Abstract
Biallelic germline mismatch repair (MMR) gene (MLH1, MSH2, MSH6, and PMS2) mutations are an extremely rare event that causes constitutional mismatch repair deficiency (CMMRD) syndrome. CMMRD is underdiagnosed and often debuts with pediatric malignant brain tumors. A high degree of clinical awareness of the CMMRD phenotype is needed to identify new cases. Immunohistochemical (IHC) assessment of MMR protein expression and analysis of microsatellite instability (MSI) are the first tools with which to initiate the study of this syndrome in solid malignancies. MMR IHC shows a hallmark pattern with absence of staining in both neoplastic and non-neoplastic cells for the biallelic mutated gene. However, MSI often fails in brain malignancies. The aim of this report is to draw attention to the peculiar IHC profile that characterizes CMMRD syndrome and to review the difficulties in reaching an accurate diagnosis by describing the case of two siblings with biallelic MSH6 germline mutations and brain tumors. Given the difficulties involved in early diagnosis of CMMRD we propose the use of the IHC of MMR proteins in all malignant brain tumors diagnosed in individuals younger than 25 years-old to facilitate the diagnosis of CMMRD and to select those neoplasms that will benefit from immunotherapy treatment.
Keywords: constitutional mismatch repair deficiency syndrome, MMR gene expression, immunohistochemistry, MSH6 gene
1. Introduction
Constitutional mismatch repair deficiency syndrome (CMMRD, MIM No. 276300) is caused by biallelic germline mutations in mismatch repair (MMR) system genes [1,2]. PMS2 is the most frequently mutated gene accounting for 60% of CMMRD cases. The remaining 40% are due to MSH6 (20–30%), MLH1 and MSH2 (10–20%) pathogenic variants [3]. A unique case of homozygous EPCAM patient has been described [4]. It is usually seen among families of individuals with Lynch syndrome (LS) who carry monoallelic mutations in one of these genes [1,2]. CMMRD is a very rare familial syndrome with about 200 cases reported since the first description in 1999 [3]. It has an autosomal recessive pattern of inheritance and affects children and adolescents who mainly develop brain tumors, gastrointestinal neoplasms, and hematological malignancies. Other LS-related neoplasms, such as endometrial and urinary tract carcinomas, can also develop. Additionally, some features of neurofibromatosis (NF) type 1, especially café-au-lait macules (CALMs) with irregular borders and different degrees of pigmentation, may be present in the absence of NF1/SPRED1 germline mutations [5,6]. The presence of multiple pilomatricomas [7], brain anomalies [8] and developmental vascular abnormalities has also been described [9]. Consanguinity of the parents is also frequently present [10,11]. Gastrointestinal lesions are age-dependent and include adenomas with or without high grade dysplasia as well as polyps resembling those seen in juvenile polyposis [12]. Other rare manifestations such as pediatric systemic lupus erythematosus have been described in 2.5% of CMMRD patients [13]. The pleiotropic CMMRD phenotype is not specific to the syndrome as these features overlap with other childhood tumor syndromes, in particular NF1. These features are used in a scoring system from the European C4CMMRD Consortium to raise clinical suspicion of CMMRD (Table 1) [3]:
Table 1.
Scoring system to determine germline testing in eligibility for CMMRD.
| Indication for CMMRD Testing in a Cancer Patient | ≥3 Points |
|---|---|
| Malignancies/premalignancies: one is mandatory; if more than one is present in the patient, add the points | |
| Carcinoma from the LS spectrum * at age <25 years | 3 points |
| Multiple bowel adenomas at age <25 years and absence of APC/MUTYH mutation(s) or a single high-grade dysplasia adenoma at age <25 years | 3 points |
| WHO grade III or IV glioma at age <25 years | 2 points |
| NHL of T-cell lineage or sPNET at age <18 years | 2 points |
| Any malignancy at age <18 years | 1 point |
| Additional features: optional; if more than one of the following is present, add the points | |
| Clinical sign of NF1 and/or ≥2 hyperpigmented and/or hypopigmented skin alterations Ø > 1 cm in the patient | 2 points |
| Diagnosis of LS in a first-degree or second-degree relative | 2 points |
| Carcinoma from LS spectrum * before the age of 60 in first-degree, second-degree, and third-degree relative | 1 point |
| A sibling with carcinoma from the LS spectrum *, high-grade glioma, sPNET or NHL | 2 points |
| A sibling with any type of childhood malignancy | 1 point |
| Multiple pilomatricomas in the patient | 2 points |
| One pilomatricoma in the patient | 1 point |
| Agenesis of the corpus callosum or non-therapy-induced cavernoma in the patient | 1 point |
| Consanguineous parents | 1 point |
| Deficiency/reduced levels of IgG2/4 and/or IgA | 1 point |
* Colorectal, endometrial, small bowel, ureter, renal pelvis, biliary tract, stomach, bladder carcinoma. Abbreviations: CMMRD, constitutional mismatch repair deficiency; LS, Lynch syndrome; NHL, non-Hodgkin’s lymphomas; sPNET, supratentorial primitive neuroectodermal tumors.
The identification of pathogenic variants in both alleles of an MMR gene is required to confirm the diagnosis. However, the presence of PMS2 pseudogenes [14] and variants of unknown significance (VUS) makes this difficult. To improve the previous consensus established [15] a new diagnostic criterion for CMMRD elaborated by an international panel of experts has recently been described to unmask this rare syndrome (Table 2) [4]. They established four definitive criteria to ensure diagnosis of the syndrome with strong evidence and three probable criteria with moderate evidence based on the combination of (a) the MMR germline results, (b) ancillary testing including the immunohistochemistry of the four MMR genes on non-neoplastic tissue, germline microsatellite instability [16], ex vivo MSI plus methylation tolerance [17], in vitro repair assay and NGS detection [18], low-level MSI in tissue [19,20] and (c) clinical manifestations.
Table 2.
CMMRD diagnostic criteria.
| Criterion | Germline result * PMS2, MSH6, MSH2, MLH1 | Positive Ancillary Testing † | Clinical Phenotype | |
|---|---|---|---|---|
| Definitive diagnosis (strong evidence of CMMRD) | 1 | Biallelic pathogenic variants (P/P) *, confirmed in trans. ‡ | Not required, unless unaffected >25 years then one required. † | Not required if under age 25 (if no malignancy over age 25, ancillary testing required). |
| 2 | Biallelic P/LP or LP/LP * variants, confirmed in trans ‡ | One required† for hallmark CMMRD. Two required† for C4CMMRD criteria. ** | Hallmark CMMRD cancer diagnosis or C4CMMRD criteria of 3 points ** (then two ancillary tests required). | |
| 3 | Heterozygous P or LP variant (±VUS * or likely benign variants). | One required †. | Hallmark CMMRD cancer diagnosis. | |
| 4 | No P or LP MMR variants (including VUS/ VUS). †† Or no testing available (i.e., deceased proband). | Two required †. | Hallmark CMMRD cancer diagnosis. | |
| Likely diagnosis (moderate evidence of CMMRD) | 5 | Biallelic P/LP * or LP/LP variants, confirmed in trans.§ | Not required. | C4CMMRD criteria of 3 points. ** |
| 6 | No P or LP MMR variants (including VUS/ VUS). †† Or no testing available (i.e., deceased proband). | One required. † | Hallmark CMMRD cancer diagnosis. | |
| 7 | Heterozygous P or LP variant or no testing available (i.e., deceased proband). | Two required. † | C4CMMRD criteria of 3 points. ** Individuals aged <18 with NF1 features (i.e., no malignancy or polyposis history). Malignancy under age 30 |
* Biallelic–impacts same gene on both parental alleles (i.e., PMS2/PMS2); P, pathogenic (ACMG C5); LP, likely pathogenic (ACMG C4); VUS (ACMG C3). Multigene panel testing is recommended to investigate overlapping conditions. Consider phenotype of individual to rule out overlapping syndromes. All families should be assessed in a specialized center for diagnosis. † Ancillary testing does not include tumor mutation burden and signature at this time. Functional testing should be published with proven high sensitivity and specificity performed in an accredited (e.g., CAP-inspected) laboratory authorized to give a clinically usable report. If discrepancy occurs among tests, multiple ancillary tests should be used to reach a more conclusive decision. ‡ In trans variants can be proven by testing parents, offspring or other relatives. If unavailable to confirm variants in trans, individual should fulfil criterion 3. § If unavailable to confirm variants in trans, individual should fulfil criterion 6. Hallmark CMMRD cancer: glioma or CNS embryonal tumors 25 years, hematological cancer (excluding Hodgkin’s lymphoma) <18 years, GI adenocarcinoma <25 years, or >10 adenomatous GI polyps <18 years (after ruling out polyposis conditions). ** C4CMMRD criteria outlined in Table 1. †† Consanguinity further supports a diagnosis of CMMRD due to a homozygous MMR gene mutation that is unidentifiable. ACMG, American College of Medical Genetics; CAP, College of American Pathologists; C4CMMRD, European Consortium Care for CMMRD; CMMRD, constitutional mismatch repair deficiency syndrome; CNS, Central Nervous System; GI, gastrointestinal; MMR, mismatch repair; NF1, neurofibromatosis type 1.
In addition, guidelines for surveillance of individuals with CMMRD have been recently proposed by the European C4CMMRD Consortium [21] and the International BMMRD Consortium [22]. Even so, identification, surveillance and care of CMMRD patients continue to be a challenge.
According to recent epidemiological studies, carriers of the pathogenic variants of MMR are relatively common (up to 1 in 279 of the general population), with PMS2 being the most frequently mutated gene [23]. In children with cancer, germline alterations have been found in up to 6% of cases, mostly affecting DNA repair genes, and the hypermutated tumors corresponded to high-grade gliomas with biallelic mutations in MSH6 and PMS2. [24]. As a result, these tumors are very sensitive to immunotherapy and resistant to standard treatment with temozolomide [25].
CMMRD remains an underdiagnosed syndrome that usually debuts with malignant brain tumors in pediatric age with some clinical features that should raise suspicion of this syndrome [26] and especially in the context of high consanguinity as shown by the study of Amaya et al [11]. In this study, the prevalence of CMMRD was evaluated among patients with malignant brain tumors younger than 18 years with a high rate of consanguinity. Among the 36 high-grade gliomas, 17 (39%) showed a lack of staining for an MMR protein, and of these 82% of cases showed the characteristic pattern of CMMRD. Patients with lack of staining in both neoplastic and normal cells also had CALM, consanguinity, and a family history of cancer. In addition, the median age at diagnosis was 12.2 years, similar to that of sporadic tumors.
