Abstract
Simple Summary
Gut microbiota is emerging as new diagnostic and prognostic marker and/or therapeutic target to improve the management of cancer. This review aims to summarize microbial signatures that have been associated with digestive and other cancers. We report the clinical relevance of these microbial markers to predict the response to cancer therapy. Among these biomarkers, colibactin-producing E. coli are prevalent in the colonic mucosa of patients with colorectal cancer and they promote colorectal carcinogenesis in several pre-clinical models. Here we discuss the promising use of colibactin-producing E. coli as a new predictive factor and a therapeutic target in colon cancer management.
Abstract
The gut microbiota is crucial for physiological development and immunological homeostasis. Alterations of this microbial community called dysbiosis, have been associated with cancers such colorectal cancers (CRC). The pro-carcinogenic potential of this dysbiotic microbiota has been demonstrated in the colon. Recently the role of the microbiota in the efficacy of anti-tumor therapeutic strategies has been described in digestive cancers and in other cancers (e.g., melanoma and sarcoma). Different bacterial species seem to be implicated in these mechanisms: F. nucleatum, B. fragilis, and colibactin-associated E. coli (CoPEC). CoPEC bacteria are prevalent in the colonic mucosa of patients with CRC and they promote colorectal carcinogenesis in susceptible mouse models of CRC. In this review, we report preclinical and clinical data that suggest that CoPEC could be a new factor predictive of poor outcomes that could be used to improve cancer management. Moreover, we describe the possibility of using these bacteria as new therapeutic targets.
Keywords: intestinal microbiota, CoPEC, colibactin, cancer, E. coli, dysbiosis, anti-cancer treatment, prognosis, biomarker, colorectal cancer
1. Introduction
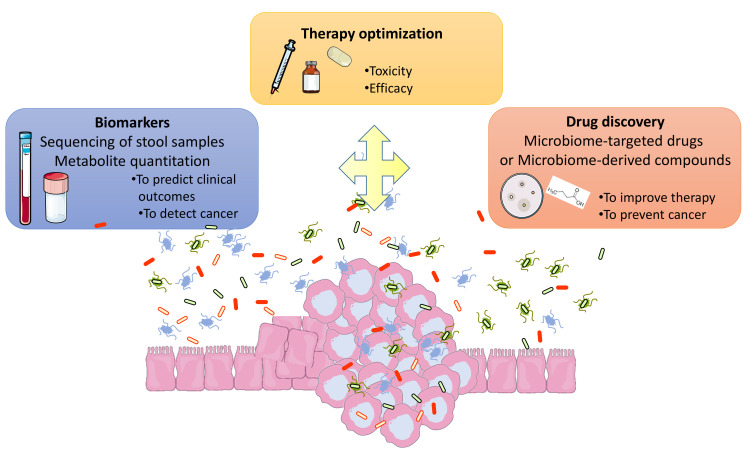
The human gut microbiota is a complex microbial ecosystem harboring more than 100 billion bacteria, archaea, viruses, parasites and fungi that maintain symbiotic interactions with the host [1,2,3]. This intricate relationship plays a crucial role in maintaining healthy homeostasis including nutrient absorption, vitamin biosynthesis, immune system development, the maintenance of epithelial mucosa integrity, and resistance against pathogens [4,5]. Given the symbiotic relationship between the host and gut microbiota, an alteration from the normal microbiota composition, referred to as dysbiosis, has been observed in several human diseases including digestive cancers and other extra-digestive malignancies [2,6,7]. Gut microbiota dysbiosis could promote carcinogenesis in various tumor pathologies [8]. Association between repeated courses of antibiotics and the development of a variety of tumors has been demonstrated [9]. Most evidence supporting a causal role for gut dysbiosis as a modulator of cancer development is related to colorectal cancer (CRC) [10,11,12,13]. Gut microbiota have also been associated with a number of other malignancies, including hepatocellular carcinoma and breast cancer via its influence on energy metabolism and obesity and other specific mechanisms (for review [8]). In addition, scientists have investigated the entire microbial community in the tumor landscape through next generation sequencing technologies [14,15]. Even if the signature of dysbiosis is not yet defined in cancers, one emerging translational application of the gut microbiome is its potential as diagnostic and/or prognostic biomarker or as a therapeutic target (cf. Figure 1). The intestinal microbiota could therefore be a new tool to improve the management of cancers. Because the colonic microbiota is mostly represented by bacteria, the bacterial community remains the most studied.
Figure 1.
Intestinal microbiome for a better management of cancer patients. New biomarkers based on microbial composition of the stool are emerging to predict clinical outcomes. Metabolites signatures and/or sequencing oral samples will also be developed as diagnostic or prognostic biomarkers. Studies about interactions between gut microbiota and treatments (surgery, chemotherapy, immunotherapy, radiotherapy) could allow to improve their efficacy and decrease some side effects (e.g., post-surgical complications, toxicity). Gut microbiota data could be use for the discovery of new and innovative therapeutic tools as small bioactive molecule which mimic benefic microbial effect or targeting procarcinogenic bacteria. Microbial intervention could be developed including prebiotics, probiotics, phages. Natural products and/or diet complementation can also be considered. These strategies will have to be adapted according to tumor characteristics and to the patient’s environment, lifestyle, host susceptibility and comorbidities.
In this review, we provide clinical data on the potential use of gut microbiota-related biomarkers for cancer screening and prognosis. We describe the results obtained for CRC and for others cancers. We next focused on colibactin-associated E. coli (CoPEC), a pro-carcinogenic bacterium that is more prevalent in aggressive form of CRC [16,17]. We discussed the preclinical and clinical data, which suggest that CoPEC could be a new prognostic biomarker in CRC management. Finally, we describe the possibility of using CoPEC as a new therapeutic target.
2. Gut Microbiota Biomarker for Cancer Screening
2.1. Intestinal Microbiota Biomarker for CRC Screening
The actual 5-year survival rate is 90% for a localized CRC, but this rapidly decrease to <15% for metastatic disease [18]. For this reason, an early, noninvasive, and accessible method for CRC screening is critical. The main screening strategy used worldwide is the fecal immunochemical test (FIT), which has a limited sensitivity of 79% for CRC diagnosis [19]. The multitarget stool DNA test, not routinely used, is very effective in detecting CRC (sensitivity 92.3%) but presents a higher false-positive rate than FIT [20]. Therefore, more accurate and noninvasive biomarkers for early CRC screening is still needed. Given the link between gut microbiota dysbiosis and CRC [21], microbial markers have recently emerged as a promising indicator in early CRC screening (cf Table 1). Known specific oncogenic gut bacteria have been assessed individually from both fecal and mucosal samples from patients with CRC, including strains of Bacteroides fragilis, Escherichia coli, Enterococcus faecalis, Streptococcus gallolyticus, and Fusobacterium nucleatum [16,17,21,22,23,24,25]. Over the last few years, many studies using sequencing methods with whole-genome shotgun (WGS) and/or 16S RNAr DNA sequencing have also highlighted several signatures (mostly in feces) between individuals with CRC and healthy control subjects [13,26,27,28,29,30]. Overall, these datasets revealed a global compositional shift in microbiota from individuals with CRC compared to healthy controls, notably with a reduced bacterial [30,31,32]. No single specific oncomicrobe has been found to be universally present in all patients with CRC. Significantly enriched and depleted fecal microorganisms have been identified between CRC and control populations. Differences were not affected when controlling for potential confounding by age, body mass index (BMI) or sex (40). These identified polymicrobial signatures could potentially be harnessed as screening biomarkers for CRC [25,30,31,32,33,34,35,36,37] (cf. Table 1). However at present, these is no consensus to define this signature. Consistently, alone or combined with other bacteria, Fusobacterium nucleatum emerged as a fecal marker in CRC both in feces and tumor samples compared to control subjects and surrounding normal tissue [30,34,35,38]. In addition, quantifying fecal Fusobacterium nucleatum abundance could improve the performance of the FIT in detecting CRC and advanced adenoma with an increase in the AUC from 0.86 to 0.95 [27,39]. Overall, the majority of the published data were obtained from fecal sample analysis. Because oral and gut microbiomes were predictive of each other and over-representations of several oral bacteria as F. nucleatum were detected in the stool from patients with CRC, microbial-biomarkers of CRC could be also investigated in oral samples [33,40]. Recently, Dohlman et al. discovered promising endogenous microbial DNA signatures in blood samples from CRC patients featuring mucosal barrier injury in CRC [41].
Table 1.
Intestinal microbiota-related biomarkers for the screening of CRC, digestive cancers and other cancers.
| Cancer | Microbiota-Related Marker | Techniques | Samples | Ref |
|---|---|---|---|---|
| CRC | Diversity | |||
| Reduction of diversity (metagenome) | Shotgun metagenomic analysis | Feces | [30,31,32] | |
| Increase of diversity | Blood | [41] | ||
| Polymicrobial signatures | ||||
| 4 validated markers including F. nucleatum, P. micra, S. moorei, P. stomatis | Shotgun metagenomic analysis | Feces | [30] | |
| 7 species including B. fragilis, F. nucleatum, Porphyromonasasaccharolytica, P. micra, P. intermedia, A. finegoldii, and T. acidaminovorans | Feces | [31] | ||
| 29 species including F. nucleatum, P. micra, S. moorei, P. stomatis, Ruminococcus torques | Feces | [35] | ||
| 16 species including P. stomatis, F. nucleatum, Parvimonasspp., P. asaccharolytica, G. morbillorum, C. symbiosum and P. micra | Feces | [37] | ||
| 22 species including F. nucleatum and B. fragilis | Shotgun metagenomic analysis and 16S rRNA gene sequencing | Feces | [34] | |
| 34 species including F. nucleatum, P. micra, S. moorei, P. stomatis | 16S rRNA gene sequencing |
Feces | [33] | |
| 12 species including F. nucleatum, B. fragilis, P. micra, P. stomatis | Tissue | [36] | ||
| Single bacteria | ||||
| F. nucleatum (metagenome, 16S rRNA gene sequencing, PCR) | Shotgun metagenomic analysis, 16S rRNA gene sequencing, PCR | Feces and Tissue | [25,30,35,36,41] | |
| Colibactin-Producing E. coli (metagenome, PCR) | Shotgun metagenomic analysis, PCR | Feces and Tissue | [16,17,22,25,35,41] | |
| Bacteroidetes fragilis (ETBF) (metagenome, PCR) | Shotgun metagenomic analysis, PCR | Feces and Tissue | [21,32,36] | |
| Enterococcus faecalis (PCR) | PCR | Feces | [23] | |
| Streptococcus gallolyticus (metagenome, PCR) | Shotgun metagenomic analysis, PCR | Tissue | [24,37] | |
| Parvimonas micra (PCR) | PCR | Feces | [25] | |
| Other digestive cancers | ||||
| Gastric cancer | Desulfovibrio, Escherichia, Faecalibacterium or Oscillospira | 16S rRNA gene sequencing |
Feces | [48] |
| Lactobacillus and Megasphaera | Feces | [49] | ||
| Hepatocellular Carcinoma | Increase of diversity (vs. cirrhosis) 30 OTUs including Klebsiella, Prevotella and Haemophilus (vs. control) |
16S rRNA gene sequencing |
Feces | [50] |
| Pancreatic ductal adenocarcinoma | Decrease of diversity | 16S rRNA gene sequencing |
Feces | [51] |
| Increase of Bacteroidetes and decrease of Firmicutes and Proteobacteria. Set of 40 genera | Feces | [51] | ||
| Set of 14 species including Akkermansia and Bacteroidales | Feces | [52] | ||
| Esophageal cancer | Bacteroides, Bifidobacterium, Streptococcus, or Lachnospira | 16S rRNA gene sequencing |
Feces | [53] |
| Other cancers | ||||
| Breast cancer | Increase of Clostridiaceae, Faecalibacterium, and Ruminococcaceae, and decrease of Dorea and Lachnospiraceae | 16S rRNA gene sequencing |
Feces | [54] |
| Breast cancer (post-menopausal) | 14 optimal species markers including Escherichia coli, Eubacterium eligens, Proteus mirabilis, and Fusobacterium varium | Shotgun metagenomic analysis | Feces | [55] |
| Lung cancer | Actinobacteria (phyla), Bifidobacterium and Enterococcus (genus) | 16S rRNA gene sequencing |
Feces | [56] |
Microbial-derived metabolite analyses in human biofluids (blood, urine, saliva, and fecal) could also be a promising and noninvasive approach for early CRC screening [42]. Although the method of sample collection is not standardized, four human studies supported the screening potential of fecal metabolic profiling using the nuclear magnetic resonance (NMR) method in CRC cohort patients [43,44,45,46]. Fecal metabolite levels such as lactate, glucose, and some amino acids were higher in CRC patients than in controls [43,45]. Moreover, the levels of SCFAs, glutamate and succinate varied significantly according to CRC stage [45]. In a recent study, Lin et al. demonstrated that fecal acetate had the highest diagnostic performance for discriminating CRC from controls [44]. Conversely, Chen et al. observed that butyrate and several butyrate-producing bacteria were depleted in CRC patients stools [47].
