Abstract
A primary factor in tumor morbidity and mortality, lung adenocarcinoma (LUAD) is known to be a major subtype of lung cancer, having the lowest survival rate among all other cancers. Using The Cancer Genome Atlas (TCGA) database the relationship between the immune infiltrate and the NUP62CL was explored and the value of the NUP62CL expression in the prognosis and diagnosis LUAD was examined. Using the logistic regression and the Wilcoxon signed‐rank test the relationship between the NUP62CL and the clinico‐pathological features was analyzed. There was a significant correlation between the clinical stage (p = 0.005), the N (p = 0.004), and the decreased expression of NUP62CL. The prognosis of LUAD with high NUP62CL expression was revealed to be worse than that with low NUP62CL expression (p < 0.001) by the Kaplan‐Meier survival analysis. The potentiality of NUP62CL to be a significant factor of prognosis for LUAD was indicated by the analyses of the multivariate and the univariate Cox regression models. In LUAD, the crucial role of recombination and maintenance of telomere as a significant pathway for NUP62CL was suggested by the Gene Set Enrichment Analysis (GSEA). To analyze the correlation between the genes and the tumor infiltrating immune cells the CIBERSORT was used. Moreover the positive correlation with the NUP62CL expression in LUAD of the infiltration level of the tumor infiltrating B lymphocytes and memory CD4+ T cells was exhibited by CIBERSORT. Therefore, NUP62CL may be a new valuable prognostic indicator for LUAD.
Keywords: GEO, immune infiltration, LUAD, NUP62CL, prognostic, TCGA
High expression of NUP62CL was associated with low OS in patients with LUAD. Cox regression model analysis indicated that NUP62CL was an important prognostic factor of LUAD. Maintaining telomere recombination may be an important signaling pathway for NUP62CL to play a role in LUAD. In addition, NUP62CL was related to the infiltration level of memory CD4+ T cells.

1. INTRODUCTION
Lung cancer is a major factor in tumor morbidity and mortality. Rebecca L. et al. established that with 1.8 million patient deaths, denoting almost 18.4% of the total (one in every five), and 2.1 million potential new diagnoses, lung cancer has shown the lowest rate of survival among all cancers. 1 , 2 Although the treatment of various therapies has made continuous progress, the 5‐year survival rate is still relatively poor. Lung cancer affords an overall five‐year relative survival of 20.3% in females and 15.5% in males, subsequent to age‐adjustment. 3 Moreover early diagnosis can significantly improve the prognosis, however, most patients when firstly diagnosis have developed to advanced stage. 2 , 4 Hence, the identification of novel immune‐related therapeutic targets and valuable biomarkers for the patients of lung cancer coupled with the further research and development of cancer treatment has become imperative today.
Non‐small cell lung cancer, or NSCLC, with an 80% incidence, is the principal type of lung cancer. 5 With adenocarcinoma being the leading subtype accounting for nearly 50% of patients afflicted with lung cancer, the primary NSCLC histological phenotypes have been known to include the large cell carcinoma, squamous cell carcinoma, and the adenocarcinoma. 6 Lung cancer comprises both cancerous and non‐cancerous cells. The knowledge about the role of these non‐cancer cells remains a mystery. The infiltrating immune cells, the vascular cells, and the stromal cells are known to be the components of the non‐cancerous cells in varied proportions. Immune cell infiltration from among these could potentially improve clinical prognosis, and thereby be effectively targeted in therapy, as indicated via recent drug trials that targeted physiological pathways regulating autoimmune responses, as also the duration and severity of disease immune responses, and immune checkpoints, to reveal how they considerably prolonged survival in cases of solid tumors, such as melanoma, NSCLC, renal cell carcinoma and triple negative breast cancer. 7 , 8 , 9 , 10 In addition, the most widely studied immune cells are tumor infiltrating lymphocytes (TIL). Previous studies have shown that the presence of TIL is potentially predictive and prognostic for specific breast cancer subtypes. As indicated by large‐scale adjuvant studies, in primary biopsy the higher TIL levels have been shown to be associated with reduced recurrence and improved overall survival (OS) particularly in cases with triple negative breast cancer (TNBC), the HER2‐positive, and the human epidermal growth factor receptor 2. 11 , 12 , 13 Amplified TIL levels were also correlated with higher pathological complete response, or PCR rates, subsequent to neoadjuvant therapy in TNBC and HER2‐positive tumor patients. 14 , 15 , 16 Nonetheless, in the case of lung adenocarcinoma the role of these tumor‐infiltrating immune cells remains significantly unknown.
