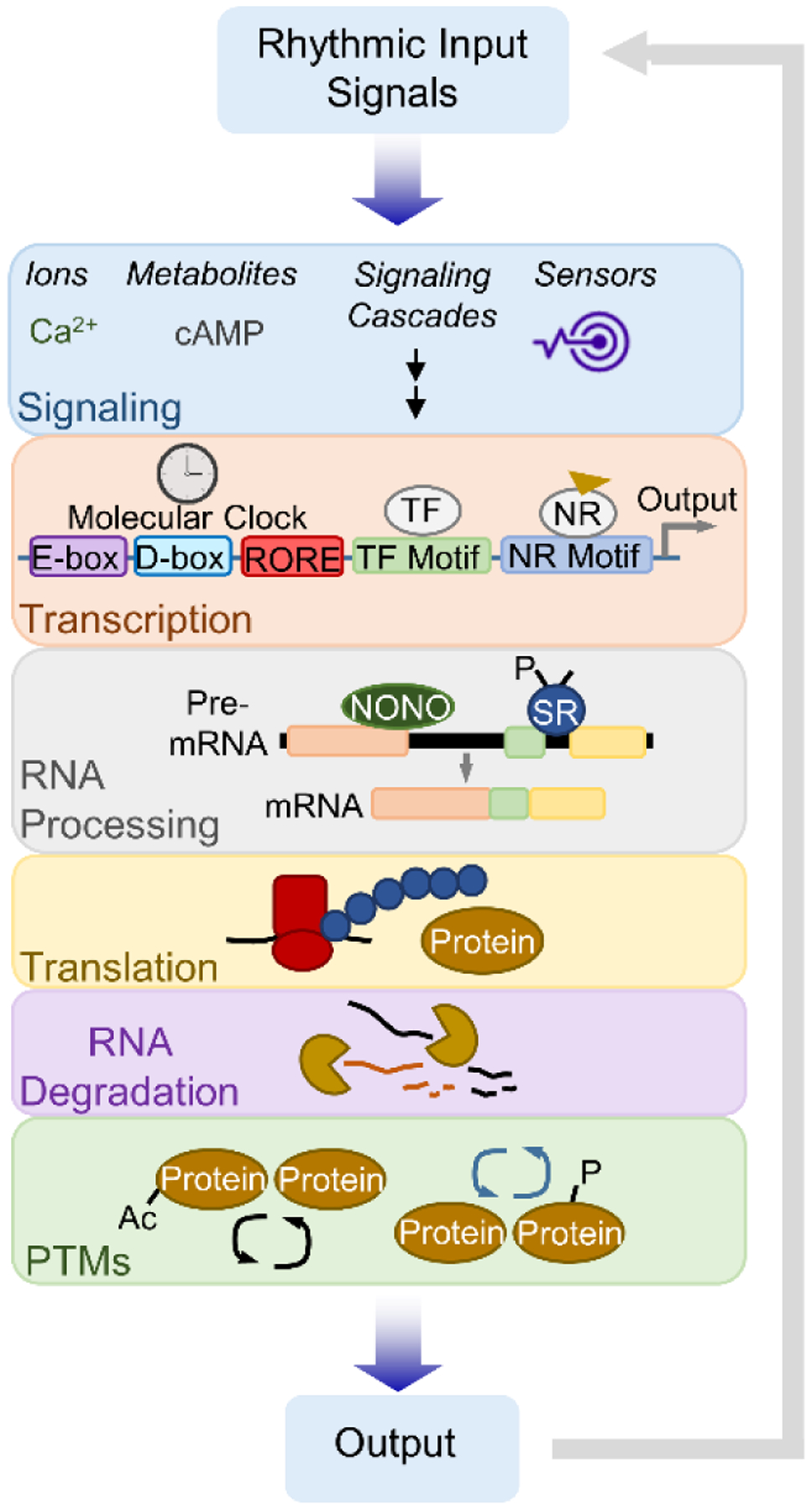
The clock can autonomously support a portion of circadian output. Nuclear receptors (NRs) regulate gene expression in tandem with clock proteins and are activated by extrinsic ligands, many of which oscillate in the bloodstream as a result of clock control in another tissue. The daily nuclear accumulation of certain transcription factors (TFs) is driven by feeding or body temperature rhythms. Systemic rhythms post-transcriptionally shape mRNA and protein oscillations. Feeding and fasting regulate RNA processing, translation and degradation while temperature fluctuations can induce alternative splicing programs that generate rhythmicity. Daily systemic energy metabolism regulates the functional status of proteins through post-translational modifications tied to metabolite levels. cAMP – cyclic adenosine monophosphate; NONO – non-POU domain-containing octamer-binding protein; SR – serine/arginine-rich splicing factor family; Ac – acetylation; P – phosphorylation.