CMMRD tumor identification requires the use of the tests applied for LS identification, namely immunohistochemical (IHC) assessment of the four MMR proteins expression and/or analysis of microsatellite instability (MSI) [15]. In CMMRD, however, these studies show some peculiar findings, knowledge of which is mandatory to avoid underdiagnosis of this rare syndrome. Specifically, loss of MMR protein immunoreaction is not only seen in tumor cells but also in normal cells. Moreover, in CMMRD brain malignancies the standard MSI analysis fails to show instability [2,7,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66] although the use of an extended Bethesda panel with 10 additional mononucleotide repeat markers has detected subtle frameshifts [61]. Nowadays, the use of MMR IHC is becoming increasingly widespread mainly due to universal LS screening of colorectal and endometrial tumors [67,68]. Its use will increase with the adoption of the tissue diagnostic approach, a revolutionary paradigm shift in cancer treatment and drug development [69]. Therefore, a good knowledge of the immunohistochemical pattern characteristic of CMMRD is crucial to avoid pitfalls in its interpretation, given the great diversity of lesions and neoplasms that may be the first manifestation of this syndrome.
The aim of this report is to draw attention to the peculiar IHC profile that characterizes CMMRD syndrome, and to review the methods and difficulties in reaching a correct diagnosis. We demonstrate this by the case of two children with biallelic MSH6 germline mutations and brain tumors, and a review of the literature.
2. Cases
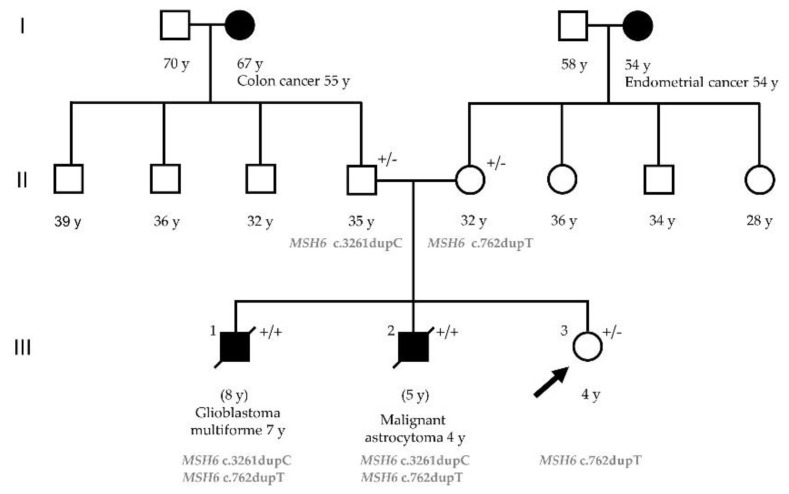
A healthy four-years-old girl was referred to our Neurofibromatosis Clinic due to a suspected family history of NF involving her two brothers. This girl showed no NF signs at that time, but her parents were worried about her future. The older brother was born with six CALMs and was considered as a potential NF type 1 patient and, consequently, was included in a NF surveillance program. At the age of seven he had complained of headaches and vomiting and been diagnosed as having a brain tumor. The latter was totally resected and found to be a glioblastoma multiforme (GBM). The patient died at the age of eight due to a spinal cord relapse of the tumor. The younger brother was born with a single CALM but was considered as an NF type 1 patient due to his older brother’s medical history. At the age of four he developed headaches and vomiting and an NF-related pilocytic astrocytoma was suspected in an MRI study. A conservative management was initially recommended. However, headaches were persistent and an MRI performed two months later showed a growing tumor. A surgical resection was carried out but, unfortunately, a complicated post-operative course led to death two weeks after surgery. The pathological diagnosis was malignant astrocytoma. There was no additional family history of NF. The parents were healthy and non-consanguineous. The paternal and maternal grand-mothers had been diagnosed with cancer, colorectal cancer at the age of 55 and endometrial cancer at 54, respectively. The family pedigree is shown in Figure 1.
Figure 1.
Pedigree of the family with biallelic MSH6 mutations. The arrow indicates the proband. Filled symbols indicate affected subject; open symbols unaffected subjects. The age at tumor onset is shown with the tumor type. +/− = MSH6 monoallelic mutation carrier (Lynch syndrome); +/+ = MSH6 biallelic mutation carrier (CMMRD).
We challenged the initial diagnosis of NF in this family due to the absence of previous family history of the disease and the paucity of NF manifestations, although a gonadal mosaicism in one of the progenitors could not be ruled out.
2.1. Immunohistochemical Analysis of MMR Proteins
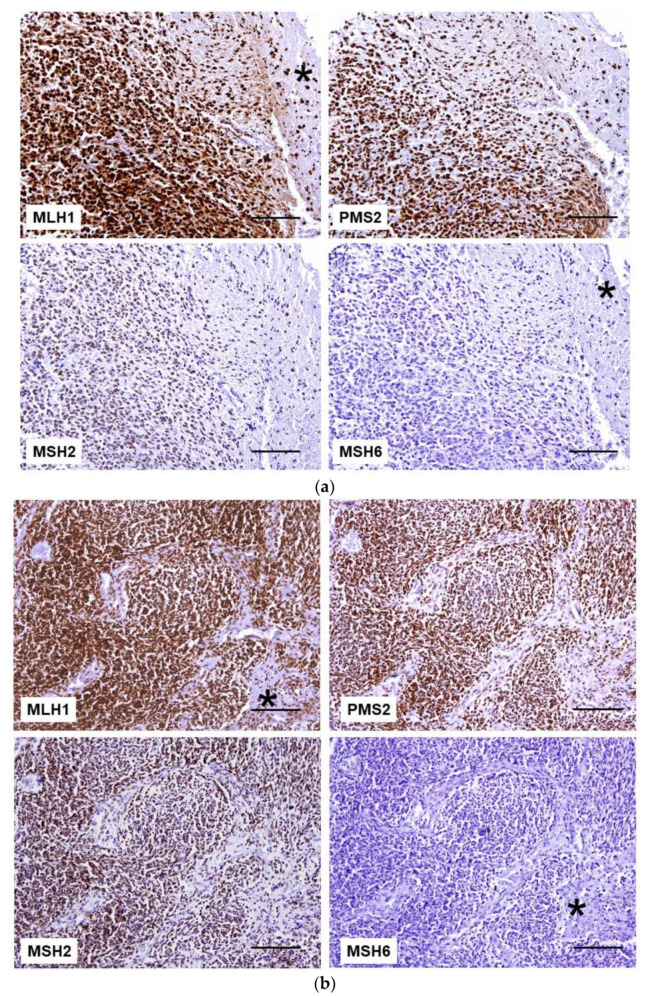
Both brain tumors showed positive nuclear immunostaining in neoplastic cells as well as in adjacent normal brain tissue with antibodies against MLH1, PMS2, and MSH2 proteins. In contrast, absence of MSH6 expression was observed in both tumor cells and the surrounding normal tissue (Figure 2). An external positive control tissue from another individual was also studied to assess the quality of the immunostaining in each case.
Figure 2.
MMR protein IHC in brain tumors. (a) Glioblastoma cells and normal brain tissue (*) retained the expression of MLH1, PMS2, and MSH2 proteins. MSH6 staining is lost in both tumor cells and normal tissue (*). (b) In malignant astrocytoma, neoplastic, stromal, and endothelial cells are immunoreactive to anti-MLH1 (*), anti-PMS2, and anti-MSH2 antibodies. In contrast, the lack of MSH6 staining is observed in all cells (*). Scale bar: 100 µm.
2.2. MSI Analysis
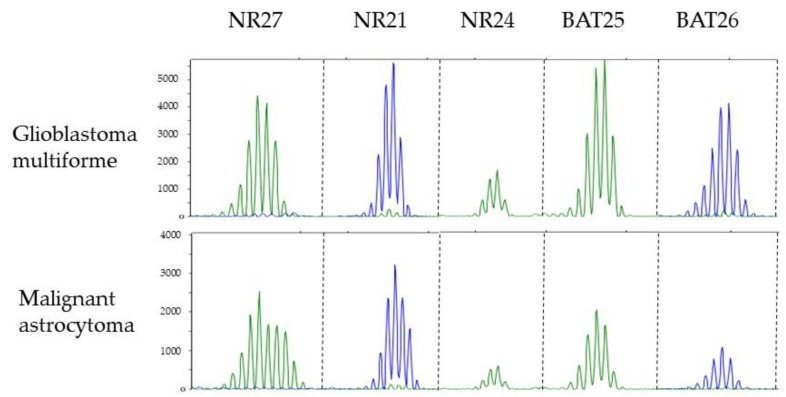
Analysis of MSI showed that none of the brain tumors were MSI-H. In the GBM case all markers showed an apparently stable profile (Figure 3), whereas in the malignant astrocytoma case only the NR27 marker showed a broader profile. This may represent a germline variant as this specific marker is known to be quasi-monomorphic [70].
Figure 3.
Electropherograms showing microsatellite profiles of five mononucleotide repeats markers (NR21, NR24, NR27, BAT25, and BAT26) in each tumor. A broader spectrum is observed in marker NR27 in the malignant astrocytoma.
2.3. Germline Mutation Analysis
Sequence analysis of the MSH6 gene in father’s blood sample displayed a heterozygous single nucleotide duplication in exon 5 (c.3261dupC) leading to a frameshift and premature termination (p.Phe1088Leufs*5). The same analysis in the mother’s sample showed a heterozygous single nucleotide duplication in exon 4 (c.762dupT) leading to a nonsense mutation, p (Glu255 *). Both variants were classified as pathogenic (class 5). Analysis of the two brothers’ tumor brain tissues confirmed that both were compound heterozygous MSH6 c.3261dupC and c.762dupT mutation carriers. Our patient (III.3 Figure 1), the four-year-old girl, was heterozygous for the c.762dupT MSH6 gene mutation. This result ruled out the diagnosis of CMMRD syndrome and she was diagnosed with LS.
2.4. Somatic Mutational Analysis
Acquired polymerase proofreading defects are characteristic of CMMRD brain tumors, leading to high mutational burden and associated with response to immune checkpoint inhibitors [25,71,72]. Therefore, the analysis of POLE and POLD1 genes in the brain tumor DNA sample from patient III-2 revealed the presence of four variants in POLD1 (Table 3). Of note, one of these variants, p.Glu318Lys affected the catalytic residue of POLD1 and is classified as pathogenic. Unfortunately, the mutational analysis was performed after the patient’s death and could not be used to guide an appropriated treatment.
Table 3.