In the mitochondrial transport, DNA repair, gene transcription, and microtubule regulation, the NUP family has been observed to play an important role. 17 , 18 , 19 , 20 , 21 The FG repeat domain of NUP98 is encoded by a chimeric gene, the NUP98–NSD1. Approximately 5% of people have t (5; 11) (q35; p15.5) translocation in human acute myeloid leukemia. The study by Gang G. Wang et al. found that the main transformation characteristic of NUP98–NSD1 is that it does not allow cell differentiation, thereby promoting self‐renewal of progenitor cells, and in experiments found that NUP98–NSD1 can enhance self‐renewal by activating Hox‐A gene and Meis1 transcription and to prevent differentiation, the potent leukemia oncogenes HoxA9 and Meis1 are co‐expressed in NUP98–NSD1 immortalized progenitor cells. Acute myeloid leukemia could be caused by NUP98–NSD1. 19 , 22 Yumei Leng et al. Showed that tumor protein MUC1‐C was related to human cancer and entered the nucleus through the way of binding with NUP62. 23 In addition, In the export of HIV retroviral RNA particles in the human cells, the NUP62 has been indicated to play a significant role. 24 Nonetheless, the involvement of NUP62CL in the development of cancer presently has not been indicated by any study so far.
The prognostic value of the NUP62CL expression in LUAD was evaluated from the data acquired from The Cancer Genome Atlas (TCGA). In the LUAD‐pathogenesis‐linked NUP62CL regulatory network, additional insights into the engagement of the biological pathways were provided by the GSEA. Moreover to study the tumor‐infiltrating immune cells and the NUP62CL correlation, along with the density of the distinct tumor‐infiltrating immune cells under various microenvironments was determined using the CIBERSORT as a recent metagene approach. Moreover, the tumor‐immunity‐interactions correlation, the mechanism involved, besides the understanding and establishing the positive role of the NUP62CL could be possible from the conclusions we drew.
2. MATERIALS AND METHODS
2.1. Data acquisition and bioinformatics analysis
This study used data from the public domain. The prognostic information of tumors and normal tissues, with the gene expression profiles of the patients of LUAD were downloaded from TCGA (https://portal.gdc.cancer.gov/) for our study, and included 535 tumor tissues. Age, total survival time, gender, clinical stage, grade, TNM stage (T, tumor; N, node; M, metastasis) were included. Subsequently, 503 data along with 522 RNA sequence data were used for further survival analysis and correlation analyses, respectively. In addition, to study the relationship between the invasive immune cells and the expression of NUP62CL the tumor tissue was segregated into high and low expression groups based on the expression of the NUP62CL.
2.2. Gene set enrichment analysis
The GSEA computational method helps ascertain statistically significant and consistent variances that exist in any two biological states of an a priori defined set of genes. 25 In this study, to investigate the potential mechanism of NUP62CL expression in the prognosis of LUAD and to explain the significant differences in the survival rates between the low and high NUP62CL expression groups, the use of GSEA was done. The nominal p value and normalized enrichment score (NES) helped assess pathway enrichment (p < 0.05, FDR < 0.25).
2.3. Evaluation of tumor infiltrating immune cells
As per normalized gene expression data, the relative proportion of 22 kinds of immune cell infiltration was inferred by CIBERSORT algorithm. In other words, deemed to be the smallest depiction of any cell type, the set of reference gene expression values was deployed using the deconvolution algorithm of CIBERSORT. From a large sample of mixed cell types, to infer the proportion of the cell types the Support Vector Regression (SVR) was used based upon these values. Standard annotation files helped compile gene expression datasets, which were then uploaded to the CIBERSORT portal (http://cibersort.stanford.edu/). The algorithm runs with 1000 default signature matrices arranged. CIBERSORT validates conclusions via Monte Carlo sampling, wherein a p value is derived to deconvolute each sample. Moreover, CIBERSORT inferred 22 kinds of immune cells. To assess the effect of NUP62CL expression, 535 samples from TCGA were used to represent all genes.
2.4. Verification analysis of NUP62CL
From the Gene Expression Omnibus (GEO) repository the GSE13213 data set was downloaded and 117 samples were collected. Using the R survival package, the relationship between NUP62CL expression and the OS was analyzed in the patients with LUAD. To analyze the 9736 tumor and the 8587 normal samples contained in the TCGA and the GTEX, using the standard processing flow, the online database GEPIA was used. 26 In the patients with LUAD, the relationship between the expression of NUP62CL and the OS was analyzed using GEPIA. The differential expression of NUP62CL under the various pathological stages was compared, while the differential expression of the NUP62CL in the normal and the tumor tissues was observed through boxplot. In addition, the protein locations could be evaluated accurately using the antibody profiles from the 32 human tissues and their protein expression profiles from the Human Protein Atlas network database (HPA) (www.proteinatlas.org). To analyze the expression of NUP62CL in the LUAD and the normal tissues, the HPA was used.
