Bacteria and phytoplankton form close associations in the ocean that are driven by the exchange of chemical compounds. The bacterial signal 2-heptyl-4-quinolone (HHQ) slows phytoplankton growth; however, the mechanism responsible remains unknown.
KEYWORDS: HHQ, Pseudoalteromonas, cell cycle, phytoplankton, quorum sensing, virus-host interactions
ABSTRACT
Interactions between phytoplankton and heterotrophic bacteria fundamentally shape marine ecosystems by controlling primary production, structuring marine food webs, mediating carbon export, and influencing global climate. Phytoplankton-bacterium interactions are facilitated by secreted compounds; however, linking these chemical signals, their mechanisms of action, and their resultant ecological consequences remains a fundamental challenge. The bacterial quorum-sensing signal 2-heptyl-4-quinolone (HHQ) induces immediate, yet reversible, cellular stasis (no cell division or mortality) in the coccolithophore Emiliania huxleyi; however, the mechanism responsible remains unknown. Using transcriptomic and proteomic approaches in combination with diagnostic biochemical and fluorescent cell-based assays, we show that HHQ exposure leads to prolonged S-phase arrest in phytoplankton coincident with the accumulation of DNA damage and a lack of repair despite the induction of the DNA damage response (DDR). While this effect is reversible, HHQ-exposed phytoplankton were also protected from viral mortality, ascribing a new role of quorum-sensing signals in regulating multitrophic interactions. Furthermore, our data demonstrate that in situ measurements of HHQ coincide with areas of enhanced micro- and nanoplankton biomass. Our results suggest bacterial communication signals as emerging players that may be one of the contributing factors that help structure complex microbial communities throughout the ocean.
IMPORTANCE Bacteria and phytoplankton form close associations in the ocean that are driven by the exchange of chemical compounds. The bacterial signal 2-heptyl-4-quinolone (HHQ) slows phytoplankton growth; however, the mechanism responsible remains unknown. Here, we show that HHQ exposure leads to the accumulation of DNA damage in phytoplankton and prevents its repair. While this effect is reversible, HHQ-exposed phytoplankton are also relieved of viral mortality, elevating the ecological consequences of this complex interaction. Further results indicate that HHQ may target phytoplankton proteins involved in nucleotide biosynthesis and DNA repair, both of which are crucial targets for viral success. Our results support microbial cues as emerging players in marine ecosystems, providing a new mechanistic framework for how bacterial communication signals mediate interspecies and interkingdom behaviors.
INTRODUCTION
Interactions between marine phytoplankton and bacteria have been shown to fundamentally shape marine ecosystems, particularly by mediating biogeochemical cycling, regulating productivity, and trophic structure (1–3). Bacterium-phytoplankton interactions are complex, often being species specific (4) or temporally ephemeral (5), and can span the spectrum from antagonistic to beneficial (6, 7). Increasingly, it is clear that these intricate interkingdom interactions are facilitated by excreted chemical compounds that mediate a suite of processes such as nutrient transfer, primary production, and shifts in community composition. Linking chemical compound identity with a mechanism of action and ecological consequences will strengthen our understanding of how these fundamental and multifaceted interactions govern marine ecosystem function.
First discovered in marine systems 4 decades ago (8), quorum sensing (QS) is a form of microbial cell-cell communication through which marine bacteria use diffusible chemical signals to facilitate coordinated and cooperative biogeochemically important behaviors (9). Recent work finds that alkylquinolone-based QS signals can modulate interspecies behavior, suggesting that these molecules may influence cellular communication at the interkingdom level (10). In particular, the alkylquinolone QS signal 2-heptyl-4-quinolone (HHQ) functions as a messenger molecule able to modulate bacterial virulence behavior, facilitating the emergence of the pathogen Pseudomonas aeruginosa within polymicrobial communities (11, 12). Trafficking of hydrophobic alkylquinolones, including HHQ, is aided by the release of outer membrane vesicles containing micromolar concentrations of alkylquinolones that are produced by P. aeruginosa and serve as signal delivery vehicles to neighboring recipient cells (13). Purified outer membrane vesicles isolated from P. aeruginosa have also been shown to possess significant antimicrobial activity, inhibiting the growth of adjacent Gram-positive bacteria (13). Additionally, HHQ has also been implicated in antagonizing fungal biofilm formation (12); downregulating eukaryotic host immune responses via the suppression of a key transcription factor, NF-κB (10); and activating receptors found to play a role in innate immune signaling in airway epithelia (14). These findings support the influence of alkylquinolones in mediating host-microbe interactions.
Recently, HHQ was isolated from marine gammaproteobacteria (Pseudomonas sp. and Pseudoalteromonas sp.) and was observed to cause significant shifts in both natural phytoplankton and microbial communities (15), including species-specific static phytoplankton growth (no growth or mortality) at nanomolar concentrations (16). Static growth in phytoplankton has been observed previously, in relation to both bacterial exudates (17, 18) and nutrient stress (19–22). However, the underlying molecular mechanism(s) by which HHQ influences phytoplankton fitness and the outcomes of ecological interactions remains unknown. For example, host physiology has been demonstrated to be an integral factor in the success of viral infection of phytoplankton, with infection success and burst size being influenced by host conditions (23). However, the role that HHQ plays in mediating microbial interactions beyond phytoplankton growth alterations has yet to be investigated.
To better understand how HHQ alters molecular function and ecological interactions in marine microbes, ultrastructural observations and diagnostic biochemical assays were integrated with transcriptomic and proteomic studies to link the persistent but reversible physiological impact of nanomolar concentrations of HHQ on a model marine phytoplankton, Emiliania huxleyi. Furthermore, we examined if HHQ could disrupt virus-induced mortality in E. huxleyi, thereby ascribing a new role for bacterial quorum-sensing signals. E. huxleyi plays a central role in mediating ocean carbon (24) and sulfur (25) cycling; thus, the results presented here emphasize the importance of considering the ecological consequences of chemically mediated bacterium-phytoplankton interactions on global primary production and biogeochemical cycles.
RESULTS AND DISCUSSION
Response to HHQ exposure.
Following 96 h of exposure to 100 ng ml−1 of HHQ, batch cultures of axenic E. huxleyi (CCMP2090) exhibited cellular stasis (no cell division or mortality) concomitant with a significant increase in forward scatter, red fluorescence, and side scatter, proxies for cell size, chlorophyll content, and cell granularity, respectively (P value of <0.01 for all comparisons by repeated-measures analysis of variance [ANOVAR]) (Fig. 1). The photosynthetic efficiency (Fv/Fm) did not change in response to long-term HHQ exposure (ANOVAR). Additionally, after only 24 h of HHQ exposure, phytoplankton cells were observed to have enlarged chloroplasts with distended thylakoid membranes containing numerous intraorganelle vesicles, abundant cytoplasmic vesicles/vacuoles, homogeneous nucleus staining lacking defined euchromatin/heterochromatin regions with disintegrated nuclear envelops, and osmium-rich puncta within and adjacent to the chloroplasts, likely indicating enhanced lipid storage (see Fig. S1 in the supplemental material). To examine if the physiological effects induced by HHQ exposure were reversible, 96-h-exposed HHQ cultures were diluted roughly ∼80-fold with f/2 medium without silica (26) to a final concentration of 1.25 ng ml−1 HHQ, a concentration demonstrated not to influence E. huxleyi growth. Cells previously exposed to HHQ showed recovery mirroring paired vehicle control cultures (ANOVAR) (Fig. S2). Taken together, HHQ-treated E. huxleyi cells appear to mirror previous studies in which cellular arrest has been observed in phytoplankton in response to bacterially derived chemical exposure (17, 18, 27–29) as well as nutrient limitation (20–22). In order to elucidate if the observed cellular stasis is mechanistically similar to those observed previously in the literature, we conducted cell cycle, transcriptomic, and proteomic analyses of HHQ-exposed E. huxleyi.
FIG 1.
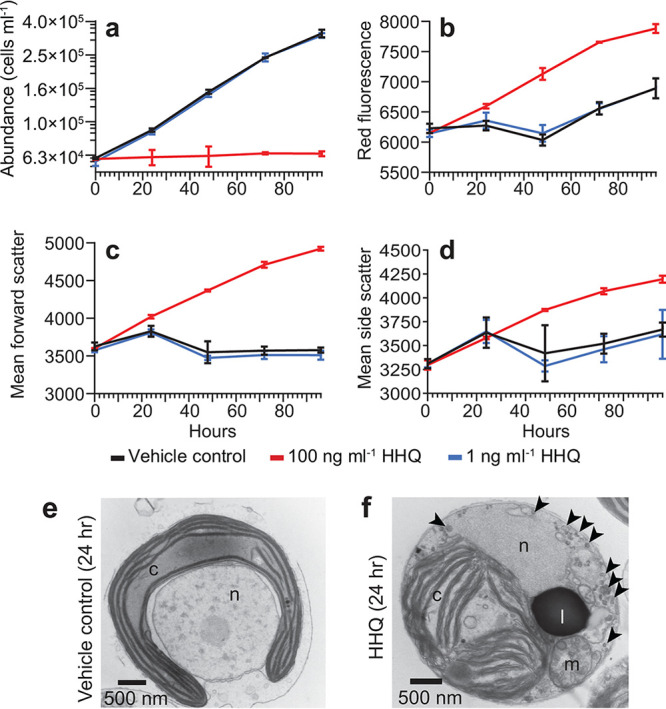
Exposure to HHQ halts cell division and alters cell morphology. (a through d) E. huxleyi cultures (n = 3) were exposed to HHQ or the vehicle control (DMSO) at the 0-h time point (T0) and monitored by flow cytometry for cell abundance (a), red fluorescence in relative fluorescence units (RFU) (695/50 nm) (a proxy for chlorophyll a intensity) (b), forward scatter (a proxy for cell size) (c), and side scatter (a proxy for cell granularity) (d) over 96 h. Means ± standard deviations are shown. In 100-ng ml−1 HHQ-exposed cells, all parameters measured were significantly different from those for the vehicle control (P < 0.05 by repeated-measures analysis of variance). Note that in panel a, data for HHQ-treated cells at 1 ng ml−1 sit directly beneath data for the vehicle control (DMSO). (e and f) Transmission electron microscopy micrographs of E. huxleyi cells exposed to the vehicle control (DMSO) (e) or 100 ng ml−1 HHQ (f) for 24 h. Subcellular structures include the chloroplast (c), lipid droplet (l), mitochondria (m), nucleus (n), and vacuoles (black arrowheads).
Composite TEM images showing representative images of E. huxleyi cells exposed to the vehicle control (DMSO) (a through f) or 100 ng ml−1 HHQ (g through l) for 24 h. In select images, subcellular structures are labeled for identification. c, chloroplast; l, lipid droplet; m, mitochondria; n, nucleus; p, pyrenoid; black arrow, Golgi apparatus; v, vacuole; white arrowheads, vesicles. Bar = 500 nm. Download FIG S1, PDF file, 0.7 MB (698.4KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
E. huxleyi cultures were examined for their ability to recover following exposure to 100 ng ml−1 of HHQ for 96 h. Following HHQ exposure, cultures were monitored for their ability to recover by measuring the growth rate (a) (each point represents the exponential growth rate over the previous 24 h), red fluorescence in RFU (a proxy for chlorophyll a intensity) (b), and forward scatter (a proxy for cell size) (c). Each symbol represents the mean from three independent replicates ± the standard deviation. Significant differences between HHQ-exposed cells and the vehicle control were assessed using ANOVAR (P < 0.05). Download FIG S2, PDF file, 0.3 MB (332.9KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evidence for S-phase arrest.
