Abstract
Background
Obesity, characterized by an increased amount of adipose tissue, is a metabolic chronic alteration which has reached pandemic proportion. Lifestyle changes are the first line therapy for obesity and a large variety of dietary approaches have demonstrated efficacy in promoting weight loss and improving obesity-related metabolic alterations. Besides diet and physical activity, bariatric surgery might be an effective therapeutic strategy for morbid obese patients. Response to weight-loss interventions is characterised by high inter-individual variability, which might involve epigenetic factors. microRNAs have critical roles in metabolic processes and their dysregulated expression has been reported in obesity.
Aim
The aim of this review is to provide a comprehensive overview of current studies evaluating changes in microRNA expression in obese patients undergoing lifestyle interventions or bariatric surgery.
Results
A considerable number of studies have reported a differential expression of circulating microRNAs before and after various dietary and bariatric surgery approaches, identifying several candidate biomarkers of response to weight loss. Significant changes in microRNA expression have been observed at a tissue level as well, with entirely different patterns between visceral and subcutaneous adipose tissue. Interestingly, relevant differences in microRNA expression have emerged between responders and non-responders to dietary or surgical interventions. A wide variety of dysregulated microRNA target pathways have also been identified, helping to understand the pathophysiological mechanisms underlying obesity and obesity-related metabolic diseases.
Conclusions
Although further research is needed to draw firm conclusions, there is increasing evidence about microRNAs as potential biomarkers for weight loss and response to intervention strategies in obesity.
Keywords: Obesity, microRNA, Weight loss, Diet, Bariatric surgery, Obesity treatment
The challenges of obesity treatment
Obesity is a common condition that has been consistently linked to an increased risk of developing a wide range of disorders, such as metabolic syndrome (MS), type 2 diabetes (T2D), cardiovascular diseases (CVD), non-alcoholic fatty liver disease (NAFLD), musculoskeletal diseases and some cancers [1–4]. Over the last decades, the prevalence of this condition has dramatically risen, reaching pandemic proportions. Globally, it has been estimated that nearly 39% of adults are overweight and 13% are obese [5]. Obesity is defined as excessive fat accumulation ensuing from a chronic imbalance between calorie intake and energy expenditure, which is linked to unhealthy diet habits and physical inactivity [6]. Excess energy is stored as triglycerides in white adipose tissue (WAT), which comprises subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT). Although adipocytes display similar functions in SAT and VAT, the latter is more markedly associated with abnormal metabolic profiles, directly contributing to insulin resistance [7, 8]. Specifically, in vitro studies have reported a more consistent release of free fatty acids (FFA) in VAT due to a higher lipolytic activity, which is able to induce insulin resistance impairing insulin signalling [9, 10]. It has been observed that VAT expresses several pro-inflammatory and anti-inflammatory mediators which are critically involved in the maintenance of immune metabolic homeostasis [11]. Furthermore, the greater number of macrophages in VAT compared to SAT leads to a more marked production of pro-inflammatory cytokines, significantly contributing to the chronic low grade inflammation typical of obesity and metabolic diseases [12, 13].
Despite the advances in therapeutic strategies, obesity management remains highly challenging. Lifestyle modification is the primary approach for weight loss and fat mass reduction. Indeed, dietary energy restriction and increased energy expenditure through physical activity are the well-known cornerstones of obesity treatment [14]. Specifically, energy restriction is crucial for the weight-loss phase, while physical activity is essential for long-term maintenance. A decrease of at least 5% of the initial body weight is considered clinically significant and can be effectively achieved by lowering the daily caloric intake by 500 kcal [15]. Of note, exercise without dietary restrictions is able to achieve only 3–5% weight loss in patients with obesity [16]. A wide variety of dietary intervention patterns, such as Mediterranean diet, low carbohydrate diet, low-fat diet and ketogenic diet (KD), which significantly differ in the proportions of macronutrients, have been extensively evaluated in clinical trials and displayed great efficacy in reducing body weight and in improving metabolic abnormalities in obese patients [17–19]. On the other hand, bariatric surgery is a highly effective strategy for weight loss and comorbidities improvement in morbidly obese patients when life style intervention fails. Several surgical procedures based on restrictive or malabsorptive approaches are currently available. Specifically, laparoscopic sleeve gastrectomy (LSG) and laparoscopic adjustable gastric banding (LAGB) significantly restrict food intake, while Roux-en-Y gastric by-pass (RYGB) is mainly malabsorptive [20]. The most frequent complications of bariatric surgery include dumping, nausea, vomiting, diarrhoea, infections, stenosis, bleeding, increased risk of alcohol abuse after surgery, and perioperative death.[21, 22].
Despite the different therapeutic options, response to weight-loss programmes is hallmarked by high inter-individual variability, which might be partly explained by epigenetic factors [23–25]. In light of this, the development of novel tools based on nutrigenetic and nutrigenomic information is of utmost importance in the development of customized approaches in obesity management [26]. Thus, great efforts are being made to identify valuable predictors of response to diet or bariatric surgery interventions. Recently microRNAs, a class of small non-coding RNAs that regulate gene expression, have gained much attention not only as regulators of biological processes but also as prognostic biomarkers in obesity management.
General aspects of microRNAs
microRNAs are small (19–25 nucleotides) non-coding single stranded RNAs that function destabilizing or depleting target mRNAs [27]. Since their discovery in Caenorhabditis elegans in 1993 they have been described in many other species, even viruses, and to date 2654 human mature microRNA sequences have been identified (miRbase version 22.1 released in October 2018) [28–31]. Their dysregulation has been described in the context of many pathological processes, spanning from cancer to neurological and metabolic diseases. Therefore, although their function has not been yet completely defined, their huge number and wide species distribution suggest a crucial role in gene regulation [32, 33].
microRNAs biogenesis, secretion and function involve many complex molecular events, not completely understood yet. microRNA genes are usually transcribed by RNA polymerase II and sometimes by RNA polymerase III. The pri-miRNA sequence is capped at the 5′ and polyadenylated at the 3′ end and recognized by the Microprocessor complex, constituted by the Di George Syndrome Critical Region 8 (DGCR8) nuclear protein and the RNAse III Drosha. This first nuclear maturation step releases a precursor microRNA, called pre-miRNA. The protein Exportin-5 is then responsible for the translocation from the nucleus to the cytoplasm, where the pre-miRNA is cleaved by the RNAse III Dicer with the production of a double strand microRNA of about 22 nucleotides in length. After loading onto Ago2, a member of the Argonaute (Ago) protein family, the microRNA is included into the RNA-induced silencing complex (RISC), a big ribonucleoprotein effector complex. The binding with Ago2 favours the most stable strand, while the passenger strand is degraded to produce a mature RISC. The main mechanisms responsible for the target mRNA silencing are mRNA degradation and translational repression. Specifically, if the sequence homology between the microRNA and its target mRNA is complete, Ago proteins degrade the target mRNA. Otherwise, if the sequence homology is only partial, there is only translational repression [34, 35].
Although the cellular compartment is the site of microRNA production and action, recent evidence demonstrated that microRNAs can also act into the extracellular compartment after secretion. One of the most intriguing hypothesis related to their function and origin sustains that extracellular microRNAs are involved in cell-to-cell communication. Circulating microRNAs have been detected in different biological fluids, such as serum, plasma and urine [36, 37]. To date, it has been demonstrated that microRNAs can be packaged into shedding vesicles and exosomes or coupled with high-density lipoproteins (HDL) and low-density lipoproteins (LDL) or Ago proteins, and actively secreted by cells. In addition, microRNAs can be passively secreted after incorporation in apoptotic bodies [38].
In addition, microRNA encapsulation into extracellular vesicles is a well-structured and defined process. The microvesicle-packed microRNAs do not simply mirror the cell of origin repertoire, since some microRNAs are preferentially exported or retained in cells [39].
Importantly, thanks to the circulating microRNA association with both protein complexes and/or to their packing into extracellular vesicles, microRNAs show high stability into the extracellular environment, rendering not only very stable extracellular molecules, but also capable of acting as diagnostic, prognostic and therapeutic biomarkers [40–42].
Moreover, microRNAs play key roles in physiological and pathological processes. They are important in fat cell formation and in the regulation of metabolic and endocrine functions. Indeed, the integral role of microRNAs in adipose tissue is emerging from studies demonstrating that the inhibition of Drosha and Dicer in human mesenchymal stem cells (MSC) inhibited the differentiation into adipocytes, conversely Dicer in 3T3-L1 cells inhibited adipogenesis [43, 44]. Other studies showed that microRNA are involved in the regulation of adipogenesis, acting as stimulators (i.e. miR-21, miR-26b, miR-30, miR-103, miR-143, miR-148, miR-181a, miR-199a, miR-378) or inhibitors (i.e. let7, miR-22, miR-125a, miR-224) of human adipocyte differentiation programmes [45–51]. As regards human obesity, several studies identified dysregulated microRNAs. In particular, many microRNAs were differentially regulated in WAT of obese subjects compared to lean human subjects [52–54]. Finally, several microRNAs are known to influence lipolytic activity [55–57] and adipokine production in fat cells [58, 59].
microRNAs and weight loss: evidence from lifestyle intervention studies
Interestingly, compelling evidence suggests that differences in the outcomes of dietary intervention might be related to epigenetic factors [25]. The role of microRNAs as biomarkers of weight loss after dietary and lifestyle interventions has been extensively investigated in a wide number of studies, exploring microRNA expression at a systemic level (white blood cells, serum and plasma) and in adipose tissue (Table 1).
Table 1.
