Abstract
pCTX-M3 is the archetypic member of the IncM incompatibility group of conjugative plasmids (recently referred to as IncM2). It is responsible for the worldwide dissemination of numerous antibiotic resistance genes, including those coding for extended-spectrum β-lactamases and conferring resistance to aminoglycosides. The IncM plasmids acquired during evolution diverse mobile genetic elements found in one or two multiple resistance regions, MRR(s), grouping antibiotic resistance genes as well as mobile genetic elements or their remnants. The IncM plasmids can be found in bacteria inhabiting various environments. The information on the structure and biology of pCTX-M3 is integrated in this review. It focuses on the functional modules of pCTX-M3 responsible for its replication, stable maintenance, and conjugative transfer, indicating that the host range of the pCTX-M3 replicon is limited to representatives of the family Enterobacteriaceae (Enterobacterales ord. nov.), while the range of recipients of its conjugation system is wide, comprising Alpha-, Beta-, and Gammaproteobacteria, and also Firmicutes.
Keywords: plasmid, IncM, blaCTX-M-3, conjugative transfer, antibiotic resistance
1. Introduction
Plasmids are DNA molecules able to replicate independently from the host-cell chromosome, widespread in bacteria and archaea, but also found in some eukaryotes. They have a modular structure comprising a backbone and a load. The backbone is composed of a replicon, regions encoding maintenance functions (oligomer resolution, partition, and post-segregational killing), and conjugative transfer genes. The plasmid load comprises all DNA fragments gathered during plasmid evolution, encoding, for example, antibiotic resistance, metabolic pathways, or virulence factors.
Plasmids are part of a mobile gene pool available for bacteria. Especially in Gram-negative bacteria, plasmids transferred during conjugation play an important role, being responsible for the bacterial ability to adapt quickly to a changing environment. Of particular importance are plasmids involved in the spreading of antimicrobial resistance in clinically relevant bacterial species. Several bacteria resistant to last resort antibiotics were listed in 2017 by the World Health Organization as those of critical priority for research and development of new antibiotics (https://www.who.int, accessed on 20 November 2020). Beside carbapenem-resistant Acinetobacter baumannii, Pseudomonas aeruginosa, and species of the Enterobacteriaceae family (recently reclassified as order Enterobacterales [1]), this priority also concerns Enterobacteriaceae species producing extended-spectrum β-lactamases (ESBL). According to the definition by D. Livermore, “an ESBL is any β-lactamase, ordinarily acquired and not inherent to a species, that can rapidly hydrolyze, or confer resistance to, oxyimino-cephalosporins (not carbapenems) or any β-lactamase mutant, within a family, that has an enhanced ability to do so” [2].
The ESBLs encountered in Europe are variants of β-lactamase types SHV, TEM, and CTX-M, with the latter widely disseminated in the 2000s [3]. The first such enzyme was detected in Munich, Germany in 1989 as conferring resistance to cefotaxime in Escherichia coli and therefore was named CTX-M for cefotaximase-Munich [4]. Since then, more than 300 CTX-M β-lactamases have been described, classified into seven groups based on their phylogeny: CTX-M-1, -M-2, -M-8, -M-9, -M-14, -M-15, and -M-25. Genes encoding enzymes of the first two groups evolved by escape of chromosomal genes from Kluyvera ascorbata, and those of groups 8 and 9 from K. georgiana [5,6]. The insertion sequence ISEcp1 was often involved in the initial mobilization of blaCTX-M ancestors from the genome of Kluyvera sp., and is frequently found in the proximity of the blaCTX-M genes on plasmids. Moreover, the ISEcp1 element can also be responsible for the high-level expression of the blaCTX-M genes [5].
The mobilization of the CTX-M-encoding genes into large conjugative plasmids has brought about their global dissemination. Around the year 2000, the CTX-M-9 and CTX-M-14 variants were the most prevalent in Spain, CTX-M-1—in Italy, CTX-M-3 – in Poland, and the CTX-M-15 variant in France and the UK ([3] and citations therein). Plasmids responsible for the spread of the blaCTX-M gene variants often belong to one of six major resistance plasmid families identified in clinically relevant Enterobacteriaceae (for review see [7,8]). Worldwide, the most frequently isolated epidemic clone was E. coli ST131 with blaCTX-M-15 located on a plasmid of the IncF incompatibility group [9].
2. Isolation of pCTX-M3
The “Polish CTX-M variant” was detected in 1996 in a hospital in Warsaw, when three Citrobacter freundii strains and one Escherichia coli strain resistant to cefotaxime and susceptible to ceftazidime were isolated from urine samples of different patients [10]. The resistance was associated with a large plasmid coding for a CTX-M variant new at that time, CTX-M-3. This plasmid could be easily transferred to E. coli via conjugation and conferred resistance to penicillins, cephalosporins, and aztreonam, as well as to the aminoglycosides, gentamicin and tobramycin, and trimethoprim-sulfamethoxazole [10]. Between 1998 and 2000 bacteria carrying this plasmid became widely disseminated in Poland and they were found in 15 hospitals in ten Polish cities; they represented eight Enterobacteriaceae species: Klebsiella pneumoniae, K. oxytoca, Enterobacter cloacae, Serratia marcescens, C. freundii, E. coli, Salmonella enterica, and Morganella morganii [11]. The nucleotide sequence of the plasmid isolated from the clinical strain C. freundii 2526, and subsequently named pCTX-M3, has been determined and deposited in the GenBank database under the accession no. AF550415.
3. Analysis of pCTX-M3 Nucleotide Sequence
The pCTX-M3 plasmid is 89,468 bps in size and carries 103 putative genes, of which 22 orfs have no homologues in public databases and are regarded as pCTX-M3-specific [12].