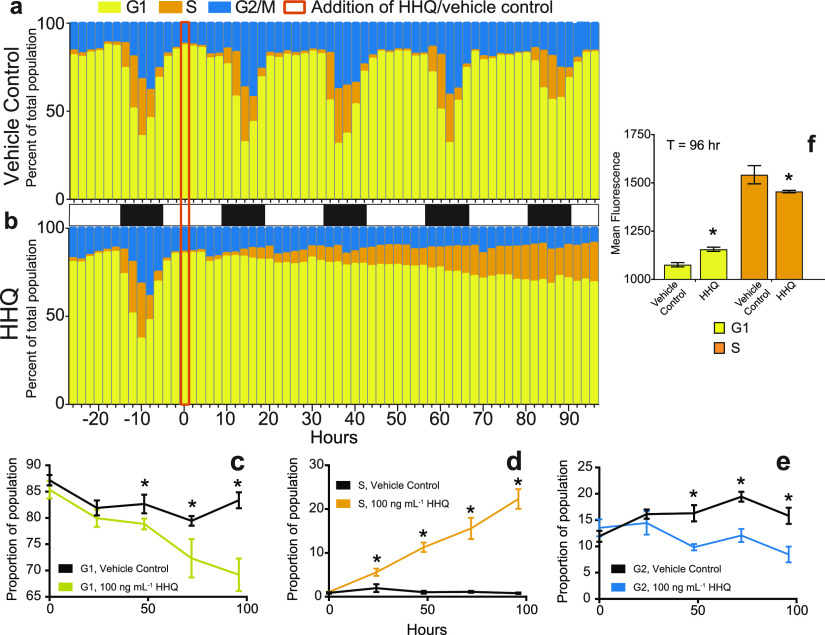
The DNA content of E. huxleyi cells following HHQ exposure was tracked for 96 h via flow cytometry, and cells treated with HHQ ceased the typical diurnal cell cycle progression within 24 h of HHQ addition (Fig. 2). Over 96 h, the proportions of HHQ-exposed cells in both G1 and G2 phases were shown to steadily decrease, whereas the proportion of cells in S phase significantly increased (P > 0.01 by ANOVAR) (Fig. 2c through e). Additionally, HHQ-exposed cells found in G1 phase demonstrated a significantly higher DNA content per cell than their paired vehicle controls (P value of <0.05 by Welch’s approximate t test) (Fig. 2f). These results suggest that HHQ-exposed cells are attempting to duplicate their genome but are stalling in early S phase.
FIG 2.
HHQ triggers stalling in S phase. (a and b) The cell cycle stage was quantified by profiling the fluorescence (575/25 nm), a proxy for DNA content, of propidium iodide-stained E. huxleyi cultures (n = 3) exposed to either the vehicle control (DMSO) (a) or 100 ng ml−1 HHQ (b) for 96 h. (c through e) The proportion of cells in each cell stage was determined from density plots of the distribution of cells with various DNA contents ranging from 2N (G1) to 4N (G2) at T0, T24, T48, T72, and T96. Cells with intermediate DNA content were denoted as S phase, as the genome replicated. Each plot represents the mean ± standard deviation for triplicate samples (P < 0.05 by ANOVAR). (f) Mean fluorescences (575/25 nm) of G1- and S-phase cells treated with the vehicle control (DMSO) or 100 ng ml−1 HHQ for 96 h and stained with propidium iodide were compared via Welch’s approximate t test (P < 0.01). As DNA replication occurs only in S phase, the increase in the mean fluorescence for HHQ-treated cells that fall within the G1 gate suggests that these cells are currently in S phase but stall early in the process of DNA synthesis and are unable to synthesize enough additional DNA to fall within the S-phase region.
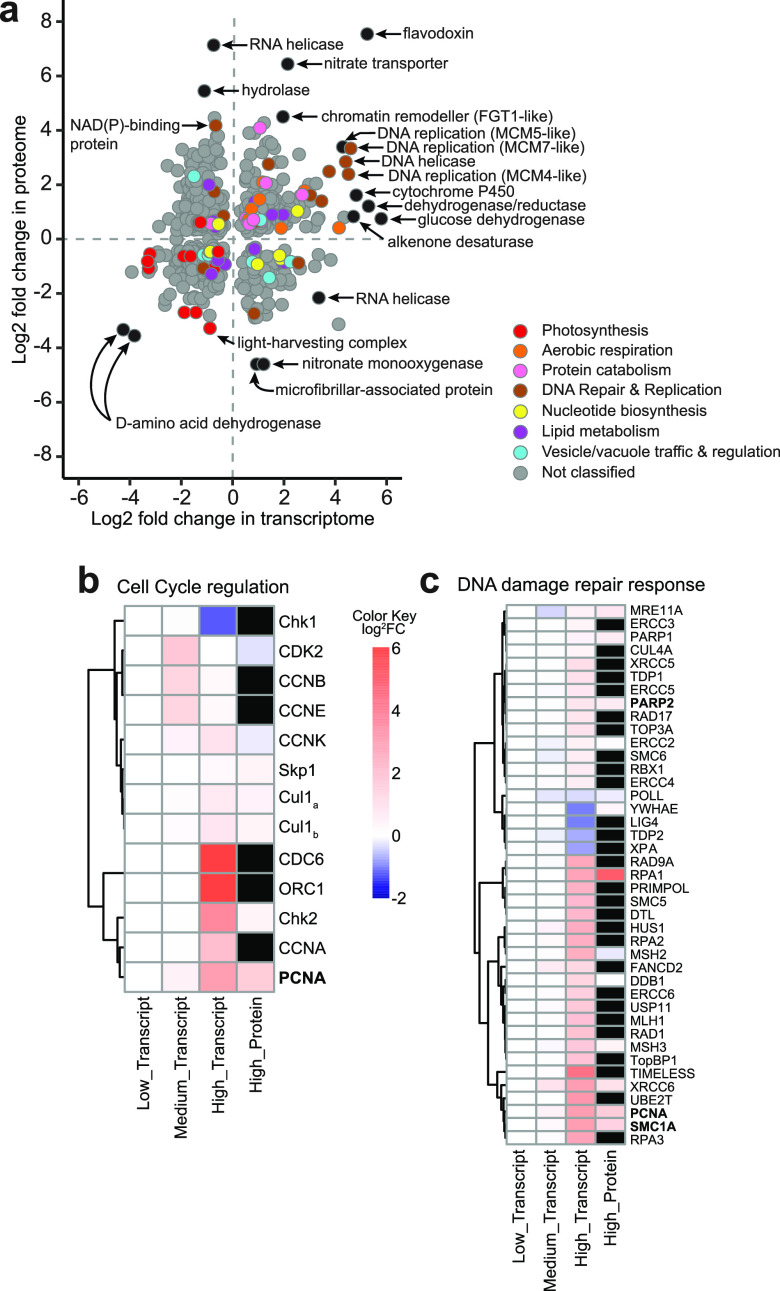
Whole-cell transcriptomic and proteomic analyses were performed on E. huxleyi cells exposed to HHQ concentrations of 1 ng ml−1 (low), 10 ng ml−1 (medium), and 100 ng ml−1 (high), with samples taken at 24 h (transcripts) and 72 h (transcripts and proteins) (Fig. S3). E. huxleyi cultures demonstrated unique transcriptomic and proteomic profiles in response to each HHQ concentration, with the greatest numbers of differentially expressed genes being found in higher-HHQ treatments, compared to the dimethyl sulfoxide (DMSO) vehicle control (Table 1; Fig. S3a and b). After 24 h of HHQ exposure, 39.8% of transcripts in high-HHQ samples were differentially expressed relative to the DMSO vehicle controls (q value of <0.05 by a Wald test) (Table 1). Similarly, after 72 h of exposure, replicate high-HHQ samples continued to appear distinct from the DMSO vehicle control samples (Fig. S3a and b), with 37.6% of transcripts (q value of <0.05 by a Wald test) and 15.9% of proteins (q value of <0.05 by Welch’s approximate t test) significantly changing in relative abundance and abundance, respectively (Table 1). When examined together, a total of 665 genes and corresponding proteins were found to significantly change in abundance at 72 h under high-HHQ treatment relative to the vehicle control (Fig. 3) (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). In general, processes associated with DNA replication and repair, aerobic respiration, and protein catabolism yielded higher relative transcript and protein abundances under high-HHQ treatment, while photosynthetic components/processes were detected at lower relative transcript and protein abundances (Fig. 3a) (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). Far fewer genes and proteins were found to be differentially expressed in the low- and medium-HHQ treatments (Table 1), which is likely related to the observed recovery of cell growth in these treatments (Fig. S3c). The growth of cells exposed to low HHQ concentrations was nearly identical to that of the DMSO control throughout the experiment. By 72 h, no genes or proteins in the low-HHQ treatment were differentially expressed compared to the control. While the medium-HHQ treatment demonstrated some growth inhibition over the first 24 h, by 72 h, the population had largely recovered (Fig. S3c). For cell populations exposed to medium HHQ concentrations at 24 h, transcripts related to cell cycle progression, cytoskeletal regulation, and mitosis demonstrated increased relative abundances compared to the control (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). By 72 h, there was no clear trend observed in functions related to differentially expressed transcripts in the medium-HHQ samples. For the purposes of elucidating the molecular target(s) of HHQ, we therefore focused our efforts on the analysis of the high-HHQ transcriptomic and proteomic data.
TABLE 1.
Summary of differentially expressed transcripts and proteins following HHQ exposurea
| HHQ concn (ng/ml) | No. of transcripts or proteins |
|||||
|---|---|---|---|---|---|---|
| 24 h |
72 h |
|||||
| Upregulated | Downregulated | Unchanged | Upregulated | Downregulated | Unchanged | |
| Transcripts | ||||||
| 1 | 13 | 20 | 31,476 | 0 | 0 | 31,549 |
| 10 | 2,702 | 1,990 | 26,817 | 382 | 159 | 31,008 |
| 100 | 5,948 | 6,605 | 18,956 | 6,166 | 5,698 | 19,685 |
| Proteins | ||||||
| 1 | NA | NA | NA | 0 | 0 | 5,528 |
| 10 | NA | NA | NA | 3 | 0 | 5,525 |
| 100 | NA | NA | NA | 628 | 375 | 4,525 |
Expression with each HHQ treatment was compared to expression with the vehicle control (DMSO) treatment (q value of <0.05 by Welch’s approximate t test). NA, not applicable.
FIG 3.
Molecular and proteomic changes as a result of HHQ exposure. (a) Comparison of log2 fold changes in transcript (x axis) and protein (y axis) expression from E. huxleyi cultures (n = 4) following exposure to 100 ng ml−1 HHQ for 72 h compared to the vehicle control (DMSO). Only shared differentially expressed transcripts (q value of <0.05 by a Wald test) and proteins (q value of <0.05 by Welch’s approximate t test) are shown for a total of 665 genes/proteins. Transcripts and proteins with similar functions are colored via gene ontology (GO) annotation according to the curated groupings shown in Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1. Genes and proteins without GO annotations or annotations outside the selected groupings are shown in gray. Selected outliers are labeled in black. (b and c) Heat maps displaying putative homologs of E. huxleyi protein-coding genes associated with cell cycle regulation (b) and the DNA damage repair response (c) after 72 h of HHQ exposure. Black boxes indicate proteins that were not detected in the proteomic analysis. Names in boldface type indicate those protein-coding genes found within the scatterplot in panel a. Dendrograms indicate hierarchical clustering based on the similarity of gene/protein expression levels. FC, fold change.