Studies that evaluated microRNA expression change after lifestyle intervention strategies
| Study | Population | Intervention | Source | Regulated miRNAs | Role/target | |
|---|---|---|---|---|---|---|
| Milagro et al. 2013 [60] | 10 obese women | 8-week hypocaloric diet | PBMC |
miR-935, miR-4772 (↑) miR-376b (↓) |
Biomarkers of weight loss | |
| Marques-Rocha et al. 2016 [61] | 40 MS | 8-week hypocaloric Mediterranean diet | WBC |
miR-155-3p (↓) let-7b (↑) |
Biomarkers of improvement in quality of diet and weight loss (155-3p) | |
| Garcia-Lacarte et al. 2018 [62] | 96 MS | 8-week hypocaloric diet | WBC | miR-612, miR-1976 (↑) | Biomarkers of high response to diet/TP53 and CD40 | |
| Garcia-Lacarte et al. 2019 [63] | 96 MS | 12-week hypocaloric diet | WBC | miR-548q, miR-1185-1 (↑) | Biomarkers of high response to diet/GSK3B | |
| Hess et al. 2020 [65] | 85 obese/overweight | 12-week hypocaloric diet | Serum |
miR-222-3p (↑) miR-122-5p, miR-193a-5p (↓) |
Biomarkers of weight loss and MS/glucose metabolism | |
| Giardina et al. 2019 [71] | 103 obese | 6-month LGI (n = 36), HGI (n = 36), LF (n = 31) diet; | Plasma |
(↓) in LGI vs HGI diet: miR-139-3p, miR-411, miR-432, miR-99b, miR-340, miR-423-5p, miR-361, let-7c (↓) in LF diet: miR-139-3p |
Association with specific dietary patterns/Lipid metabolism | |
| Assman et al. 2020 [72] |
78 obese 25 CTRL |
high-protein diet (n = 38); LF diet (n = 40) | Plasma |
LF diet: miR-130a-3p, miR-142-5p, miR-144-5p, miR-15a-5p, miR-221-3p, miR-29c-3p (↓) miR-22-3p (↑) |
Biomarkers of response to LF diet/PPAR-γ, PPAR- α, SIRT1 (miR-22-3p) | |
| Manning et al. 2019 [75] |
80 obese women 80 CTRL women |
4-week very-low-calorie diet | Plasma |
miR-126, miR-375, miR-376, miR-499, miR-642 (↑) miR-208, miR-433 (↓) |
Biomarkers of weight loss/glucose metabolism, inflammation, angiogenesis, cell death | |
| Cannataro et al. 2019 [76] | 36 obese | 6-week KD | Plasma |
Women: miR-148b-3p, miR-26a-5p, miR-520h, miR-548d-3p Both sexes: miR-30e-5p, miR-502-5p, miR-590-5p, miR-644a, let-7b-5p, miR-143-3p, miR-504-5p |
Sex differences in microRNA expression after KD/ inflammation, immunity, glucose metabolism | |
| Margolis et al. 2017 [77] | 16 elderly men | 28-day hypocaloric diet | Serum | miR-133a-3p, miR-133b (↑) | Biomarkers of reduced skeletal-muscle regeneration | |
| Parr et al. 2016 [78] | 111 obese | 16-week diet (HPHC/HPMC/CON) and physical activity | Plasma |
miR-140, miR-221, miR-223 (↑) low responders: miR-935 (↑) |
Biomarkers of response to intervention | |
| Donghui et al. 2019 [79] |
37 obese male adolescents 10 CTRL |
6-week hypocaloric diet and aerobic exercise | Serum | miR-126 (↑) | Biomarker of weight loss/endothelial function | |
| Kristensen et al. 2017 [80] | 38 obese | 15-week hypocaloric diet and physical activity | SAT |
miR-29a-3p, miR-29a-5p (↑) miR-20b-5p, miR-454-3p (↓) |
Glucose uptake, lipid metabolism, energy homeostasis | |
| Giardina et al. 2018 [84] | 8 obese | 6-month LGI (n = 3), HGI (n = 3), LF (n = 2) diet; | SAT |
LGI diet: miR-551b, miR-221, miR-378, let7a (↓) HGI diet: miR-1276, miR-132, miR-29a (↓) LF diet: miR-661, miR-1179, miR-132, miR-221, miR-29a, miR-378 (↓) |
Association with metabolic and body composition parameters | |
MS, metabolic syndrome; PBMC, peripheral blood mononuclear cells; WBC, white blood cells; GSK3B, glycogen synthase kinase-3 B; LGI, low glycaemic index; HGI, high glycaemic index; LF, low fat; CTRL, control subjects; PPAR-γ, peroxisome proliferator-activated receptor γ; PPAR-α, peroxisome proliferator-activated receptor α; SIRT1, sirtuin-1; KD, ketogenic diet; HPH, high dairy protein and carbohydrate; HPMC, high dairy protein and moderate carbohydrate; CON, low dairy protein and high carbohydrate; SAT, subcutaneous adipose tissue
microRNA expression in white blood cells
In 2013, Milagro et al., characterized microRNA expression in peripheral blood mononuclear cells (PBMC) from 10 obese women undergoing an 8-week energy-restricted diet. The population was further categorized as responder (R, > 5% body mass loss, n = 5) and non-responder (NR, < 5% body mass loss, n = 5) to the intervention. At baseline, the study revealed a different expression of five microRNAs between the two groups. Specifically, in the NR group at baseline, miR-935 and miR-4772 were up-regulated, whereas miR-223, miR-224 and miR-376b were down-regulated. Notably, miR-935, miR-4772 and miR-376b also showed a relevant association with the magnitude of weight loss, being therefore valuable candidate biomarkers for weight loss and response to diet [60]. Subsequently, Marques-Rocha et al., focused on the modulation of the expression of nine selected inflammation-related microRNAs in white blood cells of 40 patients affected by MS after an 8-week energy-restricted Mediterranean diet. Two microRNAs showed a statistically significant change after the dietary intervention; specifically, miR-155-3p strongly decreased, while let-7b was significantly up-regulated. In addition, the authors showed that let-7b, miR-125b, miR-130a, miR-132-3p and miR-422b were positively associated with the Health Eating Index (HEI) improvement, a scoring metric of diet quality, and miR-155-3p also with weight loss [61]. Another study evaluated microRNA changes in white blood cells obtained from patients with MS enrolled in the RESMENA (Metabolic Syndrome Reduction in Navarra) nutritional trial, after two different energy-restricted dietary interventions. The enrolled subjects were classified as high responders (HR) and low responders (LR), when weight loss after 8 weeks was respectively higher or lower than 8%. Microarray analyses were performed to detect microRNA methylation (31 LR vs 16 h) and expression (14 LR vs 10 h) before and after intervention. Six microRNAs (miR-1237, miR-1976, miR-642, miR-636, miR-612, miR-193B) were identified as both hypomethylated and over-expressed in HR. Notably, miR-612 and miR-1976 were the most hypomethylated and over-expressed, respectively. The bioinformatics analysis revealed TP53 and CD40 as miR-612 and miR-1976 targets, respectively. Both of them were modulated after the dietary intervention, comparing HR to LR, suggesting a role of these microRNA/gene axes in MS and obesity [62]. The same authors further reported an up-regulation of miR-548q and miR-1185-1 in HR, compared to sex and gender-matched LR. In functional assay, miR-548q and miR-1185-1 reduced glycogen synthase kinase-3 B (GSK3B) gene expression, targeting its mRNA [63]. GSK3B is known to be involved in pro-inflammatory responses by promoting the expression of pro-inflammatory cytokines such as IL-1ß, IL-6 or TNF-α [64]. Accordingly, the authors reported a negative correlation between miR-1185-1 expression and serum levels of IL-6 [63].
Circulating microRNAs
Several studies focused on the modulation of blood circulating microRNA levels after dietary interventions, comparing the effects of different dietary plans, with variable proportions of macronutrients.
Hess et al., evaluated the expression of selected serum microRNAs associated with obesity and MS in obese and overweight patients (n = 85), before and after a 12-week hypocaloric diet. The dietary intervention consisted of a 500 kcal/day restriction and a baseline randomization to fibre supplements or placebo. Circulating levels of miR-222-3p significantly increased after weight loss, while miR-122-5p and miR-193a-5p were reduced. At baseline, the concentrations of miR-122-5p and miR-193a-5p were higher in the presence of MS and positively correlated with markers of insulin resistance and body composition, such as HOMA-IR (homeostasis model assessment of insulin resistance), waist circumference and visceral fat mass [65]. Notably, the up-regulation of circulating miR-122-5p has been linked to NAFLD and its reduction after weight loss could therefore reflect an improvement in liver health [66]. As miR-122-5p and miR-193a-5p correlated with insulin resistance parameters, their reduction after intervention might mirror an improvement in glucose metabolism as well. MiR-222-3p has been previously linked to T2D and gestational diabetes (GDM), although its role in regulating glucose metabolism has yet to be elucidated [67–69]. Overall, these results suggest that specific microRNA patterns of expression might hallmark metabolically unhealthy obesity.
Ortega et al., investigated selected plasma circulating microRNAs in nine obese patients following a 14-week energy-restricted diet. However, the study revealed no significant associations between weight loss (17% reduction in body mass) and circulating microRNAs (miR-520c-3p, miR-15a, miR-590-5p, miR-126, miR-636, miR-625) [70].
Giardina et al., analysed plasma microRNA expression before and 6 months after three energy-restricted diets. Specifically, a discovery cohort of eight patients that underwent high-throughput screening was sub-grouped into three categories: moderate carbohydrate and low glycaemic index diet (LGI, n = 3), moderate carbohydrate and high glycaemic index diet (HGI, n = 3) and low fat and high glycaemic index diet (LF, n = 2). The authors identified eight differentially expressed microRNAs (miR-139-3p, miR-411, miR-432, miR-99b, miR-340, miR-423-5p, miR-361 and let-7c), whose significantly dysregulated levels were confirmed in a wider cohort of 103 patients (LGI, n = 36; HGI, n = 36; LF, n = 31). All these microRNAs were down-regulated in the LGI compared to the HGI group, with miR-361 showing the greatest reduction. Besides this, miR-139-3p was down-regulated both in the HGI and in the LF group compared to the baseline and positively correlated with both LDL-cholesterol and total cholesterol. MiR-340 was down-regulated only in the HGI group and positively correlated with triglycerides changes. MiR-432 and miR-423 were down-regulated only in the LF group and the latter had the same correlations reported for miR-139-3p. Finally, let-7c correlated with all lipid parameters [71].
