Abstract
Background
Gastric cancer (GC) is a common and deadly malignancy in the world. CircRNAs have emerged as important regulators in human diseases, including GC. In this work, we intended to explore the role of circ_CORO1C in GC progression and potential mechanism.
Methods
Quantitative real-time PCR (qRT-PCR) or Western blot assay was performed to examine the expression of circRNA coronin-like actin-binding protein 1C (circ_CORO1C), microRNA (miR)-138-5p and Krueppel-like factor 12 (KLF12) in clinical samples and cells. Cell colony formation ability and viability were measured by colony formation assay and methyl thiazolyl tetrazolium (MTT) assay, respectively. Expression of cell proliferation and epithelia-mesenchymal transition (EMT) biomarker was detected by Western blot analysis. And cell metastasis, including migration and invasion, and apoptosis were analyzed via Transwell assay and flow cytometry, respectively. Target relationship among circ_CORO1C, miR-138-5p and KLF12 was validated by dual-luciferase reporter assay. The in vivo role of circ_CORO1C was investigated by tumor xenograft assay.
Results
Circ_CORO1C and KLF12 were upregulated, while miR-138-5p was downregulated in GC tissues and cells. Circ_CORO1C knockdown suppressed colony formation ability, viability, migration, invasion and EMT in GC cells, while promoted cell apoptosis in vitro. Circ_CORO1C targeted miR-138-5p, the inhibition of which could attenuate silenced circ_CORO1C-induced inhibitory effects on GC progression. MiR-138-5p repressed the aggressive malignant behaviors of GC cells by directly targeting KLF12. Circ_CORO1C deficiency inhibited GC tumor growth in vivo.
Conclusion
Depletion of circ_CORO1C suppressed GC progression by regulating miR-138-5p/KLF12 axis, offering a potential molecular target for GC therapy.
Keywords: gastric cancer, circ_CORO1C, miR-138-5p, KLF12, tumor growth
Introduction
According to the Global Cancer Statistics 2018, Gastric cancer (GC), a frequent malignancy in the digestive system, ranks as the third lethiferous disease among cancer-related deaths in the world.1 Due to a lack of advanced diagnostic techniques, a large number of people are diagnosed until advanced stages.2 Although current therapeutic approaches for cancers have improved a lot, the 5-year survival rate of GC patients is relatively low.3 Therefore, clarifying the molecular mechanisms underlying GC progression and searching for dependable diagnostic biomarkers and treatment targets are necessary.
Circular RNAs (circRNAs) are a novel category of non-coding RNAs, featured by covalently closed continuous loop, exerting a significant role in multiple physiological and pathological processes, especially in tumorigenesis, serving as diagnosis biomarkers or therapy strategies for human cancers.4,5 Because of the special circular structure, circRNAs are more stable than linear RNAs.6 On account of the development of high-throughput sequencing technology, many circRNAs were disclosed to have a vital regulatory impact on GC development, such as circNRIP1,7 circCACTIN (ID: hsa_circ_0092303 in circBase)8 and hsa_circ_0000993.9 As for circ_CORO1C (hsa_circ_0000437), which was reported to be dysregulated in GC tissues,10 its definite role in GC progression needs to be substantiated.
MicroRNAs (miRNAs) are also non-coding RNA molecules, with approximately 22 nucleotides in length, closely implicated with animal development and disease.11 Typically, miRNAs could trigger degradation of messenger RNAs (mRNAs) and reduce their translation, involved in cellular processes during the development and progression of cancer, including GC.12 MiR-138-5p, was a well-studied miRNA, acting as a tumor suppressor in some human cancers, like prostate cancer, lung adenocarcinoma, retinoblastoma, bladder cancer.13–16 Furthermore, miR-138-5p also conferred inhibition effect on GC cell proliferation and metastasis.17 As a target candidate gene of circ_CORO1C, the role of miR-138-5p in circ_CORO1C-mediated GC development was then explored.
Krueppel-like factor 12 (KLF12) belongs to the KLF family, which is an important zinc finger transcription factor.18 KLF12 was manifested to exert diverse functions in several human cancers, serving as an oncogenic stimulus.19–21 Moreover, KLF12 had a pivotal role in poorly differentiated GC and was identified as a promising target for GC therapy.22 Here, KLF12 was estimated to be a target mRNA of miR-138-5p by miRDB. The role of circ_CORO1C/miR-138-5p/KLF12 axis in GC progression remains unknown.
Herein, we detected a dysregulated circRNA circ_CORO1C in GC tissues and cells. In addition, its functional impact on GC progression and the regulatory axis circ_CORO1C/miR-138-5p/KLF12 were firstly testified.
Materials and Methods
Collection of Clinical Specimens
Prior to the conduction of this study, we got the authorization of the Ethics Committee of Sunshine Union Hospital. GC tissues (n=50) and matched adjacent normal tissues were collected from 50 patients hospitalized at Sunshine Union Hospital in accordance with the Declaration of Helsinki. Before resection surgery, these patients or their guardians signed the written informed consent, and all patients did not undergo chemotherapy, radiotherapy or immunotherapy.
Cell Culture and Transfection
Human normal gastric epithelial cell line GES-1 (CL-0563), GC cell lines HGC27 (CL-0107), AGS (CL-0022) and MKN45 (CL-0292) were acquired from Procell (Wuhan, China). Besides, GC MKN74 cells were bought from Mingzhoubio (Ningbo, China). Cell culture was maintained in Roswell Park Memorial Institute medium (RPMI)-1640 (Gibco, Grand Island, NY, USA) added with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Gibco) in a 37°C humidified incubator containing 5% CO2.
For knockdown of circ_CORO1C, small interfering RNA specifically against circ_CORO1C (si-circ_CORO1C) was designed and synthesized by GENEWIZ (Suzhou, China), with si-NC as negative control. MiR-138-5p mimic (miR-138-5p), miR-138-5p inhibitor (anti-miR-138-5p) and their corresponding negative controls (miR-NC and anti-miR-NC) were supplied by RIBOBIO Co. Ltd. (Guangzhou, China). To upregulate KLF12, the overexpression plasmid pcDNA-KLF12 (KLF12) was constructed by introducing the full-length cDNA sequence of KLF12 into pcDNA3.1 vector (Geenseed Biotech, Guangzhou, China), with pcDNA as negative control. For transfection assay, Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) was utilized.