POLD1 variants found in the analysis of the brain tumor of individual III.2. In silico predictions based on Alamut software.
| Variant Calling | Coverage | Rs ID | MAF | |||
|---|---|---|---|---|---|---|
| Gene (transcript) | cDNA change | Predicted protein change | Allelic frequency | Read depth | (ExAC/ESP) | |
| POLD1 (NM_001256849.1) | c.952G>A | p.Glu318Lys | 0.264 | 772 | rs775232133 | 0.000010/NR |
| c.1096G>A | p.Ala366Thr | 0.235 | 267 | – | NR/NR | |
| c.2788G>A | p.Ala930Thr | 0.482 | 410 | rs144111108 | 0.00010/0.00008 | |
| c.3157C>T | p.Arg1053Cys | 0.62 | 91 | rs779208942 | 0.00004/NR | |
| In Silico Predictions | Protein Domain | |||||
| Protein Function | ||||||
| Gene (transcript) | Splicing | SIFT (score) | Mutation Taster (p-value) | Polyphen2/HumDiv (score) | Polyphen2/HumVar (score) | |
| POLD1 (NM_001256849.1) | No changes | Deleterious (0) |
Disease causing (1) | Probably damaging (1.000) | Probably damaging (0.998) | Exonuclease(catalytic residue) |
| Unconclusive | Deleterious (0.05) |
Disease causing (0.956) | Benign (0.087) | Benign (0.271) | Exonuclease | |
| No changes | Tolerated (0.13) |
Disease causing (1) | Probably damaging (1.000) | Probably damaging (0.998) | – | |
| No changes | Deleterious (0) |
Disease causing (1) | Probably damaging (0.999) | Probably damaging (0.973) | – | |
Abbreviations: NR, not reported.
3. Brain Tumors in CMMRD Syndrome
In order to compare our findings with those previously reported, the results of MMR protein expression using IHC and MSI analysis from a total of 118 reported brain tumors in patients with CMMRD syndrome have been reviewed and are specified in Table 4 [2,7,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66].
Table 4.
Clinicopathological and molecular features of brain tumors in CMMRD syndrome.
| Chronological Case Order | Reference (Number) | Case ID | Age at Diagnosis (Years) | Gender | Histology | Altered Gene | IHC | MSI (Marker) |
|---|---|---|---|---|---|---|---|---|
| 1 | Wang [2] | 1 | 18 | M | Medulloblastoma | MLH1 | NA | * MSI-H |
| 2 | 2 | 7 | F | Medulloblastoma | MLH1 | NA | NA | |
| 3 | De Rosa [27] | 1 | 14 | F | Oligodendroglioma GIII | PMS2 | NA | NA |
| 4 | 2 | 13 | F | Neuroblastoma | PMS2 | NA | NA | |
| 5 | Vikky [28] | 1 | 4 | F | Glioma | MLH1 | NA | NA |
| 6 | Bougeard [29] | 1 | 3 | M | GBM | MSH2 | NA | MSS |
| 7 | Menko [30] | 1 | 10 | M | Oligodendroglioma | MSH6 | MSH6+ | MSS |
| 8 | De Vos 2004 [14] | 1 | 8 | M | PNET | PMS2 | NA | NA |
| 9 | 2 | 14 | F | PNET | PMS2 | NA | NA | |
| 10 | De Vos 2006 [31] | 1 | 8 | NS | PNET | PMS2 | NA | NA |
| 11 | 2 | 4 | NS | PNET | PMS2 | NA | NA | |
| 12 | 3 | 15 | NS | Glioma | PMS2 | NA | NA | |
| 13 | 4 | 6 | NS | Astrocytoma | PMS2 | NA | NA | |
| 14 | 5 | 7 | NS | GBM | PMS2 | NA | NA | |
| 15 | 6 | 2 | NS | Giant cell glioblastoma | PMS2 | NA | NA | |
| 16 | Agostini [32] | 1 | 18 | M | Giant cell glioblastoma | PMS2 | PMS2- | MSS |
| 17 | Ostergaard [33] | 1 | 9 | M | Astrocytoma GIII | MSH6 | MSH6- | NA |
| 18 | 2 | 2 | F | GBM of spinal cord | MSH6 | MSH6- | NA | |
| 19 | Hegde [34] | 1 | 8 | F | GBM | MSH6 | NA | MSI-H (D2S123, D17S250, BAT25, D18S35, TP53-DI, D1S283, TP53-Penta, FGA, NR-21, NR-22, NR-24) |
| 20 | Durno [35] | 1 | 14 | F | Anaplastic astrocytoma | MLH1 | NA | NA |
| 21 | Krüger [36] | 1 | 9 | M | GBM | PMS2 | NA | NA |
| 22 | 2 | 6 | F | GBM | PMS2 | NA | NA | |
| 23 | 3 | 9 | M | GBM | PMS2 | NA | NA | |
| 24 | Gururangan [37] | 1 | 19 | M | Astrocytoma GIII | PMS2 | PMS2- | MSS |
| 25 | 2 | 11 | M | Anaplastic oligodendroglioma | PMS2 | NA | NA | |
| 26 | Auclair [38] | 1 | 7 | F | GBM | MSH6 | * MSH6– | * MSI-L |
| 27 | 2 | 19 | F | Oligodendroglioma | PMS2 | * PMS2– | * MSI-H | |
| 28 | Poley [39] | 1 | 4 | M | GBM | MLH1 | MLH1– | MSS |
| 29 | 2 | 6 | M | Oligodendroglioma | MSH6 | MSH6– | MSI-L | |
| 30 | 3 | 8 | M | Medulloblastoma | MSH6 | MSH6– | MSI-L | |
| 31 | Scott [40] | 1 | 7 | F | Medulloblastoma | MSH6 | MSH6– | NA |
| 32 | Kratz [41] | 1 | 9 | F | PNET | PMS2 | NA | NA |
| 33 | Etzler [42] | 1 | 6 | NS | Medulloblastoma | MSH6 | NA | NA |
| 34 | 2 | 9 | NS | GBM | MSH6 | MSH6– | MSS | |
| 35 | 3 | 10 | NS | GBM | PMS2 | PMS2– | MSS | |
| 36 | Senter [43] | 1 | 23 | NS | Brain tumor | PMS2 | NA | NA |
| 37 | 2 | 35 | NS | Glioma | PMS2 | NA | NA | |
| 38 | 3 | 7 | NS | Medulloblastoma | PMS2 | NA | NA | |
| 39 | Tan [44] | 1 | 8 | M | GBM | PMS2 | NA | NA |
| 40 | Toledano [45] | 1 | NS | M | Anaplastic astrocytoma GIII | MSH2 | NA | NA |
| 41 | 2 | 13 | M | Anaplastic astrocytoma GII | MSH2 | NA | NA | |
| 42 | Sjursen [46] | 1 | 10 | F | Giant cell glioblastoma | PMS2 | PMS2+ | MSI-H (BAT25, BAT26,BAT40,D2S123,D5S107, D5S346, D5S406,D13S153, D17S250) |
| 43 | Giunti [47] | 1 | 10 | M | GBM | PMS2 | NA | NA |
| 44 | 2 | 4 | F | Brain tumor | PMS2 | NA | NA | |
| 45 | Roy [48] | 1 | 8 | F | Medulloblastoma | PMS2 | NA | NA |
| 46 | Herkert [49] | 1 | 2 | M | Angiosarcoma cerebral | PMS2 | PMS2- | MSI-H (BAT25, BAT26, D2S123, D5S346, D17S250) |
| 47 | 2 | 34 | M | GBM | PMS2 | NA | NA | |
| 48 | 3 | 9 | F | Anaplastic ganglioma | PMS2 | PMS2- | MSI-H (BAT25, BAT26, D2S123, D5S346, D17S250) | |
| 49 | Ilencikova [50] | 1 | 11 | F | Fibrillar astrocitoma | MSH6 | NA | NA |
| 50 | 1 | 12 | M | Anaplastic astrocytoma GIII | MSH6 | MSH6- | MSI-L (BAT26) | |
| 51 | 2 | 10 | M | GBM | MSH6 | NA | NA | |
| 52 | Leenen [51] | 1 | 4 | F | PNET | PMS2 | PMS2- | MSS |
| 53 | 2 | 7 | M | Glioblastoma GIII | PMS2 | PMS2- | MSI-H (NR21,BAT26) | |
| 54 | Johannesma [52] | 1 | 11 | F | Papillary glioneural tumor | PMS2 | PMS2– | MSS(BAT26, BAT25, NR21, NR24, MONO 27, Penta D, Penta C) |
| 55 | Baas [8] | 1 | 3 | M | GBM | MLH1 | MLH1– | NA |
| 56 | 2 | 2 | M | Astrocytoma of spinal cord | PMS2 | §MLH1–/PMS2- | NA | |
| 57 | Lindsay [53] | 1 | 12 | M | Medulloblastoma | PMS2 | NA | NA |
| 58 | Walter [54] | 1 | 13 | F | GBM | PMS2 | NA | NA |
| 59 | Yeung [55] | 1 | 3 | M | Optic pathway glioma | PMS2 | NA | NA |
| 60 | Chmara [7] | 1 | 11 | M | Anaplastic oligodendroglioma | PMS2 | NA | NA |
| 61 | 2 | 4 | M | GBM | NA | NA | NA | |
| 62 | 3 | 9 | M | GBM | PMS2 | NA | NA | |
| 63 | 4 | 4 | F | GBM | PMS2 | NA | NA | |
| 64 | Bakry [56] | 1 | 5 | NS | Pleomorphic xantho-astrocytoma | PMS2 | PMS2– | NS |
| 65 | 2 | 8 | NS | GBM | MSH6 | MSH6– | NS | |
| 66 | 3 | 11 | NS | GBM | PMS2 | PMS2– | NS | |
| 67 | 4 | 13 | NS | GBM | PMS2 | PMS2– | NS | |
| 68 | 5 | 4 | NS | GBM | PMS2 | NA | NS | |
| 60 | 6 | 21 | NS | Oligodendroglioma | PMS2 | NA | NS | |
| 70 | 7 | 24 | NS | GBM | PMS2 | NA | NS | |
| 71 | 8 | 10 | NS | Anaplastic oligodendroglioma | MSH6 | MSH6– | NS | |
| 72 | 9 | 11 | NS | Anaplastic astrocytoma | MSH6 | MSH6– | NS | |
| 73 | 10 | 17 | NS | GBM | PMS2 | PMS2– | NS | |
| 74 | 11 | 12 | NS | GBM | MSH6 | NA | NS | |
| 75 | 12 | 8 | NS | GBM | NA | PMS2– | NS | |
| 76 | 13 | 9 | NS | GBM | NA | PMS2– | NS | |
| 77 | 14 | 4 | NS | GBM | NA | PMS2– | NS | |
| 78 | 15 | 16 | NS | Anaplastic astrocytoma | NA | PMS2– | NS | |
| 79 | 16 | 11 | NS | Pleomorphic xantho-astrocytoma | NA | PMS2– | NS | |
| 80 | Bougeard [57] | 1 | 11 | F | GBM | PMS2 | * PMS2– | *MSI-H |
| 81 | 2 | 9 | M | GBM | PMS2 | * PMS2– | *MSI-H | |
| 82 | Daou [58] | 1 | 22 | F | Anaplastic ganglioglioma | PMS2 | PMS2– | NA |
| 83 | Lavoine [59] | 1 | 21 | NS | Ganglioglioma | PMS2 | PMS2– | MSI-H |
| 84 | 2 | 40 | NS | Glioblastoma | PMS2 | NA | NA | |
| 85 | 3 | 11 | NS | Glioblastoma | PMS2 | # MLH1-/PMS2- | MSI-H | |
| 86 | 4 | 6 | NS | GBM | PMS2 | # MLH1-/PMS2- | MSS | |
| 87 | 5 | 13 | NS | GBM | PMS2 | PMS2– | NA | |
| 88 | 6 | 22 | NS | GBM | PMS2 | PMS2– | MSI-H | |
| 89 | 7 | 32 | NS | GBM | PMS2 | PMS2– | MSI-H | |
| 90 | 8 | 34 | NS | GBM | PMS2 | PMS2– | MSI-H | |
| 91 | 9 | 6 | NS | Spinal GBM | PMS2 | PMS2– | NA | |
| 92 | 10 | 6 | NS | GBM | PMS2 | NA | NA | |
| 93 | 11 | 5 | NS | Medulloblastoma | MLH1 | # MLH1–/PMS2– | MSS | |
| 94 | 12 | 5 | NS | Oligodendroglioma | MLH1 | # MLH1–/PMS2– | MSS | |
| 95 | 13 | 6 | NS | GBM | MSH6 | MSH6– | MSS | |
| 96 | 14 | 9 | NS | Astrocytoma | MSH6 | MSH6+ | MSS | |
| 97 | 15 | 14 | NS | GBM | MSH6 | # MSH2–/MSH6– | MSS | |
| 98 | Nguyen [60] | 1 | 6 | F | Medulloblastoma | MLH1 | § MLH1–/PMS2- | MSI-H (TP53, D17S250, D2S123, D5S346) |
| 99 | 2 | 5 | M | Glioma IV | MLH1 | § MLH1-/PMS2- | MSI-H (TP53, D17S250, D2S123,D5S2013) | |
| 100 | 3 | 5 | M | Brain tumor | NA | NA | NA | |
| 101 | Maletzki [61] | 1 | 11 | F | Anaplastic astrocytoma | MSH6 | NA | MSI-H (BAT25, BAT26, Cat25, Bat40, NR24, MRPL2, TP53, DAMS) |
| 102 | 2 | 10 | M | GBM | MSH6 | NA | MSI-H (BAT25, BAT26, Cat25, Bat40, NR24, TP53, DAMS) | |
| 103 | 3 | 4 | F | GBM | PMS2 | NA | MSI-H (BAT25, BAT26, Cat25, Bat40, NR21, NR24, NR27, MRPL2, TP53, DAMS) | |
| 104 | Taeubner [62] | 1 | 13 | F | Medulloblastoma | MSH6 | MSH6 + | NA |
| 105 | AlHarbi [63] | 1 | 5 | F | GBM | MSH6 | NA | NA |
| 106 | Baig [64] | 1 | 11 | M | GBM | PMS2 | NA | NA |
| 107 | 2 | 9 | F | PNET, astrocytoma | NA | NA | NA | |
| 108 | 3 | 5 | M | PNET | NA | NA | NA | |
| 109 | 4 | 1 | M | PNET | NA | NA | NA | |
| 110 | 5 | 7 | M | GBM | PMS2 | NA | NA | |
| 111 | 6 | 9 | F | PNET, astrocytoma | PMS2 | NA | NA | |
| 112 | Bush [65] | 1 | 27 | F | GBM | MSH6 | NA | NA |
| 113 | Farah [66] | 1 | 10 | M | High-grade Glioma | PMS2 | PMS2– | NA |
| 114 | 2 | 22 | F | GBM | PMS2 | PMS2– | NA | |
| 115 | 3 | 10 | M | Medulloblastoma | PMS2 | PMS2- | NA | |
| 116 | 4 | 12 | F | Brain tumor | PMS2 | PMS2- | NA | |
| 117 | Current study | 1 | 7 | M | GBM | MSH6 | MSH6- | MSS |
| 118 | 2 | 4 | M | Malignant astrocytoma | MSH6 | MSH6- | MSI-L (NR27) |
Abbreviations: IHC, immunohistochemistry; MSI, microsatellite instability; GBM, glioblastoma multiforme; NA, not analyzed; NS, not specified; MSI-H, microsatellite instability, high grade; MSI-L, microsatellite instability, low grade; MSS, microsatellite instability, stable; § MLH1, staining loss in tumor cells but not in non-neoplastic cells. # IHC, immunohistochemical pattern not specified; *, results from Lavoine et al., [59].