The expression of 86 circulating obesity-related microRNAs has been explored in obese patients, randomly assigned to a moderately high-protein diet (n = 38) or a low-fat diet (n = 40), and in normal weight controls (n = 25). Among the differentially expressed microRNAs, in obese subjects compared to controls seven microRNAs (miR-130a-3p, miR-142-5p, miR-144-5p, miR-15a-5p, miR-22-3p, miR-221-3p and miR-29c-3p) were significantly associated with response to low-fat diet and strongly discriminated between responders and non-responders. In particular, a correlation between miR-22-3p expression and the entity of weight loss has been observed [72]. Interestingly, several regulators of fatty acid metabolism, such as peroxisome proliferator-activated receptor α (PPAR-α) and sirtuin-1 (SIRT1), are confirmed targets of miR-22 [73, 74].
A global microRNA profiling has been performed in plasma samples obtained before and after weight loss from obese women undergoing a 4-week very-low-calorie diet and from age-matched lean women. The expression of ~ 800 microRNAs has been evaluated (n = 8 samples each group) and validated in a cohort of 80 samples for each group. 21 microRNAs were found to be significantly dysregulated in obese women at baseline compared to lean women, their predicted targets belonged to pathways involved in glucose metabolism, inflammation, angiogenesis and cell death. Interestingly, the expression of seven microRNAs (miR-126, miR-208, miR-375, miR-376, miR-433, miR-499, miR-642) significantly changed in obese women after weight loss becoming similar to lean women, suggesting that an acute weight loss programme might effectively reverse the marked dysregulation of circulating microRNAs in obese subjects [75].
The effects of a KD on biochemical parameters, body composition and plasma microRNA profile has been evaluated in 36 obese subjects after 6 weeks of intervention. The KD is a very low carbohydrate nutritional regimen, characterized by less than 30–50 g of carbohydrates per day. In this study a biphasic KD was carried out: in the first 3-week phase, carbohydrate content was lower than 30 g per day, while in the last three weeks it reached 120 g per day. During the subsequent 6-weeks of follow-up, serum samples were collected for a high-throughput screening analysis on 799 microRNAs. The dietary programme improved body mass index (BMI), plasma insulin and triglyceride levels in both sexes, while the effect on microRNA expression profile was dissimilar between men and women. In females, at the end of the KD programme, miR-148b-3p, miR-26a-5p, miR-30e-5p, miR-502-5p, miR-520 h, miR-548d-3p, miR-590-5p and miR-644a were significantly different compared to the baseline; while in males a change in expression was observed for miR-30e-5p, miR-502-5p, miR-590-5p and miR-644a. Interestingly three microRNAs, let-7b-5p, miR-143-3p, miR-504-5p, showed the same statistical difference between baseline and 6 weeks of KD in both sexes. Bioinformatics analysis revealed that the differentially expressed microRNA target genes were linked to cytokine signalling pathways, inflammation and immunity, nutrient metabolism, oxidative phosphorylation, PPARs functional regulation and insulin signalling pathways [76]. Margolis et al., instead focused on the modulation of selected skeletal-muscle specific microRNAs in serum (c-myomiR: miR-1, miR-133a-3p, miR-133b, miR-206) and their relationship with protein synthesis rates. In detail, 16 elderly men (mean age 64 ± 2 years) were first subjected to a first 7-day period of eucaloric weight maintenance (WM) diet, to allow adaptation to the subsequent 28-day 30% energy-restricted (ER) diet. In the ER period, the authors showed not only an up-regulation of the c-myomiR score, with significant increase of miR-133a-3p and miR-133b, but also an inverse correlation between this score and the whole body protein synthesis rate, suggesting that ER may have a negative impact on skeletal-muscle regeneration, the top predicted target process at bioinformatics analysis. However, this study has several limitations, including the small number of participants, the lack of muscle biopsy analysis and body composition evaluation, such as fat-free mass [77].
Parr et al., evaluated plasma expression of 13 selected microRNAs, previously shown to be modulated by energy restriction and with putative roles in weight loss, at baseline and after a 16-week diet and physical activity intervention. The enrolled patients (n = 111) were subjected to physical exercise and randomly assigned to three dietary groups with different macronutrient intake: high dairy protein and carbohydrate (HPHC), high dairy protein and moderate carbohydrate (HPMC), low dairy protein and high carbohydrate (CON). Circulating microRNAs were analysed in 40 patients and further divided into two subgroups on the basis of body mass reduction: “high responders”, HiRes, ≥ 10% (n = 8 HPMC, n = 5 HPHC, n = 9 CON) and “low responders”, LoRes, ≤ 5% (n = 6 HPMC, n = 9 HPHC, n = 3 CON). In accordance with the above findings reported by Milagro et al., higher levels of miR-935 were observed in LoRes compared to HiRes at baseline and miR-140, miR-221 and miR-223 expression increased after 16-week weight-loss intervention, suggesting a putative role of these microRNAs as biomarkers of variability in individual response to weight-loss interventions. Interestingly, miR-935 was the only microRNA already higher at baseline in the LoRes group whose significant difference was maintained until the end of the intervention period. However, an important limitation of this study is related to the combination of dietary regimens and physical exercise that does not allow to evaluate the impact of the single intervention on the selected circulating microRNA profile [78]. The effects of exercise combined with dietary intervention has indeed been evaluated in other studies. One of them focused on the modulation of endothelial function in a population of obese adolescents. This study was carried out on an experimental group (EG) of 37 obese male adolescents (12–18 years) and a control group (CG, n = 10) of normal weight adolescents. The intervention was based on a 6-week calorie restriction diet associated with aerobic exercise. Regression analysis indicated a positive correlation between changes in serum miR-126 levels and BMI, serum levels of endothelium-derived nitric oxide (NO) and endothelin-1 (ET-1), before and after dietary intervention in the EG respect to CG. These data suggest that miR-126 might be involved in the improvement of microvascular endothelial function measured by RHI index [79].
microRNA expression in adipose tissue
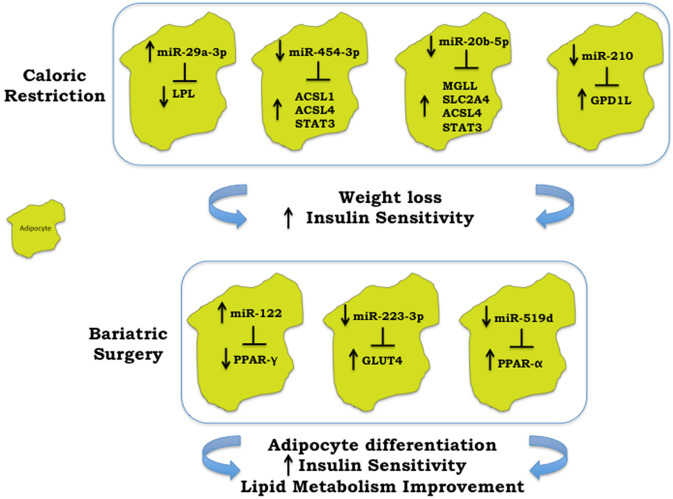
Kristensen et al., analysed microRNA expression in SAT derived from 19 obese patients undergoing a 15-week weight-loss strategy including both physical exercise and a hypocaloric dietary regimen. They identified nine differentially expressed microRNAs after intervention, but the statistically significant modulation of only three of these microRNAs was confirmed by validation on a wider cohort of patients (n = 38). In detail, miR-29a-3p and miR-29a-5p were up-regulated, while miR-20b-5p and miR-454-3p were down-regulated (the latter only had a borderline down-regulation), comparing the baseline with the post-intervention expression. In addition, the authors identified 56 predicted target genes of these four validated microRNAs and noticed a statistically significant inverse correlation between acyl-CoA synthetase long-chain family member 1 (ACSL1) with miR-454-3p; monoglyceride lipase (MGLL) and solute carrier family 2 member 4 (SLC2A4), which is the main responsible for glucose uptake into adipocytes with miR-20b-5p; lipoprotein lipase (LPL) with miR-29a-3p and finally acyl-CoA synthetase long-chain family member 4 (ACSL4) and signal transducer and activator of transcription 3 (STAT3) with both miR-454-3p and miR-20b-5p (Fig. 1). This last observation is interesting since these two microRNAs may act together in the modulation of genes, such as ACSL4 and STAT3, involved in energy homeostasis and metabolism [80]. Other studies that analysed microRNA expression in SAT have been conducted. In detail, He et al., examined possible molecular mechanisms underlying obesity through the integration of genetic obesity-associated loci derived from the largest genome-wide association study (GWAS) meta-analysis for BMI, gene expression and microRNA profiles, in adipose tissue from 200 subjects. This approach was further supported by the analysis of five publicly available studies comparing obese and non-obese subjects who underwent different dietary intervention strategies. It is worth noting that He et al., identified a molecular axis involving hypoxia/Glycerol-3-Phosphate Dehydrogenase 1 Like (GPD1L)/miR-210 that can be linked to fat accumulation and obesity. GPD1L has been recently demonstrated to be a regulator of HIF-1α stability and a validated target of miR-210, a master regulator of hypoxia. Since adipose tissue in patients is scarcely oxygenated, with a consequent relative hypoxic status, the authors hypothesized that the increase of HIF-1α expression and activity in high fat diet (HFD) models could induce an up-regulation of miR-210, with the subsequent reduction in GPD1L levels and a further repression of prolyl hydroxylases (PHDs) activity. All these events may determine the activation of a feedback loop that further promote HIF-1α accumulation, with subsequent worsening of obesity, insulin resistance and adipose tissue inflammation. Interestingly, low calorie diet was able to induce a relative reduction in the hypoxic state and of miR-210 levels, paralleled with an increase of GPD1L levels (Fig. 1) [81–83]. Giardina et al. [71] instead evaluated the microRNA profile in a randomly selected representative cohort of eight patients subjected to different dietary interventions, previously described. The 13 most differentially expressed microRNAs in SAT were further selected for validation in a wider cohort of 48 patients. The expression of miR-551b, miR-221, miR-378 and let7a was down-regulated after the LGI intervention. The expression of miR-1276, miR-132 and miR-29a was down-regulated after the HGI intervention, while the expression of miR-661, miR-1179, miR-132, miR-221, miR-29a and miR-378 was down-regulated after the LF intervention. Each dietary approach induced a significant reduction in body weight, BMI, waist circumference, fat mass, fat-free mass and HOMA-IR compared to the respective baseline values. Moreover, the expression of some microRNAs, namely miR-551b, miR-1179, miR-132, miR-221, miR-29a, miR-34a, miR-378, correlated positively with biochemical and anthropometric variables, indicating that weight loss and changes in body composition, rather than dietary composition, could be the main drivers in the modulation of microRNAs profile [84].
