Streptococcus suis is a causative agent of meningitis, polyarthritis, and polyserositis in swine, and it is a major cause of zoonotic meningitis in humans. Here, we investigate epigenetic gene regulation in S. suis by multiple phasevarions controlled by the phase-variable type III DNA methyltransferase ModS.
KEYWORDS: R-M systems, Streptococcus suis, methyltransferase, phase variation, phasevarion, veterinary pathogen
ABSTRACT
Streptococcus suis is a significant cause of bacterial meningitis in humans, particularly in Southeast Asia, and is a leading cause of respiratory and invasive disease in pigs. Phase-variable DNA methyltransferases, associated with restriction-modification (R-M) systems, are a source of epigenetic gene regulation, controlling the expression of multiple genes. These systems are known as phasevarions (phase-variable regulons) and have been characterized in many host-adapted bacterial pathogens. We recently described the presence of a Type III DNA methyltransferase in S. suis, ModS, which contains a simple sequence repeat (SSR) tract within the open reading frame of the modS gene and which differed in length between individual strains. We also observed that multiple allelic variants of the modS gene were present in a population of S. suis isolates. Here, we demonstrate that a biphasic ON-OFF switching of expression occurs in the two most common ModS alleles, ModS1 and ModS2, and that switching is dependent on SSR tract length. Furthermore, we show using single-molecule real-time (SMRT) sequencing that ModS1 and ModS2 are active methyltransferases in S. suis. ON-OFF switching of each ModS allele results in the regulation of distinct phasevarions, with the ModS2 phasevarion impacting growth patterns and antibiotic resistance. This is the first demonstration of a phase-variable Type III DNA methyltransferase in a Gram-positive organism that controls a phasevarion. Characterizing the phenotypic effects of phasevarions in S. suis is key to understanding pathogenesis and the development of future vaccines.
IMPORTANCE Streptococcus suis is a causative agent of meningitis, polyarthritis, and polyserositis in swine, and it is a major cause of zoonotic meningitis in humans. Here, we investigate epigenetic gene regulation in S. suis by multiple phasevarions controlled by the phase-variable Type III DNA methyltransferase ModS. This is the first characterized example of a Type III R-M system regulating a phasevarion in a Gram-positive organism. We demonstrate that biphasic ON-OFF switching of ModS expression results in differences in bacterial growth and antibiotic resistance. Understanding the effects of ModS phase variation is required to determine the stably expressed antigenic repertoire of S. suis, which will direct and inform the development of antimicrobial treatments and vaccines against this important pathogen.
INTRODUCTION
Streptococcus suis is an important veterinary pathogen that contributes a substantial disease burden to the swine industry (1). S. suis is also a significant pathogen in humans, causing zoonotic meningitis, most commonly associated with occupational exposure (2) and consumption of contaminated pork products (3). S. suis frequently colonizes the upper respiratory tract of pigs, where it is considered a commensal organism (4). Virulent strains of S. suis are able to adhere to and invade epithelial cells of the airway, allowing access to the bloodstream. In both pigs and humans, S. suis is able to breach the blood-brain barrier, resulting in the development of meningitis and septic shock (4).
Phase variation is the rapid and reversible switching of gene expression, and it plays an important role in the pathogenesis of many organisms, where it is usually associated with expression of surface factors such as capsules (5) and adhesins (6, 7) and with lipooligosaccharide biosynthesis (8). Randomized gene expression as a result of phase variation can complicate vaccine development, as it results in an unstable antigenic repertoire, and is potentially a major cause of vaccine escape. Many bacterial pathogens also encode cytoplasmically located phase-variable DNA methyltransferases, which are associated with restriction-modification (R-M) systems (9). Variable methyltransferase expression results in genome-wide methylation differences, resulting in differential gene regulation by epigenetic mechanisms in systems called phasevarions (phase-variable regulons) (9, 10). Many phasevarions are controlled by the ON-OFF switching of Type III DNA methyltransferases, encoded by mod genes. Phasevarions controlled by Type III mod genes have been described in Haemophilus influenzae (11, 12), Moraxella catarrhalis (13), Neisseria spp. (14), Helicobacter pylori (15), and Kingella kingae (16). The phasevarions in these human-adapted pathogens all control expression of genes involved in the pathogenesis of these organisms (17–19). All of these phase-variable mod genes contain simple sequence repeat (SSR) DNA tracts within their open reading frame. These SSR tracts are highly unstable and are prone to variation in length due to slipped-strand mispairing during DNA replication, resulting in the biphasic ON-OFF switching of gene expression; the mod gene is either in-frame and Mod is expressed (ON), or a variation in SSR tract length results in a frameshift and premature stop codon and Mod is not expressed (OFF) (20).
The methyltransferase specificity of a Type III Mod protein is dictated by the central target recognition domain (TRD) of the encoding mod gene (9, 10). This TRD differs between the alleles of individual mod genes, with the 5′ and 3′ regions of individual mod genes being highly conserved between alleles (9, 10). For example, there are 21 modA alleles present in nontypeable Haemophilus influenzae (NTHi) and Neisseria species, six modB and seven modD alleles have been identified in Neisseria gonorrhoeae and Neisseria meningitidis, and 19 modH alleles are present in H. pylori (10, 21). Our previous analysis of the restriction enzyme database REBASE demonstrated the presence of a mod gene in S. suis, which we have named modS, that contained a GAGCA(n) SSR tract (22). A follow-up study analyzing a large collection of S. suis isolates determined the presence of three distinct modS alleles, each of which methylated an adenine in distinct DNA sequences. ModS1 methylated 5′-GCG(m6)ADT-3′ (D is either A, G, or T; bold text indicates methylated adenine), ModS2 methylated 5′-VTC(m6)ATC-3′ (V is either A, G, or C), and ModS3 (present in a single strain identified in GenBank) methylated 5′-GTTC(m6)ANNNB-3′ (B is either C, G, or T; N is any nucleotide) (23). These specificities were determined through heterologous expression of the methyltransferase in Escherichia coli. While various lengths of GAGCA(n) SSR tracts were present in different strains of S. suis carrying the same modS allele, it was not determined whether modS is phase variable or whether the ModS protein is an active methyltransferase in S. suis when expressed.
In this study, we investigated phase variation of the two most prevalent modS alleles, modS1 and modS2, by enriching populations of S. suis strains for specific GAGCA(n) SSR tract lengths in each encoding modS gene. We then determined if differential protein expression occurred in enriched ON-OFF populations of S. suis by using sequential window acquisition of all theoretical mass spectra (SWATH-MS). Clinically relevant phenotypes were assessed in vitro to determine if phase variation of the ModS methyltransferase could result in relevant phenotypic changes or advantages in vivo. Studying the impact of these systems not only provides an understanding of disease pathogenesis but will also guide the development of vaccines by defining the stably expressed antigenic repertoire of S. suis.
RESULTS
ModS is a phase-variable DNA methyltransferase.