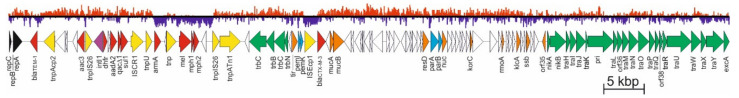
The mean G+C content in pCTX-M3 DNA is 51%, but it varies from 32% up to 65% in different sequence blocks (Figure 1). The minimal G+C value is found in two regions, one comprising armA, mel, mph1, and mph2, and the other within ISEcp1 and blaCTX-M-3, while the maximum in the sequence around orf8, a pCTX-M3-specific gene. Antibiotic resistance genes are located in two horizontally acquired regions, the first comprising blaCTX-M-3 with ISEcp1, responsible for its introduction to the plasmid, and the second, a multiple resistance region (MRR). It comprises blaTEM-1, situated next to the replicon, a type I integron with the aadA2, dfrA12, and sul1 gene cassettes, and the low-(G+C) region mentioned above, with armA coding for a 16S rRNA methylase and three putative macrolide resistance genes mel, mph1, and mph2. The MRR lies between the replicon and the trb transfer region; it is 27 kb in size and contains all the mobile elements of pCTX-M3 except for ISEcp1 [12]. Moreover, MRR has a mosaic structure with areas of high and low G+C contents (Figure 1), indicating that it arose by multiple genetic events, of which the first was acquisition of Tn1 together with blaTEM-1 [12]. It is worth mentioning that armA, mel, mph1, mph2, and the integron are located within Tn1548, a composite transposon bordered by two IS26 copies [13], also comprising the ISCR1 element regarded as a gene-capturing system (for review see [14]). It has been speculated that Tn1548 gave rise to the AbGRI3 resistance islands of Acinetobacter baumannii [15].
Figure 1.
Schematic representation of pCTX-M3 sequence. Open reading frames (ORFs) are represented by arrows indicating their orientation. Genes encoding the replicon are in black, post-segregational killing and partition systems in blue, and conjugative transfer in green. Drug resistance genes are in red, transposase genes in yellow, other genes with homologues in databases in orange, and those of known functions are subscribed. A G+C plot is presented at the top; values higher than the average are in red and lower in purple.
3.1. Replicon
pCTX-M3 replicates owing to a single replicon in 96% identical to that of pMU604 (mini-pMU607.1) of K. pneumoniae (GenBank acc. no. U27345) and comprise the repCBA region. repA codes for a replication initiator protein and repB for a small leader peptide translationally coupled with RepA [16], while the function of repC is unclear. The regulation of expression of the replication region in the pMU604 plasmid was deciphered by Athanasopoulos et al. [16] and found to be mainly post-transcriptional. The expression of repB and repA is repressed by an antisense RNA of ca. 77 nucleotides, called RNA I, whose target lies in repB, the leader region of the rep mRNA. Expression of repA is dependent on rep mRNA forming a tertiary structure called a pseudoknot, which enables translation of repA mRNA. Interaction of RNA I with its target sequesters the mRNA bases involved in the formation of the pseudoknot, and thereby prevents repA expression [16,17].
In E. coli cells pCTX-M3 is present in approx. 0.5–0.7 copies per chromosome (measured in cells of early stationary culture), while a minireplicon, comprising the repCBA region, is present in 8–10 copies per chromosome [18].
The pCTX-M3 replicon was originally classified as a member of an IncL/M incompatibility group [12]. However, in 2015 [19] three IncL/M plasmid lines, IncL, IncM1, and IncM2, were distinguished on the basis of their incRNA identity, which is 104 bases long [20] and partially overlaps the RNA I described by Athanasopoulos et al. [16]. Representatives of the IncL and IncM plasmids have been shown to be mutually compatible, unlike members of the IncM1 and IncM2 groups. pCTX-M3 was reclassified as an IncM2 group plasmid [19].
The range of hosts in which pCTX-M3 can replicate comprises Enterobacteriaceae (Enterobacterales ord. nov.) [21].
3.2. Stable Maintenance Systems
pCTX-M3 is stably maintained in bacterial populations (it is carried by 100% E. coli cells after 60 generations [18]). A stable maintenance of a plasmid in a bacterial population is ensured by a concerted action of the partition system, post-segregational killing system, resolvase, and finally by the conjugation system. The maintenance of the pCTX-M3 plasmid is ensured by the parA–parB module coding for a partition system homologous to the parM–parR-encoded system from the R1/NR1 plasmid [12], with the first gene coding for an actin-type segregation protein and the second for a DNA-binding adaptor protein (for review see [22]). In pCTX-M3 the centromere-like sequence parS is located upstream of parA, similarly to parC, the centromere in the parMR system [18]. Another component has been shown to be important in the pCTX-M3 partition system, playing a role in the stabilization of unstable vectors—the nuc gene located just downstream of parB and coding for a nuclease [18]. In IncI1 plasmids, such as R64, nuc is located within the tra region, upstream of sogL coding for primase. However, its inactivation had no effect on the conjugative transfer of this plasmid [23]. Resolvase, an enzyme resolving plasmid oligomers into monomeric forms, is probably encoded by the resD gene in pCTX-M3 [12]. A post-segregational killing (PSK) system is encoded by the pemI–pemK operon showing high similarity to that of NR1 (97% identity of PemI and 95% identity of PemK) [12]. pemK codes for a toxin, which was shown to be a sequence-specific endoribonuclease cleaving mRNAs to inhibit protein synthesis, and pemI for its antidote [24]. Deletion of pemI–pemK resulted in only a small drop in pCTX-M3 maintenance in E. coli cells (97% retention of the PSK system-deficient plasmid after 60 generations), indicating the unimportance of the PSK system for the segregational stability of pCTX-M3 and the main role of the partition system [18].
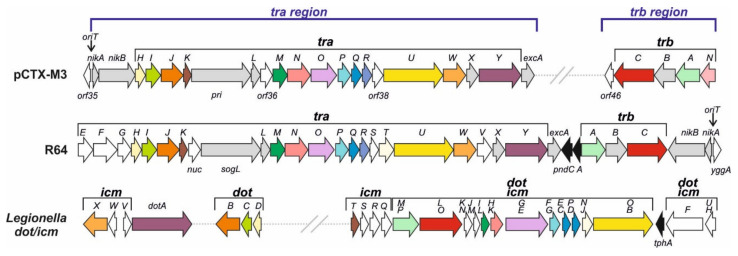
The pCTX-M3 conjugation system is encoded in two distantly located regions, tra and trb, both displaying an operon structure. It is related to that of IncI1 plasmids, ColIb-P9 and R64, in both the amino acid sequences of respective proteins (30 to 65% similarity of amino acid sequence) and in the gene syntheny (Figure 2) [12]. Nevertheless, several genes from each system are unique. The two systems also differ in the location of entire gene segments—in pCTX-M3, the oriT region (the origin of transfer sequence oriT together with nikA and nikB, coding for an accessory protein and a relaxase, respectively) is a part of the tra operon, while in the IncI1 plasmids it is located downstream of the trb cluster and the two regions are transcribed convergently. Moreover, there is no traEFG in pCTX-M3, and its tra and trb regions are separated by the IncM replicon and the 27-kb MRR.