2.5. Statistical analysis
R version.3.5.1 assayed all statistical queries. The correlation of the clinico‐pathological features with NUP62CL was evaluated through the logistic regression and the Wilcoxon signed‐rank test. To analyze the impact of the NUP62CL expression on the clinical characteristics of patients with LUAD and their OS, the Cox regression model was applied. All tests were regarded as statistically significant with the p < 0.05.
3. RESULTS
3.1. Patient characteristics
The clinical and gene expression data of 522 tumors, presented in Table 1, were downloaded from TCGA database. In our study, a total of 280 female patients and 242 male patients were analyzed. The disease cited stage wise as a patient count/percentage, was: Stage I: 279/53.45%, Stage II: 124/23.75%, Stage III: 85/16.28%, and Stage IV: 26/4.98%. The tumor included 32.95% T1 (n = 172), 53.83% T2 (n = 281), 9.00% T3 (n = 47), and 3.64% T4 (n = 19). The Node included 64.17% N0 (n = 335), 18.20% N1 (n = 95), 14.37% N2 (n = 75), and 0.38% N3 (n = 2). A total of 25 cases (4.79%) had distant metastases (Table 1).
TABLE 1.
Characteristics of LUAD patients in TCGA and GEO database
| Clinical characteristics | TCGA (522) % | GEO (117) % | ||
|---|---|---|---|---|
| Age (mean ± SD) | 65.33 ± 10.02 | 60.68 ± 10.17 | ||
| Survival time (year) | 2.11 ± 2.28 | 5.38 ± 2.40 | ||
| Gender | ||||
| Female | 280 | 53.64 | 57 | 48.72 |
| Male | 242 | 46.36 | 60 | 51.28 |
| Stage | ||||
| I | 279 | 53.45 | 79 | 67.52 |
| II | 124 | 23.75 | 13 | 11.11 |
| III | 85 | 16.28 | 25 | 21.37 |
| IV | 26 | 4.98 | 0 | 0 |
| T | ||||
| 1 | 172 | 32.95 | 54 | 46.15 |
| 2 | 281 | 53.83 | 50 | 42.74 |
| 3 | 47 | 9 | 8 | 6.84 |
| 4 | 19 | 3.64 | 5 | 4.27 |
| M | ||||
| 0 | 353 | 67.62 | 117 | 100 |
| 1 | 25 | 4.79 | 0 | 0 |
| N | ||||
| 0 | 335 | 64.17 | 87 | 74.36 |
| 1 | 95 | 18.2 | 8 | 6.84 |
| 2 | 75 | 14.37 | 22 | 18.8 |
| 3 | 2 | 0.38 | 0 | 0 |
Abbreviations: M, metastasis; N, node; T, tumor.
3.2. Association with clinico‐pathological variables and NUP62CL expression
We evaluated NUP62CL expression data across all patient characteristics derived from TCGA. Figure 1 indicated a significant correlation of NUP62CL decreased expression with clinical stage (p = 0.005) and N (p = 0.004). According to logistic regression analysis, the reduced NUP62CL expression was significantly correlated with gender (OR=0.66, p = 0.019), the clinical stage (OR = 0.30, p = 0.009, for IV vs. I), T (OR = 0.50, p = 0.041, for 3 vs. 1), N (OR = 0.49, p = 0.007, for 3 vs. 0) and M (OR = 0.37, p = 0.029, for 1 vs. 0) (Table 2). Therefore, LUAD with high NUP62CL expression presents a greater likelihood of progressing to advanced stages and metastasizing distantly, relative to the low expression group.
FIGURE 1.
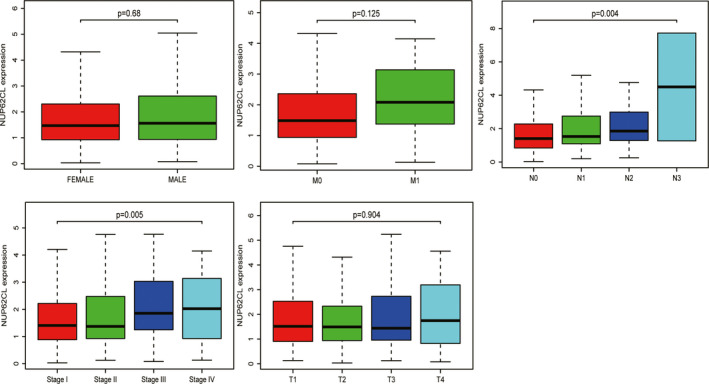
Association with NUP62CL expression and clinicopathologic characteristics, including: gender, clinical stage, and TNM stage
TABLE 2.