Fig. 1.
Caloric restriction and/or bariatric surgery modulation of adipose tissue microRNAs. a Caloric Restriction. The increase of miR-29a-3p after caloric restriction was paralleled with lipoprotein lipase (LPL) reduction, while the reduction of miR-454-3p, miR-20b-5p and miR-210 were paralleled with the increase of other pivotal players in controlling weight loss and insulin resistance. Specifically, acyl-CoA synthetase long-chain family member 1 (ACSL1) up-regulation was related with miR-454-3p down-regulation. Monoglyceride lipase (MGLL) and solute carrier family 2 member 4 (SLC2A4) were related with miR-20b-5p modulation. While acyl-CoA synthetase long-chain family member 4 (ACSL4) and signal transducer and activator of transcription 3 (STAT3) up-regulation was related with both miR-454-3p and miR-20b-5p down-regulation. The reduction of miR-210 was coupled with Glycerol-3-Phosphate Dehydrogenase 1 Like (GPD1L) increase. b Bariatric surgery. The increase of miR-122 was linked with peroxisome proliferator-activated receptor γ (PPAR-γ), while miR-223-3p and miR-519d were related with glucose transporter 4 (GLUT-4) and PPAR-α up-regulation, respectively
microRNAs and weight loss: evidence from bariatric surgery intervention studies
A wide number of studies focused on the possible relationship between microRNA expression and bariatric surgery (Table 2), highlighting that surgically induced weight loss is effective in rescuing microRNA signature [85].
Table 2.
Studies that reported changes in microRNA expression after bariatric surgery
| Study | Population | Intervention | Source | Regulated miRNAs | Role/target |
|---|---|---|---|---|---|
| Hulsmans et al. 2012 [86] |
9 obese 6 CTRL |
RYGB | PBMC | miR-181 (↑) | TLR-NFkB pathway |
| Ortega et al. 2013 [70] | 6 obese | RYGB | Plasma |
miR-16-1, miR-122, miR-140-5p, miR-193a-5p (↓) miR-221 and miR-199a-3p (↑) |
– |
| Nunez-Lopez et al. 2017 [87] | 22 obese | RYGB | Plasma |
miR-15a (↑) miR-34a, miR-122 (↓) |
Biomarkers of weight loss /glucose metabolism |
| Atkin et al. 2018 [88] | 29 T2D | RYGB | Plasma |
miR-7-5p, let-7f-5p, miR-15b-5p, miR-320c, miR-205-5p, miR-335-5p (↑) let-7i-5p (↓) |
Inflammation, adipocyte proliferation, ß-cell function, thyroid and pituitary function |
| Hubal et al. 2017 [92] | 6 obese women | RYGB | Plasma and serum adipocyte-derived exosomes | let-7a-5p, miR-16-5p | Insulin signalling |
| Bae et al. 2019 [93] |
16 obese 18 CTRL |
LSG (n = 2) RYGB (n = 14) | Serum exosomes | miR-424-5p | Biomarker of weight loss |
| Macartney-Coxson et al. 2020 [94] | 15 obese women | RYGB |
SAT VAT |
SAT: miR-23a-5p, miR-27a-5p, miR-200c-3p, miR-223-3p, miR-1246, miR-24-2-5p, miR-128, miR-421, miR-3178, miR-1224-5p, miR-221, miR-22, miR-762 (↓) VAT: miR-223-3p (↓) |
Inflammation, glucose uptake |
| Liao et al. 2018 [98] |
20 obese 8 CTRL |
LSG |
SAT VAT |
VAT: miR-122 (↑) |
PPAR-γ |
| Kurylowicz et al. 2016 [99] |
20 obese 7 CTRL |
Bariatric surgery | SAT | miR-146b-3p, miR-146b-5p, miR-223-3p, miR-223-5p, miR-941 (↑) | BMPR2, FOXP1, IGF1R |
| Ortega et al. 2015 [100] | 16 obese | RYGB | SAT | miR-155, miR-221, miR-130b (↓) | Inflammation |
| Ortega et al. 2015 [101] | 9 obese women | RYGB | SAT | miR-19a/b, miR-146a/b, miR-155, miR-193b, miR-221, miR-222, miR-223, miR-376c, miR-411 (↓) | Glucose uptake, lipid metabolism, energy homeostasis |
| Nardelli et al. 2017 [102] |
3 obese 2 CTRL |
LAGB | SAT |
miR-519d, miR-299-5p, miR-212, miR-671-3p (↓) miR-370, miR-487a (↑) |
PPAR-α (miR-519d) |
CTRL, control subjects; RYGP, Roux-en-Y gastric by-pass; PBMC, peripheral blood mononuclear cells; T2D, type 2 diabetes; SAT, subcutaneous adipose tissue; VAT, visceral adipose tissue; LSG, laparoscopic sleeve gastrectomy; BMPR2, bone morphogenic protein receptor 2; FOXP, forkhead box protein P1; IGF1R, insulin-like growth factor receptor 1; LAGB, laparoscopic adjustable gastric banding
microRNA expression in white blood cells
Changes in inflammatory toll-like receptor (TLR)/nuclear factor kB (NFkB) related microRNAs have been evaluated in circulating monocytes from obese subjects undergoing RYGB and lean controls. Microarray analysis identified 133 differentially expressed microRNAs in circulating monocytes of obese subjects (n = 9) compared with lean controls (n = 6). In addition, target prediction revealed that nine dysregulated microRNA families were associated with TLR/NFkB pathway, among them, in a larger validation cohort of 14 lean controls and 21 morbidly obese patients, only the miR-181 family resulted to be down-regulated in monocytes of obese patients and its dysregulated levels were restored after bariatric surgery [86].
Circulating microRNAs
In the context of bariatric surgery, several studies focused on the expression pattern of circulating microRNAs before and after surgical intervention. In 2013, Ortega et al., analysed the differences in the microRNA expression profile before and after RYGB in plasma samples from six morbidly obese patients. The results were validated in an independent cohort of 22 obese patients and 14 circulating microRNAs resulted significantly modulated. In particular, the authors observed a marked reduction in miR-16-1, miR-122, miR-140-5p, miR-193a-5p and an up-regulation of miR-221 and miR-199a-3p [70].
Nunez-Lopez et al., were interested in the identification of non-invasive biomarkers for metabolic changes resulting from RYGB. They recruited 22 morbidly obese subjects who had undergone RYGB surgery 1–3 months before. At baseline evaluation, subjects were divided in two subgroups: 11 subjects were subjected to a 6-month exercise-programme (EX) and the other 11 to a control health education intervention (CON). Interestingly, 94 plasma microRNAs, selected on the basis of their relationship with metabolism, were analysed. The authors identified three patterns of circulating microRNA changes in the patient cohort at the end of the intervention. Specifically, three microRNAs were modulated both in the CON and in the EX group (miR-15a increased, while miR-34a and miR-122 decreased), suggesting that their modulation may indicate a general response to weight loss induced by RYGB surgery; three microRNAs were modulated only in the CON group (miR-7 and miR-106 increased, while miR-221 decreased) and four microRNAs only in the EX group (miR-135b, miR-144 and miR-206 decreased, while miR-149 increased). Interestingly, the 10 above mentioned microRNAs significantly correlated with indices of HOMA-IR, β-cell function, body composition, plasma lipids and liver function. Moreover, miR-15a, miR-7, miR-106b and miR-135b correlated, already at the baseline, with insulin sensitivity (miR-15a), glucose effectiveness (miR-7 and miR-106b), and acute insulin response to glucose (miR-135b), suggesting their role as predictive biomarkers of cardiometabolic changes [87].
Atkin et al., analysed plasma microRNA expression in obese patients (n = 29) with T2D, undergoing RYGB. Six microRNAs (miR-7-5p, let-7f-5p, miR-15b-5p, miR-320c, miR-205-5p, miR-335-5p) significantly increased, while let-7i-5p decreased after surgery. Interestingly, a marked reduction in blood glucose and glycated haemoglobin (HbA1c) levels was observed after surgery in a high percentage of patients (more than 60%) [88]. Most of the regulated microRNAs were found to be expressed in natural killer cells, which play critical roles in obesity-induced inflammation [89]. Furthermore, the same microRNAs have been shown to be involved in different biological processes, such as adipocyte proliferation, ß-cell function, thyroid and pituitary function [90, 91].