(a) Principal-component analysis performed on the regularized-logarithm-transformed transcriptomic read count matrix. Each point represents an experimental sample, with point color indicating the treatment and shape indicating the time point. The first component explains 39% of the variance, and the second component explains 20%. (b) Principal-component analysis performed on the logarithm-transformed proteomic data matrix (protein summed peptide peak area values + 1). Each point represents an experimental sample, with point color indicating the treatment. The first component explains 29.8% of the variance, and the second component explains 16.2%. Note that protein was collected only at the 72-h time point. (c) Growth curve of E. huxleyi (CCMP2090 [axenic]) cultures exposed to 1, 10, and 100 ng ml−1 HHQ and sampled for RNA (at 24 and 72 h) and protein (at 72 h). The arrow indicates the time of HHQ addition. Download FIG S3, PDF file, 0.2 MB (261.1KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Indeed, at the physiological level, the response of E. huxleyi to HHQ parallels phosphorus (P) limitation in phytoplankton (i.e., S/G2-phase arrest, decreased growth rate, and increased chlorophyll content, forward scatter, and side scatter) (19–21, 30). However, the canonical response in P-limited cells of the upregulation of both alkaline phosphatase and phosphodiesterases (31–33) was not observed in cells exposed to HHQ, nor did we see a significant induction of acid phosphatases, pyrophosphatase, phosphorus transporters, or ATP-sulfurylase enzymes known to be induced following P limitation in HHQ-exposed cells, indicating the lack of phosphorus stress (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). Therefore, while the patterns of cell cycle arrest are similar between HHQ-treated E. huxleyi and nutrient limitation, the underlying mechanisms are distinct.
In phytoplankton, cellular arrest is often accompanied by the induction of autocatalytic or programmed cell death (PCD) responses such as increased reactive oxygen production or caspase-like activity (34), and previous findings in mammalian cells indicate that HHQ has the ability to activate PCD pathways (35). However, no evidence of PCD/apoptosis was observed in HHQ-exposed E. huxleyi cells using a series of diagnostic fluorescence assays (i.e., membrane permeabilization, caspase activity, and reactive oxygen species [ROS] and nitrous oxide [NO] production) (Fig. S4). Additionally, no transcripts or proteins associated with PCD increased in abundance with exposure to HHQ (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). The lack of PCD induction in HHQ-exposed cells may stem from E. huxleyi’s arrest in early S phase (Fig. 2d), as cellular arrest during S phase does not induce apoptotic pathways but rather curtails DNA replication, thereby dramatically extending the cell cycle (36). The transcriptomic profile of HHQ-exposed cells demonstrates an increased relative abundance of canonical transcripts facilitating the G1/S transition, including cell division control protein 6 (CDC6), origin recognition complex subunit 1 (ORC1), and cyclins A, B, E, and K (Fig. 3b). Moreover, significant increases in relative transcript abundances of DNA replication fork machinery (i.e., DNA polymerases α, ε, and δ; DNA primase; replication protein A; topoisomerases [TOPO]; the minichromosomal maintenance complex; proliferating cell nuclear antigen; and replication factor C) (Fig. 3c) 72 h after HHQ exposure suggest an intent to replicate DNA, a hallmark of S phase (37). Yet despite this observed induction of DNA replication machinery, DNA synthesis was severely curtailed following HHQ exposure (Fig. 2), suggesting that HHQ exposure interferes with the ability of E. huxleyi cells to correctly complete the DNA replication process.
Diagnostic biochemical assays for caspase activity from whole-cell lysates or purified caspase enzyme (n = 3) (comparison of the whole-cell lysate at 72 h of DMSO treatment versus the whole-cell lysate at 72 h of HHQ treatment [P value of <0.05 by Welch’s approximate t test {no significance observed}]) (a), the presence of active caspase proteases in vivo (CaspACE) (b), reactive nitrogen species (DAF-FM diacetate) (c), reactive oxygen species (CM-H2DCFDA) from DMSO- or HHQ-exposed cultures from 1 to 72 h posttreatment (d), reactive oxygen species (CM-H2DCFDA) from 72-h DMSO- or HHQ-exposed cultures that were spiked with the algicide tetrabromopyrrole (TBP) (positive control) for 1 or 2 h (e), apoptosis (Image-iT Dead stain) from DMSO- or HHQ-exposed cultures from 1 to 72 h posttreatment (f), apoptosis (Image-iT Dead stain) from 72-h DMSO- or HHQ-exposed cultures that were spiked with TBP (positive control) for 4.5 h (g), and mitochondrial membrane integrity (MitoHealth stain) (h). Bars represent the means ± standard deviations from triplicate readings. Significance in panels b through d, f, and h was based on ANOVAR followed by Dunnett’s multiple-comparison test (P value of <0.05). Significance in panels e and g was based on Student’s t test (P value of <0.05). Cells exposed to HHQ were significantly different from the control only using MitoHealth stain after 24 h of exposure. However, the two treatments were not significantly different from one another at all subsequent time points. Download FIG S4, PDF file, 0.3 MB (309.3KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Disruption of DNA replication induces DNA damage response (DDR) pathways, activating effector kinases such as Chk1 and Chk2 necessary for halting DNA synthesis and the induction of cell cycle arrest to allow time for repair (38). We observed transcripts for Chk1 and Chk2 to be differentially expressed under HHQ treatment (Fig. 3b). Furthermore, a significant decrease in relative histone transcript and protein abundances, a hallmark of DNA synthesis disruption, was observed (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1) following HHQ exposure. As DNA replication and histone production are coupled, cells experiencing DNA replication stress will remove histone transcripts (39).
Possible protein targets of HHQ.
During S phase, a cell must tightly regulate the availability of nucleotides to ensure faithful DNA replication (40). Therefore, S-phase cells rely on de novo nucleotide synthesis pathways to produce enough materials for complete genome replication (41). Several transcripts and proteins involved in de novo purine (amidophosphoribosyltransferase, trifunctional purine biosynthetic protein adenosine 3, phosphoribosylformylglycinamidine synthase, bifunctional purine biosynthesis protein, adenylosuccinate synthase, IMP dehydrogenase, and GMP synthase) and pyrimidine (carbamoyl phosphate synthase II, aspartate carbamoyltransferase, and CTP synthases) nucleotide synthesis increased in abundance with HHQ exposure (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). Increased nucleotide synthesis may indicate the need to produce the necessary materials to replenish nucleotide pools during replication. However, only partial replication of the E. huxleyi genome following HHQ exposure was observed (Fig. 2), suggesting that HHQ may disrupt nucleotide production, thereby limiting nucleotide availability.
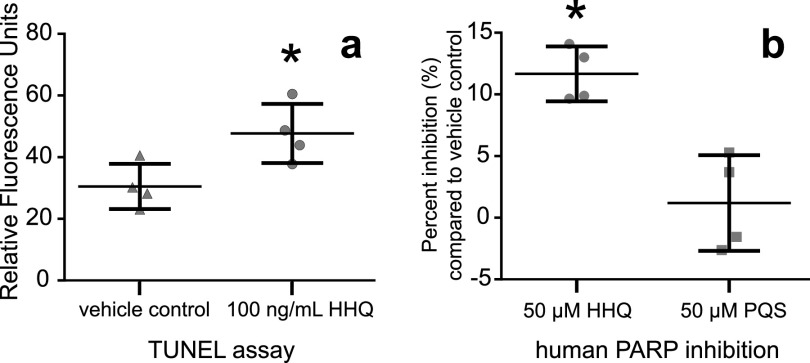
Select alkylquinolones are known to inhibit a key rate-limiting enzyme directly involved in pyrimidine synthesis, dihydroorotate dehydrogenase (DHODH) (42). DHODH inhibition in eukaryotes may induce intra-S-phase arrest due to severely diminished cellular nucleotide pools that can disrupt DNA replication, stall replication forks, and increase the frequency of genomic DNA lesions, including strand breaks, during S phase (43, 44). Indeed, after 46 h of HHQ exposure, a significant increase in DNA strand breaks was observed in culture (P = 0.032 by Welch’s approximate t test) (Fig. 4a) and was not observed when HHQ was directly exposed to genomic E. huxleyi or lambda DNA (Fig. S5). This indicates that DNA strand breaks are not caused directly by HHQ but are caused indirectly through other mechanisms. It has been previously observed that following the induction of DNA damage during S phase, cells will enter intra-S-phase arrest that drastically decreases the rate of DNA replication to allow the DDR to resolve any DNA lesions (36). With the exception of preliminary work in Chlamydomonas reinhardtii and dinoflagellates, the DDR has not been well characterized in phytoplankton (45, 46). Of the 57 mammalian DDR protein homologs in the E. huxleyi genome (E value of ≤10−20), 41 were significantly differentially expressed (at the transcript and/or protein level), of which 37 increased in relative abundance at 72 h under high-HHQ exposure (Fig. 3c), indicating that the cell is attempting to repair DNA lesions. However, DNA damage induced by the inhibition of DHODH is known to activate apoptotic pathways through the hyperactivation of the DDR (47). No apoptotic pathway activation was observed with HHQ exposure, suggesting that the DDR itself may also be impacted by HHQ.
FIG 4.
Exposure to HHQ leads to cellular DNA damage and inhibition of human PARP. (a) Cultures (n = 4) of E. huxleyi were exposed to 100 ng ml−1 HHQ or the vehicle control (DMSO) for 46 h before pigments were removed and cells were stained using an in vivo TUNEL assay to detect the presence of DNA ends, a proxy for DNA breaks. (b) Inhibition of the human PARP-1 enzyme by 50 μM HHQ and 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS). Percent PARP inhibition was measured using the PARP universal colorimetric assay kit (R&D Systems). The absorbance values for quadruplicate wells containing HHQ or PQS were compared to those of the vehicle control, and this ratio was subtracted from 100% to determine PARP inhibition. Points represent individual replicates. Asterisks indicate a significant difference between the treatment and the vehicle control (P value of <0.05 by Welch’s approximate t test).
HHQ does not directly lead to DNA strand breaks. Lambda DNA and genomic DNA isolated from E. huxleyi CCMP2090 were incubated with 100 ng ml−1 HHQ or the volumetric equivalent of DMSO for 24 h at 18°C, and the presence of DNA strand breaks was assessed by an agarose gel stained with ethidium bromide. Download FIG S5, PDF file, 0.6 MB (608.7KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A master regulator of the DDR involved in chromatin remodeling, nucleolar structure, and genome stability is poly(ADP-ribose) polymerase (PARP) (48). PARP binds to sites of DNA damage and stalls replication forks, producing negatively charged ADP-ribose polymer scaffolds that attract repair proteins (49). PARP homologs in E. huxleyi were found to be increased in both relative transcript abundance and protein abundance under HHQ treatment (Fig. 3c). Under high levels of DNA damage or if repair mechanisms are compromised, PARP can become overactivated and deplete cellular NAD+ and ATP pools, thereby initiating apoptotic pathways (50). However, no apoptotic activity was observed in E. huxleyi cells following HHQ exposure (Fig. S4), indicating that HHQ may inhibit PARP activity. Indeed, HHQ was found to significantly inhibit human PARP activity (P = 0.0002 by Welch’s approximate t test) (Fig. 4b), while a closely related alkylquinolone, 2-heptyl-3-hydroxy-4(1H)-quinolone (PQS), did not possess PARP-inhibitory activity, nor did it impact E. huxleyi growth (Fig. 4b and Fig. S6a). Interestingly, the genomes of phytoplankton species unaffected by HHQ (16) did not reveal the presence of any PARP homologs, further implicating PARPs in the response of phytoplankton to HHQ.