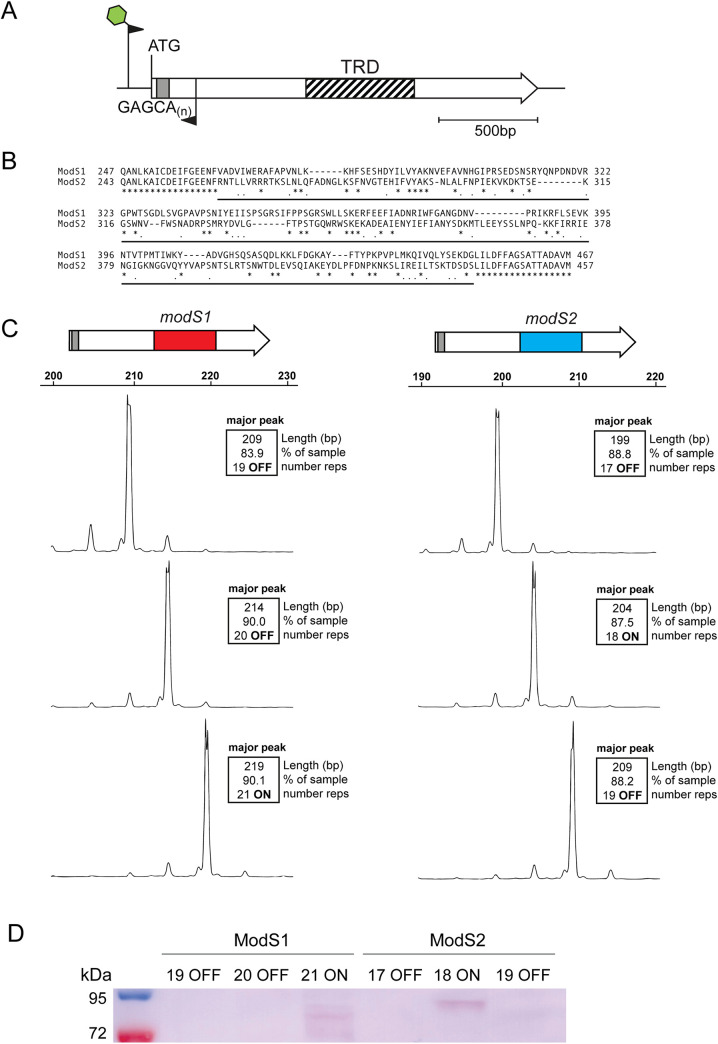
In all previous examples of phase-variable Type III mod genes, SSR tract length variation results in a biphasic ON-OFF switching of expression. In order to determine if variation in length of the GAGCA(n) repeat tract (Fig. 1A) located in the modS open reading frame led to phase-variable expression of the gene, isogenic strains were generated that contained defined lengths of this SSR tract. The isolation of three consecutive repeat tract lengths would theoretically result in one strain where the respective modS gene is ON [GAGCA(n) repeat tract length will place the gene in frame and expressed], and two strains of the triplet will be OFF [GAGCA(n) repeat tract length will place the gene out of frame and not expressed]. These enriched populations were generated in S. suis strains LSS89 (modS1) or SS1056 (modS2), as our analysis of the TRDs of these two alleles showed that they were highly variable and therefore were predicted to methylate different target sequences (Fig. 1B) as we demonstrated previously with heterologous overexpression of ModS1 and ModS2 (23). Strains were enriched for SSR tract lengths containing either 19, 20, or 21 repeats in modS1 and 17, 18, or 19 repeats in modS2. Fragment analysis of each strain confirmed enrichment of each SSR tract length to above 80% (Fig. 1C).
FIG 1.
Expression of ModS alleles. (A) The modS gene contains a variable-length GAGCA(n) simple sequence repeat (SSR) tract (gray box) near the start of the gene and a variable central target recognition domain (TRD), represented by the hatched box. The 5′ and 3′ regions of the modS gene are highly (>95% nucleotide identity) conserved (white). PCR over the SSR tract was determined by 6-carboxyfluorescein (FAM)-labeled PCR using the primers SsuT3-F-FAM and SsuT3-R and analyzed using fragment length analysis. (B) Alignment of the TRD regions of ModS1 and ModS2 showing <25% amino acid identity. An asterisk (*) represents identical amino acid residues; dots (.) represents similar amino acid residues (basic, acidic, and neutral); the TRD region is underlined. Alignments carried out in ClustalW. (C) Fragment length analysis traces of the enriched modS1 and modS2 populations of strains LSS89 and SS1056, respectively, containing three consecutive GAGCA(n) SSR tract lengths. (D) Western blot analysis using ModS antiserum demonstrates that the ModS protein is only present in S. suis populations enriched for 21 repeats in ModS1 (LSS89) and 18 repeats in ModS2 (SS1056), demonstrating phase-variable expression of this protein.
Western blotting of whole-cell lysates of these enriched strains with antiserum generated against the conserved region of all ModS proteins demonstrate that variation in SSR tract length led to a biphasic ON-OFF switching of expression (Fig. 1D; see full blot in Fig. S1 in the supplemental material). The ModS1 protein was only produced in the LSS89 strain, which contained 21 GAGCA repeats in the modS1 gene. ModS2 was only produced in the SS1056 strain, which was enriched for 18 GAGCA repeats. This matches the prediction from the annotated start codon of both genes (see Fig. S2 in the supplemental material).
Expression of ModS is dependent on simple sequence repeat (SSR) tract length. (A) Coomassie staining of whole-cell lysates from enriched strains of S. suis. (B) Western blot using anti-ModS antiserum demonstrates that ModS1 is only produced in strains enriched for 21 GAGCA repeats and that ModS2 is only produced in strains enriched for 18 repeats. The dotted line represents the region of blot presented in Fig. 1D. Download FIG S1, PDF file, 1.7 MB (1.7MB, pdf) .
Copyright © 2021 Tram et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Three open reading frames occur in the modS gene due to variation in length of the GAGCA(n) simple sequence repeat tract. Due to a frameshift downstream of the GAGCA(n) SSR tract, 19 and 20 repeats result in a premature stop codon (in bold, highlighted with an asterisk [*]), and consequently no expression of the ModS protein; 21 GAGCA(n) repeats in the SSR tract results in the gene being in-frame, and therefore ModS protein is produced. Download FIG S2, PDF file, 0.1 MB (96KB, pdf) .
Copyright © 2021 Tram et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To demonstrate methyltransferase activity of the expressed ModS protein, Pacific Biosciences (PacBio) single-molecule real-time (SMRT) sequencing was carried out on genomic DNA isolated from our triplet sets of enriched isogenic strains (19, 20, and 21 repeats for modS1 in strain LSS89; 17, 18, and 19 repeats for modS2 in strain SS1056). We detected a separate, distinct Type I methyltransferase motif in strain LSS89 compared to strain SS1056 (Table 1), with these motifs being fully methylated in all three strains of each enriched triplet set. When comparing the methylomes of enriched LSS89 strains, we detected a Type III methyltransferase motif, 5′-GCG(m6)AT, which was only present in the strain where ModS1 was expressed (21 repeats ON; Table 1). This methylation by ModS1 occurred at 2859 5′-GCG(m6)AT sites throughout the genome, and was not detected in strains enriched for 19 or 20 GCACA repeats in the modS1 gene. SS1056 enriched strains exhibited a different Type III motif, 5′-VTC(m6)ATC, which was only present when ModS2 was expressed (18 repeats ON; Table 1). This motif was methylated 4,810 times in the SS1056 genome, and again was not methylated in strains enriched for 17 or 19 repeats (OFF) in the modS2 gene. These motifs are the same as those detected previously when these ModS alleles were heterologously overexpressed in E. coli (5).
TABLE 1.
Summary of methylomes for S. suis strains LSS89 (ModS1) and SS1056 (ModS2)a
 |
Strains were enriched for GAGCA(n) SSR tracts of 19, 20, or 21 GAGCA repeats in the modS1 gene in strain LSS89 or 17, 18, or 19 GAGCA repeats in the modS2 gene of strain SS1056. Type III methyltransferase motifs are only detected in strains enriched for an ON number of repeats (ModS1 in red in strain LSS89, 21 repeats; ModS2 in blue in strain SS1056, 18 repeats) and match the protein expression detected in Fig. 1. Bold text indicates methylated adenine. In ModS2, V represents A, G, or C. Percentage (%) values represent motifs detected/motifs present. Full methylome data are presented in Data Set S1 in the supplemental material.
All single-molecule real-time (SMRT) sequencing methylome data summarized in Table 1 for our enriched modS1 and modS2 populations of strains LSS89 and SS1056, respectively, containing three consecutive GAGCA(n) SSR tract lengths. Download Data Set S1, XLSX file, 0.01 MB (13.3KB, xlsx) .
Copyright © 2021 Tram et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Biphasic switching of ModS1 and ModS2 results in the expression of distinct phasevarions.