Hubal et al., explored plasma and serum adipocyte-derived-exosomal microRNAs in six obese women before (T0) and 1-year (T1) after RYGB. Interestingly, 29 differentially expressed microRNAs between T0 and T1 were significantly correlated to improvement in the HOMA-IR. Pathway analysis identified the Insulin Receptor Signalling as one of the most enriched pathways. Moreover, two of the above mentioned microRNAs (let-7a-5p, miR-16-5p) correlated not only to the Insulin Receptor Signalling, showing the highest number of targets in this pathway, but also to branched chain amino acids (BCAAs) levels, which are strictly connected to insulin dysregulation. These findings indicate that changes in circulating adipocyte-derived exosomal microRNAs may be connected with post-surgery improvements in glucose homeostasis and insulin resistance [92].
Other authors explored circulating microRNAs in serum exosomes obtained from obese patients (n = 16), before and 6 months after bariatric surgery, and from healthy subjects (n = 18). Nine exosomal and 32 circulating microRNAs displayed higher and lower expression, respectively, in obese patients after intervention compared to baseline. Remarkably, among the nine up-regulated microRNAs, the levels of miR-424-5p before surgery positively correlated with weight loss after intervention [93]. Due to their high stability, exosomal microRNAs are novel candidate biomarkers for several pathological conditions. Since a considerable number of exosomal microRNAs decreased after bariatric surgery, in parallel with the reduction in fat mass, it can be speculated that most circulating exosomal microRNAs derive from adipose tissue.
microRNA expression in adipose tissue
Although mounting evidence has focused on circulating microRNA profiling to individuate candidate biomarkers of response to weight loss after dietary intervention or bariatric surgery, a debate is still on-going on whether circulating microRNA dysregulation directly reflects changes at a tissue or cellular level. In light of this, several studies focused on the differences in microRNA expression profile between SAT and VAT, before and after bariatric surgery.
Recently, microRNA expression has been explored in SAT and VAT of obese women (n = 15) collected both during RYGB and then after 17 months, at the time of a second surgery for other purposes. Several microRNAs, namely miR-23a-5p, miR-27a-5p, miR-200c-3p, miR-223-3p, miR-1246, miR-24-2-5p, miR-128, miR-421, miR-3178, miR-1224-5p, miR-221, miR-22 and miR-762, were significantly down-regulated after surgery in SAT, while only miR-223-3p showed significant down-regulation in VAT [94]. Selected targets of miR-223-3p, which have been found to be over-expressed in obesity [95], have been evaluated. In both tissues, NLRP3 (NACHT, LRRand PYD domains-containing protein 3) and leptin (LEP) mRNAs were significantly down-regulated, while glucose transporter 4 (GLUT-4) was up-regulated after surgery. Furthermore, the GLUT-4 mRNA positively correlated to miR-223-3p expression in SAT (Fig. 1) [94]. Remarkably, the NLRP3 consists of a set of intracellular sensors and receptors, known as inflammasome, recognized to act as key contributors to the chronic inflammatory state observed in obesity [96].
Liao et al., compared the microRNA expression pattern in SAT and VAT in 20 patients undergoing LSG and 8 normal weight subjects undergoing laparoscopic cholecystectomy. SAT and VAT from obese patients were characterized by 18 differentially expressed microRNAs: 12 were significantly up-regulated and 6 down-regulated in VAT. Conversely, these differences were less marked in non-obese subjects. Among the differentially expressed microRNAs, miR-122 was the most up-regulated in VAT from both obese and non-obese subjects in respect to SAT, and its over-expression was confirmed by RT-qPCR in a wider cohort of patients. By over-expressing this microRNA in fully differentiated mouse 3T3-L1 cells, the authors identified the peroxisome proliferator-activated receptor γ (PPAR-γ) as the most significantly altered pathway. Moreover, the over-expression of miR-122 induced a significant reduction in PPAR-γ mRNA, while its inhibition led to a significant increase in PPAR-γ mRNA expression levels, suggesting that miR-122 regulates PPAR-γ, one of the most important players in adipocyte differentiation (Fig. 1) [97, 98].
Kurylowicz et al., instead compared microRNA profile before and after bariatric surgery only in SAT. In detail, they analysed the microRNA profile in VAT and SAT from 10 obese patients (O), VAT and SAT from 7 normal weight subjects (N) and SAT from 10 obese patients about 2-years after surgery-induced weight loss (PO). By focusing on the differences in microRNA expression between SAT samples before and after weight loss (SAT-O vs SAT-PO), the authors identified 58 over and 3 under-expressed microRNAs in SAT-O compared with SAT-PO. Five of them, namely miR-146b-3p, miR-146b-5p, miR-223-3p, miR-223-5p and miR-941, were also under-expressed in SAT-N in respect to SAT-O. However, there were also differences in the microRNA expression pattern between SAT-PO and SAT-N samples. In particular, 79 microRNAs were differentially expressed: 42 were under-expressed, whereas 37 over-expressed in SAT-PO in respect to SAT-N, indicating that obesity may determine persistent miRNome changes. Finally, the authors reported that several targets with well-known roles in obesity, such as the bone morphogenic protein receptor 2 (BMPR2), the forkhead box protein P1 (FOXP1) and the insulin-like growth factor receptor 1 (IGF1R), comprised the highest number of binding sites in the differentially expressed microRNAs [99].
The SAT microRNA profile of obese subjects was also analysed by other authors. In 2015, Ortega et al. evaluated the microRNA profile changes in SAT from obese subjects after laparoscopic RYGB. The first study enrolled 16 obese patients in which 15 microRNAs showed significant modification after surgery-induced weight loss. Among them, miR-155, miR-221 and miR-130b displayed decreased expression after treatment [100]. The same authors identified significant reduction in 11 microRNAs (miR-19a/b, miR-146a/b, miR-155, miR-193b, miR-221, miR-222, miR-223, miR-376c, miR-411) in SAT samples of nine morbidly obese women after surgery-induced weight loss. Interestingly, three of them, namely miR-155, miR-221 and miR-222, were associated with obesity-related inflammation and weight loss, suggesting that microRNA modulation after weight loss may underline improvement in patients’ inflammatory status [101]. Subsequently, in 2017 Nardelli et al., analysed microRNA expression pattern in SAT obtained from three severely obese women before (T0) and three years after LAGB (T1), and from two lean subjects, undergoing laparoscopic cholecystectomy. Specifically, at T0, 43 microRNAs resulted significantly up-regulated and 15 down-regulated between obese and lean subjects. Of the above cited microRNAs, four resulted down-regulated (miR-519d, miR-299-5p, miR-212, miR-671-3p) and two up-regulated (miR-370, miR-487a) also in T1 compared to T0. MiR-370, miR-487a and miR-519d dysregulation was also confirmed by validation assays. Interestingly, the level of PPAR-α protein, a validated target of miR-519d [53], increased at T1 compared to T0, suggesting that miR-519d targets PPAR-α in SAT and may be involved in the improvement of lipid metabolism and SAT function after surgery (Fig. 1) [102].
Overall, in a meta-analysis of 17 studies evaluating the differential expression of microRNA before and after bariatric surgery both in humans and in animal models, several microRNAs were reported to be consistently down-regulated (miR-93-5p, miR-106b-5p, let-7b-5p, let-7i-5p, miR-16-5p, miR-19b-3p, miR-92a-3p, miR-222-3p, miR-142-3p, miR-140-5p, miR-155-5p, miR-320-3p) or up-regulated (miR-7-5p and miR-320c), with high concordance between studies [103]. However, it is worth highlighting that marked differences between the selected studies have emerged (i.e. population, type of intervention, follow-up length, collected samples) and further research is therefore needed to draw firm conclusions.
Conclusions and future perspectives
Over the last 20 years, obesity and obesity-related disorders have rapidly become a public health concern worldwide. Recently, microRNAs have gained much attention as epigenetic modulators in obesity, helping understand the pathophysiological mechanisms underlying this condition. Several microRNAs have been found to be differentially regulated in obesity and their expression was significantly modified by different weight-loss approaches, such as diet, physical activity and bariatric surgery. Specifically, the above mentioned studies explored microRNAs expression in several compartments, such as blood cells, serum, plasma, adipose tissue, and the effect of a wide variety of currently available intervention strategies for obesity treatment has been evaluated. In particular, dietary interventions with marked differences in macronutrient composition have been compared, highlighting different effects on microRNA expression profile. These findings suggest that weight loss obtained through entirely different dietary strategies might be mirrored by profound changes in microRNA signature. As regards bariatric surgery, it should be highlighted that the vast majority of the available intervention studies have focused on the effect of the malabsorptive procedure RYGB, whereas data on other widely adopted surgical approaches, such as LSG and LAGB, are still limited. Future research is therefore needed to address this issue to possibly compare the effect of different surgical interventions on circulating and tissue microRNAs expression. Of note, the assessment of microRNA profile in adipose tissue before and after obesity treatment approaches gave critical information about the metabolic changes occurring in response to weight loss. Indeed, the definition of the mechanisms through which microRNAs regulate adipose tissue metabolism is of utmost interest, as it can lead to significant improvement in obesity treatment. Importantly, diverse patterns of microRNA expression between VAT and SAT have been observed in several studies, suggesting that weight loss might differently modify the metabolic profile of these tissues.
Furthermore, substantial differences in microRNAs expression have emerged between responders and non-responders to dietary and surgical interventions. Considering the consistent variability in individual response to weight-loss interventions, circulating microRNAs might be valuable biomarkers of efficacy, possibly helping in the differentiation between responders and non-responders. Remarkably, a wide variety of dysregulated microRNA target pathways, some of them crucially involved in glucose and lipid metabolism, energy homeostasis, inflammation, immunity, endothelial function, have also been identified, helping understand the pathophysiological mechanisms underlying obesity and obesity-related metabolic diseases.