Figure 2.
Gene organization of the transfer regions of pCTX-M3 and R64 and the dot/icm region of Legionella pneumophila. ORFs homologous in all systems are indicated by coloring. ORFs homologous in both plasmids are shown in grey, and those unique to one system only are shown in white. Gene group designation is indicated in brackets and individual genes are marked with appropriate letters. ORFs unrelated to the conjugation system are shown as black arrows.
The oriTpCTX-M3 sequence lies upstream of nikA and comprises two pairs of inverted repeats and the nic site ACATCTTG↓T similar to that of the IncI1 plasmids [25]. Moreover, pCTX-M3 can mobilize plasmids bearing oriT of the IncI1 plasmid ColIb-P9, and ColIb-P9 can mobilize plasmids bearing oriTpCTX-M3, albeit both with low efficiency [12].
pCTX-M3 can be transferred during bacterial conjugation not only on solid supports, but also in liquid. Interestingly, in contrast to the IncI1 plasmids which require type IV pili for conjugation in liquid media, pCTX-M3 does not encode additional pili [12,26], similarly to other IncM and IncL plasmids [27].
3.2.1. pCTX-M3 Conjugative Transfer System
The conjugative transfer can be regarded as a rolling circle (RC) replication coupled with a type IV protein transport system (T4SS) by a dedicated coupling protein (CP) [28]. During conjugation of bacteria other than Streptomyces, a plasmid or an integrative conjugative element (ICE) is transported as single-stranded DNA (ssDNA) complexed with specific proteins. The DNA transfer occurs after a physical contact between the donor and the recipient cell is established thanks to the activity of the MPF (mating pair formation) system, a secretion machinery for DNA–protein complexes, homologous with the type IV secretion system (T4SS). During bacterial conjugation the plasmid DNA is processed by the relaxosome complex (also called DTR—DNA transfer and replication). Its action involves the nicking of one strand of the plasmid DNA at a specific sequence, the nic site, located within the origin of transfer (oriT). The free 3’OH end of the nicked strand then serves as a primer for the synthesis of a new DNA strand in a process resembling RC replication. The relaxosome complex is composed of the relaxase, which is an ssDNA transesterase, and auxiliary proteins [29]. The relaxase remains covalently bound to the 5’ end of the ssDNA and is transported to the recipient cell. The DNA segment entering the recipient cell the earliest is termed the plasmid leading region. The coupling protein (CP) bringing together the action of the DTR and MPF systems is an ATPase, and it is also involved in targeting the transported plasmid DNA in the form of a nucleoprotein to the MPF system [30]. In the recipient cell, the ssDNA is circularized and then copied to form dsDNA [29].
The MPF systems of conjugative plasmids are evolutionarily related to the T4SS which transports virulence proteins of some pathogenic Gram-negative bacteria into eukaryotic cells [31]. The prototypical T4SS is the VirB/VirD4 transporter transferring the oncogenic T-DNA from Agrobacterium tumefaciens into plant cells. This T4SS consists of 11 proteins (VirB1 to VirB11), and VirD4 being the CP (for review, see [32,33]). The translocation channel comprises the VirB3 and VirB6–VirB10 proteins. Three components form the core channel complex in the outer membrane, also called the outer membrane complex (OMC): VirB9, the pore-forming protein; VirB7, a small lipoprotein; and VirB10, spanning both the inner and the outer membrane. The inner membrane complex (IMC) comprises the VirB3, VirB6, and VirB8 proteins. Interactions of the IMC with OMC and with the ATPases VirB4 and VirB11 result in the formation of a pore. The extracellular structure important for establishing the contact between the donor and recipient cells, the T-pilus, consists of VirB2, the major subunit, and VirB5, the minor component localized at the pilus tip. Additionally required for the T-pilus assembly is VirB3, the least-characterized MPF component. Finally, VirB1 shows homology to a lytic transglycosylase that cleaves peptidoglycan [34,35]. Three cytoplasmic ATPases, VirB4, VirB11, and the coupling protein VirD4, provide energy for the system.
The T4SSs of Gram-negative bacteria fall into two large phylogenetic groups, IVA and IVB [36]. The VirB/VirD4 transporter of A. tumefaciens is the prototype of type IVA systems (T4ASS) [32]. The type IVB group (T4BSS) comprises homologues of the Dot/Icm transporter of virulence proteins of Legionella pneumophila, the causative agent of Legionnaires’ disease, composed of approximately 27 proteins [34,37]. Most of the Dot/Icm proteins share homology with the elements of conjugation system of the IncI1 plasmid R64 [38] (Figure 2).
The homology between the proteins involved in R64 conjugative transfer and the components of the VirB/VirD4 transporter of A. tumefaciens is rather low [39]. The R64 conjugative transfer system is encoded by 22 genes from the tra region, traE–traY and the nuc gene, plus the trbA–C genes. Altogether, 16 of them have been shown to be indispensable for conjugation [23]. The conjugative system of pCTX-M3 also comprises tra and trb genes; however, as already mentioned, the transfer regions of these two plasmids do differ. Nevertheless, the pCTX-M3 proteins, similarly to that of R64, display some homology to a number of Dot/Icm proteins (Figure 2). Results of our bioinformatic analysis (for details see [21]) suggest that the T4SS of the pCTX-M3 conjugation system is composed of:
The transmembrane core subcomplex. Its OMC comprises TraN, TraI, and TraH, the homologues of Legionella DotH, DotC, and DotD, respectively. These Legionella proteins localize to the outer membrane forming the pore, similar to that formed by the VirB7–VirB9 proteins; by combining with the inner membrane proteins DotF and DotG they form the transmembrane core subcomplex [40]. In pCTX-M3, the DotF and DotG homologues are TraP and TraO; the latter is also a distant homologue of VirB10 [21,41];
The pilus—the TraR and TraQ proteins are distant homologues of VirB2, the major pilus subunit;
The VirB11 and VirB4 ATPases—TraJ is a homologue of VirB11 and TraU is a distant homologue of Legionella DotO and Agrobacterium VirB4, the only component common to all T4SSs of Gram-negative and Gram-positive bacteria, and archaea [39];
The coupling protein—the putative CP is TrbC. The CP subcomplex may also comprise TrbA, a DotM homologue;
The entry exclusion system—TraY and ExcA of pCTX-M3 are close homologues of respective R64 proteins [42].