Of the 118 patients in the series, the gender was specified in 77, 43 (56%) males and 34 (44%) females.
Regarding the distribution of tumors by age, 72 (61%) appeared in the first decade of life, 26 (22%) between 11 and 15 years, 6 (5%) between 16 and 20, 7 (6%) between 21 and 24 and 6 (5%) over 25 years.
The most frequently altered gene was PMS2 with 68 (63%) cases, followed by MSH6 (27, 25%), MLH1 (10, 9%) and MSH2 (3, 3%). In 10 cases there is no data available on the altered gene.
MMR IHC was not performed in 59 tumors. Among the 59 cases analyzed, loss of PMS2 expression was the most frequent finding with 31 (52.5%), MSH6 was negative in 14 (23.7%), MLH1 and PMS2 negative in seven (11.8%), MLH1 negative in two (3%), MSH2 and MSH6 negative in one (1.5%). PMS2 was positive in one (1.5%) (case 42) and MSH6 positive in three (5%) (cases 7, 96, and 104). The sensitivity of MMR IHC for identifying patients affected by CMMRD was 93.2%, as it showed loss of expression in at least one protein in 55 of the 59 brain neoplasms analyzed. In 54 cases it was possible to compare the result of germline analysis with MMR IHC and a good agreement was observed in 47 (87%). The remainder seven (13%) with non-concordant results were as follow: three mutated MHS6 with positive MSH6 (cases 7, 96, and 104), two mutated PMS2 with negative MLH1 and PMS2 (cases 85 and 86), one mutated MSH6 with negative MSH2 and MSH6 (case 97) and one mutated PMS2 with positive PMS2 (case 42).
MSI analysis was performed in 56 (47%) tumors, but in 16 the results were not specified [48]. The results of the 40 tumors with available data showed that MSI-H was the most frequent finding with 19 (47.5%) followed by MSS with 16 (40%) and MSI-L with five (12.5%).
4. Discussion
The identification of individuals with CMMRD syndrome remains a challenge and a high degree of clinical awareness of its features and diagnostic pitfalls is required.
The cases presented here illustrate how a multidisciplinary approach, with the participation of pathologists, dermatologists, geneticists, pediatricians, gastroenterologists, and neurooncologists, is needed to ensure an early diagnosis of CMMRD in brain tumor patients as well as to provide the best treatment. MMR deficient high-grade gliomas are resistant to standard treatments with combination of temozolomide and radiation but sensitive to immunotherapy. In addition, the overall survival of patients with CMMRD and malignant brain tumors is poor and worse than seen in the same population with no predisposition to hereditary cancer [26]. For example, today, according to C4CMMRD criteria, [3] the clinical suspicion of CMMRD should have been raised in the older sibling, who with more than 5 CALMs met the suspicion of NF1 at the time of diagnosis of GBM. The clinical presentation of the second sibling would be even more obvious by having a suspected NF1 skin lesion, the background of the sibling with a glioma, and in both parental lines second-degree relatives with LS-associated carcinomas before the age of 60 years. At that point, reaching a score superior to three points in the C4CMMRD criteria, further analysis should be done to confirm a diagnosis of CMMRD. The finding of a somatic pathogenic variant in POLD1 in the high-grade glioma of this patient suggested the hypermutant nature of the tumor that characterizes brain malignancies developed in CMMRD.
IHC study of MMR is one of the ancillary tests included, since in CMMRD at least one protein must be totally negative in normal tissue. For that reason, any unusual immunostaining pattern of MMR protein in neoplastic and normal cell should be a clue to guide the germ-line analysis. Herein, in the two new cases of CMMRD by biallelic MSH6 mutations with brain neoplasm, the IHC study of MMR proteins was crucial to arrive at the correct molecular diagnosis. The characteristic pattern of expression seen in CMMRD, namely lack of nuclear expression in both the tumor and the normal cells with MSH6 protein, was crucial. To avoid the pitfall of interpreting the lack of staining in all cells as a failure of the technique, it is important to include an external positive control from another individual in each determination. MMR IHC is a useful tool for identifying CMMRD patients with brain neoplasms with a sensitivity of 93% based on the cases reviewed in Table 4, showing loss of expression in at least one protein in 55 of the 59 tumors analyzed. However, unusual results of MMR protein expression have also been found. Tumors from patients with a homozygous MLH1 mutation displayed negative MLH1 staining only in neoplastic cells, with loss of PMS2 in both neoplastic and normal cells [60]. Baas et al described a spinal cord astrocytoma from a biallelic PMS2 mutation carrier with loss of MLH1 in tumor cells, probably as a result of somatic alteration, with lack of PMS2 staining in tumor and normal tissue [8]. In addition, MSH6 and PMS2 could retain expression in tumors in the case of biallelic germline missense mutations because of residual protein expression [29,45,58,61]. This is observed in four tumors in the series reviewed (cases 7, 42, 96, and 104) and could explain the lower correlation between the loss of protein expression with the mutated gene in brain tumors compared to what occurs in other types of neoplasms such as colon and endometrial carcinoma. Another widely available assay to guide germline diagnosis of CMMRD is tumor MSI analysis, but CMMRD brain tumors were often MSS or MSI-L. The two brain neoplasms herein reported were MSS and the other MSI-L due to minor alterations with the mononucleotide NR27 marker. NR27 is one of the five markers of the quasi-monomorphic panel (NR21, NR27, NR24, BAT25 and BAT26) described by Buhard et al [70] and different from those validated in colorectal cancer due to its better specificity, which allows the avoidance of normal cells as a control [73]. However, this panel was not as efficient in detecting MSI in CMMRD tumors [52]. In contrast, dinucleotide repeat markers have been shown to be most useful in CMMRD-associated brain tumors, although they are insensitive to MSH6 deficiency. In fact, this was well illustrated by Nguyen et al., analyzing different tumors of CMMRD patients with mononucleotide and dinucleotide repeat markers [60]. One sarcoma and two brain tumors were MSI-H only in dinucleotide repeat markers while one colorectal cancer was unstable in all types of marker. Of the 19 MSI-H brain tumors, the markers used in the analysis were detailed in 10 cases and in nine of these the markers were dinucleotide repeats [26,38,41,52,53]. Bakry et al., described 16 brain tumors and only one was MSI-H but the markers used were not specified [56]. Another way to enhance the efficiency of MSI analysis has been to extend the number of non-coding mononucleotide repeat markers. Using this approach with 10 markers, Maletzki et al demonstrated MSI-H in 3/3 brain tumors analyzed (Table 4) [61]. Even so, MSI in the identification of CMMRD patients with brain tumors showed a low sensitivity of 47.5%, which was not enough to unmask half of the patients affected by the syndrome. Another possible option that would allow the identification of CMMRD in brain tumors would be sequencing to determine MSI, mutation burden and signatures, and other alterations characteristic of these tumors, such as POLE or POLD1 variants. In fact, a recent report has revealed novel signatures that are uniquely attributed to mismatch repair deficiency by using exome- and genome-wide microsatellite instability analysis [74]. In this study different microsatellite (MS)-mutated loci, lack of recurrently mutated MS-loci and lack of long MS-indels have been identified as the main differences between childhood and adult MMR-deficient cancers. These differences may explain why conventional electrophoretic MSI assays, based on the detection of indels of >3 bases in a small number of MS analyzed, were unable to detect MSI in pediatric tumors. Germline analysis confirms the suspicion of CMMRD only if pathogenic variants in both alleles of an MMR gene are found. Unfortunately, this approach is not always definitive due to the detection of VUS and the presence of pseudogenes of PMS2 [14], the most frequent gene affected in CMMRD patients. In CMMRD, MSI may be detected in DNA from all normal cells, which is a hallmark of the syndrome. Following this principle, different techniques have been developed to detect low-frequency microsatellite length variants. Ingham et al [16] described the germline MSI (gMSI), a PCR-based assay with the DNA from the peripheral blood leukocytes (PBLs) detecting dinucleotide repeats that are not sensitive to loss of MSH6 activity. Bodo et al. described an ex vivo MSI (evMSI) using mononucleotide repeats but requiring a long process of culturing of lymphoid precursor cells as well as analysis of alkylation tolerance [17]. Recently, Gallon et al [19] perfected the strategy with the use of short (7–12 bp) mononucleotide repeats to detect low-frequency microsatellite length variants in PBLs. Their smMIP and sequencing-based assay can also be used as a screening test as it is an inexpensive test suitable for high throughput detection of at-risk populations. A promising new approach to detecting highly sensitive MSI, hsMSI, based on a panel of unstable monomorphic markers allows the identification of gMSI in normal cells in PBLs and minimally invasive samples showing robust results in the diagnosis of CMMRD [20]. Both next generation sequencing-based approaches do not have the limitations of the previous tests and provide an accurate test for CMMRD regardless of the affected gene.