Directing clock output genes
Systematic analyses of genome-wide datasets collected over the circadian cycle have conceptualized how the clock controls rhythmic gene expression. Key findings are that 1) BMAL1 positioning on chromatin is mostly cell-autonomous; 2) BMAL1 binding sites have little overlap between tissues and are distinguishable by the state of nucleosome depletion; 3) clock protein binding sites overlap heavily with lineage-specific TFs; and 4) rhythmic promoter-enhancer interactions are associated with rhythmic genes (8, 122–124). Thus, local clocks are poised but require signals from the whole network for complete temporal control of transcription (53). Many extrinsic rhythmic signals impinge on TFs and epigenetic regulators that interact with the clock machinery (21, 24, 77, 122, 125).
An important level of control is provided by nuclear receptors (NRs), which can operate as a point of metabolic integration for the clock. NRs regulate the expression of specific sets of genes in response to ligands emanating from extrinsic sources. Their circadian function depends on at least four sources of rhythmic information. One, amounts of ligand often oscillate in the serum, resulting in cyclic NR activity. To illustrate, the hypothalamic-pituitary-adrenal axis controls the serum oscillation of corticosterone and circadian glucocorticoid receptor activation in several tissues (16, 96, 97). Two, many NRs are rhythmically expressed and have nuclear protein levels that rise and fall over the course of a day, therefore activity depends on the phase relationship between receptor and ligand (126, 127). Three, some NRs and other cooperating TFs continue to oscillate in clock-disrupted Cry1/2-null mice, pointing to control by behavior or systemic cues (126). Four, several examples exist of NRs interacting with clock proteins at specific times during the day, resulting in circadian regulation of distinct sets of genes (128). CRY1 and CRY2, for example, interact broadly with various NRs, co-occupy thousands of genomic binding sites and function as co-repressors of gene expression (129). NR function is a striking example of the complexity of circadian regulation as it relates to systemic physiology.
A point to consider is that molecular oscillations can exist in the absence of the essential clock gene Bmal1. Skin fibroblasts and liver from Bmal1−/− mice exhibit large scale oscillations of transcripts, proteins and protein phosphorylation sites that persist under free-running conditions (i.e. no extrinsic forces present), are temperature compensated and can be entrained (synchronized), suggesting the presence of a non-canonical mechanism that meets the criteria of a molecular clock (130). However, the physiological relevance of this phenomenon is unclear, since almost all transcripts and proteins that oscillate in Bmal1−/− do not oscillate in wild type (53, 54, 130). One possibility is that these non-canonical rhythms arise under conditions of BMAL1 disruption, and therefore may be relevant in pathological contexts.
Concluding Remarks
In modern society, aligning our internal time with geophysical time is challenging. Misalignment, wherein eating and sleeping patterns conflict with the natural light-dark cycle, is associated with metabolic syndrome, cardiovascular disease, neurological conditions and cancer. Shift workers are particularly vulnerable to misalignment, and reports of hospital nurses and police officers confirm that internal desynchrony is a consequence of an inverted work schedule (131, 132). And just as circadian disruption can induce pathologies, pathologies can induce circadian disruption. In mice, bouts of intermittent hypoxia – similar to obstructive sleep apnea in humans – shift the phase of the clocks in the liver, kidney and lung such that inter-organ timing is discombobulated (133). It has become urgent that we uncover the molecular underpinnings of the relationship between the circadian clock and disease.
Studies in mice demonstrate a direct cause and effect relationship between desynchrony and metabolic dysfunction. Mice with deletion of Bmal1 in the SCN retain peripheral clock synchrony under a light-dark cycle, yet become desynchronized under constant darkness, concomitant with fat accumulation and impaired glucose utilization. When synchrony is reestablished by restricting food intake to a 12 hr window, body weight and glucose metabolism are restored (55). These results suggest it is not merely the presence of a restorative signal that is essential, but rather its rhythmicity. The physiological glucocorticoid rhythm is not obesogenic, but lack of oscillation and its flattening to a persistent signal of the same magnitude triggers adipocyte differentiation and fat accumulation (134).
