To the Editor
Cutaneous T-cell lymphomas (CTCL) are a heterogeneous group of lymphoid malignancies derived from skin-homing T cells. Mycosis fungoides (MF) is the most common form of CTCL, and Sezary syndrome (SS) is an aggressive variant with varying levels of clonal lymphocytes in the blood. The variable presentation and lack of definitive diagnostic markers make CTCL diagnosis challenging. While the biology of these malignancies is not fully understood, some microbes, particularly viruses, have been hypothesized to play roles in malignant T cell transformation in CTCL (Berger et al., 2002, Mirvish et al., 2013, van der Loo et al., 1979). However, high throughput sequencing approaches have failed to consistently detect viral sequences in the skin or peripheral blood of CTCL patients (Anderson et al., 2018, Dereure et al., 2013). Infections are common in advanced stage patients, and antibiotic treatment results in skin improvement and decreased disease activity (Lindahl et al., 2019). Hence, to better understand the spectrum of microbial involvement in CTCL, we performed a comprehensive evaluation of the skin microbiome in a cohort of MF and SS patients as compared to healthy controls.
In this pilot study, we used shotgun metagenomic sequencing to investigate microbial communities at pre-determined, matched skin sites in four MF patients (stages IA to IIIA), two SS patients (stage IVA1), and ten age- and sex-matched healthy volunteers (HVs) (Table S1). The study was approved by the Institutional Review Boards of Johns Hopkins and the National Institute of Arthritis and Musculoskeletal and Skin Diseases, NIH. Subjects provided written informed consent and underwent skin preparatory regimens. Pre-moistened swabs were used to collect samples from the nares, lower back and thigh skin (sites of CTCL predilection), and air controls (see Supplementary Materials and Methods for details on patient recruitment and sampling). DNA was isolated, and libraries were created for metagenomic sequencing (Oh et al., 2014).
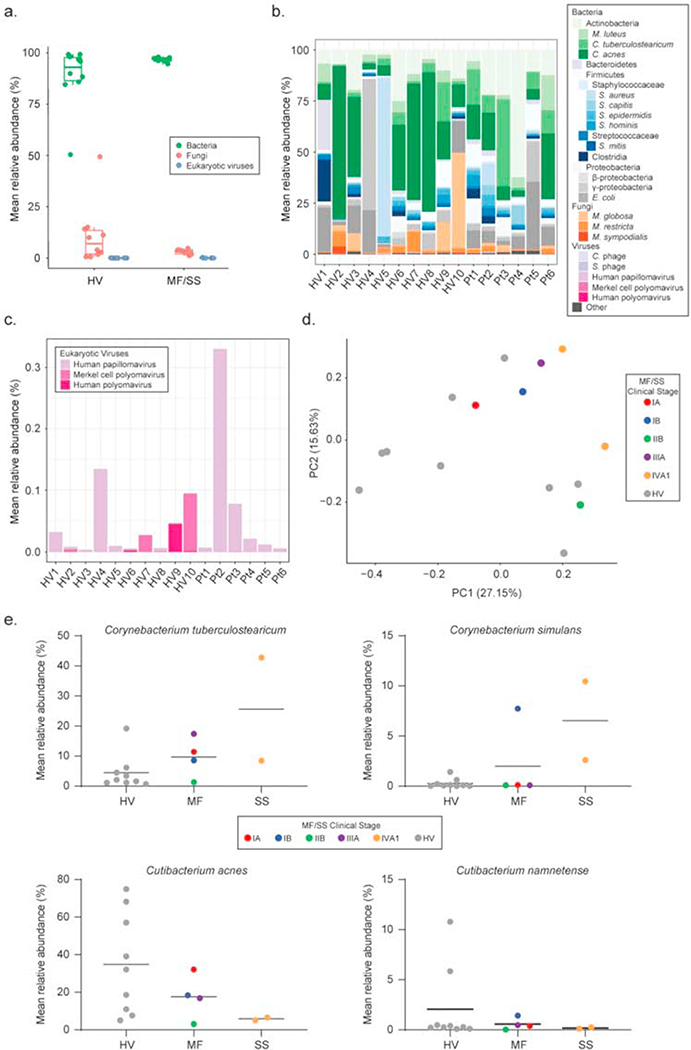
Bacterial, fungal, and viral communities were investigated by mapping microbial reads to a multi-kingdom reference database (Table S2). Analyses focused on comparing the microbiomes between lesional patient skin and HV skin from the lower back and thigh. Bacteria predominated microbial communities at all sites (Figure 1a, S1a, S2a, Table S3). Of the most abundant taxa across kingdoms (Figure 1b, S1b, S2b), less than 0.5% of metagenomic reads mapped to eukaryotic viruses (predominantly Papillomaviridae and Polyomaviridae) in MF/SS and HV lower backs (0.09% ± 0.1% vs. 0.05% ± 0.05%) and thighs (0.08% ± 0.1% vs. 0.07% ± 0.08%) with no discernible differences regardless of sampling area (Figure 1c, S1c, S2c, Table S4). Similarly, fungal abundances did not differ significantly between HVs and MF/SS (Figure 1b, S1b, Table S5); Shannon diversity was comparable (Figure S3a, S3b).