In view of all the aforesaid data and in accordance with the recent consensus of diagnostic criteria [4], we propose the use of MMR IHC in malignant brain tumors diagnosed at age younger than 25 years-old to identify those with CMMRD that may have gone undetected due to few accompanying key manifestations or lack of family history of LS spectrum neoplasms. Cases reviewed of the literature in this study showed that the majority of brain tumors occurred before the age of 16 years (98/117, 84%) and 95% of cases occurred up to the age of 25 years (111/117). Only six (5%) patients were older than 25 years (27, 32, 34 (×2), 35, 40). However, given the controversy in considering “MMR-deficiency” in adult brain tumors as a predictor of treatment [75,76,77], and because these neoplasms are not included in the LS tumor spectrum, we do not believe it is appropriate to consider universal screening by IHC in brain tumors in adults older than 25 years.
Of all the available ancillary tests, we believe that MMR IHC is the most suitable for use as a screening method because it is a simple, fast, efficient, and economical technique, fully implemented in pathology departments, which allows the suspicion of CMMRD with a high sensitivity (93.2%) when MMR proteins are not expressed in tumor cells or normal adjacent cells. It is crucial for the pathologist to know the expression pattern that characterizes CMMRD to avoid misinterpretation.
To integrate MMR IHC screening into the established consensus clinical guidelines [3,4], we believe that the best strategy to follow would be to screen for CMMRD using both the MMR IHC in brain malignancy neoplasms in younger than 25 years of age and the C4CMMRD clinical scoring system [3] to select patients for germline genetic testing, those with an MMR-deficient tumor or with a score of 3 or more points; and that the final diagnosis would be made according to the recent guidelines of the International CMMRD Consortium [4]. Thus, patients excluded from MMR IHC screening due to presenting with brain tumor over the age of 25 years would be identified as CMMRD using the existing clinical scoring system of the C4CMMRD consortium based on the set of clinical features that make up their phenotype. This is true for all six patients over 25 years of age in the series reviewed (Table 4). In addition, those brain tumors retaining PMS2 or MSH6 expression were also identified applying the clinical scoring system (Table 5).
Table 5.
Phenotypic features of patients who did not meet criteria for MMR IHC screening or had IHC false-negative results.
| Case ID | Reference (Number) | Reason for Non-Selection by IHC Screening |
Malignancies (Age at Diagnosis) | Others (Age at Diagnosis) |
Points * |
|---|---|---|---|---|---|
| 37 | Senter [43] | Age at brain tumor diagnosis | Glioma (35) Rectum (24) Endometrial (35) | Brother: CRC (26), glioblastoma (34) Brother: Glioma (24) | 5 |
| 46 | Herkert [49] | Age at brain tumor diagnosis | GBM (34) CRC (21) Duodenal (32) Jejunal (×2) (34) | CR polyps (11) GI polyps with dysplasia (21 and 32), congenital asplenia, left isomerism, ventricle septum defect LS family |
6 |
| 84 | Levoine [59] | Age at brain tumor diagnosis | Glioblastoma (40)CRC (22 and 32) | Polyps (38) CALMs LS family | 5 |
| 89 | Levoine [59] | Age at brain tumor diagnosis | GBM (32) CRC (20)Gastric (32) | Polyps (20) | 3 |
| 90 | Levoine [59] | Age at brain tumor diagnosis | GBM (34) CRC (22 and 25) Endometrial (35) Small-bowel (36) | Polyps (22) Consanguinity LS family | 4 |
| 112 | Bush [65] | Age at brain tumor diagnosis | GBM (27) | Multiple polyps (23) CALMs Sister: 2 adenomas and 1 with high-grade dysplasia (21) breast cancer (29) LS family | 3 |
| 42 | Sjursen [46] | PMS2+ | Giant cell glioblastoma (10) CRC (20) Duodenal (26) Ileal (30 and 36) Endometrial (31) Jejunal (42) Basal cell carcinoma several times |
Polyps Sister: CRC (16)Father: Gastric cancer (64) | 4 |
| 7 | Menko [30] | MSH6+ | Oligodendroglioma (10) CRC (12) | CALMs Consanguinity | 8 |
| 96 | Levoine [59] | MSH6+ | Astrocytoma (9) | CALMs | 4 |
| 104 | Taeubner [62] | MSH6+ | Medulloblastoma (13) | CALMs Other skin lesions reminiscent of NF1 | 4 |
Abbreviations: CR, colorectal; CRC, colorectal carcinoma; GI, gastrointestinal; GBM, glioblastoma multiforme; CALMs, café-au-lait macules, LS, Lynch syndrome; *, according to C4CMMRD clinical scoring system [3]; NF1, neurofibromatosis type 1.
At the same time, it will ensure the selection of an effective treatment, as MSI-H tumor cells are resistant to O6-methylating agents such as temozolomide [78], but may be good candidates for immunotherapy treatments [79,80].
5. Material and Methods
5.1. Immunohistochemical Analysis of MMR Proteins
Formalin-fixed, paraffin-embedded tissue sections representative of both brain tumors were studied using standard IHC techniques. The mouse primary antibodies used were the following: anti-hMLH1 (clone G168-15, PharMingen, San Diego, CA, USA), anti-hMSH2 (clone G219-1129, PharMingen, San Diego, CA, USA), anti-hMSH6 (clone 44, Transduction Laboratories, San Diego, CA, USA), and anti-PMS2 (clone A16-4, PharMingen, San Diego, CA, USA). Nuclear immunoreaction in lymphocytes, normal brain or endothelial cells served as positive control. A separate tissue section was used as an additional positive control since internal positive control staining is not observed in CMMRD tissues.
5.2. MSI Analysis
Tumor genomic DNA from the two brain tumor patients was obtained from paraffin-embedded tissue. Briefly, 5 mm hematoxylin-stained paraffin sections were deparaffinized with xylene and rehydrated with ethanol. Tissue was manually macro-dissected for tumor cell enrichment. Finally, DNA was isolated using QIAamp DNAmicrokit (Qiagen GmbH, Hilden, Germany). Five mononucleotide microsatellites (NR21, NR24, NR27, BAT25 and BAT26) were studied using the primers and conditions previously described by Buhard et al. [70]. Fragments were analyzed on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems/Thermo Fisher Scientific, Waltham, MA, USA).
5.3. Germline MSH6 Mutational Analysis
MSH6 germline point mutation analysis was performed on genomic DNA isolated from peripheral blood leucocytes from patients’ parents using PCR and direct sequencing of the whole coding sequence and intron–exon boundaries. The identified MSH6 mutations were subsequently analyzed in brain tumor DNA from their affected sons and blood sample from the healthy daughter. Variant classification was determined according to InSIGHT classification guidelines [81].
5.4. Targeted Next Generation Sequencing Analysis
The brain tumor DNA of patient III-2 was analyzed using a targeted Next Generation Sequencing custom panel, previously described in [82], which included the coding region of POLE and POLD1 genes, among other genes (APC, AXIN2, BMPR1A; BUB1, BUB1B, BUB3, CDH1, CHEK2, ENG, EPCAM, EXO1, FAN1, MLH1, MLH3, MSH2, MSH3, MSH6, MUTYH, PMS1, PMS2, POLD1, POLE, PTEN, SMAD4, STK11 and TP53) and somatic hotspot mutations (design size: 319 Kb). Tumor from patient III-1 did not fulfill quality requirements to be analyzed. Briefly, capture of the target regions was performed using HaloPlex Target Enrichment kit 1-500 kb (Agilent Technologies, Santa Clara, CA, USA), according to the HaloPlex Target Enrichment System-Fast Protocol Version B. Library concentrations were normalized to 0.44 nM. Pooled libraries were sequenced in a MiSeq (Illumina, San Diego, CA, USA) with paired-end 250 bp reads plus an 8-base index read, using MiSeq Reagent Kit v3. Agilent SureCall application was used to trim, align and call variants. Variant filtering was performed based on Phred quality ≥30, alternative allele ratio ≥0.20, read depth ≥30x in the analyzed FFPE sample. Identified variants were then filtered against common single-nucleotide polymorphisms (MAF>1 according to the Exome Aggregation Consortium (ExAC) and the NHLBI GO Exome Sequencing Project (ESP) data). Pathogenicity assessment of the identified variants was aided by Alamut software v2.9.0. (Sophia Genetics SA, Saint Sulpice, Switzerland). The size of the panel used prevented the analysis of tumor mutational burden or mutational signatures.
6. Conclusions
Recognizing the lack of MMR protein expression in both tumor cells and normal tissue as a pattern of CMMRD-related IHC is of crucial importance. The IHC approach is particularly relevant in brain malignancies in which MSI analysis may fail to reveal an MSI-H pattern using a panel of mononucleotide repeat markers. Therefore, we propose the use of MMR IHC in malignant brain tumors diagnosed below 25 years of age to unmask those suffering from CMMRD that may have gone undetected due to the few key manifestations or the lack of family history of LS spectrum neoplasms.