A deletion analysis of individual genes from the tra and trb regions of pCTX-M3 revealed that all but three of them are important for conjugative transfer both in liquid media and on solid surface [21]. It is worth mentioning that deletion of traI or traO in IncI1 plasmids only diminished the conjugative transfer on solid support, and deletion of traH had no influence [23]. In contrast, the deletion of homologues of any of these three genes in pCTX-M3 abolished the transfer, suggesting different compositions of the core transmembrane subcomplexes encoded by these two plasmids.
The three genes found to be dispensable for the pCTX-M3 conjugative transfer are: orf36, located in the middle of the tra region, orf46 from the trb region, and orf35 from the leading region. Interestingly, a pCTX-M3 derivative without orf35 or orf36 used as a helper plasmid demonstrated an increased mobilization efficiency [21]. Deletion of orf35 resulted in a 100-fold increase in mobilization efficiency while the effect of orf36 deletion was dependent on the recipient species: with A. tumefaciens and E. coli as recipients, the increase was ca. 10- and 5-fold, respectively. An analysis of transcript levels of the 5′-most genes of the tra region of pCTX-M3 lacking orf35 led to a conclusion that the pCTX-M3 tra operon, which encodes both the relaxase complex and T4SS, is subject to orf35-dependent repression: expression of nikA, nikB, and traH was elevated ca. 40-, 23-, and 80-fold [21]. Deletion of orf36 did not change the level of transcripts of nikA or nikB, while that of traH was increased 120-fold, suggesting that the T4SS-encoding genes are controlled by an additional promoter subject to orf36-dependent repression. However, the overproduction of the Orf36 protein was not succeeded [43]. It should be noted here that within orf36 but on the opposite strand two other orfs were detected, one coding for an H-NS-like protein and the other an Hha-like protein, both members of the nucleoid-associated proteins (NAP) family. Plasmid-encoded NAPs participate in transcriptional silencing of horizontally acquired genes to prevent a fitness decrease in the host cell [44]. These two NAP-encoding orfs can also be identified in other plasmids of the IncL and IncM groups, especially when automatically annotated. They were first described in the R446 plasmid by Tietze and Tschäpe in 1994 [45] who were looking for an E. coli mutant that was insensitive to an M-specific phage. Taking into account that orf36 and NAP-encoding genes are located in the center of the tra region, the mode of regulation of the conjugative transfer genes in pCTX-M3 requires further studies.
Not only conjugative plasmids can be transferred during conjugation. Plasmids carrying an oriT sequence at least compatible with the MPF encoded by a plasmid co-residing in the host cell can be mobilized to conjugative transfer. The plasmid supplying the MPF is then regarded as a helper plasmid, and the plasmid being transferred with its help is called a mobilizable plasmid.
pCTX-M3 acting as a helper plasmid can mobilize the oriTpCTX-M3-bearing plasmids into recipients such as A. tumefaciens, Cupriavidus necator (previously Ralstonia eutropha), and Pseudomonas putida, representatives of Alpha-, Beta-, and Gammaproteobacteria, respectively [21], and also into the Gram-positive bacteria Bacillus subtilis and Lactococcus lactis [25]. pCTX-M3 itself, when transferred, cannot be established in any these bacteria [21,25]. These indicate that the range of hosts of the conjugative transfer system of pCTX-M3 is much broader than the host range of its replicon, which is restricted to Enterobacteriaceae (Enterobacterales ord. nov.).
4. Evolution of pCTX-M3-Related IncM Plasmids
At the end of the 20th century, 47% of strains of the plant pathogen Erwinia amylovora, the causative agent of fire blight, isolated in Lebanon carried a ca. 60-kb plasmid of the IncL/M group, pEL60 [46]. Its backbone is almost identical to that of pCTX-M3, e.g., the identity of the repCBA genes is 93%. The main difference between these plasmids is the 29-kb sequence present in pCTX-M3, organized in two regions with antibiotic resistance genes: first, downstream of pemK, with ISEcp1 introducing blaCTX-M-3, and the second, the MRR, separating repA and the trb region. Although pEL60 appeared to be the pCTX-M3 ancestor, it was classified as an IncM1 plasmid [19]. Surprisingly, the nucleotide sequence of a plasmid isolated from Enterobacter hormaechei in an Australian hospital, in 2019, pCP15_002 (GenBank acc. no. CP042490), is over 96% identical to that of pEL60. The plasmid carries no antibiotic resistance genes, but its entire sequence can be found in pCTX-M3: it is 98–99% identical to the pCTX-M3 backbone, with the rep region identical in 100%. Therefore, pC15_002 could be considered the actual pCTX-M3 ancestor, still circulating in the environment.
Interestingly, another plasmid, pCTX-M360, classified as IncM2, isolated from a multidrug-resistant K. pneumoniae strain from a patient in an intensive care unit in China in 2003, comprised a pEL60-like backbone (the rep region of 100% identity to that of pCTX-M3) with the ISEcp1-blaCTX-M-3 module inserted downstream of pemK, and with an insertion of Tn2 downstream of repA, introducing the blaTEM-1 gene [47]. Remnants of a similar transposon, Tn1, also with blaTEM-1, can be found within the MRR of pCTX-M3 [12]. Therefore, pCTX-M360 could be viewed as an intermediate between the ancestor, which probably was pEL60-like (or pC15_002-like), and pCTX-M3, and the insertion of Tn1 would then represent an early event in the evolutionary sequence leading to pCTX-M3.
Yet another plasmid, pNDM-HK, with blaNDM-1 encoding the New Delhi metallo-β-lactamase, isolated from E. coli of clinical origin in Hong Kong in 2009, can be considered a member of the pCTX-M3 family, and is also classified in the IncM2 subgroup [48]. It does not contain the ISEcp1-blaCTX-M-3 module, and within its MRR the module with the blaNDM-1 gene replaces the integron fragment located downstream of one of the IS26 copies in pCTX-M3 [48]. Additionally, pNDM-OM, another plasmid with the blaNDM-1 gene, isolated from a clinical K. pneumoniae strain in the Sultanate of Oman in 2010, is identical to pNDM-HK, lacking only two insertion sequences present in the latter [49]; pNDM-OM was also classified as belonging to the IncM2-subgroup.