Figure 1. Bacteria predominate in skin microbiomes of both MF/SS patients and healthy adults, but bacterial communities differ between patients and controls.
(a) Relative abundances of lower back skin microbes classified by kingdom in HVs (n=10) and MF/SS patients (n=6). Boxplot with each dot corresponding to one subject and representing the mean relative abundance (%) of one lower back sample per subject (lesional skin for patients; healthy skin for HVs). For each box, the central line represents the median, lower and upper edges represent the 1st and 3rd quartile, and whiskers represent values up to 1.5 times the interquartile range. Shotgun metagenomics microbial reads were mapped to a multi-kingdom reference database containing 2356 bacterial, 395 fungal, 4695 viral and 67 archaeal reference genomes using Bowtie2. (b) Mean relative abundances (%) of the major microbial taxa detected across kingdoms in lower back skin samples of HVs compared to MF/SS. In the barplot, colors represent distinct taxa. Due to the unusually high relative abundance (>75%) of the plant Gammaproteobacteria (Xanthomonas campestris) on the skin of HV4, this subject was removed from downstream analyses. (c) Relative abundances (%) of eukaryotic viruses on the skin of HVs and MF/SS patients. (d) Principle coordinates analysis (PCoA) of lower back skin bacterial communities in HVs and MF/SS. The distance between samples was measured using Bray-Curtis dissimilarity index. Each dot corresponds to one lower back sample per subject. HVs (n=9) are represented by gray dots. MF/SS patients are color-coded by clinical stage IA (n=1), IB (n=1), IIB (n=1), IIIA (n=1), IVA1 (n=2). Clustering of dots in the PCoA indicates higher similarity between skin bacterial communities. (e) Mean relative abundances (%) of common cutaneous bacteria (Corynebacterium spp. and Cutibacterium spp.) on the lower back skin of HVs (n=9) compared to MF (n=4) and SS (n=2) patients. Each dot corresponds to one subject and represents the relative abundance in one representative lower back sample. Dots are color-coded as in (d). Black horizontal lines represent the mean. HV, healthy volunteer; MF, mycosis fungoides; SS, Sezary syndrome.
Given the low viral and fungal relative abundances, we focused on bacterial communities in MF/SS and HVs. We performed principle coordinates analysis (PCoA) using Bray-Curtis dissimilarity index which demonstrated separation of HVs and MF/SS bacterial skin communities on both lower backs and thighs (Figure 1d, S1d). Superimposing MF/SS stages on the PCoA showed greatest separation between HVs and stage IVA1 patients, suggesting stage IV patients’ skin microbiomes are the most distinct from controls.