Acknowledgments
We are indebted to the patients, their families and the Spanish Association of Neurofibromatosis.
Author Contributions
Conceptualization, M.P. and E.M.; Data curation, C.C. and I.B.; Formal analysis, C.C., C.S., A.M.M.-M., J.D.V., E.D. and E.M.; Investigation, C.C., C.S., A.M.M.-M., M.P., E.D. and E.M.; Methodology, C.S., A.M.M.-M. and E.D.; Supervision, M.P., M.E. and E.M.; Writing–original draft, C.C.; Writing–review & editing, C.C., C.S., A.M.M.-M., I.B., M.P., E.D., M.E. and E.M. All authors have read and agreed to the published version of the manuscript.
Funding
We thank CERCA Programme/Generalitat de Catalunya for institutional support. The research leading to these results has received funding from the Secretariat for Universities and Research of the Ministry of Business and Knowledge of the Government of Catalonia (2017SGR1080 and 2017SGR1282); the Spanish Ministry of Economy and Competitiveness and co-funded by FEDER funds -a way to build Europe- (grant SAF2015-68016-R and PID2019-111254RB-I00), CIBERONC, MCIU/AEI/FEDER, UE (RTI2018-094049-B-I00 and SAF2014-55000); “la Caixa” Banking Foundation (LCF/PR/GN18/51140001) and Olga Torres Foundation.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and complies with Regulation (EU) 2016/679 of 27 April 2016 on the protection of natural persons with regard to the processing of personal data and on the free movement of such data and Spanish Organic Law 3/2018 of 5 December on the Protection of Personal Data and Guarantee of Digital Rights. This project (PI-20-314) was approved by the Ethics Committee of Hospital Universitari Germans Trias i Pujol (9 October 2020).
Informed Consent Statement
Informed consent was obtained from subjects involved in the study.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ricciardone M.D., Ozçelik T., Cevher B., Ozdağ H., Tuncer M., Gürgey A., Uzunalimoğlu O., Cetinkaya H., Tanyeli A., Erken E., et al. Human MLH1 deficiency predispones to hematological malignancy and neurofibromatosis type 1. Cancer Res. 1999;59:290–293. [PubMed] [Google Scholar]
- 2.Wang Q., Lasset C., Desseigne F., Frappaz D., Bergeron C., Navarro C., Ruano E., Puisieux A. Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res. 1999;59:294–297. [PubMed] [Google Scholar]
- 3.Wimmer K., Kratz C.P., Vasen H.F., Caron O., Colas C., Entz-Werle N., Gerdes A.M., Goldberg Y., Ilencikova I., Muleris M., et al. EU-consortium care for CMMRD (C4CMMRD). Diagnostic criteria for constitutional mismatch repair deficiency syndrome: Suggestions of the European consortium ‘care for CMMRD’ (C4CMMRD) J. Med. Genet. 2014;51:355–365. doi: 10.1136/jmedgenet-2014-102284. [DOI] [PubMed] [Google Scholar]
- 4.Aronson M., Colas C., Shuen A., Hampel H., Foulkes W.D., Feldman H.B., Goldberg Y., Muleris M., Schneider K.W., McGee R.B., et al. Diagnostic criteria for constitutional mismatch repair deficiency (CMMRD): Recommendations from the international consensus working group. J. Med. Genet. 2021:2020–107627. doi: 10.1136/jmedgenet-2020-107627. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q., Montmain G., Ruano E., Upadhyaya M., Dudley S., Liskay R.M., Thibodeau S.N., Puisieux A. Neurofibromatosis type 1 gene as a mutational target in a mismatch repair-deficient cell type. Hum. Genet. 2003;112:117–123. doi: 10.1007/s00439-002-0858-4. [DOI] [PubMed] [Google Scholar]
- 6.Brems H., Chmara M., Sahbatou M., Denayer E., Taniguchi K., Kato R., Somers R., Messiaen L., Schepper S.D., Fryns J.P., et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat. Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- 7.Chmara M., Wernstedt A., Wasag B., Peeters H., Renard M., Beert E., Brems H., Giner T., Bieber I., Hamm H., et al. Multiple pilomatricomas with somatic CTNNB1 mutations in children with constitutive mismatch repair deficiency. Genes Chromosomes Cancer. 2013;52:656–664. doi: 10.1002/gcc.22061. [DOI] [PubMed] [Google Scholar]
- 8.Baas A.F., Gabbett M., Rimac M., Kansikas M., Raphael M., Nievelstein R.A., Nicholls W., Offerhaus J., Bodmer D., Wernstedt A., et al. Agenesis of the corpus callosum and gray matter heterotopia in three patients with constitucional mismatch repair deficiency syndrome. Eur. J. Hum. Genet. 2013;21:55–61. doi: 10.1038/ejhg.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiran S.I., Ben-Sira L., Elhasid R., Roth J., Tabori U., Yalon M., Constantini S., Dvir R. Multiple brain developmental venous anomalies as a marker for constitutional mismatch repair deficiency syndrome. Am. J. Neuroradiol. 2018;39:1943–1946. doi: 10.3174/ajnr.A5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baris H.N., Barnes-Kedar I., Toledano H., Halpern M., Hershkovitz D., Lossos A., Lerer I., Peretz T., Kariv R., Cohen S., et al. Constitutional mismatch repair deficiency in israel: High proportion of founder mutations in MMR genes and consanguinity. Pediatr. Blood Cancer. 2016;63:418–427. doi: 10.1002/pbc.25818. [DOI] [PubMed] [Google Scholar]
- 11.Amayiri N., Tabori U., Campbell B., Bakry D., Aronson M., Durno C., Rakopoulos P., Malkin D., Qaddoumi I., Musharbash A., et al. High frequency of mismatch repair deficiency among pediatric high grade gliomas in Jordan. Int. J. Cancer. 2016;138:380–385. doi: 10.1002/ijc.29724. [DOI] [PubMed] [Google Scholar]
- 12.Levi Z., Kariv R., Barnes-Kedar I., Goldberg Y., Half E., Morgentern S., Eli B., Baris H.N., Vilkin A., Belfer R.G., et al. The gastrointestinal manifestation of constitutional mismatch repair deficiency syndrome: From a single adenoma to polyposis-like phenotype and early onset cancer. Clin. Genet. 2015;88:474–478. doi: 10.1111/cge.12518. [DOI] [PubMed] [Google Scholar]
- 13.Toledano H., Orenstein N., Sofrin E., Ruhrman-Shahar N., Amarilyo G., Basel-Salmon L., Shuldiner A.R., Smirin-Yosef P., Aronson M., Al-Tarrah H., et al. Paediatric systemic lupus erythematosus as a manifestation of constitutional mismatch repair deficiency. J. Med. Genet. 2020;57:505–508. doi: 10.1136/jmedgenet-2019-106303. [DOI] [PubMed] [Google Scholar]
- 14.Vos M.D., Hayward B.E., Picton S., Sheridan E., Bonthron D.T. Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am. J. Hum. Genet. 2004;74:954–964. doi: 10.1086/420796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suerink M., Ripperger T., Messiaen L., Menko F.H., Bourdeaut F., Colas C., Jongmans M., Goldberg Y., Nielsen M., Muleris M., et al. Constitutional mismatch repair deficiency as a differential diagnosis of neurofibromatosis type 1: Consensus guidelines for testing a child without malignancy. J. Med. Genet. 2019;56:53–62. doi: 10.1136/jmedgenet-2018-105664. [DOI] [PubMed] [Google Scholar]
- 16.Ingham D., Diggle C.P., Berry I., Bristow C.A., Hayward B.E., Rahman N., Markham A.F., Sheridan E.G., Bonthron D.T., Carr I.M. Simple detection of germline microsatellite instability for diagnosis of constitutional mismatch repair cancer syndrome. Hum. Mutat. 2013;34:847–852. doi: 10.1002/humu.22311. [DOI] [PubMed] [Google Scholar]
- 17.Bodo S., Colas C., Buhard O., Collura A., Tinat J., Lavoine N., Guilloux A., Chalastanis A., Lafitte P., Coulet F., et al. Diagnosis of constitutional mismatch repair-deficiency syndrome based on microsatellite instability and lymphocyte tolerance to methylating agents. Gastroenterology. 2015;149:1017–1029. doi: 10.1053/j.gastro.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Shuen A.Y., Lanni S., Panigrahi G.B., Edwards M., Yu L., Campbell B.B., Mandel A., Zhang C., Zhukova N., Alharbi M., et al. Functional repair assay for the diagnosis of constitutional mismatch repair deficiency from non-neoplastic tissue. J. Clin. Oncol. 2019;37:461–470. doi: 10.1200/JCO.18.00474. [DOI] [PubMed] [Google Scholar]
- 19.Gallon R., Mühlegger B., Wenzel S.S., Sheth H., Hayes C., Aretz S., Dahan K., Foulkes W., Kratz C.P., Ripperger T., et al. A sensitive and scalable microsatellite instability assay to diagnose constitutional mismatch repair deficiency by sequencing of peripheral blood leukocytes. Hum. Mutat. 2019;40:649–655. doi: 10.1002/humu.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.González-Acosta M., Marín F., Puliafito B., Bonifaci N., Fernández A., Navarro M., Salvador H., Balaguer F., Iglesias S., Velasco A., et al. High-sensitivity microsatellite instability assessment for the detection of mismatch repair defects in normal tissue of biallelic germline mismatch repair mutation carriers. J. Med. Genet. 2020;57:269–273. doi: 10.1136/jmedgenet-2019-106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasen H.F., Ghorbanoghli Z., Bourdeaut F., Cabaret O., Caron O., Duval A., Entz-Werle N., Goldberg Y., Ilencikova D., Kratz C.P., et al. Guidelines for surveillance of individuals with constitucional mismatch repair-deficiency proposed by the European Consortium “Care for CMMR-D” (C4CMMR-D) J. Med. Genet. 2014;51:283–293. doi: 10.1136/jmedgenet-2013-102238. [DOI] [PubMed] [Google Scholar]