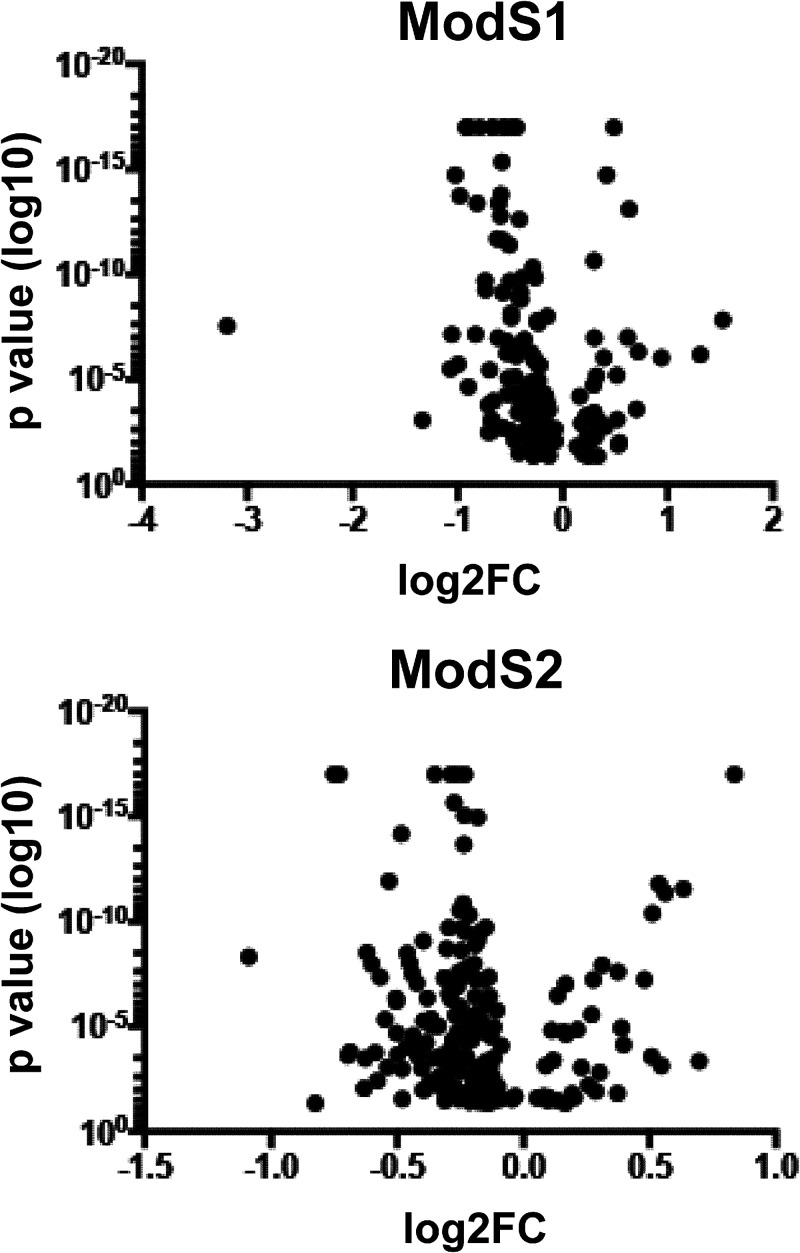
Quantitative proteomics was carried out to determine if distinct protein expression profiles occurred as a result of ModS expression in S. suis, i.e., if these phase-variable methyltransferases control phasevarions. The expression profiles of an ON-OFF ModS1 pair from strain LSS89 (21 repeats ON and 19 repeats OFF) and an ON-OFF ModS2 pair from strain SS1056 (18 repeats ON and 17 repeats OFF) were assessed using SWATH-MS. Both strain pairs exhibited unique changes in protein expression dependent on phase variation of their respective modS gene, with SWATH-MS covering ∼23% of total proteins in LSS89 strains and ∼22% of total proteins in SS1056 strains. Significant differences in protein abundance between the ON and OFF populations are represented as volcano plots in Fig. 2. In strain LSS89, ModS1 ON resulted in higher expression of several proteins involved in a range of cellular processes compared to strains where ModS1 was OFF. Multiple transcriptional regulators were upregulated by expression of ModS1, including the replication initiator protein DnaA (24) and a YlbF/YmcA-type protein (25), as well as transcription factors and repressors. Many ribosomal subunits were upregulated, as well as those involved in general metabolism, such as a nitroreductase (26), glucokinase (27), and a protein involved in alkylphosphonate utilization (28) (Table 2). Conversely, in strains where ModS1 was not expressed, proteins such as ribosomal proteins, several components of ABC transporters, and the fatty acid binding protein DegV (29) were all increased in expression (Table 2).
FIG 2.
Volcano plot demonstrating changes to protein expression as a result of ModS. SWATH-MS proteomics demonstrated a coverage of 450 of 1,964 identified proteins (∼23%) in the LSS89 ModS1 ON-OFF strain pair, and 411 of 1,905 (∼22%) identified proteins in the SS1056 ModS2 ON-OFF strain pair. The x axis indicates relative fold difference in protein abundance in ON compared to OFF; the y axis indicates statistical significance.
TABLE 2.
Differentially regulated proteins (>1.5-fold) in the ModS1 phasevarion
| GenPept accession no. | Protein | Fold changea | P |
|---|---|---|---|
| Downregulated in modS1 ON | |||
| WP_002940030.1 | 30S ribosomal protein S12 | 2.87 | 1.43 × 10−8 |
| WP_002938143.1 | ABC transporter permease | 2.48 | 6.54 × 10−7 |
| WP_009909754.1 | 50S ribosomal protein L27 | 1.92 | 9.04 × 10−7 |
| WP_014638653.1 | ABC transporter substrate-binding protein | 1.65 | 4.81 × 10−7 |
| WP_002936253.1 | DegV family protein | 1.62 | 2.53 × 10−4 |
| WP_023369127.1 | Sugar ABC transporter substrate-binding protein | 1.55 | 7.86 × 10−14 |
| WP_004195491.1 | 50S ribosomal protein L23 | 1.54 | 9.68 × 10−8 |
| Upregulated in modS1 ON | |||
| WP_002936659.1 | 50S ribosomal protein L29 | 9.14 | 2.76 × 10−8 |
| WP_002940438.1 | Amino acid ABC transporter ATP-binding protein | 2.1 | 8.50 × 10−4 |
| WP_012027640.1 | Ribosome biogenesis GTPase Der | 2.08 | 3.12 × 10−6 |
| WP_002942806.1 | CsbD family protein | 2.03 | 6.94 × 10−8 |
| WP_014636557.1 | Transcriptional repressor | 1.99 | 1.78 × 10−15 |
| WP_002936247.1 | HU family DNA-binding protein | 1.97 | 1.81 × 10−6 |
| WP_002936483.1 | 50S ribosomal protein L7/L12 | 1.89 | 1.82 × 10−14 |
| WP_002938891.1 | Acyl carrier protein | 1.88 | ≤1.00 × 10−17 |
| WP_002936048.1 | DUF1846 domain-containing protein | 1.86 | ≤1.00 × 10−17 |
| WP_014736429.1 | Nitroreductase family protein | 1.77 | 2.17 × 10−5 |
| WP_014735259.1 | 30S ribosomal protein S10 | 1.76 | ≤1.00 × 10−17 |
| WP_002940682.1 | 30S ribosomal protein S16 | 1.73 | 6.91 × 10−8 |
| WP_002934959.1 | DUF1149 family protein | 1.67 | 4.09 × 10−14 |
| WP_023368958.1 | Chromosomal replication initiator protein DnaA | 1.66 | ≤1.00 × 10−17 |
| WP_023369777.1 | dTDP-4-dehydrorhamnose reductase | 1.64 | 1.96 × 10−10 |
| WP_002936660.1 | 50S ribosomal protein L16 | 1.62 | 5.40 × 10−10 |
| WP_004194840.1 | DUF2829 domain-containing protein | 1.6 | 1.60 × 10−4 |
| WP_012027178.1 | PTS sugar transporter subunit IIA | 1.6 | 3.27 × 10−3 |
| WP_002936656.1 | 50S ribosomal protein L24 | 1.59 | 3.52 × 10−6 |
| WP_002937025.1 | Response regulator transcription factor | 1.59 | 2.89 × 10−3 |
| WP_011922043.1 | DivIVA domain-containing protein | 1.59 | 8.40 × 10−4 |
| WP_002938966.1 | YlbF/YmcA family competence regulator | 1.58 | ≤1.00 × 10−17 |
| WP_012775074.1 | Alkylphosphonate utilization protein | 1.54 | 8.72 × 10−4 |
| WP_023370402.1 | ROK family glucokinase | 1.53 | ≤1.00 × 10−17 |
| WP_002936622.1 | 30S ribosomal protein S13 | 1.51 | 1.00 × 10−4 |
| WP_023369315.1 | Glycine cleavage system protein H | 1.5 | 1.94 × 10−12 |
Fold change presented as modS1 ON versus modS1 OFF.