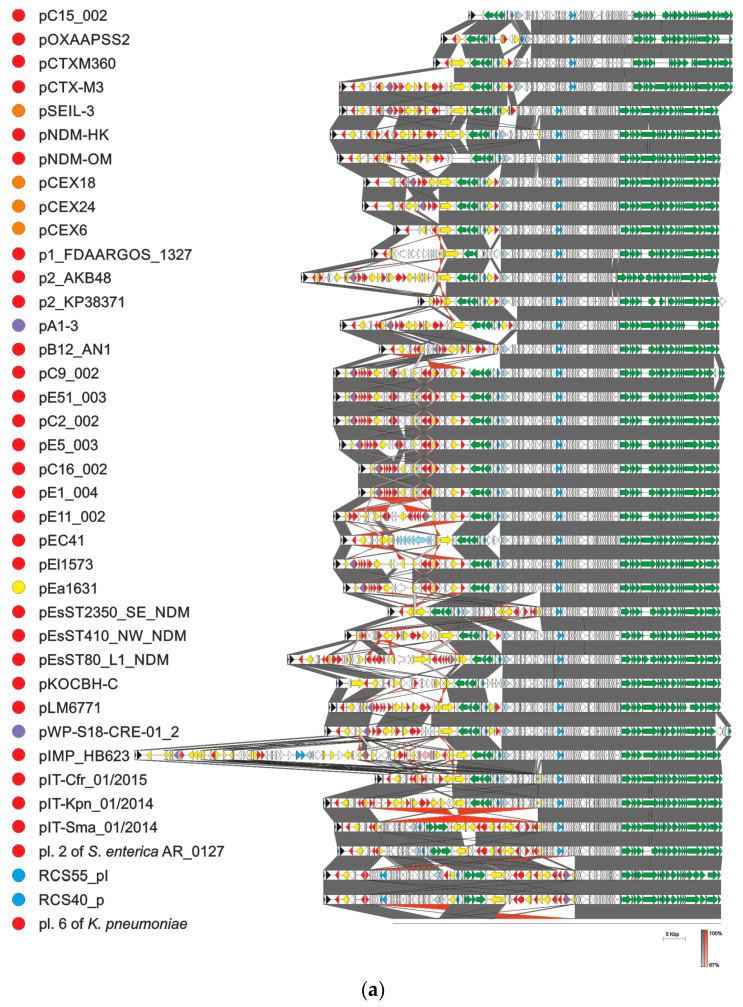
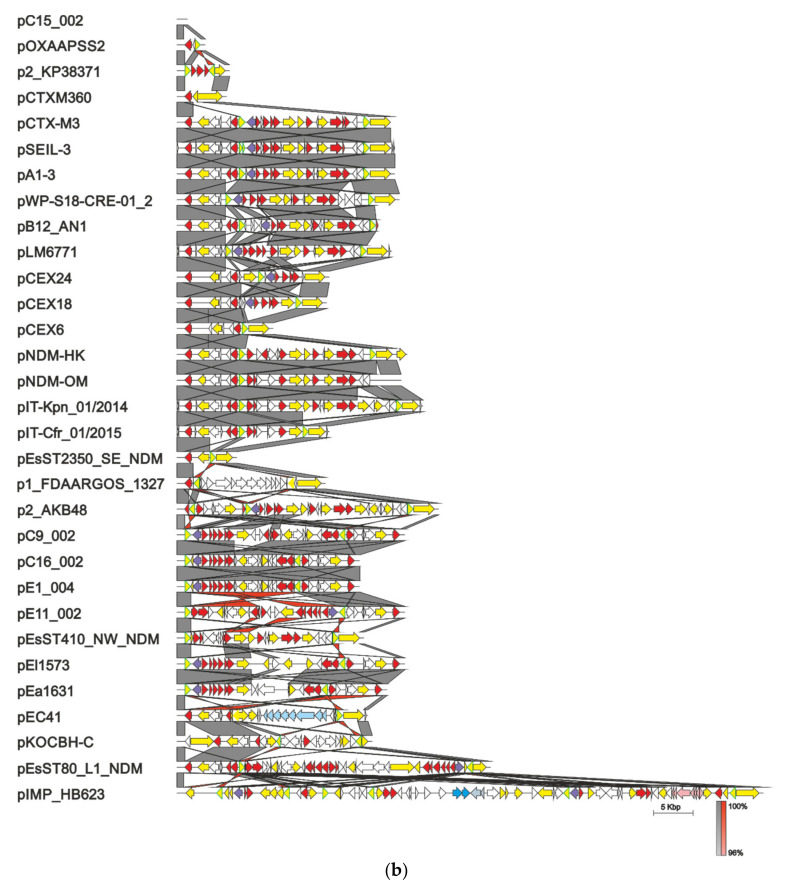
All known plasmids with 100% identity of their replicon region to the corresponding sequence of pCTX-M3 (1658 bp, starting 74 bp upstream of repC through repA, comprising repCBA and the RNA I-encoding sequence), found in the GenBank database (as of 25 March 2021), should be considered members of the pCTX-M3 family. They were found in Enterobacteriaceae strains (Enterobacterales ord. nov.) associated with health care (K. pneumoniae, E. coli, C. freundii, Serratia marcescens, Enterobacter cloacae, E. hormaechei, S. enterica subsp. enterica serovar Senftenberg, K. michiganensis, Enterobacter kobei, and Leclercia adecarboxylata). Some were also isolated from farm animals (from K. pneumonia, E. coli, E. cloacae, and S. enterica subsp. enterica serovar Havana), and additionally, a few originated from bacteria from municipal wastewater (K. pneumonia and E. cloacae), and one from Klebsiella aerogenes (pEa1631) from wildlife. Altogether, these environments indicate a vast diversity of reservoirs of the pCTX-M3 family plasmids. The 39 members of this family are shown in Figure 3a to the exclusion of sequences coming from the same survey and differing only in single nucleotides. As described above in detail for pCTX-M3, in most family members the modules comprising genes conferring antibiotic resistance can be found in two locations: the MRR between the rep region and the trb genes, and the shorter one downstream of pemK. The MRRs of the pCTX-M3 family plasmids, shown in Figure 3b, comprise a striking variety of genes conferring resistance to:
Figure 3.