NUP62CL expression associated with clinical pathological characteristics (logistic regression)
| Clinical characteristics | Total (N) | Odds ratio in EMID1 expression | p‐value |
|---|---|---|---|
| Gender (male vs. female) | 522 | 0.66 (0.46, 0.93) | 0.019 |
| Age (≥65 vs. <65) | 522 | 0.75 (0.52, 1.06) | 0.105 |
| Stage (IV. vs. I) | 305 | 0.30 (0.11, 0.71) | 0.009 |
| T (3 vs. 1) | 219 | 0.50 (0.25, 0.96) | 0.041 |
| M (1 vs. 0) | 378 | 0.37 (0.14, 0.87) | 0.029 |
| N (3 vs. 0) | 337 | 0.49 (0.28, 0.81) | 0.007 |
Abbreviations: M, metastasis; N, node; T, tumor.
3.3. Survival analysis and multivariate analysis of NUP62CL
The prognosis of the low expression of NUP62CL was shown to be better than that of the high expression of NUP62CL (p < 0.001) by the Kaplan‐Meier survival analysis, as illustrated in Figure 2A. The high expression of NUP62CL having a significant relationship to the low OS of the LUAD patients was indicted by the Univariate analysis. Other clinicopathological variables, including stage, T, N, were also associated with low survival in patients with LUAD. The probability of the NUP62CL being an independent prognostic factor for LUAD was demonstrated by the Multivariate analysis (Table 3, Figure 2B,C).
FIGURE 2.
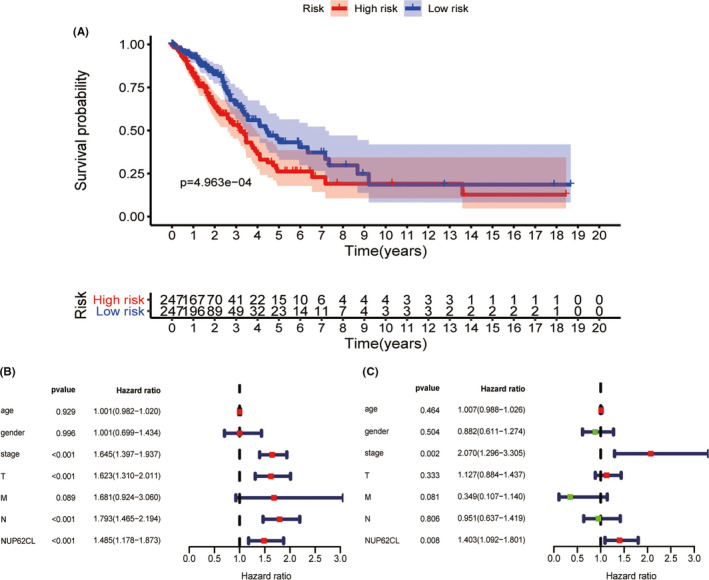
A, Survival analysis of NUP62CL by Kaplan Meier survival curve. B, Univariate analysis of NUP62CL. C, Multivariate survival model of NUP62CL. M, metastasis; N, node; T, tumor
TABLE 3.
A, Associations with overall survival and clinicopathologic characteristics in TCGA patients using Cox regression. B, Multivariate survival model after variable selection
| Clinicopathologic variable | HR | HR.95L | HR.95H | p‐value |
|---|---|---|---|---|
| (A) | ||||
| Age | 1.00 | 0.98 | 1.02 | 0.929 |
| Gender | 1.00 | 0.70 | 1.43 | 0.996 |
| Stage | 1.64 | 1.40 | 1.94 | <0.001 |
| T | 1.62 | 1.31 | 2.01 | <0.001 |
| M | 1.68 | 0.92 | 3.06 | 0.089 |
| N | 1.79 | 1.46 | 2.19 | <0.001 |
| NUP62CL | 1.49 | 1.18 | 1.87 | 0.001 |
| (B) | ||||
| Stage | 2.07 | 1.30 | 3.31 | 0.002 |
| T | 1.13 | 0.88 | 1.44 | 0.333 |
| N | 0.95 | 0.64 | 1.42 | 0.806 |
| NUP62CL | 1.40 | 1.09 | 1.80 | 0.008 |
Abbreviations: M, metastasis; N, node; T, tumor.
3.4. Identification of a NUP62CL linked signaling pathway enabled by GSEA
The Gene Set Enrichment Analysis (GSEA) between the high and the low expression data of NUP62CL was conducted to identify the significant signaling pathways activated in the Lung adenocarcinoma. Table 4 displays the details of the significant variances (FDR < 0.25, NOM p < 0.05) in the enrichment of MSigDB Collection (c5.all.v7.0.symbols.gmt), presented by GSEA. The differentially enriched high expression phenotypes in the NUP62CL have been exhibited in the Figure 3A as, DNA dependent DNA replication maintenance of fidelity, g2 DNA damage checkpoint, replication fork processing, telomere maintenance via recombination, site of DNA damage, positive regulation of double strand break repair, mitotic recombination, regulation of sister chromatid cohesion, centriolar satellite, mitotic g2m transition checkpoint.
TABLE 4.