Extended validation studies are required to define CRC signatures of dysbiosis. Future investigations should consider the heterogeneity of CRC. Because microbiota differ with gender and life style, these parameters have to be considered [1,32]. Moreover different clinical studies have suggested that the dysbiotic signature of CRC might be different according to the tumor characteristics (KRAS, BRAF mutations, and DNA mismatch repair MMR alterations) and/or tumor sidedness (proximal or distal location) [57,58,59,60,61,62,63,64].
2.2. Intestinal Microbiota Biomarker for Other Cancers Screening
While this has been relatively well studied in CRC, there are also emerging data suggesting gut microbiota dysbiosis as a potential noninvasive tool for early diagnosis of a variety of cancers (cf. Table 1). Even if 70% of gastric cancer (GC) is associated with Helicobacter pylori infection, this bacterium is not a relevant screening marker. Indeed, only 1–4% of H. pylori infected individuals will eventually develop GC. Recently, two clinical studies explored the correlation between fecal bacteria markers and GC diagnosis/occurrence compared to healthy people [48,49]. Liu et al. interestingly identified Desulfovibrio, Escherichia, Faecalibacterium, or Oscillospira as fecal biomarkers to predict GC with a precision of 90% or above [48]. Moreover, it has been suggested that the gut microbiota could play a role in the carcinogenesis of hepatocellular carcinoma (HCC) through the gut–liver axis [65,66]. Ren et al. [50] collected 419 human samples and found that fecal microbial diversity was increased in early HCC versus cirrhosis. A core set of 30 OTUs was sufficient to identify early HCC. Moreover, clinical studies of oral, fecal, and pancreatic microbiota composition have been reported in relation to pancreatic ductal adenocarcinoma (PDAC) [52,67,68]. Researchers have attempted to identify the fecal microbiota of patients with PDAC when compared to control patients. At the phylum level, a significant increase in Bacteroidetes abundance and a decrease in Firmicutes and Proteobacteria abundances were seen in the fecal microbiota of patients with PDAC versus healthy controls [51]. A recent clinical study provided sequencing data on the gut microbiota composition of esophageal cancer (EC) patients and screened out the optimal potential microbiota biomarkers for EC screening. The authors found that Lachnospira had higher accuracy in EC diagnosis [53].
In addition to these studies on digestive tract cancers, intestinal microbiota dysbiosis has been described in patients with cancer in other locations. Growing evidence has demonstrated a gut microbiota imbalance in breast cancer (BC) [69]. Several studies identified a variable gut microbiota composition in women with BC with elevated levels of Clostridiaceae, Faecalibacterium, and Ruminococcaceae, and a decrease in the levels of Dorea and Lachnospiraceae compared to paired healthy controls [54], confirming that the gut microbiome could be used as a relevant biomarker in BC investigation. Zhu et al. used shotgun metagenomic analysis and showed that postmenopausal women with BC have a different fecal microbiota composition than healthy controls [55]. Moreover, although the role of the lung microbiome in lung cancer (LC) has been investigated, very few studies have evaluated the gut microbiota involvement in LC development. Zhuang et al. [56] compared the gut microbiota of 30 LC patients to 30 healthy controls and found a higher abundance of the bacterial phylum Actinobacteria and genus Bifidobacterium in controls, while patients with LC showed elevated levels of Enterococcus, suggesting their use as possible biomarkers of LC.
Currently, intestinal microbiome biomarkers at the metagenomic or metabolomics level are still not being used in clinical practice for cancer screening. Further prospective investigations are required to confirm whether these microbial biomarkers can successfully identify patients with an increased risk of cancer and provide early intervention.
3. Gut Microbiota Biomarkers Predicting Prognosis and/or Treatment Response
3.1. Biomarker to Predict Cancer Therapy Efficacy
Currently, an emerging approach is to consider the influence of the gut microbiota on cancer therapy efficacy, thereby raising the possibility of using gut microbial community as a predictive biomarker for cancer treatments. Evidence from recent preclinical and clinical studies suggests that the gut microbiota may influence the efficacy of antitumor therapeutic strategies, especially immunotherapy and chemotherapy, as well as radiotherapy and surgery. Specific mechanisms have been reported for digestive cancers, but also for other cancer, mostly through an integration of metagenomic shotgun sequencing and 16S rRNA gene sequencing of patient stool samples [70]. Herein, we review the clinical relevance of observational patient cohorts supporting the use of gut bacteria related biomarkers to predict the response to cancer therapy.
3.1.1. Immunotherapy
A prime example of the gut microbiota influencing therapeutic responses is through immunotherapy. Over the last few years, immunotherapy has revolutionized the therapeutic approach to cancer. Immune checkpoint inhibitors (ICIs), including anti-PD-1 and anti-CTLA-4 have become the most promising treatment, with prolonged responses and improved survival in both solid tumors (for example: melanoma and renal cell carcinoma, non-small cell lung cancer, and mismatch-repair deficient colorectal cancer) and lymphomas [71,72,73,74]. However, immunotherapy only benefits a subset of patients. Preclinical mouse models showed that the gut microbiota modulates tumor response to ICIs [75,76]. Human gut metagenomic analysis revealed that responder patients had different microbiota compositions than non-responders. For example, Chaput et al. [77] revealed, in a cohort of 26 patients with metastatic melanoma treated with ipilimumab (anti-CTLA-4), that patients whose baseline gut microbiota was enriched with Faecalibacterium and other Firmicutes had longer progression-free survival and overall survival. A recent clinical cohort reported that antibiotic administration could inhibit the clinical benefit of anti-PD-1 in patients with metastatic renal or lung cancer. The authors also revealed correlations between clinical responses to anti-PD-1 and the relative fecal abundance of Akkermansia muciniphila in patients at the time of diagnosis [72]. Moreover, in 2018, two studies published in Science confirmed that the gut microbiota composition influences clinical response to anti-PD-1 therapy in melanoma patients [71,73]. Gopalakrishnan et al. [71] used the metagenomics WGS approach on stool sample from 43 metastatic melanoma patients to observe that Fecalibacterium spp. were overrepresented in responder patients while Bacteroides thetaiotaomicron, Escherichia coli, and Anaerotruncus colihominis were enriched in non-responders. Similarly, Matson et al. [73] showed that bacterial species such as Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium were more abundant in the feces from responder patients than non-responders. Both studies demonstrated that the « responder » phenotype could be transferred into germ-free or antibiotic-treated mouse models via fecal microbiota transplant (FMT), leading to a greater efficacy of ICI therapy while FMT from the non-responders profile failed to. More recently, Baruch et al. and Davar et al. provided the first-in-human proof-of-concept evidence for the effectiveness of FMT to affect immunotherapy response in metastatic melanoma patients [78,79]. Based on these published reports, the clinical relevance of the gut microbiota as a novel biomarker of ICI response must be validated in additional prospective clinical studies.
3.1.2. Chemotherapy-Radiotherapy
The Gut microbiota may shape responses to chemo and/or radiotherapy. Alexander et al. have proposed in their review the ‘TIMER’ mechanistic framework (from Translocation, Immunomodulation, Metabolism, Enzymatic degradation, and Reduced diversity and ecological variation) to explain how gut bacteria influence chemotherapy effects on the host [80]. Numerous preclinical studies have demonstrated the involvement of the gut microbiome in the efficacy of a range of chemotherapies such as cyclophosphamide [81], oxaliplatin [74,82] or gemcitabine [83] in CRC and other cancer types. More recently, a metagenome association study in lung cancer patients showed that Streptococcus mutans and Enterococcus casseliflavus were linked to better chemotherapy outcomes [84]. Clinical evidence-based research on the role of potential gut microbial biomarkers for predicting patient’s response to chemotherapy is scarce. In 2020, two Asian cohort studies [85,86] aimed to evaluate the predictive value of the gut microbiome in terms of the response after preoperative concurrent chemoradiation (nCCRT) in patients with rectal cancer. Jang et al. [85], after analyzing 45 fecal samples, showed that Bacteroidales (Bacteroidaceae, Rikenellaceae, Bacteroides) were relatively more abundant in patients with a noncomplete response than in those with complete response. Additionally, the authors found that Duodenibacillus massiliensis was associated with a complete response. Yi et al. [86] identified potential microbial biomarkers for predicting the response to nCCRT in locally advanced rectal cancer: butyrate-producing bacteria, including Roseburia, Dorea, and Anaerostipes were overrepresented in responders, whereas Coriobacteriaceae and Fusobacterium were markedly higher in non-responders. Although gut dysbiosis could be a relevant biomarker to predict radiotherapy-induced mucositis [87,88], the direct impact on the patients’ response to radiotherapy has not yet been investigated yet. Recently Guo et al. described in mice that radiation survivors could be identified by specific bacterial and metabolite profiles (Lachnosspiraceae and Enterococcaceae increases and downstream metabolites such as propionate and tryptophan pathways) [89]. These data suggest that some microbiota-associated markers could be identified to predict radiation injury. Further clinical studies are needed to identify the microbial populations involved in radioresistance.