Quantitative Real-Time PCR (qRT-PCR)
To isolate total RNA from clinical samples or cells, TRIzol Reagent (Invitrogen) was applied based on the user’s manual. After determination of purity and concentration, 1 μg RNA was subjected to reverse-transcription and qPCR using M-MLV Reverse Transcriptase (Invitrogen) and SYBR Premix ExTaq kit (TaKaRa, Otsu, Japan), respectively, to evaluate the expression levels of circ_CORO1C, CORO1C and KLF12, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous control. The expression of miR-138-5p was analyzed utilizing miRNA First-Strand cDNA Synthesis Kit (Agilent, Santa Clara, CA, USA) and all-in-one miRNA RT-qPCR Detection Kit (GeneCopoeia Inc., Rockville, MD, USA), normalized to housekeeping gene U6. Relative expression levels were calculated utilizing 2–ΔΔCt method.23 Primers used in this assay were exhibited in Table 1.
Table 1.
Primers Used for qRT-PCR Assay
| Forward Primer (5ʹ→3ʹ) | Reverse Primer (5ʹ→3ʹ) | |
|---|---|---|
| circ_CORO1C | AATGTGAGATTGCCAGAA | GTCAGGGTCATAGAAAGG |
| CORO1C | TCCTCCCTCTGCACAAGACT | GGATCTGCCATACCATGACC |
| KLF12 | CGGCAGTCAGAGTCAAAACAG | CGGCTTCCATATCGGGATAGT |
| GAPDH | AATCCCATCACCATCTTCC | CATCACGCCACAGTTTCC |
| miR-138-5p | GCGAGCTGGTGTTGTGAATC | AGTGCAGGGTCCGAGGTATT |
| U6 | CGCTTCACGAATTTGCGTGTCAT | GCTTCGGCAGCACATATACTAAAAT |
RNase R Digestion
To confirm the circular structure of circ_CORO1C, RNase R treatment was employed. Total RNA (5 μg) isolated from HGC-27 and MKN45 cells was treated with RNase R (Geneseed, Guangzhou, China) or not (Mock) at 37°C for 30 min. Afterwards, qRT-PCR was used to detect the expression of circ_CORO1C and CORO1C.
Colony Formation Assay
To assess the plate clone formation capacity of GC cells, transfected HGC-27 and MKN45 cells were plated into 6-well plates with an initial density of 500 cells per well. After routine culture for 2 weeks, generated colonies were immobilized by methanol, stained with 0.1% crystal violet at indoor temperature and counted exploiting Image J software (NIH, Bethesda, MD, USA). Colony formation rate indicates the percentage of the number of colonies among the number of seeded cells.
Methyl Thiazolyl Tetrazolium (MTT) Assay
MTT assay was employed to examine the cell viability of GC cells. After transfection, 3 × 103 HGC-27 and MKN45 cells were plated into 96-well plates. At 0 d, 1 d, 2 d or 3 d post seeding, 10 μL 0.5 mg/mL MTT solution (Beyotime Institute of Biotechnology, Haimen, China) was pipetted into each well. After incubation for another 4 h, the original medium was replaced by 150 μL dimethyl sulfoxide (DMSO). 15 min later, the optical density (OD) of each well was recorded at the wavelength of 490 nm exploiting a microplate reader (BioTek Instruments Inc., Winooski, VT, USA).
Western Blot Analysis
Clinical samples or cells were lysed in Radio-Immunoprecipitation Assay (RIPA) buffer mixed with protease and phosphates inhibitors (CWBIO, Beijing, China) for protein isolation. After quantified with a bicinchoninic acid assay (BCA) Kit (Beyotime Institute of Biotechnology), protein samples were subjected to separation through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Subsequently, membranes were blocked with 5% fat-free milk, then incubated with primary antibody and corresponding secondary antibody. Protein bands were treated with an enhanced chemiluminescence (ECL) Kit (Millipore) for visualization and analyzed by Image J software. Antibodies used in this assay were purchased from Abcam (Shanghai, China): anti-Ki67 (ab92742, 1:2000 dilution), anti-E-cadherin (ab40772; 1:1500 dilution), anti-N-cadherin (ab18203, 1:2000 dilution), anti-vimentin (ab193555, 1:1000 dilution), anti-KLF12 (ab221602, 1:1000 dilution), anti-GAPDH (ab128915, 1:3000 dilution) and Goat Anti-Rabbit IgG H&L (HRP; ab205718, 1:5000 dilution).
Transwell Assay
This assay was employed to assess the migrated and invaded abilities of GC cells with the help of Transwell chamber (8 μm pores; Millipore). For migration, 1 × 104 HGC-27 and MKN45 cells in RPMI-1640 medium without FBS were titled into the upper chamber. Meanwhile, RPMI-1640 medium harboring 10% FBS was added into the bottom chamber. Following 24-h incubation, cells were subjected to fixation, staining and counting under a microscope (amplification: 100 ×).
For invasion assay, 5 × 104 GC cells were initially seeded in the upper chamber enveloped with Matrigel (R&D Systems, Inc., Minneapolis, MN, USA). The remaining procedures were the same as the migration assay.
Flow Cytometry
Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (Beyotime Institute of Biotechnology) was used to determine the cell apoptosis of GC cells utilizing as per the guideline of the manufacturer. In brief, transfected HGC-27 and MKN45 cells were harvested and incubated with Annexin V-FITC and propidium iodide (PI) in dark. In the end, apoptotic GC cells were observed using a flow cytometer (CoulterEpics, Miami, FL, USA).
Target Gene Prediction and Dual-Luciferase Reporter Assay
Starbase 3.0 (http://starbase.sysu.edu.cn/index.php) and miRDB (http://www.mirdb.org/index.html) were applied to forecast the possible targets of circ_CORO1C and miR-138-5p, respectively. Results showed that both circ_CORO1C and 3ʹ-untranslated region (3ʹUTR) of KLF12 contained binding sites with miR-138-5p.