Signaling pathways most significantly correlated with NUP62CL expression based on their normalized enrichment score (NES) and p‐value
| GO name | NES | NOM p‐val | FDR q‐val |
|---|---|---|---|
| Positive | |||
| DNA dependent DNA replication maintenance of fidelity | 2.288 | 0.000 | 0.002 |
| G2 DNA damage checkpoint | 2.329 | 0.000 | 0.003 |
| Replication fork processing | 2.257 | 0.000 | 0.005 |
| Telomere maintenance via recombination | 2.168 | 0.000 | 0.033 |
| Site of DNA damage | 2.117 | 0.004 | 0.069 |
| Positive regulation of double strand break repair | 2.102 | 0.000 | 0.076 |
| Mitotic recombination | 2.091 | 0.000 | 0.078 |
| Regulation of sister chromatid cohesion | 2.066 | 0.000 | 0.090 |
| Centriolar satellite | 2.070 | 0.000 | 0.097 |
| Mitotic g2m transition checkpoint | 2.047 | 0.002 | 0.098 |
Gene sets with NOM p < 0.05 and FDR q‐val < 0.25 are considered as significant.
Abbreviations: FDR, false discovery rate; NES, normalized enrichment score; NOM, nominal.
FIGURE 3.
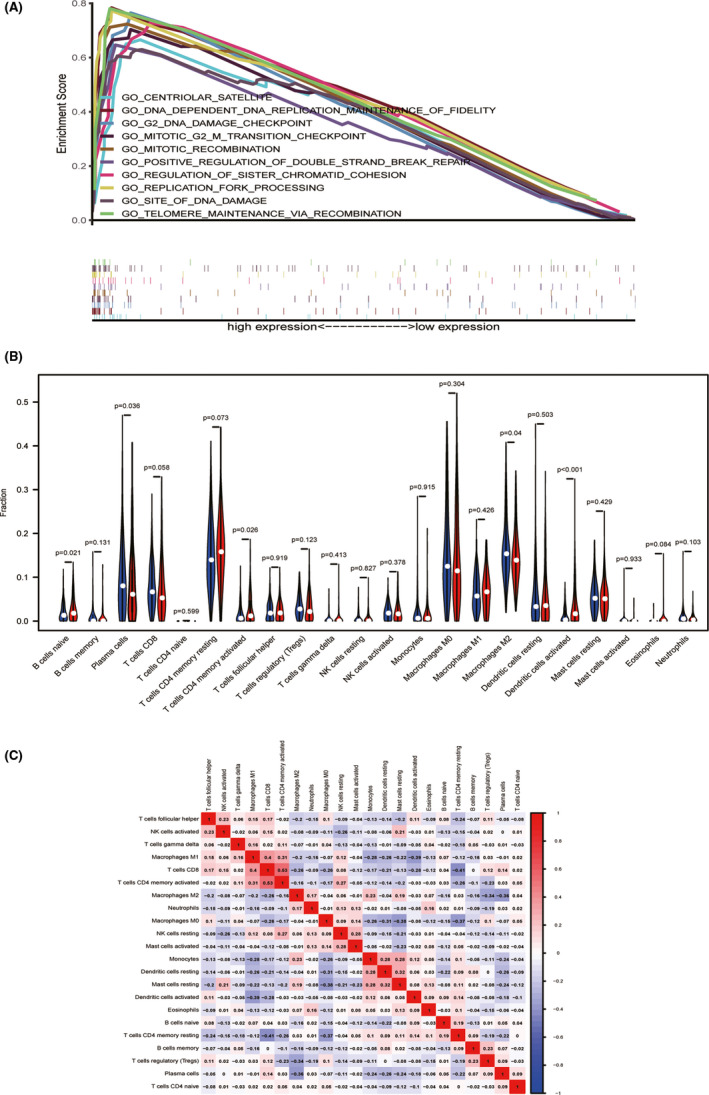
A, GO term analysis showed 10 positive correlation groups. B, The proportion of 22 immune cell subtypes in high and low NUP62CL expression group. Red indicated high expression and blue indicated low expression. C, Heatmap of 22 immune infiltration cells in LUAD samples
3.5. Relationship between the tumor‐infiltrating immune cells and the NUP62CL expression
To investigate whether NUP62CL expression is associated with immune infiltration in lung adenocarcinoma, to explore the gene expression profiles of the samples downloaded and to infer the density of the 22 immune cells the CIBERSORT resource was deployed in this study. The total 535 tumor samples were segregated according to the NUP62CL expression into two parts as, 268 samples of high expression and 267 samples of low expression. Subsequently, using CIBERSORT the relative proportion of the 22 kinds of immune cell infiltration in these tumor samples were estimated. The result of the relative proportion of these immune cell infiltration is shown in Figure 3B. In the high expression group, B cells naïve (p = 0.021), T cells CD4+ memory activated (p = 0.026) and the activated (p < 0.001) dendritic cells were found to be significantly increased. However, the Macrophages M2 (p = 0.04) and the Plasma cells (p = 0.036) were observed to have been significantly increased in the low expression group. Moreover a weak to moderate correlation was presented by the diverse tumor‐infiltrating immune cells subgroups (Figure 3C).