3.1.3. Surgery
To the best of our knowledge, a key role of gut microbiota in surgical outcomes has mainly been demonstrated in CRC. In particular, anastomotic leaks (AL) constitute the most life-threatening postoperative complications following colorectal procedures (ranging from 2 to 19%) despite a consistent improvement of surgical techniques and perioperative care [90]. The potential role of gut microbiota on the pathogenesis of AL following CRC surgery is increasingly obvious [91,92]. Previous works on animal models have suggested the causal role of specific microorganisms presenting with high collagenolytic properties including Enterococcus faecalis and Pseudomonas aeruginosa [93]. These specific bacterial strains possess high collagenase activity and activate matrix metalloproteinase (MMP-9), thus leading to tissue breakdown and AL through collagen-degrading ability. Shogan et al., found in humans undergoing colonic resection that their anastomotic tissues were still colonized with E. faecalis and other bacterial strains with collagen-degrading/MMP-9-activating properties. However, the available literature reporting the relationship between gut microbiota and AL in human patients with CRC is very scarce. The presence of E. faecalis in the drain fluid of patients with colorectal AL suggests its potential use as a fast screening tool for AL prevention in the early postoperative phase [94]. Based on data from a pilot study [95], Van Praagh et al. investigated the mucosa-associated microbiota composition in AL depending on the placement (or not) of a bioresorbable sheath (C-seal) into the anastomosis [96]. In non-C-seal patients, AL was significantly associated with a lower microbial diversity and a higher abundance of the mucin-degrading microbiome families Lachnospiraceae and Bacteroidaceae compared to matched patients with no AL. The authors indicated that, in non-C-seal patients, a gut microbiota composition consisting of ≥60% Lachnospiraceae and Bacteroidaceae was a predictive factor for AL. More recently, Palmisano et al. [97] demonstrated an increase in preoperative aggressive bacteria Acinetobacter lwoffii and Hafnia alvei and a low abundances of protective bacteria in five patients with AL after colonic resection. All of this clinical evidence strengthened the hypothesis that a peculiar gut microbiota composition could represent a risk factor for AL occurrence and could be a good biomarkers to predict AL. Surgical site infection (SSI: superficial, deep, or organ/space) and postoperative ileus (POI) are other common complications following surgical resection of CRC. Currently, very few clinical studies have linked gut microbiota with SSI or POI-related bacteria in CRC patients after surgery. Only one recent clinical study reported Faecalibacterium in mucosal samples from CRC patients as a possible predictive biomarker of POI [98]. Ohigashi et al. is the only clinical report to identify causative fecal bacteria enriched in SSI after CRC surgery: S. aureus, P. aeruginosa, and Enterococcus spp. [99]. Altogether, further clinical investigations to identify microbial signatures associated with a high risk of postoperative complications following colorectal surgery are needed. Prospective clinical studies are ongoing including our METABIOTE study [100].
3.2. Biomarker Predicting Prognosis and Long-Term Outcomes in CRC
Several studies have also demonstrated that specific microbial markers may serve as a prognostic indicator of cancer survival especially in CRC. The most studied bacteria related to CRC’ patient survival is Fusobacterium nucleatum. Flanagan et al. found a shortened disease-free survival in CRC patients related to higher levels of F. nucleatum [101]. The mucosal amount of F. nucleatum was associated with an advanced stage, proximal tumor location and shorter patient survival by Mima et al. [63]. In addition, a high abundance of F. nucleatum was associated with molecular alterations such as CIMP high, wild-type p53, MSI-high and BRAF mutation in colon tumor tissue [102]. The presence of enterotoxigenic Bacteroides fragilis in the colonic mucosa was also associated with a higher CRC stage. Wei and coworkers [102] concluded that the abundance of F. nucleatum or B. fragilis was a prognostic biomarker of poor survival. Moreover, Bonnet et al. [16] observed an increased level of mucosa-associated and internalized E. coli in CRC compared to normal tissue patients. A relationship between poor CRC prognostic indicators (tumor-node-metastasis stage) and colonization of mucosa by E. coli was reported. Pathogenic E. coli strains producing a genotoxin named « colibactin » (CoPEC) were more prevalent in the mucosa of CRC patients with stages III/IV than stage I tumors. More validations studies are needed before using these biomarkers as prognosis factors in daily clinical practice.
In conclusion, preclinical tumor models and observational cohorts of cancer patients are providing accumulating evidence that the composition of the gut microbiota seems to play a decisive role in the response of cancer patients to anticancer therapeutics such as chemotherapy, radiotherapy or ICIs. These studies mostly used high-throughput bacterial 16S rRNA gene sequencing and metagenomics approaches but using metabolome analysis in this context appears to be a promising method. Future challenges should be to develop interventional approaches by modulating the gut microbiota composition to safely boost the clinical efficacy of these anticancer treatments [103,104]. This new therapeutic strategy named “oncobiotic” is expected to strengthen the oncological arsenal [105,106,107]. Different strategies is developing ad oral administration of live microorganisms (bacteria and/or phages), probiotic, prebiotic, fecal microbiota transfer, or bacterial metabolites. They will be tested alone or in association with classical treatment and be adapted and personalized to the gut dysbiosis determined for each patient [105]. Further clinical research is needed to confirm the use of oncobiotics to target the microbiome and improve cancer treatment.
4. Colibactin-Producing E. coli in the Intestinal Microbiota
Colibactin is a bacterial genotoxin and was first identified in 2006 by Nougayrède et al. in the E. coli meningitis strain IHE3034 [108]. Colibactin-producing E. coli (CoPEC) has been isolated from the intestinal microbiota as commensal bacteria [109,110,111,112,113], in infectious diseases such as septicemia [110,114], newborn meningitis [112], urinary tract infections [106], and colorectal neoplasia tissues [22,115].
Several studies found that CoPEC was more common in the gut microbiota of 55 to 67% of CRC patients, while they were found in 18 to 21% of healthy subjects [17,22,115,116]. A study in Japan did not find an increased prevalence of pks genes in CRC patients (43%) when compared to healthy patients (46%) [117], but in their study, bacterial DNA was isolated by colonic lavage followed by PCR amplification, as opposed to the direct isolation of mucosa-associated E. coli from healthy and CRC patients as reported previously. Recently, a study by Iyadorai et al., reporting lower levels of colonization, confirmed this higher prevalence of CoPEC in CRC patient mucosa [118]. In addition, it was demonstrated that genes in the pks island were significantly more prevalent in tissue than blood, suggesting that E. coli strains expressing this gene are associated with CRC tissues [41]. Our team also reported that colonization of the intestinal mucosa by CoPEC was more prevalent in patients with stage III or IV CRC than in those with stage I CRC, suggesting a possible link with cancer aggressiveness [16]. In this study, we showed that the sex and the age of the patients with colon cancer did not influence the colonization by associated and internalized E. coli. Watanabe et al. observed that the prevalence of CoPEC was positively associated with male sex in healthy Japanese individuals [119]. The correlation between the presence of CoPEC and colorectal tumors has led to intensive research to unveil the possible pro-carcinogenic mechanisms of colibactin.
4.1. Pks Genomic Island and Colibactin Structure
Colibactin is synthesized from the pks genomic island which is a 54-kb nonribosomal peptide synthetase–polyketide synthase (NRPS–PKS) gene cluster that encodes the enzymes responsible for its biosynthesis [120]. Colibactin is a secondary metabolite that undergoes a prodrug activation mechanism. It involves the installation of a structural motif at the N-terminus, which is removed at the final stage of biosynthesis [121,122]. Notably, the pks island was identified in extraintestinal pathogenic E. coli but has also been found in other members of the family Enterobacteriaceae (Klebsiella pneumoniae, Enterobacter aerogenes, and Citrobacter koseri). The pks genomic island phylogeny indicated horizontal acquisition/transmission and the possibility of exchange between compatible E. coli subtypes [123].
Because of its high instability, the structure of colibactin has remained unknown for a long time. Since colibactin cannot be purified directly from a CoPEC culture media, a commonly employed strategy is to compare bacterial metabolites in pks+-E. coli and pks−-E. coli deficient for colibactin production. In pks−-E. coli, precolibactins accumulate, allowing their identification by mass spectrometry. However, the purification and characterization of precolibactins remains a challenge, and the compounds isolated so far have been obtained in extremely small quantities. The structure of colibactin has only been revealed recently. Since it was demonstrated that colibactin-producing bacteria cross-link DNA, the group of Crawford and colleagues used DNA as a probe to isolate colibactin from bacterial cultures and elucidated the structure of the colibactin residue when bound to two nucleobases [124]. Colibactin biosynthesis is not described in this paper since our team and others have extensively reviewed the process previously [125,126].
4.2. Pro-Carcinogenic Activity of CoPEC in CRC
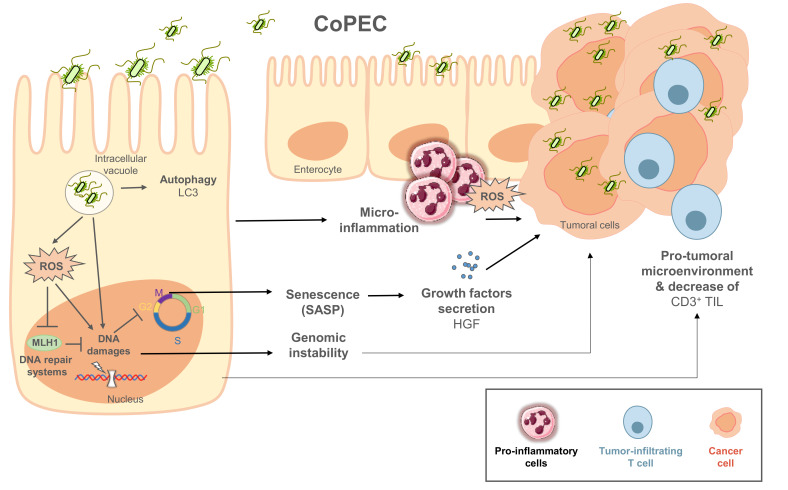
Colibactin has been reported to cause DNA double-strand breaks in vitro [108]. Since this first publication, several teams have reported the induction of DNA double-strand breaks, in vitro and in vivo, as well as chromosomal instability through ROS production and cell cycle arrest, suggesting the potential role of colibactin in CRC development [22,61,108,113,127,128,129,130]. The pro-carcinogenic activity of CoPEC has been demonstrated in several preclinical animal models. CoPEC promoted colorectal cancer formation in multiple intestinal neoplasia (ApcMin/+) mice [16], in AOM-DSS-treated mice [131], in IL-10-deficient (IL-10−/−) mice treated with AOM [22], and in Apcmin/+/IL-10−/− mice [132]. Recently, we showed that autophagy in infected epithelial cell could control the pro-carcinogenic activity of CoPEC [133]. However, the precise mechanisms of colibactin pro-carcinogenic activity is still under investigation (cf. Figure 2).
Figure 2.
Pro-carcinogenic activity of CoPEC in colonic mucosa. Escape of CoPEC from autophagy in infected epithelial cells could lead colibactin to alkylate DNA, and further to cause DNA damage and then cell cycle arrest. In addition, CoPEC induces cellular oxidative stress, leading to inhibition of the DNA repair protein MLH1. All of these mechanisms participate in genomic instability in infected epithelial cells. In addition, CoPEC induced senescence of infected epithelial cells, accompanied by secretion of inflammatory mediators and growth, promoting the proliferation of adjacent uninfected cells. They could also affect the tumor immune microenvironment, even at distances from the tumor site through the reduction of tumor-infiltrating CD3+ T-cells.