To validate the target binding, a fragment of circ_CORO1C or KLF12 3ʹUTR containing the estimated binding position was inserted into the pGL4 vector (Promega Corp., Madison, WI, USA) to synthesize wild type luciferase reporter plasmid circ_CORO1C-wt or KLF12-wt. The mutant type ones (circ_CORO1C-mut and KLF12-mut) were supplied by RIBOBIO Co. Ltd. HGC-27 and MKN45 cells were transfected with a constructed luciferase reporter and miR-NC or miR-138-5p for 48 h, followed by luciferase intensity determination using Dual-Luciferase Reporter Assay System (Promega Corp.).
Tumor Xenograft Assay
Animal experiments were performed after approval of the permission from the Ethics Committee of Sunshine Union Hospital according to the Guide for the Care and Use of Laboratory Animals (GB/T 35892–2018). Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines. Short hairpin RNA (shRNA) against circ_CORO1C (sh-circ_CORO1C) and matched negative control (sh-NC) were constructed and provided by Genomeditech (Shanghai, China). 5-week-old male BALB/C nude mice bought from Beijing Laboratory Animal Center (Beijing, China) were injected with MKN45 cells (2 × 106) stably expressing sh-circ_CORO1C or sh-NC (n=5). 1 week after injection, the size of generated tumors was measured once a week with the following formula: 0.5 × length × width2. 4 weeks later, all mice were sacrificed, and tumor tissues were taken out for weigh, qRT-PCR and Western blot assays.
Statistical Analysis
Data generated from 3 independent experiments were processed using GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA) and shown as mean ± standard deviation. Statistical methods included Student’s t-test or one-way analysis of variance (ANOVA) with Tukey’s tests. Correlation among the expression of circ_CORO1C, miR-138-5p and KLF12 in GC tissues was analyzed via Pearson correlation analysis. P-value < 0.05 was identified as statistically significant.
Results
Dysregulation of circ_CORO1C in GC Tissues and Cells
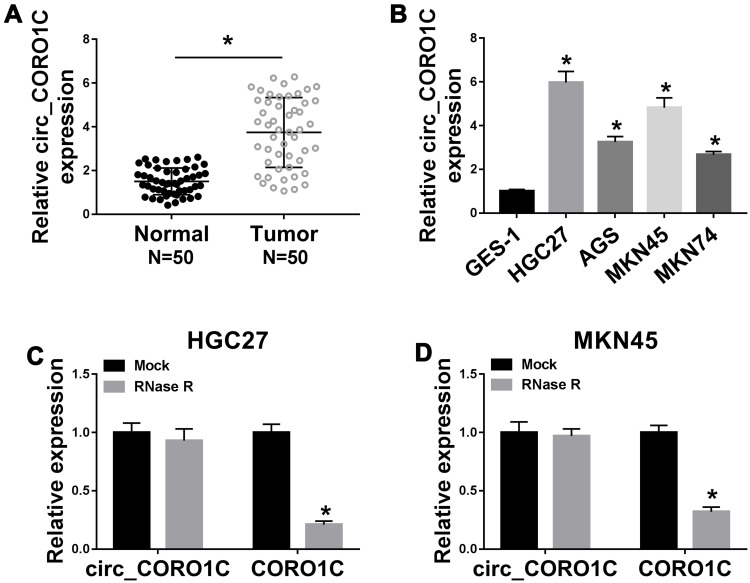
To identify the role of circ_CORO1C in GC development, the expression of circ_CORO1C in GC tissues and cell lines was detected. As exhibited in Figure 1A, circ_CORO1C was apparently upregulated in GC tissues relative to normal tissues. Besides, circ_CORO1C was also abundant in GC cells (HGC-27, AGS, MKN45 and MKN74) when compared to GES-1 cells (Figure 1B). Then, the stability of circ_CORO1C in GC cells was evaluated. Following RNase R treatment, the expression of CORO1C, rather than circ_CORO1C, was significantly declined in HGC-27 and MKN45 cells, suggesting that circ_CORO1C had a loop structure (Figure 1C and D). Furthermore, the circRNA ring structure of circ_CORO1C was also verified using divergent primers (Supplementary Fig 1). In short, circ_CORO1C was upregulated in GC tissues and cells.
Figure 1.
Dysregulation of circ_CORO1C in GC tissues and cells. (A) QRT-PCR assay for the relative expression of circ_CORO1C in GC tissues (n=50) and matched adjacent normal tissues. (B) QRT-PCR assay for the relative expression of circ_CORO1C in GES-1, HGC-27, AGS, MKN45 and MKN74 cells. (C and D) QRT-PCR assay for the relative expression of circ_CORO1C and CORO1C in HGC-27 and MKN45 cells digested with RNase R or not (Mock). *P <0.05.