Impact of alkylquinolones on phytoplankton growth and protein homology modeling. (a) Effect of PQS on E. huxleyi growth. Shown is a dose-response curve of E. huxleyi (strain CCMP2090 [axenic]) in response to PQS. Each symbol represents the mean from three independent replicates ± the standard deviation. Protein homology modeling of the E. huxleyi PARP-1-like homolog (GenBank accession number XP_005783504.1) to the crystal structure of human PARP-1 (PDB accession number 2RD6) with the PARP inhibitor veliparib was performed. (b) The sequence of the E. huxleyi PARP-like homolog (GenBank accession number XP_005783504.1) was aligned to the sequence of human PARP-1 (PDB accession number 2RD6). Two key amino acid residues (red), Tyr and Glu, are strictly conserved between human PARP and the E. huxleyi protein. (c and d) The active site of the structure under PDB accession number 2RD6 shows the binding of the inhibitor veliparib. Regions in the structure under PDB accession number 2RD6 with the greatest homology to the E. huxleyi PARP-like homolog are shown in pink, with the remainder of the protein chain in green. The region comprising the active site for small-molecule binding is highly conserved between the two proteins, with two key binding interactions observed: the nearly coplanar arrangement of a tyrosine side chain phenyl to the inhibitor and a water-mediated hydrogen bond of the basic nitrogen atom of the inhibitor to a glutamate side chain carboxyl. Download FIG S6, PDF file, 0.9 MB (908KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inhibition of PARP activity in the presence of DNA damage drastically reduces the effectiveness of the DDR and is known to induce cellular arrest in the S phase (51). Together, our observations of prolonged S-phase arrest (Fig. 2), the upregulation of the DDR in HHQ-exposed cultures (Fig. 3c), the conserved nature of the mammalian and E. huxleyi PARP catalytic sites (Fig. S6b through d), and the chemical structural similarities of HHQ to known inhibitors of both PARP and DHODH with core benzimidazole moieties (52) collectively suggest that HHQ may function simultaneously to inhibit both PARP and DHODH activity in E. huxleyi. Additional experiments using E. huxleyi enzymes are needed to fully characterize whether PARP and DHODH are molecular targets of HHQ.
HHQ impacts on energy production.
To facilitate DNA synthesis and repair, the cell requires large ATP pools (53). In HHQ-exposed cells, the increased relative transcript abundance of enzymes in the tricarboxylic acid (TCA) cycle (i.e., isocitrate dehydrogenase, α-ketoglutarate dehydrogenase, succinate dehydrogenase, fumarase, and malate dehydrogenase) (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1) may signal the overproduction of reducing equivalents for ATP production via oxidative phosphorylation. Additionally, the increase in the relative transcript abundance of metabolic efficiency controllers, sirtuin-like deacetylases (54), observed following HHQ treatment (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1) may be a direct result of PARP inhibition. Sirtuins compete with PARPs for NAD+, and the expression of deacetylases is dependent on NAD+ availability (55). PARP inhibition is known to drastically increase cellular NAD+ pools, thereby promoting sirtuin expression and activity (56). Increased sirtuin activity in HHQ-exposed cells may also explain the increase in the relative transcript abundance of manganese superoxide dismutase (Mn-SOD) (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1), an antioxidant enzyme that protects the cell from ROS-induced damage, as sirtuins are known to induce the production of Mn-SOD proteins (57). Finally, increased relative transcript abundance of the tryptophan-mediated de novo NAD+ synthesis pathway was also observed, potentially in an attempt to increase NAD+ availability (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). Taken together, these results suggest that HHQ exposure promotes increased energy production in E. huxleyi, which can fuel various cellular biosynthesis and repair pathways while staving off the induction of PCD.
Increased cellular demand for ATP would necessitate the induction of glycolytic enzymes. However, following HHQ treatment, there was a significant decrease in the relative transcript abundance of hexokinase (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1), the first step in glycolysis, consistent with previous work demonstrating that alkylquinolones suppress the induction of this glycolytic enzyme through direct targeting of transcription factor hypoxia-inducible factor 1 (HIF-1) protein degradation via proteasomal pathways (58). Furthermore, we observed a shift to the Entner-Doudoroff glycolytic pathway in HHQ-treated cells (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1), which can conserve amino acid resources due to a low protein demand in comparison to other pathways (59). Moreover, we observed increases in relative transcript abundances leading to the production of aspartate (i.e., the TCA cycle, the aspartate-arginosuccinate shunt, glutamic oxaloacetic transaminase [GOT], and C4-like photosynthesis) in parallel with decreases in transcripts for aspartate utilization pathways, with the exception of nucleotide synthesis (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). Aspartate is known to rescue cells from S-phase arrest by fueling de novo nucleotide synthesis (60).
HHQ impacts on photosynthesis and redox.
HHQ-induced cell cycle arrest in E. huxleyi did not significantly alter the photosynthetic energy conversion efficiency; however, the majority of light-harvesting complexes and transcripts of the Calvin cycle decreased in relative abundance under HHQ exposure (Fig. 3a). These findings parallel those described previously for the diatom Phaeodactylum tricornutum undergoing chemically mediated cell cycle arrest (61). In plants, the coordinated downregulation of transcripts involved in photosynthesis, electron transport, and the Calvin cycle is thought to allow for the reallocation of resources toward defense against bacterial and viral pathogens (62). However, a decrease in transcript abundance does not always correlate with a loss of protein function, as photosynthetic proteins have a long functional half-life in the cell, with the exception of ferredoxin (Fd) and ferredoxin NADP+ oxidoreductase (FNR), both of which are involved in maintaining the cellular redox state following pathogen infection (62). Together, both ferredoxin and the isofunctional flavodoxin (Fld) participate in electron shuttling, preventing electron misrouting that can lead to ROS accumulation and restoring chloroplast redox homeostasis under environmental stress (63). Indeed, the genes and proteins with the most significant differential expression levels under HHQ exposure in E. huxleyi were Fd (58-fold increase in transcript and 3-fold increase in protein abundances), FNR (85-fold increase in transcript abundance), and Fld (38-fold increase in transcript and 186-fold increase in protein abundances) (Fig. 3a) (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1), which may explain the observed lack of ROS production (Fig. S4). Additional reduction systems, including FAD/NAD(P)-binding oxidoreductase, ferredoxin nitrite reductase (Fd-NR), and glutathione reductase (GR), in HHQ-treated E. huxleyi cells were also significantly induced, which could ameliorate NADPH buildup (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). Moreover, increased relative expression of vitamin B6 (VitB6) transcripts following HHQ treatment could protect against oxidative stress in chloroplasts (64), while increased relative expression levels of transcripts encoding proline oxidase (POX), pyrroline-5-carboxylate reductase (P5CR), and manganese superoxide dismutase (Mn-SOD) could explain the lack of mitochondrial ROS toxicity (Fig. S4) (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1). Together, these results suggest that HHQ-exposed E. huxleyi uniformly decreased the relative abundance of photosynthetic gene transcripts in support of a coordinated induction of defense responses aimed at maintaining cellular redox homeostasis without debilitating photosynthetic capacity.
Consequences of HHQ-induced cellular stasis.
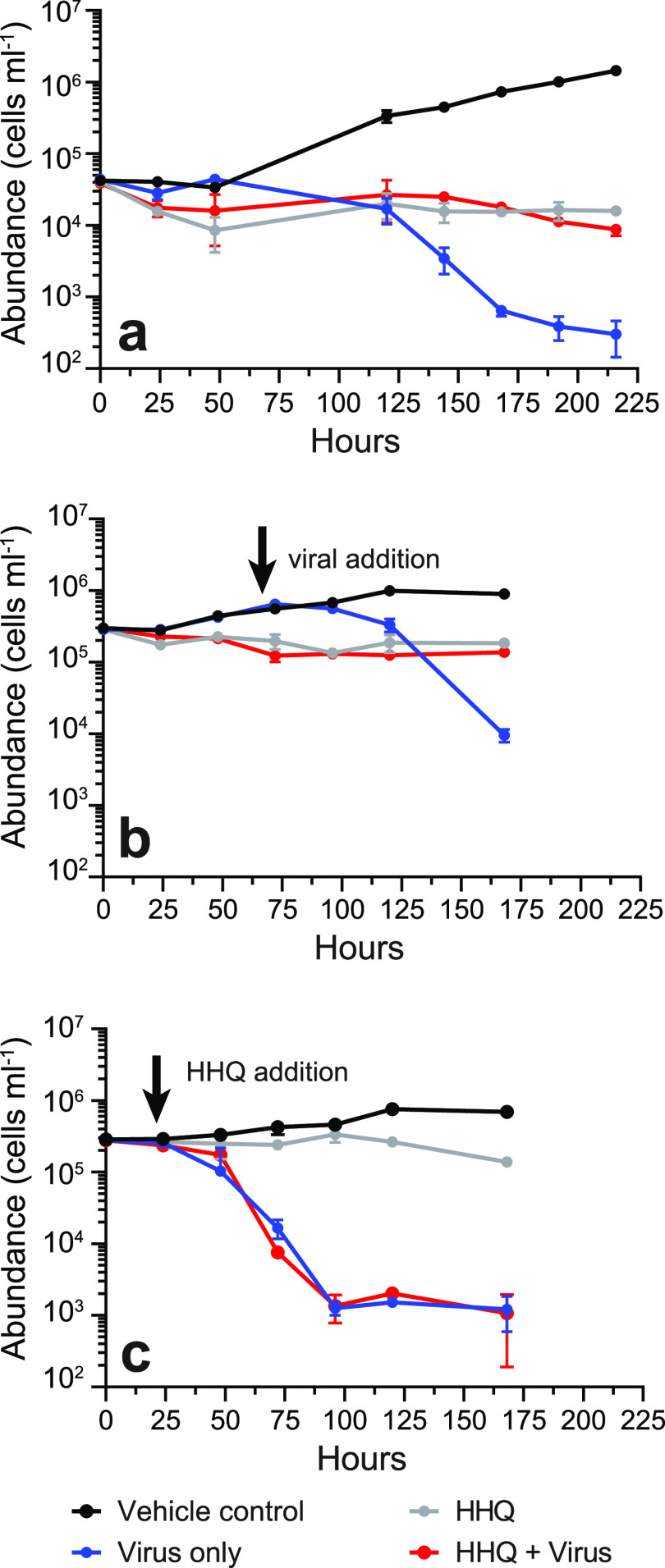
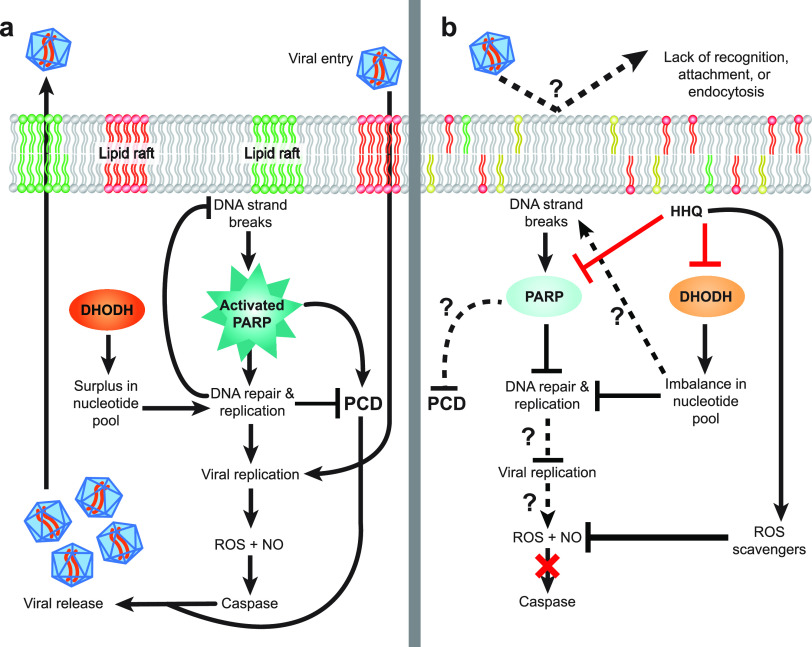
Given that viral replication requires the hijacking of host replication machinery and HHQ exposure inhibited DNA replication and repair in E. huxleyi, the impact of HHQ on host-virus dynamics was investigated. When E. huxleyi cells were exposed to HHQ and Emiliania huxleyi virus (EhV) strain 207 concurrently, virus-induced cellular death was significantly reduced (P value of <0.0001 by ANOVAR) (Fig. 5a). This outcome was observed regardless of whether viruses were added simultaneously with HHQ (Fig. 5a) or 72 h after HHQ treatment (Fig. 5b). However, if HHQ addition was delayed even by 24 h, virus-induced mortality occurred in E. huxleyi (Fig. 5c). These results indicate the possibility that HHQ exposure early in viral infection critically impacts the effectiveness of the virus. There are numerous mechanisms by which HHQ may inhibit virus-induced mortality of E. huxleyi (Fig. 6). For example, HHQ may impact the entry of the virus into the cell. Significant morphological restructuring occurred following 24 h of HHQ exposure, which may prevent viral recognition, attachment, and/or endocytosis. Previous work has demonstrated that within 24 h of E. huxleyi viral infection, the virus requires the induction of host DNA replication machinery (65). Thus, HHQ may either inhibit the virus’ ability to manipulate DNA replication or acquire necessary nucleotides for transcription, thereby stalling infection success. HHQ may also stall the induction of ROS production, which has been demonstrated previously to be necessary for successful E. huxleyi viral infection (66). In the transcriptomic and proteomic data presented here, significant upregulation of a variety of antioxidants, including Fd, FNR, Fld, Fd-NR, GR, Mn-SOD, POX, and VitB6, may counteract virus-induced remodeling of the host antioxidant network essential for viral replication. Likewise, the expression and activation of caspase and metacaspase proteases during infection are critical for enabling virus-induced lysis in E. huxleyi (67). However, these proteases were not upregulated and did not show activity in HHQ-exposed cells (Fig. S4) (see Supplemental Data File 1 at https://doi.org/10.6084/m9.figshare.14414285.v1), further suggesting that HHQ exposure may disrupt these critical processes in viral infection. Attenuation of viral mortality would theoretically permit increased survival of phytoplankton and allow bacteria to continue to take advantage of coordinated nutrient exchange, common between bacteria and phytoplankton (68). Thus, the impacts of HHQ exposure on phytoplankton may have ecological consequences beyond shifts in algal physiology, to impacts on large-scale biogeochemical cycles.