Current data support a model in which colibactin alkylates DNA via a cyclopropane ring conjugated to α,β-unsaturated imine, leading to the formation of adenine-colibactin adducts and DNA crosslinks [124,134]. The induction of double-strand breaks has been reported to involve a copper-mediated oxidative cleavage [135]. In addition to DNA damage, CoPEC induces cellular oxidative stress in vitro, leading to decreased expression of DNA repair proteins MLH1 and MSH2, and thus increased genomic instability [61]. Recently, important studies have provided strong evidence regarding the etiological role of colibactin in the first step of human colorectal carcinogenesis [130,136,137]. Dziubańska-Kusibab et al. reported that colibactin-induced DNA double-strand breaks were enriched for an AT-rich hexameric sequence motif associated with distinct DNA-shape characteristics. The exact double-strand-break loci corresponded with mutational hot spots in previously identified cancer genomes [136,138]. In a second work, Iftekhar et al. investigated the transformation potential of a short-term infection with CoPEC using primary murine colon epithelial cells [130]. Infected organoids show characteristics of CRC cells (e.g., increased proliferation, impaired differentiation, several mutations in genes related to p53-signalling pathways). Finally, Pleguezuelos-Manzano et al. identified a unique mutational signature caused by exposure to CoPEC that is enriched in human CRC tumors and metastases [137]. After chronic exposure of human organoid cultures to human CRC-derived CoPEC or the corresponding colibactin-deficient E. coli strain, the authors used WGS on the organoids to show an increase in single-base substitutions and single T deletions at T homopolymers in colibactin-exposed cultures when compared to the mutant. After examination of the WGS datasets, the same mutational signature was detected in a subset of 5876 human cancer genomes from two independent cohorts, predominantly in CRC. Moreover, both mutational signatures were positively correlated with each other, suggesting their common origin. Therefore, to link colibactin-mutational signatures with oncogenic mutations, Clevers and his team showed that colibactin signatures were found in 2.4% of CRC driver mutations, with 5.3% of APC mutations having the highest rate. This molecular fingerprint provides a direct link between colibactin exposure and the DNA damage patterns that drive CRC development [139].
In addition to its genotoxic effect, it was demonstrated that CoPEC infection induces senescence of infected epithelial cells accompanied by the secretion of inflammatory mediators and growth factors (senescence associated secretory phenotype profile-SASP), thus promoting the proliferation of adjacent uninfected cells [131]. Dalmasso et al. demonstrated that cellular senescence was the consequence of the induction of miR-20a-5p expression, which targets SENP1, and leads to alteration of p53 SUMOylation [140]. In addition, CoPEC could also affect the tumor immune microenvironment, even distant from the tumor site. Indeed, we observed in both CRC patients and ApcMin/+ mice colonized by CoPEC a reduced density of tumor-infiltrating CD3+ T-cells [141]. We noticed a significant decrease in antitumor T-cells in the MLNs of CoPEC-infected mice when compared to controls, suggesting that CoPEC could induce a pro-carcinogenic immune microenvironment through a reduction of the antitumor immune system. To our knowledge, the pro-carcinogenic effect of CoPEC has only been studied in CRC. However, the mechanisms described above, for which mediators act at distances from sites of infection, suggest that they could play a promoting role in other types of cancers. In addition to the role of CoPECs in colorectal carcinogenesis, different studies suggest that these bacteria may be diagnostic and prognostic factors in CRC patients.
4.3. CoPEC Detection as a Diagnostic or Prognostic Marker for Colorectal Cancers
Preclinical and clinical studies described that CoPEC has been associated with aggressive CRC and could be a poor prognostic factor. Specifically, a higher E. coli colonization rate and a higher prevalence of CoPEC are found in patients with TNM stage III or IV tumors (UICC TNM Classification, 8th Edition, 2017) [16]. Eklof et al. showed that the prevalence of CoPEC was progressively increased in the adenoma-carcinoma sequence [17]. They described that the presence of the pks island and F. nucleatum detection could predict cancer with a specificity of 63.1% and a sensitivity of 84.6%, suggesting the potential value of these microbial parameters for CRC diagnosis. In addition, we observed that CoPEC was more frequently identified in the microsatellite stable (MSS) CRC phenotype than the MSI (microsatellite instability) phenotype which is associated with better long-term outcomes [61]. Moreover, CoPEC gut colonization might contribute to modulating the immunotherapy efficacy in a preclinical model [141]. Indeed, our study showed that colonization by CoPEC led to a decreased response to anti-PD-1 immunotherapy in MC38-grafted mice. This work is the first to show that CoPEC can influence treatment efficacy. All of these data suggest that the presence of CoPEC in the intestinal microbiota could be a marker of poor prognosis of patients with CRC. In a clinical study, a high level of E. coli was shown in the feces of non-responders versus responders to anti PD-1 treatment of melanoma patients suggesting that E. coli and CoPEC could be predictive markers of cancers outcome [71]. Two clinical studies are in progress to evaluate the prognostic value of CoPEC in CRC and rectal cancer. First, the METABIOTE project evaluates the impact of new prognostic tools such as the gut microbiota, including the CoPEC prevalence and body composition profile, on surgical and oncologic results in a prospective cohort of 300 patients ([100]; ClinicalTrials.gov Identifier: NCT03843905). Second, the MICARE project aims to determine the CoPEC predictive value of a poor response to neoadjuvant treatment of rectal Cancers (ClinicalTrials.gov Identifier: NCT04103567). Extended studies are also required to determine the possible interaction between CoPEC and anticancer therapies in CRC and other cancers.
5. Targeting CoPEC in CRC Therapy
In the field of targeting gut microbiota for colon cancer therapy, two objectives have been distinguished: the first is the anti-tumor effect reducing activity of the pro-carcinogenic microorganisms, while another aspect is the increase of the CRC treatment efficacy eliminating the targeted bacteria. In regard to CoPEC targeting, most of the publications have focused on its anti-tumor effect.
Several strategies have been applied to target colibactin and inhibit its activity. Using docking approach, our team has discovered in 2016 two boron-based compounds that are ligands of the serine peptidase ClbP and prevent the genotoxicity induced by CoPEC in a preclinical mouse model [128]. Extensive additional data regarding the specificity of these compounds and possible side effects on host health are required. Oswald and his team, which first reported the existence of colibactin, reported that high iron intake could decrease the synthesis of colibactin [142,143], suggesting that iron could be a good candidate to regulate CoPEC virulence and carcinogenic factors. However, this observation suggests the complexity of targeting the intake of an essential nutrient with a controversial role in carcinogenesis. They also discovered a new interplay between the synthesis of the genotoxin colibactin and the polyamine spermidine by showing that endogenous spermidine synthase SpeE is required for full genotoxic activity of CoPEC [144]. Since polyamines are abundant in cancer tissue and are associated with cell proliferation, the relationship between spermidine levels in the colonic lumen and the pro-carcinogenic activity of CoPEC has to be investigated in order to find a possible way to inhibit a possible deleterious synergy in patients. The same team recently discovered that polyphosphate kinase (PPK) is required for the promoter activity of clbB, one of the genes in the pks island, and therefore for the production of colibactin [145]. They demonstrated that 5-aminosalicylic acid, a commonly prescribed drug and inhibitor of PPK, can inhibit the activity of the clbB promoter and thus reduce colibactin production, leading to reduced genotoxic activity in HeLa cells. In addition, all these strategies developed to eliminate CoPEC colonization and/or activity could improve the efficacy of CRC therapies. We have recently demonstrated in a preclinical model that gut colonization with a CoPEC strain could strongly reduce the efficiency of anti-PD-1 therapy [141]. Extensive studies are required to explore the role of CoPEC in cancer treatment outcomes and to find new strategies to target their activities to improve therapy efficiency.
The addition of natural compounds to the diet could appear to be an innovative alternative to reduce CoPEC activity. Decreased expression of clbB gene in the presence of the Sub-MIC concentrations of cinnamon essential oil and cinnamaldehyde has been observed in CoPEC isolated from CRC compared to untreated isolates [146]. Extracts of Terminalia catappa, Psidium guajava, and Sandoricum koetjape, as well as their metabolites tannin and quercetin, have been demonstrated to downregulate the expression of several clb genes in a CoPEC strain and protect eukaryotic epithelial cells from infection and DNA damage in vitro [147]. In a clinical study performed on healthy Japanese individuals, the prevalence of CoPEC was negatively associated with the intake of green tea [119]. Recently, impact of prebiotics (inulin and galacto-saccharide) on the genotoxicity of CoPEC, was studied in vitro. Unexpected results were obtained with an increase of DNA damage induced by the CoPEC after the treatment [148]. Preclinical studies are needed to confirm these data. Even if probiotic and/or an oncobiotic strategy has not yet been evaluated in vivo to reduce CoPEC colonization and/or activity, it could be an interesting approach in combination with CRC reference therapies.
These works highlight several promising approaches to reduce CoPEC activity in CRC patients harboring these bacteria and achieve promising outcomes in patient therapy. Preclinical and clinical studies will be necessary to develop and validate these innovative therapies targeting the CoPEC. With the current data, it is likely that the first clinical application of CoPEC detection will be the search for these bacteria in the feces as predictive biomarkers in order to better monitor CRC patients after treatment and prevent recurrences.
6. Conclusions
Among the multiple factors that may eventually result in carcinogenesis, gut microbiota dysbiosis has been described with increased attention. The gut microbiota could then lead to innovative and curative medical proposals for CRC and other cancers in addition to existing methods. Among these microbial factors, CoPEC emerges as an effective therapeutic target and predictive marker for CRC prognosis and long-term outcomes.
Acknowledgments
The authors acknowledge Medical Art Servier.