High fat diet disrupts the temporal coordination of metabolic pathways between tissues, as revealed by simultaneous profiling of the circadian metabolome in brain, body and serum of mice (62). Moreover, lack of a discernable feeding-fasting rhythm ablates oscillation of key metabolic genes (95, 118). By contrast, the powerful and durable effect of food and food timing on the clock can be harnessed to bolster the system. There is recent evidence that some patients with metabolic syndrome benefit from time restricted eating (135). Deciphering the circuitry of communication between clocks has the potential to transform our understanding of physiology and metabolism, and may hold therapeutic promise to identify innovative, non-invasive strategies to fight disease and improve human wellbeing.
Figure Caption 0. Cellular to Organismal Timekeeping: Communication Between Clocks.
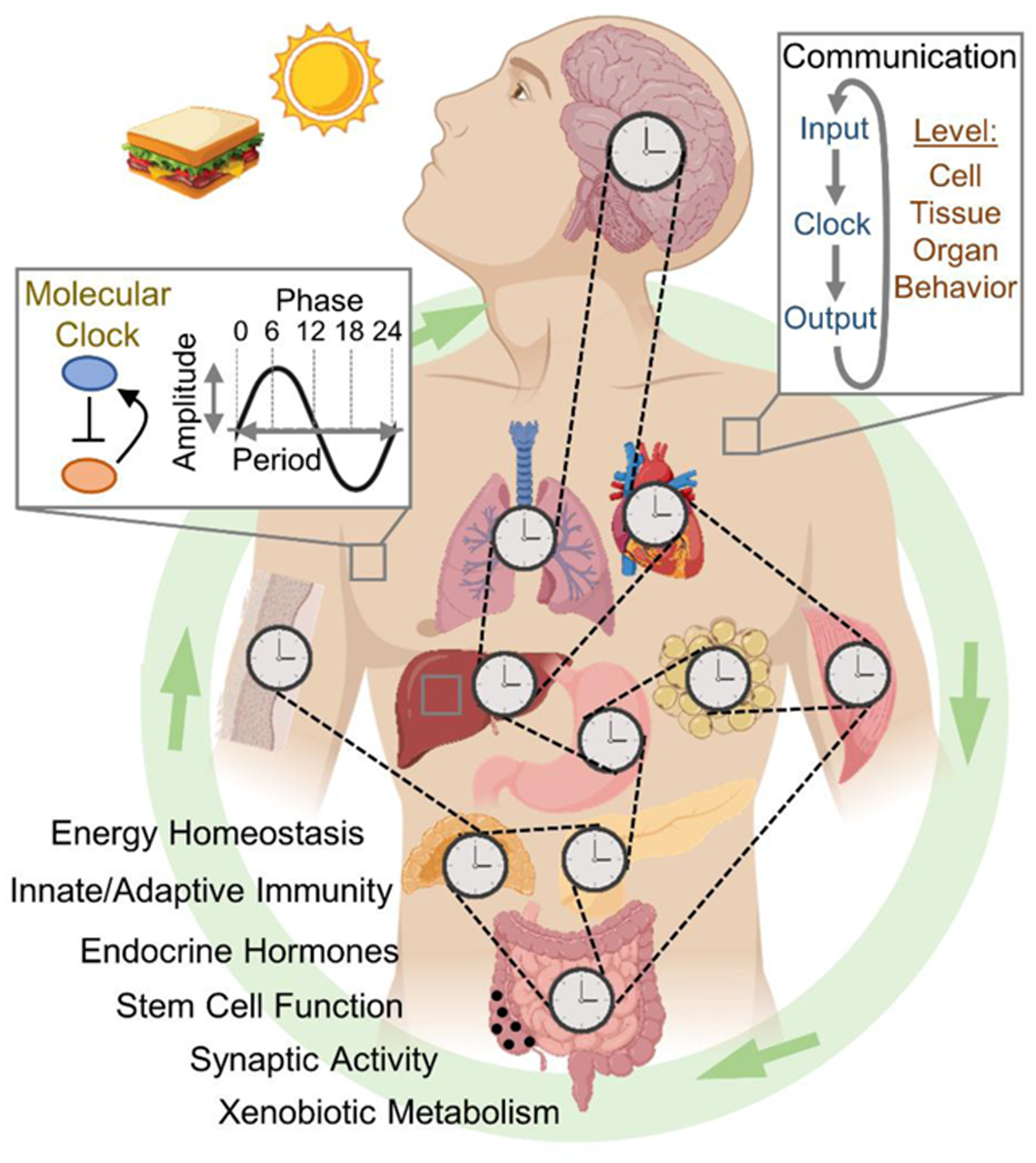
The mammalian circadian clock is a coupled network of cell and tissue clocks. Light and food are predominant cues, pushing and pulling on the phase, enhancing or attenuating the amplitude and activating or inhibiting functional rhythms. In a tissue-specific manner, clocks receive input signals and convert them into timed functional outputs, many of which in turn act as inputs, effectively connecting the network.