FIG 5.
HHQ can inhibit E. huxleyi virus-induced mortality. The abundance (cells per milliliter) of E. huxleyi over time (hours) after being exposed to either the vehicle control (DMSO), HHQ (100 ng ml−1), EhV 207, or HHQ plus EhV 207 (MOI = 80) was determined. (a) HHQ and the virus were added together. (b) HHQ was added at T0, and the virus was added after 72 h. (c) Virus was added at T0, and HHQ addition was delayed for 24 h (n = 3 for each treatment in each experiment). Means ± standard deviations are shown.
FIG 6.
Proposed model for the role of HHQ in influencing viral success in E. huxleyi. (a) During the infection of a phytoplankton cell, viruses are recognized via specific surface receptors and will enter the cell via endocytosis through distinct lipid rafts. Once inside the cell, the virus hijacks host replication machinery to produce additional viral particles. This replication is dependent on functional de novo nucleotide synthesis enzymes, such as dihydroorotate dehydrogenase (DHODH), to provide the cell with sufficient nucleotide materials. Likewise, functional DNA repair, often mediated by poly(ADP-ribose) polymerase (PARP), is necessary to ensure that replication can continue. Successful viral replication then generates intracellular reactive oxygen species (ROS) and nitric oxide (NO) signaling, which in turn activates caspase proteases, allowing the release of replicated viral particles via programmed cell death (PCD)-induced cell lysis. (b) In HHQ-exposed phytoplankton cells, virus-induced mortality was not observed, but the mechanism by which HHQ impacts viral cycling remains unclear. HHQ may directly inhibit (shown as red lines) the activity of DHODH and PARP, which would prevent the production of viral particles via the collapse of DNA replication machinery. HHQ may also indirectly impact parts of the virus cycle (shown as dotted lines) by changing host physiology to disrupt recognition, nucleotide production, ROS production, caspase activation, or PCD.
Summary.
Our laboratory findings demonstrate that a quorum-sensing signal produced by a marine bacterium significantly, but reversibly, leads to DNA lesions and cell cycle arrest in a eukaryotic phytoplankter, which can influence interkingdom virus-host interactions. In the eastern tropical South Pacific, >1-ng liter−1 surface concentrations of HHQ were found to correlate with areas of enhanced phytoplankton biomass (Fig. S7). These low concentrations of HHQ in bulk seawater are not surprising, as N-acyl homoserine lactones (69), vitamins (70), and other highly labile, trafficked compounds fundamental for growth and signaling are often found in low concentrations in bulk seawater. We anticipate that the primary abiotic sink for HHQ will be photooxidation by sunlight; however, the rate of photooxidation in seawater will strongly depend on a myriad of factors (e.g., depth, day length, and dissolved organic matter [DOM] concentrations, etc.). Previous work showed that the half-life of quinoline, the parent compound of HHQ, varied between 8 and 53 days using solar simulations (71). Furthermore, these measured bulk concentrations likely do not represent the effective concentration that a marine microbial cell would experience in the phycosphere (72).
Detection of HHQ in the marine environment. (a) Cruise track of the U.S. GEOTRACES GP16 cruise in 2013 in the eastern southern tropical Pacific Ocean. (b) Composite figure showing the detection of HHQ in environmental samples. Gray bars indicate the concentrations of HHQ from six stations along the cruise track. Circles indicate the percent relative abundances of picoplankton (yellow) and micro- and nanoplankton (red). Previously, HHQ was isolated from laboratory cultures of both marine Pseudoalteromonas and Pseudomonas spp. (c and d) Extracted ion chromatograms for HHQ ([M + H]+ = m/z 244.17) detected by high-resolution (450K) orbitrap liquid chromatography-electrospray ionization mass spectrometry (LC-ESIMS) for the standard (c) and environmental (d) samples. (e) HHQ confirmation via tandem mass spectrometry (MS/MS) analysis in environmental samples compared to an authentic standard. High-mass-accuracy high-energy collisional dissociation (HCD) fragmentation spectra (35% collision energy) of the [M + H]+ ion were collected at 38 min. Download FIG S7, PDF file, 0.4 MB (381.5KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Previous work has demonstrated that HHQ can significantly alter natural microbial community composition and growth rates (15), and here, we find that detectable in situ HHQ concentrations correlated with enhanced phytoplankton biomass. Together, these findings suggest that alkylquinolone signaling may play a significant role in structuring complex microbial communities, ultimately influencing primary production and biogeochemical cycles. In addition, our findings highlight the functional duality of bacterial cues that serve as diffusive messengers used as a communication tool in microbial communities but also as chemical mediators of marine microbial interactions.
MATERIALS AND METHODS
General cultivation conditions.
For all experiments, axenic Emiliania huxleyi (CCMP2090, non-lith forming) (from the National Center for Marine Algae and Microbiota, East Boothbay, ME) was grown in natural seawater-based f/2 medium without silica (26). Cultures were maintained on a 14-h/10-h light (80 ± 5 μmol photons m−2 s−1)/dark cycle at 18°C, with a salinity of 35. These conditions are referred to here as general culturing conditions. Strain purity was confirmed using f/2 MM and f/2 MB purity test broths and visually confirmed by epifluorescence microscopy (73). Cultures (20 ml) were transferred weekly to maintain exponentially growing cultures.
Phytoplankton cells were enumerated by a hemocytometer or using a flow cytometer (Guava; Millipore). Via the flow cytometer, cell abundance was determined by using species-specific settings, including their forward scatter, side scatter, and red fluorescence (695/50-nm) emission characteristics for evaluating chlorophyll intensity. All samples were run at 0.24 μl s−1 for 3 min, either live or fixed with glutaraldehyde (0.5% final concentration). A correction factor was applied to fixed cell abundances to account for cell loss due to preservation.
Growth experiments.
The HHQ concentration resulting in 50% growth inhibition (IC50) was determined using triplicate, 2- or 20-ml cultures of E. huxleyi (∼100,000 cells ml−1) exposed to HHQ (between 0.25 and 512 ng ml−1), PQS (0.5 to 530 μg ml−1), or the vehicle control (0.1% DMSO) for 72 h. Growth rates were calculated using an exponential growth equation and were plotted against the HHQ concentration to determine the IC50 at 72 h postexposure as described previously (16). Concentrations of DMSO below 0.5% (vol/vol) have no impact on axenic E. huxleyi growth. DMSO was used as the solvent vehicle for HHQ and PQS.
To examine the impacts of HHQ, triplicate flasks of 30-ml cultures of E. huxleyi (∼50,000 cells ml−1) were exposed to either 1 or 100 ng ml−1 HHQ or a vehicle (0.1% DMSO) control. The experiment mixture was sampled daily for 96 h to monitor E. huxleyi abundance, forward scatter, side scatter, red fluorescence (695/50 nm), and photosynthetic efficiency (Fv/Fm). Fv/Fm was measured using a fluorescence induction and relaxation (FIRe) system (Satlantic). Samples were dark adapted for 30 min, and photosystem II kinetics were measured from the average of 10 iterations of an 80-μs single-turnover event and 1,000 ms of weak modulated light.
To measure recovery, after 96 h of HHQ exposure, triplicate 2-ml aliquots of an HHQ-exposed culture were transferred into 198 ml of fresh medium, effectively diluting HHQ to 1 ng ml−1. The same dilution was made with the vehicle control treatment, and the experiment mixture was sampled daily for E. huxleyi growth rate, forward scatter, side scatter, and red fluorescence (695/50 nm).
To investigate viral infection dynamics, triplicate 50-ml cultures were prepared for the following treatments: E. huxleyi (∼40,000 cells ml−1) plus the vehicle control (0.1% DMSO), E. huxleyi plus EhV 207 (3.2 × 106 EhV particles ml−1), E. huxleyi plus HHQ (100 ng ml−1), and E. huxleyi plus HHQ and EhV 207. The multiplicity of infection (MOI) was 80, to ensure successful viral infection potential. Samples were taken daily to monitor E. huxleyi abundance.
For all growth experiments, excluding the IC50 calculation, significant differences between treatments were determined by comparing abundances over time using ANOVAR, followed by Dunnett’s multiple-comparison test (74). All data were tested to ensure that they passed the assumptions for normality and sphericity prior to running the ANOVAR.
Physiological assays.
Propidium iodide (PI) was used to quantitatively discriminate cell cycle stages in HHQ-exposed phytoplankton cultures over 122 h. Three replicate 2-liter cultures at ∼33,000 cells ml−1 were dosed with either 100 ng ml−1 HHQ or the vehicle control (0.002% DMSO). Fixed cells were enumerated every 24 h via flow cytometry. Every 2 h, approximately 106 cells were subsampled, pelleted, and washed twice via centrifugation at 3,214 × g for 15 min at 18°C. The dry cell pellets were resuspended in 1 ml of ice-cold liquid chromatography-mass spectrometry (LC-MS)-grade methanol, transferred to microcentrifuge tubes, and stored at −80°C. For reading, methanol-fixed cells were centrifuged at 16,000 × g for 10 min at 4°C, methanol was removed, and pellets were resuspended in 1 ml of 1× Dulbecco’s phosphate-buffered saline (DPBS) before repelleting by centrifugation at 16,000 × g for 10 min at 4°C. The pellet was resuspended in 0.5 ml of FxCycle PI/RNase solution (Thermo Fisher), incubated for 30 min in the dark, and then measured via flow cytometry (583/26-nm emission).
Diagnostic fluorescent dye assays were used to measure indicators of cell stress and programmed cell death (PCD) following HHQ treatment. Intercellular reactive oxygen species (ROS) and nitric oxide (NO) production, mitotoxicity, cytotoxicity, and caspase protease levels and activity in whole-cell lysates were measured in E. huxleyi (starting cell concentration of ∼100,000 cells ml−1) following HHQ treatment (70 ng ml−1 or 100 ng ml−1) at various time points up to 72 h postexposure. See Text S1 in the supplemental material for detailed protocols.