Author Contributions
Writing—original draft preparation, J.V., R.V. and M.B.; Writing—review and editing, J.V., R.V., N.B. and M.B.; Supervision, N.B. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge the support received from the Ministère de la Recherche et de la Technologie, Inserm and Université Clermont-Auvergne (UMR1071), INRAE (USC-2018), the CPER 2016, La ligue contre le cancer 63/03 and by the French government IDEX-ISITE initiative (Grant number: 16-IDEX-0001-CAP 20-25) of the University of Clermont Auvergne.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shanahan F., Ghosh T.S., O’Toole P.W. The Healthy Microbiome-What Is the Definition of a Healthy Gut Microbiome? Gastroenterology. 2021;160:483–494. doi: 10.1053/j.gastro.2020.09.057. [DOI] [PubMed] [Google Scholar]
- 2.Thursby E., Juge N. Introduction to the Human Gut Microbiota. Biochem. J. 2017;474:1823–1836. doi: 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tremaroli V., Bäckhed F. Functional Interactions between the Gut Microbiota and Host Metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 4.Garrett W.S., Gordon J.I., Glimcher L.H. Homeostasis and Inflammation in the Intestine. Cell. 2010;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human Nutrition, the Gut Microbiome and the Immune System. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwabe R.F., Jobin C. The Microbiome and Cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y.-J., Li S., Gan R.-Y., Zhou T., Xu D.-P., Li H.-B. Impacts of Gut Bacteria on Human Health and Diseases. Int. J. Mol. Sci. 2015;16:7493–7519. doi: 10.3390/ijms16047493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helmink B.A., Khan M.A.W., Hermann A., Gopalakrishnan V., Wargo J.A. The Microbiome, Cancer, and Cancer Therapy. Nat. Med. 2019;25:377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 9.Boursi B., Mamtani R., Haynes K., Yang Y.-X. Recurrent Antibiotic Exposure May Promote Cancer Formation--Another Step in Understanding the Role of the Human Microbiota? Eur. J. Cancer. 2015;51:2655–2664. doi: 10.1016/j.ejca.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy B.S., Weisburger J.H., Narisawa T., Wynder E.L. Colon Carcinogenesis in Germ-Free Rats with 1,2-Dimethylhydrazine and N-Methyl-n’-Nitro-N-Nitrosoguanidine. Cancer Res. 1974;34:2368–2372. [PubMed] [Google Scholar]
- 11.Li Y., Kundu P., Seow S.W., de Matos C.T., Aronsson L., Chin K.C., Kärre K., Pettersson S., Greicius G. Gut Microbiota Accelerate Tumor Growth via C-Jun and STAT3 Phosphorylation in APCMin/+ Mice. Carcinogenesis. 2012;33:1231–1238. doi: 10.1093/carcin/bgs137. [DOI] [PubMed] [Google Scholar]
- 12.Wang X., Yang Y., Huycke M.M. Microbiome-Driven Carcinogenesis in Colorectal Cancer: Models and Mechanisms. Free Radic. Biol. Med. 2017;105:3–15. doi: 10.1016/j.freeradbiomed.2016.10.504. [DOI] [PubMed] [Google Scholar]
- 13.Wong S.H., Yu J. Gut Microbiota in Colorectal Cancer: Mechanisms of Action and Clinical Applications. Nat. Rev. Gastroenterol. Hepatol. 2019;16:690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 14.Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J. Meta-Analysis of Gut Microbiome Studies Identifies Disease-Specific and Shared Responses. Nat. Commun. 2017;8:1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saus E., Iraola-Guzmán S., Willis J.R., Brunet-Vega A., Gabaldón T. Microbiome and Colorectal Cancer: Roles in Carcinogenesis and Clinical Potential. Mol. Aspects Med. 2019;69:93–106. doi: 10.1016/j.mam.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonnet M., Buc E., Sauvanet P., Darcha C., Dubois D., Pereira B., Déchelotte P., Bonnet R., Pezet D., Darfeuille-Michaud A. Colonization of the Human Gut by E. Coli and Colorectal Cancer Risk. Clin. Cancer Res. 2014;20:859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 17.Eklöf V., Löfgren-Burström A., Zingmark C., Edin S., Larsson P., Karling P., Alexeyev O., Rutegård J., Wikberg M.L., Palmqvist R. Cancer-Associated Fecal Microbial Markers in Colorectal Cancer Detection. Int. J. Cancer. 2017;141:2528–2536. doi: 10.1002/ijc.31011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 19.Imperiale T.F., Gruber R.N., Stump T.E., Emmett T.W., Monahan P.O. Performance Characteristics of Fecal Immunochemical Tests for Colorectal Cancer and Advanced Adenomatous Polyps: A Systematic Review and Meta-Analysis. Ann. Intern. Med. 2019;170:319–329. doi: 10.7326/M18-2390. [DOI] [PubMed] [Google Scholar]
- 20.Kisiel J.B., Limburg P.J. Colorectal Cancer Screening With the Multitarget Stool DNA Test. Am. J. Gastroenterol. 2020;115:1737–1740. doi: 10.14309/ajg.0000000000000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobhani I., Tap J., Roudot-Thoraval F., Roperch J.P., Letulle S., Langella P., Corthier G., Tran Van Nhieu J., Furet J.P. Microbial Dysbiosis in Colorectal Cancer (CRC) Patients. PLoS ONE. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arthur J.C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J.M., Fan T.-J., Campbell B.J., Abujamel T., Dogan B., Rogers A.B., et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balamurugan R., Rajendiran E., George S., Samuel G.V., Ramakrishna B.S. Real-Time Polymerase Chain Reaction Quantification of Specific Butyrate-Producing Bacteria, Desulfovibrio and Enterococcus Faecalis in the Feces of Patients with Colorectal Cancer. J. Gastroenterol. Hepatol. 2008;23:1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- 24.Abdulamir A.S., Hafidh R.R., Bakar F.A. Molecular Detection, Quantification, and Isolation of Streptococcus Gallolyticus Bacteria Colonizing Colorectal Tumors: Inflammation-Driven Potential of Carcinogenesis via IL-1, COX-2, and IL-8. Mol. Cancer. 2010;9:249. doi: 10.1186/1476-4598-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Löwenmark T., Löfgren-Burström A., Zingmark C., Eklöf V., Dahlberg M., Wai S.N., Larsson P., Ljuslinder I., Edin S., Palmqvist R. Parvimonas Micra as a Putative Non-Invasive Faecal Biomarker for Colorectal Cancer. Sci. Rep. 2020;10:15250. doi: 10.1038/s41598-020-72132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C., Nielsen T., Pons N., Levenez F., Yamada T., et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang Q., Chiu J., Chen Y., Huang Y., Higashimori A., Fang J., Brim H., Ashktorab H., Ng S.C., Ng S.S.M., et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin. Cancer. Res. 2017;23:2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 28.Ternes D., Karta J., Tsenkova M., Wilmes P., Haan S., Letellier E. Microbiome in Colorectal Cancer: How to Get from Meta-Omics to Mechanism? Trends Microbiol. 2020;28:401–423. doi: 10.1016/j.tim.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Yachida S., Mizutani S., Shiroma H., Shiba S., Nakajima T., Sakamoto T., Watanabe H., Masuda K., Nishimoto Y., Kubo M., et al. Metagenomic and Metabolomic Analyses Reveal Distinct Stage-Specific Phenotypes of the Gut Microbiota in Colorectal Cancer. Nat. Med. 2019;25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 30.Yu J., Feng Q., Wong S.H., Zhang D., Liang Q.Y., Qin Y., Tang L., Zhao H., Stenvang J., Li Y., et al. Metagenomic Analysis of Faecal Microbiome as a Tool towards Targeted Non-Invasive Biomarkers for Colorectal Cancer. Gut. 2017;66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 31.Dai Z., Coker O.O., Nakatsu G., Wu W.K.K., Zhao L., Chen Z., Chan F.K.L., Kristiansen K., Sung J.J.Y., Wong S.H., et al. Multi-Cohort Analysis of Colorectal Cancer Metagenome Identified Altered Bacteria across Populations and Universal Bacterial Markers. Microbiome. 2018;6:70. doi: 10.1186/s40168-018-0451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng Q., Liang S., Jia H., Stadlmayr A., Tang L., Lan Z., Zhang D., Xia H., Xu X., Jie Z., et al. Gut Microbiome Development along the Colorectal Adenoma-Carcinoma Sequence. Nat. Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 33.Baxter N.T., Ruffin M.T., Rogers M.A.M., Schloss P.D. Microbiota-Based Model Improves the Sensitivity of Fecal Immunochemical Test for Detecting Colonic Lesions. Genome Med. 2016;8:37. doi: 10.1186/s13073-016-0290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeller G., Tap J., Voigt A.Y., Sunagawa S., Kultima J.R., Costea P.I., Amiot A., Böhm J., Brunetti F., Habermann N., et al. Potential of Fecal Microbiota for Early-Stage Detection of Colorectal Cancer. Mol. Syst. Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirbel J., Pyl P.T., Kartal E., Zych K., Kashani A., Milanese A., Fleck J.S., Voigt A.Y., Palleja A., Ponnudurai R., et al. Meta-Analysis of Fecal Metagenomes Reveals Global Microbial Signatures That Are Specific for Colorectal Cancer. Nat. Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osman M.A., Neoh H.-M., Ab Mutalib N.-S., Chin S.-F., Mazlan L., Raja Ali R.A., Zakaria A.D., Ngiu C.S., Ang M.Y., Jamal R. Parvimonas Micra, Peptostreptococcus Stomatis, Fusobacterium Nucleatum and Akkermansia Muciniphila as a Four-Bacteria Biomarker Panel of Colorectal Cancer. Sci. Rep. 2021;11:2925. doi: 10.1038/s41598-021-82465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas A.M., Manghi P., Asnicar F., Pasolli E., Armanini F., Zolfo M., Beghini F., Manara S., Karcher N., Pozzi C., et al. Metagenomic Analysis of Colorectal Cancer Datasets Identifies Cross-Cohort Microbial Diagnostic Signatures and a Link with Choline Degradation. Nat. Med. 2019;25:667–678. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X., Zhu X., Cao Y., Fang J.-Y., Hong J., Chen H. Fecal Fusobacterium Nucleatum for the Diagnosis of Colorectal Tumor: A Systematic Review and Meta-Analysis. Cancer Med. 2019;8:480–491. doi: 10.1002/cam4.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong S.H., Kwong T.N.Y., Chow T.-C., Luk A.K.C., Dai R.Z.W., Nakatsu G., Lam T.Y.T., Zhang L., Wu J.C.Y., Chan F.K.L., et al. Quantitation of Faecal Fusobacterium Improves Faecal Immunochemical Test in Detecting Advanced Colorectal Neoplasia. Gut. 2017;66:1441–1448. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding T., Schloss P.D. Dynamics and Associations of Microbial Community Types across the Human Body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dohlman A.B., Arguijo Mendoza D., Ding S., Gao M., Dressman H., Iliev I.D., Lipkin S.M., Shen X. The Cancer Microbiome Atlas: A Pan-Cancer Comparative Analysis to Distinguish Tissue-Resident Microbiota from Contaminants. Cell Host Microbe. 2021;29:281–298.e5. doi: 10.1016/j.chom.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nannini G., Meoni G., Amedei A., Tenori L. Metabolomics Profile in Gastrointestinal Cancers: Update and Future Perspectives. World J. Gastroenterol. 2020;26:2514–2532. doi: 10.3748/wjg.v26.i20.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monleón D., Morales J.M., Barrasa A., López J.A., Vázquez C., Celda B. Metabolite Profiling of Fecal Water Extracts from Human Colorectal Cancer. NMR Biomed. 2009;22:342–348. doi: 10.1002/nbm.1345. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y., Ma C., Bezabeh T., Wang Z., Liang J., Huang Y., Zhao J., Liu X., Ye W., Tang W., et al. 1 H NMR-Based Metabolomics Reveal Overlapping Discriminatory Metabolites and Metabolic Pathway Disturbances between Colorectal Tumor Tissues and Fecal Samples. Int. J. Cancer. 2019;145:1679–1689. doi: 10.1002/ijc.32190. [DOI] [PubMed] [Google Scholar]
