Abstract
The calcineurin B-like protein (CBL) and CBL-interacting protein kinase (CIPK) play important roles in plant signal transduction and response to abiotic stress. Plants of Medicago genus contain many important forages, and their growth is often affected by a variety of abiotic stresses. However, studies on the CBL and CIPK family member and their function are rare in Medicago. In this study, a total of 23 CBL and 58 CIPK genes were identified from the genome of Medicago sativa as an important forage crop, and Medicaog truncatula as the model plant. Phylogenetic analysis suggested that these CBL and CIPK genes could be classified into five and seven groups, respectively. Moreover, these genes/proteins showed diverse exon-intron organizations, architectures of conserved protein motifs. Many stress-related cis-acting elements were found in their promoter region. In addition, transcriptional analyses showed that these CBL and CIPK genes exhibited distinct expression patterns in various tissues, and in response to drought, salt, and abscisic acid treatments. In particular, the expression levels of MtCIPK2 (MsCIPK3), MtCIPK17 (MsCIPK11), and MtCIPK18 (MsCIPK12) were significantly increased under PEG, NaCl, and ABA treatments. Collectively, our study suggested that CBL and CIPK genes play crucial roles in response to various abiotic stresses in Medicago.
Keywords: Medicago sativa, Medicago truncatula, CBL genes, CIPK genes, abiotic stresses, ABA, defense compounds, expression profiling
1. Introduction
Plants are exposed to various adverse environment stresses during their growth and development process, including high salinity, drought, cold, and pathogens. Plants have evolved complex signal transduction pathways to cope with these fluctuating environments during their life cycle [1]. Calcium, an important second messenger in plant cell signaling, involves various signal transduction pathways [2]. Calcium is able to convert external signals into cytoplasmic information to further drive the response to specific stimuli [3]. Calcium signals are mainly perceived by Ca2+ sensors, including calmodulins (CAMs), calmodulin-like proteins (CMLs), calcium-dependent protein kinases (CDPKs), and the plant-specific calcineurin B-like proteins (CBLs) [4]. The CBLs belong to a unique group of calcium sensors in plants that can be specifically targeted by CBL-interacting protein kinases (CIPKs) to transduce the perceived calcium signal upon various stimulations. CBLs and CIPKs together form the Ca2+-mediated CBL-CIPK network [5].
CBL proteins share some signature motifs or domains. CBL proteins contain an important structural component as calcium binding sites, which can capture Ca2+ ions through four EF hand domains, and the number of amino acids spaced between the four EF-hand domains is conservative [6,7]. Furthermore, serine residue in the PFPF motif, is necessary for CIPKs to phosphorylate CBLs [8]. There are two important domains in the C-terminal. The conserved NAF motif is responsible for the interaction with Ca2+ bound CBLs, thus activating targeted CIPKs [9,10]. Another important domain is the protein phosphotase interaction (PPI) domain, which can target the special member of the protein phosphotase 2C [11].
CBL and CIPK genes have been identified in many plant species, including Arabidopsis thaliana [2], Oryza sativa [12], Brassica napus [1], and Ananas comosus [13]. Many studies have demonstrated that the CBL and CIPK proteins can function properly. For example, the over-expression of AtCBL5 confer salt tolerance in the transgenic Arabidopsis plants [14]. AtCIPK8 itself can regulate the low-affinity phase of the nitrate primary response [15]. Meanwhile, the CBL and CIPK can work together as a network to widely regulate various metabolism processes in plant response to abiotic stress and nutrient signaling cascades [16,17,18], which included, but were not limited to, the transport of sodium (Na+), potassium (K+), nitrate (NO3−) cross plasma membrane and vacuole membrane, and in auxin and ABA signal transduction [19]. Meanwhile, the CBL-CIPK network can transfer the kinase to the plasma membrane and activate the plasma membrane-localized Na+/H+ antiporter AtNHX7 (SOS1) and the vacuolar H+-ATPase to improve salt tolerance in roots of Arabidopsis (AtCBL4-AtCIPK24 pathway) [3,20,21]. Moreover, AtCBL10 can target AtCIPK24, and this interaction is related to the vacuolar compartments and can protect shoots from salt stress [22].
Alfalfa (M. sativa) is a perennial legume plant, which is one of the important forage crops. Alfalfa had high yield (representing about 2600 kg per hectare) and high protein content (with about 15–20% of crude protein) [23,24]. However, the genetic analysis on the tetraploid alfalfa genome is complicated due to the general intolerance of inbreeding. On the other hand, alfalfa is not tolerant to many abiotic stresses in the ecosystem [24]. M. truncatula emerged as model legumes for fundamental research in the 1990s [25]. As a reference species, M. truncatula has many advantages, such as short life cycle, easy control of diploid inheritance, small genome, easy transformation, high genetic diversity, and homology with legumes that have much larger genomes [25]. Abundant genome and genetic resources are available for M. truncatula, which is also valuable for the comparable studies of its close relative alfalfa.
Therefore, in the present study, we investigated the CBL and CIPK genes closely related to abiotic stress through comparative analysis between M. sativa and M. truncatula. We identified CBL and CIPK genes from both plant species, and analyzed their multiple sequence alignment, phylogenetic relationship, gene structure, protein motifs, and cis-acting elements. Furthermore, the expression profiles of CBLs and CIPKs of M. sativa and M. truncatula in response to various abiotic stresses were analyzed. Our comparative studies on CBL and CIPK genes from M. sativa and M. truncatula will provide a basis for future investigation on abiotic stress responses medicated by CBLs and CIPKs, and will facilitate the functional characterization of individual CBLs and CIPKs genes in responses to stresses and developmental signals.
2. Results
2.1. Identification of CBL-CIPK Genes in the M. sativa and M. truncatula Genome
Based on domain confirmation and homology search, a total of 10 CBLs and 26 CIPKs from M. sativa, and 13 CBLs and 32 CIPKs from M. truncatula, were identified, respectively. Their sequence were obtained from the M. sativa and M. truncatula genome database and further analyzed. Physico-chemical properties of CBL and CIPK genes, including TIGR locus, homologous gene, molecular weights, isoelectric points, possible subcellular localization were listed in Table 1. The encoded protein length of Ms/Mt CBL and Ms/Mt CIPK genes range from 173–336/191–257 aa and 207–519/237–518 aa. We also found the corresponding MW of Ms/Mt CBL and Ms/Mt CIPK ranged from 19.49–38.24/21.89–29.50 kDa and 25.00–58.00/23.36–57.89 kDa.
Table 1.
Properties of the predicted CBL proteins in M. sativa and M. truncatula.
| Gene Name | TIGR Locus | Homologous Gene | Chromosome Location | pI | MW | Protein Length | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| MsCBL1 | MsG0280007543.01.T01 | MtCBL5 | Chr2: 16946249–16952230 | 6.2 | 38,240.38 | 336 | Extracellular |
| MsCBL2 | MsG0380014586.01.T01 | MtCBL6 | Chr3: 56818145–56821980 | 4.9 | 19,486.31 | 173 | Plasma membrane |
| MsCBL3 | MsG0380016524.01.T01 | MtCBL7 | Chr3: 84678194–84682482 | 4.9 | 38,012.36 | 334 | Plasma membrane |
| MsCBL4 | MsG0480023701.01.T01 | MtCBL9 | Chr4: 88741570–88745444 | 4.55 | 24,633.18 | 217 | Plasma membrane |
| MsCBL5 | MsG0580024593.01.T01 | MtCBL7 | Chr5: 6687437–6692205 | 4.77 | 30,599.99 | 265 | Plasma membrane |
| MsCBL6 | MsG0580030002.01.T01 | MtCBL10 | Chr5: 104956837–104960723 | 5.02 | 29,541.79 | 256 | Plasma membrane |
| MsCBL7 | MsG0880043002.01.T01 | MtCBL12 | Chr8: 17451660–17454557 | 4.82 | 27,195.04 | 238 | Plasma membrane |
| MsCBL8 | MsG0880046091.01.T01 | MtCBL7 | Chr8: 68926482–68929407 | 4.99 | 20,024.43 | 173 | Plasma membrane |
| MsCBL9 | MsG0880046710.01.T01 | MtCBL8 | Chr8: 77208147–77210234 | 4.79 | 26,744.53 | 253 | Extracellular |
| MsCBL10 | MsG0880047760.01.T01 | MtCBL9 | Chr8: 90474978–90479379 | 5.64 | 31,753.31 | 273 | Plasma membrane |
| MtCBL1 | MtrunA17_Chr1g0151621 | MsCBL3 | Chr1: 4424275–4430607 | 4.68 | 28,348.38 | 249 | Plasma membrane |
| MtCBL2 | MtrunA17_Chr2g0290751 | MsCBL6 | Chr2: 10098104–10100632 | 4.8 | 24,076.53 | 211 | Plasma membrane |
| MtCBL3 | MtrunA17_Chr2g0290761 | MsCBL6 | Chr2: 10120014–10122541 | 5.02 | 24,212.82 | 211 | Plasma membrane |
| MtCBL4 | MtrunA17_Chr2g0290771 | MsCBL6 | Chr2: 10131045–10133055 | 4.44 | 21,889.77 | 191 | Plasma membrane |
| MtCBL5 | MtrunA17_Chr2g0290781 | MsCBL6 | Chr2: 10140913–10143707 | 4.76 | 27,082.64 | 238 | Plasma membrane |
| MtCBL6 | MtrunA17_Chr3g0102821 | MsCBL6 | Chr3: 27325210–27329496 | 4.7 | 25,954.5 | 226 | Plasma membrane |
| MtCBL7 | MtrunA17_Chr4g0055421 | MsCBL5 | Chr4: 49038532–49043130 | 4.74 | 24,483.05 | 213 | Plasma membrane |
| MtCBL8 | MtrunA17_Chr4g0063121 | MsCBL9 | Chr4: 54777738–54779483 | 4.54 | 23,911.69 | 214 | Plasma membrane |
| MtCBL9 | MtrunA17_Chr4g0076551 | MsCBL6 | Chr4: 64369731–64374408 | 4.75 | 25,972.49 | 227 | Plasma membrane |
| MtCBL10 | MtrunA17_Chr5g0446931 | MsCBL6 | Chr5: 43351610–43355760 | 4.71 | 26,064.69 | 226 | Plasma membrane |
| MtCBL11 | MtrunA17_Chr8g0341941 | MsCBL3 | Chr8: 5383017–5389089 | 4.78 | 29,496.87 | 257 | Plasma membrane |
| MtCBL12 | MtrunA17_Chr8g0346911 | MsCBL7 | Chr8: 9926428–9929518 | 4.99 | 27,094.98 | 236 | Plasma membrane |
| MtCBL13 | MtrunA17_Chr8g0388331 | MsCBL3 | Chr8: 46203348–46207118 | 4.97 | 24,028.73 | 210 | Plasma membrane |
In addition, the isoelectric point (pI) of the Ms/Mt CBL and Ms/Mt CIPK ranges from 4.55–6.20/4.44–5.02 and 4.65–9.29/5.69–9.44, respectively (Table 1 and Table 2). The corresponding homologous CBL and CIPK genes of M. sativa and M. truncatula were identified in parallel by sequence alignment. The predicted subcellular locations suggested that most of the CBL and CIPK proteins from M. sativa and M. truncatula were located in the plasma membrane or extracellular (Table 1 and Table 2).