Depletion of circ_CORO1C Repressed GC Cell Proliferation and Metastasis, While Promoted Cell Apoptosis
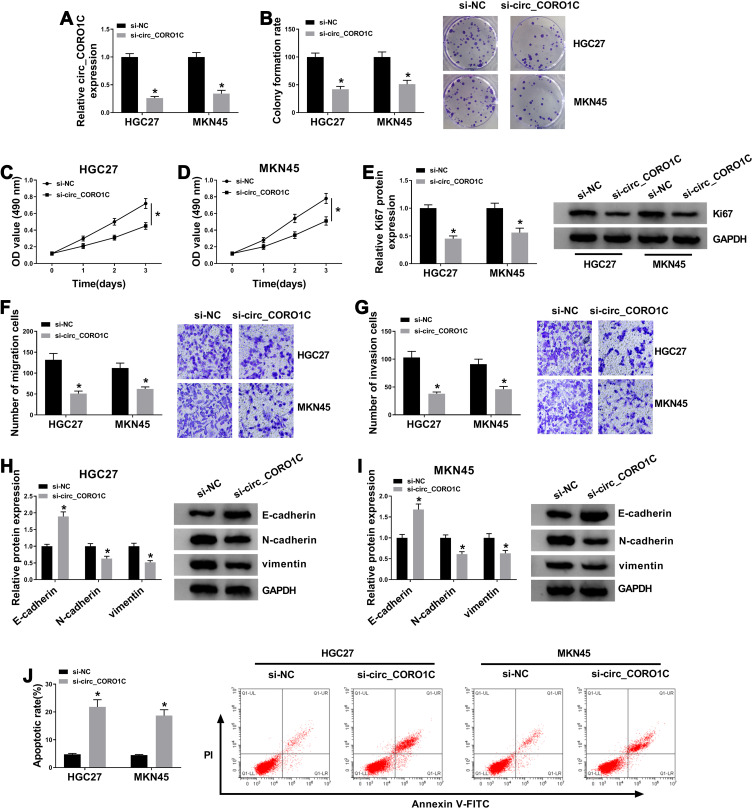
Subsequently, loss-of-function assays were performed to explore the effects of circ_CORO1C on cellular behaviors of GC cells. Circ_CORO1C knockdown cells were successfully constructed by transfecting siRNA against circ_CORO1C, and cells transfected with si-NC served as control (Figure 2A). Following colony formation assay showed that circ_CORO1C knockdown reduced the colony formation ability of GC cells (Figure 2B). Depletion of circ_CORO1C also inhibited the cell viability of HGC-27 and MKN45 cells (Figure 2C and D). Western blot assay showed that circ_CORO1C knockdown also reduced the protein level of cell proliferation biomarker Ki67 (Figure 2E). Transwell assay suggested that circ_CORO1C deficiency remarkably repressed cell migration (Figure 2F) and invasion (Figure 2G) of HGC-27 and MKN45 cells. Western blot assay was also conducted to clarify the role of circ_CORO1C in the epithelia-mesenchymal transition (EMT) process. Obviously, depletion of circ_CORO1C triggered the upregulation of E-cadherin and the downregulation of N-cadherin and vimentin (Figure 2H and I). Flow cytometry witnessed circ_CORO1C knockdown-induced elevated apoptotic rate of GC cells (Figure 2J). Taken together, depletion of circ_CORO1C hampered proliferation and metastasis, while facilitated cell apoptosis of GC cells.
Figure 2.
Depletion of circ_CORO1C repressed GC cell proliferation and metastasis, while promoted cell apoptosis. HGC-27 and MKN45 cells were transfected with si-NC or si-circ_CORO1C. (A) QRT-PCR assay for the relative expression of circ_CORO1C in transfected cells. (B) Colony formation assay for the colony formation ability of transfected GC cells. (C and D) MTT assay for the cell viability of transfected cells. (E) Western blot assay for the protein level of Ki67 in transfected cells. (F and G) Transwell assay for the migration and invasion of transfected cells. (H and I) Western blot assay for the protein levels of E-cadherin, N-cadherin and vimentin in transfected cells. (J) Flow cytometry for the apoptotic rate of transfected cells. *P <0.05.
Circ_CORO1C Acted as a Sponge of miR-138-5p
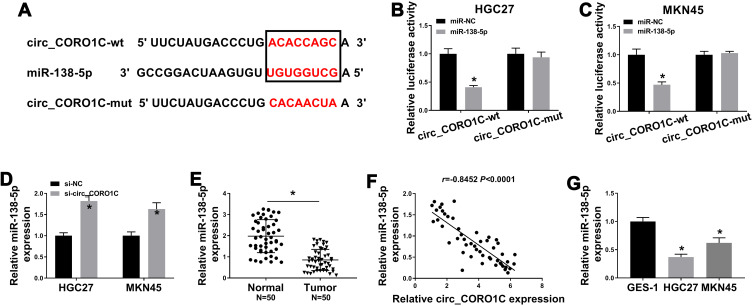
Mechanically, circRNAs were reported to functioned by sponging miRNAs.24 Thus, we searched the target miRNA of circ_CORO1C using Starbase 3.0, let-7a-5p, miR-138-5p, miR-379-3p, miR-411-3p and miR-448 were estimated to be potential candidates. Additionally, the expression level of miR-138-5p was highest in GC cells with circ_CORO1C depletion among these 5 miRNAs (Supplementary Fig 2), so it was selected for further investigation. The complementary binding sites between circ_CORO1C and miR-138-5p are exhibited in Figure 3A. Dual-luciferase reporter assay was used to confirm the target relationship. Obviously, introduction of miR-138-5p significantly reduced the luciferase intensity of circ_CORO1C-wt (more than 50%) in HGC-27 and MKN45 cells, rather than circ_CORO1C-mut (Figure 3B and C). Additionally, we found that miR-138-5p expression in HGC-27 and MKN45 cells was upregulated by circ_CORO1C inhibition (Figure 3D). The expression of miR-138-5p in GC tissues was lower than that in normal tissues (Figure 3E), which was negatively correlated with circ_CORO1C expression (r= −0.8452, P <0.0001) (Figure 3F). The downregulation of miR-138-5p was observed in HGC-27 and MKN45 cells, with respect to GES-1 cells (Figure 3G). Collectively, circ_CORO1C could target miR-138-5p in GC cells.
Figure 3.
Circ_CORO1C acted as a sponge of miR-138-5p. (A) The binding sites between circ_CORO1C and miR-138-5p, as well as the mutant. (B and C) Dual-luciferase reporter assay for the luciferase intensity of circ_CORO1C-wt and circ_CORO1C-mut in HGC-27 and MKN45 cells transfected with miR-NC or miR-138-5p. (D) QRT-PCR assay for the relative expression of miR-138-5p in HGC-27 and MKN45 cells transfected with si-NC or si-circ_CORO1C. (E) QRT-PCR assay for the relative expression of miR-138-5p in GC tissues (n=50) and matched adjacent normal tissues. (F) Pearson correlation analysis for the correlation between expression of circ_CORO1C and miR-138-5p in GC tissues. (G) QRT-PCR assay for the relative expression of miR-138-5p in GES-1, HGC-27 and MKN45 cells. *P <0.05.