Acknowledgements
This work is dedicated to the memory of Paolo Sassone-Corsi (1956-2020), a great scientist, mentor and human. We would like to thank all members of the Sassone-Corsi lab for conceptual discussions and careful revision of this manuscript.
Funding:
The P.S.-C. lab is supported by NIH grants R21DK114652, R21AG053592 and a Challenge Grant of the Novo Nordisk Foundation (NNF-202585). K.B.K is supported by the NIDDK grant F32DK121425.
Footnotes
Competing interests: The authors have no competing interests.
References
- 1.Meyers SR, Malinverno A, Proterozoic Milankovitch cycles and the history of the solar system. Proc Natl Acad Sci U S A 115, 6363–6368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills JN, Minors DS, Waterhouse JM, The circadian rhythms of human subjects without timepieces or indication of the alternation of day and night. J Physiol 240, 567–594 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitmore D, Foulkes NS, Sassone-Corsi P, Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 404, 87–91 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Moore RY, Eichler VB, Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res 42, 201–206 (1972). [DOI] [PubMed] [Google Scholar]
- 5.Schibler U, Sassone-Corsi P, A web of circadian pacemakers. Cell 111, 919–922 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Yoo SH et al. , PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamia KA, Storch KF, Weitz CJ, Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A 105, 15172–15177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koike N et al. , Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aryal RP et al. , Macromolecular Assemblies of the Mammalian Circadian Clock. Mol Cell 67, 770–782 e776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Partch CL, Green CB, Takahashi JS, Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24, 90–99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papazyan R, Zhang Y, Lazar MA, Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol 23, 1045–1052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McIntosh BE, Hogenesch JB, Bradfield CA, Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol 72, 625–645 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Hennig S et al. , Structural and functional analyses of PAS domain interactions of the clock proteins Drosophila PERIOD and mouse PERIOD2. PLoS Biol 7, e94 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghuram S et al. , Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol 14, 1207–1213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin L et al. , Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Oishi K et al. , Genome-wide expression analysis reveals 100 adrenal gland-dependent circadian genes in the mouse liver. DNA Res 12, 191–202 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P, Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A 99, 7728–7733 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buhr ED, Yoo SH, Takahashi JS, Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330, 379–385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U, Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol 12, 1574–1583 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Gerber A et al. , Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell 152, 492–503 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Nakahata Y et al. , The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bass J, Lazar MA, Circadian time signatures of fitness and disease. Science 354, 994–999 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Hirano A, Fu YH, Ptacek LJ, The intricate dance of post-translational modifications in the rhythm of life. Nat Struct Mol Biol 23, 1053–1060 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Asher G et al. , SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Adamovich Y, Ladeuix B, Golik M, Koeners MP, Asher G, Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1alpha. Cell Metab 25, 93–101 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Nagoshi E et al. , Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Welsh DK, Logothetis DE, Meister M, Reppert SM, Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Webb B, Angelo N, Huettner JE, Herzog ED, Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc Natl Acad Sci U S A 106, 16493–16498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noguchi T, Wang LL, Welsh DK, Fibroblast PER2 circadian rhythmicity depends on cell density. J Biol Rhythms 28, 183–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmal C, Herzog ED, Herzel H, Measuring Relative Coupling Strength in Circadian Systems. J Biol Rhythms 33, 84–98 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guenthner CJ et