Table 2.
Properties of the predicted CIPK proteins in M. sativa and M. truncatula.
| Gene Name | TIGR Locus | Homologous Gene | Chromosome Location | pI | MW | Protein Length | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| MsCIPK1 | MsG0180000513.01.T01 | MtCIPK1 | Chr1: 7219758–7220941 | 8.93 | 42,516.31 | 376 | Plasma membrane |
| MsCIPK2 | MsG0180000521.01.T01 | MtCIPK1 | Chr1: 7356600–7357829 | 8.99 | 46,413.89 | 410 | Plasma membrane |
| MsCIPK3 | MsG0180001670.01.T01 | MtCIPK2 | Chr1: 24938893–24940191 | 8.85 | 48,636.26 | 433 | Plasma membrane |
| MsCIPK4 | MsG0180004166.01.T01 | MtCIPK3 | Chr1: 74311950–74313263 | 8.86 | 48,883.24 | 438 | Plasma membrane |
| MsCIPK5 | MsG0180005611.01.T01 | MtCIPK4 | Chr1: 93961932–93963092 | 8.09 | 44,450.35 | 387 | Extracellular |
| MsCIPK6 | MsG0280006904.01.T01 | MtCIPK5 | Chr2: 7792269–7793606 | 8.87 | 51,100.97 | 446 | Plasma membrane |
| MsCIPK7 | MsG0280006906.01.T01 | MtCIPK6 | Chr2: 7819514–7821070 | 6.56 | 57,894.45 | 519 | Plasma membrane |
| MsCIPK8 | MsG0280011471.01.T01 | MtCIPK9 | Chr2: 84572890–84574260 | 9.15 | 51,553.44 | 457 | Plasma membrane |
| MsCIPK9 | MsG0380014945.01.T01 | MtCIPK15 | Chr3: 62899825–62901207 | 8.58 | 52,512.51 | 461 | Plasma membrane |
| MsCIPK10 | MsG0380014950.01.T01 | MtCIPK16 | Chr3: 62944869–62946200 | 8.45 | 50,073.61 | 444 | Plasma membrane |
| MsCIPK11 | MsG0380015546.01.T01 | MtCIPK17 | Chr3: 71653648–71654991 | 8.94 | 51,591.75 | 448 | Plasma membrane |
| MsCIPK12 | MsG0480018187.01.T01 | MtCIPK18 | Chr4: 1337477–1338790 | 8.58 | 50,031.2 | 438 | Plasma membrane |
| MsCIPK13 | MsG0480019352.01.T01 | MtCIPK19 | Chr4: 18884311–18885931 | 8.55 | 56,612.39 | 504 | Plasma membrane |
| MsCIPK14 | MsG0580027748.01.T01 | MtCIPK19 | Chr5: 67945209–67946826 | 8.36 | 56,525.14 | 503 | Plasma membrane |
| MsCIPK15 | MsG0580028307.01.T01 | MtCIPK24 | Chr5: 78145226–78153759 | 6.37 | 51,712.85 | 453 | Plasma membrane |
| MsCIPK16 | MsG0580028588.01.T01 | MtCIPK25 | Chr5: 82921999–82923423 | 9.05 | 53,886.01 | 475 | Plasma membrane |
| MsCIPK17 | MsG0580028590.01.T01 | MtCIPK26 | Chr5: 82973828–82975942 | 7.14 | 49,482.95 | 443 | Plasma membrane |
| MsCIPK18 | MsG0580029551.01.T01 | MtCIPK27 | Chr5: 98438716–98444881 | 6.45 | 53,945 | 475 | Plasma membrane |
| MsCIPK19 | MsG0780039388.01.T01 | MtCIPK28 | Chr7: 63273062–63274447 | 8.02 | 52,396.38 | 462 | Plasma membrane |
| MsCIPK20 | MsG0780039391.01.T01 | MtCIPK29 | Chr7: 63319880–63321148 | 8.95 | 47,583.98 | 423 | Plasma membrane |
| MsCIPK21 | MsG0880042910.01.T01 | MtCIPK30 | Chr8: 15785566–15788993 | 9.03 | 52,865.88 | 464 | Plasma membrane |
| MsCIPK22 | MsG0880042931.01.T01 | MtCIPK30 | Chr8: 16193214–16197233 | 8.99 | 50,776.14 | 448 | Plasma membrane |
| MsCIPK23 | MsG0880042932.01.T01 | MtCIPK31 | Chr8: 16202211–16203469 | 4.65 | 23,360.42 | 207 | Plasma membrane |
| MsCIPK24 | MsG0880046785.01.T01 | MtCIPK20 | Chr8: 78103239–78112793 | 9.29 | 46,753.34 | 410 | Plasma membrane |
| MsCIPK25 | MsG0880047252.01.T01 | MtCIPK21 | Chr8: 83803682–83805677 | 9.18 | 33,282.91 | 296 | Plasma membrane |
| MsCIPK26 | MsG0880047603.01.T01 | MtCIPK23 | Chr8: 88629631–88630695 | 8.59 | 40,180.45 | 355 | Plasma membrane |
| MtCIPK1 | MtrunA17_Chr1g0150321 | MsCIPK2 | Chr1: 3363660–3365695 | 6.51 | 57,997.54 | 518 | Plasma membrane |
| MtCIPK2 | MtrunA17_Chr1g0166101 | MsCIPK3 | Chr1: 15531311–15533332 | 8.71 | 48,428.7 | 435 | Plasma membrane |
| MtCIPK3 | MtrunA17_Chr1g0188031 | MsCIPK4 | Chr1: 37868206–37870202 | 7.6 | 49,372.84 | 440 | Plasma membrane |
| MtCIPK4 | MtrunA17_Chr1g0205421 | MsCIPK5 | Chr1: 50641418–50642779 | 9.44 | 28,820.94 | 243 | Plasma membrane |
| MtCIPK5 | MtrunA17_Chr2g0284011 | MsCIPK6 | Chr2: 4993431–4995206 | 8.77 | 50,920.75 | 445 | Plasma membrane |
| MtCIPK6 | MtrunA17_Chr2g0284031 | MsCIPK7 | Chr2: 5004740–5007006 | 8.92 | 53,811.89 | 474 | Plasma membrane |
| MtCIPK7 | MtrunA17_Chr2g0302431 | MsCIPK22 | Chr2: 19969671–19976812 | 7.56 | 36,077.73 | 320 | Plasma membrane |
| MtCIPK8 | MtrunA17_Chr2g0304111 | MsCIPK18 | Chr2: 21918963–21927365 | 8.71 | 44,042.03 | 391 | Plasma membrane |
| MtCIPK9 | MtrunA17_Chr2g0333821 | MsCIPK8 | Chr2: 51523898–51527942 | 7.99 | 52,575.69 | 462 | Plasma membrane |
| MtCIPK10 | MtrunA17_Chr3g0091281 | MsCIPK11 | Chr3: 12548682–12549395 | 8.9 | 48,125.84 | 423 | Plasma membrane |
| MtCIPK11 | MtrunA17_Chr3g0091291 | MsCIPK11 | Chr3: 12550018–12550736 | 8.44 | 57,285.85 | 510 | Plasma membrane |
| MtCIPK12 | MtrunA17_Chr3g0091311 | MsCIPK11 | Chr3: 12582134–12582970 | 8.95 | 47,560 | 422 | Plasma membrane |
| MtCIPK13 | MtrunA17_Chr3g0091411 | MsCIPK11 | Chr3: 12692385–12693326 | 9.27 | 30,232.13 | 257 | Plasma membrane |
| MtCIPK14 | MtrunA17_Chr3g0091421 | MsCIPK11 | Chr3: 12693793–12694524 | 8.93 | 52,506.01 | 457 | Plasma membrane |
| MtCIPK15 | MtrunA17_Chr3g0107011 | MsCIPK9 | Chr3: 30606080–30609128 | 9.05 | 52,009.48 | 447 | Plasma membrane |
| MtCIPK16 | MtrunA17_Chr3g0107051 | MsCIPK10 | Chr3: 30640386–30641717 | 9.22 | 47,462.77 | 420 | Plasma membrane |
| MtCIPK17 | MtrunA17_Chr3g0114511 | MsCIPK11 | Chr3: 35974723–35976525 | 8.67 | 50,488.22 | 446 | Plasma membrane |
| MtCIPK18 | MtrunA17_Chr4g0001331 | MsCIPK12 | Chr4: 900468–902885 | 9.27 | 28,716.18 | 243 | Plasma membrane |
| MtCIPK19 | MtrunA17_Chr4g0013131 | MsCIPK14 | Chr4: 10571450–10575122 | 9.1 | 49,421.81 | 435 | Plasma membrane |
| MtCIPK20 | MtrunA17_Chr4g0063881 | MsCIPK24 | Chr4: 55303349–55314602 | 8.53 | 49,753.07 | 436 | Plasma membrane |