Circ_CORO1C Acted as Oncogenic Role in GC Cells by Directly Targeting miR-138-5p
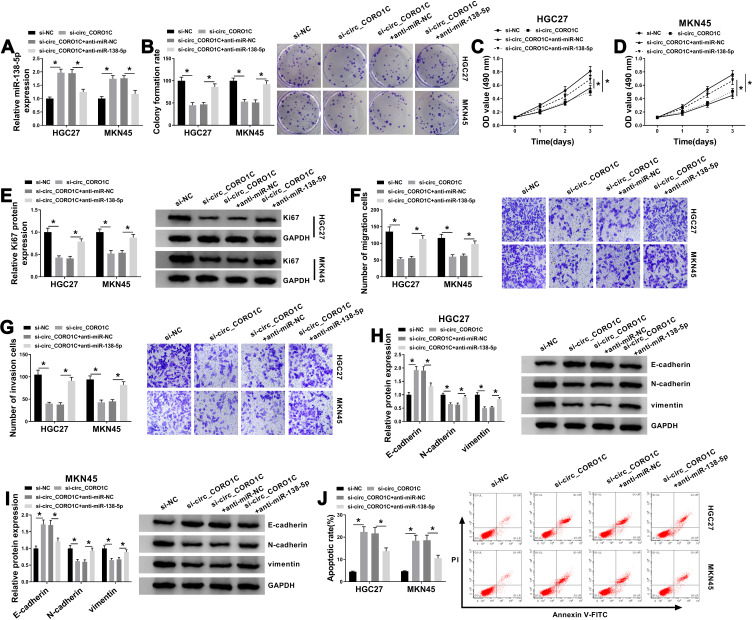
In view of circ_CORO1C targeted miR-138-5p in GC cells, we then conducted a series of rescue experiments to investigate the co-effect of circ_CORO1C and miR-138-5p on cellular behaviors. Using qRT-PCR assay, it was found that circ_CORO1C deficiency-induced miR-138-5p upregulation was ameliorated by anti-miR-138-5p (Figure 4A). Following functional assays demonstrated that si-circ_CORO1C repressed colony formation ability (Figure 4B), cell viability (Figure 4C and D), cell proliferation (Figure 4E), migration (Figure 4F) and invasion (Figure 4G), and upregulated the expression of E-cadherin while downregulated N-cadherin and vimentin (Figure 4H and I), as well as promoted apoptotic (Figure 4J). These effects were all partially reversed by anti-miR-138-5p. In sum, circ_CORO1C affected the growth and metastasis of GC cells by directly targeting miR-138-5p.
Figure 4.
Circ_CORO1C exerted its oncogenic role in GC cells by directly targeting miR-138-5p. HGC-27 and MKN45 cells were transfected with si-NC, si-circ_CORO1C, si-circ_CORO1C+anti-miR-NC or si-circ_CORO1C+anti-miR-138-5p. (A) QRT-PCR assay for the relative expression of miR-138-5p in transfected cells. (B) Colony formation assay for the colony formation ability of transfected GC cells. (C and D) MTT assay for the cell viability of transfected cells. (E) Western blot assay for the protein level of Ki67 in transfected cells. (F and G) Transwell assay for the migration and invasion of transfected cells. (H and I) Western blot assay for the protein levels of E-cadherin, N-cadherin and vimentin in transfected cells. (J) Flow cytometry for the apoptotic rate of transfected cells. *P <0.05.
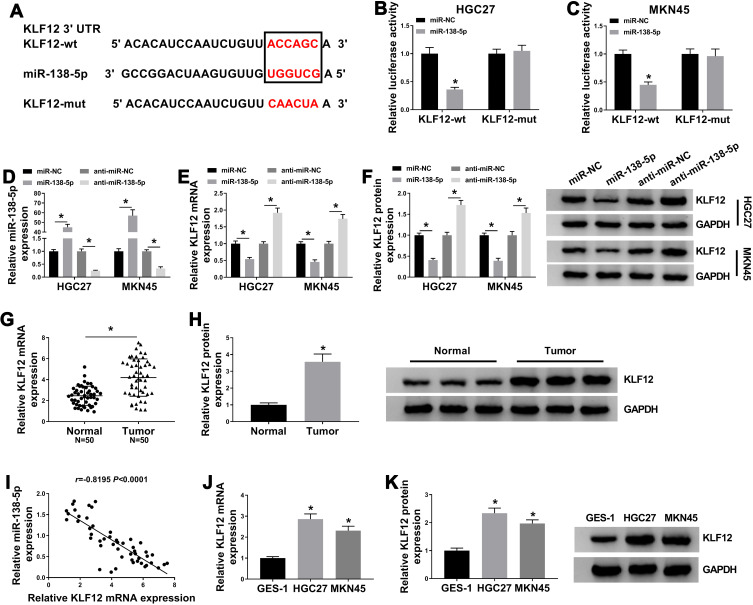
KLF12 Was a Direct Target of miR-138-5p
To unveil the interaction between KLF12 and miR-138-5p, miRDB was used. Results showed that KLF12 was a downstream gene of miR-138-5p (Figure 5A). Dual-luciferase reporter assay disclosed that miR-138-5p induced about 60% reduction of the luciferase intensity in KLF12-wt, while no obvious influence was observed in KLF12-mut (Figure 5B and C). By using qRT-PCR assay, we found that miR-138-5p mimic efficiently upregulated miR-138-5p expression, while miR-138-5p inhibitor caused opposite effect (Figure 5D). Moreover, miR-138-5p mimic repressed the mRNA and protein expression KLF12; on the contrary, anti-miR-138-5p upregulated KLF12 expression (Figure 5E and F). In addition, mRNA and protein levels of KLF12 were upregulated in GC tissues (Figure 5G and H), as well as in GC cells (Figure 5J and K), in contrast to the corresponding control. KLF12 mRNA expression in GC tissues was inversely correlated with miR-138-5p expression (r= −0.8195, P <0.0001) (Figure 5I). Therefore, miR-138-5p targeted KLF12 in GC cells.
Figure 5.