It should be highlighted that some discrepancies emerged between studies. Nevertheless, these conflicting results might be attributed to several factors, such as heterogeneity of the included populations, different samples collected (plasma, serum, adipose tissue), type of intervention adopted (diet composition, physical activity interference, surgical procedure), variability in the analytic methods for microRNA profiling. As regards the latter, in future studies a strong effort should be made in analysing microRNAs through absolute quantification and RNA identification. Indeed, applying standardized techniques for RNA extraction and performing highly multiplexed single molecule counting would be a crucial advance in analytical methodologies.
Acknowledgements
This review did not receive any specific grant from funding agencies in the public, commercial, or no-for-profit sectors.
Funding
Open access funding provided by Università degli Studi di Roma La Sapienza within the CRUI-CARE Agreement..
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
For this type of study formal consent in not required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
G. Catanzaro and T. Filardi equally contributed to the manuscript.
References
- 1.Anastasi E, Filardi T, Tartaglione S, Lenzi A, Angeloni A, Morano S. Linking type 2 diabetes and gynecological cancer: an introductory overview. Clin Chem Lab Med. 2018;56(9):1413–1425. doi: 10.1515/cclm-2017-0982. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2015 Obesity Collaborators Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med. 2017;377(1):13–27. doi: 10.1056/NEJMoa1614362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein S. Clinical obesity issues from an internist's perspective. Obes Res. 2002;10(Suppl 1):87S–88S. doi: 10.1038/oby.2002.198. [DOI] [PubMed] [Google Scholar]
- 4.Kopelman PG. Obesity as a medical problem. Nature. 2000;404(6778):635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization (2020) Obesity and overweight: fact sheets. https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight. Accessed 16 Jul 2020
- 6.Galgani J, Ravussin E. Energy metabolism, fuel selection and body weight regulation. Int J Obes (Lond) 2008;32(Suppl 7):S109–119. doi: 10.1038/ijo.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Despres JP. Abdominal obesity as important component of insulin-resistance syndrome. Nutrition. 1993;9(5):452–459. [PubMed] [Google Scholar]
- 8.Jensen MD. Visceral fat: culprit or canary? Endocrinol Metab Clin North Am. 2020;49(2):229–237. doi: 10.1016/j.ecl.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Ostman J, Arner P, Engfeldt P, Kager L. Regional differences in the control of lipolysis in human adipose tissue. Metabolism. 1979;28(12):1198–1205. doi: 10.1016/0026-0495(79)90131-8. [DOI] [PubMed] [Google Scholar]
- 10.Roden M, Price TB, Perseghin G, Petersen KF, Rothman DL, Cline GW, Shulman GI. Mechanism of free fatty acid-induced insulin resistance in humans. J Clin Invest. 1996;97(12):2859–2865. doi: 10.1172/JCI118742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santangelo C, Filardi T, Perrone G, Mariani M, Mari E, Scazzocchio B, Masella R, Brunelli R, Lenzi A, Zicari A, Morano S. Cross-talk between fetal membranes and visceral adipose tissue involves HMGB1-RAGE and VIP-VPAC2 pathways in human gestational diabetes mellitus. Acta Diabetol. 2019;56(6):681–689. doi: 10.1007/s00592-019-01304-x. [DOI] [PubMed] [Google Scholar]
- 12.Harman-Boehm I, Bluher M, Redel H, Sion-Vardy N, Ovadia S, Avinoach E, Shai I, Kloting N, Stumvoll M, Bashan N, Rudich A. Macrophage infiltration into omental versus subcutaneous fat across different populations: effect of regional adiposity and the comorbidities of obesity. J Clin Endocrinol Metab. 2007;92(6):2240–2247. doi: 10.1210/jc.2006-1811. [DOI] [PubMed] [Google Scholar]
- 13.Giannattasio S, Corinaldesi C, Colletti M, Di Luigi L, Antinozzi C, Filardi T, Scolletta S, Basili S, Lenzi A, Morano S, Crescioli C. The phosphodiesterase 5 inhibitor sildenafil decreases the proinflammatory chemokine IL-8 in diabetic cardiomyopathy: in vivo and in vitro evidence. J Endocrinol Invest. 2019;42(6):715–725. doi: 10.1007/s40618-018-0977-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen MD, Ryan DH, Apovian CM, Ard JD, Comuzzie AG, Donato KA, Hu FB, Hubbard VS, Jakicic JM, Kushner RF, Loria CM, Millen BE, Nonas CA, Pi-Sunyer FX, Stevens J, Stevens VJ, Wadden TA, Wolfe BM, Yanovski SZ, Jordan HS, Kendall KA, Lux LJ, Mentor-Marcel R, Morgan LC, Trisolini MG, Wnek J, Anderson JL, Halperin JL, Albert NM, Bozkurt B, Brindis RG, Curtis LH, DeMets D, Hochman JS, Kovacs RJ, Ohman EM, Pressler SJ, Sellke FW, Shen WK, Smith SC, Jr, Tomaselli GF, American College of Cardiology, American Heart Association Task Force on Practice G. Obesity S 2013 AHA/ACC/TOS guideline for the management of overweight and obesity in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and The Obesity Society. Circulation. 2014;129(25 Suppl 2):S102–138. doi: 10.1161/01.cir.0000437739.71477.ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCafferty BJ, Hill JO, Gunn AJ. Obesity: scope, lifestyle interventions, and medical management. Tech Vasc Interv Radiol. 2020;23(1):100653. doi: 10.1016/j.tvir.2020.100653. [DOI] [PubMed] [Google Scholar]
- 16.Chin SH, Kahathuduwa CN, Binks M. Physical activity and obesity: what we know and what we need to know. Obes Rev. 2016;17(12):1226–1244. doi: 10.1111/obr.12460. [DOI] [PubMed] [Google Scholar]
- 17.Foster D, Sanchez-Collins S, Cheskin LJ. Multidisciplinary team-based obesity treatment in patients with diabetes: current practices and the state of the science. Diabetes Spectr. 2017;30(4):244–249. doi: 10.2337/ds17-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trumbo P, Schlicker S, Yates AA, Poos M, Food, Nutrition Board of the Institute of Medicine TNA Dietary reference intakes for energy, carbohydrate, fiber, fat, fatty acids, cholesterol, protein and amino acids. J Am Diet Assoc. 2002;102(11):1621–1630. doi: 10.1016/s0002-8223(02)90346-9. [DOI] [PubMed] [Google Scholar]
- 19.Yannakoulia M, Poulimeneas D, Mamalaki E, Anastasiou CA. Dietary modifications for weight loss and weight loss maintenance. Metabolism. 2019;92:153–162. doi: 10.1016/j.metabol.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Arterburn DE, Courcoulas AP. Bariatric surgery for obesity and metabolic conditions in adults. BMJ. 2014;349:g3961. doi: 10.1136/bmj.g3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abell TL, Minocha A. Gastrointestinal complications of bariatric surgery: diagnosis and therapy. Am J Med Sci. 2006;331(4):214–218. doi: 10.1097/00000441-200604000-00008. [DOI] [PubMed] [Google Scholar]
- 22.Svensson PA, Anveden A, Romeo S, Peltonen M, Ahlin S, Burza MA, Carlsson B, Jacobson P, Lindroos AK, Lonroth H, Maglio C, Naslund I, Sjoholm K, Wedel H, Soderpalm B, Sjostrom L, Carlsson LM. Alcohol consumption and alcohol problems after bariatric surgery in the Swedish obese subjects study. Obes (Silver Spring) 2013;21(12):2444–2451. doi: 10.1002/oby.20397. [DOI] [PubMed] [Google Scholar]
- 23.Bouchard L, Rabasa-Lhoret R, Faraj M, Lavoie ME, Mill J, Perusse L, Vohl MC. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr. 2010;91(2):309–320. doi: 10.3945/ajcn.2009.28085. [DOI] [PubMed] [Google Scholar]
- 24.Campion J, Milagro FI, Martinez JA. Individuality and epigenetics in obesity. Obes Rev. 2009;10(4):383–392. doi: 10.1111/j.1467-789X.2009.00595.x. [DOI] [PubMed] [Google Scholar]
- 25.Milagro FI, Campion J, Cordero P, Goyenechea E, Gomez-Uriz AM, Abete I, Zulet MA, Martinez JA. A dual epigenomic approach for the search of obesity biomarkers: DNA methylation in relation to diet-induced weight loss. FASEB J. 2011;25(4):1378–1389. doi: 10.1096/fj.10-170365. [DOI] [PubMed] [Google Scholar]
- 26.Goni L, Cuervo M, Milagro FI, Martinez JA. Future perspectives of personalized weight loss interventions based on nutrigenetic, epigenetic, and metagenomic data. J Nutr. 2015;146(4):905S–912S. doi: 10.3945/jn.115.218354. [DOI] [PubMed] [Google Scholar]
- 27.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 28.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 29.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20(1):5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 30.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 31.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15(12):2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 34.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Guarino E, Delli Poggi C, Grieco GE, Cenci V, Ceccarelli E, Crisci I, Sebastiani G, Dotta F. Circulating microRNAs as biomarkers of gestational diabetes mellitus: updates and perspectives. Int J Endocrinol. 2018;2018:6380463. doi: 10.1155/2018/6380463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 39.Guduric-Fuchs J, O'Connor A, Camp B, O'Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 41.Guay C, Regazzi R. Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat Rev Endocrinol. 2013;9(9):513–521. doi: 10.1038/nrendo.2013.86. [DOI] [PubMed] [Google Scholar]