- 22.Durno C.A., Sherman P.M., Aronson M., Malkin D., Hawkins C., Bakry D., Bouffet E., Gallinger S., Pollett A., Campbell B., et al. Phenotypic and genotypic characterisation of biallelic mismatch repair deficiency (BMMR-D) syndrome. Eur. J. Cancer. 2015;51:977–983. doi: 10.1016/j.ejca.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Win A.K., Jenkins M.A., Dowty J.G., Antoniou A.C., Lee A., Giles G.G., Buchanan D.D., Clendenning M., Rosty C., Ahnen D.J., et al. Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2017;26:404–412. doi: 10.1158/1055-9965.EPI-16-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gröbner S.N., Worst B.C., Weischenfeldt J., Buchhalter I., Kleinheinz K., Rudneva V.A., Johann P.D., Balasubramanian G.P., Segura-Wang M., Brabetz S., et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 25.Shlien A., Campbell B.B., Borja R.D., Alexandrov L.B., Merico D., Wedge D., Van Loo P., Tarpey P.S., Coupland P., Behjati S., et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat. Genet. 2015;47:257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 26.Guerrini-Rousseau L., Varlet P., Colas C., Andreiuolo F., Bourdeaut F., Dahan K., Devalck C., Faure-Conter C., Genuardi M., Goldberg Y., et al. Constitutional mismatch repair deficiency-associated brain tumors: Report from the European C4CMMRD consortium. Neurooncol. Adv. 2019;1:vdz033. doi: 10.1093/noajnl/vdz033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosa M.D., Fasano C., Panariello L., Scarano M.I., Belli G., Iannelli A., Ciciliano F., Izzo P. Evidence for a recessive inheritance of Turcot’s syndrome caused by compound heterozygous mutations within the PMS2 gene. Oncogene. 2000;19:1719–1723. doi: 10.1038/sj.onc.1203447. [DOI] [PubMed] [Google Scholar]
- 28.Vilkki S., Tsao J.L., Loukola A., Pöyhönen M., Vierimaa O., Herva R., Aaltonen L.A., Shibata D. Extensive somatic microsatellite mutations in normal human tissue. Cancer Res. 2001;61:4541–4544. [PubMed] [Google Scholar]
- 29.Bougeard G., Charbonnier F., Moerman A., Martin C., Ruchoux M.M., Drouot N., Frébourg T. Early onset brain tumor and lymphoma in MSH2-deficient children. Am. J. Hum. Genet. 2003;72:213–216. doi: 10.1086/345297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menko F.H., Kaspers G.L., Meijer G.A., Claes K., Van Hagen J.M., Gille J.J. A homozygous MSH6 mutation in a child with café-au-lait spots, oligodendroglioma and rectal cancer. Fam. Cancer. 2004;3:123–127. doi: 10.1023/B:FAME.0000039893.19289.18. [DOI] [PubMed] [Google Scholar]
- 31.Vos M.D., Hayward B.E., Charlton R., Taylor G.R., Glaser A.W., Picton S., Cole T.R., Maher E.R., McKeown C.M.E., Mann J.R., et al. PMS2 mutations in childhood cancer. J. Natl. Cancer Inst. 2006;98:358–361. doi: 10.1093/jnci/djj073. [DOI] [PubMed] [Google Scholar]
- 32.Agostini M., Tibiletti M.G., Lucci-Cordisco E., Chiaravalli A., Morreau H., Furlan D., Boccuto L., Pucciarelli S., Capella C., Boiocchi M., et al. Two PMS2 mutations in a Turcot syndrome family with small bowel cancers. Am. J. Gastroenterol. 2005;100:1886–1891. doi: 10.1111/j.1572-0241.2005.50441.x. [DOI] [PubMed] [Google Scholar]
- 33.Ostergaard J.R., Sunde L., Okkels H. Neurofibromatosis von Recklinghausen type I phenotype and early onset of cancers in siblings compound heterozygous for mutations in MSH6. Am. J. Med. Genet. A. 2005;139:96–105. doi: 10.1002/ajmg.a.30998. [DOI] [PubMed] [Google Scholar]
- 34.Hegde M.R., Chong B., Blazo M.E., Chin L.H.E., Ward P.A., Chintagumpala M.M., Kim J.Y., Plon S.E., Richards C.S. A homozygous mutation in MSH6 causes Turcot syndrome. Clin. Cancer Res. 2005;11:4689–4693. doi: 10.1158/1078-0432.CCR-04-2025. [DOI] [PubMed] [Google Scholar]
- 35.Durno C., Aronson M., Bapat B., Cohen Z., Gallinger S. Family history and molecular features of children, adolescents, and young adults with colorectal carcinoma. Gut. 2005;54:1146–1150. doi: 10.1136/gut.2005.066092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krüger S., Kinzel M., Walldorf C., Gottschling S., Bier A., Tinschert S., Von Stackelberg A., Henn W., Görgens H., Boue S., et al. Homozygous PMS2 germline mutations in two families with early-onset haematological malignancy, brain tumours, HNPCC-associated tumours, and signs of neurofibromatosis type 1. Eur. J. Hum. Genet. 2008;16:62–72. doi: 10.1038/sj.ejhg.5201923. [DOI] [PubMed] [Google Scholar]
- 37.Gururangan S., Frankel W., Broaddus R., Clendenning M., Senter L., McDonald M., Eastwood J., Reardon D., Vredenburgh J., Quinn J., et al. Multifocal anaplastic astrocytoma in a patient with hereditary colorectal cancer, transcobalamin II deficiency, agenesis of the corpus callosum, mental retardation, and inherited PMS2 mutation. Neuro Oncol. 2008;10:93–97. doi: 10.1215/15228517-2007-037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auclair J., Leroux D., Desseigne F., Lasset C., Saurin J.C., Joly M.O., Pinson S., Xu X.L., Montmain G., Ruano E., et al. Novel biallelic mutations in MSH6 and PMS2 genes, gene conversion as a likely cause of PMS2 gene inactivation. Hum. Mutat. 2007;28:1084–1090. doi: 10.1002/humu.20569. [DOI] [PubMed] [Google Scholar]
- 39.Poley J.W., Wagner A., Hoogmans M.M., Menko F.H., Tops C., Kros J.M., Reddingius R.E., Meijers-Heijboer H., Kuipers E.J., Dinjens W.N.M. Rotterdam initiative on gastrointestinal hereditary tumors. Biallelic germline mutations of mismatch-repair genes: A possible cause for multiple pediatric malignancies. Cancer. 2007;109:2349–2356. doi: 10.1002/cncr.22697. [DOI] [PubMed] [Google Scholar]
- 40.Scott R.H., Mansour S., Pritchard-Jones K., Kumar D., MacSweeney F., Rahman N. Medulloblastoma, acute myelocytic leukemia and colonic carcinomas in a child with biallelic MSH6 mutations. Nat. Clin. Pract. Oncol. 2007;4:130–134. doi: 10.1038/ncponc0719. [DOI] [PubMed] [Google Scholar]
- 41.Kratz C.P., Niemeyer C.M., Jüttner E., Kartal M., Weninger A., Schmitt-Graeff A., Kontny U., Lauten M., Utzolino S., Rädecke J., et al. Childhood T-cell non-Hodgkin’s lymphoma, colorectal carcinoma and brain tumor in association with café-au-lait spots caused by a novel homozygous PMS2 mutation. Leukemia. 2008;22:1078–1080. doi: 10.1038/sj.leu.2405008. [DOI] [PubMed] [Google Scholar]
- 42.Etzler J., Peyrl A., Zatkova A., Schildhaus H.-U., Ficek A., Merkelbach-Bruse S., Kratz C.P., Attarbaschi A., Hainfellner J.A., Yao S., et al. RNA-based mutation analysis identifies an unusual MSH6 splicing defect and circumvents PMS2 pseudogene interference. Hum. Mutat. 2008;29:299–305. doi: 10.1002/humu.20657. [DOI] [PubMed] [Google Scholar]
- 43.Senter L., Clendenning M., Sotamaa K., Hampel H., Green J., Potter J.D., Lindblom A., Lagerstedt K., Thibodeau S.N., Lindor N.M., et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan T.Y., Orme L.M., Lynch E., Croxford M.A., Dow C., Dewan P.A., Lipton L. Biallelic PMS2 mutations and a distinctive childhood cancer syndrome. J. Pediatr. Hematol. Oncol. 2008;30:254–257. doi: 10.1097/MPH.0b013e318161aa20. [DOI] [PubMed] [Google Scholar]
- 45.Toledano H., Goldberg Y., Kedar-Barnes I., Baris H., Porat R.M., Shochat C., Bercovich D., Pikarsky E., Lerer I., Yaniv I., et al. Homozygosity of MSH2 c.1906G→C germline mutation is associated with childhood colon cancer, astrocytoma and signs of Neurofibromatosis type I. Fam. Cancer. 2009;8:187–194. doi: 10.1007/s10689-008-9227-3. [DOI] [PubMed] [Google Scholar]
- 46.Sjursen W., Bjørnevoll I., Engebretsen L.F., Fjelland K., Halvorsen T., Myrvold H.E. A homozygote splice site PMS2 mutation as cause of Turcot syndrome gives rise to two different abnormal transcripts. Fam. Cancer. 2009;8:179–186. doi: 10.1007/s10689-008-9225-5. [DOI] [PubMed] [Google Scholar]
- 47.Giunti L., Cetica V., Ricci U., Giglio S., Sardi I., Paglierani M., Andreucci E., Sanzo M., Forni M., Buccoliero A.M., et al. Type A microsatellite instability in pediatric gliomas as an indicator of Turcot syndrome. Eur. J. Hum. Genet. 2009;17:919–927. doi: 10.1038/ejhg.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy S., Raskin L., Raymond V.M., Thibodeau S.N., Mody R.J., Gruber S.B. Pediatric duodenal cancer and biallelic mismatch repair gene mutations. Pediatr. Blood Cancer. 2009;53:116–120. doi: 10.1002/pbc.21957. [DOI] [PubMed] [Google Scholar]
- 49.Herkert J.C., Niessen R.C., Olderode-Berends M.J., Veenstra-Knol H.E., Vos Y.J., Van der Klift H.M., Scheenstra R., Tops C.M.J., Karrenbeld A., Peters F.T.M., et al. Paediatric intestinal cancer and polyposis due to bi-allelic PMS2 mutations: Case series, review and follow-up guidelines. Eur. J. Cancer. 2011;47:965–982. doi: 10.1016/j.ejca.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 50.Ilencikova D., Sejnova D., Jindrova J., Babal P. High-grade brain tumors in siblings with biallelic MSH6 mutations. Pediatr. Blood Cancer. 2011;57:1067–1070. doi: 10.1002/pbc.23217. [DOI] [PubMed] [Google Scholar]
- 51.Leenen C.H., Geurts-Giele W.R., Dubbink H.J., Reddingius R., Van den Ouweland A.M., Tops C.M.J., Van de Klift H.M., Kuipers E.J., Van Leerdam M.E., Dinjens W.N.M., et al. Pitfalls in molecular analysis for mismatch repair deficiency in a family with biallelic pms2 germline mutations. Clin. Genet. 2011;80:558–565. doi: 10.1111/j.1399-0004.2010.01608.x. [DOI] [PubMed] [Google Scholar]
- 52.Johannesma P.C., Van der Klift H.M., Van Grieken N.C., Troost D., Riele H.T., Jacobs M.A., Postma T.J., Heideman D.A., Tops C.M., Wijnen J.T., et al. Childhood brain tumours due to germline bi-allelic mismatch repair gene mutations. Clin. Genet. 2011;80:243–255. doi: 10.1111/j.1399-0004.2011.01635.x. [DOI] [PubMed] [Google Scholar]