KLF12 was a direct target of miR-138-5p. (A) The binding sites between miR-138-5 and KLF12, as well as the mutant. (B and C) Dual-luciferase reporter assay for the luciferase intensity of KLF12-wt and KLF12-mut in HGC-27 and MKN45 cells transfected with miR-NC or miR-138-5p. (D) QRT-PCR assay for the relative expression of miR-138-5p in HGC-27 and MKN45 cells transfected with miR-NC, miR-138-5p, anti-miR-NC or anti-miR-138-5p. (E and F) QRT-PCR and Western blot assays for the mRNA and protein levels of KLF12 in HGC-27 and MKN45 cells transfected with miR-NC, miR-138-5p, anti-miR-NC or anti-miR-138-5p. (G) QRT-PCR for the mRNA level of KLF12 in GC tissues (n=50) and matched adjacent normal tissues. (H) Western blot assay for the protein level of KLF12 in GC tissues and matched adjacent normal tissues. (I) Pearson correlation analysis for the correlation between expression of KLF12 mRNA and miR-138-5p in GC tissues. (J and K) QRT-PCR and Western blot assays for the mRNA and protein levels of KLF12 in GES-1, HGC-27 and MKN45 cells. *P <0.05.
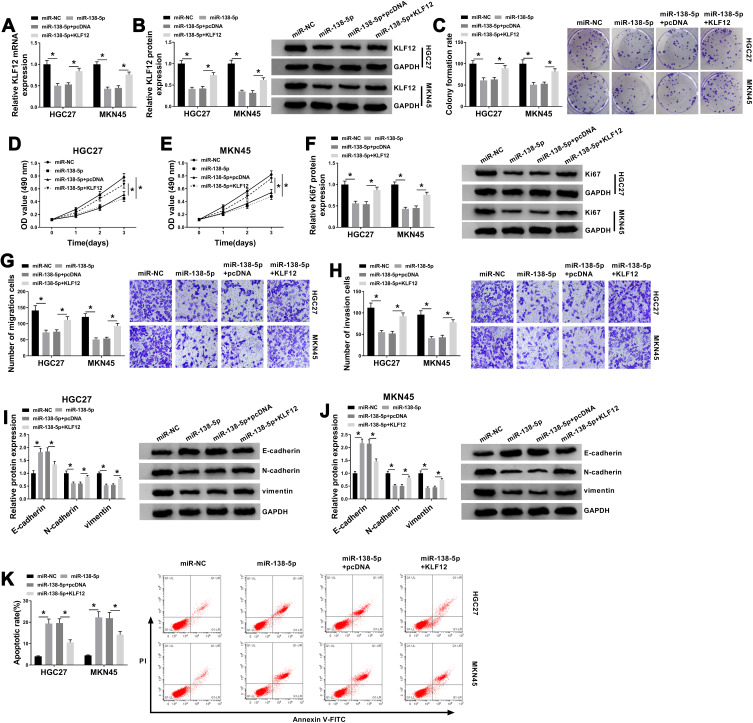
KLF12 Attenuated miR-138-5p-Induced GC Cell Proliferation and Metastasis Inhibition and Cell Apoptosis Promotion
We then explored the effects of miR-138-5p and KLF12 on the cellular behaviors. As exhibited in Figure 6A and B, miR-138-5p mimic obviously reduced the mRNA and protein levels of KLF12, which was relieved by introduced KLF12. Additionally, upregulated miR-138-5p inhibited colony formation ability (Figure 6C), cell viability (Figure 6D and E), proliferation (Figure 6F), migration (Figure 6G) and invasion (Figure 6H), and upregulated E-cadherin while downregulated N-cadherin and vimentin (Figure 6I and J), as well as increased apoptotic rate (Figure 6K) in GC cells, all these effects were weakened by the introduction of KLF12. Collectively, miR-138-5p suppressed the growth and metastasis of GC cells by downregulating KLF12 expression.
Figure 6.
KLF12 could attenuate miR-138-5p-induced GC cell proliferation and metastasis inhibition and cell apoptosis promotion. HGC-27 and MKN45 cells transfected with miR-NC, miR-138-5p, miR-138-5p+pcDNA or miR-138-5p+KLF12. (A and B) QRT-PCR and Western blot assays for the mRNA and protein levels of KLF12 in transfected cells. (C) Colony formation assay for the colony formation ability of transfected GC cells. (D and E) MTT assay for the cell viability of transfected cells. (F) Western blot assay for the protein level of Ki67 in transfected cells. (G and H) Transwell assay for the migration and invasion of transfected cells. (I and J) Western blot assay for the protein levels of E-cadherin, N-cadherin and vimentin in transfected cells. (K) Flow cytometry for the apoptotic rate of transfected cells. *P <0.05.
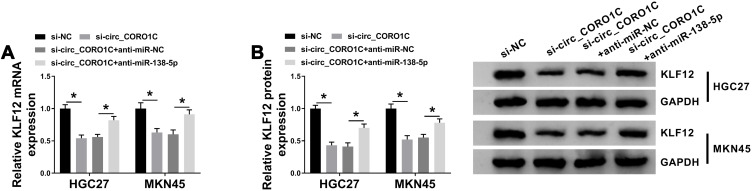
Circ_CORO1C Positively Regulated KLF12 by Sponging miR-138-5p in GC Cells
Based on the above results, the effect of circ_CORO1C on KLF12 expression in HGC-27 and MKN45 cells was analyzed. Results from qRT-PCR and Western blot showed that circ_CORO1C knockdown downregulated KLF12 expression at mRNA and protein levels, while anti-miR-138-5p largely restored it (Figure 7A and B). Thus, circ_CORO1C positively regulated KLF12 expression by absorbing miR-138-5p in GC cells.
Figure 7.
Circ_CORO1C positively regulated KLF12 by sponging miR-138-5p in GC cells. HGC-27 and MKN45 cells transfected with si-NC, si-circ_CORO1C, si-circ_CORO1C+anti-miR-NC or si-circ_CORO1C+anti-miR-138-5p. (A and B) QRT-PCR and Western blot assays for the mRNA and protein levels of KLF12 in transfected cells. *P <0.05.