- 42.Faruq O, Vecchione A. microRNA: diagnostic perspective. Front Med (Lausanne) 2015;2:51. doi: 10.3389/fmed.2015.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujimoto Y, Nakagawa Y, Shingyouchi A, Tokushige N, Nakanishi N, Satoh A, Matsuzaka T, Ishii KA, Iwasaki H, Kobayashi K, Yatoh S, Suzuki H, Yahagi N, Urayama O, Yamada N, Shimano H. Dicer has a crucial role in the early stage of adipocyte differentiation, but not in lipid synthesis, in 3T3-L1 cells. Biochem Biophys Res Commun. 2012;420(4):931–936. doi: 10.1016/j.bbrc.2012.03.110. [DOI] [PubMed] [Google Scholar]
- 44.Oskowitz AZ, Lu J, Penfornis P, Ylostalo J, McBride J, Flemington EK, Prockop DJ, Pochampally R. Human multipotent stromal cells from bone marrow and microRNA: regulation of differentiation and leukemia inhibitory factor expression. Proc Natl Acad Sci USA. 2008;105(47):18372–18377. doi: 10.1073/pnas.0809807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y, Du J, Zhang P, Zhao X, Li Q, Jiang A, Jiang D, Tang G, Jiang Y, Wang J, Li X, Zhang S, Zhu L. MicroRNA-125a-5p mediates 3T3-L1 preadipocyte proliferation and differentiation. Molecules. 2018;23:2. doi: 10.3390/molecules23020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol. 2009;23(6):925–931. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Son YH, Ka S, Kim AY, Kim JB. Regulation of adipocyte differentiation via MicroRNAs. Endocrinol Metab (Seoul) 2014;29(2):122–135. doi: 10.3803/EnM.2014.29.2.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi C, Zhang M, Tong M, Yang L, Pang L, Chen L, Xu G, Chi X, Hong Q, Ni Y, Ji C, Guo X. miR-148a is associated with obesity and modulates adipocyte differentiation of mesenchymal stem cells through Wnt signaling. Sci Rep. 2015;5:9930. doi: 10.1038/srep09930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ouyang D, Xu L, Zhang L, Guo D, Tan X, Yu X, Qi J, Ye Y, Liu Q, Ma Y, Li Y. MiR-181a-5p regulates 3T3-L1 cell adipogenesis by targeting Smad7 and Tcf7l2. Acta Biochim Biophys Sin (Shanghai) 2016;48(11):1034–1041. doi: 10.1093/abbs/gmw100. [DOI] [PubMed] [Google Scholar]
- 50.Huang S, Wang S, Bian C, Yang Z, Zhou H, Zeng Y, Li H, Han Q, Zhao RC. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21(13):2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L, Hou J, Ye L, Chen Y, Cui J, Tian W, Li C, Liu L. MicroRNA-143 regulates adipogenesis by modulating the MAP2K5-ERK5 signaling. Sci Rep. 2014;4:3819. doi: 10.1038/srep03819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortega FJ, Moreno-Navarrete JM, Pardo G, Sabater M, Hummel M, Ferrer A, Rodriguez-Hermosa JI, Ruiz B, Ricart W, Peral B, Fernandez-Real JM. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE. 2010;5(2):e9022. doi: 10.1371/journal.pone.0009022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinelli R, Nardelli C, Pilone V, Buonomo T, Liguori R, Castano I, Buono P, Masone S, Persico G, Forestieri P, Pastore L, Sacchetti L. miR-519d overexpression is associated with human obesity. Obesity (Silver Spring) 2010;18(11):2170–2176. doi: 10.1038/oby.2009.474. [DOI] [PubMed] [Google Scholar]
- 54.Arner E, Mejhert N, Kulyte A, Balwierz PJ, Pachkov M, Cormont M, Lorente-Cebrian S, Ehrlund A, Laurencikiene J, Heden P, Dahlman-Wright K, Tanti JF, Hayashizaki Y, Ryden M, Dahlman I, van Nimwegen E, Daub CO, Arner P. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012;61(8):1986–1993. doi: 10.2337/db11-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin YY, Chou CF, Giovarelli M, Briata P, Gherzi R, Chen CY. KSRP and MicroRNA 145 are negative regulators of lipolysis in white adipose tissue. Mol Cell Biol. 2014;34(12):2339–2349. doi: 10.1128/MCB.00042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorente-Cebrian S, Mejhert N, Kulyte A, Laurencikiene J, Astrom G, Heden P, Ryden M, Arner P. MicroRNAs regulate human adipocyte lipolysis: effects of miR-145 are linked to TNF-alpha. PLoS ONE. 2014;9(1):e86800. doi: 10.1371/journal.pone.0086800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Li C, Li H, Song Y, Zhao Y, Zhai L, Wang H, Zhong R, Tang H, Zhu D. miR-378 activates the pyruvate-PEP futile cycle and enhances lipolysis to ameliorate obesity in mice. EBioMedicine. 2016;5:93–104. doi: 10.1016/j.ebiom.2016.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lorente-Cebrian S, Gonzalez-Muniesa P, Milagro FI, Martinez JA. MicroRNAs and other non-coding RNAs in adipose tissue and obesity: emerging roles as biomarkers and therapeutic targets. Clin Sci (Lond) 2019;133(1):23–40. doi: 10.1042/CS20180890. [DOI] [PubMed] [Google Scholar]
- 59.Xu LL, Shi CM, Xu GF, Chen L, Zhu LL, Zhu L, Guo XR, Xu MY, Ji CB. TNF-alpha, IL-6, and leptin increase the expression of miR-378, an adipogenesis-related microRNA in human adipocytes. Cell Biochem Biophys. 2014;70(2):771–776. doi: 10.1007/s12013-014-9980-x. [DOI] [PubMed] [Google Scholar]
- 60.Milagro FI, Miranda J, Portillo MP, Fernandez-Quintela A, Campion J, Martinez JA. High-throughput sequencing of microRNAs in peripheral blood mononuclear cells: identification of potential weight loss biomarkers. PLoS ONE. 2013;8(1):e54319. doi: 10.1371/journal.pone.0054319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marques-Rocha JL, Milagro FI, Mansego ML, Zulet MA, Bressan J, Martinez JA. Expression of inflammation-related miRNAs in white blood cells from subjects with metabolic syndrome after 8 wk of following a Mediterranean diet-based weight loss program. Nutrition. 2016;32(1):48–55. doi: 10.1016/j.nut.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 62.Garcia-Lacarte M, Martinez JA, Zulet MA, Milagro FI. Implication of miR-612 and miR-1976 in the regulation of TP53 and CD40 and their relationship in the response to specific weight-loss diets. PLoS ONE. 2018;13(8):e0201217. doi: 10.1371/journal.pone.0201217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Lacarte M, Mansego ML, Zulet MA, Martinez JA, Milagro FI. miR-1185-1 and miR-548q are biomarkers of response to weight loss and regulate the expression of GSK3B. Cells. 2019;8:12. doi: 10.3390/cells8121548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beurel E. Regulation by glycogen synthase kinase-3 of inflammation and T cells in CNS diseases. Front Mol Neurosci. 2011;4:18. doi: 10.3389/fnmol.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hess AL, Larsen LH, Udesen PB, Sanz Y, Larsen TM, Dalgaard LT. Levels of circulating miR-122 are associated with weight loss and metabolic syndrome. Obes (Silver Spring) 2020;28(3):493–501. doi: 10.1002/oby.22704. [DOI] [PubMed] [Google Scholar]
- 66.Cai C, Lin Y, Yu C. Circulating miRNAs as novel diagnostic biomarkers in nonalcoholic fatty liver disease: a systematic review and meta-analysis. Can J Gastroenterol Hepatol. 2019;2019:2096161. doi: 10.1155/2019/2096161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi Z, Zhao C, Guo X, Ding H, Cui Y, Shen R, Liu J. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERalpha expression in estrogen-induced insulin resistance. Endocrinology. 2014;155(5):1982–1990. doi: 10.1210/en.2013-2046. [DOI] [PubMed] [Google Scholar]
- 68.Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E, Xifra G, Martinez C, Ricart W, Rieusset J, Rome S, Karczewska-Kupczewska M, Straczkowski M, Fernandez-Real JM. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37(5):1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 69.Filardi T, Catanzaro G, Mardente S, Zicari A, Santangelo C, Lenzi A, Morano S, Ferretti E. Non-coding RNA: role in gestational diabetes pathophysiology and complications. Int J Mol Sci. 2020;21:11. doi: 10.3390/ijms21114020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ortega FJ, Mercader JM, Catalan V, Moreno-Navarrete JM, Pueyo N, Sabater M, Gomez-Ambrosi J, Anglada R, Fernandez-Formoso JA, Ricart W, Fruhbeck G, Fernandez-Real JM. Targeting the circulating microRNA signature of obesity. Clin Chem. 2013;59(5):781–792. doi: 10.1373/clinchem.2012.195776. [DOI] [PubMed] [Google Scholar]
- 71.Giardina S, Hernandez-Alonso P, Diaz-Lopez A, Salas-Huetos A, Salas-Salvado J, Bullo M. Changes in circulating miRNAs in healthy overweight and obese subjects: effect of diet composition and weight loss. Clin Nutr. 2019;38(1):438–443. doi: 10.1016/j.clnu.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 72.Assmann TS, Riezu-Boj JI, Milagro FI, Martinez JA. Circulating adiposity-related microRNAs as predictors of the response to a low-fat diet in subjects with obesity. J Cell Mol Med. 2020;24(5):2956–2967. doi: 10.1111/jcmm.14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gurha P, Wang T, Larimore AH, Sassi Y, Abreu-Goodger C, Ramirez MO, Reddy AK, Engelhardt S, Taffet GE, Wehrens XH, Entman ML, Rodriguez A. microRNA-22 promotes heart failure through coordinate suppression of PPAR/ERR-nuclear hormone receptor transcription. PLoS ONE. 2013;8(9):e75882. doi: 10.1371/journal.pone.0075882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kurylowicz A, Owczarz M, Polosak J, Jonas MI, Lisik W, Jonas M, Chmura A, Puzianowska-Kuznicka M. SIRT1 and SIRT7 expression in adipose tissues of obese and normal-weight individuals is regulated by microRNAs but not by methylation status. Int J Obes (Lond) 2016;40(11):1635–1642. doi: 10.1038/ijo.2016.131. [DOI] [PubMed] [Google Scholar]