- 53.Lindsay H., Jubran R.F., Wang L., Kipp B.R., May W.A. Simultaneous colonic adenocarcinoma and medulloblastoma in a 12-year-old with biallelic deletions in PMS2. J. Pediatr. 2013;163:601–603. doi: 10.1016/j.jpeds.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walter A.W., Ennis S., Best H., Vaughn C.P., Swensen J.J., Openshaw A., Gripp K.W. Constitutional mismatch repair deficiency presenting in childhood as three simultaneous malignancies. Pediatr. Blood Cancer. 2013;60:E135–E136. doi: 10.1002/pbc.24613. [DOI] [PubMed] [Google Scholar]
- 55.Yeung J.T., Pollack I.F., Shah S., Jaffe R., Nikiforova M., Jakacki R.I. Optic pathway glioma as part of a constitutional mismatch-repair deficiency syndrome in a patient meeting the criteria for neurofibromatosis type 1. Pediatr. Blood Cancer. 2013;60:137–139. doi: 10.1002/pbc.24254. [DOI] [PubMed] [Google Scholar]
- 56.Bakry D., Aronson M., Durno C., Rimawi H., Farah R., Alharbi Q.K., Alharbi M., Shamvil A., Ben-Shachar S., Mistry M., et al. Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: Report from the constitutional mismatch repair deficiency consortium. Eur. J. Cancer. 2014;50:987–996. doi: 10.1016/j.ejca.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 57.Bougeard G., Olivier-Faivre L., Baert-Desurmont S., Tinat J., Martin C., Bouvignies E., Vasseur S., Huet F., Couillault G., Vabres P., et al. Diversity of the clinical presentation of the MMR gene biallelic mutations. Fam. Cancer. 2014;13:131–135. doi: 10.1007/s10689-013-9676-1. [DOI] [PubMed] [Google Scholar]
- 58.Daou B., Zanello M., Varlet P., Brugieres L., Jabbour P., Caron O., Lavoine N., Dhermain F., Willekens C., Beuvon F., et al. An unusual case of constitutional mismatch repair deficiency syndrome with anaplastic ganglioglioma, colonic adenocarcinoma, osteosarcoma, acute myeloid leukemia, and signs of neurofibromatosis type 1: Case report. Neurosurgery. 2015;77:E145–E152. doi: 10.1227/NEU.0000000000000754. [DOI] [PubMed] [Google Scholar]
- 59.Lavoine N., Colas C., Muleris M., Bodo S., Duval A., Entz-Werle N., Coulet F., Cabaret O., Andreiuolo F., Charpy C., et al. Constitutional mismatch repair deficiency syndrome: Clinical description in a French cohort. J. Med. Genet. 2015;52:770–778. doi: 10.1136/jmedgenet-2015-103299. [DOI] [PubMed] [Google Scholar]
- 60.Nguyen A., Bougeard G., Koob M., Chenard M.P., Schneider A., Maugard C., Entz-Werle N. MSI detection and its pitfalls in CMMRD syndrome in a family with a bi-allelic LH1 mutation. Fam. Cancer. 2016;15:571–577. doi: 10.1007/s10689-016-9894-4. [DOI] [PubMed] [Google Scholar]
- 61.Maletzki C., Huehns M., Bauer I., Ripperger T., Mork M.M., Vilar E., Klöcking S., Zettl H., Prall F., Linnebacher M. Frameshift mutational target gene analysis identifies similarities and differences in constitutional mismatch repair-deficiency and Lynch syndrome. Mol. Carcinog. 2017;56:1753–1764. doi: 10.1002/mc.22632. [DOI] [PubMed] [Google Scholar]
- 62.Taeubner J., Wimmer K., Muleris M., Lascols O., Colas C., Fauth C., Brozou T., Felsberg J., Riemer J., Gombert M., et al. Diagnostic challenges in a child with early onset desmoplastic medulloblastoma and homozygous variants in MSH2 and MSH6. Eur. J. Hum. Genet. 2018;26:440–444. doi: 10.1038/s41431-017-0071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.AlHarbi M., Mobark N.A., AlMubarak L., Aljelaify R., AlSaeed M., Almutairi A., Alqubaishi F., Hussain M.E., Balbaid A.A.O., Marie A.S., et al. Durable response to nivolumab in a pediatric patient with refractory glioblastoma and constitutional biallelic mismatch repair deficiency. Oncologist. 2018;23:1401–1406. doi: 10.1634/theoncologist.2018-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baig S.M., Fatima A., Tariq M., Khan T.N., Ali Z., Faheem M., Mahmood H., Killela P., Waitkus M., He Y., et al. Hereditary brain tumor with a homozygous germline mutation in PMS2: Pedigree analysis and prenatal screening in a family with constitutional mismatch repair deficiency (CMMRD) syndrome. Fam. Cancer. 2019;18:261–265. doi: 10.1007/s10689-018-0112-4. [DOI] [PubMed] [Google Scholar]
- 65.Bush L., Aronson M., Tabori U., Campbell B.B., Bedgood R.B., Jasperson K. Delineating a new feature of constitutional mismatch repair deficiency (CMMRD) syndrome: Breast cancer. Fam. Cancer. 2019;18:105–108. doi: 10.1007/s10689-018-0088-0. [DOI] [PubMed] [Google Scholar]
- 66.Farah R.A., Maalouf F., Chahine N.A., Farhat H., Campbell B., Zhukova N., Durno C., Aronson M., Hawkins C., Bouffet E., et al. Ongoing issues with the management of children with constitutional mismatch repair deficiency syndrome. Eur. J. Med. Genet. 2019;62:103706. doi: 10.1016/j.ejmg.2019.103706. [DOI] [PubMed] [Google Scholar]
- 67.Giardiello F.M., Allen J.I., Axilbund J.E., Boland C.R., Burke C.A., Burt R.W., Church J.M., Dominitz J.A., Johnson D.A., Kaltenbach T., et al. Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US Multi-society Task Force on colorectal cancer. Am. J. Gastroenterol. 2014;109:1159–1179. doi: 10.1038/ajg.2014.186. [DOI] [PubMed] [Google Scholar]
- 68.Watkins J.C., Yang E.J., Muto M.G., Feltmate C.M., Berkowitz R.S., Horowitz N.S., Syngal S., Yurgelun M.B., Chittenden A., Hornick J.L., et al. Universal screening for mismatch-repair deficiency in endometrial cancers to identify patients with Lynch syndrome and Lynch-like syndrome. Int. J. Gynecol. Pathol. 2017;36:115–127. doi: 10.1097/PGP.0000000000000312. [DOI] [PubMed] [Google Scholar]
- 69.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buhard O., Cattaneo F., Wong Y.F., Yim S.F., Friedman E., Flejou J.F., Duval A., Hamelin R. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J. Clin. Onco. 2006;24:241–245. doi: 10.1200/JCO.2005.02.7227. [DOI] [PubMed] [Google Scholar]
- 71.Hodel K.P., Borja R.D., Henninger E.E., Campbell B.B., Ungerleider N., Light N., Wu T., LeCompte K.G., Goksenin A.Y., Bunnell B.A., et al. Explosive mutation accumulation triggered by heterozygous human Pol ε proofreading-deficiency is driven by suppression of mismatch repair. Elife. 2018;7:e32692.:e32692. doi: 10.7554/eLife.32692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campbell B.B., Light N., Fabrizio D., Zatzman M., Fuligni F., Borja R.D., Davidson S., Edwards M., Elvin J.A., Hodel K.P., et al. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171:1042–1056. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bacher J.W., Flanagan L.A., Smalley R.L., Nassif N.A., Burgart L.J., Halberg R.B., Megid W.M.A., Thibodeau S.N. Development of a fluorescent multiplex assay for detection of MSI-High tumors. Dis. Markers. 2004;20:237–250. doi: 10.1155/2004/136734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung J., Maruvka Y.E., Sudhaman S., Kelly J., Haradhvala N.J., Bianchi V., Edwards M., Forster V.J., Nunes N.M., Galati M.A., et al. DNA Polymerase and mismatch repair exert distinct microsatellite instability signatures in normal and malignant human cells. Cancer Discov. 2020;11:1–16. doi: 10.1158/2159-8290.CD-20-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Indraccolo S., Lombardi G., Fassan M., Pasqualini L., Giunco S., Marcato R., Gasparini A., Candiotto C., Nalio S., Fiduccia P., et al. Genetic, epigenetic, and immunologic profiling of MMR-deficient relapsed glioblastoma. Clin. Cancer Res. 2019;25:1828–1837. doi: 10.1158/1078-0432.CCR-18-1892. [DOI] [PubMed] [Google Scholar]
- 76.Balana C., Vaz M.A., Sepúlveda J.M., Mesia C., Barco S.D., Pineda E., Muñoz-Langa J., Estival A., Las Peñas R.D., Fuster J., et al. A phase II randomized, multicenter, open-label trial of continuing adjuvant temozolomide beyond 6 cycles in patients with glioblastoma (GEINO 14-01) Neuro Oncol. 2020;22:1851–1861. doi: 10.1093/neuonc/noaa107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lombardi G., Barresi V., Indraccolo S., Simbolo M., Fassan M., Mandruzzato S., Simonelli M., Caccese M., Pizzi M., Fassina A., et al. Pembrolizumab activity in recurrent high-grade gliomas with partial or complete loss of mismatch repair protein expression: A monocentric, observational and prospective pilot study. Cancers. 2020;12:2283. doi: 10.3390/cancers12082283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fedier A., Fink D. Mutations in DNA mismatch repair genes: Implications for DNA damage signaling and drug sensitivity. Int. J. Oncol. 2004;24:1039–1047. doi: 10.3892/ijo.24.4.1039. [DOI] [PubMed] [Google Scholar]
- 79.Tabori U., Hansford J.R., Achatz M.I., Kratz C.P., Plon S.E., Frebourg T., Brugières L. Clinical management and tumor surveillance recommendations of inherited mismatch repair deficiency in childhood. Clin. Cancer Res. 2017;23:e32–e37. doi: 10.1158/1078-0432.CCR-17-0574. [DOI] [PubMed] [Google Scholar]
- 80.Bouffet E., Larouche V., Campbell B.B., Merico D., Borja R.D., Aronson M., Durno C., Krueger J., Cabric V., Ramaswamy V., et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 2016;34:2206–2211. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- 81.Thompson B.A., Spurdle A.B., Plazzer J.P., Greenblatt M.S., Akagi K., Al-Mulla F., Bapat B., Bernstein I., Capellá G., Dunnen J.T.D., et al. Application of a 5-tiered scheme for standardized classification of 2360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vargas-Parra G.M., González-Acosta M., Thompson B.A., Gómez C., Fernández S., Dámaso E., Pons T., Morak M., Valle J.D., Iglesias S., et al. Elucidating the molecular basis of MSH2-deficient tumors by combined germline and somatic analysis. Int. J. Cancer. 2017;141:1365–1380. doi: 10.1002/ijc.30820. [DOI] [PubMed] [Google Scholar]