| MtCIPK21 | MtrunA17_Chr4g0069301 | MsCIPK18 | Chr4: 59217861–59224659 | 8.85 | 51,572.49 | 460 | Plasma membrane |
| MtCIPK22 | MtrunA17_Chr4g0072831 | MsCIPK24 | Chr4: 61783649–61786108 | 9.05 | 27,643.06 | 237 | Cytoplasmic |
| MtCIPK23 | MtrunA17_Chr4g0074111 | MsCIPK26 | Chr4: 62835031–62837115 | 9.34 | 25,004.1 | 214 | Plasma membrane |
| MtCIPK24 | MtrunA17_Chr5g0428031 | MsCIPK15 | Chr5: 29699497–29710190 | 9.26 | 50,104.11 | 444 | Plasma membrane |
| MtCIPK25 | MtrunA17_Chr5g0432471 | MsCIPK16 | Chr5: 33183949–33187482 | 8.97 | 50,856.92 | 446 | Plasma membrane |
| MtCIPK26 | MtrunA17_Chr5g0432491 | MsCIPK17 | Chr5: 33195433–33198761 | 6.82 | 50,243.62 | 441 | Plasma membrane |
| MtCIPK27 | MtrunA17_Chr5g0441011 | MsCIPK18 | Chr5: 39527588–39534318 | 6.72 | 50,665.32 | 446 | Plasma membrane |
| MtCIPK28 | MtrunA17_Chr7g0244521 | MsCIPK19 | Chr7: 33731410–33735051 | 8.92 | 52,017.29 | 453 | Plasma membrane |
| MtCIPK29 | MtrunA17_Chr7g0244561 | MsCIPK20 | Chr7: 33768481–33770580 | 5.69 | 52,186.74 | 465 | Plasma membrane |
| MtCIPK30 | MtrunA17_Chr8g0346071 | MsCIPK21 | Chr8: 9057047–9061702 | 8.58 | 53,028.89 | 465 | Plasma membrane |
| MtCIPK31 | MtrunA17_Chr8g0346081 | MsCIPK21 | Chr8: 9065393–9070388 | 9.22 | 50,033.79 | 443 | Plasma membrane |
| MtCIPK32 | MtrunA17_Chr8g0379091 | MsCIPK7 | Chr8: 39906567–39908530 | 8.93 | 52,494.6 | 465 | Plasma membrane |
2.2. Multiple Sequence Alignment, Phylogenetic Analysis and Classification of CBL and CIPK Genes
Sequence alignments of the multiple amino acids between MsCBLs and MtCBLs indicated that the sequences of MsCBLs and MtCBLs are highly conserved (Figure S1): all the CBLs containing four EF hand motifs and one PFPF motif, which are similar to the CBLs from Arabidopsis [6]. Among them, MsCBL2 had no EF hand 4 motif and PFPF motif, MtCBL3 had no PFPF motif, and MsCBL8 had no EF hand 2 and 3 motifs.
Similarly, the alignment results showed that all the CIPKs contain an N-terminal catalytic kinase domain and a C terminal regulatory domain, which are jointed by a variable domain (Figure S2). Correspondingly, the multiple amino acids of CIPK includes four domains: ATP binding site motif, activation loop motif, NAF motif and PPI motif [9]. Among them, MsCIPK23, MsCIPK26, and MtCIPK11 do not have ATP binding site motif. MsCIPK24 and MtCIPK22 do not have the activation loop motif, and they are homologous genes. MsCIPK10, 11, 12, 13, 14 do not have NAF motif, although they have been reported to be necessary to mediate interactions between CBL and CIPK proteins. In addition, MsCIPK15, 23, and MtCIPK10, 11, 12, 13, 14, 22 do not contain PPI motif.
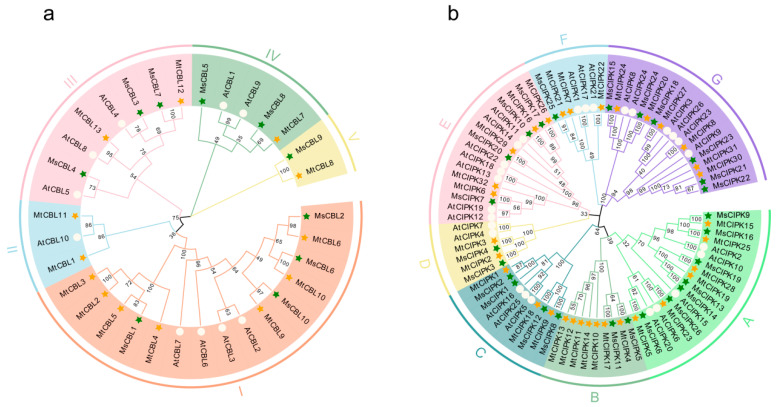
To investigate the evolutionary relationships of CBL and CIPK between M. truncatula and M. sativa and proteins from other plant species, a phylogenetic tree was constructed using the full amino acid sequences of CBL and CIPK family proteins from M. sativa, M. truncatula, and A. thaliana. It was shown that all of the CBLs could be classified into five distinct groups (I to V, Figure 1a), and all CIPKs into seven distinct groups (group A to G, Figure 1b) based on their sequence similarity, respectively. All of the CBL and CIPK genes from M. sativa and M. truncatula have their corresponding homologous genes.
Figure 1.
The phylogenetic analysis of CBL (a) and CIPK (b) proteins families across Medicago and Arabidopsis. Full length protein sequences of CBLs and CIPKs were constructed using MEGA-X based on the Neighbor-Joining (NJ) method; bootstrap was 1000 replicates. Subfamilies are highlighted with different colors. The green solid pentagrams, orange solid pentagrams, and hollow circles represent CBL and CIPK proteins from M. sativa (Ms), M. truncatula (Mt), and A. thaliana (At), respectively.
Most CBL and CIPK genes in M. sativa and M. truncatula have homologous genes in Arabidopsis. The largest cluster for CBL was group I with 15 members, and the smallest cluster was group V with two members. However, there was no CBL member from Arabidopsis in group V, only a pair of homologous genes MsCBL9 and MtCBL8. In addition, the largest group of CIPK are group A and G with both 18 members, and the smallest group is D, with only 6 CIPK members. In group B, 9 CIPK members of M. sativa and M. truncatula were closely related, but no AtCIPK members was found in this group.
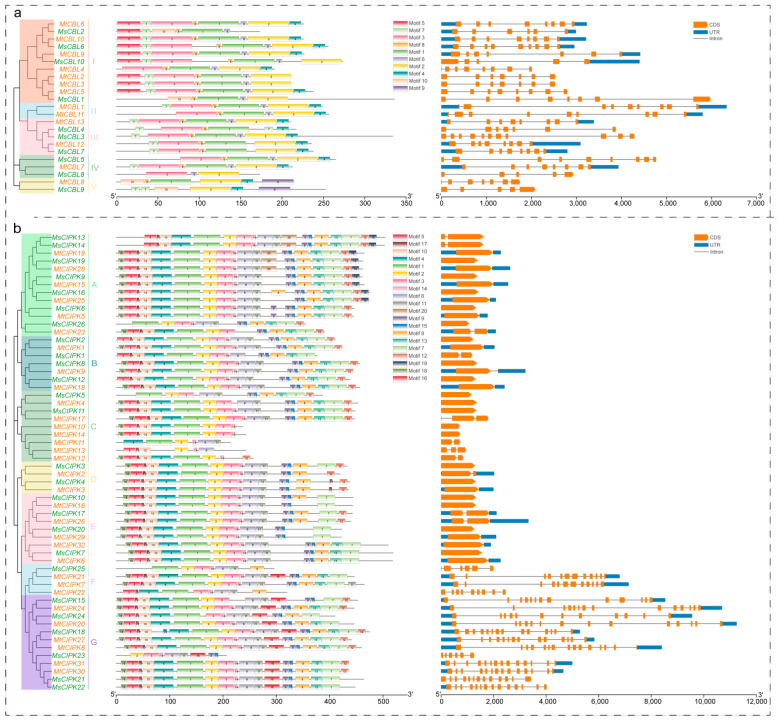
2.3. Analyses of Conserved Motif and Gene Structure
Conserved motifs and intron/exon organization were analyzed in order to further investigate the structural features of CBLs and CIPKs of M. sativa and M. truncatula (Figure 2a). In the CBL proteins, motifs 1–3, 5, 7–8 corresponded to the conserved domain EF hand, while motif 4 corresponded to the conserved domain PFPF motif. Motif 1, 5, 8, 9 of CIPK proteins play key roles, which correspond to activation loop, ATP binding site, PPI motif, and NAF motif, respectively. The conserved NAF motif mediates CBL and CIPK physical interactions [9], indicating that the identified CIPKs may be functionally linked in the CBL and CIPK network. Moreover, the deletion of NAF motif gene was only found in group C, which may be related to classification and function. However, they all have motif 1 of the activation loop or motif 5 of the ATP binding site domain. Therefore, all the 8 CIPKs identified in this study showed conserved features of the CIPKs family, which is consistent with the motif analysis of CIPKs in Arabidopsis [2].