In strain SS1056, ModS2 ON-OFF switching resulted in varied expression of a distinct set of proteins (Table 3). ModS2 expression increased the levels of proteins involved in amino acid metabolism such as cysteine synthase and aminopeptidase D (30). Proteins involved in general metabolism, such as dihydroxyacetone kinase (31), also showed increased expression in ModS2 ON. The DNA biosynthesis enzymes ribonucleotide-diphosphate reductase and ribonucleotide-triphosphate reductase (32) were increased in expression in ModS2 ON, as was an acyl carrier protein involved in fatty acid biosynthesis (33). Three proteins were upregulated in strains that did not express ModS2 (OFF), namely, an ATP-dependent Clp protease, the nucleotide exchange factor GrpE, and a glyoxalase/bleomycin resistance/extradiol dioxygenase family protein involved in resistance to antimicrobials.
TABLE 3.
Differentially regulated proteins (>1.5-fold) in the ModS2 phasevarion
| GenPept accession no. | Protein | Fold changea | P |
|---|---|---|---|
| Downregulated in modS2 ON | |||
| WP_002935876.1 | Glyoxalase/bleomycin resistance/extradiol dioxygenase family protein | 1.79 | ≤1.00 × 10−17 |
| WP_044673510.1 | ATP-dependent Clp protease ATP-binding subunit | 1.62 | 4.35 × 10−4 |
| WP_024387015.1 | Nucleotide exchange factor GrpE | 1.55 | 2.81 × 10−12 |
| Upregulated in modS2 ON | |||
| WP_012028296.1 | 50S ribosomal protein L20 | 2.13 | 4.82 × 10−9 |
| WP_012027287.1 | Peptidylprolyl isomerase | 1.77 | 4.40 × 10−2 |
| WP_002938891.1 | Acyl carrier protein | 1.68 | ≤1.00 × 10−17 |
| WP_044754372.1 | Cysteine synthase A | 1.66 | ≤1.00 × 10−17 |
| WP_002936486.1 | 50S ribosomal protein L10 | 1.62 | 2.35 × 10−4 |
| WP_044680965.1 | Class 1b ribonucleoside-diphosphate reductase subunit alpha | 1.61 | 1.54 × 10−4 |
| WP_044771061.1 | Dihydroxyacetone kinase subunit L | 1.55 | 8.68 × 10−3 |
| WP_024384331.1 | Aminopeptidase P family protein | 1.54 | 3.24 × 10−4 |
| WP_044681922.1 | Ribonucleoside-triphosphate reductase | 1.54 | 2.97 × 10−9 |
| WP_079394259.1 | Amino acid ABC transporter substrate-binding protein | 1.52 | 1.07 × 10−8 |
| WP_044674536.1 | Winged helix-turn-helix transcriptional regulator | 1.5 | 1.70 × 10−4 |
Fold change presented as modS2 ON versus modS2 OFF.
ModS switching results in differences in growth.
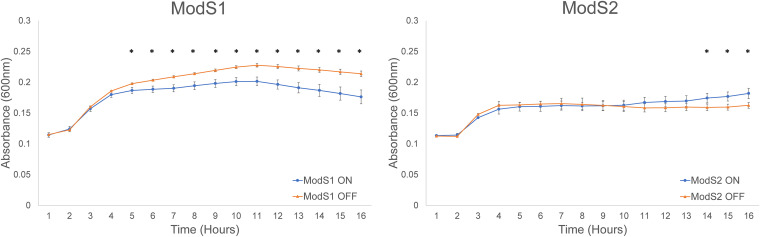
Our proteomic analysis of ModS switching demonstrated several proteins that could potentially impact growth rate exhibited differential regulation in both ModS alleles. These included proteins involved in general metabolic enzymes, DNA transcription factors and repressors, and biosynthesis enzymes (Tables 2 and 3). In order to determine if these altered protein expression levels could affect S. suis growth rates, we conducted standard growth curves of our enriched ModS ON-OFF pairs in rich medium. These growth curves showed a small but repeatable (n = 3) difference in growth rate for both strain pairs (Fig. 3). Starting at the same optical density at 600 nm (OD600), when ModS1 was ON, growth was to a lower final OD (P < 0.05), and conversely, when ModS2 was ON, growth was to a higher final OD (P < 0.05).
FIG 3.
Growth curves of S. suis populations enriched for modS1 and modS2. ON-OFF strain pairs for ModS1 (strain LSS89) and ModS2 (SS1056) were grown in rich medium (THB-Y broth) for 18 h with shaking. Statistically significant differences (P < 0.05) in absorbance at each time point are indicated by asterisks, assessed using Student’s t test.
ModS2 phase variation results in differences in antibiotic resistance.
SWATH-MS also demonstrated differential expression of an antibiotic resistance protein in ModS2 (GenPept accession no. WP_002935876.1), annotated as a glyoxalase/bleomycin resistance/extradiol dioxygenase family protein, which exhibited higher expression in strains which did not express ModS2 (OFF) compared to ModS2 ON. We therefore carried out a MIC analysis using several beta-lactam antibiotics (ampicillin, penicillin, and amoxicillin) and a glycopeptide antibiotic (vancomycin) (Table 4). Although there was no change in MIC to amoxicillin, vancomycin, and penicillin, there was a 2-fold increase in resistance to ampicillin when ModS2 was OFF compared to ModS2 ON (0.32 μg/ml versus 0.16 μg/ml).
TABLE 4.
MICs of ModS2 ON versus ModS2 OFF enriched S. suis strains
| Drug | MIC (μg/ml) for S. suis: |
|
|---|---|---|
| ModS2 ON | ModS2 OFF | |
| Ampicillin | 0.16 | 0.31 |
| Vancomycin | 1.25 | 1.25 |
| Amoxicillin | 0.16 | 0.16 |
| Penicillin | 0.63 | 0.63 |
DISCUSSION
Over the last 15 years, phasevarions (phase-variable regulons) controlled by ON-OFF switching of Type III mod genes have emerged as an important gene regulation strategy in a diverse range of host-adapted pathogens (9, 12, 13, 15, 16, 21). In all of these examples, phase-variable ON-OFF switching of mod expression occurs via changes in the length of locus-encoded SSR tracts. In this study, using a combination of strain enrichment, Western blotting, and SMRT sequencing, we conclusively demonstrate that S. suis contains multiple allelic variants of a new phase-variable Type III mod gene, modS, with this representing the first example of a phasevarion controlled by biphasic ON-OFF switching of a Type III DNA methyltransferase in a Gram-positive organism (Fig. 1). Utilizing an antiserum that recognizes all ModS alleles with equal affinity (9, 34), it was found that both ModS alleles were only expressed in strains with specific SSR tract lengths. ModS1 was only present in strains with 21 GAGCA repeats, and ModS2 was only expressed in strains with 18 GAGCA repeats. In both cases, the repeat length placed the modS gene in the correct reading frame and demonstrates that the expression of the ModS protein is dependent on the SSR tract length.
SMRT sequencing was used to determine methyltransferase activity and specificity of ModS1 and ModS2. LSS89 strains of S. suis enriched for ModS1 expression (21 GACAC repeats ON) exhibited methylation of the adenine in 5′-GCGm6AT motifs, which was not seen in strains in which ModS1 was OFF (19 or 20 GACAC repeats). ModS2-expressing strains (SS1056) of S. suis exhibited methylation of a Type III Mod motif of 5′-VTCm6ATC motifs (where V can be either A, G, or C) when ModS2 was predicted to be ON (18 GACAC repeats). This confirmed our previous findings when we overexpressed ModS1 and ModS2 using a recombinant E. coli system (23). SMRT sequencing also demonstrates that both ModS1 and ModS2 are active methyltransferases in S. suis, methylating at thousands of sites in their respective genomes (in strain LSS89, 2,861 5′-GCGm6AT sites are present; in SS1056, 4,812 5′-TCm6ATC sites are present; see Data Set S1 in the supplemental material).