We then further investigated specific taxa contributing to differences in bacterial communities between MF, SS, and HV skin. Given the association with Staphylococcus aureus colonization and infection in CTCL (Krejsgaard et al., 2014, Lindahl et al., 2019) and reported staphylococcal-corynebacterial interactions (Ramsey et al., 2016), we compared these and other common cutaneous bacteria. Staphylococcus aureus abundances were low in most MF, SS, and HV skin samples (Figure S4a, S4b). One HV and one MF patient had higher S. aureus relative abundances on the skin. Commensal staphylococci (S. capitis, S. epidermidis, and S. hominis) trended higher in MF (3.8% ± 3.9%, 2.7% ± 2.1%, and 1.8% ± 2.4%, respectively) versus HV lower back skin (0.6% ± 0.6%, 1.4% ± 1.1%, and 0.8% ± 1.2%, respectively) (Figure S4). Two Corynebacterium species (C. tuberculostearicum and C. simulans) were increased on MF and SS skin, with highest mean relative abundances in SS patients (C. tuberculostearicum on lower back: 25.6% ± 24.3% (SS) vs. 4.4% ± 5.8% (HV); C. simulans on lower back: 6.5% ± 5.5% (SS) vs. 0.3% ± 0.5% (HV)) (Figure 1e, S1e). MF and SS skin also displayed lower relative abundances of Cutibacterium acnes and Cutibacterium namnetense as compared to HV skin. These bacterial shifts were not statistically significant, likely due to the small number of patients. However, comparing HV to MF to SS skin, we observed increasing trends in the mean relative abundances of Corynebacterium species and decreasing trends in Cutibacterium species, suggesting bacterial shifts may correlate with disease stage or treatment status (Figure 1e, S1e).
Our findings suggest eukaryotic DNA viruses are negligible components of the skin microbiome in our MF/SS cohort. These results corroborate and extend previous reports suggesting CTCL is unlikely to originate from infection by a directly oncogenic DNA virus (Dulmage et al., 2015). However, other mechanisms by which viral pathogens can affect neoplastic transformation must be further explored including 1) indirect viral tumorigenesis by transient exposure to viral genomes (‘hit-and-run’ oncogenesis) (Niller et al., 2011), 2) integration of retroviruses or DNA viral elements into human host DNA, and 3) antigenic stimulation of T cells in the peripheral blood.
Our MF/SS patients showed no marked differences in skin viral or fungal communities as compared to age-matched HVs sampled at consistent sites. Nonetheless, we observed bacterial community shifts including higher relative abundances of Corynebacterium spp. and lower relative abundances of Cutibacterium spp. in MF/SS skin. Several staphylococcal and corynebacterial species trended higher in MF/SS skin and would be important to examine in larger studies. Interestingly, relative abundances of C. tuberculostearicum were high (>25% on average) in stage IVA1 patients. Advanced stage patients may be at increased risk of infection from impaired skin integrity and immune dysregulation (Axelrod et al., 1992), and C. tuberculostearicum has been shown to upregulate proinflammatory responses in human skin cells, suggesting a potential link to cutaneous inflammation (Altonsy et al., 2020).
Recently, Salava et al. reported some differences in bacterial communities between clinically unaffected and affected patient skin without including HV controls (Salava et al., 2020). Due to potential absence of uninvolved contralateral skin in advanced stage patients, reports of lymphoma involvement in normal-appearing skin in CTCL patients (Pujol et al., 2000), and different skin microbiomes in anatomically distinct but adjacent sites, we studied control samples from age-matched HVs sampled at comparable anatomical sites.
The separation of bacterial communities between stage IVA1 patients and HVs is notable, and it is intriguing that bacterial shifts appear to correlate with disease stage. Whether these findings are driven by disease severity or other unrelated variables, including systemic treatments, is unknown. Further investigation with larger and ideally multi-center CTCL cohorts is warranted to validate these findings and to evaluate the relationship between disease stage and the skin microbiome.
Supplementary Material
ACKNOWLEDGEMENTS
This publication was supported by the Intramural Research Programs of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (CPH, MAM, JP, J-HJ, HHK) and the National Human Genome Research Institute (CPH, XH, JAS). The study utilized the computational resources of the NIH HPC Biowulf Cluster (http://hpc.nih.gov). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The authors thank other members of the Segre and Kong labs for their underlying efforts and the healthy volunteers and patients with cutaneous T-cell lymphoma for their contributions.