pCTX-M3 family plasmids. (a) Alignment of pCTX-M3 and related plasmids; (b) Alignment of diverse multiple resistance regions. The ORFs are represented as arrows, rep genes are in black, drug resistance genes in red, mercury-resistance operon in pink, ferric citrate transport genes in pale blue, conjugative transfer genes in green, transposase genes in yellow (IS26 transposase genes are outlined in green in panel b), integron integrase-encoding genes in violet, parAB and pemIK in blue, mucA and mucB in grey, and others in white. Plasmid nucleotide sequences were taken from GenBank at following accession numbers: pC15_002—CP042490, pOXAPSS2—KU159086, pCTXM360—EU938349, pCTX-M3—AF550415, pSEIL-3—MN380440, pNDM-HK—HQ451074, pNDM-OM—JX988621, pCEX18—LC556221, pCEX24—LC556214, pCEX6—LC556219, p1_FDAARGOS_1327—CP069833, p2_AKB48—CP044337, p2_KP38371—CP014298, pA1-3—LC508263, pB12AN_1—CP026156, pC9_002—CP042532, pE51_003—CP042537, pC2_002—CP042522, pE5_003—CP042574, pC16_002—CP042580, pE1_004—CP042509, pE11_002—CP042526, pEC41—MW548582, pEl1573—JX101693, pEa1631—MG516907, pEsST2350_SE_NDM—CP031322, pEsST410_NW_NDM—CP031235, pEsST80_L1_NDM—CP031216, pKOCBH-C—CP035217, pLM6771—KX009507, pWP5-S18-CRE-01_2—AP022128, pIMP_HB623—KM877517, pIT-Cfr_01/2015—MH722216, pIT-Kpn_01/2014—MH722217, pIT-Sma_01/2014—MH722219, pl. 2 of S. enterica subsp. enterica serovar Senftenberg AR_0127—CP032193, RCS55_pl—LT985387, RCS40_pl—LT985241, and plasmid 6 K. pneumoniae—LT968692. All sequences were rotated or inverted and rotated to begin with repC. Plasmid schemes are drawn to scale. Original annotations are used. Color dots in front of the plasmid name denote the source of bacteria from which the plasmid was isolated: red—humans, orange—farm animals, yellow—wildlife, blue—humans or animals, and purple—wastewater. Sequence homology is color-coded according to scale below the alignment.
β-lactam antibiotics—blaTEM-1, usually located in proximity of the rep region, blaIMP-4 located within the integron, and also blaNDM-1, blaDHA-1, blaOXA-16, blaOXA-48, and blaIMP-34;
aminoglycosides—armA (nbrB) imported into the plasmid with ISCR1 as a part of a composite transposon flanked by two IS26 elements, aac(6’)-Ib4 related to IS26 and aac(3)-IId located within an integron, also aac(6’)-Ib-cr, aadA1, aadA16, aadA2, and strAB;
macrolides—mel, mph1, and mph2, frequently ISCR1 related;
chloramphenicol—catB3 located within the integron;
quinolones—qnrB2 and aac(6’)-Ib-cr;
sulphonamides and trimethoprim—sul1 and dfrA12, located within an integron, and also dfrA14 or dfrA27;
quaternary ammonium compounds—qacE and qacG2;
mercury—the mer operon;
fosfomycin—fosC2;
bleomycin—bleMBL.
The second location harbors blaCTX-M-3 (blaCTX-M-15 in one case) accompanied by ISEcp1. Only in pEsST2350_SE_NDM, a complex structure with multiple transposase genes and antibiotic resistance genes was found in this region.
All plasmids but one, pIMP_HB623 from an E. cloacae clinical strain from China, range in size from 66 to 98.5 kb. pIMP_HB623 of 133.2 kb carries a novel combination of genes conferring resistance to imipenem and fosfomycin within its 73-kb MRR [50]. This is a complex structure apparently generated by multiple recombination events. In four of the pCTX-M3-family plasmids the trb region is inverted: plasmid 2 of S. enterica subsp. enterica serovar Senftenberg from the USA, plasmid 6 of K. pneumoniae isolated in a Chinese hospital, and two plasmids, RCS55_pI and RCS40_p, isolated from E. coli strains in France. The inverted sequence blocks are bordered by two IS26s. Apart from this inversion, RCS55_pI and RCS40_p are 99% identical to pCTX-M3 along the entire sequence. Interestingly, pB12AN_1 isolated from a clinical K. pneumoniae strain in China lacks the trb genes, indicating a loss of self-transfer abilities. In the vast majority of the pCTX-M3 family plasmids the IS26 insertion sequences or their remnants are present at the borders of the sequence blocks that differentiate their MRR regions (Figure 3b). As in other plasmids, IS26 elements are important players in the plasmid evolution of the pCTX-M3 family by participating in the acquisition of antibiotic resistance genes [51,52]. It is also worth mentioning that some of the plasmids shown in Figure 3 were accompanied in the host cell by plasmids of other incompatibility groups facilitating further recombination events.
5. Taxonomy of IncL/M Plasmids
Since the 1970s numerous other plasmids classified as IncM, IncL, or IncL/M have been isolated from clinically relevant bacteria from the family Enterobacteriaceae (Enterobacterales ord. nov.). Their characteristic feature is one or two MRR(s) grouping antibiotic resistance genes as well as mobile genetic elements or their remnants. The problem of their classification was well presented in papers introducing new classification strategies in 2015 [19] and 2021 [53]. Initially, as discussed by Carattoli et al. [19], two groups were distinguished within 17 IncL/M plasmids—IncL and IncM, shown to be compatible with each other. Then, two subgroups were further distinguished among the IncM plasmids, IncM1 and IncM2; they were incompatible but phylogenetically well separated. Recently, the phylogenetic relations of 148 plasmids from the former IncL/M group were analyzed by Blackwell et al. [53]. By comparing the nucleotide sequences of 70 plasmid backbones they distinguished five subgroups within the IncL group, L1–L5. Plasmids recognized previously by Carattoli et al. [19] as members of the IncM1 and IncM2 groups were divided into five subgroups of IncM1, a single subgroup of IncM2, and additionally, four other subgroups were separated, IncM3–IncM6, [53]. While pEL60 became the unique member of the IncM3 subgroup, pCTX-M3, pCTXM360, pNDM-HK, and pNDM-OM remained the members of the IncM2 subgroup.