SWATH-MS proteomic analysis confirmed that the altered methylation resulting from ModS phase variation results in a change in the expression of numerous proteins in distinct phasevarions. These changes in protein expression were seen across many families of protein, including those involved in central metabolism, gene regulation, and transporters. Although phase variation can be a frequent cause of vaccine escape across bacterial species (20), our investigation shows that no proteins currently investigated as vaccine targets are subject to variation by ModS1 or ModS2 in the strains tested (35, 36). By randomly switching expression of different sets of genes, phasevarions allow generation of multiple phenotypic states within a bacterial population. This gives the population an extra contingency strategy for survival, as some phenotypes may be more beneficial under certain environmental conditions, or give certain individual bacteria a particular advantage, allowing survival of the population through selection bottlenecks. Phase-variable switching of ModS resulted in differences in growth rate, but it is unclear whether this is a result of an alteration of regulatory genes due to ModS switching, or another, as-yet-uncharacterized effect of ModS phase variation. When strain pairs were started at the same seeding density, although the final differences in final OD between enriched ON and OFF strain pairs were small, they are statistically significant (P < 0.05) and could have an effect on both long-term carriage and disease, particularly if one population has an advantage resulting from differential protein expression over time. ModS2 phase variation also resulted in a difference in antibiotic resistance. A protein repressed by ModS2 expression, annotated as a glyoxalase, bleomycin resistance, and extradiol dioxygenase family protein, has been described as involved in resistance to beta-lactam and glycopeptide antibiotics (37), which are commonly used to treat bacterial infections in humans as well as commercial pig farms (38). The MIC of S. suis was assessed against three commonly used beta-lactams (ampicillin, amoxicillin, and penicillin) and a glycopeptide from the same antibiotic class as bleomycin (vancomycin). A strain in which ModS2 was OFF, where the expression of this resistance protein was increased, showed a small (2-fold) increase in MIC toward ampicillin compared to that of the isogenic strain in which ModS2 was ON. Although small, there may be a cumulative effect, particularly as many beta-lactam antibiotics are extensively used prophylactically in commercial pig farms (39–42). This has the potential to result in a long-term increase in ampicillin resistance in S. suis populations, although this would need to be studied in a model system and is beyond the scope of this work.
Although many of the changes in protein expression determined by our SWATH-MS analysis of enriched ON-OFF population in each ModS allele were small (1.5-fold to 2-fold), SWATH-MS is a technique which quantitates protein expression based on abundance (43). Differential expression of proteins will therefore only identify differences in highly expressed proteins, and expression differences of proteins beyond the limit of detection will be missed. We recognize that this is a limitation of our approach, but a SWATH-MS proteomics approach has been previously used to characterize phasevarion-mediated changes to protein expression (13, 44), with similar small differences in expression also reported in these phasevarions. Therefore, while we have determined that both ModS1 and ModS2 affect the expression of small distinct sets of proteins, it is likely that further changes to gene and protein expression remain undetected. In order to thoroughly characterize each phasevarion, further analysis will be needed. It will also be important to characterize the phasevarions controlled by other modS alleles; in previous work we showed that a third modS allele—modS3—was present in S. suis. The ModS3 allele methylated a distinct sequence to ModS1 and ModS2 (23) and is therefore highly likely to control a different phasevarion. The presence of additional modS alleles should also be determined and studied, in order to detail all gene and protein expression changes mediated by phasevarions in S. suis.
This characterization of ModS is the first described instance of a phasevarion in a Gram-positive organism controlled by a Type III methyltransferase. Different alleles of ModS methylate distinct motifs and result in distinct phasevarions. These distinct ModS phasevarions result in gross differences in growth rate and, in the case of ModS2, differential resistance to antibiotics. Both of these phenotypes could affect disease and pathology, and this remains to be studied using both in vitro models and, ideally, an in vivo challenge using the natural host (pigs). A thorough understanding of phase variation of gene expression, and in particular of phasevarions, is required in order to determine the stably expressed antigenic repertoire of a bacterial species. The prevalence of phase-variable methyltransferases across the bacterial domain demonstrates that phasevarions are a widespread contingency strategy (22, 45, 46) and that characterization of these systems is imperative in order to rationally design effective vaccines that only target stably expressed antigens in the organisms where they are present.
MATERIALS AND METHODS
Bacterial strains and growth.
S. suis strains LSS89 and SS1056 (47) used in this study were grown in Todd-Hewitt broth (THB; Oxoid) supplemented with 2% yeast extract (THB-Y) or on THB-Y plates (THB-Y supplemented with 1.5% wt/vol agar). Cultures were incubated overnight at 37°C.
Strain enrichment.
Fragment length analysis of the SSR tract in each modS allele was conducted to determine the length of the GAGCA(n) SSR tract. A PCR was performed using a fluorescently labeled forward primer SsuT3-F-FAM (5′-FAM-CAT CAA AAA CGG CTT GAC AGC C) and the reverse primer SsuT3-R (5′-GCA ATG TTG TCT GAT AAA ACA TCT TTT G), as described previously (23). DNA fragment length analysis was carried out at the Australian Genome Research Facility (AGRF; Brisbane, Australia). This technique was used to enrich populations of S. suis strains for defined SSR lengths through a combination of fragment length analysis and subculturing. S. suis populations of strain LSS89 (encoding modS1) were enriched for 19, 20, and 21 GAGCA repeats, and strain SS1056 (encoding modS2) were enriched for 17, 18, and 19 GAGCA repeats. Populations containing >80% of each single tract length were considered to be enriched and were used in subsequent studies.
SDS-PAGE and Western blot analysis.
Cell lysates of each enriched strain were prepared from two 25-ml overnight liquid cultures of S. suis cells grown in THB-Y broth and pooled. The absorbance of each culture was assessed, and samples were then normalized by centrifugation and resuspension to a calculated OD600 of 30.0 in 1 ml of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% Triton X-100, 0.5% SDS, and 50 mM Tris HCl [pH 8]). Samples were sonicated twice for 30 s at a probe intensity (amplitude) of 15 μm, on ice. Lysates were clarified by centrifugation at 14,000 × g for 20 min to remove cellular debris.
Supernatants were prepared for SDS-PAGE using BOLT 4× loading dye containing 100 mM dithiothreitol (Sigma) and boiled for 30 min, then loaded on precast 4 to 12% BOLT Bis-Tris gel (Thermo Fisher) and run for 45 min at 165 V in morpholinepropanesulfonic acid (MOPS) 1× running buffer according to the manufacturer’s instructions (Thermo Fisher). Proteins were visualized by Coomassie brilliant blue staining. Twice the amount of lysate was loaded onto gels for use in Western blotting, and proteins were transferred to preequilibrated polyvinylidene difluoride (PVDF) membranes (Bio-Rad) at 20 V for 60 min using the BOLT transfer system (Thermo Fisher). Following transfer, membranes were blocked with 10% skim milk in Tris-buffered saline with Tween 20 (TBST) for 60 min. Membranes were incubated with a 1:1,000 dilution of anti-ModS antiserum (raised as detailed below) overnight at 4°C, followed by a 1:10,000 anti-mouse alkaline phosphatase secondary antibody (Sigma). Blots were developed using nitro-blue tetrazolium (NBT)/5-bromo-4-chloro-3′-indolyphosphate (BCIP) (Roche) according to the manufacturer’s instructions.
Construction of a TRD-less ModS protein.