3.6. Verification analysis
To determine whether the prognosis of NUP62CL is reliable, we used GEO database, GEPIA database and HPA database to verify NUP62CL. GSE13213 was downloaded from GEO database and included 117 sample information (Table 1). The high OS in patients with LUAD was found to be associated with the low expression of NUP62CL from the analysis of the data (Figure 4A). From the GEPIA database, it was found that the expression of NUP62CL was significantly higher in the tumor tissues than in the normal group (Figure 4B). Late pathological stage and low OS (p < 0.001) were found to be correlated significantly with the high expression of NUP62CL (Figure 4C,D). Higher incidence of the expression of NUP62CL in tumor tissue than in the normal tissue was indicated by the immunohistochemistry analysis from HPA (Figure 4E).
FIGURE 4.
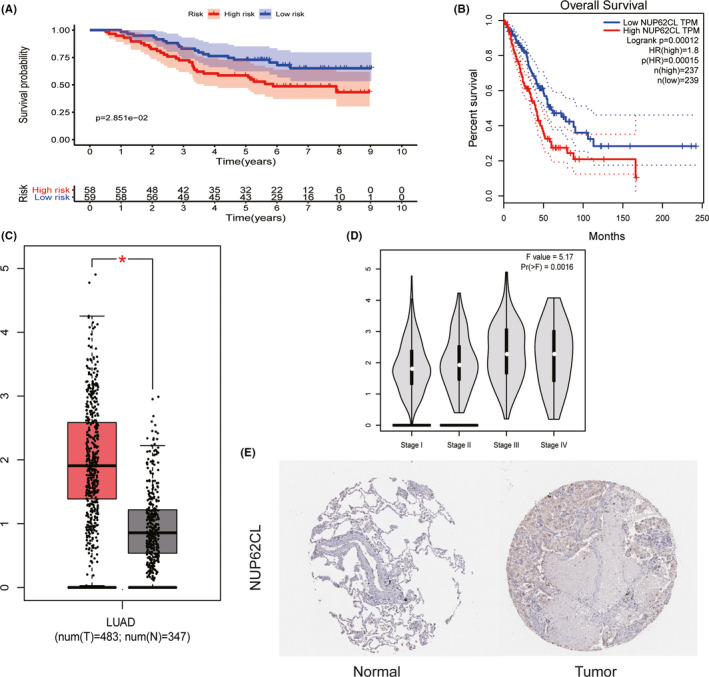
A, The expression level and overall survival of NUP62CL in GEO database. B, The expression level and overall survival of NUP62CL in GEPIA. C, The expression level of NUP62CL in normal and tumor tissues obtained from GEPIA. D, Differential expression of NUP62CL in different pathological stage of GEPIA. E, The expression of NUP62CL protein in lung by HPA immunohistochemistry
4. DISCUSSION
Lung cancer is a leading morbidity and mortality factor, with adenocarcinoma as the primary subtype. In‐depth research on lung adenocarcinoma is imperative to improve the prognosis and the survival rate of the patients with lung cancer. 27 Possibly, our study might be the first to suggest the association with cancer of the NUP62CL expression and its potential to be a biomarker in the lung adenocarcinoma prognosis.
Our study established an increased NUP62CL expression in LUAD as correlating with overall survival and poor prognosis, as well as advanced clinic‐pathological aspects, such as clinical stage, tumor, node, metastasis. To explore further the functionality of the NUP62CL in LUAD and to specify the following as differentially enriched in its high expression phenotype the GSEA was deployed by the study, DNA‐dependent DNA replication maintenance of fidelity, g2 DNA damage checkpoint, replication fork processing, telomere maintenance via recombination, site of DNA damage, positive regulation of double strand break repair, mitotic recombination, regulation of sister chromatid cohesion, centriolar satellite, mitotic g2m transition checkpoint. Moreover, a significant correlation was established between the immune infiltration levels and the NUP62CL through CIBERSORT in case of the LUAD afflicted patients, including B cells naive, T cells CD4 memory activated, Dendritic cells activated, Plasma cells and Macrophages M2. The potentiality of the NUP62CL to be an independent prognostic biomarker in LUAD was suggested by the results of this study.