Figure 2.
Analyses on phylogenetic relationships, motifs, and gene structure of CBL and CIPK genes from M. sativa and M. truncatula. The groups and its color of the phylogenetic tree are consistent with those in Figure 1. The motifs were indicated in different colored boxes with different numbers and the sequence information for each motif is provided in Figures S1 and S2. Exon–intron structure and conserved domain of CBL and CIPK genes. Blue boxes indicate 5′- and 3′- untranslated regions; orange boxes indicate exons; black lines indicate introns. (a,b) Phylogenetic relationships, motifs, and gene structure of CBL genes (a) and CIPK genes (b).
The gene structure of CBLs and CIPKs was analyzed, and it showed that each CBL has multiple introns. However, CIPK family members were clustered into an intron-rich clade (≥5 introns per gene) and an intron-poor clade (≤2 introns per gene), the intron-rich CIPK genes were clustered in subgroup F and G, while the intron-poor genes are presented in the other five subgroups (A–E). The structural differences in the CIPKs may allow CIPK genes to function differently because of the functional domains determine the function of genes [9].
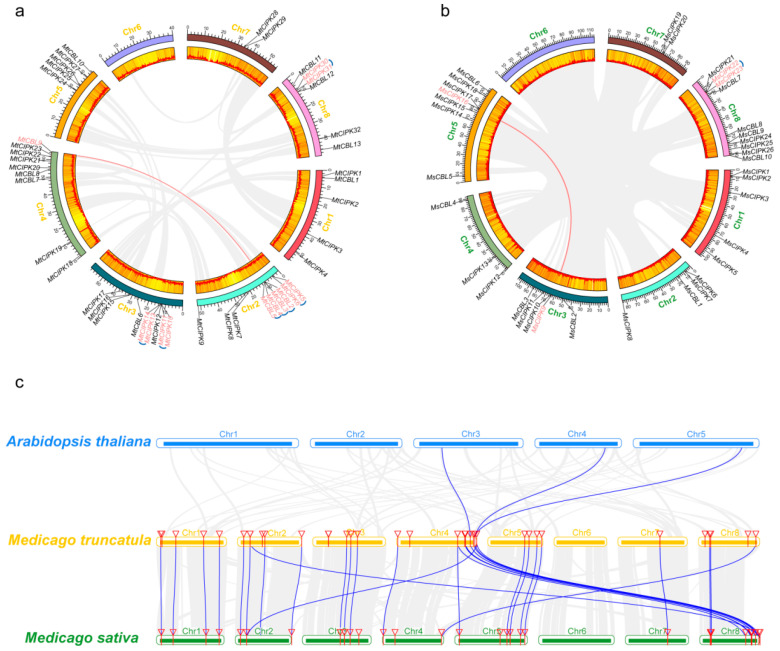
2.4. Analysis of Collinearity and Chromosome Location of CBL and CIPK Genes
To further investigate the evolutionary mechanism of CBL and CIPK gene family, both tandem and segmental duplication events were analyzed. Overall, the distribution of CBL and CIPKs are not even in both M. truncatula and M. sativa. MtCBLs were distributed on six out of all eight chromosomes. There are four MtCBL genes in chromosome 2, three in chromosome 4 and 8, and one in chromosome 1, 3, 5; but none in chromosome 6 or 7 (Figure 3a). Unlike M. truncatula, CBL genes in M. sativa distributed only on 5 chromosomes, but not on chromosomes 1, 6, and 7 (Figure 3b). Additionally, four, two, two, one, and one CBLs genes were on chromosomes 8, 3, 5, 2, and 4 of M. sativa, respectively (Figure 3b).
Figure 3.
Chromosome distributions of CBLs and CIPKs in M. truncatula and M. sativa. The chromosomal location and interchromosomal relationships of CBL and CIPK genes. The tandem duplicated genes are marked by blue arc trajectory and segmentally duplicated genes are connected by red curves. The chromosomal location and interchromosomal relationships of M. truncatula (a) and M. sativa (b). (c) Synteny analysis of CBL and CIPK genes between M. sativa, M. truncatula and representative plant species (Arabidopsis). Gray lines in the background indicate the collinear blocks within M. truncatula, and M. sativa/Arabidopsis, and the blue lines highlight the syntenic CBL and CIPK gene pairs.
For the CIPK genes, except chromosome 6 that did not have any gene for both M. truncatula and M. sativa (Figure 3b), all the other chromosomes have two to eight CIPKs for both M. truncatula and M. sativa (Figure 3b). In comparison, chromosome 3 of M. truncatula, and chromosome 8 of M. sativa had the most CIPK genes (Figure 3).
In addition, one CBL gene pair and one CIPK gene pair could be identified as segmental duplication events, two for MtCBL, four for MtCIPK, and one for MsCIPK were found to be tandem duplication in M. truncatula and M. sativa (Figure 3a,b). All above results inferred that both segmental duplication and tandem duplication events played an important driving force for the evolution of CBL genes, and the later played a predominant role. It is worth mentioning that although M. sativa and M. truncatula each had a pair of duplication genes, the number of tandem duplication genes in M. truncatula is far more than that of M. sativa.
Furthermore, three comparative syntenic maps of M. sativa and M. truncatula associated with the representative plant species Arabidopsis were constructed to illustrate the evolution relationship of CBL and CIPK genes family (Figure 3c). Notably, three orthologous pairs and 28 orthologous pairs were found between M. truncatula and Arabidopsis, and M. truncatula and M. sativa, respectively (Supplementary File S1). There were two genes in M. truncatula which showed a collinear relationship with Arabidopsis and M. sativa, respectively. MtCBL7 was collinear with MsCBL5 and MsCBL8, and MsCBL10 was collinear with MtCBL2 and MtCBL9. Correspondingly, MtCIPK21 and MtCIPK23 were found to be associated with two collinear gene pairs in Arabidopsis and M. sativa, which may play a special role in the growth and development of Medicago.
Moreover, to better understand the evolutionary selection pressure during the formation of CBL and CIPK gene families, the Ka/Ks values of CBL and CIPK gene pairs were analyzed for both M. sativa and M. truncatula (Supplementary File S1). Among all the CBL and CIPK gene pairs that are repeated in tandem and in segments, as well as the M. sativa and M. truncatula orthologous gene pairs, except for a homologous gene pairs (MtCBL8-MsCBL9), the Ka/Ks values of the remaining gene pairs are all less than 1. Taken together, these results indicated that the CBL and CIPK genes of M. sativa and M. truncatula may have undergone strong purification selection pressure during evolution.
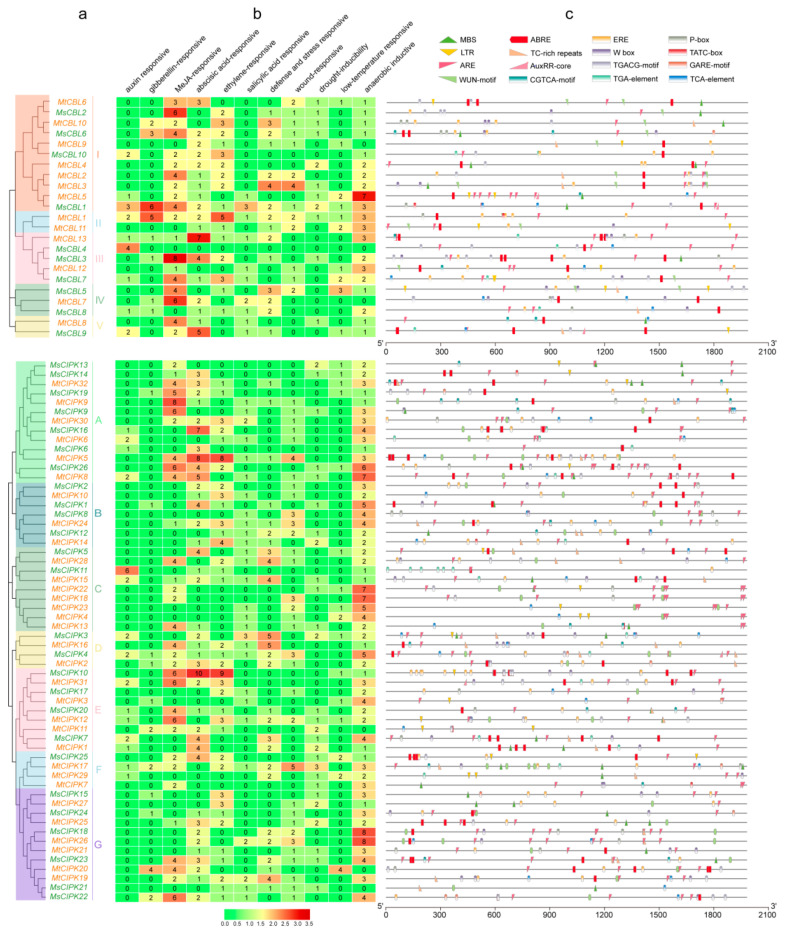
2.5. Analyses of the Cis-Acting Element and Location of CBL and CIPK Genes
The cis-acting element is important for the binding of transcription factors, which control the expression of many target genes. Here, we focused on hormones and abiotic stress related cis-acting elements, including auxin responsive (AuxRE-core), gibberellin-responsive (GARE-motif, P-box, TATC-box), MeJA-responsive (TGACG-motif, CGTCA-motif), abscisic acid-responsive (ABRE), ethylene-responsive (ERE), salicylic acid responsiveness (TCA-element), defense and stress responsiveness (TC-rich repeats, W-box), wound responses (WUN motif), MYB binding site involved in drought-inducibility (MBS), low temperature-responsive (LTR), and anaerobic induction (ARE) (Figure 4 and Supplementary File S2).