al. , Circadian rhythms of Per2::Luc in individual primary mouse hepatocytes and cultures. PLoS One 9, e87573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lande-Diner L, Stewart-Ornstein J, Weitz CJ, Lahav G, Single-cell analysis of circadian dynamics in tissue explants. Mol Biol Cell 26, 3940–3945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izumo M et al. , Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. Elife 3, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hastings MH, Maywood ES, Brancaccio M, The Mammalian Circadian Timing System and the Suprachiasmatic Nucleus as Its Pacemaker. Biology (Basel) 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crosio C, Cermakian N, Allis CD, Sassone-Corsi P, Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci 3, 1241–1247 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Hastings MH, Maywood ES, Brancaccio M, Generation of circadian rhythms in the suprachiasmatic nucleus. Nat Rev Neurosci 19, 453–469 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Brancaccio M, Patton AP, Chesham JE, Maywood ES, Hastings MH, Astrocytes Control Circadian Timekeeping in the Suprachiasmatic Nucleus via Glutamatergic Signaling. Neuron 93, 1420–1435 e1425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tso CF et al. , Astrocytes Regulate Daily Rhythms in the Suprachiasmatic Nucleus and Behavior. Curr Biol 27, 1055–1061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brancaccio M et al. , Cell-autonomous clock of astrocytes drives circadian behavior in mammals. Science 363, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barca-Mayo O et al. , Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat Commun 8, 14336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morioka N et al. , The induction of Per1 expression by the combined treatment with glutamate, 5-hydroxytriptamine and dopamine initiates a ripple effect on Bmal1 and Cry1 mRNA expression via the ERK signaling pathway in cultured rat spinal astrocytes. Neurochem Int 90, 9–19 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Park JS et al. , Differential Phase Arrangement of Cellular Clocks along the Tonotopic Axis of the Mouse Cochlea Ex Vivo. Curr Biol 27, 2623–2629 e2622 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gould PD et al. , Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noguchi T, Wang CW, Pan H, Welsh DK, Fibroblast circadian rhythms of PER2 expression depend on membrane potential and intracellular calcium. Chronobiol Int 29, 653–664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang SL, Yue Z, Arnold DM, Artiushin G, Sehgal A, A Circadian Clock in the Blood-Brain Barrier Regulates Xenobiotic Efflux. Cell 173, 130–139 e110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rash JE et al. , Connexin36 vs. connexin32, “miniature” neuronal gap junctions, and limited electrotonic coupling in rodent suprachiasmatic nucleus. Neuroscience 149, 350–371 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sengiku A et al. , Circadian coordination of ATP release in the urothelium via connexin43 hemichannels. Sci Rep 8, 1996 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrenko V et al. , Pancreatic alpha- and beta-cellular clocks have distinct molecular properties and impact on islet hormone secretion and gene expression. Genes Dev 31, 383–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamajuku D et al. , Real-time monitoring in three-dimensional hepatocytes reveals that insulin acts as a synchronizer for liver clock. Sci Rep 2, 439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams J et al. , Epithelial and stromal circadian clocks are inversely regulated by their mechano-matrix environment. J Cell Sci 131, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Neill JS, Hastings MH, Increased coherence of circadian rhythms in mature fibroblast cultures. J Biol Rhythms 23, 483–488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guan D et al. , The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koronowski KB et al. , Defining the Independence of the Liver Circadian Clock. Cell 177, 1448–1462 e1414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Welz PS et al. , BMAL1-Driven Tissue Clocks Respond Independently to Light to Maintain Homeostasis. Cell 178, 1029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kolbe I, Leinweber B, Brandenburger M, Oster H, Circadian clock network desynchrony promotes weight gain and alters glucose homeostasis in mice. Mol Metab 30, 140–151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Husse J, Leliavski A, Tsang AH, Oster H, Eichele G, The light-dark cycle controls peripheral rhythmicity in mice with a genetically ablated suprachiasmatic nucleus clock. FASEB J 28, 4950–4960 