6. Conclusions
pCTX-M3 is a representative of a large family of low copy-number plasmids that are segregationally stable. Its conjugation system enables transfer of plasmids to a broad range of recipients of Alpha-, Beta-, and Gammaproteobacteria, and even to some Firmicutes, while it replicates only in Enterobacterales (ord. nov.). The reservoir of the pCTX-M3 family plasmids, which confer resistance to a wide variety of antibiotics, was found worldwide in diverse environments. As a part of a bacterial horizontal gene pool, such plasmids are involved in the exchange and spread of antibiotic resistance genes and finally, when disseminated in relevant bacteria, they pose serious problems for the health of humans and animals, compromising the efficiency of therapies. Therefore, it is an urgent need to understand the regulation of their conjugation system, aiming to develop agents which could counteract or control the conjugative transfer of plasmids.
7. Patents
Dmowski, M.; Kern-Zdanowicz, I. The helper plasmid, bacterial strain and the broad host range system for plasmid mobilization and uses thereof. PL230884. 10 September 2012.
Acknowledgments
I thank Michał Dmowski (Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland) for critical reading of the manuscript.
Funding
The project was funded by the National Science Centre, grant N N401 534640.
Conflicts of Interest
The author declares no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Adeolu M., Alnajar S., Naushad S., Gupta R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morgane. Int. J. Syst. Evol. Microbiol. 2016;66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 2.Cornaglia G., Garau J., Livermore D.M. Living with ESBLs. Clin. Microbiol. Infect. 2007;14:1–2. doi: 10.1111/j.1198-743X.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 3.Livermore D.M., Canton R., Gniadkowski M., Nordmann P., Rossolini G.M., Arlet G., Ayala J., Coque T.M., Kern-Zdanowicz I., Luzzaro F., et al. CTX-M: Changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 2007;59:165–174. doi: 10.1093/jac/dkl483. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind A., Schweighart S., Grimm H. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990;18:294–298. doi: 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez M.M., Power P., Radice M., Vay C., Famiglietti A., Galleni M., Ayala J.A., Gutkind G. Chromosome-Encoded CTX-M-3 from Kluyvera ascorbata: A Possible Origin of Plasmid-Borne CTX-M-1-Derived Cefotaximases. Antimicrob. Agents Chemother. 2004;48:4895–4897. doi: 10.1128/AAC.48.12.4895-4897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson A.B., Silverman M., Boyd D.A., Mcgeer A., Willey B.M., Daneman N., Mulvey M.R. Identification of a Progenitor of the CTX-M-9 Group of Extended-Spectrum beta-Lactamases from Kluyvera georgiana Isolated in Guyana. Antimicrob. Agents Chemother. 2005;49:2112–2115. doi: 10.1128/AAC.49.5.2112-2115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carattoli A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Woodford N. Successful, multiresistant bacterial clones. J. Antimicrob. Chemother. 2008;61:233–234. doi: 10.1093/jac/dkm474. [DOI] [PubMed] [Google Scholar]
- 10.Gniadkowski M., Schneider I., Pałucha A., Jungwirth R., Mikiewicz B., Bauernfeind A. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: Identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 1998;42:827–832. doi: 10.1128/AAC.42.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baraniak A., Fiett J., Sulikowska A., Hryniewicz W., Gniadkowski M. Countrywide Spread of CTX-M-3 Extended-Spectrum β-Lactamase-Producing Microorganisms of the Family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 2002;46:151–159. doi: 10.1128/AAC.46.1.151-159.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gołębiewski M., Kern-Zdanowicz I., Zienkiewicz M., Adamczyk M., Żylinska J., Baraniak A., Gniadkowski M., Bardowski J., Cegłowski P. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 2007;51:3789–3795. doi: 10.1128/AAC.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granier S.A., Hidalgo L., San Millan A., Escudero J.A., Gutierrez B., Brisabois A., Gonzalez-Zorn B. ArmA Methyltransferase in a Monophasic Salmonella enterica Isolate from Food. Antimicrob. Agents Chemother. 2011;55:5262–5266. doi: 10.1128/AAC.00308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toleman M.A., Bennett P.M., Walsh T.R. ISCR elements: Novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackwell G.A., Holt K.E., Bentley S.D., Hsu L.Y., Hall R.M. Variants of AbGRI3 carrying the armA gene in extensively antibiotic-resistant Acinetobacter baumannii from Singapore. J. Antimicrob. Chemother. 2017;72:dkw542. doi: 10.1093/jac/dkw542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Athanasopoulos V., Praszkier J., Pittard A.J. The replication of an IncL/M plasmid is subject to antisense control. J. Bacteriol. 1995;177:4730–4741. doi: 10.1128/JB.177.16.4730-4741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Athanasopoulos V., Praszkier J., Pittard A.J. Analysis of elements involved in pseudoknot-dependent expression and regulation of the repA gene of an IncL/M plasmid. J. Bacteriol. 1999;181:1811–1819. doi: 10.1128/JB.181.6.1811-1819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mierzejewska J., Kulińska A., Jagura-Burdzy G. Functional analysis of replication and stability regions of broad-host-range conjugative plasmid CTX-M3 from the IncL/M incompatibility group. Plasmid. 2007;57:95–107. doi: 10.1016/j.plasmid.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Carattoli A., Seiffert S.N., Schwendener S., Perreten V., Endimiani A. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS ONE. 2015;10:e0123063. doi: 10.1371/journal.pone.0123063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osborn A.M., Fernanda M., Tatley S., Steyn L.M., Pickup R.W., Saunders J.R. Mosaic plasmids and mosaic replicons: Evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology. 2000;146:2267–2275. doi: 10.1099/00221287-146-9-2267. [DOI] [PubMed] [Google Scholar]
- 21.Dmowski M., Gołębiewski M., Kern-Zdanowicz I. Characteristics of the conjugative transfer system of the IncM plasmid pCTX-M3 and identification of its putative regulators. J. Bacteriol. 2018;200 doi: 10.1128/JB.00234-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salje J., Gayathri P., Löwe J. The ParMRC system: Molecular mechanisms of plasmid segregation by actin-like filaments. Nat. Rev. Microbiol. 2010;8:683–692. doi: 10.1038/nrmicro2425. [DOI] [PubMed] [Google Scholar]