Figure 4.
Putative cis-elements and transcription factor binding sites in the promoter regions of CBL and CIPK genes from M. sativa and M. truncatula. (a) The groups and color are indicated as in Figure 1. (b) The colors and numbers of the grid indicated the numbers of different cis-acting elements in these CBL and CIPK genes. (c) The colored block represented different types of cis-acting elements and their locations in each CBL and CIPK gene.
Our results showed that the promoter of CBLs and CIPKs genes contained various cis-acting elements with different number. In particular, many of them have stress-related cis-acting elements including ABRE, W-box, and MBS repeats (Figure 4). Among them, MtCBL3, 6, MsCBL3, 9 have at least three ABRE repeat elements (Figure 4b,c). In addition, MtCIPK5, MsCIPK6, MsCIPK10 contain seven, eight, and ten W-box repeat elements, respectively (Figure 4b,c) and MtCIPK15, 16, 19, 28, MsCIPK5, 7, contain at least three W-box repeat elements. Moreover, only MtCIPK7 contained three MBS elements, and all the other genes had less than three MBS elements (Figure 4b,c). These genes are likely inducible by various stresses.
2.6. Analysis of the Expression Levels of CBL and CIPK Genes from Microarray Data
It is well known that CBL-CIPK complexes play important roles in the response of plants to external stimuli from environment [20,21,26]. Therefore, we retrieved genechip data of M. truncatula from the MtGEA web server (root under culture salinity, root under hydroponic salinity, root and shoot under drought treatment), and analyzed gene expression profiles in M. truncatula. One representative probe was selected for each gene, and the expression level of the representative probe is relatively close to the average value of multiple probes (Supplementary File S3).
To further screen the stress responsive genes under different stress treatments, the average change folds of each gene under individual treatment (total change folds for all treatments/number of treatments) of the MtCBL and MtCIPK genes in each treatment under different stress treatments was calculated (Figure 5 and Supplementary File S3). In addition, we also counted the number of treatments for each gene that were up-regulated by more than 2 times, which is consistent with the results of the above genes (Figure 5). Our expression analysis indicated that many CBL and CIPK genes are induced by these stresses (Figure 5).
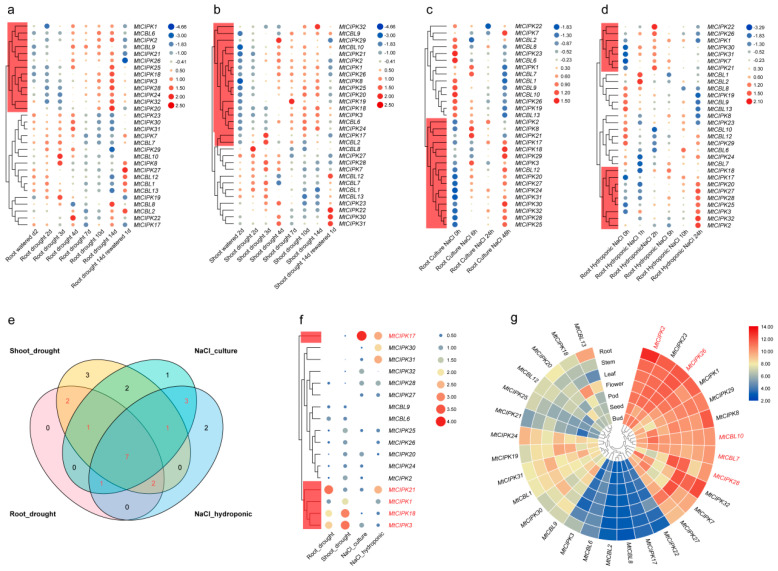
Figure 5.
MtCBL and MtCIPK gene expression profiles under different treatments, different times, and different tissues retrieved from microarray data. Each column represents one treatment, each row represents one gene, and each member is normalized in the same column. The relative expression levels are log2-transformed and visualized for heatmap. The color of circles from blue to red shows the expression level from negative to positive values after normalization. The red box indicates genes with increased expression. (a) Gene expression of root under different drought treatment time. (b) Gene expression of shoot under different drought treatment time. (c) Gene expression of root under different NaCl-culture time. (d) Gene expression of root under different NaCl-hydroponics time. (e) Venn diagram of gene expression levels after four different treatments. (f) Gene expression levels of common differentially expressed genes after different treatments in the Figure 5e. (g) Expression profiles of genes in 7 different tissues retrieved from microarray data.
Under drought treatment, 13 genes (MtCIPK1, MtCBL6, MtCIPK2, etc.) and 18 genes (MtCIPK32, MtCBL9, MtCBL29, etc.) were high induced in roots or in shoots, respectively (Figure 5a,b). In details, the expression levels of these genes were significantly increased under drought stress, but decreased after re-watering. It is worth noting that MtCBL6 and MtCBL9 were the only two CBL genes that were highly induced upon drought treatment, whether in roots or in stems (Figure 5a,b).
In addition, the gene expression levels were investigated in the roots that were treated with NaCl under culture and hydroponic condition, and it was found that 16 genes (MtCIPK2, MtCIPK8, MtCIPK21, etc., Figure 5c) and 18 genes (MtCIPK22, MtCIPK26, MtCIPK18, etc., Figure 5d) were highly induced under these two conditions, respectively. Notably, all of them were CIPK genes except one CBL (MtCBL12) gene that were highly induced under the culture NaCl treatment condition.
Moreover, Venn diagram, showing the differential expression level of all genes under four different treatments (Figure 5a–d), demonstrated that 17 genes were highly expressed under both drought and NaCl treatment (in red in Figure 5e). In particular, MtCIPK1, 3, 17, 18, 21 showed relatively higher expression level than the other genes under four different treatments (Figure 5f).
Due to the close relationship between gene expression level and gene function, we also investigated the expression profiles of CBLs and CIPKs in seven tissues of M. truncatula (root, stem, leaf, flower, pod, seed, and bud) with available microarray data (Figure 5g). Remarkably, 12 genes (MtCIPK1, 2, 7, 8, 23, 26, 27, 28, 29, 32, and MtCBL7, 10) showed relatively high expression level in various tissues. On the contrary, 6 genes (MtCBL2, 6, 8, and MtCIPK3, 17, 22) were expressed at relatively low level in different tissues (Figure 5g). Besides, the expression levels of the remaining 12 genes (MtCIPK1, 9, 12, 13 and MtCIPK18–21, 24, 25, 30, and 31) showed different pattern in various tissues. Altogether, the expression profiles of CBLs and CIPKs genes varied greatly, suggesting that they may be functional in different tissues.
2.7. Validation of the Expression Profile of Stress-Responsive CBL and CIPK Genes by qPCR Analysis
In order to verify the expression profiles of MtCBL and MtCIPK genes from microarray data, qPCR verification were carried out with ten selected genes. Among them, three MtCIPK genes (MtCIPK2, 26, 28) and two MtCBL genes (MtCBL7, 10) were selected among the highly expressed genes as representatives (Figure 5g). In addition to these five genes, the other five genes (MtCIPK1, 3, 17, 18, 21) were also selected for qPCR verification because they were highly inducible under stresses (Figure 5f). In addition, their homology genes from M. sativa, as shown in Table 1 and Table 2, were also selected as gene pairs for qPCR verification for comparison purpose.
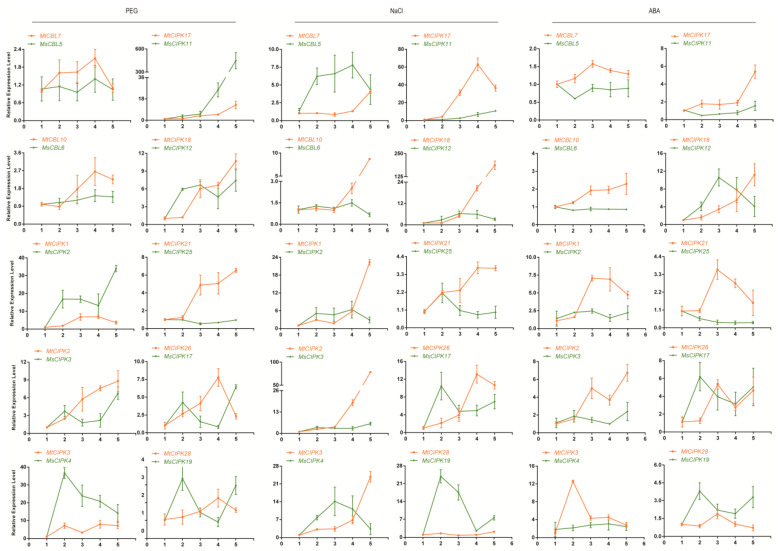
The expression of these twenty genes were analyzed under two stresses (NaCl and PEG) and ABA treatments (Figure 6). Overall, most genes were strongly induced by multiple treatments. In more details, under PEG stress, all of these genes except MsCIPK25, were significantly up-regulated to different levels, although their patterns are not the same for M. truncatula or M. sativa at different time points (Figure 6, left). Under NaCl treatment, many of the genes were up-regulation at different level and time points, but MtCIPK1–3, 8 and MtCBL10 particularly were significantly up-regulated at 72 h (Figure 6, middle). Upon ABA treatment, the fold changes of all genes were not more than 15 folds, with many of them around 6 folds (Figure 6, right). However, the expression levels of MsCBL5, 6, MsCIPK2, 4, 11, 25 did not show any significant changes, indicating they were not induced by ABA treatment (Figure 6, right).