Abbreviations:
- CTCL
cutaneous T-cell lymphoma
- MF
mycosis fungoides
- SS
Sezary syndrome
- HV
healthy volunteer
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
DATA AVAILABILITY STATEMENT:
All sequencing data were deposited and are available at the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA642893.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- Altonsy MO, Kurwa HA, Lauzon GJ, Amrein M, Gerber AN, Almishri W, et al. Corynebacterium tuberculostearicum, a human skin colonizer, induces the canonical nuclear factor-kappaB inflammatory signaling pathway in human skin cells. Immun Inflamm Dis 2020;8:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson ME, Nagy-Szakal D, Jain K, Patrone CC, Frattini MG, Lipkin WI, et al. Highly Sensitive Virome Capture Sequencing Technique VirCapSeq-VERT Identifies Partial Noncoding Sequences but no Active Viral Infection in Cutaneous T-Cell Lymphoma. J Invest Dermatol 2018;138:1671–73. [DOI] [PubMed] [Google Scholar]
- Axelrod PI, Lorber B, Vonderheid EC. Infections Complicating Mycosis Fungoides and Sézary Syndrome. JAMA 1992;267:1354–58. [PubMed] [Google Scholar]
- Berger CL, Hanlon D, Kanada D, Dhodapkar M, Lombillo V, Wang N, et al. The growth of cutaneous T-cell lymphoma is stimulated by immature dendritic cells. Blood 2002;99:2929–39. [PubMed] [Google Scholar]
- Dereure O, Cheval J, Du Thanh A, Pariente K, Sauvage V, Claude Manuguerra J, et al. No evidence for viral sequences in mycosis fungoides and Sezary syndrome skin lesions: a high-throughput sequencing approach. J Invest Dermatol 2013;133:853–5. [DOI] [PubMed] [Google Scholar]
- Dulmage BO, Feng H, Mirvish E, Geskin L. Black cat in a dark room: The absence of a directly oncogenic virus does not eliminate the role of an infectious agent in cutaneous T-cell lymphoma pathogenesis. Br J Dermatol 2015;172:1449–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejsgaard T, Willerslev-Olsen A, Lindahl LM, Bonefeld CM, Koralov SB, Geisler C, et al. Staphylococcal enterotoxins stimulate lymphoma-associated immune dysregulation. Blood 2014;124:761–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl LM, Willerslev-Olsen A, Gjerdrum LMR, Nielsen PR, Blumel E, Rittig AH, et al. Antibiotics inhibit tumor and disease activity in cutaneous T-cell lymphoma. Blood 2019;134:1072–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirvish JJ, Pomerantz RG, Falo LD, Geskin LJ. Role of infectious agents in cutaneous T-cell lymphoma: Facts and controversies. Clin Dermatol 2013;31:423–31. [DOI] [PubMed] [Google Scholar]
- Niller HH, Wolf H, Minarovits J. Viral hit and run-oncogenesis: genetic and epigenetic scenarios. Cancer Lett 2011;305:200–17. [DOI] [PubMed] [Google Scholar]
- Oh J, Byrd AL, Deming C, Conlan S, NISC Comparative Sequencing Program, Kong HH, et al. Biogeography and individuality shape function in the human skin metagenome. Nature 2014;514:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol RM, Gallardo F, Llistosella E, Blanco A, Bernado L, Bordes R, et al. Invisible mycosis fungoides: A diagnostic challenge. J Am Acad Dermatol 2000;42:324–8. [DOI] [PubMed] [Google Scholar]
- Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP. Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species. Front Microbiol 2016;7:1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salava A, Deptula P, Lyyski A, Laine P, Paulin L, Vakeva L, et al. Skin microbiome in cutaneous T cell lymphoma by 16S and whole genome shotgun sequencing. [e-pub ahead of print]. J Invest Dermatol; doi: 10.1016/j.jid.2020.03.951 (accessed 23 June 2020). [DOI] [PubMed] [Google Scholar]
- van der Loo EM, van Muijen GN, van Vloten WA, Beens W, Scheffer E, Meijer CJ. C-type virus-like particles specifically localized in Langerhans cells and related cells of skin and lymph nodes of patients with mycosis fungoides and Sezary’s syndrome. A morphological and biochemical study. Virchows Arch B Cell Pathol Incl Mol Pathol 1979;31:193–203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.