- 23.Komano T., Yoshida T., Narahara K., Furuya N. The transfer region of Incl1 plasmid R64: Similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 2000;35:1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., Zhang Y., Zhu L., Suzuki M., Inouye M. Interference of mRNA Function by Sequence-specific Endoribonuclease PemK*. J. Biol. Chem. 2004;279 doi: 10.1074/jbc.M314284200. [DOI] [PubMed] [Google Scholar]
- 25.Dmowski M., Kern-Zdanowicz I. A novel mobilizing tool based on the conjugative transfer system of the IncM Plasmid pCTX-M3. Appl. Environ. Microbiol. 2020;86:1–18. doi: 10.1128/AEM.01205-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida T., Furuya N., Ishikura M., Isobe T., Haino-fukushima K., Ogawa T., Komano T. Purification and Characterization of Thin Pili of IncI1 Plasmids ColIb-P9 and R64: Formation of PilV-Specific Cell Aggregates by Type IV Pili. J. Bacteriol. 1998;180:2842–2848. doi: 10.1128/JB.180.11.2842-2848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradley D.E. Determination of pili by conjugative bacterial drug resistance plasmids of incompatibility groups B, C, H, J, K, M, V, and X. J. Bacteriol. 1980;141:828–837. doi: 10.1128/JB.141.2.828-837.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llosa M., Gomis-Rüth F.X., Coll M., de la Cruz F. Bacterial conjugation: A two-step mechanism for DNA transport. Mol. Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 29.Zechner E.L., Lang S., Schildbach J.F. Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. B Biol. Sci. 2012;367:1073–1087. doi: 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koraimann G., Wagner M.A. Social behavior and decision making in bacterial conjugation. Front. Cell. Infect. Microbiol. 2014;4:54. doi: 10.3389/fcimb.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallden K., Rivera-Calzada A., Waksman G. Type IV secretion systems: Versatility and diversity in function. Cell. Microbiol. 2010;12:1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitzschke A., Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 2010;29:1021–1032. doi: 10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fronzes R., Schäfer E., Wang L., Saibil H.R., Orlova E.V., Waksman G. Structure of a Type IV Secretion System Core Complex. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christie P.J. The mosaic type IV secretion systems. EcoSal Plus. 2016;7 doi: 10.1128/ecosalplus.ESP-0020-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhatty M., Laverde Gomez J.A., Christie P.J. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grohmann E., Christie P.J., Waksman G., Backert S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol. Microbiol. 2018;107:455–471. doi: 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segal G., Feldman M., Zusman T. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 2005;29:65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Nagai H., Kubori T. Type IVB secretion systems of Legionella and other gram-negative bacteria. Front. Microbiol. 2011;2:1–12. doi: 10.3389/fmicb.2011.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guglielmini J., Bertrand N., Abby S.S., Garcillan-Barcia M.P., de la Cruz F., Rocha E.P.C. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or. Nucleic Acids Res. 2014;42:5715–5727. doi: 10.1093/nar/gku194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vincent C.D., Friedman J.R., Jeong K.C., Sutherland M.C., Vogel J.P. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol. Microbiol. 2012;85:378–391. doi: 10.1111/j.1365-2958.2012.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guglielmini J., Quintais L., Garcillán-Barcia M.P., de la Cruz F., Rocha E.P.C. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakuma T., Tazumi S., Furuya N., Komano T. ExcA proteins of IncI1 plasmid R64 and IncIγ plasmid R621a recognize different segments of their cognate TraY proteins in entry exclusion. Plasmid. 2013;69:138–145. doi: 10.1016/j.plasmid.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Wiśniewska M., Kern-Zdanowicz I. Attempts to overproduce the product of the orf36 gene of the pCTX-M3 plasmid of Enterobacteriaceae. Institute of Biochemistry and Biophysics, Polish Academy of Sciences; Warsaw, Poland: 2017. [Google Scholar]
- 44.Dillon S.C., Dorman C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 45.Tietze E., Tschäpe H. Temperature-dependent expression of conjugation pili by IncM plasmid-harbouring bacteria: Identification of plasmid-encoded regulatory functions. J. Basic Microbiol. 1994;34:105–116. doi: 10.1002/jobm.3620340206. [DOI] [PubMed] [Google Scholar]
- 46.Foster G.C., McGhee G.C., Jones A.L., Sundin G.W. Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 2004;70:7539–7544. doi: 10.1128/AEM.70.12.7539-7544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu W., Luo L., Wang J., Zhuang X., Zhong L., Liao K., Zeng Y., Lu Y. Complete nucleotide sequence of pCTX-M360, an intermediate plasmid between pEL60 and pCTX-M3, from a multidrug-resistant Klebsiella pneumoniae strain isolated in China. Antimicrob. Agents Chemother. 2009;53:5291–5293. doi: 10.1128/AAC.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ho P.L., Lo W.U., Yeung M.K., Lin C.H., Chow K.H., Ang I., Tong A.H.Y., Bao J.Y.-J., Lok S., Lo J.Y.C. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS ONE. 2011;6:e17989. doi: 10.1371/journal.pone.0017989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonnin R.A., Nordmann P., Carattoli A., Poirel L. Comparative Genomics of IncL/M-Type Plasmids: Evolution by Acquisition of Resistance Genes and Insertion Sequences. Antimicrob. Agents Chemother. 2013;57:674–676. doi: 10.1128/AAC.01086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y., Lo W.-U., Lai E.L., Chow K.-H., Ho P.-L. Complete Sequence of the Multidrug-Resistant IncL/M Plasmid pIMP-HB623 Cocarrying blaIMP-34 and fosC2 in an Enterobacter cloacae Strain Associated with Medical Travel to China. Antimicrob. Agents Chemother. 2015;59 doi: 10.1128/AAC.00375-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Partridge S.R., Thomas L.C., Ginn A.N., Wiklendt A.M., Kyme P., Iredell J.R. A novel gene cassette, aacA43, in a plasmid-borne class 1 integron. Antimicrob. Agents Chemother. 2011;55:2979–2982. doi: 10.1128/AAC.01582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harmer C.J., Hall R.M. IS26-Mediated Formation of Transposons Carrying Antibiotic Resistance Genes. mSphere. 2016;1 doi: 10.1128/mSphere.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blackwell G.A., Doughty E.L., Moran R.A. Evolution and dissemination of L and M plasmid lineages carrying antibiotic resistance genes in diverse Gram-negative bacteria. Plasmid. 2021;113:102528. doi: 10.1016/j.plasmid.2020.102528. [DOI] [PubMed] [Google Scholar]