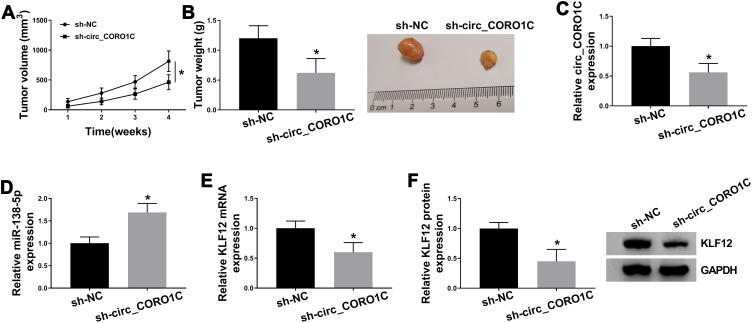
Silencing of circ_CORO1C Inhibited Tumor Growth in vivo
To explore the role of circ_CORO1C in vivo, a tumor xenograft model was constructed by injecting MKN45 cells stably expressing sh-circ_CORO1C or sh-NC. As shown in Figure 8A and B and Supplementary Fig 3, the volume and weight of tumors generated in sh-circ_CORO1C group were significantly decreased in contrast to those in sh-NC group. In addition, circ_CORO1C (Figure 8C) and KLF12 (Figure 8E and F) were downregulated, while miR-138-5p (Figure 8D) was upregulated in sh-circ_CORO1C group relative to sh-NC group. The above results suggested that circ_CORO1C knockdown could inhibit tumor growth in vivo.
Figure 8.
Silencing of circ_CORO1C inhibited tumor growth in vivo. 5-week-old male BALB/C nude mice were subcutaneously injected with MKN45 cells stably expressing sh-circ_CORO1C or sh-NC (n=5). (A) Tumor growth curve of nude mice. (B) Tumor weight in nude mice at 4 weeks post injection. (C and D) QRT-PCR assay for the expression of circ_CORO1C (C) and miR-138-5p (D) in generated tumors. (E and F) QRT-PCR and Western blot assays for the mRNA (E) and protein (F) levels of KLF12 in generated tumors. *P <0.05.
Discussion
Recently, more and more circRNAs associated with GC have been identified, followed by clarification of their biological functions and action mechanisms.25 In fact, circRNAs take part in the regulation of tumorigenic properties of GC cells and are correlated with several clinical characteristics of GC patients.26 In our study, circ_CORO1C was identified as an oncogenic circRNA in GC, the knockdown of which inhibited GC cell proliferation and metastasis, while promoted cell apoptosis in vitro, as well as blocked tumor growth in vivo. Furthermore, circ_CORO1C might exert oncogenic role through miR-138-5p/KLF12 axis.
As far as we know, few research focused on circ_CORO1C. There was a report that mentioned the dysregulation of circ_CORO1C in GC tissues.10 Herein, 50 pairs of GC tissues and normal tissues were collected, and the upregulation of circ_CORO1C in GC tissues relative to normal tissues was observed. Circ_CORO1C was also highly expressed in GC cells, implying its oncogenic potency in GC. Loss-of-function assays suggested that circ_CORO1C knockdown repressed aggressive malignant behaviors of GC cells and tumor growth in mice model.
Functionally, circRNAs could act as sponges of miRNAs, indirectly affecting the expression and function of downstream mRNAs.27,28 Here, we utilized Starbase 3.0 to predict the target miRNA of circ_CORO1C, and miR-138-5p was found to have binding sites with circ_CORO1C, which was confirmed by dual-luciferase reporter assay. Pang et al corroborated that miR-138 was connected to advanced TNM stage and lymph node metastasis, and miR-138 blocked aggressive development of GC.17 MiR-138-5p was downregulated in GC, which could confer anti-proliferation and anti-metastasis influence on GC cells by targeting EGFR.29 Additionally, miR-138-5p was sponged by circular RNA circNRIP1, involved in hypoxia-induced 5-FU resistance of GC cells.30 Similarly, downregulation of miR-138-5p was detected in GC tissues and cells. Apart from this, interference of miR-138-5p largely weakened the inhibition effect of circ_CORO1C knockdown on GC development, indicating that circ_CORO1C exerted its oncogenic role by absorbing miR-138-5p.
It is widely accepted that circRNA could function as competing endogenous RNA (ceRNA) of miRNAs, indirectly affecting the expression and function of mRNAs.31 In our study, KLF12 was firstly identified as a target gene of miR-138-5p, validated by dual-luciferase reporter assay. As reported previously, KLF12 was targeted by miR-137 and miR-200a-3p, two tumor suppressor genes in GC.32,33 Furthermore, KLF12 was upregulated in GC, which could participate in lncRNA TTN-AS1 or HOTAIR-mediated GC progression.34,35 Here, we also revealed the upregulation of KLF12 in GC. Functionally, overexpressed KLF12 attenuated the suppressive effect on GC cell proliferation and metastasis mediated by miR-138-5p. It was also found that circ_CORO1C positively regulated KLF12 expression by antagonizing miR-138-5p. In sum, circ_CORO1C exerted oncogenic role in GC via regulating miR-138-5p/KLF12 axis.
Taken together, we firstly uncover the oncogenic role of circ_CORO1C in GC progression. Depletion of circ_CORO1C could repress GC cell proliferation, metastasis and tumorigenicity through miR-138-5p/KLF12 axis. Our findings suggested that circ_CORO1C might be applied for GC therapy.
Acknowledgment
Yongqiang Fan and Min Liu are co-first authors.
Funding Statement
No funding was received.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of Sunshine Union Hospital. Written informed consent was obtained from all enrolled patients.