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Tahara Y et al. , In vivo monitoring of peripheral circadian clocks in the mouse. Curr Biol 22, 1029–1034 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Saini C et al. , Real-time recording of circadian liver gene expression in freely moving mice reveals the phase-setting behavior of hepatocyte clocks. Genes Dev 27, 1526–1536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Vinne V, Swoap SJ, Vajtay TJ, Weaver DR, Desynchrony between brain and peripheral clocks caused by CK1delta/epsilon disruption in GABA neurons does not lead to adverse metabolic outcomes. Proc Natl Acad Sci U S A 115, E2437–E2446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pando MP, Morse D, Cermakian N, Sassone-Corsi P, Phenotypic rescue of a peripheral clock genetic defect via SCN hierarchical dominance. Cell 110, 107–117 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Hamaguchi Y, Tahara Y, Hitosugi M, Shibata S, Impairment of Circadian Rhythms in Peripheral Clocks by Constant Light Is Partially Reversed by Scheduled Feeding or Exercise. J Biol Rhythms 30, 533–542 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Dyar KA et al. , Atlas of Circadian Metabolism Reveals System-wide Coordination and Communication between Clocks. Cell 174, 1571–1585 e1511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masri S et al. , Lung Adenocarcinoma Distally Rewires Hepatic Circadian Homeostasis. Cell 165, 896–909 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cooper SJ, From Claude Bernard to Walter Cannon. Emergence of the concept of homeostasis. Appetite 51, 419–427 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Perrin L et al. , Human skeletal myotubes display a cell-autonomous circadian clock implicated in basal myokine secretion. Mol Metab 4, 834–845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gamble KL, Berry R, Frank SJ, Young ME, Circadian clock control of endocrine factors. Nat Rev Endocrinol 10, 466–475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Benitah SA, Welz PS, Circadian Regulation of Adult Stem Cell Homeostasis and Aging. Cell Stem Cell 26, 817–831 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Godinho-Silva C et al. , Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature 574, 254–258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Acosta-Galvan G et al. , Interaction between hypothalamic dorsomedial nucleus and the suprachiasmatic nucleus determines intensity of food anticipatory behavior. Proc Natl Acad Sci U S A 108, 5813–5818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buijs FN et al. , Suprachiasmatic Nucleus Interaction with the Arcuate Nucleus; Essential for Organizing Physiological Rhythms. eNeuro 4, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orozco-Solis R et al. , The Circadian Clock in the Ventromedial Hypothalamus Controls Cyclic Energy Expenditure. Cell Metab 23, 467–478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Astiz M, Heyde I, Oster H, Mechanisms of Communication in the Mammalian Circadian Timing System. Int J Mol Sci 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo H, Brewer JM, Champhekar A, Harris RB, Bittman EL, Differential control of peripheral circadian rhythms by suprachiasmatic-dependent neural signals. Proc Natl Acad Sci U S A 102, 3111–3116 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saini C, Morf J, Stratmann M, Gos P, Schibler U, Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev 26, 567–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U, System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5, e34 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adamovich Y et al. , Oxygen and Carbon Dioxide Rhythms Are Circadian Clock Controlled and Differentially Directed by Behavioral Signals. Cell Metab 29, 1092–1103 e1093 (2019). [DOI] [PubMed] [Google Scholar]
- 77.Peek CB et al. , Circadian Clock Interaction with HIF1alpha Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab 25, 86–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Asher G, Sassone-Corsi P, Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161, 84–92 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Bruning F et al. , Sleep-wake cycles drive daily dynamics of synaptic phosphorylation. Science 366, (2019). [DOI] [PubMed] [Google Scholar]
- 80.Noya SB et al. , The forebrain synaptic transcriptome is organized by clocks but its proteome is driven by sleep. Science 366, (2019). [DOI] [PubMed] [Google Scholar]
- 81.Noguchi T, Ikeda M, Ohmiya Y, Nakajima Y, A dual-color luciferase assay system reveals circadian resetting of cultured fibroblasts by co-cultured adrenal glands. PLoS One 7, e37093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barnea M, Chapnik N, Genzer Y, Froy O, The circadian clock machinery controls adiponectin expression. Mol Cell Endocrinol 399, 284–287 (2015). [DOI] [PubMed] [Google Scholar]