The modS1 gene was amplified from S. suis strain LSS89, excluding the CAGCA(n) SSR tract, using the primers SsuT3-oE-F (5′-AGTCAG CATATG AGC AGA GCA AAG CAA AAG CTT GGA GAA TAC ACT CAA G; underlined text represents NdeI site for cloning) and SsuT3-oE-R (5′-AGTCAG GGATCC CTA CAC CAC CTT CAC TTT GGT ACC; underlined text represents BamHI site for cloning) with KOD Hot Start DNA polymerase (Merck Millipore) according to the manufacturer’s instructions. The resulting product was then cloned into the NdeI-BamHI site of the expression vector pET15b (Novagen) with the gene in-frame with the N-terminal His tag to generate vector pET15b:SsuT3-His_tag. The TRD coding region of modS was then removed, and the conserved 5′ and 3′ coding regions were fused. This was generated by using the pET15b::SsuT3-His_tag vector as a template via inverse PCR with the primers SsuT3-TRD-remove-F (5′-CCCA AAT ATC TCA TCA CAG ATA GCT TTG AGG TTG G) and SsuT3-TRD-remove-R (5′-GCT GTT ATG CAG CTG AAT GCA GAA GAT GG) binding either side of the TRD, using KOD Hot Start DNA polymerase (Merck Millipore) according to the manufacturer’s instructions. His-tagged TRD-less ModS was overexpressed in E. coli BL21 using isopropyl β-d-1-thiogalactopyranoside (IPTG) induction (0.5 mM) overnight at 37°C with 200 rpm shaking, and protein purified using Talon resin (TaKaRa Bio) using standard protocols in 50 mM phosphate buffer (pH 7.4) containing 300 mM NaCl.
Generation of ModS antiserum.
A cohort of five 6- to 8-week-old female BALB/c mice (Animal Resources Centre, WA, Australia) were immunized subcutaneously with 25 μg of recombinant TRD-less ModS protein in 25 μl phosphate-buffered saline (PBS), mixed with 25 μl Freund’s adjuvant (Freund’s complete adjuvant [FCA] on day 0 and Freund’s incomplete adjuvant [FIA] subsequently on days 14, 21, 28, and 42; Merck, Darmstadt, Germany). Terminal bleeds were collected on day 58, and serum was separated via centrifugation. Preimmune (naive) serum was collected from cohorts prior to immunization via tail bleed. Serum was stored in 50% glycerol at −20°C. This antiserum recognizes all ModS alleles with equal affinity, as it was raised against the conserved regions of ModS shared by all alleles. All animal work was carried out according to the Australian Code for the Care and Use of Animals for Scientific Purposes, with approval from the Griffith University Animal Ethics Committee (GLY/16/19/AEC).
Single-molecule real-time (SMRT) sequencing and methylome analysis.
Genomic DNA from our enriched triplet sets of S. suis strains LSS89 (modS1 19, 20, and 21 repeats) and SS1056 (modS2 17, 18, and 19 repeats) was prepared from an overnight culture in THB-Y broth, and high-molecular-weight genomic DNA was isolated using the Sigma GenElute kit (Sigma-Aldrich) according to the manufacturer's instructions. SMRT sequencing and methylome analysis was carried out as previously described (48, 49). Briefly, DNA was sheared to an average length of approximately 5 to 10 kb (genomic DNA) using g-Tubes (Covaris, Woburn, MA), and SMRTbell template sequencing libraries were prepared using sheared DNA. DNA was end repaired, then ligated to hairpin adapters. Incompletely formed SMRTbell templates were degraded with a combination of exonuclease III (New England Biolabs; Ipswich, MA) and exonuclease VII (USB; Cleveland, OH). Primer was annealed and samples were sequenced on the PacBio Sequel system (Menlo Park, CA) using standard protocols for long insert libraries. SMRT sequencing and methylome analysis was carried out at SNPSaurus (University of Oregon, OR, USA).
SWATH-MS proteomics.
Overnight cultures of each S. suis strain (107 CFU/ml) were harvested, lysed in guanidium buffer (6 M guanidium chloride, 50 mM Tris-HCl [pH 8], and 10 mM dithiothreitol) and incubated at 30°C for 30 min with shaking (500 rpm). Cysteines of the total protein were alkylated by addition of acrylamide to a final concentration of 25 mM and incubated at 30°C for 60 min with shaking (500 rpm). Concentration of samples was assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher). A 100-μg aliquot of the protein was then precipitated by addition of 1:1 methanol:acetone at −20°C overnight. The protein was pelleted at 18,000 × g for 10 min, and supernatant was removed before the pellet was resuspended in 50 μl trypsin reaction buffer,1 μg trypsin (New England Biolabs) was added, and the suspension incubated overnight at 37°C. Tryptic-digested peptides were then desalted and purified using a ZipTip (Millipore) per the manufacturer’s instructions. SWATH-MS was performed as previously described (50). Briefly, tryptic peptides were analyzed by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) using a Prominence nanoLC system (Shimadzu) and a TripleTOF 5600 mass spectrometer with a NanoSpray III interface (Sciex). Peptides were separated on a Vydac Everest reversed-phase C18 high-performance liquid chromatography (HPLC) column at a flow rate of 1 μl/min. A gradient of 10% to 60% buffer B over 45 min, with buffer A (1% acetonitrile and 0.1% formic acid) and buffer B (80% acetonitrile and 0.1% formic acid) was used. A mass spectrometry (MS)-time of flight (TOF) scan was performed from an m/z range of 350 to 1,800 for 0.5 s, followed by information-dependent acquisition of MS/MS of the top 20 peptides from m/z 40 to 1,800 for 0.05 s per spectrum, with automated CE (capillary electrophoresis) selection. Identical LC conditions were used for SWATH-MS. SWATH-MS of triplicate biological replicates was performed with the same MS-TOF scan, followed by high-sensitivity information-independent acquisition with m/z isolation windows with 1 m/z window overlap each for 0.1 s across an m/z range of 400 to 1,250. Collision energy was automatically assigned by Analyst software (AB Sciex) based on m/z window ranges. Proteins were identified by searching against S. suis Lss89 and SS1056 genomes (NCBI accession no. GCA_900059105.1 and GCA_900051945.1, respectively) and common contaminants with standard settings using ProteinPilot 5.0.1 (AB Sciex). False-discovery-rate analysis was performed on all searches. ProteinPilot search results were used as ion libraries for SWATH analyses. The abundance of proteins was measured automatically using PeakView (AB Sciex) with standard settings. Comparison of protein relative abundance was performed based on protein intensities or ion intensities using a linear mixed-effects model with the MSstats package in R. Proteins with ≥1.5-fold changes in abundance and with adjusted P values greater than ≤0.05 were considered differentially expressed.
Growth curves and MIC assays.
Growth curves were conducted by dilution of overnight cultures to an OD600 of 0.1. These cultures were then used to seed a 96-well plate in triplicate. Plates were then incubated at 37°C with shaking in a Tecan Infinite 200 Pro plate reader, with absorbance readings (OD600) being taken every 15 min.
The MIC was measured by broth microdilution in triplicate experiments based on CLSI guidelines as described previously (51). Briefly, an overnight culture of S. suis was diluted to an OD600 of 0.1 and subcultured grown to the mid-log phase for 3 h at 37°C. Mid-log-phase cultures were diluted to an OD600 of 0.2, and 50 μl of each culture was added to 96-well plates containing serially diluted antibiotic concentrations (5 to 0.08 μg/ml), and plates were grown at 37°C with 5% CO2 for 24 h. The MIC (mg/liter) was determined as the last dilution at which turbidity was observed following overnight growth, with all assays being performed in triplicate.
Data availability.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (52) partner repository under data set identifier PXD023726.
All data for growth curves presented in Fig. 3. Download Data Set S2, XLSX file, 0.01 MB (14.8KB, xlsx) .
Copyright © 2021 Tram et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Amanda Nouwens and Peter Josh in the SCMB MS facility, University of Queensland, for conducting SWATH-MS proteomic analysis. We thank Allison Banse and Eric Johnson at SNPsaurus (University of Oregon, OR, USA) for conducting SMRT sequencing and methylome analysis.
This work was funded by an Australian Research Council Discovery Project DP180100976 to J.M.A. and P.J.B. and by Australian National Health and Medical Research Council (NHMRC) Principal Research Fellowship 1138466 to M.P.J.