Consent for Publication
Patients agree to participate in this work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Coccolini F, Montori G, Ceresoli M, et al. Advanced gastric cancer: what we know and what we still have to learn. World J Gastroenterol. 2016;22(3):1139–1159. doi: 10.3748/wjg.v22.i3.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.RaFiei E, Mohammadian-Hafshejani A, Towhidi F, Makhsosi BR, Salehiniya H. Lack of any relationship of stomach cancer incidence and mortality with development in Asia. Asian Pac J Cancer Prev. 2016;17(8):3777–3783. [PubMed] [Google Scholar]
- 4.Xu Z, Yan Y, Zeng S, et al. Circular RNAs: clinical relevance in cancer. Oncotarget. 2018;9(1):1444–1460. doi: 10.18632/oncotarget.22846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11(1):21. doi: 10.1186/s13045-018-0569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi: 10.1080/15476286.2016.1220473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20. doi: 10.1186/s12943-018-0935-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Song X, Chen X, et al. Circular RNA CircCACTIN promotes gastric cancer progression by sponging MiR-331-3p and regulating TGFBR1 expression. Int J Biol Sci. 2019;15(5):1091–1103. doi: 10.7150/ijbs.31533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong S, Wang J, Hou J, et al. Circular RNA hsa_circ_0000993 inhibits metastasis of gastric cancer cells. Epigenomics. 2018;10(10):1301–1313. doi: 10.2217/epi-2017-0173 [DOI] [PubMed] [Google Scholar]
- 10.Vidal AF, Ribeiro-Dos-Santos AM, Vinasco-Sandoval T, et al. The comprehensive expression analysis of circular RNAs in gastric cancer and its association with field cancerization. Sci Rep. 2017;7(1):14551. doi: 10.1038/s41598-017-15061-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi: 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- 12.Shrestha S, Hsu SD, Huang WY, et al. A systematic review of microRNA expression profiling studies in human gastric cancer. Cancer Med. 2014;3(4):878–888. doi: 10.1002/cam4.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H, Xiong Y, Wu Z, et al. MIR-138-5P inhibits the progression of prostate cancer by targeting FOXC1. Mol Genet Genomic Med. 2020;8(4):e1193. doi: 10.1002/mgg3.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu D, Gu L, Li Z, Jin W, Lu Q, Ren T. MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathol Res Pract. 2019;215(5):861–872. doi: 10.1016/j.prp.2019.01.029 [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Yao YJ, Zheng F, et al. Mir-138-5p acts as a tumor suppressor by targeting pyruvate dehydrogenase kinase 1 in human retinoblastoma. Eur Rev Med Pharmacol Sci. 2017;21(24):5624–5629. doi: 10.26355/eurrev_201712_14005 [DOI] [PubMed] [Google Scholar]
- 16.Yang R, Liu M, Liang H, et al. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Mol Cancer. 2016;15(1):82. doi: 10.1186/s12943-016-0569-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pang L, Li B, Zheng B, Niu L, Ge L. miR-138 inhibits gastric cancer growth by suppressing SOX4. Oncol Rep. 2017;38(2):1295–1302. doi: 10.3892/or.2017.5745 [DOI] [PubMed] [Google Scholar]
- 18.Lam VC, Folkersen L, Aguilar OA, Lanier LL. KLF12 regulates mouse NK cell proliferation. J Immunol. 2019;203(4):981–989. doi: 10.4049/jimmunol.1900396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SH, Park YY, Cho SN, Margalit O, Wang D, DuBois RN. Krüppel-like factor 12 promotes colorectal cancer growth through early growth response protein 1. PLoS One. 2016;11(7):e0159899. doi: 10.1371/journal.pone.0159899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Pu J, Zhang Y, et al. DANCR contributed to hepatocellular carcinoma malignancy via sponging miR-216a-5p and modulating KLF12. J Cell Physiol. 2019;234(6):9408–9416. doi: 10.1002/jcp.27625 [DOI] [PubMed] [Google Scholar]
- 21.Song P, Yin SC. Long non-coding RNA 319 facilitates nasopharyngeal carcinoma carcinogenesis through regulation of miR-1207-5p/KLF12 axis. Gene. 2019;680:51–58. doi: 10.1016/j.gene.2018.09.032 [DOI] [PubMed] [Google Scholar]
- 22.Nakamura Y, Migita T, Hosoda F, et al. Krüppel-like factor 12 plays a significant role in poorly differentiated gastric cancer progression. Int J Cancer. 2009;125(8):1859–1867. doi: 10.1002/ijc.24538 [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 24.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 25.Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. 2020;77(9):1661–1680. doi: 10.1007/s00018-019-03345-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang X, Wen J, Sun M, Yuan Y, Xu Q. CircRNAs and its relationship with gastric cancer. J Cancer. 2019;10(24):6105–6113. doi: 10.7150/jca.32927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Qi X, Liu L, et al. Emerging epigenetic regulation of circular RNAs in human cancer. Mol Ther Nucleic Acids. 2019;16:589–596. doi: 10.1016/j.omtn.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu X, Song M, Wang F. Circulating circular RNAs as biomarkers of cancer. Non-Coding RNA Invest. 2019;3:8. doi: 10.21037/ncri.2019.02.01 [DOI] [Google Scholar]
- 29.Wang Y, Zhang H, Ge S, et al. Effects of miR‑138‑5p and miR‑204‑5p on the migration and proliferation of gastric cancer cells by targeting EGFR. Oncol Rep. 2018;39(6):2624–2634. doi: 10.3892/or.2018.6389 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Xu G, Li M, Wu J, Qin C, Tao Y, He H. Circular RNA circNRIP1 sponges microRNA-138-5p to maintain hypoxia-induced resistance to 5-fluorouracil through HIF-1α-dependent glucose metabolism in gastric carcinoma. Cancer Manag Res. 2020;12:2789–2802. doi: 10.2147/CMAR.S246272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281. doi: 10.18632/oncotarget.19154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du Y, Chen Y, Wang F, Gu L. miR-137 plays tumor suppressor roles in gastric cancer cell lines by targeting KLF12 and MYO1C. Tumour Biol. 2016;37(10):13557–13569. doi: 10.1007/s13277-016-5199-3 [DOI] [PubMed] [Google Scholar]
- 33.Jia C, Zhang Y, Xie Y, et al. miR-200a-3p plays tumor suppressor roles in gastric cancer cells by targeting KLF12. Artif Cells Nanomed Biotechnol. 2019;47(1):3697–3703. doi: 10.1080/21691401.2019.1594857 [DOI] [PubMed] [Google Scholar]
- 34.Dong MM, Peng SJ, Yuan YN, Luo HP. LncRNA TTN-AS1 contributes to gastric cancer progression by acting as a competing endogenous RNA of miR-376b-3p. Neoplasma. 2019;66(4):564–575. doi: 10.4149/neo_2018_180927N721 [DOI] [PubMed] [Google Scholar]
- 35.Xun J, Wang C, Yao J, Gao B, Zhang L. Long non-coding RNA HOTAIR modulates KLF12 to regulate gastric cancer progression via PI3K/ATK signaling pathway by sponging miR-618. Onco Targets Ther. 2019;12:10323–10334. doi: 10.2147/OTT.S223957 [DOI] [PMC free article] [PubMed] [Google Scholar]