Figure 6.
Quantification of gene expression levels of selected CBL and CIPK genes from M. sativa and M. truncatula under PEG, NaCl, and ABA stress using qPCR. Data are average of three independent biological samples ±SE and vertical bars indicate standard deviation. The value 1–5 of X axis for M. sativa represent treatment period of 0, 4, 6, 8, and 24 h, respectively. The value 1–5 of X-axis for M. truncatula represent treatment period of 0, 2, 24, 48, 72 h, respectively.
3. Discussion
The signaling system composed of CBLs and CIPKs as a calcium sensors is a key regulatory node in stress signaling pathways in plant. Significantly, regulation of membrane transport processes seems to be an emerging theme in the function of CBL-CIPK signaling machinery [19]. Extensive evidence confirmed the idea that CBL-CIPK proteins network regulates abiotic stress in many species, but little is known in Medicago. In the current study, a search for CBL and CIPK genes brought about the identification of 10 MsCBL, 13 MtCBL, 26 MsCBL, and 32 MtCBL genes in M. sativa and M. truncatula, respectively.
Multiple sequence alignments showed that most of the Medicago CBL proteins contained four EF hand motifs, which are necessary for CBL proteins to bind Ca2+, and the linker between each EF hand motif is absolutely conserved among CBL proteins [27]. Among them, MsCBL8 had no EF hand 2 and 3 motifs, and the phylogenetic analysis of MsCBL8 did indicate that it has high homology with AtCBL1 and AtCBL9 protein (Figure S1), suggesting that it might have specific functions and needs further study. Most CIPK proteins have been demonstrated to contain two domains: An N-terminal kinase catalytic domain (NAF motif) and a C-terminal regulatory domain (PPI motif) (Figure S2). However, MsCIPK10, 11, 12, 13, 14 do not have NAF motif or PPI motif, and the phylogenetic analysis of them clustered together. Therefore, they may perform special functions because they contain the ATP binding site domain and activation loop domain, belonging to the CIPK family. Phylogenetic analysis showed that CBLs and CIPKs had an independent Medicago cluster without Arabidopsis members (cluster V for CBL, cluster B for CIPK). Other clustering patterns were highly consistent with Arabidopsis [2]. Therefore, these members play important roles in the evolution of CBLs and CIPKs families, respectively.
Conserved motif identification shows that most CIPKs identified have a NAF domain in the C-terminal region (Figure 2b). It can be seen that they are close phylogenetically, and the types and positions of their motifs are more similar. Some motifs only exist in part of the CBL or CIPK amino acid sequence. For example, motif 5 belongs to the EF hands domain of CBL protein that only appears in one group I (Figure 2a), and these findings suggested that these motifs may be related to the functional diversity of CBL proteins. Gene structures like intron/exon organizations often reflect the evolution within some gene families [28,29,30]. Gene structure analysis showed that CBLs have more introns, among them, MsCBL3 have 10 introns, while MsCBL8, 9, 10 have 4 introns (Figure 2a). The phylogenetic relationship analysis showed that they belong to different groups, and the functions of them might be different, as shown in Figure 2b. Interestingly, CIPKs can be divided into two types: Poor introns (A–E) and rich introns (F,G). Similar patterns of intron-rich/poor CIPK family members have also been observed in model plants, Arabidopsis, rice, poplar, and soybean [12,31,32]. Significantly, there are abundant introns in CIPK proteins of green algae [33], moss, and ferns, and the intron deletion of CIPK first occurs in seed plants, which may be due to the species evolution caused by intron deletion in intron-rich members [2].
Duplication and divergence play an important role in expansion and evolution of gene families [34,35]. We found one (3.8%) segmental duplication and tandem duplication among 26 MsCIPKs, and one (10%) segmental duplication in ten MtCBLs, three (30%) tandem duplication in ten MtCBLs and four (12.5%) tandem duplication among thirty-two MtCIPKs (Figure 3). It shows that gene duplication is the main evolutionary force for the expansion of CBL and CIPK gene families in Medicago. Based on gene expression, combined with phylogenetic analysis and collinearity analysis, it is valuable to understand the functions of CBL and CIPK genes in special physiological processes. For example, MtCIPK31 has the highest expression level in seeds and flowers, but is relatively low in other tissues, which is consistent with its tandem duplication MtCIPK30 (Figure 5g), it is worth noting that there is a collinearity between MsCIPK21 and MtCIPK30. Interesting, their closest ortholog gene was AtCIPK9 in Arabidopsis, which is expressed predominantly in root, where it regulates K+-homeostasis under low-K+ stress in Arabidopsis [36,37]. This result indicated that MtCIPK30 and MtCIPK31 may share similar function in regulate K+-homeostasis under abiotic stress in Medicago. Another orthologous pair MsCIPK22 and MsCIPK23 were homologies of OsCIPK9 that were induced by multiple stresses, including cold, drought, PEG, and ABA treatments [38], suggesting the potential function of MsCIPK22 and MsCIPK23 in participating abiotic stress.
High-salinity or drought soil is the most serious abiotic stress [39]. It is urgent to improve the salinity and drought tolerance of alfalfa to increase yield. The transcript data from genechip under NaCl and drought treatments suggested that the expressions of several CBL and CIPK genes were highly induced or drastically changed (Figure 5). Simultaneously, the expression pattern of all ten genes were verified to be up-regulated under several treatments in M. truncatula by qPCR analyses (Figure 6) and suggested that these ten genes are involved in plant abiotic stress and hormone induction. Correspondingly, three gene pairs (MtCIPK2-MsCIPK3, MtCIPK17-MsCIPK11, and MtCIPK18-MsCIPK12) were significantly up-regulated under PEG, NaCl, and ABA treatments, and their expression patterns were the same in M. sativa and M. truncatula. By identifying the cis-acting elements bound by specific transcription factors, it is possible to reveal the transcriptional regulatory mechanism and gene expression patterns of the plant environmental adaptation. Unsurprisingly, all these gene pairs contain at least two of abscisic acid-responsive (ABRE), defense and stress responsiveness (TC-rich repeats, W-box), and MYB binding site involved in drought inducible (MBS) (except for the MtCIPK18 gene). Notably, MsCIPK3 gene contains more defense and stress responsiveness elements. Consequently, these genes may play key roles in enhancing the stress resistance of M. sativa and M. truncatula.
Previous studies have confirmed that CBL, CIPK, or CBL-CIPK complex function in regulating the complex process of plant growth and responding to environmental stress [18,40]. AtCIPK7 regulates cold stress responses via its interaction with AtCBL1 [41]. However, the homologous genes MtCIPK2 and MsCIPK3 exhibited high expression under several stresses, indicating that these two genes are involved in other pathways except cold stress. AtCIPK25 participates in the development of root meristem through auxin and cytokinin signaling [42]. The closely related genes of MtCIPK18 and MsCIPK12 may also play a similar role in resisting environmental adversity. AtCBL1 was reported to interact with AtCIPK1, involved in ABA-dependent stress responses [43]. Moreover, AtCBL1/9 can interact with AtCIPK23 to regulate both K+ and NO3− uptake in plants [44]. The ortholog gene MtCBL7 (AtCBL9) was up-regulated by PEG, NaCl, and ABA stress in M. truncatula (Figure 6). Interesting, AtCBL9–AtCIPK1 complex are also involved in ABA independent stress responses [43]. OsCIPK1 from rice was speculated to connect ABA and cold signaling pathway [40]. The orthologous gene MtCIPK21 (AtCIPK1) was up-regulated by almost all kinds of stresses in M. truncatula, and ABA-responsive element ABRE was found in the promoter region of MtCIPK21 (Figure 4). These results suggested the potential roles of CBL and CIPK genes in M. sativa and M. truncatula under abiotic stresses-resistance and hormone-resistance. Meanwhile, these homologous genes of M. sativa and M. truncatula did not all exhibit the same expression pattern under different stress treatments. It is possible that these genes have changed to varying degrees during the species evolution.