Telomerase expression is regulated by receptor cells, so telomere sequence will wear out gradually in each round of DNA replication. Finally, this loss leads to the failure of chromosome ends to distinguish themselves from DNA double strand breaks, and to produce DNA damage reactions that prevent further proliferation of cells. Telomere wear limits the number of times cells enter the replication cycle, and constitutes the internal source of cell death. Therefore, proper telomere function can prevent tumor. 28 On the contrary, telomere dysfunction can trigger the fusion of chromosome ends, which leads to genomic instability and promotes tumor development. 29 Moreover, most cancer cells reactivate telomerase, leading to the avoidance of growth arrest and thus infinite proliferation. 28 Therefore, we speculate that NUP62CL can reactivate telomerase and promote the development of LUAD. However, the mechanism between melanin pathway and lung adenocarcinoma needs further study.
The independent prediction of the survival in cancer and the sentinel lymph node status could be enabled by the tumor infiltrating lymphocyte as a primary prognostic biomarker in the tumor progression. 30 , 31 The NUP62CL regulated tumor immunology was conclusively indicated by the positive correlation with memory CD4+ T cells and the significant aspect of our study entailing the NUP62CL expression regarding the immune infiltration levels in LUAD. Cornelia Voigt et al. found that memory CD4+ T cells can produce and release IL‐22 through the regulation of IL‐1 in cancer cells, thus promoting the growth of tumor. 32 Therefore, we believed that the increased expression of NUP62CL can regulate the production of IL‐22 by memory CD4+ T cells, thus promoting the development of LUAD. The tumor infiltrating B lymphocytes play an important role in the tumor development and exist in all stages of cancer as an indispensable part of the tumor microenvironment. The importance of the B cells infiltrating tumors as regulators of lung cancer progression was indicated by Wang et al. 33 on one hand, by promoting the T cell response and secreting the tumor‐specific antibodies, the tumor infiltrating B lymphocytes could exert anti‐tumor immunity; on the other hand, B cells can also develop into an immunosuppressive phenotype that secretes IL‐10, thereby promoting tumor development. Our research results showed that highly expressed tumor infiltrating B lymphocytes may be an important factor in promoting the development of LUAD. Previous studies have found that tumor‐infiltrating T cells and B cells are very close, and the functional interaction between them can enhance local immune activation, thereby contributing to a better prognosis for cancer patients. 34 However, in LUAD, whether tumor‐infiltrating T cells and B cells also have such a role before to promote the development of tumors, this needs more research to explore.
Nonetheless, there were several shortcomings in this study. First, the inevitable missing of certain useful information due to the less number of samples of the clinical data types. Second, the signal mechanism at the cytological level was not analyzed by the study. Nonetheless, an expanded clinical sampling in future research would be needed to validate our conclusions.
To conclude, the mechanism of the NUP62CL, the survival outcomes, and the relationship between clinico‐pathologic variables and the NUP62CL in cases of LUAD were explored and assessed by our study. Telomere maintenance via recombination could potentially constitute the key NUP62CL‐regulated pathways in LUAD. Moreover, the key pathway of NUP62CL regulation in lung adenocarcinoma needs further validation. Overall, with reference to LUAD, we analyzed 22 immune cell subsets and ascertained NUP62CL significantly correlated with tumor‐infiltrating memory CD4+ T cells and tumor infiltrating B lymphocytes, primarily as potential prognostic biomarkers for progression. Therefore, NUP62CL may be a promising prognostic biomarker of lung adenocarcinoma.
CONFLICTS OF INTEREST
The authors declare that they have no conflict of interest.
AUTHORS CONTRIBUTIONS
Shiqi Ren and Wei Wang conceived and designed the experiments, performed the experiments, prepared figures and/or tables, and approved the final draft. Chenlin Zhang and Yidan Sun analysed the data, prepared figures and/or tables, and approved the final draft. Mengjing Sun, Yingjing Wang, Xiaojing Zhang performed the experiments, prepared figures and/or tables, and approved the final draft. Ziheng Wang conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft. Bin lu conceived and designed the experiments, analysed the data, and approved the final draft. Lanlan Yao conceived and designed the experiments, analysed the data, authored or reviewed drafts of the paper, and approved the final draft.
Shiqi Ren and Wei Wang contributed equally to this work.
Contributor Information
Bing Lu, Email: bing.lu66@outlook.com.