- 83.Chen S et al. , Angptl8 mediates food-driven resetting of hepatic circadian clock in mice. Nat Commun 10, 3518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.LeSauter J, Hoque N, Weintraub M, Pfaff DW, Silver R, Stomach ghrelin-secreting cells as food-entrainable circadian clocks. Proc Natl Acad Sci U S A 106, 13582–13587 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Landgraf D et al. , Oxyntomodulin regulates resetting of the liver circadian clock by food. Elife 4, e06253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma X, Zhou Z, Chen Y, Wu Y, Liu Y, RBP4 functions as a hepatokine in the regulation of glucose metabolism by the circadian clock in mice. Diabetologia 59, 354–362 (2016). [DOI] [PubMed] [Google Scholar]
- 87.Bookout L et al. , FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nat Med 19, 1147–1152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramanathan C et al. , mTOR signaling regulates central and peripheral circadian clock function. PLoS Genet 14, e1007369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crosby P et al. , Insulin/IGF-1 Drives PERIOD Synthesis to Entrain Circadian Rhythms with Feeding Time. Cell 177, 896–909 e820 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mendez-Ferrer S, Lucas D, Battista M, Frenette PS, Haematopoietic stem cell release is regulated by circadian oscillations. Nature 452, 442–447 (2008). [DOI] [PubMed] [Google Scholar]
- 91.He W et al. , Circadian Expression of Migratory Factors Establishes Lineage-Specific Signatures that Guide the Homing of Leukocyte Subsets to Tissues. Immunity 49, 1175–1190 e1177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.de Juan A et al. , Artery-Associated Sympathetic Innervation Drives Rhythmic Vascular Inflammation of Arteries and Veins. Circulation 140, 1100–1114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Man K, Loudon A, Chawla A, Immunity around the clock. Science 354, 999–1003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Petrenko V, Dibner C, Cell-specific resetting of mouse islet cellular clocks by glucagon, glucagon-like peptide 1 and somatostatin. Acta Physiol (Oxf) 222, e13021 (2018). [DOI] [PubMed] [Google Scholar]
- 95.Greenwell BJ et al. , Rhythmic Food Intake Drives Rhythmic Gene Expression More Potently than the Hepatic Circadian Clock in Mice. Cell Rep 27, 649–657 e645 (2019). [DOI] [PubMed] [Google Scholar]
- 96.Sotak M et al. , Peripheral circadian clocks are diversely affected by adrenalectomy. Chronobiol Int 33, 520–529 (2016). [DOI] [PubMed] [Google Scholar]
- 97.Su Y et al. , Effects of adrenalectomy on daily gene expression rhythms in the rat suprachiasmatic and paraventricular hypothalamic nuclei and in white adipose tissue. Chronobiol Int 32, 211–224 (2015). [DOI] [PubMed] [Google Scholar]
- 98.Wang D et al. , Effects of feeding time on daily rhythms of neuropeptide and clock gene expression in the rat hypothalamus. Brain Res 1671, 93–101 (2017). [DOI] [PubMed] [Google Scholar]
- 99.Breit A et al. , Insulin-like growth factor-1 acts as a zeitgeber on hypothalamic circadian clock gene expression via glycogen synthase kinase-3beta signaling. J Biol Chem 293, 17278–17290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McDearmon EL et al. , Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 314, 1304–1308 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chavan R et al. , Liver-derived ketone bodies are necessary for food anticipation. Nat Commun 7, 10580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fustin JM, Karakawa S, Okamura H, Circadian Profiling of Amino Acids in the SCN and Cerebral Cortex by Laser Capture Microdissection-Mass Spectrometry. J Biol Rhythms 32, 609–620 (2017). [DOI] [PubMed] [Google Scholar]
- 103.Paschos GK et al. , Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med 18, 1768–1777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kettner NM et al. , Circadian Dysfunction Induces Leptin Resistance in Mice. Cell Metab 22, 448–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zeng W et al. , Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell 163, 84–94 (2015). [DOI] [PubMed] [Google Scholar]
- 106.Kalsbeek A et al. , The suprachiasmatic nucleus generates the diurnal changes in plasma leptin levels. Endocrinology 142, 2677–2685 (2001). [DOI] [PubMed] [Google Scholar]
- 107.Yi X et al. , Ventromedial arcuate nucleus communicates peripheral metabolic information to the suprachiasmatic nucleus. Endocrinology 147, 283–294 (2006). [DOI] [PubMed] [Google Scholar]