Additional methodological information. Download Text S1, PDF file, 0.2 MB (241.9KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
E. huxleyi DNA integrity was examined using a modified protocol for the Click-iT terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) Alexa Fluor 488 imaging assay kit (Thermo Fisher). Four replicate E. huxleyi cultures (∼250,000 cells ml−1) were assayed according to the manufacturer’s protocol and sampled after 46 h of HHQ exposure, with tagged cells being enumerated via flow cytometry (512/18-nm emission). See Text S1 in the supplemental material for detailed protocols.
Transmission electron microscopy (TEM).
Replicate 20-ml cultures of exponentially growing E. huxleyi cells (∼100,000 cells ml−1) were exposed to either 100 ng ml−1 HHQ or the vehicle control (0.2% DMSO) for 24 h. Samples were concentrated by filtration on a 0.45-μm polycarbonate filter, transitioned out of f/2 medium via three sequential washes with 10 ml of 0.2 M sodium cacodylate buffer (pH 7.2), and then fixed in 2% glutaraldehyde in 0.2 M sodium cacodylate buffer (pH 7.2). Samples were postfixed in 2.0% osmium tetroxide for 1 h at room temperature and rinsed in double-distilled water (ddH2O) prior to en bloc staining with 2% uranyl acetate. After dehydration through a graded ethanol series, the cells were infiltrated and embedded in Embed-812 (Electron Microscopy Sciences). Thin sections were stained with uranyl acetate and lead citrate and examined with a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera and AMT Advantage NanoSprint500 software.
Transcriptomic and proteomic analyses.
A large-scale culturing experiment was performed with axenic E. huxleyi cells treated with either three concentrations of HHQ (1 ng ml−1, 10 ng ml−1, and 100 ng ml−1) or the vehicle control (0.002% DMSO) for 72 h. Following HHQ/DMSO exposure, 400-ml subsamples were taken from each quadruplicate 2-liter bottle at both 24 and 72 h for total RNA isolation, and an additional 1,200-ml subsample was taken at 72 h for total protein isolation. Total RNA and protein were isolated and quantified as described in Text S1 in the supplemental material.
For transcriptome sequencing (RNA-seq) analysis, the Kapa stranded mRNA-Seq library preparation kit (Kapa Biosystems) was used to prepare library samples, and the samples were sequenced on the NextSeq platform (Illumina) to generate 75-bp paired-end reads. Low-quality reads and adaptor sequences were trimmed using Trimmomatic (V0.38) (75). Transcript abundances were determined using Salmon (V0.12.0) (76) and the Ensembl (77) gene predictions for E. huxleyi CCMP1516 (the nonaxenic form of CCMP2090 [ftp://ftp.ensemblgenomes.org/pub/protists/release-41/fasta/emiliania_huxleyi/cdna/]) as a transcript target index (k-mer size = 23). Normalization and determination of significantly differentially abundant transcripts were performed using the DESeq2 R package (V1.22.1) (78). Tests for differential expression were carried out with the Wald test using a negative binomial generalized linear model. Logarithmic fold change (LFC) estimates were shrunken using the apeglm package (V1.6.0) (79) within DESeq2. The resulting P values were adjusted using the Benjamini-Hochberg (BH) procedure (80) (see Text S1 in the supplemental material).
For proteomic analysis, proteins were solubilized in urea, reduced, alkylated, and trypsin digested as described previously (81). The resulting peptide samples were desalted with a minicentrifugal C18 column according to the manufacturer’s instructions (Nest Group). Peptides were chromatographically separated (precolumn, 3-cm by 100-μm internal diameter [ID]; analytical column, 30-cm by 75-μm ID; resin, 3-μm C18-AQ) with a nanoAcquity ultraperformance liquid chromatography (UPLC) system (2 to 35% acetonitrile [ACN] and 0.1% [vol/vol] formic acid; 250 nl min−1 for 90 min) directly in line with a Fusion Lumos Orbitrap Tribrid mass spectrometer (Thermo Fisher Scientific) operated in the data-independent acquisition (DIA) mode according to methods described previously (82). To generate a peptide spectral library, 1 μg of a pooled sample containing equal parts from each peptide digest was analyzed with six gas-phase fractions covering m/z 400 to 1,000 in increments of 100 m/z (4 m/z staggered MS2 windows and 2 m/z overlap). Each bioreplicate was then quantified in single DIA analyses (MS1, m/z 400 to 1,000; 8 m/z staggered MS2 windows and 4 m/z overlap).
In order to generate absolute abundance measurements of detected proteins, raw MS data files were processed using msconvert (ProteoWizard) for demultiplexing and peak picking. EncyclopeDIA (V0.7.4) was used to (i) search the resulting fragmentation spectra against the UniProt E. huxleyi CCMP1516 protein and contaminant database (10.0-ppm precursor, fragment, and library tolerances), (ii) provide peptide-level area under the curve (AUC) data, and (iii) generate quantitative reports of identified peptides and proteins for each HHQ MS experiment (1% false discovery rate). Significant changes (q < 0.05) in protein abundances between HHQ treatments and the vehicle control were calculated as log2 fold changes between treatments. Complete details of protein sample preparations, chromatographic separations, mass spectrometry detection, and quantification can be found in Text S1 in the supplemental material.
Proteomic data were matched to the transcriptomic data utilizing the corresponding NCBI accession numbers. As many of the genes and proteins were uncharacterized, potential homologs of known proteins of interest were identified by querying the amino acid sequences of selected human proteins against the translated E. huxleyi (CCMP2090) genome, utilizing a significance threshold of an E value of <1 × 10−20. Combined data were visualized utilizing the ggplot2 and pheatmap packages in R.
PARP inhibition and homology modeling.
To examine the impact of alkylquinolone exposure on mammalian PARP activity, an inhibition assay was performed using the PARP universal colorimetric assay kit (R&D Systems) according to the manufacturer’s instructions. Human PARP enzyme (0.5 U) was exposed to 50 μM HHQ (n = 4), 50 μM PQS (n = 4), or the vehicle control (0.25% DMSO) (n = 4) for 15 min prior to the addition of PARP activity buffer. See Text S1 in the supplemental material for a detailed protocol.
The E. huxleyi sequence under GenBank accession number XP_005783504.1 was aligned to the Protein Data Bank (PDB) database to determine the closest structural homolog with a small molecular inhibitor, veliparib, in the active site that could lend insight into HHQ binding.
Detection of HHQ in environmental samples.
Seawater samples were collected along a cruise track from Manta, Ecuador, to Tahiti from October to December 2013 (U.S. GEOTRACES EPZT GP16) as described previously (83). Briefly, seawater was collected at a 3-m depth by a tow fish and pumped at a flow rate of 250 ml min−1 through a 0.2-μm filter and a polytetrafluoroethylene column packed with 20 g of polystyrene resin (Bondesil ENV; Agilent). Each sample represents an integrated average of 400 to 600 liters of water across a wide region. Samples were frozen onboard at −20°C. Prior to analysis, thawed columns were rinsed with 500 ml of 18.2 MΩ-cm ultrahigh-purity water (qH2O) and eluted with 250 ml of LC-MS-grade methanol. The extracts were concentrated by rotary evaporation and brought up in a final volume of 6 ml of qH2O that was stored at −20°C. The organic extracts were separated by a high-pressure liquid chromatography system (Dionex Ultimate 3000) coupled to an Orbitrap Fusion MS instrument (Thermo Scientific). The specific methodology can be found in Text S1 in the supplemental material.
Data availability.
Sequences from this study have been deposited in the Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE131846. The raw mass spectrometry proteomics data and subsequent spectral libraries have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under accession number PXD011560 (https://www.ebi.ac.uk/pride/archive/projects/PXD011560).
ACKNOWLEDGMENTS
We acknowledge the support from the Electron Microscopy Resource Laboratory at the University of Pennsylvania for TEM sample processing. We thank Kay Bidle for thoughtful feedback on a previous draft of the manuscript and the viral cultures, Vinayak Agarwal for homology modeling support and discussions, Bradley Moore for tetrabromopyrrole, and Katie Barott for flow cytometry support. We thank members of the Whalen laboratory, including Ellysia Overton, Yongjie Gao, Carlotta Pazzi, Megan Coolahan, Shreya Kishore, and Lucy Zhao, for assistance in phytoplankton sampling for RNA and protein isolation and constructive discussions. We thank the Georgia Genomics and Bioinformatics Core Facility for RNA sequencing.
Funding for this work was supported by an NSF grant (OCE-1657808) awarded to K.E.W. and E.L.H. K.E.W. was also supported by a faculty research grant from Haverford College as well as funding from the Koshland Integrated Natural Science Center and Green Fund at Haverford College. E.L.H. was also supported by a Sloan Foundation research fellowship. B.L.N. was supported by an NSF grant (OCE-1633939). M.C.M. was supported by an NIH training grant (T32 HG000035). Mass spectrometry was partially supported by the University of Washington Proteomics Resource (UWPR95794). D.R. was supported by funding through the Gordon and Betty Moore Foundation (grant 6000), a Simons Collaboration for Ocean Processes and Ecology grant (329108), and an NSF grant (OCE-1736280). R.B. was supported by an NSF graduate research fellowship and an NSF grant (OCE-1829761).
We declare no competing interests.