- 45.Lin Y., Ma C., Liu C., Wang Z., Yang J., Liu X., Shen Z., Wu R. NMR-Based Fecal Metabolomics Fingerprinting as Predictors of Earlier Diagnosis in Patients with Colorectal Cancer. Oncotarget. 2016;7:29454–29464. doi: 10.18632/oncotarget.8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le Gall G., Guttula K., Kellingray L., Tett A.J., Ten Hoopen R., Kemsley E.K., Savva G.M., Ibrahim A., Narbad A. Metabolite Quantification of Faecal Extracts from Colorectal Cancer Patients and Healthy Controls. Oncotarget. 2018;9:33278–33289. doi: 10.18632/oncotarget.26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W., Liu F., Ling Z., Tong X., Xiang C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients with Colorectal Cancer. PLoS ONE. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S., Dai J., Lan X., Fan B., Dong T., Zhang Y., Han M. Intestinal Bacteria Are Potential Biomarkers and Therapeutic Targets for Gastric Cancer. Microb. Pathog. 2021;151:104747. doi: 10.1016/j.micpath.2021.104747. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., Shen J., Shi X., Du Y., Niu Y., Jin G., Wang Z., Lyu J. Gut Microbiome Analysis as a Predictive Marker for the Gastric Cancer Patients. Appl. Microbiol. Biotechnol. 2021;105:803–814. doi: 10.1007/s00253-020-11043-7. [DOI] [PubMed] [Google Scholar]
- 50.Ren Z., Li A., Jiang J., Zhou L., Yu Z., Lu H., Xie H., Chen X., Shao L., Zhang R., et al. Gut Microbiome Analysis as a Tool towards Targeted Non-Invasive Biomarkers for Early Hepatocellular Carcinoma. Gut. 2019;68:1014–1023. doi: 10.1136/gutjnl-2017-315084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren Z., Jiang J., Xie H., Li A., Lu H., Xu S., Zhou L., Zhang H., Cui G., Chen X., et al. Gut Microbial Profile Analysis by MiSeq Sequencing of Pancreatic Carcinoma Patients in China. Oncotarget. 2017;8:95176–95191. doi: 10.18632/oncotarget.18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Half E., Keren N., Reshef L., Dorfman T., Lachter I., Kluger Y., Reshef N., Knobler H., Maor Y., Stein A., et al. Fecal Microbiome Signatures of Pancreatic Cancer Patients. Sci. Rep. 2019;9:16801. doi: 10.1038/s41598-019-53041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng Y., Tang D., Hou P., Shen W., Li H., Wang T., Liu R. Dysbiosis of Gut Microbiota in Patients with Esophageal Cancer. Microb Pathog. 2021;150:104709. doi: 10.1016/j.micpath.2020.104709. [DOI] [PubMed] [Google Scholar]
- 54.Goedert J.J., Jones G., Hua X., Xu X., Yu G., Flores R., Falk R.T., Gail M.H., Shi J., Ravel J., et al. Investigation of the Association between the Fecal Microbiota and Breast Cancer in Postmenopausal Women: A Population-Based Case-Control Pilot Study. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/djv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu J., Liao M., Yao Z., Liang W., Li Q., Liu J., Yang H., Ji Y., Wei W., Tan A., et al. Breast Cancer in Postmenopausal Women Is Associated with an Altered Gut Metagenome. Microbiome. 2018;6:136. doi: 10.1186/s40168-018-0515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhuang H., Cheng L., Wang Y., Zhang Y.-K., Zhao M.-F., Liang G.-D., Zhang M.-C., Li Y.-G., Zhao J.-B., Gao Y.-N., et al. Dysbiosis of the Gut Microbiome in Lung Cancer. Front. Cell Infect Microbiol. 2019;9:112. doi: 10.3389/fcimb.2019.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mima K., Cao Y., Chan A.T., Qian Z.R., Nowak J.A., Masugi Y., Shi Y., Song M., da Silva A., Gu M., et al. Fusobacterium Nucleatum in Colorectal Carcinoma Tissue According to Tumor Location. Clin. Transl. Gastroenterol. 2016;7:e200. doi: 10.1038/ctg.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Viljoen K.S., Dakshinamurthy A., Goldberg P., Blackburn J.M. Quantitative Profiling of Colorectal Cancer-Associated Bacteria Reveals Associations between Fusobacterium Spp., Enterotoxigenic Bacteroides Fragilis (ETBF) and Clinicopathological Features of Colorectal Cancer. PLoS ONE. 2015;10:e0119462. doi: 10.1371/journal.pone.0119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim K., Castro E.J.T., Shim H., Advincula J.V.G., Kim Y.-W. Differences Regarding the Molecular Features and Gut Microbiota Between Right and Left Colon Cancer. Ann. Coloproctol. 2018;34:280–285. doi: 10.3393/ac.2018.12.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richard M.L., Liguori G., Lamas B., Brandi G., da Costa G., Hoffmann T.W., Pierluigi Di Simone M., Calabrese C., Poggioli G., Langella P., et al. Mucosa-Associated Microbiota Dysbiosis in Colitis Associated Cancer. Gut Microbes. 2018;9:131–142. doi: 10.1080/19490976.2017.1379637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gagnière J., Bonnin V., Jarrousse A.-S., Cardamone E., Agus A., Uhrhammer N., Sauvanet P., Déchelotte P., Barnich N., Bonnet R., et al. Interactions between Microsatellite Instability and Human Gut Colonization by Escherichia Coli in Colorectal Cancer. Clin. Sci. (Lond.) 2017;131:471–485. doi: 10.1042/CS20160876. [DOI] [PubMed] [Google Scholar]
- 62.Karakas Y., Dizdar O. Tumor Sidedness and Prognosis in Colorectal Cancer: Is Microbiome the Missing Link? JAMA Oncol. 2017;3:1000. doi: 10.1001/jamaoncol.2017.0034. [DOI] [PubMed] [Google Scholar]
- 63.Mima K., Nishihara R., Qian Z.R., Cao Y., Sukawa Y., Nowak J.A., Yang J., Dou R., Masugi Y., Song M., et al. Fusobacterium Nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flemer B., Lynch D.B., Brown J.M.R., Jeffery I.B., Ryan F.J., Claesson M.J., O’Riordain M., Shanahan F., O’Toole P.W. Tumour-Associated and Non-Tumour-Associated Microbiota in Colorectal Cancer. Gut. 2017;66:633–643. doi: 10.1136/gutjnl-2015-309595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta H., Youn G.S., Shin M.J., Suk K.T. Role of Gut Microbiota in Hepatocarcinogenesis. Microorganisms. 2019;7:121. doi: 10.3390/microorganisms7050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou A., Tang L., Zeng S., Lei Y., Yang S., Tang B. Gut Microbiota: A New Piece in Understanding Hepatocarcinogenesis. Cancer Lett. 2020;474:15–22. doi: 10.1016/j.canlet.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 67.Thomas R.M., Jobin C. Microbiota in Pancreatic Health and Disease: The next Frontier in Microbiome Research. Nat. Rev. Gastroenterol. Hepatol. 2020;17:53–64. doi: 10.1038/s41575-019-0242-7. [DOI] [PubMed] [Google Scholar]
- 68.Li Q., Jin M., Liu Y., Jin L. Gut Microbiota: Its Potential Roles in Pancreatic Cancer. Front. Cell Infect. Microbiol. 2020;10:572492. doi: 10.3389/fcimb.2020.572492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laborda-Illanes A., Sanchez-Alcoholado L., Dominguez-Recio M.E., Jimenez-Rodriguez B., Lavado R., Comino-Méndez I., Alba E., Queipo-Ortuño M.I. Breast and Gut Microbiota Action Mechanisms in Breast Cancer Pathogenesis and Treatment. Cancers. 2020;12:2465. doi: 10.3390/cancers12092465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Villéger R., Lopès A., Carrier G., Veziant J., Billard E., Barnich N., Gagnière J., Vazeille E., Bonnet M. Intestinal Microbiota: A Novel Target to Improve Anti-Tumor Treatment? Int. J. Mol. Sci. 2019;20:4584. doi: 10.3390/ijms20184584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gopalakrishnan V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., et al. Gut Microbiome Modulates Response to Anti-PD-1 Immunotherapy in Melanoma Patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Routy B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy against Epithelial Tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 73.Matson V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.-L., Luke J.J., Gajewski T.F. The Commensal Microbiome Is Associated with Anti-PD-1 Efficacy in Metastatic Melanoma Patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koulouridi A., Messaritakis I., Gouvas N., Tsiaoussis J., Souglakos J. Immunotherapy in Solid Tumors and Gut Microbiota: The Correlation-A Special Reference to Colorectal Cancer. Cancers. 2020;13:43. doi: 10.3390/cancers13010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sivan A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.-L., et al. Commensal Bifidobacterium Promotes Antitumor Immunity and Facilitates Anti-PD-L1 Efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vétizou M., Pitt J.M., Daillère R., Lepage P., Waldschmitt N., Flament C., Rusakiewicz S., Routy B., Roberti M.P., Duong C.P.M., et al. Anticancer Immunotherapy by CTLA-4 Blockade Relies on the Gut Microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chaput N., Lepage P., Coutzac C., Soularue E., Le Roux K., Monot C., Boselli L., Routier E., Cassard L., Collins M., et al. Baseline Gut Microbiota Predicts Clinical Response and Colitis in Metastatic Melanoma Patients Treated with Ipilimumab. Ann. Oncol. 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 78.Davar D., Dzutsev A.K., McCulloch J.A., Rodrigues R.R., Chauvin J.-M., Morrison R.M., Deblasio R.N., Menna C., Ding Q., Pagliano O., et al. Fecal Microbiota Transplant Overcomes Resistance to Anti-PD-1 Therapy in Melanoma Patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baruch E.N., Youngster I., Ben-Betzalel G., Ortenberg R., Lahat A., Katz L., Adler K., Dick-Necula D., Raskin S., Bloch N., et al. Fecal Microbiota Transplant Promotes Response in Immunotherapy-Refractory Melanoma Patients. Science. 2021;371:602–609. doi: 10.1126/science.abb5920. [DOI] [PubMed] [Google Scholar]