4. Materials and Methods
4.1. Identification of CBL-CIPK Genes in the Medicago Genome
The genome sequences and deduced protein sequences of CBLs and CIPKs were downloaded from the M. sativa and M. truncatula genome website (http://www.medicagogenome.org/, accessed on 15 November 2020) [45]. The first step in determining the CBL (EF-hand calcium-binding domain (PS50222)) and CIPK (NAF domain (PF03822)) family is to download Hidden Markov Model (HMM) profiles from Pfam protein family database (https://pfam.xfam.org/, accessed on 16 November 2020). Secondly, the CBL and CIPK gene sequences from Arabidopsis were downloaded from TAIR (https://www.arabidopsis.org/, accessed on 16 November 2020). The CBLs and CIPKs sequences from Arabidopsis were used as a query (p < e−5) to search the protein sequence of M. sativa and M. truncatula. The identified sequences were further verified, and redundant sequences were removed from the list. Further, output putative CBL and CIPK protein sequences were submitted to InterProScan (https://www.ebi.ac.uk/interpro/search/sequence-search, accessed on 20 November 2020), CDD (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 20 November 2020), Pfam (https://pfam.xfam.org/, accessed on 22 November 2020), and SMART (http://smart.embl-heidelberg.de/, accessed on 22 November 2020) to confirm the conserved CBL and CIPK domain. Finally, all CBL and CIPK candidate genes were obtained and assigned based on their locations on chromosome, including 10 MsCBL genes, 13 MtCBL genes, 26 MsCIPK genes, and 32 MtCIPK genes. Correspondingly, ExPASy (https://web.expasy.org/compute_pi/, accessed on 24 November 2020) was used to determine the isoelectric point (pI) and molecular weight (MW) of CBL and CIPK proteins. Finally, subcellular localization of CBL and CIPK genes were predicted by using the Softberry Home Page (http://linux1.softberry.com/berry.phtm, accessed on 24 November 2020).
4.2. Sequence Analyses and Structural Characterization of Medicago CBL and CIPK Genes
Sequence alignment analysis of CBL and CIPK domain sequences were carried out by using jalview v2.10.5 (Solvusoft Corporation, Las Vegas, NV, USA). Conserved motifs in CBL and CIPK protein sequences were identified by the MEME program (MEME-Suite version 5.1.0, http://meme-suite.org/, accessed on 2 December 2020) with default settings, except the motif number of CBL was set as 10 and 20 for CIPK, and the width of minimum and maximum motif were set as 10 and 200, respectively. The visualization of exon–intron positions and conserved motifs was executed through Amazing Optional Gene Viewer software (TBtools v1.068, South China Agricultural University, Guangzhou, Guangdong, China) [46].
4.3. Phylogenetic Analysis and Classification of the CBL and CIPK Genes
The phylogenetic relationship of CBL-CIPK proteins among M. sativa, M. truncatula, and Arabidopsis were analyzed with identified CBL-CIPK amino acid sequences from M. sativa, M. truncatula, and Arabidopsis. The phylogenetic trees were constructed by using MEGA-X (Mega Limited, Auckland, New Zealand) [47] using the Neighbor-Joining (NJ) method with default parameters with bootstrap value of 1000. Finally, EvolView (https://evolgenius.info/evolview-v2/, accessed on 5 December 2020) was used to modify the evolutionary tree.
4.4. Analysis of Collinearity and Chromosome Location
The chromosomal localization of all CBLs and CIPKs of M. sativa and M. truncatula were obtained from the corresponding website. Multiple collinear Scan toolkit (TBtools v1.068 (mcscanx), South China Agricultural University, Guangzhou, Guangdong, China) was used to analyze the gene duplication events with default parameters [48]. All the CBL and CIPK genes were similarly mapped to eight M. sativa and M. truncatula chromosomes, respectively, followed by the analysis on their intraspecific synteny relationship in M. sativa and M. truncatula using Amazing Gene Location software (TBtools), respectively [46]. In order to exhibit the interspecific synteny relationship among M. sativa, M. truncatula, and the representative model plant species (Arabidopsis), the syntenic maps were constructed using the Dual Systeny Plotter software (TBtools) [46]. The simple Ka/Ks calculator software is used to calculate non synchronous (Ka) and synchronous (Ks) values of CBL and CIPK gene pairs (TBtools) [46].
4.5. Analyses of the Cis-Acting Elements and Location of CBL and CIPK Genes in Medicago
The cis-acting elements in the 2000 bp upstream sequences of the coding region of CBL and CIPK genes were analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 10 December 2020) [49]. TBtools was used to visualize the cis-acting elements CBL and CIPK genes of Medicago.
4.6. Analysis of the Expression Levels of CBL and CIPK Genes from Genechip Data
For the analysis of the expression level of CBL and CIPK genes from microarray data, we download all the genechip data from M. truncatula Gene Expression Atlas (https://Mtgea.noble.org/v3/, accessed on 12 December 2020)), which has been developed as a compendium or “atlas” of gene expression profiles for the M. truncatula genes. The selected genechip data covered the roots and shoots, from plants subjected to drought stress, salt stress, and ABA stresses, and specific cell and tissue types. Amazing HeatMap software was used to generate the heatmap (TBtools) [46].
4.7. Plant Materials and Treatments
The M. sativa (Zhongmu No.1, Institute of Animal Science of CAAS, Beijing, China) and M. truncatula (cv. Jemalong A17, Institute of Animal Science of CAAS, Beijing, China) plants used in this study was stored at the Institute of Animal Science of CAAS. The roots, stems, leaves, flowers, pods (20-day old pods), and seeds (20-day old seeds) of mature M. sativa and M. truncatula plants, were collected separately for RNA extraction and used for qPCR analysis. Meanwhile, the expression patterns of CBL and CIPK genes were also investigated under different stress and hormone treatments (PEG, NaCl and ABA). The material and method used for different treatments were the same for M. sativa as for M. truncatula as previously reported by Yang et al. [50].
4.8. Analysis of the Gene Expression by qRT-PCR
Total RNA extraction, first strand cDNA synthesis, and qPCR procedures and methods were the same as previously reported by Yang et al. [50]. Each reaction was performed in biological triplicates and the data from qPCR was analyzed using 2−ΔΔCT method. The results were analyzed by means ± standard deviation (SD). The primer sequences used in this study were shown in details in Table S1.
5. Conclusions
The CBL-CIPK network is well-recognized as one of the resistance mechanisms. This study analyzed the CBL and CIPK genes on a genome-wide scale in M. sativa and M. truncatula. A total of 10 MsCBLs, 13 MtCBLs, 26 MsCIPKs, and 32 MtCIPKs were identified, respectively, in M. sativa and M. truncatula. These genes show high similarity in amino acid sequence, motif compositions, and conservative gene structure. In addition, phylogenetic analysis and collinearity analysis on CBL and CIPK in different species revealed their evolutionary patterns and predicted their functions in complex environments. Moreover, the expression profile of CBL and CIPK genes in different tissues and treatments of Medicago were analyzed and were verified by qPCR. It was found that most of the genes were highly expressed, especially MtCIPK2-MsCIPK3, MtCIPK17-MsCIPK11, and MTCIPK18-MsCIPK12. These gene pairs showed the same expression pattern in M. sativa and M. truncatula, and they may play an important role in response of plant stresses. This study compares the CBL-CIPK family of M. sativa and M. truncatula to provide new ideas for understanding the evolutionary relationship between the same genus and related crops, which also provides a comprehensive information basis for better characterizing the role of the CBL-CIPK family in plant stress resistance.
Abbreviations
CBL: calcineurin B-like protein; CIPK: CBL-interacting protein kinase; MW: Molecular weight; PI: Isoelectric point; qPCR: Quantitative polymerase chain reaction; aa: Amino acid; ABA: Abscisic acid; PEG: Polyethylene glycol.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22094634/s1, Figure S1: Alignment of multiple CBL amino acid sequences from M. sativa and M. truncatula. The sequence conservation is shown as a percentage bar-score below. The sequence logo is shown at the bottom, summarizing the occurrence of given amino acids at specific positions. Four well-conserved EF-hand motif and the PFPF motif were boxed. Figure S2: Alignment of multiple CIPK amino acid sequences from M. sativa and M. truncatula. The sequence conservation is shown as a percentage bar-score below. The sequence logo is shown at the bottom, summarizing the occurrence of given amino acids at specific positions. The ATP binding site, the activation loop, the NAF motif and PPI motifs for CIPK proteins were boxed. Table S1: List of primers and their sequences used in this study. Supplementary File S1. All tandem and segmental duplicated CBL and CIPK gene pairs in Medicago, and the orthologous gene pairs between Medicago and Arabidopsis; Supplementary File S2. List of all identified cis-acting elements in all CBL and CIPK genes found in Medicago; Supplementary File S3. Detailed information on the expression levels of CBL and CIPK gene genes retrieved from microarray data for M. truncatula, including eight different tissues and treatments in response to drought and NaCl treatments.