REFERENCES
- 1.Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat Rev Microbiol 5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 2.Buchan A, LeCleir GR, Gulvik CA, Gonzalez JM. 2014. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12:686–698. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 3.Cole JJ. 1982. Interactions between bacteria and algae in aquatic ecosystems. Annu Rev Ecol Syst 13:291–314. doi: 10.1146/annurev.es.13.110182.001451. [DOI] [Google Scholar]
- 4.Fukami K, Nishimura S, Ogusa M, Asada M, Nishijima T. 1997. Continuous culture with deep seawater of a benthic food diatom Nitzschia sp. Hydrobiologia 358:245–249. doi: 10.1023/A:1003141104693. [DOI] [Google Scholar]
- 5.Danger M, Oumarou C, Benest D, Lacroix G. 2007. Bacteria can control stoichiometry and nutrient limitation of phytoplankton. Funct Ecol 21:202–210. doi: 10.1111/j.1365-2435.2006.01222.x. [DOI] [Google Scholar]
- 6.Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, Thingstad F. 1983. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10:257–263. doi: 10.3354/meps010257. [DOI] [Google Scholar]
- 7.Mayali X, Azam F. 2004. Algicidal bacteria in the sea and their impact on algal blooms. J Eukaryot Microbiol 51:139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x. [DOI] [PubMed] [Google Scholar]
- 8.Nealson KH, Platt T, Hastings JW. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol 104:313–322. doi: 10.1128/JB.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hmelo LR. 2017. Quorum sensing in marine microbial environments. Annu Rev Mar Sci 9:257–281. doi: 10.1146/annurev-marine-010816-060656. [DOI] [PubMed] [Google Scholar]
- 10.Reen FJ, McGlacken GP, O’Gara F. 2018. The expanding horizon of alkyl quinolone signalling and communication in polycellular interactomes. FEMS Microbiol Lett 365:fny076. doi: 10.1093/femsle/fny076. [DOI] [PubMed] [Google Scholar]
- 11.Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reen FJ, Mooij MJ, Holcombe LJ, McSweeney CM, McGlacken GP, Morrissey JP, O’Gara F. 2011. The Pseudomonas quinolone signal (PQS), and its precursor HHQ, modulate interspecies and interkingdom behaviour. FEMS Microbiol Ecol 77:413–428. doi: 10.1111/j.1574-6941.2011.01121.x. [DOI] [PubMed] [Google Scholar]
- 13.Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 14.Freund JR, Mansfield CJ, Doghramji LJ, Adappa ND, Palmer JN, Kennedy DW, Reed DR, Jiang P, Lee RJ. 2018. Activation of airway epithelial bitter taste receptors by Pseudomonas aeruginosa quinolones modulates calcium, cyclic-AMP, and nitric oxide signaling. J Biol Chem 293:9824–9840. doi: 10.1074/jbc.RA117.001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whalen KE, Becker JW, Schrecengost AM, Gao YJ, Giannetti N, Harvey EL. 2019. Bacterial alkylquinolone signaling contributes to structuring microbial communities in the ocean. Microbiome 7:93. doi: 10.1186/s40168-019-0711-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey EL, Deering RW, Rowley DC, El Gamal A, Schorn M, Moore BS, Johnson MD, Mincer TJ, Whalen KE. 2016. A bacterial quorum-sensing precursor induces mortality in the marine coccolithophore, Emiliania huxleyi. Front Microbiol 7:59. doi: 10.3389/fmicb.2016.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pokrzywinski KL, Tilney CL, Warner ME, Coyne KJ. 2017. Cell cycle arrest and biochemical changes accompanying cell death in harmful dinoflagellates following exposure to bacterial algicide IRI-160AA. Sci Rep 7:45102. doi: 10.1038/srep45102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Tol HM, Amin SA, Armbrust EV. 2017. Ubiquitous marine bacterium inhibits diatom cell division. ISME J 11:31–42. doi: 10.1038/ismej.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKew BA, Metodieva G, Raines CA, Metodiev MV, Geider RJ. 2015. Acclimation of Emiliania huxleyi (1516) to nutrient limitation involves precise modification of the proteome to scavenge alternative sources of N and P. Environ Microbiol 17:4050–4062. doi: 10.1111/1462-2920.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parpais J, Marie D, Partensky F, Morin P, Vaulot D. 1996. Effect of phosphorus starvation on the cell cycle of the photosynthetic prokaryote Prochlorococcus spp. Mar Ecol Prog Ser 132:265–274. doi: 10.3354/meps132265. [DOI] [Google Scholar]
- 21.Rokitta S, Von Dassow P, Rost B, John U. 2016. P- and N-depletion trigger similar cellular responses to promote senescence in eukaryotic phytoplankton. Front Mar Sci 3:109. doi: 10.3389/fmars.2016.00109. [DOI] [Google Scholar]
- 22.Vaulot D, Olson RJ, Merkel S, Chisholm SW. 1987. Cell-cycle response to nutrient starvation in 2 phytoplankton species, Thalassiosira weissflogii and Hymenomonas carterae. Mar Biol 95:625–630. doi: 10.1007/BF00393106. [DOI] [Google Scholar]
- 23.Mojica KD, Brussaard CP. 2014. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol Ecol 89:495–515. doi: 10.1111/1574-6941.12343. [DOI] [PubMed] [Google Scholar]
- 24.Balch WM, Holligan PM, Kilpatrick KA. 1992. Calcification, photosynthesis and growth of the bloom-forming coccolithophore, Emiliania huxleyi. Cont Shelf Res 12:1353–1374. doi: 10.1016/0278-4343(92)90059-S. [DOI] [Google Scholar]
- 25.Simo R. 2001. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol Evol 16:287–294. doi: 10.1016/s0169-5347(01)02152-8. [DOI] [PubMed] [Google Scholar]
- 26.Guillard RRL. 1975. Culture of phytoplankton for feeding marine invertebrates, p 29–60. In Smith WL, Chaney MH (ed), Culture of marine invertebrate animals. Plenum Press, New York, NY. [Google Scholar]
- 27.Barak-Gavish N, Frada MJ, Ku C, Lee PA, DiTullio GR, Malitsky S, Aharoni A, Green SJ, Rotkopf R, Kartvelishvily E, Sheyn U, Schatz D, Vardi A. 2018. Bacterial virulence against an oceanic bloom-forming phytoplankter is mediated by algal DMSP. Sci Adv 4:eaau5716. doi: 10.1126/sciadv.aau5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bramucci AR, Case RJ. 2019. Phaeobacter inhibens induces apoptosis-like programmed cell death in calcifying Emiliania huxleyi. Sci Rep 9:5215. doi: 10.1038/s41598-018-36847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segev E, Wyche TP, Kim KH, Petersen J, Ellebrandt C, Vlamakis H, Barteneva N, Paulson JN, Chai L, Clardy J, Kolter R. 2016. Dynamic metabolic exchange governs a marine algal-bacterial interaction. Elife 5:e17473. doi: 10.7554/eLife.17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shemi A, Schatz D, Fredricks HF, Van Mooy BA, Porat Z, Vardi A. 2016. Phosphorus starvation induces membrane remodeling and recycling in Emiliania huxleyi. New Phytol 211:886–898. doi: 10.1111/nph.13940. [DOI] [PubMed] [Google Scholar]
- 31.Dyhrman ST. 2016. Nutrients and their acquisition: phosphorus physiology in microalgae, p 155–183. In Borowitzka M, Beardall J, Raven J (ed), The physiology of microalgae: developments in applied phycology, vol 6. Springer, Cham, Switzerland. [Google Scholar]
- 32.Dyhrman ST, Ruttenberg KC. 2006. Presence and regulation of alkaline phosphatase activity in eukaryotic phytoplankton from the coastal ocean: implications for dissolved organic phosphorus remineralization. Limnol Oceanogr 51:1381–1390. doi: 10.4319/lo.2006.51.3.1381. [DOI] [Google Scholar]
- 33.Xu Y, Wahlund TM, Feng L, Shaked Y, Morel FMM. 2006. A novel alkaline phosphatase in the coccolithophore Emiliania huxleyi (Prymnesiophyceae) and its regulation by phosphorus. J Phycol 42:835–844. doi: 10.1111/j.1529-8817.2006.00243.x. [DOI] [Google Scholar]
- 34.Bidle KD. 2015. The molecular ecophysiology of programmed cell death in marine phytoplankton. Annu Rev Mar Sci 7:341–375. doi: 10.1146/annurev-marine-010213-135014. [DOI] [PubMed] [Google Scholar]
- 35.Holban AM, Bleotu C, Chifiriuc MC, Bezirtzoglou E, Lazar V. 2014. Role of Pseudomonas aeruginosa quorum sensing (QS) molecules on the viability and cytokine profile of human mesenchymal stem cells. Virulence 5:303–310. doi: 10.4161/viru.27571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao HX, Poovey CE, Privette AA, Grant GD, Chao HY, Cook JG, Purvis JE. 2017. Orchestration of DNA damage checkpoint dynamics across the human cell cycle. Cell Syst 5:445–459.e5. doi: 10.1016/j.cels.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Limas JC, Cook JG. 2019. Preparation for DNA replication: the key to a successful S phase. FEBS Lett 593:2853–2867. doi: 10.1002/1873-3468.13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciardo D, Goldar A, Marheineke K. 2019. On the interplay of the DNA replication program and the intra-S phase checkpoint pathway. Genes (Basel) 10:94. doi: 10.3390/genes10020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christopher A, Hameister H, Corrigall H, Ebenhoh O, Muller B, Ullner E. 2016. Modelling robust feedback control mechanisms that ensure reliable coordination of histone gene expression with DNA replication. PLoS One 11:e0165848. doi: 10.1371/journal.pone.0165848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pai CC, Kearsey SE. 2017. A critical balance: dNTPs and the maintenance of genome stability. Genes (Basel) 8:57. doi: 10.3390/genes8020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lane AN, Fan TW. 2015. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res 43:2466–2485. doi: 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Seyedsayamdost MR. 2017. Synergy and target promiscuity drive structural divergence in bacterial alkylquinolone biosynthesis. Cell Chem Biol 24:1437–1444.e3. doi: 10.1016/j.chembiol.2017.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnould S, Rodier G, Matar G, Vincent C, Pirot N, Delorme Y, Berthet C, Buscail Y, Noel JY, Lachambre S, Jarlier M, Bernex F, Delpech H, Vidalain PO, Janin YL, Theillet C, Sardet C. 2017. Checkpoint kinase 1 inhibition sensitises transformed cells to dihydroorotate dehydrogenase inhibition. Oncotarget 8:95206–95222. doi: 10.18632/oncotarget.19199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fairus AKM, Choudhary B, Hosahalli S, Kavitha N, Shatrah O. 2017. Dihydroorotate dehydrogenase (DHODH) inhibitors affect ATP depletion, endogenous ROS and mediate S-phase arrest in breast cancer cells. Biochimie 135:154–163. doi: 10.1016/j.biochi.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Li C, Wong JTY. 2019. DNA damage response pathways in dinoflagellates. Microorganisms 7:191. doi: 10.3390/microorganisms7070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vlcek D, Sevcovicova A, Sviezena B, Galova E, Miadokova E. 2008. Chlamydomonas reinhardtii: a convenient model system for the study of DNA repair in photoautotrophic eukaryotes. Curr Genet 53:1–22. doi: 10.1007/s00294-007-0163-9. [DOI] [PubMed] [Google Scholar]
- 47.Liu LC, Dong Z, Lei Q, Yang J, Hu HR, Li Q, Ji YC, Guo LY, Zhang YL, Liu YL, Cui HJ. 2017. Inactivation/deficiency of DHODH induces cell cycle arrest and programed cell death in melanoma. Oncotarget 8:112354–112370. doi: 10.18632/oncotarget.19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jagtap P, Szabo C. 2005. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov 4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 49.Wei HT, Yu XC. 2016. Functions of PARylation in DNA damage repair pathways. Genomics Proteomics Bioinformatics 14:131–139. doi: 10.1016/j.gpb.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu SW, Wang HM, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. 2002. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 51.Noel G, Godon C, Fernet M, Giocanti N, Megnin-Chanet F, Favaudon V. 2006. Radiosensitization by the poly(ADP-ribose) polymerase inhibitor 4-amino-1,8-naphthalimide is specific of the S phase of the cell cycle and involves arrest of DNA syntheses. Mol Cancer Ther 5:564–574. doi: 10.1158/1535-7163.MCT-05-0418. [DOI] [PubMed] [Google Scholar]
- 52.Abdullah I, Chee CF, Lee YK, Thunuguntla SSR, Reddy KS, Nellore K, Antony T, Verma J, Mun KW, Othman S, Subramanya H, Rahman NA. 2015. Benzimidazole derivatives as potential dual inhibitors for PARP-1 and DHODH. Bioorg Med Chem 23:4669–4680. doi: 10.1016/j.bmc.2015.05.051. [DOI] [PubMed] [Google Scholar]