- 80.Alexander J.L., Wilson I.D., Teare J., Marchesi J.R., Nicholson J.K., Kinross J.M. Gut Microbiota Modulation of Chemotherapy Efficacy and Toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017;14:356–365. doi: 10.1038/nrgastro.2017.20. [DOI] [PubMed] [Google Scholar]
- 81.Viaud S., Saccheri F., Mignot G., Yamazaki T., Daillère R., Hannani D., Enot D.P., Pfirschke C., Engblom C., Pittet M.J., et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A., Molina D.A., Salcedo R., Back T., Cramer S., et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geller L.T., Barzily-Rokni M., Danino T., Jonas O.H., Shental N., Nejman D., Gavert N., Zwang Y., Cooper Z.A., Shee K., et al. Potential Role of Intratumor Bacteria in Mediating Tumor Resistance to the Chemotherapeutic Drug Gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao Z., Fei K., Bai H., Wang Z., Duan J., Wang J. Metagenome Association Study of the Gut Microbiome Revealed Biomarkers Linked to Chemotherapy Outcomes in Locally Advanced and Advanced Lung Cancer. Thorac. Cancer. 2021;12:66–78. doi: 10.1111/1759-7714.13711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jang B.-S., Chang J.H., Chie E.K., Kim K., Park J.W., Kim M.J., Song E.-J., Nam Y.-D., Kang S.W., Jeong S.-Y., et al. Gut Microbiome Composition Is Associated with a Pathologic Response After Preoperative Chemoradiation in Patients with Rectal Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2020;107:736–746. doi: 10.1016/j.ijrobp.2020.04.015. [DOI] [PubMed] [Google Scholar]
- 86.Yi Y., Shen L., Shi W., Xia F., Zhang H., Wang Y., Zhang J., Wang Y., Sun X., Zhang Z., et al. Gut Microbiome Components Predict Response to Neoadjuvant Chemoradiotherapy in Patients with Locally Advanced Rectal Cancer: A Prospective, Longitudinal Study. Clin. Cancer Res. 2020 doi: 10.1158/1078-0432.CCR-20-3445. [DOI] [PubMed] [Google Scholar]
- 87.Al-Qadami G., Sebille Y.V., Le H., Bowen J. Gut Microbiota: Implications for Radiotherapy Response and Radiotherapy-Induced Mucositis. Expert Rev. Gastroenterol. Hepatol. 2019;13:485–496. doi: 10.1080/17474124.2019.1595586. [DOI] [PubMed] [Google Scholar]
- 88.Liu J., Liu C., Yue J. Radiotherapy and the Gut Microbiome: Facts and Fiction. Radiat. Oncol. 2021;16:9. doi: 10.1186/s13014-020-01735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo H., Chou W.-C., Lai Y., Liang K., Tam J.W., Brickey W.J., Chen L., Montgomery N.D., Li X., Bohannon L.M., et al. Multi-Omics Analyses of Radiation Survivors Identify Radioprotective Microbes and Metabolites. Science. 2020;370 doi: 10.1126/science.aay9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stormark K., Krarup P.-M., Sjövall A., Søreide K., Kvaløy J.T., Nordholm-Carstensen A., Nedrebø B.S., Kørner H. Anastomotic Leak after Surgery for Colon Cancer and Effect on Long-Term Survival. Colorectal. Dis. 2020;22:1108–1118. doi: 10.1111/codi.14999. [DOI] [PubMed] [Google Scholar]
- 91.Koliarakis I., Athanasakis E., Sgantzos M., Mariolis-Sapsakos T., Xynos E., Chrysos E., Souglakos J., Tsiaoussis J. Intestinal Microbiota in Colorectal Cancer Surgery. Cancers. 2020;12:3011. doi: 10.3390/cancers12103011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bartolini I., Risaliti M., Ringressi M.N., Melli F., Nannini G., Amedei A., Muiesan P., Taddei A. Role of Gut Microbiota-Immunity Axis in Patients Undergoing Surgery for Colorectal Cancer: Focus on Short and Long-Term Outcomes. World J. Gastroenterol. 2020;26:2498–2513. doi: 10.3748/wjg.v26.i20.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shogan B.D., Belogortseva N., Luong P.M., Zaborin A., Lax S., Bethel C., Ward M., Muldoon J.P., Singer M., An G., et al. Collagen Degradation and MMP9 Activation by Enterococcus Faecalis Contribute to Intestinal Anastomotic Leak. Sci. Transl. Med. 2015;7:286ra68. doi: 10.1126/scitranslmed.3010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Komen N., Slieker J., Willemsen P., Mannaerts G., Pattyn P., Karsten T., de Wilt H., van der Harst E., van Leeuwen W., Decaestecker C., et al. Polymerase Chain Reaction for Enterococcus Faecalis in Drain Fluid: The First Screening Test for Symptomatic Colorectal Anastomotic Leakage. The Appeal-Study: Analysis of Parameters Predictive for Evident Anastomotic Leakage. Int. J. Colorectal. Dis. 2014;29:15–21. doi: 10.1007/s00384-013-1776-8. [DOI] [PubMed] [Google Scholar]
- 95.van Praagh J.B., de Goffau M.C., Bakker I.S., Harmsen H.J.M., Olinga P., Havenga K. Intestinal Microbiota and Anastomotic Leakage of Stapled Colorectal Anastomoses: A Pilot Study. Surg. Endosc. 2016;30:2259–2265. doi: 10.1007/s00464-015-4508-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Praagh J.B., de Goffau M.C., Bakker I.S., van Goor H., Harmsen H.J.M., Olinga P., Havenga K. Mucus Microbiome of Anastomotic Tissue During Surgery Has Predictive Value for Colorectal Anastomotic Leakage. Ann. Surg. 2019;269:911–916. doi: 10.1097/SLA.0000000000002651. [DOI] [PubMed] [Google Scholar]
- 97.Palmisano S., Campisciano G., Iacuzzo C., Bonadio L., Zucca A., Cosola D., Comar M., de Manzini N. Role of Preoperative Gut Microbiota on Colorectal Anastomotic Leakage: Preliminary Results. Updates Surg. 2020;72:1013–1022. doi: 10.1007/s13304-020-00720-x. [DOI] [PubMed] [Google Scholar]
- 98.Jin Y., Geng R., Liu Y., Liu L., Jin X., Zhao F., Feng J., Wei Y. Prediction of Postoperative Ileus in Patients With Colorectal Cancer by Preoperative Gut Microbiota. Front. Oncol. 2020;10:526009. doi: 10.3389/fonc.2020.526009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ohigashi S., Sudo K., Kobayashi D., Takahashi T., Nomoto K., Onodera H. Significant Changes in the Intestinal Environment after Surgery in Patients with Colorectal Cancer. J. Gastrointest. Surg. 2013;17:1657–1664. doi: 10.1007/s11605-013-2270-x. [DOI] [PubMed] [Google Scholar]
- 100.Veziant J., Poirot K., Chevarin C., Cassagnes L., Sauvanet P., Chassaing B., Robin F., Godfraind C., Barnich N., Pezet D., et al. Prognostic Value of a Combination of Innovative Factors (Gut Microbiota, Sarcopenia, Obesity, Metabolic Syndrome) to Predict Surgical/Oncologic Outcomes Following Surgery for Sporadic Colorectal Cancer: A Prospective Cohort Study Protocol (METABIOTE) BMJ Open. 2020;10:e031472. doi: 10.1136/bmjopen-2019-031472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Flanagan L., Schmid J., Ebert M., Soucek P., Kunicka T., Liska V., Bruha J., Neary P., Dezeeuw N., Tommasino M., et al. Fusobacterium Nucleatum Associates with Stages of Colorectal Neoplasia Development, Colorectal Cancer and Disease Outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 102.Wei Z., Cao S., Liu S., Yao Z., Sun T., Li Y., Li J., Zhang D., Zhou Y. Could Gut Microbiota Serve as Prognostic Biomarker Associated with Colorectal Cancer Patients’ Survival? A Pilot Study on Relevant Mechanism. Oncotarget. 2016;7:46158–46172. doi: 10.18632/oncotarget.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Daillère R., Derosa L., Bonvalet M., Segata N., Routy B., Gariboldi M., Budinská E., De Vries I.J.M., Naccarati A.G., Zitvogel V., et al. Trial Watch: The Gut Microbiota as a Tool to Boost the Clinical Efficacy of Anticancer Immunotherapy. Oncoimmunology. 2020;9:1774298. doi: 10.1080/2162402X.2020.1774298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Woelk C.H., Snyder A. Modulating Gut Microbiota to Treat Cancer. Science. 2021;371:573–574. doi: 10.1126/science.abg2904. [DOI] [PubMed] [Google Scholar]
- 105.Grajeda-Iglesias C., Durand S., Daillère R., Iribarren K., Lemaitre F., Derosa L., Aprahamian F., Bossut N., Nirmalathasan N., Madeo F., et al. Oral Administration of Akkermansia Muciniphila Elevates Systemic Antiaging and Anticancer Metabolites. Aging (Albany NY) 2021;13 doi: 10.18632/aging.202739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zitvogel L., Daillère R., Roberti M.P., Routy B., Kroemer G. Anticancer Effects of the Microbiome and Its Products. Nat. Rev. Microbiol. 2017;15:465–478. doi: 10.1038/nrmicro.2017.44. [DOI] [PubMed] [Google Scholar]
- 107.Zitvogel L., Ma Y., Raoult D., Kroemer G., Gajewski T.F. The Microbiome in Cancer Immunotherapy: Diagnostic Tools and Therapeutic Strategies. Science. 2018;359:1366–1370. doi: 10.1126/science.aar6918. [DOI] [PubMed] [Google Scholar]
- 108.Nougayrède J.-P., Homburg S., Taieb F., Boury M., Brzuszkiewicz E., Gottschalk G., Buchrieser C., Hacker J., Dobrindt U., Oswald E. Escherichia Coli Induces DNA Double-Strand Breaks in Eukaryotic Cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 109.Dubois D., Delmas J., Cady A., Robin F., Sivignon A., Oswald E., Bonnet R. Cyclomodulins in Urosepsis Strains of Escherichia Coli. J. Clin. Microbiol. 2010;48:2122–2129. doi: 10.1128/JCM.02365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnson J.R., Johnston B., Kuskowski M.A., Nougayrede J.-P., Oswald E. Molecular Epidemiology and Phylogenetic Distribution of the Escherichia Coli Pks Genomic Island. J. Clin. Microbiol. 2008;46:3906–3911. doi: 10.1128/JCM.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Putze J., Hennequin C., Nougayrède J.-P., Zhang W., Homburg S., Karch H., Bringer M.-A., Fayolle C., Carniel E., Rabsch W., et al. Genetic Structure and Distribution of the Colibactin Genomic Island among Members of the Family Enterobacteriaceae. Infect Immun. 2009;77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nowrouzian F.L., Oswald E. Escherichia Coli Strains with the Capacity for Long-Term Persistence in the Bowel Microbiota Carry the Potentially Genotoxic Pks Island. Microb. Pathog. 2012;53:180–182. doi: 10.1016/j.micpath.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 113.Payros D., Secher T., Boury M., Brehin C., Ménard S., Salvador-Cartier C., Cuevas-Ramos G., Watrin C., Marcq I., Nougayrède J.-P., et al. Maternally Acquired Genotoxic Escherichia Coli Alters Offspring’s Intestinal Homeostasis. Gut Microbes. 2014;5:313–325. doi: 10.4161/gmic.28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Micenková L., Beňová A., Frankovičová L., Bosák J., Vrba M., Ševčíková A., Kmeťová M., Šmajs D. Human Escherichia Coli Isolates from Hemocultures: Septicemia Linked to Urogenital Tract Infections Is Caused by Isolates Harboring More Virulence Genes than Bacteraemia Linked to Other Conditions. Int. J. Med. Microbiol. 