- 108.Mure LS et al. , Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB, A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 111, 16219–16224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Granados-Fuentes D, Tseng A, Herzog ED, A circadian clock in the olfactory bulb controls olfactory responsivity. J Neurosci 26, 12219–12225 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Preussner M et al. , Body Temperature Cycles Control Rhythmic Alternative Splicing in Mammals. Mol Cell 67, 433–446 e434 (2017). [DOI] [PubMed] [Google Scholar]
- 112.Erion R, King AN, Wu G, Hogenesch JB, Sehgal A, Neural clocks and Neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Barber F, Erion R, Holmes TC, Sehgal A, Circadian and feeding cues integrate to drive rhythms of physiology in Drosophila insulin-producing cells. Genes Dev 30, 2596–2606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dyar KA et al. , Transcriptional programming of lipid and amino acid metabolism by the skeletal muscle circadian clock. PLoS Biol 16, e2005886 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Solanas G et al. , Aged Stem Cells Reprogram Their Daily Rhythmic Functions to Adapt to Stress. Cell 170, 678–692 e620 (2017). [DOI] [PubMed] [Google Scholar]
- 116.Vollmers C et al. , Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 106, 21453–21458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Atger F et al. , Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci U S A 112, E6579–6588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Su Y, Foppen E, Zhang Z, Fliers E, Kalsbeek A, Effects of 6-meals-a-day feeding and 6-meals-a-day feeding combined with adrenalectomy on daily gene expression rhythms in rat epididymal white adipose tissue. Genes Cells 21, 6–24 (2016). [DOI] [PubMed] [Google Scholar]
- 119.de Goede P et al. , Differential effects of diet composition and timing of feeding behavior on rat brown adipose tissue and skeletal muscle peripheral clocks. Neurobiol Sleep Circadian Rhythms 4, 24–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Benegiamo G et al. , The RNA-Binding Protein NONO Coordinates Hepatic Adaptation to Feeding. Cell Metab 27, 404–418 e407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thaiss CA et al. , Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations. Cell 167, 1495–1510 e1412 (2016). [DOI] [PubMed] [Google Scholar]
- 122.Shostak A, Brunner M, Help from my friends-cooperation of BMAL1 with noncircadian transcription factors. Genes Dev 33, 255–257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mermet J et al. , Clock-dependent chromatin topology modulates circadian transcription and behavior. Genes Dev 32, 347–358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beytebiere JR et al. , Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes Dev 33, 294–309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Masri S et al. , Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 158, 659–672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang J et al. , Nuclear Proteomics Uncovers Diurnal Regulatory Landscapes in Mouse Liver. Cell Metab 25, 102–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang X et al. , Nuclear receptor expression links the circadian clock to metabolism. Cell 126, 801–810 (2006). [DOI] [PubMed] [Google Scholar]
- 128.Qu M, Duffy T, Hirota T, Kay SA, Nuclear receptor HNF4A transrepresses CLOCK:BMAL1 and modulates tissue-specific circadian networks. Proc Natl Acad Sci U S A 115, E12305–E12312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kriebs A et al. , Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity. Proc Natl Acad Sci U S A 114, 8776–8781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ray S et al. , Circadian rhythms in the absence of the clock gene Bmal1. Science 367, 800–806 (2020). [DOI] [PubMed] [Google Scholar]
- 131.Resuehr D et al. , Shift Work Disrupts Circadian Regulation of the Transcriptome in Hospital Nurses. J Biol Rhythms 34, 167–177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Koshy A, Cuesta M, Boudreau P, Cermakian N, Boivin DB, Disruption of central and peripheral circadian clocks in police officers working at night. FASEB J 33, 6789–6800 (2019). [DOI] [PubMed] [Google Scholar]
- 133.Manella G et al. , Hypoxia induces a time- and tissue-specific response that elicits intertissue circadian clock misalignment. Proc Natl Acad Sci U S A 117, 779–786 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bahrami-Nejad Z et al. , A Transcriptional Circuit Filters Oscillating Circadian Hormonal Inputs to Regulate Fat Cell Differentiation. Cell Metab 27, 854–868 e858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wilkinson MJ et al. , Ten-Hour Time-Restricted Eating Reduces Weight, Blood Pressure, and Atherogenic Lipids in Patients with Metabolic Syndrome. Cell Metab, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