- 53.Milanese C, Bombardieri CR, Sepe S, Barnhoorn S, Payan-Gomez C, Caruso D, Audano M, Pedretti S, Vermeij WP, Brandt RMC, Gyenis A, Wamelink MM, de Wit AS, Janssens RC, Leen R, van Kuilenburg ABP, Mitro N, Hoeijmakers JHJ, Mastroberardino PG. 2019. DNA damage and transcription stress cause ATP-mediated redesign of metabolism and potentiation of anti-oxidant buffering. Nat Commun 10:4887. doi: 10.1038/s41467-019-12640-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang HC, Guarente L. 2014. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab 25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canto C, Menzies KJ, Auwerx J. 2015. NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab 22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. 2011. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab 13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webster BR, Lu Z, Sack MN, Scott I. 2012. The role of sirtuins in modulating redox stressors. Free Radic Biol Med 52:281–290. doi: 10.1016/j.freeradbiomed.2011.10.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Legendre C, Reen FJ, Mooij MJ, McGlacken GP, Adams C, O’Gara F. 2012. Pseudomonas aeruginosa alkyl quinolones repress hypoxia-inducible factor 1 (HIF-1) signaling through HIF-1α degradation. Infect Immun 80:3985–3992. doi: 10.1128/IAI.00554-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R. 2013. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci U S A 110:10039–10044. doi: 10.1073/pnas.1215283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel D, Menon D, Bernfeld E, Mroz V, Kalan S, Loayza D, Foster DA. 2016. Aspartate rescues S-phase arrest caused by suppression of glutamine utilization in KRas-driven cancer cells. J Biol Chem 291:9322–9329. doi: 10.1074/jbc.M115.710145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim J, Brown CM, Kim MK, Burrows EH, Bach S, Lun DS, Falkowski PG. 2017. Effect of cell cycle arrest on intermediate metabolism in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci U S A 114:E8007–E8016. doi: 10.1073/pnas.1711642114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bilgin DD, Zavala JA, Zhu J, Clough SJ, Ort DR, DeLucia EH. 2010. Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33:1597–1613. doi: 10.1111/j.1365-3040.2010.02167.x. [DOI] [PubMed] [Google Scholar]
- 63.Lodeyro AF, Ceccoli RD, Pierella Karlusich JJ, Carrillo N. 2012. The importance of flavodoxin for environmental stress tolerance in photosynthetic microorganisms and transgenic plants. Mechanism, evolution and biotechnological potential. FEBS Lett 586:2917–2924. doi: 10.1016/j.febslet.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 64.Havaux M, Ksas B, Szewczyk A, Rumeau D, Franck F, Caffarri S, Triantaphylides C. 2009. Vitamin B6 deficient plants display increased sensitivity to high light and photo-oxidative stress. BMC Plant Biol 9:130. doi: 10.1186/1471-2229-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ku C, Sheyn U, Sebe-Pedros A, Ben-Dor S, Schatz D, Tanay A, Rosenwasser S, Vardi A. 2020. A single-cell view on alga-virus interactions reveals sequential transcriptional programs and infection states. Sci Adv 6:eaba4137. doi: 10.1126/sciadv.aba4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheyn U, Rosenwasser S, Ben-Dor S, Porat Z, Vardi A. 2016. Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean. ISME J 10:1742–1754. doi: 10.1038/ismej.2015.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bidle KD, Haramaty L, Ramos JBE, Falkowski P. 2007. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc Natl Acad Sci U S A 104:6049–6054. doi: 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cooper MB, Smith AG. 2015. Exploring mutualistic interactions between microalgae and bacteria in the omics age. Curr Opin Plant Biol 26:147–153. doi: 10.1016/j.pbi.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 69.Krupke A, Hmelo LR, Ossolinski JE, Mincer TJ, Van Mooy BA. 2016. Quorum sensing plays a complex role in regulating the enzyme hydrolysis activity of microbes associated with sinking particles in the ocean. Front Mar Sci 3:55. doi: 10.3389/fmars.2016.00055. [DOI] [Google Scholar]
- 70.Sanudo-Wilhelmy SA, Gomez-Consarnau L, Suffridge C, Webb EA. 2014. The role of B vitamins in marine biogeochemistry. Annu Rev Mar Sci 6:339–367. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 71.Kochany J, Maguire RJ. 1994. Photodegradation of quinoline in water. Chemosphere 28:1097–1110. doi: 10.1016/0045-6535(94)90328-X. [DOI] [Google Scholar]
- 72.Seymour JR, Amin SA, Raina JB, Stocker R. 2017. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol 2:17065. doi: 10.1038/nmicrobiol.2017.65. [DOI] [PubMed] [Google Scholar]
- 73.Becker JW, Brandon ML, Rappe MS. 2007. Cultivating microorganisms from dilute aquatic environments: melding traditional methodology with new cultivation techniques and molecular methods, p 399–406. In Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA (ed), Manual of environmental microbiology, 9th ed. ASM Press, Washington, DC. [Google Scholar]
- 74.Dunnett CW. 1955. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50:1096–1121. doi: 10.1080/01621459.1955.10501294. [DOI] [Google Scholar]
- 75.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kersey PJ, Allen JE, Allot A, Barba M, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Grabmueller C, Kumar N, Liu Z, Maurel T, Moore B, McDowall MD, Maheswari U, Naamati G, Newman V, Ong CK, Paulini M, Pedro H, Perry E, Russell M, Sparrow H, Tapanari E, Taylor K, Vullo A, Williams G, Zadissia A, Olson A, Stein J, Wei S, Tello-Ruiz M, Ware D, Luciani A, Potter S, Finn RD, Urban M, Hammond-Kosack KE, Bolser DM, De Silva N, Howe KL, Langridge N, Maslen G, Staines DM, Yates A. 2018. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res 46:D802–D808. doi: 10.1093/nar/gkx1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu A, Ibrahim JG, Love MI. 2019. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35:2084–2092. doi: 10.1093/bioinformatics/bty895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 81.Nunn BL, Slattery KV, Cameron KA, Timmins-Schiffman E, Junge K. 2015. Proteomics of Colwellia psychrerythraea at subzero temperatures—a life with limited movement, flexible membranes and vital DNA repair. Environ Microbiol 17:2319–2335. doi: 10.1111/1462-2920.12691. [DOI] [PubMed] [Google Scholar]
- 82.Searle BC, Pino LK, Egertson JD, Ting YS, Lawrence RT, MacLean BX, Villen J, MacCoss MJ. 2018. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat Commun 9:5128. doi: 10.1038/s41467-018-07454-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Boiteau RM, Mende DR, Hawco NJ, McIlvin MR, Fitzsimmons JN, Saito MA, Sedwick PN, DeLong EF, Repeta DJ. 2016. Siderophore-based microbial adaptations to iron scarcity across the eastern Pacific Ocean. Proc Natl Acad Sci U S A 113:14237–14242. doi: 10.1073/pnas.1608594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Composite TEM images showing representative images of E. huxleyi cells exposed to the vehicle control (DMSO) (a through f) or 100 ng ml−1 HHQ (g through l) for 24 h. In select images, subcellular structures are labeled for identification. c, chloroplast; l, lipid droplet; m, mitochondria; n, nucleus; p, pyrenoid; black arrow, Golgi apparatus; v, vacuole; white arrowheads, vesicles. Bar = 500 nm. Download FIG S1, PDF file, 0.7 MB (698.4KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
E. huxleyi cultures were examined for their ability to recover following exposure to 100 ng ml−1 of HHQ for 96 h. Following HHQ exposure, cultures were monitored for their ability to recover by measuring the growth rate (a) (each point represents the exponential growth rate over the previous 24 h), red fluorescence in RFU (a proxy for chlorophyll a intensity) (b), and forward scatter (a proxy for cell size) (c). Each symbol represents the mean from three independent replicates ± the standard deviation. Significant differences between HHQ-exposed cells and the vehicle control were assessed using ANOVAR (P < 0.05). Download FIG S2, PDF file, 0.3 MB (332.9KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(a) Principal-component analysis performed on the regularized-logarithm-transformed transcriptomic read count matrix. Each point represents an experimental sample, with point color indicating the treatment and shape indicating the time point. The first component explains 39% of the variance, and the second component explains 20%. (b) Principal-component analysis performed on the logarithm-transformed proteomic data matrix (protein summed peptide peak area values + 1). Each point represents an experimental sample, with point color indicating the treatment. The first component explains 29.8% of the variance, and the second component explains 16.2%. Note that protein was collected only at the 72-h time point. (c) Growth curve of E. huxleyi (CCMP2090 [axenic]) cultures exposed to 1, 10, and 100 ng ml−1 HHQ and sampled for RNA (at 24 and 72 h) and protein (at 72 h). The arrow indicates the time of HHQ addition. Download FIG S3, PDF file, 0.2 MB (261.1KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Diagnostic biochemical assays for caspase activity from whole-cell lysates or purified caspase enzyme (n = 3) (comparison of the whole-cell lysate at 72 h of DMSO treatment versus the whole-cell lysate at 72 h of HHQ treatment [P value of <0.05 by Welch’s approximate t test {no significance observed}]) (a), the presence of active caspase proteases in vivo (CaspACE) (b), reactive nitrogen species (DAF-FM diacetate) (c), reactive oxygen species (CM-H2DCFDA) from DMSO- or HHQ-exposed cultures from 1 to 72 h posttreatment (d), reactive oxygen species (CM-H2DCFDA) from 72-h DMSO- or HHQ-exposed cultures that were spiked with the algicide tetrabromopyrrole (TBP) (positive control) for 1 or 2 h (e), apoptosis (Image-iT Dead stain) from DMSO- or HHQ-exposed cultures from 1 to 72 h posttreatment (f), apoptosis (Image-iT Dead stain) from 72-h DMSO- or HHQ-exposed cultures that were spiked with TBP (positive control) for 4.5 h (g), and mitochondrial membrane integrity (MitoHealth stain) (h). Bars represent the means ± standard deviations from triplicate readings. Significance in panels b through d, f, and h was based on ANOVAR followed by Dunnett’s multiple-comparison test (P value of <0.05). Significance in panels e and g was based on Student’s t test (P value of <0.05). Cells exposed to HHQ were significantly different from the control only using MitoHealth stain after 24 h of exposure. However, the two treatments were not significantly different from one another at all subsequent time points. Download FIG S4, PDF file, 0.3 MB (309.3KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
HHQ does not directly lead to DNA strand breaks. Lambda DNA and genomic DNA isolated from E. huxleyi CCMP2090 were incubated with 100 ng ml−1 HHQ or the volumetric equivalent of DMSO for 24 h at 18°C, and the presence of DNA strand breaks was assessed by an agarose gel stained with ethidium bromide. Download FIG S5, PDF file, 0.6 MB (608.7KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Impact of alkylquinolones on phytoplankton growth and protein homology modeling. (a) Effect of PQS on E. huxleyi growth. Shown is a dose-response curve of E. huxleyi (strain CCMP2090 [axenic]) in response to PQS. Each symbol represents the mean from three independent replicates ± the standard deviation. Protein homology modeling of the E. huxleyi PARP-1-like homolog (GenBank accession number XP_005783504.1) to the crystal structure of human PARP-1 (PDB accession number 2RD6) with the PARP inhibitor veliparib was performed. (b) The sequence of the E. huxleyi PARP-like homolog (GenBank accession number XP_005783504.1) was aligned to the sequence of human PARP-1 (PDB accession number 2RD6). Two key amino acid residues (red), Tyr and Glu, are strictly conserved between human PARP and the E. huxleyi protein. (c and d) The active site of the structure under PDB accession number 2RD6 shows the binding of the inhibitor veliparib. Regions in the structure under PDB accession number 2RD6 with the greatest homology to the E. huxleyi PARP-like homolog are shown in pink, with the remainder of the protein chain in green. The region comprising the active site for small-molecule binding is highly conserved between the two proteins, with two key binding interactions observed: the nearly coplanar arrangement of a tyrosine side chain phenyl to the inhibitor and a water-mediated hydrogen bond of the basic nitrogen atom of the inhibitor to a glutamate side chain carboxyl. Download FIG S6, PDF file, 0.9 MB (908KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detection of HHQ in the marine environment. (a) Cruise track of the U.S. GEOTRACES GP16 cruise in 2013 in the eastern southern tropical Pacific Ocean. (b) Composite figure showing the detection of HHQ in environmental samples. Gray bars indicate the concentrations of HHQ from six stations along the cruise track. Circles indicate the percent relative abundances of picoplankton (yellow) and micro- and nanoplankton (red). Previously, HHQ was isolated from laboratory cultures of both marine Pseudoalteromonas and Pseudomonas spp. (c and d) Extracted ion chromatograms for HHQ ([M + H]+ = m/z 244.17) detected by high-resolution (450K) orbitrap liquid chromatography-electrospray ionization mass spectrometry (LC-ESIMS) for the standard (c) and environmental (d) samples. (e) HHQ confirmation via tandem mass spectrometry (MS/MS) analysis in environmental samples compared to an authentic standard. High-mass-accuracy high-energy collisional dissociation (HCD) fragmentation spectra (35% collision energy) of the [M + H]+ ion were collected at 38 min. Download FIG S7, PDF file, 0.4 MB (381.5KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional methodological information. Download Text S1, PDF file, 0.2 MB (241.9KB, pdf) .
Copyright © 2021 Pollara et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequences from this study have been deposited in the Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE131846. The raw mass spectrometry proteomics data and subsequent spectral libraries have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under accession number PXD011560 (https://www.ebi.ac.uk/pride/archive/projects/PXD011560).