Author Contributions
Experiments were performed by W.D., J.Y., L.M. and Q.S., W.D. and J.Y. analyzed the data. W.D. drafted the manuscript. Y.P. supervised the experiments and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by the National Nature Science Foundation of China (U1906201 and 31802118), and Fundamental Research Funds for Central Non-profit Scientific Institution (2020-YWF-ZX-07). The funding body played no role in the design of the study, analysis, and interpretation of data, and in writing the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data in the present study are available in the public database as referred in the Material and Method part.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang H.F., Yang B., Liu W.Z., Li H.W., Wang L., Wang B.Y., Deng M., Liang W.W., Deyholos M.K., Jiang Y.Q. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.) BMC Plant Biol. 2014;8:1471–2229. doi: 10.1186/1471-2229-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolukisaoglu U.N., Weinl S., Blazevic D., Batistic O., Kudla J.R. Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134:43–58. doi: 10.1104/pp.103.033068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kudla J., Batistic O., Hashimoto K. Calcium signals: The lead currency of plant information processing. Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan S., Lan W., Chul S.L. Potassium nutrition, sodium toxicity, and calcium signaling: Connections through the CBL-CIPK network. Plant Biol. 2009;12:339–346. doi: 10.1016/j.pbi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Deng X., Hu W., Wei S., Zhou S., Zhang F., Han J., Chen L., Li Y., Feng J., Fang B., et al. TaCIPK29, a CBL-interacting protein kinase gene from wheat, confers salt stress tolerance in transgenic tobacco. PLoS ONE. 2013;8:e69881. doi: 10.1371/journal.pone.0069881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagae M., Nozawa A., Koizumi N., Sano H., Hashimoto H., Sato M., Shimizu T. The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. Biochem. Mol. Biol. 2003;278:42240–42246. doi: 10.1074/jbc.M303630200. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez-Barrena M.J., Martınez-Ripoll M., Zhu J.K., Albert A. The structure of the Arabidopsis thaliana SOS3: Molecular mechanism of sensing calcium for salt stress response. J. Mol. Biol. 2005;345:1253–1264. doi: 10.1016/j.jmb.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 8.Du W., Lin H., Chen S., Wu Y., Zhang J., Fuglsang A.T., Palmgren M.G., Wu W., Guo Y. Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis. Plant Physiol. 2011;156:2235–2243. doi: 10.1104/pp.111.173377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albrecht V.A., Ritz O., Linder S., Harter K., Kudla J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001;20:1051–1063. doi: 10.1093/emboj/20.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y., Halfter U., Ishitani M., Zhu J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13:1383–1399. doi: 10.1105/TPC.010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohta M., Guo Y., Halfter U., Zhu J.K. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Plant Biol. 2003;100:11771–11776. doi: 10.1073/pnas.2034853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X.F., Gu Z.M., Liu F., Ma B.J., Zhang H.S. Molecular analysis of rice CIPKs involved in both biotic and abiotic stress responses. Rice Sci. 2011;18:1–9. doi: 10.1016/S1672-6308(11)60001-2. [DOI] [Google Scholar]
- 13.Aslam M., Fakher B., Jakada B.H., Zhao L., Cao S., Cheng Y., Qin Y. Genome-wide identification and expression profiling of CBL-CIPK gene family in pineapple (Ananas comosus) and the role of AcCBL1 in abiotic and biotic stress response. Biomolecules. 2019;9:293. doi: 10.3390/biom9070293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong Y.H., Sung S.J., Kim B.G., Pandey G.K., Cho J.S., Kim K.N., Luan S. Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol. Cells. 2010;29:159–165. doi: 10.1007/s10059-010-0025-z. [DOI] [PubMed] [Google Scholar]
- 15.Hu H.C., Wang Y.Y., Tsay Y.F. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009;57:264–278. doi: 10.1111/j.1365-313X.2008.03685.x. [DOI] [PubMed] [Google Scholar]
- 16.Li R.F., Zhang J.W., Wei J.H., Wang H.Z., Wang Y.Z., Ma R.C. Functions and mechanisms of the CBL-CIPK signaling system in plant response to abiotic stress. Prog. Nat. Sci. 2009;19:667–676. doi: 10.1016/j.pnsc.2008.06.030. [DOI] [Google Scholar]
- 17.Weinl S., Kudla J. The CBL-CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytol. 2009;184:517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- 18.Yu Q.Y., An L.J., Li W.L. The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014;33:203–214. doi: 10.1007/s00299-013-1507-1. [DOI] [PubMed] [Google Scholar]
- 19.Luan S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Batelli G., Verslues P.E., Agius F., Qiu Q.S., Fujii H., Pan S.Q., Schumaker K.S., Grillo S., Zhu J.K. SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 2007;27:7781–7790. doi: 10.1128/MCB.00430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu Q.S., Guo Y., Dietrich M.A., Schumaker K.S., Zhu J.K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quan R.D., Lin H.X., Mendoza I., Zhang Y.G., Cao W.H., Yang Y.Q., Shang M., Chen S.Y., Pardo J.M., Guo Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007;19:1415–1431. doi: 10.1105/tpc.106.042291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loubna F., Pascal D., Jean A., Alain G., Didier L., Romain K., Florence L., Louis-Philippe V., Laurent B. Concentration and selective separation of bioactive peptides from an alfalfa white protein hydrolysate by electrodialysis with ultrafiltration membranes. J. Membr. Sci. 2009;329:60–67. [Google Scholar]
- 24.Li X.H., Brummer E.C. Applied genetics and genomics in alfalfa breeding. Agronomy. 2012;2:40–61. doi: 10.3390/agronomy2010040. [DOI] [Google Scholar]
- 25.Barker D.G., Bianchl S., Blondon F., Dattde Y., Duc G., Essad S., Flament P., Gallusci P., Gdnier G., Guy P., et al. Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol. Biol. Report. 1990;8:40–49. doi: 10.1007/BF02668879. [DOI] [Google Scholar]
- 26.Kim B.G., Waadt R., Cheong Y.H., Pandey G.K., Dominguez-Solis J.R., Schultke S., Lee S.C., Kudla J., Luan S. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 2007;52:473–484. doi: 10.1111/j.1365-313X.2007.03249.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H.C., Yin W.L., Xia X.L. Calcineurin B-Like family in Populus: Comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regul. 2008;56:129–140. doi: 10.1007/s10725-008-9293-4. [DOI] [Google Scholar]
- 28.Boudet N., Aubourg S., Toffano-Nioche C., Kreis M., Lecharny A. Evolution of intron/exon structure of DEAD helicase family genes in Arabidopsis, Caenorhabditis, and Drosophila. Genome Res. 2015;11:2101–2114. doi: 10.1101/gr.200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J.Y., Chen N.N., Chen F., Cai B., Santo S.D., Tornielli G.B., Pezzotti M., Cheng Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera) BMC Genom. 2014;15:281. doi: 10.1186/1471-2164-15-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang N., Zheng Y., Xin H.P., Fang L.C., Li S.H. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013;32:61–75. doi: 10.1007/s00299-012-1340-y. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J.K., Chen F., Liu J.Y., Chen X.L., Hewezi T., Cheng Z.M. Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Sci. Rep. 2016;6:28225. doi: 10.1038/srep28225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy S.W., Penny D. Patterns of intron loss and gain in plants: Intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol. Biol. Evol. 2007;24:171–181. doi: 10.1093/molbev/msl159. [DOI] [PubMed] [Google Scholar]
- 33.Kleist T.J., Spencley A.L., Luan S. Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front. Plant Sci. 2014;5:e187. doi: 10.3389/fpls.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vision T.J., Brown D.G., Tanksley S.D. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 35.Hughes A.L. The evolution of functionally novel proteins after gene duplication. R. Soc. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 36.Lara A., Rodenas R., Andres Z., Martinez V., Quintero F.J., Nieves-Cordones M., Botella M.A., Rubio F. Arabidopsis K+ transporter HAK5-mediated high-affinity root K+ uptake is regulated by protein kinases CIPK1 and CIPK9. J. Exp. Bot. 2020;71:5053–5060. doi: 10.1093/jxb/eraa212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L.L., Ren H.M., Chen L.Q., Wang Y., Wu W.H. A protein kinase CIPK9 interacts with calcium sensor CBL3 and regulates K+ homeostasis under low-K+ stress in Arabidopsis. Plant Physiol. 2013;161:266–277. doi: 10.1104/pp.112.206896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang Y., Huang Y.M., Xiong L.Z. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 2007;144:1416–1428. doi: 10.1104/pp.107.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon M.C., Rhoades J.D., Draper J.H., Scardaci S.C., Spyres M.D. Assessment of salt tolerance in rice cultivars in response to salinity problems in california Crop Ecology. Prod. Manag. 1998;38:394–398. [Google Scholar]
- 40.Kanwar P., Sanyal S.K., Tokas I., Yadav A.K., Pandey A., Kapoor S., Pandey G.K. Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium. 2014;56:81–95. doi: 10.1016/j.ceca.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Huang C., Ding S., Zhang H., Du H., An L. CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci. 2011;181:57–64. doi: 10.1016/j.plantsci.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 42.Meena M.K., Vishwakarma N.K., Tripathi V., Chattopadhya D. CBL-interacting protein kinase 25 contributes to root meristem development. Soc. Exp. Biol. 2018;70:133–147. doi: 10.1093/jxb/ery334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Angelo C.D., Weinl S., Batistic O., Pandey G.K., Cheong Y.H., Schultke S.S., Albrecht V., Ehlert B., Schulz B., Harter K., et al. Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 2006;48:857–872. doi: 10.1111/j.1365-313X.2006.02921.x. [DOI] [PubMed] [Google Scholar]
- 44.Cheong Y.H., Pandey G.K., Grant J.J., Batistic O., Li L., Kim B.G., Lee S.C., Kudla J., Luan S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007;52:223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- 45.Young N.D., Debelle F., Oldroyd G.E., Geurts R., Cannon S.B., Udvardi M.K., Benedito V.A., Mayer K.F., Gouzy J., Schoof H., et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C.J., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y.P., Tang H.B., DeBarry J.D., Tan X., Li J.P., Wang X.Y., Lee T.H., Jin H.Z., Marler B., Guo H., et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Peer Y.V.D., Rouze P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J.F., Ma L., Jiang W.B., Yao Y., Tang Y.H., Pang Y.Z. Comprehensive identification and characterization of abiotic stress and hormone responsive glycosyl hydrolase family 1 genes in Medicago truncatula. Plant Physiol. Biochem. 2021;158:21–33. doi: 10.1016/j.plaphy.2020.11.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in the present study are available in the public database as referred in the Material and Method part.