- 75.Manning P, Munasinghe PE, Bellae Papannarao J, Gray AR, Sutherland W, Katare R. Acute weight loss restores dysregulated circulating microRNAs in individuals who are obese. J Clin Endocrinol Metab. 2019;104(4):1239–1248. doi: 10.1210/jc.2018-00684. [DOI] [PubMed] [Google Scholar]
- 76.Cannataro R, Perri M, Gallelli L, Caroleo MC, De Sarro G, Cione E. Ketogenic diet acts on body remodeling and microRNAs expression profile. Microrna. 2019;8(2):116–126. doi: 10.2174/2211536608666181126093903. [DOI] [PubMed] [Google Scholar]
- 77.Margolis LM, Rivas DA, Pasiakos SM, McClung JP, Ceglia L, Fielding RA. Upregulation of circulating myomiR following short-term energy restriction is inversely associated with whole body protein synthesis. Am J Physiol Regul Integr Comp Physiol. 2017;313(3):R298–R304. doi: 10.1152/ajpregu.00054.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parr EB, Camera DM, Burke LM, Phillips SM, Coffey VG, Hawley JA. Circulating microRNA responses between 'High' and 'Low' responders to a 16-Wk diet and exercise weight loss intervention. PLoS ONE. 2016;11(4):e0152545. doi: 10.1371/journal.pone.0152545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Donghui T, Shuang B, Xulong L, Meng Y, Yujing G, Yujie H, Juan L, Dongsheng Y. Improvement of microvascular endothelial dysfunction induced by exercise and diet is associated with microRNA-126 in obese adolescents. Microvasc Res. 2019;123:86–91. doi: 10.1016/j.mvr.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 80.Kristensen MM, Davidsen PK, Vigelso A, Hansen CN, Jensen LJ, Jessen N, Bruun JM, Dela F, Helge JW. miRNAs in human subcutaneous adipose tissue: effects of weight loss induced by hypocaloric diet and exercise. Obes (Silver Spring) 2017;25(3):572–580. doi: 10.1002/oby.21765. [DOI] [PubMed] [Google Scholar]
- 81.He H, Sun D, Zeng Y, Wang R, Zhu W, Cao S, Bray GA, Chen W, Shen H, Sacks FM, Qi L, Deng HW. A systems genetics approach identified GPD1L and its molecular mechanism for obesity in human adipose tissue. Sci Rep. 2017;7(1):1799. doi: 10.1038/s41598-017-01517-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kelly TJ, Souza AL, Clish CB, Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol. 2011;31(13):2696–2706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Magenta A, Greco S, Gaetano C, Martelli F. Oxidative stress and microRNAs in vascular diseases. Int J Mol Sci. 2013;14(9):17319–17346. doi: 10.3390/ijms140917319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giardina S, Hernandez-Alonso P, Salas-Salvado J, Rabassa-Soler A, Bullo M. Modulation of human subcutaneous adipose tissue microRNA profile associated with changes in adiposity-related parameters. Mol Nutr Food Res. 2018;62:2. doi: 10.1002/mnfr.201700594. [DOI] [PubMed] [Google Scholar]
- 85.Arner P, Kulyte A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015;11(5):276–288. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 86.Hulsmans M, Sinnaeve P, Van der Schueren B, Mathieu C, Janssens S, Holvoet P. Decreased miR-181a expression in monocytes of obese patients is associated with the occurrence of metabolic syndrome and coronary artery disease. J Clin Endocrinol Metab. 2012;97(7):E1213–1218. doi: 10.1210/jc.2012-1008. [DOI] [PubMed] [Google Scholar]
- 87.Nunez Lopez YO, Coen PM, Goodpaster BH, Seyhan AA. Gastric bypass surgery with exercise alters plasma microRNAs that predict improvements in cardiometabolic risk. Int J Obes (Lond) 2017;41(7):1121–1130. doi: 10.1038/ijo.2017.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Atkin SL, Ramachandran V, Yousri NA, Benurwar M, Simper SC, McKinlay R, Adams TD, Najafi-Shoushtari SH, Hunt SC. Changes in blood microRNA expression and early metabolic responsiveness 21 days following bariatric surgery. Front Endocrinol (Lausanne) 2018;9:773. doi: 10.3389/fendo.2018.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wensveen FM, Jelencic V, Valentic S, Sestan M, Wensveen TT, Theurich S, Glasner A, Mendrila D, Stimac D, Wunderlich FT, Bruning JC, Mandelboim O, Polic B. NK cells link obesity-induced adipose stress to inflammation and insulin resistance. Nat Immunol. 2015;16(4):376–385. doi: 10.1038/ni.3120. [DOI] [PubMed] [Google Scholar]
- 90.Giroud M, Karbiener M, Pisani DF, Ghandour RA, Beranger GE, Niemi T, Taittonen M, Nuutila P, Virtanen KA, Langin D, Scheideler M, Amri EZ. Let-7i-5p represses brite adipocyte function in mice and humans. Sci Rep. 2016;6:28613. doi: 10.1038/srep28613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Latreille M, Hausser J, Stutzer I, Zhang Q, Hastoy B, Gargani S, Kerr-Conte J, Pattou F, Zavolan M, Esguerra JL, Eliasson L, Rulicke T, Rorsman P, Stoffel M. MicroRNA-7a regulates pancreatic beta cell function. J Clin Invest. 2014;124(6):2722–2735. doi: 10.1172/JCI73066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hubal MJ, Nadler EP, Ferrante SC, Barberio MD, Suh JH, Wang J, Dohm GL, Pories WJ, Mietus-Snyder M, Freishtat RJ. Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obes (Silver Spring) 2017;25(1):102–110. doi: 10.1002/oby.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bae YU, Kim Y, Lee H, Kim H, Jeon JS, Noh H, Han DC, Ryu S, Kwon SH. Bariatric surgery alters microRNA content of circulating exosomes in patients with obesity. Obes (Silver Spring) 2019;27(2):264–271. doi: 10.1002/oby.22379. [DOI] [PubMed] [Google Scholar]
- 94.Macartney-Coxson D, Danielson K, Clapham J, Benton MC, Johnston A, Jones A, Shaw O, Hagan RD, Hoffman EP, Hayes M, Harper J, Langston MA, Stubbs RS. MicroRNA profiling in adipose before and after weight loss highlights the role of miR-223-3p and the NLRP3 inflammasome. Obes (Silver Spring) 2020;28(3):570–580. doi: 10.1002/oby.22722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deiuliis JA, Syed R, Duggineni D, Rutsky J, Rengasamy P, Zhang J, Huang K, Needleman B, Mikami D, Perry K, Hazey J, Rajagopalan S. Visceral adipose microRNA 223 is upregulated in human and murine obesity and modulates the inflammatory phenotype of macrophages. PLoS ONE. 2016;11(11):e0165962. doi: 10.1371/journal.pone.0165962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rheinheimer J, de Souza BM, Cardoso NS, Bauer AC, Crispim D. Current role of the NLRP3 inflammasome on obesity and insulin resistance: a systematic review. Metabolism. 2017;74:1–9. doi: 10.1016/j.metabol.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 97.Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4(4):611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 98.Liao CH, Wang CY, Liu KH, Liu YY, Wen MS, Yeh TS. MiR-122 marks the differences between subcutaneous and visceral adipose tissues and associates with the outcome of bariatric surgery. Obes Res Clin Pract. 2018;12(6):570–577. doi: 10.1016/j.orcp.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 99.Kurylowicz A, Wicik Z, Owczarz M, Jonas MI, Kotlarek M, Swierniak M, Lisik W, Jonas M, Noszczyk B, Puzianowska-Kuznicka M. NGS reveals molecular pathways affected by obesity and weight loss-related changes in miRNA levels in adipose tissue. Int J Mol Sci. 2017;19:1. doi: 10.3390/ijms19010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ortega FJ, Mercader JM, Moreno-Navarrete JM, Nonell L, Puigdecanet E, Rodriquez-Hermosa JI, Rovira O, Xifra G, Guerra E, Moreno M, Mayas D, Moreno-Castellanos N, Fernandez-Formoso JA, Ricart W, Tinahones FJ, Torrents D, Malagon MM, Fernandez-Real JM. Surgery-induced weight loss is associated with the downregulation of genes targeted by microRNAs in adipose tissue. J Clin Endocrinol Metab. 2015;100(11):E1467–1476. doi: 10.1210/jc.2015-2357. [DOI] [PubMed] [Google Scholar]
- 101.Ortega FJ, Moreno M, Mercader JM, Moreno-Navarrete JM, Fuentes-Batllevell N, Sabater M, Ricart W, Fernandez-Real JM. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clin Epigenetics. 2015;7:49. doi: 10.1186/s13148-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nardelli C, Iaffaldano L, Pilone V, Labruna G, Ferrigno M, Carlomagno N, Dodaro CA, Forestieri P, Buono P, Salvatore F, Sacchetti L. Changes in the microRNA profile observed in the subcutaneous adipose tissue of obese patients after laparoscopic adjustable gastric banding. J Obes. 2017;2017:6754734. doi: 10.1155/2017/6754734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Langi G, Szczerbinski L, Kretowski A. Meta-analysis of differential miRNA expression after bariatric surgery. J Clin Med. 2019;8:8. doi: 10.3390/jcm8081220. [DOI] [PMC free article] [PubMed] [Google Scholar]