REFERENCES
- 1.Goyette-Desjardins G, Auger JP, Xu J, Segura M, Gottschalk M. 2014. Streptococcus suis, an important pig pathogen and emerging zoonotic agent—an update on the worldwide distribution based on serotyping and sequence typing. Emerg Microbes Infect 3:e45. doi: 10.1038/emi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breton J, Mitchell WR, Rosendal S. 1986. Streptococcus suis in slaughter pigs and abattoir workers. Can J Vet Res 50:338–341. [PMC free article] [PubMed] [Google Scholar]
- 3.Susilawathi NM, Tarini NMA, Fatmawati NND, Mayura PIB, Suryapraba AAA, Subrata M, Sudewi AAR, Mahardika GN. 2019. Streptococcus suis-associated meningitis, Bali, Indonesia, 2014–2017. Emerg Infect Dis 25:2235–2242. doi: 10.3201/eid2512.181709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dutkiewicz J, Zając V, Sroka J, Wasiński B, Cisak E, Sawczyn A, Kloc A, Wójcik-Fatla A. 2018. Streptococcus suis: a re-emerging pathogen associated with occupational exposure to pigs or pork products. Part II—pathogenesis. Ann Agric Environ Med 25:186–203. doi: 10.26444/aaem/85651. [DOI] [PubMed] [Google Scholar]
- 5.Tzeng YL, Thomas J, Stephens DS. 2016. Regulation of capsule in Neisseria meningitidis. Crit Rev Microbiol 42:759–772. doi: 10.3109/1040841X.2015.1022507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atack JM, Winter LE, Jurcisek JA, Bakaletz LO, Barenkamp SJ, Jennings MP. 2015. Selection and counterselection of Hia expression reveals a key role for phase-variable expression of Hia in infection caused by nontypeable Haemophilus influenzae. J Infect Dis 212:645–653. doi: 10.1093/infdis/jiv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elango D, Schulz BL. 2020. Phase-variable glycosylation in nontypeable Haemophilus influenzae. J Proteome Res 19:464–476. doi: 10.1021/acs.jproteome.9b00657. [DOI] [PubMed] [Google Scholar]
- 8.Phillips ZN, Brizuela C, Jennison AV, Staples M, Grimwood K, Seib KL, Jennings MP, Atack JM. 2019. Analysis of invasive nontypeable Haemophilus influenzae isolates reveals selection for the expression state of particular phase-variable lipooligosaccharide biosynthetic genes. Infect Immun 87:e00093-19. doi: 10.1128/IAI.00093-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seib KL, Srikhanta YN, Atack JM, Jennings MP. 2020. Epigenetic regulation of virulence and immunoevasion by phase-variable restriction-modification systems in bacterial pathogens. Annu Rev Microbiol 74:655–671. doi: 10.1146/annurev-micro-090817-062346. [DOI] [PubMed] [Google Scholar]
- 10.Phillips ZN, Husna AU, Jennings MP, Seib KL, Atack JM. 2019. Phasevarions of bacterial pathogens—phase-variable epigenetic regulators evolving from restriction-modification systems. Microbiology (Reading) 165:917–928. doi: 10.1099/mic.0.000805. [DOI] [PubMed] [Google Scholar]
- 11.Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. 2005. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci U S A 102:5547–5551. doi: 10.1073/pnas.0501169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atack JM, Srikhanta YN, Fox KL, Jurcisek JA, Brockman KL, Clark TA, Boitano M, Power PM, Jen FE, McEwan AG, Grimmond SM, Smith AL, Barenkamp SJ, Korlach J, Bakaletz LO, Jennings MP. 2015. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat Commun 6:7828. doi: 10.1038/ncomms8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blakeway LV, Power PM, Jen FE, Worboys SR, Boitano M, Clark TA, Korlach J, Bakaletz LO, Jennings MP, Peak IR, Seib KL. 2014. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J 28:5197–5207. doi: 10.1096/fj.14-256578. [DOI] [PubMed] [Google Scholar]
- 14.Seib KL, Jen FE, Scott AL, Tan A, Jennings MP. 2017. Phase variation of DNA methyltransferases and the regulation of virulence and immune evasion in the pathogenic Neisseria. Pathog Dis 75:ftx080. doi: 10.1093/femspd/ftx080. [DOI] [PubMed] [Google Scholar]
- 15.Srikhanta YN, Gorrell RJ, Steen JA, Gawthorne JA, Kwok T, Grimmond SM, Robins-Browne RM, Jennings MP. 2011. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One 6:e27569. doi: 10.1371/journal.pone.0027569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srikhanta YN, Fung KY, Pollock GL, Bennett-Wood V, Howden BP, Hartland EL. 2017. Phasevarion-regulated virulence in the emerging pediatric pathogen Kingella kingae. Infect Immun 85:e00319-17. doi: 10.1128/IAI.00319-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brockman KL, Branstool MT, Atack JM, Robledo-Avila F, Partida-Sanchez S, Jennings MP, Bakaletz LO. 2017. The ModA2 phasevarion of nontypeable Haemophilus influenzae regulates resistance to oxidative stress and killing by human neutrophils. Sci Rep 7:3161. doi: 10.1038/s41598-017-03552-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VanWagoner TM, Atack JM, Nelson KL, Smith HK, Fox KL, Jennings MP, Stull TL, Smith AL. 2016. The modA10 phasevarion of nontypeable Haemophilus influenzae R2866 regulates multiple virulence-associated traits. Microb Pathog 92:60–67. doi: 10.1016/j.micpath.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauntlett JC, Nilsson HO, Fulurija A, Marshall BJ, Benghezal M. 2014. Phase-variable restriction/modification systems are required for Helicobacter pylori colonization. Gut Pathog 6:35. doi: 10.1186/s13099-014-0035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips ZN, Tram G, Seib KL, Atack JM. 2019. Phase-variable bacterial loci: how bacteria gamble to maximise fitness in changing environments. Biochem Soc Trans 47:1131–1141. doi: 10.1042/BST20180633. [DOI] [PubMed] [Google Scholar]
- 21.Srikhanta YN, Dowideit SJ, Edwards JL, Falsetta ML, Wu HJ, Harrison OB, Fox KL, Seib KL, Maguire TL, Wang AH, Maiden MC, Grimmond SM, Apicella MA, Jennings MP. 2009. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog 5:e1000400. doi: 10.1371/journal.ppat.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atack JM, Yang Y, Seib KL, Zhou Y, Jennings MP. 2018. A survey of type III restriction-modification systems reveals numerous, novel epigenetic regulators controlling phase-variable regulons; phasevarions. Nucleic Acids Res 46:3532–3542. doi: 10.1093/nar/gky192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atack JM, Weinert LA, Tucker AW, Husna AU, Wileman TM, F Hadjirin N, Hoa NT, Parkhill J, Maskell DJ, Blackall PJ, Jennings MP. 2018. Streptococcus suis contains multiple phase-variable methyltransferases that show a discrete lineage distribution. Nucleic Acids Res 46:11466–11476. doi: 10.1093/nar/gky913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama T, Kasho K, Kawakami H. 2017. The DnaA cycle in Escherichia coli: activation, function and inactivation of the initiator protein. Front Microbiol 8:2496. doi: 10.3389/fmicb.2017.02496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carabetta VJ, Tanner AW, Greco TM, Defrancesco M, Cristea IM, Dubnau D. 2013. A complex of YlbF, YmcA and YaaT regulates sporulation, competence and biofilm formation by accelerating the phosphorylation of Spo0A. Mol Microbiol 88:283–300. doi: 10.1111/mmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boddu RS, Perumal O, K D. 2020. Microbial nitroreductases: a versatile tool for biomedical and environmental applications. Biotechnol Appl Biochem doi: 10.1002/bab.2073. [DOI] [PubMed] [Google Scholar]
- 27.Romero-Rodriguez A, Ruiz-Villafan B, Rocha-Mendoza D, Manzo-Ruiz M, Sanchez S. 2015. Biochemistry and regulatory functions of bacterial glucose kinases. Arch Biochem Biophys 577–578:1–10. doi: 10.1016/j.abb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Schowanek D, Verstraete W. 1990. Phosphonate utilization by bacterial cultures and enrichments from environmental samples. Appl Environ Microbiol 56:895–903. doi: 10.1128/AEM.56.4.895-903.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulze-Gahmen U, Pelaschier J, Yokota H, Kim R, Kim SH. 2003. Crystal structure of a hypothetical protein, TM841 of Thermotoga maritima, reveals its function as a fatty acid-binding protein. Proteins 50:526–530. doi: 10.1002/prot.10305. [DOI] [PubMed] [Google Scholar]
- 30.Asano Y, Lübbehüsen TL. 2000. Enzymes acting on peptides containing d-amino acid. J Biosci Bioeng 89:295–306. doi: 10.1016/s1389-1723(00)88949-5. [DOI] [PubMed] [Google Scholar]