2017;307:182–189. doi: 10.1016/j.ijmm.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 115.Buc E., Dubois D., Sauvanet P., Raisch J., Delmas J., Darfeuille-Michaud A., Pezet D., Bonnet R. High Prevalence of Mucosa-Associated E. Coli Producing Cyclomodulin and Genotoxin in Colon Cancer. PLoS ONE. 2013;8:e56964. doi: 10.1371/journal.pone.0056964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nakatsu G., Li X., Zhou H., Sheng J., Wong S.H., Wu W.K.K., Ng S.C., Tsoi H., Dong Y., Zhang N., et al. Gut Mucosal Microbiome across Stages of Colorectal Carcinogenesis. Nat. Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shimpoh T., Hirata Y., Ihara S., Suzuki N., Kinoshita H., Hayakawa Y., Ota Y., Narita A., Yoshida S., Yamada A., et al. Prevalence of Pks-Positive Escherichia Coli in Japanese Patients with or without Colorectal Cancer. Gut Pathog. 2017;9:35. doi: 10.1186/s13099-017-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iyadorai T., Mariappan V., Vellasamy K.M., Wanyiri J.W., Roslani A.C., Lee G.K., Sears C., Vadivelu J. Prevalence and Association of Pks+ Escherichia Coli with Colorectal Cancer in Patients at the University Malaya Medical Centre, Malaysia. PLoS ONE. 2020;15:e0228217. doi: 10.1371/journal.pone.0228217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Watanabe D., Murakami H., Ohno H., Tanisawa K., Konishi K., Tsunematsu Y., Sato M., Miyoshi N., Wakabayashi K., Watanabe K., et al. Association between Dietary Intake and the Prevalence of Tumourigenic Bacteria in the Gut Microbiota of Middle-Aged Japanese Adults. Sci. Rep. 2020;10:15221. doi: 10.1038/s41598-020-72245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Healy A.R., Herzon S.B. Molecular Basis of Gut Microbiome-Associated Colorectal Cancer: A Synthetic Perspective. J. Am. Chem. Soc. 2017;139:14817–14824. doi: 10.1021/jacs.7b07807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bian X., Fu J., Plaza A., Herrmann J., Pistorius D., Stewart A.F., Zhang Y., Müller R. In Vivo Evidence for a Prodrug Activation Mechanism during Colibactin Maturation. Chembiochem. 2013;14:1194–1197. doi: 10.1002/cbic.201300208. [DOI] [PubMed] [Google Scholar]
- 122.Brotherton C.A., Wilson M., Byrd G., Balskus E.P. Isolation of a Metabolite from the Pks Island Provides Insights into Colibactin Biosynthesis and Activity. Org. Lett. 2015;17:1545–1548. doi: 10.1021/acs.orglett.5b00432. [DOI] [PubMed] [Google Scholar]
- 123.Suresh A., Shaik S., Baddam R., Ranjan A., Qumar S., Jadhav S., Semmler T., Ghazi I.A., Wieler L.H., Ahmed N. Evolutionary Dynamics Based on Comparative Genomics of Pathogenic Escherichia Coli Lineages Harboring Polyketide Synthase (Pks) Island. mBio. 2021;12 doi: 10.1128/mBio.03634-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xue M., Kim C.S., Healy A.R., Wernke K.M., Wang Z., Frischling M.C., Shine E.E., Wang W., Herzon S.B., Crawford J.M. Structure Elucidation of Colibactin and Its DNA Cross-Links. Science. 2019;365 doi: 10.1126/science.aax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Faïs T., Delmas J., Barnich N., Bonnet R., Dalmasso G. Colibactin: More Than a New Bacterial Toxin. Toxins. 2018;10:151. doi: 10.3390/toxins10040151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wernke K.M., Xue M., Tirla A., Kim C.S., Crawford J.M., Herzon S.B. Structure and Bioactivity of Colibactin. Bioorg. Med. Chem. Lett. 2020;30:127280. doi: 10.1016/j.bmcl.2020.127280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cuevas-Ramos G., Petit C.R., Marcq I., Boury M., Oswald E., Nougayrède J.-P. Escherichia Coli Induces DNA Damage in Vivo and Triggers Genomic Instability in Mammalian Cells. Proc. Natl. Acad. Sci. USA. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cougnoux A., Delmas J., Gibold L., Faïs T., Romagnoli C., Robin F., Cuevas-Ramos G., Oswald E., Darfeuille-Michaud A., Prati F., et al. Small-Molecule Inhibitors Prevent the Genotoxic and Protumoural Effects Induced by Colibactin-Producing Bacteria. Gut. 2016;65:278–285. doi: 10.1136/gutjnl-2014-307241. [DOI] [PubMed] [Google Scholar]
- 129.Shine E.E., Xue M., Patel J.R., Healy A.R., Surovtseva Y.V., Herzon S.B., Crawford J.M. Model Colibactins Exhibit Human Cell Genotoxicity in the Absence of Host Bacteria. ACS Chem. Biol. 2018;13:3286–3293. doi: 10.1021/acschembio.8b00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Iftekhar A., Berger H., Bouznad N., Heuberger J., Boccellato F., Dobrindt U., Hermeking H., Sigal M., Meyer T.F. Genomic Aberrations after Short-Term Exposure to Colibactin-Producing E. Coli Transform Primary Colon Epithelial Cells. Nat. Commun. 2021;12:1003. doi: 10.1038/s41467-021-21162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cougnoux A., Dalmasso G., Martinez R., Buc E., Delmas J., Gibold L., Sauvanet P., Darcha C., Déchelotte P., Bonnet M., et al. Bacterial Genotoxin Colibactin Promotes Colon Tumour Growth by Inducing a Senescence-Associated Secretory Phenotype. Gut. 2014;63:1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 132.Tomkovich S., Yang Y., Winglee K., Gauthier J., Mühlbauer M., Sun X., Mohamadzadeh M., Liu X., Martin P., Wang G.P., et al. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res. 2017;77:2620–2632. doi: 10.1158/0008-5472.CAN-16-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lucas C., Salesse L., Hoang M.H.T., Bonnet M., Sauvanet P., Larabi A., Godfraind C., Gagnière J., Pezet D., Rosenstiel P., et al. Autophagy of Intestinal Epithelial Cells Inhibits Colorectal Carcinogenesis Induced by Colibactin-Producing Escherichia Coli in ApcMin/+ Mice. Gastroenterology. 2020;158:1373–1388. doi: 10.1053/j.gastro.2019.12.026. [DOI] [PubMed] [Google Scholar]
- 134.Wilson M.R., Jiang Y., Villalta P.W., Stornetta A., Boudreau P.D., Carrá A., Brennan C.A., Chun E., Ngo L., Samson L.D., et al. The Human Gut Bacterial Genotoxin Colibactin Alkylates DNA. Science. 2019;363 doi: 10.1126/science.aar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li Z.-R., Li J., Cai W., Lai J.Y.H., McKinnie S.M.K., Zhang W.-P., Moore B.S., Zhang W., Qian P.-Y. Macrocyclic Colibactin Induces DNA Double-Strand Breaks via Copper-Mediated Oxidative Cleavage. Nat. Chem. 2019;11:880–889. doi: 10.1038/s41557-019-0317-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dziubańska-Kusibab P.J., Berger H., Battistini F., Bouwman B.A.M., Iftekhar A., Katainen R., Cajuso T., Crosetto N., Orozco M., Aaltonen L.A., et al. Colibactin DNA-Damage Signature Indicates Mutational Impact in Colorectal Cancer. Nat. Med. 2020;26:1063–1069. doi: 10.1038/s41591-020-0908-2. [DOI] [PubMed] [Google Scholar]
- 137.Pleguezuelos-Manzano C., Puschhof J., Rosendahl Huber A., van Hoeck A., Wood H.M., Nomburg J., Gurjao C., Manders F., Dalmasso G., Stege P.B., et al. Mutational Signature in Colorectal Cancer Caused by Genotoxic Pks+ E. Coli. Nature. 2020;580:269–273. doi: 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee-Six H., Olafsson S., Ellis P., Osborne R.J., Sanders M.A., Moore L., Georgakopoulos N., Torrente F., Noorani A., Goddard M., et al. The Landscape of Somatic Mutation in Normal Colorectal Epithelial Cells. Nature. 2019;574:532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 139.Arthur J.C. Microbiota and Colorectal Cancer: Colibactin Makes Its Mark. Nat. Rev. Gastroenterol. Hepatol. 2020;17:317–318. doi: 10.1038/s41575-020-0303-y. [DOI] [PubMed] [Google Scholar]
- 140.Dalmasso G., Cougnoux A., Delmas J., Darfeuille-Michaud A., Bonnet R. The Bacterial Genotoxin Colibactin Promotes Colon Tumor Growth by Modifying the Tumor Microenvironment. Gut Microbes. 2014;5:675–680. doi: 10.4161/19490976.2014.969989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lopès A., Billard E., Casse A.H., Villéger R., Veziant J., Roche G., Carrier G., Sauvanet P., Briat A., Pagès F., et al. Colibactin-Positive Escherichia Coli Induce a Procarcinogenic Immune Environment Leading to Immunotherapy Resistance in Colorectal Cancer. Int. J. Cancer. 2020;146:3147–3159. doi: 10.1002/ijc.32920. [DOI] [PubMed] [Google Scholar]
- 142.Tronnet S., Garcie C., Brachmann A.O., Piel J., Oswald E., Martin P. High Iron Supply Inhibits the Synthesis of the Genotoxin Colibactin by Pathogenic Escherichia Coli through a Non-Canonical Fur/RyhB-Mediated Pathway. Pathog. Dis. 2017;75 doi: 10.1093/femspd/ftx066. [DOI] [PubMed] [Google Scholar]
- 143.Tronnet S., Garcie C., Rehm N., Dobrindt U., Oswald E., Martin P. Iron Homeostasis Regulates the Genotoxicity of Escherichia Coli That Produces Colibactin. Infect Immun. 2016;84:3358–3368. doi: 10.1128/IAI.00659-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chagneau C.V., Garcie C., Bossuet-Greif N., Tronnet S., Brachmann A.O., Piel J., Nougayrède J.-P., Martin P., Oswald E. The Polyamine Spermidine Modulates the Production of the Bacterial Genotoxin Colibactin. mSphere. 2019;4 doi: 10.1128/mSphere.00414-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tang-Fichaux M., Chagneau C.V., Bossuet-Greif N., Nougayrède J.-P., Oswald É., Branchu P. The Polyphosphate Kinase of Escherichia Coli Is Required for Full Production of the Genotoxin Colibactin. mSphere. 2020;5 doi: 10.1128/mSphere.01195-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kosari F., Taheri M., Moradi A., Hakimi Alni R., Alikhani M.Y. Evaluation of Cinnamon Extract Effects on ClbB Gene Expression and Biofilm Formation in Escherichia Coli Strains Isolated from Colon Cancer Patients. BMC Cancer. 2020;20:267. doi: 10.1186/s12885-020-06736-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kaewkod T., Tobe R., Tragoolpua Y., Mihara H. Medicinal Plant Extracts Protect Epithelial Cells from Infection and DNA Damage Caused by Colibactin-Producing Escherichia Coli, and Inhibit the Growth of Bacteria. J. Appl. Microbiol. 2021;130:769–785. doi: 10.1111/jam.14817. [DOI] [PubMed] [Google Scholar]
- 148.Oliero M., Calvé A., Fragoso G., Cuisiniere T., Hajjar R., Dobrindt U., Santos M.M. Oligosaccharides Increase the Genotoxic Effect of Colibactin Produced by Pks+ Escherichia Coli Strains. BMC Cancer. 2021;21:172. doi: 10.1186/s12885-021-07876-8. [DOI] [PMC free article] [PubMed] [Google Scholar]