Lanlan Yao, Email: ntlyyll@163.com.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Cancer J Clin. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 3. Eberle A, Jansen L, Castro F, et al. Lung cancer survival in germany: a population‐based analysis of 132,612 lung cancer patients. Lung Cancer. 2015;90:528‐533. [DOI] [PubMed] [Google Scholar]
- 4. Cronin KA, Lake AJ, Scott S, et al. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer. 2018;124:2785‐2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Osmani L, Askin F, Gabrielson E, Li QK. Current who guidelines and the critical role of immunohistochemical markers in the subclassification of non‐small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao C, Fillmore CM, Hammerman PS, Kim CF, Kwok‐Kin W. Non‐small‐cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535‐546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti‐pd‐1 antibody in cancer. N Engl J Med. 2012;366:2443‐2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti‐pd‐l1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455‐2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nanda R, Chow LQ, Dees EC, et al. Pembrolizumab in patients with advanced triple‐negative breast cancer: phase Ib KEYNOTE‐012 Study. J Clin Oncol. 2016;34:2460‐2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ansell SM, Lesokhin AM, Borrello I, et al. PD‐1 blockade with nivolumab in relapsed or refractory hodgkin's lymphoma. N Engl J Med. 2015;372:311‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loi S, Sirtaine N, Piette F, et al. Prognostic and predictive value of tumor‐infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node‐positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin‐based chemotherapy: Big 02–98. J Clin Oncol. 2013;31:860‐867. [DOI] [PubMed] [Google Scholar]
- 12. Loi S, Michiels S, Salgado R, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Eur Phys J D. 2014;41:485‐492. [DOI] [PubMed] [Google Scholar]
- 13. Salgado R, Denkert C, Campbell C, et al. Tumor‐infiltrating lymphocytes and associations with pathological complete response and event‐free survival in HER2‐positive early‐stage breast cancer treated with lapatinib and trastuzumab: a secondary analysis of the neoaltto trial. JAMA Oncol. 2015;1:448‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carsten D, Sibylle L, Aurelia N, et al. Tumor‐associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105. [DOI] [PubMed] [Google Scholar]
- 15. Ono M, Tsuda H, Yamamoto S, et al. Tumor‐infiltrating lymphocytes are correlated with response to neoadjuvant chemotherapy in triple‐negative breast cancer. Breast Cancer Res Treat. 2012;132:793‐805. [DOI] [PubMed] [Google Scholar]
- 16. Denkert C, von Minckwitz G, Brase JC, et al. Tumor‐infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2‐positive and triple‐negative primary breast cancers. J Clin Oncol. 2015;33:983‐991. [DOI] [PubMed] [Google Scholar]
- 17. Joseph J, Dasso M. The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Letters. 2008;582(2):190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho KI, Cai Y, Yi H, Yeh A, Aslanukov A, Ferreira PA. Association of the kinesin‐binding domain of RanBP2 to KIF5B and KIF5C determines mitochondria localization and function. Traffic. 2010;8:1722‐1735. [DOI] [PubMed] [Google Scholar]
- 19. Kasper LH, Brindle PK, Schnabel CA, Pritchard CEJ, Cleary ML, van Deursen JMA. CREB binding protein interacts with nucleoporin‐specific FG repeats that activate transcription and mediate NUP98‐HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764‐776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98–NSD1 links H3K36 methylation to Hox‐A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804‐812. [DOI] [PubMed] [Google Scholar]
- 21. Palancade B, Liu X, Garcia‐Rubio M, Aguilera A, Zhao X, Doye V. Nucleoporins prevent DNA damage accumulation by modulating Ulp1‐dependent sumoylation processes. Mol Biol Cell. 2007;18:2912‐2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98‐NSD1 links H3K36 methylation to Hox‐A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804‐812. [DOI] [PubMed] [Google Scholar]
- 23. Leng Y, Cao C, Ren J, et al. Nuclear import of the MUC1‐C oncoprotein is mediated by nucleoporin Nup62. J Biol Chem. 2007;282:19321‐19330. [DOI] [PubMed] [Google Scholar]
- 24. Monette A, Pante N, Mouland AJ. HIV‐1 remodels the nuclear pore complex. J Cell Biol. 2011;193:619‐631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang Z, Li C, Kang B, Ge G, Cheng L, Gepia ZZ. A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Larson SR, Xiaotun Z, Ruth D, et al. Characterization of osteoblastic and osteolytic proteins in prostate cancer bone metastases. Prostate. 2013;73:932‐940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Apte MS, Cooper JP. Life and cancer without telomerase: Alt and other strategies for making sure ends (don’t) meet. Crit Rev Biochem Mol Biol. 2017;52:57‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharpless NE, DePinho RA. Telomeres, stem cells, senescence, and cancer. J Clin Invest. 2004;113(2):160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohtani H. Focus on TILS: prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. 2007;7:4. [PMC free article] [PubMed] [Google Scholar]
- 31. Azimi F, Scolyer RA, Rumcheva P, et al. Tumor‐infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30:2678‐2683. [DOI] [PubMed] [Google Scholar]
- 32. Voigt C, May P, Gottschlich A, Markota A, Kobold S. Cancer cells induce interleukin‐22 production from memory CD4+ T cells via interleukin‐1 to promote tumor growth. Proc Natl Acad Sci USA. 2017;114:201705165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang SS, Liu W, Ly D, Xu H, Qu L, Zhang L. Tumor‐infiltrating B cells: their role and application in anti‐tumor immunity in lung cancer. Cell Mol Immunol. 2019;16:6‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garnelo M, Tan A, Her Z, et al. Interaction between tumour‐infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2017;66:342‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]