- 31.Wei D, Wang M, Jiang B, Shi J, Hao J. 2014. Role of dihydroxyacetone kinases I and II in the dha regulon of Klebsiella pneumoniae. J Biotechnol 177:13–19. doi: 10.1016/j.jbiotec.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 32.Torrents E. 2014. Ribonucleotide reductases: essential enzymes for bacterial life. Front Cell Infect Microbiol 4:52. doi: 10.3389/fcimb.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan DI, Vogel HJ. 2010. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem J 430:1–19. doi: 10.1042/BJ20100462. [DOI] [PubMed] [Google Scholar]
- 34.Gawthorne JA, Beatson SA, Srikhanta YN, Fox KL, Jennings MP. 2012. Origin of the diversity in DNA recognition domains in phasevarion associated modA genes of pathogenic Neisseria and Haemophilus influenzae. PLoS One 7:e32337. doi: 10.1371/journal.pone.0032337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brockmeier SL, Loving CL, Nicholson TL, Wang J, Peters SE, Weinert L, Chaudhuri R, Seilly DJ, Langford PR, Rycroft A, Wren BW, Maskell DJ, Tucker AW. 2017. Use of proteins identified through a functional genomic screen to develop a protein subunit vaccine that provides significant protection against virulent Streptococcus suis in pigs. Infect Immun 86:e00559-17. doi: 10.1128/IAI.00559-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segura M. 2015. Streptococcus suis vaccines: candidate antigens and progress. Expert Rev Vaccines 14:1587–1608. doi: 10.1586/14760584.2015.1101349. [DOI] [PubMed] [Google Scholar]
- 37.dos Santos DF, Istvan P, Noronha EF, Quirino BF, Kruger RH. 2015. New dioxygenase from metagenomic library from Brazilian soil: insights into antibiotic resistance and bioremediation. Biotechnol Lett 37:1809–1817. doi: 10.1007/s10529-015-1861-x. [DOI] [PubMed] [Google Scholar]
- 38.Yongkiettrakul S, Maneerat K, Arechanajan B, Malila Y, Srimanote P, Gottschalk M, Visessanguan W. 2019. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet Res 15:5. doi: 10.1186/s12917-018-1732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lugsomya K, Chatsuwan T, Niyomtham W, Tummaruk P, Hampson DJ, Prapasarakul N. 2018. Routine prophylactic antimicrobial use is associated with increased phenotypic and genotypic resistance in commensal Escherichia coli isolates recovered from healthy fattening pigs on farms in Thailand. Microb Drug Resist 24:213–223. doi: 10.1089/mdr.2017.0042. [DOI] [PubMed] [Google Scholar]
- 40.Kouadio IK, Guessennd N, Dadie A, Koffi E, Dosso M. 2018. Comparative study of the impact of the administration of amoxicillin and Algo-Bio® alternative substance to antibiotics, on the level of selection of resistant Enterobacteriaceae in the digestive flora of piglets. J Glob Antimicrob Resist 13:161–164. doi: 10.1016/j.jgar.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Callens B, Persoons D, Maes D, Laanen M, Postma M, Boyen F, Haesebrouck F, Butaye P, Catry B, Dewulf J. 2012. Prophylactic and metaphylactic antimicrobial use in Belgian fattening pig herds. Prev Vet Med 106:53–62. doi: 10.1016/j.prevetmed.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Lekagul A, Tangcharoensathien V, Yeung S. 2019. Patterns of antibiotic use in global pig production: a systematic review. Vet Anim Sci 7:100058. doi: 10.1016/j.vas.2019.100058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krasny L, Bland P, Kogata N, Wai P, Howard BA, Natrajan RC, Huang PH. 2018. SWATH mass spectrometry as a tool for quantitative profiling of the matrisome. J Proteomics 189:11–22. doi: 10.1016/j.jprot.2018.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brockman KL, Azzari PN, Branstool MT, Atack JM, Schulz BL, Jen FE, Jennings MP, Bakaletz LO. 2018. Epigenetic regulation alters biofilm architecture and composition in multiple clinical isolates of nontypeable Haemophilus influenzae. mBio 9:e01682-18. doi: 10.1128/mBio.01682-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atack JM, Guo C, Litfin T, Yang L, Blackall PJ, Zhou Y, Jennings MP. 2020. Systematic analysis of REBASE identifies numerous Type I restriction-modification systems with duplicated, distinct hsdS specificity genes that can switch system specificity by recombination. mSystems 5:e00497-20. doi: 10.1128/mSystems.00497-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atack JM, Guo C, Yang L, Zhou Y, Jennings MP. 2020. DNA sequence repeats identify numerous Type I restriction-modification systems that are potential epigenetic regulators controlling phase-variable regulons; phasevarions. FASEB J 34:1038–1051. doi: 10.1096/fj.201901536RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinert LA, Chaudhuri RR, Wang J, Peters SE, Corander J, Jombart T, Baig A, Howell KJ, Vehkala M, Valimaki N, Harris D, Chieu TT, Van Vinh Chau N, Campbell J, Schultsz C, Parkhill J, Bentley SD, Langford PR, Rycroft AN, Wren BW, Farrar J, Baker S, Hoa NT, Holden MT, Tucker AW, Maskell DJ, BRaDP1T Consortium . 2015. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat Commun 6:6740. doi: 10.1038/ncomms7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark TA, Murray IA, Morgan RD, Kislyuk AO, Spittle KE, Boitano M, Fomenkov A, Roberts RJ, Korlach J. 2012. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res 40:e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray IA, Clark TA, Morgan RD, Boitano M, Anton BP, Luong K, Fomenkov A, Turner SW, Korlach J, Roberts RJ. 2012. The methylomes of six bacteria. Nucleic Acids Res 40:11450–11462. doi: 10.1093/nar/gks891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peak IR, Chen A, Jen FE, Jennings C, Schulz BL, Saunders NJ, Khan A, Seifert HS, Jennings MP. 2016. Neisseria meningitidis lacking the major porins PorA and PorB is viable and modulates apoptosis and the oxidative burst of neutrophils. J Proteome Res 15:2356–2365. doi: 10.1021/acs.jproteome.5b00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peak IR, Jennings CD, Jen FE, Jennings MP. 2014. Role of Neisseria meningitidis PorA and PorB expression in antimicrobial susceptibility. Antimicrob Agents Chemother 58:614–616. doi: 10.1128/AAC.02506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaino JA. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of ModS is dependent on simple sequence repeat (SSR) tract length. (A) Coomassie staining of whole-cell lysates from enriched strains of S. suis. (B) Western blot using anti-ModS antiserum demonstrates that ModS1 is only produced in strains enriched for 21 GAGCA repeats and that ModS2 is only produced in strains enriched for 18 repeats. The dotted line represents the region of blot presented in Fig. 1D. Download FIG S1, PDF file, 1.7 MB (1.7MB, pdf) .
Copyright © 2021 Tram et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Three open reading frames occur in the modS gene due to variation in length of the GAGCA(n) simple sequence repeat tract. Due to a frameshift downstream of the GAGCA(n) SSR tract, 19 and 20 repeats result in a premature stop codon (in bold, highlighted with an asterisk [*]), and consequently no expression of the ModS protein; 21 GAGCA(n) repeats in the SSR tract results in the gene being in-frame, and therefore ModS protein is produced. Download FIG S2, PDF file, 0.1 MB (96KB, pdf) .
Copyright © 2021 Tram et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All single-molecule real-time (SMRT) sequencing methylome data summarized in Table 1 for our enriched modS1 and modS2 populations of strains LSS89 and SS1056, respectively, containing three consecutive GAGCA(n) SSR tract lengths. Download Data Set S1, XLSX file, 0.01 MB (13.3KB, xlsx) .
Copyright © 2021 Tram et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All data for growth curves presented in Fig. 3. Download Data Set S2, XLSX file, 0.01 MB (14.8KB, xlsx) .
Copyright © 2021 Tram et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (52) partner repository under data set identifier PXD023726.
All data for growth curves presented in Fig. 3. Download Data Set S2, XLSX file, 0.01 MB (14.8KB, xlsx) .
Copyright © 2021 Tram et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.